Abstract
A large-scale, genome-wide association study was performed to identify genetic variations influencing serum bilirubin levels using 8841 Korean individuals. Significant associations were observed at UGT1A1 (rs11891311, P = 4.78 × 10−148) and SLCO1B3 (rs2417940, P = 1.03 × 10−17), which are two previously identified loci. The two single-nucleotide polymorphisms (SNPs) were replicated (rs11891311, P = 3.18 × 10−15) or marginally significant (rs2417940, P = 8.56 × 10−4) in an independent cohort of 1096 individuals. In a conditional analysis adjusted for the top UGT1A1 variant (rs11891311), another variant in UGT1A1 (rs4148323, P = 1.22 × 10−121) remained significant; this suggests that in UGT1A1 at least two independent genetic variations influence the bilirubin levels in the Korean population. The protein coding variant rs4148323, which is monomorphic in European-derived populations, may be specifically associated with serum bilirubin levels in Asians (P = 2.56 × 10−70). The SLCO1B3 variant (rs2417940, P = 1.67 × 10−18) remained significant in a conditional analysis for the top UGT1A1 variant. Interestingly, there were significant differences in the associated variations of SLCO1B3 between Koreans and European-derived populations. While the variant rs2417940 at intron 7 of SLCO1B3 was more significantly associated in Koreans, variants rs17680137 (P = 0.584) and rs2117032 (P = 2.76 × 10−5), two of the top-ranked SNPs in European-derived populations, did not reach the genome-wide significance level. Also, variants in SLCO1B1 did not reach genome-wide significance in Koreans. Our result supports the idea that there are considerable ethnic differences in genetic association of bilirubin levels between Koreans and European-derived populations.
INTRODUCTION
Bilirubin is a breakdown product of normal heme catabolism. Heme, which is primarily derived from the hemoglobin of red blood cells, is converted to biliverdin by heme oxygenases and is then reduced to bilirubin (1). Bilirubin is taken into the liver by the solute carrier organic anion transporter family (2) and is glucuronidated by UDP-glycosyltransferase in hepatocytes (3). The addition of one or two molecules of glucuronic acid increases the solubility of bilirubin. Once bilirubin is conjugated with glucuronic acid it is actively secreted into the bile (4).
The UDP-glycosyltransferase 1 family, polypeptide A1 (UGT1A1) and solute carrier organic anion transporter family enzymes SLCO1B1 and SLCO1B3 are responsible for glucuronidation and cellular uptake of bilirubin, respectively, and play an important role in regulating the bilirubin levels (5–7). UGT1A1 is the major determinant of serum bilirubin levels, and a genetic defect of UGT1A1 leading to complete or partial inactivation of the enzyme causes three forms of nonhemolytic, unconjugated hyperbilirubinemia: Crigler–Najjar syndrome type I and II and Gilbert syndrome (8–11). Recently, two genome-wide association studies (GWASs) based on the European-derived populations reported a strong association of UGT1A1, SLCO1B1 and SLCO1B3 with serum bilirubin levels (12,13). They confirmed that there is a substantial contribution from UGT1A1, SLCO1B1 and SLCO1B3 to bilirubin levels.
Here, we performed a GWAS to identify genetic variants associated with serum bilirubin levels in the Korean population. This is the first large-scale GWAS in an Asian population, which included 9937 Korean participants. As a result of focusing on the Asian population, we observed significant associations of UGT1A1 and SLCO1B3 with total serum bilirubin levels. It is important to note that the associated single-nucleotide polymorphisms (SNPs) were different from those in European-derived populations, which supports the idea of considerable ethnic genetic differences between Koreans and European-derived populations.
RESULTS
Genome-wide association of total serum bilirubin levels
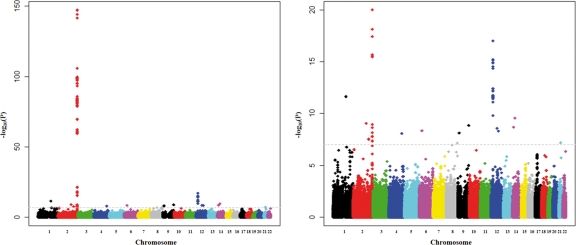
A genome-wide association of total serum bilirubin levels was tested in 8841 individuals from the Korea Association REsource (KARE, stage I) project (for detailed characteristics of the study population, see Materials and Methods and Supplementary Material, Table S1). As shown in Figure 1, significant associations were observed at two previously reported loci, UGT1A1 on 2q37 (12,13) and SLCO1B3 on 12p12 (12).
Figure 1.
Genome-wide association plot for total serum bilirubin levels (stage I). The negative common logarithms of P-value in linear regression analysis adjusted for age and sex are shown. Full results are shown in the left panel, and results with P ≥ 10–20 are shown in the right panel.
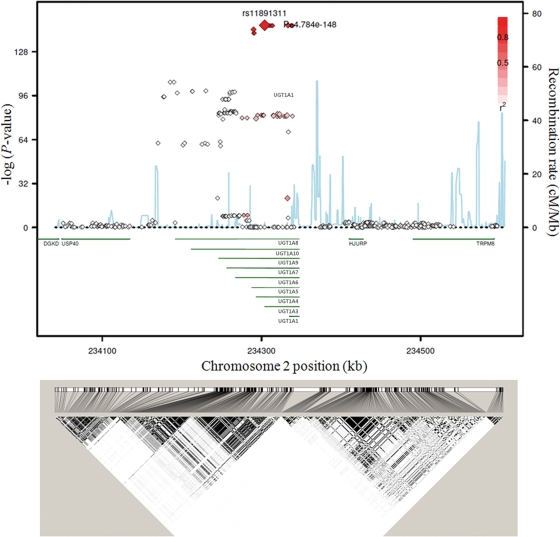
At the UGT1A1 locus, a group of 12 tightly linked SNPs [linkage disequilibrium (LD) r2 > 0.92] spanning a 50 kb genomic region showed the strongest association (P < 10−148, Fig. 2 and Supplementary Material, Table S2). The associated 50 kb region covers the UGT1A1 promoter region, which contains the TATAA box polymorphism UGT1A1*28 (rs8175347). Among the 12 SNPs, rs11891311 showed the most significant association with total serum bilirubin levels (P = 4.78 × 10−148, Table 1). The minor allele T of rs11891311 was associated with an increase in total serum bilirubin levels. The second and third most significant variants were rs887829 (P = 5.37 × 10−148) and rs6742078 (P = 7.19 × 10−148), which are located in the core promoter and intron 1 of UGT1A1, respectively (Supplementary Material, Table S2). Interestingly, the two SNPs were identified as the most significant SNPs by two independent GWASs (12,13).
Figure 2.
Association of UGT1A with total serum bilirubin levels (stage I). The association, recombination rate and linkage disequilibrium (LD) of a 540 kb genomic region (chromosome 2; 234 050 873–234 596 096) around the UGT1A cluster are plotted. The red-filled diamonds represents LD with the top SNP, rs11891311 and the blue line indicates the recombination rate.
Table 1.
Association with total serum bilirubin levels
| SNP | Chr | Positiona | Gene | Population | MAF | β | Associationb (P) |
|---|---|---|---|---|---|---|---|
| rs11891311 | 2 | 234 304 049 | UGT1A1 | Stage I (n = 8841) | 0.121 | 0.184 | 4.78 × 10−148 |
| Stage II (n = 1096) | 0.109 | 0.166 | 3.18 × 10−15 | ||||
| Combined (n = 9937) | 0.120 | 0.182 | 8.68 × 10−157 | ||||
| rs4148323 | 2 | 234 333 883 | UGT1A1 | Stage I (n = 8841) | 0.189 | 0.107 | 2.56 × 10−70 |
| Stage II (n = 1096) | 0.191 | 0.136 | 3.99 × 10−16 | ||||
| Combined (n = 9937) | 0.189 | 0.110 | 5.07 × 10−82 | ||||
| rs2417940 | 12 | 20 909 142 | SLCO1B3 | Stage I (n = 8841) | 0.212 | 0.050 | 1.03 × 10−17 |
| Stage II (n = 1096) | 0.154 | 0.073 | 8.56 × 10−04 | ||||
| Combined (n = 9937) | 0.207 | 0.048 | 2.42 × 10−17 |
The most significant SNPs for each locus are listed. Chr, chromosome; MAF, minor allele frequency.
aChromosome position (Genome Build 36.3).
bLinear regression analysis adjusted for age and sex.
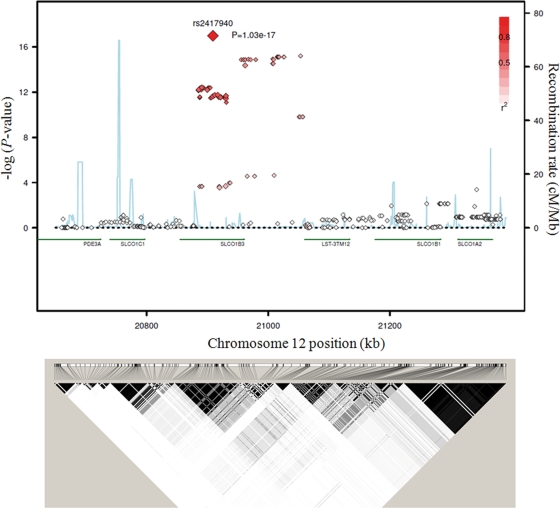
SLCO1B3 was the second most significant locus (Fig. 1 and Supplementary Material, Table S2). A strong association was observed at the genomic region that includes intron 2 through downstream of SLCO1B3 (Fig. 3). The rs2417940 variant at intron 7 of SLCO1B3 was the most significantly associated with total serum bilirubin levels at this locus (P = 1.03 × 10−17, Table 1). Meanwhile, rs17680137 (P = 0.584) and rs2117032 (P = 2.76 × 10−5), which were identified as the top-ranked SNPs in the SardiNIA study (12), did not reach genome-wide significance level (Supplementary Material, Table S3).
Figure 3.
Association of SLCO1B3 with total serum bilirubin levels (stage I). The association, recombination rate and linkage disequilibrium (LD) of a 720 kb genomic region (chromosome 12; 20 660 255–21 381 648) around SLCO1B3 are plotted. The red-filled diamonds represents LD with the top SNP, rs2417940, and the blue line indicates the recombination rate.
None of the SLCO1B1 SNPs in the Korean population reached a genome-wide significance level (Supplementary Material, Table S3). SLCO1B3 and SLCO1B1 are closely located and found at 12p12 of the human genome, but they belong to different LD blocks (Fig. 3). Among three SNPs at 12p12, which were found to have genome-wide significance in Johnson et al.'s study (13), two of them (rs4149056 and rs2417873) were filtered out by low SNP call rate (SNP call rate ≤ 95%), and rs4149000 was not significant (P = 0.013) in the Korean population (Supplementary Material, Table S3).
Replication in the second cohort
To confirm our results, we analyzed an additional data set of 1096 Korean individuals (stage II KIOM data set). We first tested whether there were significant stratification/batch effects between the two populations. Genomic control inflation scores were 1.031 (stage I, KARE) and 1.032 (stage II, KIOM), suggesting that there were no significant stratification/batch effects in the data sets. The most significant SNP in stage I, rs11891311, was strongly associated in the second (P = 3.18 × 10−15), as well as the combined population (P = 8.68 × 10−157; Table 1). rs2417940 of SLCO1B3 showed a suggestive association (P = 8.56 × 10−4) in the second data set. In addition to SNPs in UGT1A1 and SLCO1B3, several SNPs, such as rs2501324 (CCDC19, 2.33 × 10−12) at chromosome 1, rs10901296 (intergenic region, P = 1.41 × 10−9) at chromosome 9 and rs17096653 (intergenic region, P = 2.00 × 10−9) at chromosome 14, showed a significant association in stage I, but the associations were not replicated in the stage II population. For a full list of significant variants in stage I, see Supplementary Material, Table S2.
In summary, the association of UGT1A1 and SLCO1B3 with total serum bilirubin levels was confirmed in the Korean population.
GWA results are conditional on UGT1A1 (rs11891311)
UGT1A1 is the rate-limiting enzyme of bilirubin metabolism, and its association with bilirubin levels is well established. At the UGT1A1 locus, rs11891311 is one of the most significant SNPs, and a great portion of the variation in total serum bilirubin levels may be caused by rs11891311. To determine whether additional loci remained significant genome-wide after accounting for the UGT1A1 effect, we performed a conditional analysis by including the genotype of rs11891311 in the linear regression model as covariates, along with age and sex. The rs4148323 variant, which encodes a nonsynonymous change of glycine to arginine in the first exon of UGT1A1 (also known as UGT1A1*6 rs4148323), showed a remarkable association with serum bilirubin levels (stage I, P = 1.22 × 10−121; stage II, P = 6.83 × 10−24; combined, P = 3.08 × 10−139, Table 2 and Supplementary Material, Table S4). SLCO1B3 also remained significant (rs2417940; stage I, P = 1.67 × 10−18; stage II, P = 5.98 × 10−4; combined, P = 4.77 × 10−18; Table 2 and Supplementary Material, Table S4). Interestingly, the association of rs4148323 (Gly71Arg, UGT1A1*6) and rs2417940 was enhanced, not decreased, after adjustment (Tables 1 and 2). Thus, rs11891311 and rs4148323 at the UGT1A1 locus and rs2417940 at SLCO1B3 are independently associated with total serum bilirubin levels in the Korean population.
Table 2.
Conditional analysis of rs11891311
| SNP | Chr | Positiona | Gene | Population | MAF | β | Associationb (P) |
|---|---|---|---|---|---|---|---|
| rs4148323 | 2 | 234 333 883 | UGT1A1 | Stage I (n = 8841) | 0.189 | 0.137 | 1.22 × 10−121 |
| Stage II (n = 1096) | 0.191 | 0.166 | 6.83 × 10−24 | ||||
| Combined (n = 9937) | 0.189 | 0.140 | 3.08 × 10−139 | ||||
| rs2417940 | 12 | 20 909 142 | SLCO1B3 | Stage I (n = 8841) | 0.212 | 0.049 | 1.67 × 10−18 |
| Stage II (n = 1096) | 0.154 | 0.074 | 5.98 × 10−04 | ||||
| Combined (n = 9937) | 0.207 | 0.047 | 4.77 × 10−18 |
The most significant SNP for each locus is listed. Chr, chromosome; MAF, minor allele frequency.
aChromosome position (Genome Build 36.3).
bLinear regression analysis adjusted for age, sex and rs11891311.
GWA results conditional on SLCO1B3 (rs2417940)
Besides the UGT1A1 locus, the most significant association was observed at rs2417940 located in intron 7 of SLCO1B3 (combined P = 2.42 × 10−17, Table 1). We also found a suggestive association at the rs2117032 variant, which was the top SNP of SLCO1B3 in the SardiNIA study, although it did not reach a genome-wide significance level. The LD between rs2417940 and rs2117032 was 0.28 of r2 in Koreans. To test whether the association of rs2117032 is attributable to rs2417940, we performed conditional analysis on rs2417940. When adjusted for rs2417940, rs2117032 did not show a significant association (stage I, P = 0.793). Accordingly, rs2417940 may account for the association of rs2117032 in Koreans.
DISCUSSION
This is the largest GWAS of serum bilirubin levels in an Asian population. We confirmed the large impacts of UGT1A1 and SLCO1B3 on bilirubin levels, as well as the considerable ethnic genetic differences in bilirubin metabolism between Korean and European-derived populations.
We revealed that there are at least two independent genetic variations that have an effect on the bilirubin levels at the UGT1A1 locus. First, the association of the UGT1A1 locus appears to be mainly due to the functional promoter polymorphism UGT1A1*28. Along with the two previous GWASs performed in European-derived populations (12,13), we found a significant association with the promoter region of UGT1A1. The three SNPs rs887829, rs6742078 and rs11891311, which were strongly linked to each other (r2 > 0.96, Supplementary Material, Table S4) and located close to UGT1A1*28, showed the most significant association. Interestingly, Johnson et al. (13) reported that rs6742078 was in high LD (r2 = 0.88) with UGT1A1*28. Thus, the major association of the UGT1A1 promoter region may be derived from the functional promoter polymorphism UGT1A1*28 in which the longer TATAA element results in a 5-fold reduction in promoter activity (10). Also, the nonsynonymous variant rs4148323 (Gly71Arg, UGT1A1*6) of UGT1A1 accounted for variation of bilirubin levels that was not explained by the promoter SNP alone. The rs4148323 variant encodes a nonsynonymous change from glycine to arginine, and the enzymatic activity of arginine allele is reduced to 47% of the wild-type glycine allele (14). We found a significant association at rs4148323, even after adjustment for rs11891311 (Table 2 and Supplementary Material, Table S4). Therefore, the association of rs4148323 is independent of regulatory variations of UGT1A1. A previous Japanese study reporting the independence of UGT1A1*6 (rs4148323) from UGT1A1*28 also supports this finding (15).
In contrast, in both GWASs based on European-derived population, the association of the UGT1A1 locus was attributed solely to genetic variants at the promoter region. When adjusted for the top SNPs, rs887829 (12) or rs6742078 (13), none of the UGT1A1 locus SNPs remained significant. The top SNP accounts for ∼16.7–18.1% of the variation of serum bilirubin levels in the European-derived population (13). In contrast, the two independent and major modulators rs11891311 and rs4148323 accounted for only 10.47% (7.18 and 3.29%, respectively) of the variation in Koreans. The difference in allele frequency between Asians and European-derived population may, in part, explain the ethnic difference in the genetic association. The minor allele frequencies (MAFs) of rs4148323 are 0.189 and 0.163 in the Korean and HapMap Asian (JPT + CHB) populations, respectively, while rs4148323 is monomorphic in European-derived population. Meanwhile, the minor alleles of rs11891311, rs887829 and rs6742078 are more frequent in European-derived population (MAF = 0.283–0.308) than in Koreans (0.121–0.123).
SLCO1B1 and SLCO1B3 are organic anion transporter genes closely located at 12p12 of the human genome. The two previous GWASs reported dissimilar results; that is, either SLCO1B1 (13) or SLCO1B3 (12) was associated with total serum bilirubin levels. Similar to the SardiNIA study, we found a significant association at SLCO1B3, but not at SLCO1B1 (Fig. 3). The most significant association was observed at rs2417940, which is located in intron 7 of SLCO1B3 (combined P = 2.42 × 10−17, Table 1), and it was responsible for 0.635% of the variation in the bilirubin level. We also found a suggestive association of rs2117032 (2.76 × 10−5), the top SNP at SLCO1B3 in the SardiNIA study, although it did not reach the genome-wide significance level. The LD between rs2417940 and rs2117032 was 0.28 of r2 in Koreans. In conditional analysis adjusted for rs2417940, rs2117032 did not show significant association (stage I, P = 0.793), suggesting that rs2417940 may account for the association of rs2117032 in Koreans. Another top SNP, rs17680137, also showed a substantial ethnic difference. Its MAF is 0.00373 and 0.293 in Korean and European-derived populations (SardiNIA stage 1), respectively, and it is associated with bilirubin levels in only the European-derived population (Supplementary Material, Table S3). Several previous studies reported the association of SLCO1B1 with bilirubin levels in both Asian and European-derived populations (13,16,17). However, SNPs of SLCO1B1, including rs4149000, did not reach genome-wide significance in Koreans (Supplementary Material, Table S3).
In conclusion, a GWAS including 9937 Korean individuals confirmed the association of UGT1A1 and SLCO1B3 with total serum bilirubin levels in the Korean population. SLCO1B1, a locus previously identified in European-derived populations, did not reach genome-wide significance in the Korean population. The associated variations in this study differed considerably from those seen in European-derived population, even though the variations were located at the same gene, which reflects the ethnic difference of bilirubin genetics. These ethnic differences are reflected in the high Akey's FST measures in UGT1A1, SLCO1B1 and SLCO1B3 loci among different populations (Supplementary Material, Table S5). Our result emphasizes the importance of replication of GWAS in diverse populations.
MATERIALS AND METHODS
Sample description
Two cohorts (KARE, stage I and KIOM, stage II) were studied (Supplementary Material, Table S1). Details of Korea Association REsource (KARE, stage I) population were previously described (18). Briefly, the KARE data set included 8841 participants from the Ansung and Ansan areas. The KIOM (stage II) data set included 1096 individuals who visited the Korean Institute of Oriental Medicine (KIOM) and collaborative hospitals for health examinations between July 2006 and February 2009. Consent was obtained from all participants, and the Institutional Review Board of KIOM approved the study. A blood sample was obtained from each individual and used for routine blood tests, including measuring the total serum bilirubin levels. The stage I population (52.21 ± 8.92) was older than the stage II population (48.35 ± 15.68), and male individuals were more frequent in stage I (47.30%) than stage II (38.87%). Total serum bilirubin levels were different between populations with stage I (0.60 ± 0.33 mg/dl) having lower levels than stage II (0.75 ± 0.32 mg/dl). Consistent with the previous study (13), the bilirubin levels were significantly higher in males than that in females in both populations.
Genotyping and imputation
Genotyping and quality control of the KARE data set was previously described (18). Briefly, 10 004 participants from the Ansung and Ansan cohorts were genotyped using the Affymetrix Genome-Wide Human SNP Array 5.0. After standard quality control procedures [sample call rate ≥96%, SNP call rate ≥95%, MAF ≥1% and Hardy-Weinberg equilibrium (HWE) P ≥ 10−6], genotypes of 8841 individuals for 352 228 autosomal SNPs were used in subsequent analyses. The KIOM data set was genotyped and processed in the same way. After checking DNA quality, 1225 individuals were genotyped using the Affymetrix Genome-Wide Human SNP Array 5.0. We applied the same SNP filtering criteria described above and selected 1096 individuals for subsequent analyses.
Untyped HapMap SNPs were imputed with PLINK (version 1.06) using filtered haplotypes (HapMap release 23, 2 100 739 SNPs filtered from 3.99 million SNPs of JPT + CHB release 23) of 90 individuals from the Japanese and Chinese populations as a reference panel (downloaded from the PLINK web site), which lead to an imputed data set of ∼2.1 million SNPs (19). After imputation, 2 100 739 SNPs were again filtered based on the following criteria: imputation quality (PLINK information content metric ≥0.8) and MAF ≥1%, leading to a data set of 1 227 049 SNPs for the association analysis (Supplementary Material, Table S6). As both KARE (stage I) and KIOM (stage II) data sets were genotyped using the same Affymetrix SNP 5.0 array platform, the two SNP data sets were combined to a data set of 9937 individuals in the joint analysis.
Statistical analysis
The association with serum bilirubin levels was tested by linear regression analysis adjusted for age and sex using PLINK software (19). We used WGAViewer (version 1.26G) (20) to create Manhattan plots, Haploview (version 1.4) (21) to calculate LD, and SNAP (22) to annotate the proxy of the top SNP, respectively. Association with P ≤ 10−7 was considered to be genome-wide significant. Akey's FST measures among different populations were obtained from the SNP@Evolution site (23).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by a grant (to Y.S.K.) from the Ministry of Science and Technology of Korea (M10643020001-08N4302-00210) and Research Initiative program of Korea Research Institute of Bioscience and Biotechnology. The Korea Association Resource (KARE) project was supported by a grant from the Ministry for Health, Welfare and Family Affairs (4845-301-430-260-00); and a grant from the Korea National Institute of Health, Korea Center for Disease Control (4845-301-430-210-13), Republic of Korea.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Young-Kyu Park at Korea Research Institute of Bioscience and Biotechnology and Dr Sangsoo Kim at Soongsil University for their helpful comments.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Alexandra Brito M., Silva R.F., Brites D. Bilirubin toxicity to human erythrocytes: a review. Clin. Chim. Acta. 2006;374:46–56. doi: 10.1016/j.cca.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 2.Hagenbuch B., Gui C. Xenobiotic transporters of the human organic anion transporting polypeptides (OATP) family. Xenobiotica. 2008;38:778–801. doi: 10.1080/00498250801986951. doi:10.1080/00498250801986951. [DOI] [PubMed] [Google Scholar]
- 3.Ritter J.K., Crawford J.M., Owens I.S. Cloning of two human liver bilirubin UDP-glucuronosyltransferase cDNAs with expression in COS-1 cells. J. Biol. Chem. 1991;266:1043–1047. [PubMed] [Google Scholar]
- 4.Jedlitschky G., Leier I., Buchholz U., Hummel-Eisenbeiss J., Burchell B., Keppler D. ATP-dependent transport of bilirubin glucuronides by the multidrug resistance protein MRP1 and its hepatocyte canalicular isoform MRP2. Biochem. J. 1997;327(Pt 1):305–310. doi: 10.1042/bj3270305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jansen P.L., Bosma P.J., Chowdhury J.R. Molecular biology of bilirubin metabolism. Prog. Liver Dis. 1995;13:125–150. [PubMed] [Google Scholar]
- 6.Sorich M.J., Miners J.O., McKinnon R.A., Smith P.A. Multiple pharmacophores for the investigation of human UDP-glucuronosyltransferase isoform substrate selectivity. Mol. Pharmacol. 2004;65:301–308. doi: 10.1124/mol.65.2.301. doi:10.1124/mol.65.2.301. [DOI] [PubMed] [Google Scholar]
- 7.Zhang W., He Y.J., Gan Z., Fan L., Li Q., Wang A., Liu Z.Q., Deng S., Huang Y.F., Xu L.Y., et al. OATP1B1 polymorphism is a major determinant of serum bilirubin level but not associated with rifampicin-mediated bilirubin elevation. Clin. Exp. Pharmacol. Physiol. 2007;34:1240–1244. doi: 10.1111/j.1440-1681.2007.04798.x. doi:10.1111/j.1440-1681.2007.04798.x. [DOI] [PubMed] [Google Scholar]
- 8.Mackenzie P.I., Owens I.S., Burchell B., Bock K.W., Bairoch A., Belanger A., Fournel-Gigleux S., Green M., Hum D.W., Iyanagi T., et al. The UDP glycosyltransferase gene superfamily: recommended nomenclature update based on evolutionary divergence. Pharmacogenetics. 1997;7:255–269. doi: 10.1097/00008571-199708000-00001. doi:10.1097/00008571-199708000-00001. [DOI] [PubMed] [Google Scholar]
- 9.Lin J.P., Cupples L.A., Wilson P.W., Heard-Costa N., O'Donnell C.J. Evidence for a gene influencing serum bilirubin on chromosome 2q telomere: a genomewide scan in the Framingham study. Am. J. Hum. Genet. 2003;72:1029–1034. doi: 10.1086/373964. doi:10.1086/373964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bosma P.J., Chowdhury J.R., Bakker C., Gantla S., de Boer A., Oostra B.A., Lindhout D., Tytgat G.N., Jansen P.L., Oude Elferink R.P., et al. The genetic basis of the reduced expression of bilirubin UDP-glucuronosyltransferase 1 in Gilbert's syndrome. N. Engl. J. Med. 1995;333:1171–1175. doi: 10.1056/NEJM199511023331802. doi:10.1056/NEJM199511023331802. [DOI] [PubMed] [Google Scholar]
- 11.Maruo Y., Wada S., Yamamoto K., Sato H., Yamano T., Shimada M. A case of anorexia nervosa with hyperbilirubinaemia in a patient homozygous for a mutation in the bilirubin UDP-glucuronosyltransferase gene. Eur. J. Pediatr. 1999;158:547–549. doi: 10.1007/s004310051143. doi:10.1007/s004310051143. [DOI] [PubMed] [Google Scholar]
- 12.Sanna S., Busonero F., Maschio A., McArdle P.F., Usala G., Dei M., Lai S., Mulas A., Piras M.G., Perseu L., et al. Common variants in the SLCO1B3 locus are associated with bilirubin levels and unconjugated hyperbilirubinemia. Hum. Mol. Genet. 2009;18:2711–2718. doi: 10.1093/hmg/ddp203. doi:10.1093/hmg/ddp203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson A.D., Kavousi M., Smith A.V., Chen M.H., Dehghan A., Aspelund T., Lin J.P., van Duijn C.M., Harris T.B., Cupples L.A., et al. Genome-wide association meta-analysis for total serum bilirubin levels. Hum. Mol. Genet. 2009;18:2700–2710. doi: 10.1093/hmg/ddp202. doi:10.1093/hmg/ddp202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jinno H., Tanaka-Kagawa T., Hanioka N., Saeki M., Ishida S., Nishimura T., Ando M., Saito Y., Ozawa S., Sawada J. Glucuronidation of 7-ethyl-10-hydroxycamptothecin (SN-38), an active metabolite of irinotecan (CPT-11), by human UGT1A1 variants, G71R, P229Q, and Y486D. Drug. Metab. Dispos. 2003;31:108–113. doi: 10.1124/dmd.31.1.108. doi:10.1124/dmd.31.1.108. [DOI] [PubMed] [Google Scholar]
- 15.Urawa N., Kobayashi Y., Araki J., Sugimoto R., Iwasa M., Kaito M., Adachi Y. Linkage disequilibrium of UGT1A1 *6 and UGT1A1 *28 in relation to UGT1A6 and UGT1A7 polymorphisms. Oncol. Rep. 2006;16:801–806. [PubMed] [Google Scholar]
- 16.Huang C.S., Huang M.J., Lin M.S., Yang S.S., Teng H.C., Tang K.S. Genetic factors related to unconjugated hyperbilirubinemia amongst adults. Pharmacogenet. Genomics. 2005;15:43–50. doi: 10.1097/01213011-200501000-00007. doi:10.1097/01213011-200501000-00007. [DOI] [PubMed] [Google Scholar]
- 17.Ieiri I., Suzuki H., Kimura M., Takane H., Nishizato Y., Irie S., Urae A., Kawabata K., Higuchi S., Otsubo K., et al. Influence of common variants in the pharmacokinetic genes (OATP-C, UGT1A1, and MRP2) on serum bilirubin levels in healthy subjects. Hepatol. Res. 2004;30:91–95. doi: 10.1016/j.hepres.2004.07.005. doi:10.1016/j.hepres.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 18.Cho Y.S., Go M.J., Kim Y.J., Heo J.Y., Oh J.H., Ban H.J., Yoon D., Lee M.H., Kim D.J., Park M., et al. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat. Genet. 2009;41:527–534. doi: 10.1038/ng.357. doi:10.1038/ng.357. [DOI] [PubMed] [Google Scholar]
- 19.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. doi:10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ge D., Zhang K., Need A.C., Martin O., Fellay J., Urban T.J., Telenti A., Goldstein D.B. WGAViewer: software for genomic annotation of whole genome association studies. Genome Res. 2008;18:640–643. doi: 10.1101/gr.071571.107. doi:10.1101/gr.071571.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. doi:10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 22.Johnson A.D., Handsaker R.E., Pulit S.L., Nizzari M.M., O'Donnell C.J., de Bakker P.I. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. doi:10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng F., Chen W., Richards E., Deng L., Zeng C. SNP@Evolution: a hierarchical database of positive selection on the human genome. BMC Evol. Biol. 2009;9:221. doi: 10.1186/1471-2148-9-221. doi:10.1186/1471-2148-9-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.