Abstract
KCNN4 channels that provide the driving force for cAMP- and Ca2+-induced anion secretion are present in both apical and basolateral membranes of the mammalian colon. However, only a single KCNN4 has been cloned. This study was initiated to identify whether both apical and basolateral KCNN4 channels are encoded by the same or different isoforms. Reverse transcriptase-PCR (RT-PCR), real-time quantitative-PCR (RT-QPCR), and immunofluorescence studies were used to clone and identify tissue-specific expression of KCNN4 isoforms. Three distinct KCNN4 cDNAs that are designated as KCNN4a, KCNN4b, and KCNN4c encoding 425, 424, and 395 amino acid proteins, respectively, were isolated from the rat colon. KCNN4a differs from KCNN4b at both the nucleotide and the amino acid level with distinct 628 bp at the 3′-untranslated region and an additional glutamine at position 415, respectively. KCNN4c differs from KCNN4b by lacking the second exon that encodes a 29 amino acid motif. KCNN4a and KCNN4b/c are identified as smooth muscle- and epithelial cell-specific transcripts, respectively. KCNN4b and KCNN4c transcripts likely encode basolateral (40 kDa) and apical (37 kDa) membrane proteins in the distal colon, respectively. KCNN4c, which lacks the S2 transmembrane segment, requires coexpression of a large conductance K+ channel β-subunit for plasma membrane expression. The KCNN4 channel blocker TRAM-34 inhibits KCNN4b- and KCNN4c-mediated 86Rb (K+ surrogate) efflux with an apparent inhibitory constant of 0.6 ± 0.1 and 7.8 ± 0.4 μM, respectively. We conclude that apical and basolateral KCNN4 K+ channels that regulate K+ and anion secretion are encoded by distinct isoforms in colonic epithelial cells.
Keywords: smooth muscle, epithelial cells, apical membrane, basolateral membrane, potassium secretion
clotrimazole (CLT, an antifungal inhibitor)-sensitive, Ca2+-activated intermediate conductance K+ (IK) channels, known as KCNN4, are present in the hematopoietic system and in organs that secrete fluids such as pancreas, lung, salivary glands, and the colon (4, 12, 19, 21, 31, 52). KCNN4 channels are important for a wide array of different cell functions. They are involved in regulating membrane potential and Ca2+ signaling in vascular smooth muscle, endothelial cells, lymphocytes, and macrophages (11, 18, 45, 51) and are important in maintaining cell volume as well as K+ and Cl− secretion in epithelial cells (21, 28, 53). Functional studies with patch-clamp studies and CLT-sensitive K+ conductance in mucosal sheets have provided evidence for the presence of KCNN4 channels in basolateral membranes of colonic epithelial cells (7, 10, 41, 42). In recent studies, we have demonstrated CLT-sensitive K+ secretion as evidence for the additional presence of KCNN4 channels in apical membranes (21). Furthermore, KCNN4 channel function has also been shown in both apical and basolateral membranes of the human bronchial (16HBE14o) and pancreatic duct adenocarcinoma epithelial cell lines (5, 53).
Several KCNN4 orthologs have been cloned from various species and tissues (20–22, 31, 36, 52, 54). Immunofluorescence studies have localized KCNN4-like proteins in both apical and basolateral membranes of intestinal epithelial cells (9, 14, 17, 21). However, it is not known whether both apical and basolateral membrane KCNN4 channels are encoded by the same or distinct KCNN4 transcripts. Since antibodies against the NH2-terminal end or the COOH-terminal region of KCNN4 localized KCNN4-like proteins in both apical and basolateral membranes, we predicted that if apical and basolateral membrane KCNN4 channels are encoded by distinct isoforms, these isoforms would likely manifest differences in the coding regions of KCNN4 cDNA (14, 21). Thus we designed primers flanking the coding sequence of KCNN4 cDNA for RT-PCR analyses of rat colonic mRNA. In this study we demonstrate that: 1) three distinct KCNN4 cDNAs are present in the rat colon, designated as KCNN4a, KCNN4b, and KCNN4c, that encode 425, 424, and 395 deduced amino acid proteins, respectively; 2) KCNN4a-specific transcripts are expressed in the smooth muscle, whereas KCNN4b and KCNN4c specific transcripts are expressed in epithelial cells; 3) KCNN4b and KCNN4c transcripts encode basolateral (40 kDa) and apical (37 kDa) membrane KCNN4 channels in colonic epithelial cells, respectively; 4) KCNN4c, but not KCNN4b, transcripts are transcriptionally regulated during cell differentiation in IEC-6 cells (a rat intestinal crypt epithelial cell line); 5) in vitro-expressed KCNN4c proteins requires chaperon protein for plasma membrane expression in oocytes; and 6) in vitro-expressed KCNN4b and KCNN4c exhibit TRAM-34 (KCNN4 channel inhibitor)-sensitive 86Rb efflux with apparent inhibitory constant (Ki) of 0.6 ± 0.1 and 7.8 ± 0.4 μM in oocytes, respectively. Our results demonstrate that multiple KCNN4 isoforms are expressed in apical and basolateral membranes of colonic epithelial cells to regulate different cell functions in physiological and pathophysiological conditions.
MATERIALS AND METHODS
Animals and membrane preparations.
Apical and basolateral membranes were prepared from isolated colonocytes and scraped mucosa, respectively, from both the proximal and distal colon of normal nonfasting male Sprague-Dawley rats (200–250 g). All the experimental protocols used in this study were approved by the Yale University Institutional Animal Care and Use Committees. Normal human colon tissue samples were obtained from Yale University tissue bank. Apical membranes were purified by the method of Stieger et al. (49), as described previously (38). Basolateral membranes were prepared from scraped mucosa by the sucrose density gradient centrifugation technique of Biber et al. (6), as described previously (39).
Isolation of smooth muscle.
Hand-dissected rat distal colonic circular and longitudinal smooth muscles that are devoid of serosal membrane, mucosal, and submucosal cells were used for RNA extraction.
Reverse transcriptase-polymerase chain reaction (amplification of KCNN4 isoforms).
Total RNA from scraped mucosa (distal colon, proximal colon, cecum, ileum, jejunum, duodenum, and stomach), distal colonic smooth muscle, and pre- (90%) and post- (10 days) confluent IEC-6 cells were isolated using TRIzol reagent (Invitrogen, Carlsbad, CA). mRNA was purified from total RNA by using Oligotex Mini mRNA Kit (Qiagen, Valencia, CA). Reverse transcriptase-polymerase chain reaction (RT-PCR) was performed in a Thermal Cycler (Bio-Rad, Hercules, CA) using the OneStep RT-PCR kit (Qiagen). Full-length KCNN4 cDNAs were amplified from distal colon mRNA by using KCNN4-specific primers Set-I (see Table 1) that spans between 28 and 1450 bp in KCNN4 cDNA (accession no. AF156554) (21). In RT-PCR, following 30 min reverse transcription reaction at 50°C, the reaction mixture was heated for 15 min at 95°C to activate HotStarTaq DNA Polymerase and to inactivate the reverse transcriptases. The PCR reaction was then performed for 30 cycles at 95°C for 30 s, 58°C for 30 s, and 72°C for 3 min and final extension at 72°C for 10 min.
Table 1.
Primers used to amplify the full-length and specific fragments of KCNN4a, KCNN4b, and KCNN4c splice variants
| Primer Set | Primer Sequence | Primer Location in KCNN4, (Accession No. F156554) | Isoform | Expected Size cDNA Fragment, bp |
|---|---|---|---|---|
| I | Sense: 5′-CCCAGGGTGAGCAGAAACA-3′ | 28-45 | KCNN4a | 2054 |
| Antisense 5′-GTCCTGAGTAGAATGTAGATCGA-3′ | 1450-1428 | KCNN4b | 1423 | |
| KCNN4c | 1336 | |||
| II | Sense: 5′-GAGCTGGTGACTGGCCT-3′ | 153-168 | KCNN4a | 263 |
| Antisense 5′-CGTTGTCAGTCATGAACAGCT-3′ | 416-396 | KCNN4b | 263 | |
| KCNN4c | 176 | |||
| III | Sense: 5′-CACCTTGCCCTGGAGAAGA-3′ | 1301-1319 | KCNN4a | 780 |
| Antisense: 5′-GTCCTGAGTAGAATGTAGATCGA-3′ | 1450-1428 | KCNN4b | 149 | |
| KCNN4c | 149 | |||
| IV | Sense: 5′-CACCTTGCCCTGGAGAAGA-3′ | 1301-1319 | KCNN4a | None |
| Antisense: 5′-CCACTTGTGTCCTGGCATCT-5′ | 1561-1542 | KCNN4b | 260 | |
| KCNN4c | 260 |
cDNA fragments that span between 153 and 416 bp, 1301 and 1450, and 1301 and 1428 of KCNN4 were RT-PCR amplified by using rat distal colon mRNA and primer Set-II, Set-III and Set-IV, respectively (Table 1 and Fig. 1) (accession no. AF156554) (21). For RT-PCR, following 30 min reverse transcription reaction at 50°C, the reaction mixture was heated for 15 min at 95°C to activate HotStarTaq DNA Polymerase and to inactivate the reverse transcriptases. Then the PCR reaction was performed for 30 cycles at 95°C for 45 s, 59°C for 45 s, and 72°C for 1 min and final extension at 72°C for 10 min. Rat GAPDH-specific fragment was amplified as internal control using commercial primers (New England Biolabs, Ipswich, MA). The veracity of the RT-PCR products subcloned into TOPO cloning vector (Invitrogen) was confirmed by sequencing (Keck Biotechnology Resource, Yale University, New Haven, CT). Results presented represent RT-PCR analysis performed with mRNA obtained from a single animal. All RT-PCR analyses were repeated at least five times with mRNA obtained from different animals.
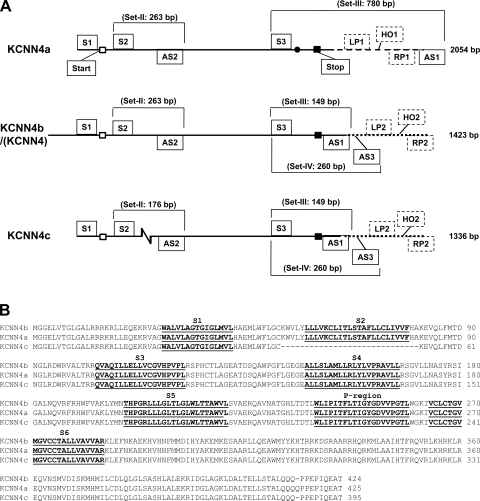
Fig. 1.
Diagrammatic alignment of cDNA sequence and primary structures of deduced amino acid sequence alignment of KCNN4a, KCNN4b/(KCNN4), and KCNN4c isoforms. A: full-length KCNN4a, KCNN4b/(KCNN4), and KCNN4c cDNAs were amplified using primer set-I. The number of base pairs (bp) amplified is given on the right. An additional codon is marked by the closed circle in the open reading frame of KCNN4a. The zigzag break denotes 87 bp deletions in KCNN4c. Open and closed squares, start and stop codons, respectively. Dashed lines, 3′-untranslated region of KCNN4a. Dotted lines, untranslated regions of KCNN4b and KCNN4c added to mark the real-time quantitative PCR (RTQ-PCR) primers. Reverse transcriptase-PCR (RT-PCR) primers (solid boxes): Sense (S) and antisense (AS) primers used to amplify full-length coding sequences of KCNN4 (KCNN4a, KCNN4b, and KCNN4c) isoforms, and isoform-specific cDNA fragments were designed from published rat KCNN4 cDNA sequence (accession no. AF156554) (21). Primer set-I (S1/AS1) that spans between 28 and 1450 bp of KCNN4 cDNA-amplified 2054 bp (KCNN4a), 1423 bp (KCNN4b), and 1336 bp (KCNN4c); set-II (S2/AS2) that spans between 153 and 416 bp KCNN4 cDNA amplified 263 bp (KCNN4a/b) and 176 bp (KCNN4c) fragments; set-III (S3/AS1) that spans between 1301 and 1450 bp of KCNN4 cDNA amplified 780 bp (KCNN4a) and 149 bp (KCNN4b/c) fragments; and set-IV (S3/AS3) that spans between 1301 and 1542 bp of KCNN4 cDNA amplified 260 bp (KCNN4b/c) fragment. The primer set and fragment sizes (bp) amplified are marked on each isoform. RTQ-PCR primers (perforated boxes): left (LP1) and right (RP1) primers that span between 1408 and 1494 and hybridizing oligo (HO1) that span between 1444 and 1468 of KCNN4a (accession no. EU872449) were used to quantify KCNN4a-specific transcripts. Left (LP2) and right (RP2) primers that span between 1478 and 1564, and hybridizing oligo that span between 1515 and 1538 of KCNN4 (accession no. AF156554) were used to quantify KCNN4b/KCNN4c-specific transcripts. B: amino acid sequences of KCNN4a, KCNN4b, and KCNN4c isoforms were aligned with the computer program PILEUP (GCG) by using default parameters. Gaps are represented by dashes. Dark lines indicate putative membrane spanning domains (MSD) (S1-S6) in addition to the pore-forming region (P-region). The entire second exon that encodes 29 amino acids (K53-A81) including the second MSD plus five exofacial and two endofacial amino acids in KCNN4a and KCNN4b is spliced out in KCNN4c. The National Center for Biotechnology Information (NCBI) accession numbers for the nucleotide sequences of KCNN4a, KCNN4b and KCNN4c are EU872449, EU872450, and EU872451, respectively.
Real-time quantitative PCR.
Real-time quantitative PCR (RTQ-PCR) analyses were performed by a two-step method using total RNA from colonic smooth muscle and mucosa of rat distal colon and 10-day postconfluent IEC-6 cells. In brief, first-strand cDNA was synthesized from total-RNA using SuperScript III and random hexamers (Invitrogen). The first-strand cDNA template (5 ng) and TaqMan Universal PCR master mix along with custom-designed KCNN4a-specific (sense 5′-TTGGTCTCTGTGTCCCTGTG-3′; antisense 5′-TGTCCAGAGATGGGAAGACA-3′) and KCNN4b/c-specific (sense 5′-GGCCACATAGCTGCCTGTTA; antisense 5′-TCCTTGAGCTCAGTCCTTCG-3′) primers (500 nM each) and 100 nM TaqMan probes (100 nM) (KCNN4a: 5′-FAM-TACACTCCTTTCCTGGGCTTTGGCA-TAMRA-3′ and KCNN4b/c: 5′-FAM-TCAGGACCCACAGAAGAATCAGGCT-TAMRA-3′) were used for RT-QPCR according to the manufacturer's instructions (Applied Biosystems, Foster City, CA). Rat β-actin amplified using TaqMan Gene Expression Assay ID Rn00667869_ml from Applied Biosystems served as an endogenous control. Threshold cycle (Ct) values of KCNN4a, KCNN4b and KCNN4c transcripts were normalized to the endogenous control. Differential expression of KCNN4a, KCNN4b/c was calculated according to the 2−ΔΔCt method (30). To ensure identical PCR efficiency between different amplicons, RTQ-PCR analysis was performed by using serially diluted cDNA to demonstrate that the efficiency of amplification of targets and the reference are equal. This study does not distinguish abundance between KCNN4b- and KCNN4c-specific transcripts, as specific primers could not be designed. RTQ-PCR analyses were performed with six different RNA preparations from different rat colonic tissues and IEC-6 cells.
Generation of polyclonal antibody to KCNN4 (anti-KCNN4-abc).
Anti-KCNN4-abc polyclonal antibody was custom produced by Antibody Solutions (Mountain View, CA) against KHTRRKDSRAARRHQRK, a peptide derived from rat colonic KCNN4-deduced amino acids 325–341 (accession no. AF156554) (21). A New Zealand White rabbit was inoculated with a keyhole limpet hemocyanin peptide conjugate mixed with an equal volume of Freund's adjuvant. The animal was injected in three subcutaneous dorsal sites with a total of 0.1 mg per injection. The injection protocol was repeated at weeks 2, 6, and 8. After 10 wk, terminal bleeds were taken. The serum was collected from clotted blood by centrifugation, and an ELISA was used to assess anti-peptide serum titer. The serum was then affinity purified over an immobilized peptide column.
Confocal laser scanning microscopy.
Rat distal colonic segments fixed in phosphate-buffered saline (PBS) containing 4% (vol/vol) paraformaldehyde plus 0.01% (vol/vol) Triton X-100 (PBS-T) were permeablized with 0.2% Triton X-100 and blocked in 0.5% (wt/vol) BSA in PBS-T. To test for the presence of KCNN4 and zonula occludens-1 (ZO-1), parallel samples were labeled with polyclonal anti-KCNN4-abc (1:50 dilution) and monoclonal ZO-1 (1:100 dilution), respectively (48). Bound antibodies were detected with secondary AlexaFluor-546 conjugates (Invitrogen) at 1:500 dilutions. Colons were mounted in vectashield with DAPI (Vector, Burlingame, CA), and image stacks with an axial spacing of 1 μm were collected on a Zeiss LSM 510 confocal laser-scanning microscope.
Immunoelectron microscopy.
Rat distal and normal human colon were fixed in 4% paraformaldehyde in 0.25 M Na-HEPES, pH 7.4, for 1 h, followed by overnight fixation in 8% paraformaldehyde at 4°C. Samples were stained by adding a few drops of a 1% toluidine blue (vol/vol) and 1% borate solution (wt/vol), rinsed several times in PBS to remove excess stain, and embedded in 10% bovine gelatin in PBS (wt/vol). Blocks of gelatin containing colonic segments were infiltrated with 2.3 M sucrose in PBS and frozen in liquid nitrogen. Frozen 75-nm sections were cut on a Leica UltraCut ultramicrotome with a FCS cryoattachment at −180°C and collected on formvar- and carbon-coated nickel grids using a 1:1 mixture of 2% methyl cellulose and 2.3 M sucrose in PBS (29). After free aldehydes had been quenched with 0.1 M NH4Cl, the grids were incubated in 1% fish skin gelatin in PBS (PBS-FSG) (wt/vol), followed by sequential staining with rabbit polyclonal anti-KCNN4-abc antibody (1:20), goat anti-rabbit IgG (1:100), and 10-nm Protein A-gold (Dept. of Cell Biology, Utrecht University, The Netherlands) in PBS-FSG. After intermediate fixation (1% glutaraldehyde for 5 min) and quenching, the sections were exposed sequentially to rabbit anti-KCNN4-abc (1:50) and 15-nm Protein A-gold conjugate. The sections were then fixed with 1% glutaraldehyde, incubated with a mixture of 1.8% methyl cellulose and 0.5% uranyl acetate, air-dried, and visualized in a Tecnai 12 Biotwin electron microscope. Images were acquired either at ×400 primary magnification (cryosemithin section, bar = 20 μm) by light microscopy or at ×21,000 primary magnification (cryothin sections, bar = 200 nm) by electron microscopy.
Oocyte expression.
PCR-amplified KCNN4b, KCNN4c, and large conductance K+ (BK) β1-subunit cDNAs subcloned into pGH19 vector were linearized with NotI. Capped cRNAs were prepared from these templates by using mMESSAGE mMACHINE T7 Kit (Ambion, Austin, TX). cRNAs transcribed from respective plasmid were dissolved in PBS at a final concentration of 0.4 μg/μl. Defolliculated Xenopus oocytes (stage V–VI) (NASCO, Fort Atkinson, WI) were injected with 50 nl of cRNA using a motor-driven injector (Drummond, PA). The cRNA-injected oocytes were incubated in Barth's medium [in mM: 84 NaCl, 1 KCl, 2.4 NaHCO3, 0.82 MgSO4, 0.33 Ca(NO3)2, 0.41 CaCl2, and 7.5 Tris·HCl (pH 7.4)] supplemented with penicillin G (100 U/ml) and streptomycin (100 μg/ml) at 18°C. Control oocytes were injected with 50 nl PBS alone. Three to five days after PBS/cRNA injection, the oocytes were used for immunofluoresence and 86Rb efflux studies.
Indirect immunofluorescence microscopy.
Cryostat sections of frozen oocytes were processed for indirect immunofluorescence microscopy. Sections incubated overnight with anti-KCNN4-abc antibody (1:20) and washed with PBS (5 min × 2) were incubated with FITC-conjugated antibodies for 10 min and washed with PBS (5 min × 2). Specimens dehydrated in 95% ethanol were mounted with Mowiol 4.88 (Hoechst, Frankfurt, Germany) containing p-phenylenediamine (1 mg/ml). Fluorescence images were acquired using a ×20 lens on a standard Nikon light microscope.
86Rb efflux.
The PBS/cRNA-injected oocytes were incubated overnight in medium containing 86Rb (1 μCi/ml Barth's medium). After overnight incubation, oocytes washed (PBS × 3) free of external 86Rb were placed in microcentrifuge tubes (6 oocytes/tube). Efflux studies were initiated by adding 100 μl K+-free Barth's medium [in mM: 84 NaCl, 1 NMG-Cl, 2.4 NaHCO3, 0.82 MgSO4, 0.33 Ca(NO3)2, 0.41 CaCl2, and 7.5 Tris·HCl (pH 7.4)]. 86Rb efflux was stopped at different times by adding ice-cold PBS (500 μl). Oocytes were washed three times with ice-cold PBS and placed individually in scintillation vials and dissolved in 100 μl 1 N NaOH. Retained 86Rb was measured by using a scintillation counter. For control studies (0 time), washed oocytes were directly placed in vials and processed for radioactive counting. At 0 time, PBS and BK β1 cRNA-injected oocytes retained 690 ± 43 and 701 ± 38 cpm/oocyte, respectively, whereas KCNN4b- and KCNN4c/BKβ1-expressing oocytes retained 839 ± 106 and 872 ± 96 cpm/oocyte, respectively. Results are presented as means ± SE of percent maximum from six oocytes.
Western blot.
Isolated colonocytes from rat proximal and distal colon and IEC-6 cells were resuspended (1:20) in ice-cold lysis buffer [50 mM Tris, pH 8.0, 0.5% SDS, 1 mM PMSF, 4 μg/ml Pepstatin A, and 1 tablet of Complete protease inhibitor/50 ml solution (Roche Applied Science, Indianapolis, IN)]. After homogenization with a tight-fit Teflon homogenizer, the homogenate was centrifuged for 15 min at 2,000 g, and 16-μl supernatants were mixed with equal volume of Laemmli buffer heated for 5 min at 95°C. The heated aliquots were immediately placed in liquid nitrogen and stored at −80°C. Frozen samples were heated at 40°C for 1–2 min and 10-μl samples (∼20 μg proteins) were loaded. The proteins were separated by two-phase Tricine polyacrylamide gel electrophoresis (10%T/6%C resolving layer, 4%T/3%C stacking layer) and transferred onto nitrocellulose membrane (Hybond ECL, Amersham Pharmacia Biotech, Piscataway, NJ) in buffer containing 10 mM 3′-(cyclohexylamino)-1-propanesulfonic acid (adjusted to pH 11) and 10% methanol. The blot was blocked overnight in TBST with 5% fat-free milk powder at 4°C. After blocking was completed, the blot was incubated for 2 h at room temperature in blocking buffer containing primary antibody [anti-KCNN4-abc (1:3,000) or anti-β-actin (1:2,500)], washed three times with TBST containing 5% milk powder, incubated for 1 h at room temperature in blocking buffer containing a 1:5,000 dilution of horseradish peroxidase-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch, West Grove, PA), and again washed three times with TBST. Immune complexes were detected on film by using enhanced chemiluminescence (Amersham Pharmacia Biotech, Piscataway, NJ). Immunoblots were also performed on colonic homogenate treated with deglycosylase (Enzymatic Protein Deglycosylation kit; Sigma, St. Louis, MO) and phosphatase (Calf Intestinal alkaline phosphatase; Sigma) enzymes.
Statistics.
Results are presented as means ± SE. Statistical analyses were performed using paired t-test and Bonferroni's one-way ANOVA post hoc test. P < 0.05 was considered to be statistically significant.
RESULTS
RT-PCR analyses of rat colonic mRNA were performed using primers set-I (Table 1) flanking the coding sequence of KCNN4 that was expected to amplify a single cDNA of ∼1.4 kb. Interestingly, analyses of the RT-PCR products indicated the presence of three cDNAs of ∼2.0, 1.4, and 1.3 kb. Sequence analyses revealed that all these three cDNAs are highly homologous to KCNN4 (Fig. 1A). Therefore, we designated these 2.0-, 1.4-, and 1.3-kb cDNAs as KCNN4a, KCNN4b, and KCNN4c, respectively (Fig. 1A). The KCNN4b nucleotide sequence is identical to the rat colonic KCNN4 sequence (also known as rIK1; comparison not shown) (21). KCNN4c differs from KCNN4b by a deletion of the entire second exon that consists of 87 bp (Fig. 1A). KCNN4a differs from KCNN4b by the presence of an additional codon in the open reading frame at position 1362 and addition of a 628-bp fragment at the 3′-untranslated region (Fig. 1A). Similar overlapping cDNAs were also isolated from a rat colonic cDNA library (Invitogen) (data not shown). These sequences were not compared with other published KCNN4 sequences as they reported only coding sequences (36, 52, 54).
The deduced amino acid sequences of KCNN4a, KCNN4b, and KCNN4c cDNAs are compared in Fig. 1B. KCNN4a, KCNN4b, and KCNN4c cDNAs encode 425, 424, and 395 deduced amino acid proteins, respectively (Fig. 1B). Amino acid comparisons indicate that KCNN4b is identical to KCNN4 (also known as rIK1) (comparison not shown) (21). As shown in Fig. 1B, KCNN4a differs from KCNN4b by the addition of an extra amino acid at position 415, whereas KCNN4c differs from KCNN4b by the deletion of a 29 amino acid motif that is encoded by the second exon. This 29 amino acid motif contains the entire predicted second membrane spanning domain (MSD) (22 amino acids) plus five exofacial and two endofacial amino acids in KCNN4a and KCNN4b isoforms.
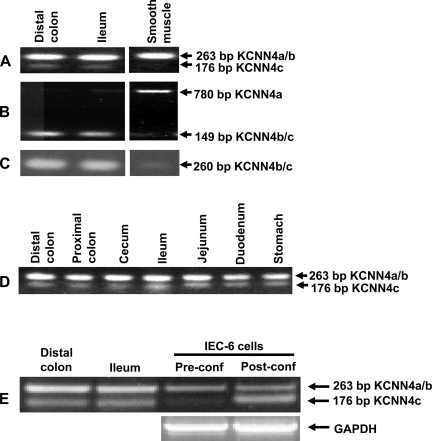
To establish that KCNN4a, KCNN4b, and KCNN4c transcripts are truly expressed and do not represent RT-PCR artifacts, additional RT-PCR analyses were performed by amplifying smaller fragments. In these studies, the primer set-II, set-III, and set-IV were employed to amplify KCNN4a-, KCNN4b-, and KCNN4c-specific fragments (Table 1; Figs. 1A and 2). The primer set-II should amplify a 263-bp fragment from KCNN4a and/or KCNN4b (KCNN4a/b), whereas it should amplify only a 176-bp fragment from KCNN4c transcripts (Fig. 2, A, D, and E). The primer set-III should amplify a 780-bp fragment from KCNN4a, whereas it should amplify only a 149-bp fragment from KCNN4b and/or KCNN4c (KCNN4b/c) transcripts (Fig. 2B). The primer set-IV should amplify only a 260-bp fragment from KCNN4b/c but not from KCNN4a transcripts (Fig. 2C). Amplification of 263-, 176-, 149-, and 260-bp fragments suggest that both KCNN4b- and KCNN4c-, but not KCNN4a-, specific transcripts are expressed in mucosal cells from the rat ileum and distal colon (Fig. 2, A–C). Since SMIK1 cDNA that encodes 425 amino acids (i.e., similar to KCNN4a) was cloned from vascular smooth muscle (36), we also examined whether KCNN4a-, KCNN4b-, and KCNN4c-specific transcripts are expressed in colonic smooth muscle cells. The strong amplification of 263 and 780-bp fragments, and the absence of the amplification of 176-, 149-, and 260-bp fragments suggest that KCNN4a, but not KCNN4b and KCNN4c, is the predominant transcript expressed in colonic smooth muscle (Fig. 2, A–C). Both KCNN4b- and KCNN4c-specific transcripts are also expressed in mucosal cells of the stomach, duodenum, jejunum, ileum and cecum (Fig. 2D). The expression of KCNN4b- and KCNN4c-specific transcripts is consistent with the demonstration that KCNN4-like proteins are present in both apical and basolateral membranes of epithelial cells of different parts of the digestive tract (14).
Fig. 2.
Expression of KCNN4a, KCNN4b, and KCNN4c isoform-specific transcripts in smooth muscle and epithelial cells. KCNN4a, KCNN4b, and KCNN4c-specific mRNA fragments were amplified by RT-PCR analyses as described in materials and methods. A: RT-PCR amplification of 263 and 176 bp fragments by primer set-II (see Table 1 and legend to Fig. 1A) suggest the expression of KCNN4a/b- and KCNN4c-specific transcripts in distal colon and ileal epithelial cells, respectively. Amplification of 263 bp, but not 176 bp, fragment suggests that only KCNN4a/b-specific transcripts are expressed in colonic smooth muscle cells. B: amplification of 780 and 149 bp fragments by primer set-III (see Table 1 and legend to Fig. 1A) indicate the expression of KCNN4a- and KCNN4b/c-specific transcripts in colonic smooth muscle and epithelial cells, respectively. C: amplification of 260-bp fragment by primer set-IV (see Table 1 and legend to Fig. 1A) indicates the expression of KCNN4b/c-specific transcripts in colonic epithelial but not in smooth muscle cells. D: the 263- and 176-bp fragments amplified by primer set-II are present in epithelial cells from all intestinal segments. D: the 263- and 176-bp fragments amplified by primer set-II are present in IEC-6 cells. Amplification of rat GAPDH specific segment was used as internal control (E).
The traces of the 780-bp fragment amplified in ileal and colonic mucosa could be a cross contamination from nonepithelial cells, as the RNA used in this study was isolated from scraped mucosa (Fig. 2B). Therefore, to establish that KCNN4b- and KCNN4c-, but not KCNN4a, specific transcripts are the epithelial-cell-specific ones, RT-PCR analyses were performed in mRNA isolated from IEC-6 cells (Fig. 2E). Similar to rat ileal mucosa, the amplification of 263- and 176-bp fragments indicate that both KCNN4b- and KCNN4c-specific transcripts are expressed in differentiated (i.e., postconfluent) IEC-6 cells (Fig. 2E). It should be emphasized that although equal amounts of internal control (GAPDH) and 263-bp fragments are amplified from both pre- and postconfluent IEC-6 cells, the amplification of the 176-bp fragment is virtually absent in preconfluent cells (Fig. 2E). In contrast to ileal and colonic mucosa, the KCNN4a-transcript-specific fragment (i.e., 780-bp fragment amplified by primer set-III) is not amplified in IEC-6 cells (data not shown). These observations 1) establish that amplified KCNN4a-, KCNN4b-, and KCNN4c-specific cDNAs do not represent RT-PCR artifacts but are truly expressed; 2) indicate that KCNN4a may be smooth muscle specific, whereas KCNN4b and KCNN4c are epithelial cell-specific isoforms; and 3) suggest that KCNN4c is transcriptionally and/or posttrancriptionally regulated during cell differentiation.
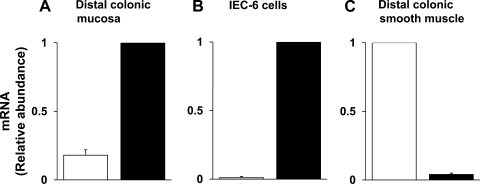
RTQ-PCR analyses were performed to establish the relative abundance of KCNN4a- and KCNN4b/c-specific transcripts in distal colonic mucosal cells, IEC-6 cells, and smooth muscle cells. As shown in Fig. 3A, although both KCNN4a- and KCNN4b/c-specific transcripts are expressed, the relative abundance of KCNN4b/c transcripts is 5.6-fold higher than that of KCNN4a transcripts in mucosa of rat distal colon. In contrast, only KCNN4b/c-, but not KCNN4a-, specific transcripts are expressed in IEC-6 cells (Fig. 3B), whereas only KCNN4a-, but not KCNN4b/c-, specific transcripts are expressed in colonic smooth muscle (Fig. 3C). These observations establish that KCNN4a transcripts are predominantly expressed in smooth muscle, whereas KCNN4b/c transcripts are present in ileal and colonic epithelial cells. Further studies, therefore, focused to characterize KCNN4b and KCNN4c transcripts.
Fig. 3.
RTQ-PCR analyses of KCNN4a- and KCNN4b/c-specific mRNA. RT-QPCR analyses were performed as described in materials and methods. A: KCNN4a- (open bars) and KCNN4b/c (closed bars)-specific mRNAs are present in 1:5 ratios in distal colonic epithelial cells. B: KCNN4b/c-, but not KCNN4a-, specific mRNA is expressed in IEC-6 cells. C: KCNN4a-, but not KCNN4b/c-, specific mRNA is present in colonic smooth muscle.
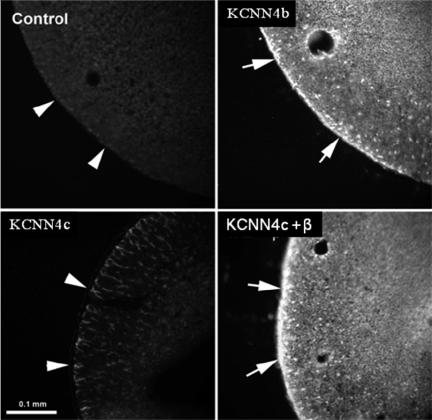
To examine whether KCNN4c that lack second MSD would express functional proteins, KCNN4c cDNA was transiently expressed in HEK293 cells. In patch-clamp studies, a few inside-out patches obtained from HEK293 cells transfected with KCNN4c cDNA exhibited channel activity with a conductance of 16 pS, but due to the low density of channels and patch instability, the Ca2+ dependence of the currents could not be determined (data not shown). Therefore, oocyte expression system was used to characterize KCNN4b and KCNN4c functions. In initial studies, immunofluorescence studies were performed to identify the plasma membrane expression of KCNN4 proteins in KCNN4 isoform-specific cRNA-injected oocytes. Anti-KCNN4-abc antibody detected KCNN4 proteins on the plasma membranes of KCNN4b cRNA-injected oocytes (Fig. 4; KCNN4b). KCNN4 proteins are not present in PBS-injected oocytes (Fig. 4; control). This observation establishes that anti-KCNN4-abc antibody recognizes KCNN4 proteins and that KCNN4 protein are not endogenously expressed in oocytes. In contrast to KCNN4b cRNA-injected oocytes, KCNN4 proteins are predominantly localized only in the cytoplasm of KCNN4c cRNA-injected oocytes (Fig. 4, KCNN4c). This observation suggests that KCNN4c proteins, which lack the second MSD (i.e., a 29-amino acid motif deletion), might require either physiological stimulation of K+ channel (i.e., K+ secretion) or a chaperone for plasma membrane delivery.
Fig. 4.
Indirect immunofluorescence imaging of KCNN4 protein in cRNA-injected oocytes. Anti-KCNN4-abc antibody localizes KCNN4 proteins on the plasma membrane of KCNN4b cRNA-injected oocytes (KCNN4b). KCNN4 protein is localized only in the cytoplasm of KCNN4c cRNA-injected oocytes (KCNN4c). Plasma membrane targeting of KCNN4c protein is substantially enhanced in oocytes coinjected with KCNN4c plus BKβ1-subunit cRNAs (KCNN4c + β). No KCNN4-specific fluorescence is detectable in water-injected oocytes (Control). Plasma membranes are indicated by arrowheads. Fluorescence images were acquired using a ×20 lens on a standard Nikon light microscope. Bar = 0.1 mm.
Incubation of KCNN4c cRNA-injected oocytes in K+-free medium that would stimulate K+ secretion did not enhance the plasma membrane expression of KCNN4c proteins, suggesting that plasma membrane expression of KCNN4c proteins may require chaperone protein(s) (data not shown). Since β-subunits served as chaperones for plasma membrane delivery of H-K-ATPase and Na-K-ATPase α-subunits, and BK channels with seven membrane-spanning domains have a β-subunit, the effect of coinjection of β-subunit cRNAs was examined on KCNN4c protein expression in oocytes (15, 33–35, 43, 44). Coinjection of KCNN4c and rat BK β1-subunit cRNAs enhanced the KCNN4 protein expression on the plasma membranes of oocytes (Fig. 4, KCNN4c + β). Coinjection of KCNN4c with H-K-ATPase and Na-K-ATPase β-subunits did not enhance plasma membrane expression of KCNN4c proteins in oocytes (data not shown). KCNN4-specific proteins were also not detected in oocytes injected with cRNA injected with BK β-subunit alone (data not shown). These observations indicate that KCNN4c proteins that lack the second MSD requires chaperone proteins for their plasma membrane expression.
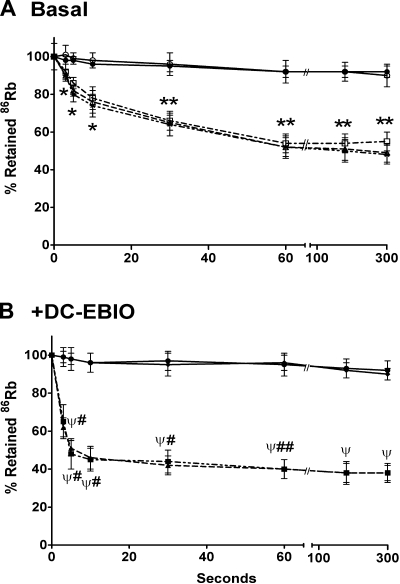
To identify whether the expressed KCNN4b and KCNN4c proteins exhibit K+ channel functions, 86Rb efflux studies were performed in oocytes injected with KCNN4b and KCNN4c plus BK β1-subunit (KCNN4c+β) cRNAs, respectively. As shown in Fig. 5A, both KCNN4b and KCNN4c+β cRNA-injected oocytes exhibit 86Rb effluxes that reach equilibrium at 60 s. To identify whether the 86Rb effluxes occur via KCNN4 channels, the effect of 100 μM 5,6-dichloro-1-ethyl-1,3-dihydro-2H-benzimidazole-2-one (DC-EBIO; a KCNN4 channel opener) was examined on 86Rb effluxes in KCNN4b and KCNN4c+β cRNA-injected oocytes (Fig. 5B) (47). The presence of DC-EBIO in the incubation medium significantly enhanced the 86Rb effluxes compared with that under basal condition and reached equilibrium within 10 s in both KCNN4b and KCNN4c+β-injected oocytes (Fig. 5B). The small conductance K+ channel variant SK3–1C, which does not produce functional proteins, has been shown to suppress SK1, SK2, and SK3 channels by intracellular sequestration of their proteins (26). Therefore, to identify whether KCNN4c proteins modulate KCNN4b function, 86Rb effluxes were measured in oocytes coinjected with KCNN4b plus KCNN4c (KCNN4b+c) cRNAs. As shown in Fig. 5A, the rates of 86Rb effluxes in KCNN4b+c cRNA-coinjected oocytes were identical to that of KCNN4b and KCNN4c+β cRNA-injected oocytes. Both PBS-injected and BKβ1 cRNA-injected oocytes exhibit minimal rate of 86Rb efflux in the presence and absence of DC-EBIO (Fig. 5, A and B). These observations suggest that KCNN4c does not modulate KCNN4b function and that in vitro-expressed KCNN4b and KCNN4c isoforms exhibit KCNN4 K+ channel function.
Fig. 5.
Time course of 86Rb efflux in KCNN4 isoform-specific cRNA-injected oocytes. 86Rb loading and effluxes were performed as described in materials and methods. A: basal 86Rb effluxes were measured by placing the oocytes in K+-free medium. Under basal conditions, oocytes injected with cRNA for KCNN4b (open triangles), KCNN4c + BKβ1 (open squares), and KCNN4b + KCNN4c (stars) exhibited a low but significant rate of 86Rb effluxes and reached equilibrium at 1 min. 86Rb effluxes were very low in PBS- (open circles) and cRNA of BKβ1- (closed circles) injected oocytes. *P < 0.05 compared with respective time point in PBS/BKβ1-injected oocytes; **P < 0.001 compared with respective time point in PBS-injected oocytes. B: 5,6-dichloro-1-ethyl-1,3-dihydro-2H-benzimidazole-2-one (DC-EBIO)-enhanced 86Rb effluxes were measured by placing the oocytes in K+-free medium containing 100 μM DC-EBIO. The presence of DC-EBIO greatly enhanced the initial rate of 86Rb effluxes in both KCNN4b (closed squares) and KCNN4c + BKβ1 (closed triangles) cRNA-injected oocytes and reached equilibrium within 1 min. 86Rb effluxes were not significantly altered by DC-EBIO in PBS (closed circles) and cRNA of BKβ1 (closed diamonds) injected oocytes. ΨP < 0.001 compared with respective time point in PBS/BKβ1-injected oocytes; #P < 0.001 compared with respective time point in Fig. 6A; ##P < 0.05 compared with respective time point in Fig. 6A.
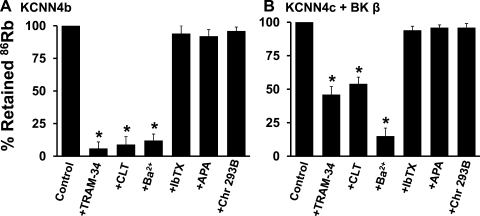
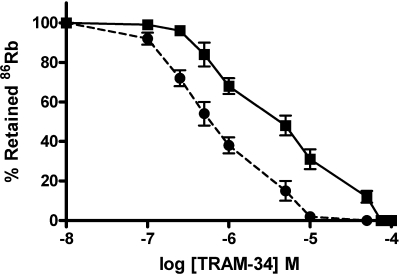
To establish that KCNN4b and KCNN4c exhibit KCNN4 K+ channel function, the effect of K+ channel blockers were examined on 86Rb effluxes in KCNN4b- and KCNN4c+β-specific cRNA-injected oocytes, respectively (Fig. 6). Since DC-EBIO-enhanced KCNN4 K+ channels have been shown resistant to TRAM-34, the effect of K+ channel blockers were examined on basal 86Rb effluxes (17). The initial rate of basal 86Rb efflux was linear up to 5 s in both KCNN4b- and KCNN4c+β cRNA-injected oocytes. Therefore, the inhibition characteristics of KCNN4b and KCNN4c+β-mediated 86Rb effluxes were performed at 5 s. Initial rate of 86Rb efflux was significantly inhibited by KCNN4 channel-specific blockers TRAM-34 (94%) and CLT (91%) and nonspecific K+ channel blocker Ba2+ (88%) in KCNN4b cRNA-injected oocytes (Fig. 6A). TRAM-34, CLT, and Ba2+ also significantly inhibited the initial rate of 86Rb efflux by 54%, 46%, and 85% in KCNN4c cRNA-injected oocytes, respectively (Fig. 6B). Although the magnitude of inhibition of 86Rb effluxes by Ba2+ is similar in both KCNN4b- and KCNN4c cRNA-injected oocytes, the magnitude of inhibition of 86Rb efflux by TRAM-34 and CLT is substantially lower in KCNN4c cRNA-injected oocytes (Fig. 6, A and B). This observation suggests that KCNN4c may be resistant to CLT and TRAM-34. Thus the inhibition kinetics of TRAM-34 was examined on both KCNN4b and KCNN4c+β cRNA-injected oocytes. As shown in Fig. 7, increasing TRAM-34 concentrations progressively inhibited the initial rate of 86Rb effluxes in both KCNN4b- and KCNN4c+β-specific cRNA-injected oocytes. Hill plot analyses of these data yielded apparent half-maximal Ki for TRAM-34 of 0.6 ± 0.1 and 7.8 ± 0.4 μM for KCNN4b and KCNN4c+β cRNA-injected oocytes, respectively. 86Rb effluxes were not inhibited by the BK channel blocker iberiotoxin, the SK channel blocker apamin, or the KCNQ channel blocker chromanol 293B in both KCNN4b and KCNN4c+β cRNA-injected oocytes (Fig. 6, A and B). These observations establish that KCNN4b and KCNN4c exhibit KCNN4 K+ channel function with distinct sensitivities to TRAM-34.
Fig. 6.
Effect of K+ channel blockers on initial rate of DC-EBIO-enhanced 86Rb efflux in KCNN4b- and KCNNc + BKβ1 cRNA-injected oocytes. 86Rb loading and effluxes were performed as described in materials and methods. 86Rb efflux was measured for 5 s by placing the KCNN4b (A) and KCNN4c + BKβ1 (B) cRNA-injected oocytes in K+-free medium and considered 100% (Control). 86Rb efflux was also measured in the presence of 5 μM TRAM-34, 1 μM clotrimazol (CLT), 5 mM Ba2+, 100 nM iberiotoxin (IbTX), 10 nM apamin (APA), and 50 μM chromanol 293B (Chr 293B). When Ba2+ was present, Na+ concentration was adjusted to maintain osmolarity. When compared with KCNN4 cRNA-injected oocytes, PBS and BKβ1 cRNA-injected oocytes exhibited <2% 86Rb efflux (data not shown). *P < 0.001 compared with control.
Fig. 7.
Effect of TRAM-34 concentrations on DC-EBIO-enhanced 86Rb efflux in KCNN4b (circles) and KCNNc + BK β1 (squares) cRNA-injected oocytes. 86Rb loading and effluxes were performed as described in materials and methods. 86Rb efflux was measured for 5 s by placing the oocytes in K+-free medium containing varying concentrations of TRAM-34 (0–100 μM). Hill plot analyses of the data yielded half-maximal inhibitory concentration (Ki) for TRAM-34 of ∼0.6 and 5 μM for KCNN4b- and KCNN4c-mediated 86Rb effluxes, respectively.
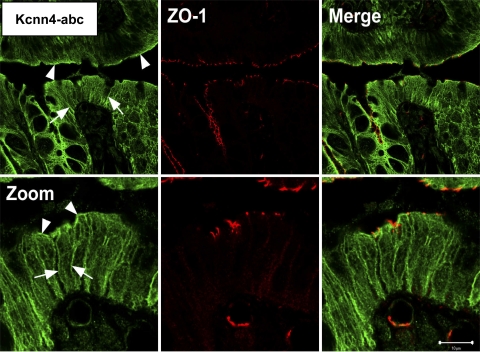
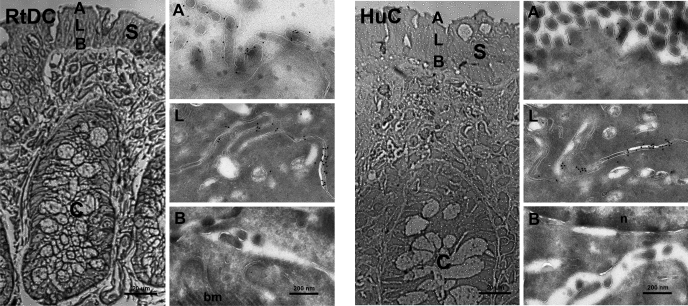
Immunological studies were performed by using the anti-KCNN4-abc antibody to identify the membrane-specific localization of KCNN4a and KCNN4b proteins in rat colonic epithelial cells. Similar to earlier observations, immunofluorescence studies performed with a confocal scanning microscope indicate that anti-KCNN4-abc antibody, which detects in vitro-expressed KCNN4 proteins in oocytes (Fig. 5), localized KCNN4-like proteins in both apical and lateral membranes in rat distal colon (Fig. 8) (14, 21). Immunogold labeling studies were performed with electron microscopy to confirm the observations with confocal microscopy that KCNN4-like proteins are present only in apical and lateral membranes of normal rat distal colon. As shown in Fig. 9, in rat distal colon the anti-KCNN4-abc antibody reacted with protein granules on the apical (Fig. 9; RtDC-A) and lateral (Fig. 9, RtDC-L) but not on basement (Fig. 9, RtDC B) membranes of rat distal colon. To identify whether apical and lateral expression of KCNN4-like proteins is unique only to the rat colon or common to other colonic epithelial cells, an immunogold labeling study was also performed in the human colon (Fig. 9, HuC). Similar to the rat colon, in human colonic epithelial cells anti-KCNN4-abc antibody also localized KCNN4-like protein granules on both the apical (Fig. 9, HuC-A) and lateral (Fig. 9, HuC-L) membranes but not on the basement (Fig. 9, HuC-B) membranes. It is to be noted here that a higher number of KCNN4-like protein granules are present in lateral membranes compared with that in apical membranes of both rat and human colonic epithelial cells (Figs. 9, RtDC and HuC). These observations establish that KCNN4-like proteins are present in both apical and lateral membranes of colonic epithelial cells.
Fig. 8.
Immunofluoresence localization of KCNN4-like proteins in normal rat distal colon. The anti-KCNN4-abc antibody recognizes the COOH-terminal region of KCNN4 proteins, which is common to KCNN4a, KCNN4b, and KCNN4c isoforms, localizes KCNN4-like proteins (green) in both apical and lateral membranes of both surface, and crypt epithelial cells in rat distal colon (left). Zonula occludens-1 (ZO-1) (red) counterstains for junctional complexes of epithelial cells using an anti-ZO1 monoclonal antibody, indicating the apical most end of the lateral membrane (middle). Merge (orange) shows overlay of both proteins (left). Arrow-heads indicate positive apical staining, whereas arrows indicate positive lateral membrane staining of anti-KCNN4-abc. Zoom = ×4; scale bar is 40 μm (top) and 10 μm (bottom). Similar observations were obtained with tissues from three rats.
Fig. 9.
Immunogold labeling of KCNN4 channels in apical (A), lateral (L), and basal (B) plasma membranes in cryothin sections of normal rat distal colon (RtDC) and normal human colon (HuC) of surface epithelium. Cryosemithin sections were prepared and trypan blue stained for orientation (left column of each panel). Tissue specimens were then ultratrimmed for cryothin sectioning focusing on small areas of interest in the surface epithelium to characterize the plasma membrane domain-specific localization of KCNN4 proteins on the ultrastructural level. Cryothin sections were immunolabeled with anti-KCNN4-abc and detected with a secondary donkey anti-rabbit 10-nm gold-labeled antibody. Anti-KCNN4-abc localized KCNN4-like proteins in A, and L plasma membrane domains of rat colonic epithelial cells, but not in B membranes of surface epithelial enterocytes sectioned. These high-resolution immunogold electron microscopy results are consistent with and confirm our observations made by confocal microscopy. S, surface epithelium; C, crypts; bm, basement membrane/collagen. Images were acquired either at ×400 primary magnification (cryosemithin section, bar = 20 μm) by light microscopy or at ×21,000 primary magnification (cryothin sections, bar = 200 nm) by electron microscopy. Similar results were obtained with two and three different human and rat tissues, respectively.
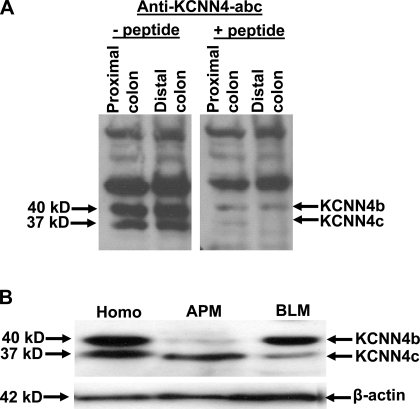
Western blot analyses were performed to identify whether the anti-KCNN4-abc antibody that localizes KCNN4-like proteins in apical and lateral membranes of colon would detect the same or different size proteins in colonic epithelial cells. Anti-KCNN4-abc antibody detects 37-, 40-, 50-, and 120-kDa proteins in epithelial cell homogenates of both proximal and distal colon (Fig. 10A, left). Incubation of anti-KCNN4-abc with 50-fold excess peptide before incubation blocked the detection of 37- and 40-kDa proteins but not 50- and 120-kDa proteins (Fig. 10A; right). Thus the 37- and 40-kDa, but not the 50- and 120-kDa, proteins detected by anti-KCNN4-abc antibody represent KCNN4-specific proteins. To establish whether the anti-KCNN4-abc antibody-specific 37- and 40-kDa proteins are present on the same or different membranes, Western blot analyses were performed on apical and basolateral membranes isolated from colonic epithelial cells of the rat distal colon (Fig. 10B). The anti-KCNN4-abc antibody that detects 37- and 40-kDa proteins in mucosal homogenate, recognizes 37- and 40-kDa proteins in apical and basolateral membranes of colonic epithelial cells (Fig. 10B). It is possible that the anti-KCNN4-abc-specific 37- and 40-kDa proteins could represent posttranslational modifications of the same transcript. However, immunoblot analyses of mucosal homogenate treated with deglycosylase and alkaline phosphatase argue against this possibility as either the mobility or the quantity of 40- and 37-kDa proteins is altered by these enzyme treatments (data not shown). These observations 1) indicate that KCNN4 channel proteins of apical (37 kDa) and basolateral (40 kDa) membranes of colonic epithelial cells are encoded by different transcripts; and 2) suggest that KCNN4b and KCNN4c transcripts may encode basolateral (40 kDa) and apical (37 kDa) KCNN4 channels, as 37- and 47-kDa proteins correspond to the expected size proteins for the deduced amino acid sequences of KCNN4c (395 amino acids) and KCNN4b (424 amino acids), respectively.
Fig. 10.
Detection of KCNN4b- and KCNN4c-specific proteins in apical and basolateral membranes of the rat distal colon. Western blot analyses were performed using anti-KCNN4-abc antibody as described in materials and methods. A: anti-KCNN4-abc polyclonal antibody detects 37, 40, 50, and 120 kDa proteins in proximal and distal colonic mucosal homogenate (left). Both 37- and 40-kDa proteins detected by the anti-KCNN4-abc antibody were eliminated when the antibody was preincubated with a 50-fold molar excess of competitor peptide corresponding to the epitope recognition sequence before blotting in both proximal and distal colon (right). B: anti-KCNN4-abc antibody detects 37- and 40-kDa proteins in colonic epithelial cell homogenate (Homo), whereas it detects 37- and 40kDa proteins in apical (APM) and basolateral (BLM) membranes, respectively.
DISCUSSION
The KCNN4 channels have been shown to regulate a variety of cell functions, such as volume regulation, cell proliferation, Ca2+ signaling, and the membrane potential that provides the driving force for Cl secretion, as well as transcellular K+ transport (1, 11, 18, 21, 28, 45, 50, 52, 53). KCNN4 channels are characterized as Ca2+-dependent, CLT-sensitive K+ currents with a conductance of 16 pS (16, 32). Ion flux studies and electrophysiological studies provided evidence for the presence of KCNN4 channels in both apical and basolateral membranes of colonic epithelial cells, respectively (16, 21). Basolateral KCNN4 channels are generally believed to maintain cell hyperpolarization to provide the driving force for cAMP- and Ca2+-induced Cl− secretions, whereas apical KCNN4 channels demonstrated to mediate Ca2+-induced K+ secretion (10, 21, 41). Immunofluorescence studies have confirmed the localization of KCNN4-like proteins in both apical and basolateral membranes of epithelial cells of intestine and colon (14, 17, 21). However, since only KCNN4 had been cloned, it is not known whether KCNN4 or other isoform(s) encode KCNN4 channels in apical and basolateral membranes of epithelial cells (21, 36, 52, 54).
The present study establishes that apical and basolateral KCNN4 channels are encoded by different transcripts in colonic epithelial cells. This conclusion is supported by the following observations: 1) two distinct KCNN4 splice variants KCNN4b and KCNN4c that encode 424 and 395 amino acid proteins are present in epithelial cells, respectively (Figs. 1–3); 2) in vitro-expressed KCNN4b and KCNN4c isoforms that exhibited KCNN4 K+ channel activities manifest differential sensitivities to TRAM-34, as KCNN4b- and KCNN4c-mediated 86Rb effluxes are inhibited by TRAM-34 with apparent half-maximal Ki of 0.6 ± 0.1 and 7.8 ± 0.4 μM, respectively (Figs. 6 and 7); and 3) the anti-KCNN4-abc antibody, which recognized in vitro-expressed KCNN4 proteins on the plasma membranes of oocytes and localized KCNN4-like proteins on both apical and basolateral membranes of the rat and human colon, detected 37- and 40-kDa proteins in apical and basolateral membrane isolated from the rat distal colon, respectively (Figs. 4, 8, 9, and 10). The 37- and 40-kDa proteins detected by anti-KCNN4-abc correspond to the expected size proteins for the deduced amino acid sequences of KCNN4c (395 amino acids) and KCNN4b (424 amino acids), respectively. Therefore, it is likely that the KCNN4 K+ channel of apical and basolateral membranes are encoded by KCNN4c and KCNN4b transcripts in rat colonic epithelial cells, respectively.
The KCNN4 channels have been shown to regulate different cell functions in epithelial and nonepithelial cells (9, 21, 28, 46, 53). The KCNN4 cloned from nonepithelial [erythropoid (MIK1) and smooth muscles (SMIK1)] and epithelial [colon (rIK1 and rSK4)] cells encode 425 and 424 amino acid proteins, respectively (21, 22, 36, 52, 54). Although these previous studies did not appreciate the single amino acid polymorphism present in epithelial and nonepithelial KCNN4, the present study provides compelling evidence that the KCNN4a and KCNN4b that encode 425 and 424 amino acid proteins, respectively, are distinct and separate transcripts. This conclusion is supported by the observations that KCNN4a and KCNN4b cDNAs consist of different 3′-untranslated regions and that the KCNN4a- and KCNN4b-specific transcripts exhibit tissue-specific expression patterns, as the former predominantly expressed in smooth muscle, whereas the latter present only in epithelial cells (Figs. 1–3). The KCNN4a and KCNN4b transcripts that exhibit tissue-specific expression patterns might be involved in regulating distinct cell functions, as the KCNN4a would regulate cell proliferation in smooth muscle, whereas KCNN4b would regulate agonist-induced K+ and anion secretion in epithelial cells (3, 50).
The KCNN4b and KCNN4c are distinct transcripts and are expressed only in epithelial cells (Fig. 3). KCNN4c differs from KCNN4b by the deletion of the second exon that encodes a 29 amino acid motif (Fig. 1). Since the deleted 29 amino acid consists of the predicted second MSD in KCNN4a and KCNN4b isoforms, the number of transmembrane helices and the membrane topology of KCNN4c with five predicted MSDs differ from KCNN4a and KCNN4b that have six MSDs. Although K+ channels and other transport proteins with both an even and odd number of MSDs can express functional proteins (8, 33, 40), proteins with an even number of MSD are considered the preferable topology (27). Therefore, it is not known whether KCNN4a/KCNN4b and KCNN4c isoforms have different membrane topology or the same membrane topology that differs from the predicted MSD, since MSD established by biochemical methods have been shown to differ from the predicted MSDs (2, 23–25).
Although KCNN4c-specific transcripts are expressed, it may not encode a functional plasma membrane protein. It is also possible that similar to the small conductance K+ channel splice variant SK3–1c, which modulates SK1, SK2, and SK3 channel function by sequestering their proteins in cytoplasm, KCNN4c might also modulate KCNN4 channel function in the same fashion in epithelial cells (26). However, the detection of a 37-kDa protein that is equivalent to the size of 395 deduced amino acids of KCNN4c suggests the expression of KCNN4c proteins on the apical membranes of the colon (Fig. 10). In vitro expression studies further support this conclusion, as anti-KCNN4-abc localized KCNN4 proteins on the plasma membranes of KCNN4c cRNA-injected oocytes and that the KCNN4c cRNA-injected oocytes exhibited KCNN4 K+ channels functions (Figs. 4–7). Although in vitro-expressed KCNN4b proteins readily localized on the plasma membranes of oocytes (Fig. 4, KCNN4b), the KCNN4c proteins required coexpression of BK β-subunit for its plasma membrane expression in oocytes (Fig. 4, KCNN4c+β). The BK β1-subunit, which functions as a Ca2+ sensor for BK channel (37), serves as a chaperone for plasma membrane expression of KCNN4c protein is a novel observation. This is the first study to demonstrate that BK β1-subunit interacts with KCNN4c protein that lacks the second transmembrane domain. Although BK β1-subunit-specific transcripts and proteins are expressed (Rajendran, VM, unpublished observations), it is not known whether KCNN4c proteins use BK β1-subunit or distinct protein(s) as chaperone for their plasma membrane expression in colonic epithelial cells.
The detection of 37- and 40-kDa proteins in colonic epithelial cells by the anti-KCNN4-abc antibody differ from that of recent studies, which used an antibody (IK38/6) raised against an NH2-terminal peptide of mouse KCNN4 (14, 17). In these studies, Furness et al. (14) have detected only a 40-kDa protein in rat colon, whereas Halm et al. (17) have detected 41- and 48-kDa proteins in hamster colon. This discrepancy with the absence of a 37-kDa protein with IK38/6 antibody may be due to different resolution used for protein separation (14, 17). However, studies that have identified a 48-kDa protein in hamster colon (17) and KCNN4 splice variant named rKCNN4c that encodes 396 amino acids (reported accession no. AB292803) suggest the existence of multiple KCNN4 isoforms in colonic epithelial cells. As discussed, the KCNN4c and KCNN4b transcripts encode apical (37 kDa) and basolateral (40 kDa) proteins in colonic epithelial cells, respectively. This conclusion is further supported by the observation that in vitro-expressed KCNN4b and KCNN4c K+ channels exhibit different sensitivity to TRAM-34, as they are inhibited with apparent Ki for TRAM-34 of 0.6 ± 0.1 and 7.8 ± 0.4 μg, respectively (Fig. 7). Although these results differ from that of human T lymphocytes KCNN4 that was shown inhibited by 0.5 μM TRAM-34 (55), it is consistent with the results reported with colonic epithelium that have shown 100 μM TRAM-34 to inhibit varying degree of short-circuit current (Isc; a measure of anion secretion) and tissue conductance induced by different agonists in guinea pig and rat colon (17). Although a high concentration of TRAM-34 was shown to inhibit different K+ channels (55), the low TRAM-34 sensitivity exerted by KCNN4c K+ channel might be due to either the lack of second MSD or the associated chaperone protein. Thus the low dose (0.5 μM) TRAM-34-resistant KCNN4c localized on the apical membrane might have provided the driving force for agonist-induced anion secretion seen in epithelial cells of the guinea pig and rat colon (17). In addition, K+ secretion mediated by apical KCNN4 channel might also contribute to the high K+ concentration present in diarrheal stool (13).
In summary, this study isolated three distinct KCNN4 (KCNN4a, KCNN4b, and KCNN4c) splice variants and identified that KCNN4a expressed predominantly in smooth muscle, whereas KCNN4c and KCNN4b are present in apical and basolateral membranes of colonic epithelial cells, respectively. In addition, this study also 1) identified that KCNN4c isoform lacks the predicted second MSD and that KCNN4c protein requires a chaperone protein for its plasma membrane expression; and 2) demonstrated that in vitro-expressed KCNN4c is relatively resistant to KCNN4 K+ channel-specific inhibitor TRAM-34. We conclude from these results that apical and basolateral KCNN4 channels encoded by KCNN4c and KCNN4b transcripts, respectively, may regulate different cell functions under physiological and pathophysiological conditions. The basolateral KCNN4 channels would provide the driving force for agonist-induced anion (Cl− and HCO3−) secretion, whereas apical KCNN4 channels would regulate K+ secretion and contribute to high K+ concentration seen in diarrheal stools. In addition, apical and basolateral KCNN4 channels would also regulate anion secretion and K+ absorption, respectively.
GRANTS
This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases Grant (DK-018777).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
ACKNOWLEDGEMENTS
Present address of C. Barmeyer: Dept. of Gastroenterology, Infectious Diseases & Rheumatology, Charité - Campus Benjamin Franklin, Berlin, Germany.
REFERENCES
- 1.Albaqumi M, Srivastava S, Li Z, Zhdnova O, Wulff H, Itani O, Wallace DP, Skolnik EY. KCa3.1.potassium channels are critical for cAMP-dependent chloride secretion and cyst growth in autosomal-dominant polycystic kidney disease. Kidney Int 74: 740–749, 2008 [DOI] [PubMed] [Google Scholar]
- 2.Bakos E, Evers R, Szakacs G, Tusnady GE, Welker E, Szabo K, de Haas M, van Deemter L, Borst P, Varadi A, Sarkadi B. Functional multidrug resistance protein (MRP1) lacking the N-terminal transmembrane domain. J Biol Chem 273: 32167–32175, 1998 [DOI] [PubMed] [Google Scholar]
- 3.Barrett KE, Keely SJ. Chloride secretion by the intestinal epithelium: molecular basis and regulatory aspects. Annu Rev Physiol 62: 535–572, 2000 [DOI] [PubMed] [Google Scholar]
- 4.Begenisich T, Nakamoto T, Ovitt CE, Nehrke K, Brugnara C, Alper SL, Melvin JE. Physiological roles of the intermediate conductance, Ca2+-activated potassium channel Kcnn4. J Biol Chem 279: 47681–47687, 2004 [DOI] [PubMed] [Google Scholar]
- 5.Bernard K, Bogliolo S, Soriani O, Ehrenfeld J. Modulation of calcium-dependent chloride secretion by basolateral SK4-like channels in a human bronchial cell line. J Membr Biol 196: 15–31, 2003 [DOI] [PubMed] [Google Scholar]
- 6.Biber J, Rechkemmer G, Bodmer M, Schroder P, Haase W, Murer H. Isolation of basolateral membranes from columnar cells of the proximal colon of the guinea pig. Biochim Biophys Acta 735: 1–11, 1983 [DOI] [PubMed] [Google Scholar]
- 7.Bleich M, Riedemann N, Warth R, Kerstan D, Leipziger J, Hor M, Driessche WV, Greger R. Ca2+ regulated K+ and non-selective cation channels in the basolateral membrane of rat colonic crypt base cells. Pflügers Arch 432: 1011–1022, 1996 [DOI] [PubMed] [Google Scholar]
- 8.Breitwieser GE. Mechanisms of K+ channel regulation. J Membr Biol 152: 1–11, 1996 [DOI] [PubMed] [Google Scholar]
- 9.Chen MX, Gorman SA, Benson B, Singh K, Hieble JP, Michel MC, Tate SN, Trezise DJ. Small and intermediate conductance Ca(2+)-activated K+ channels confer distinctive patterns of distribution in human tissues and differential cellular localisation in the colon and corpus cavernosum. Naunyn Schmiedebergs Arch Pharmacol 369: 602–615, 2004 [DOI] [PubMed] [Google Scholar]
- 10.Devor DC, Singh AK, Bridges RJ, Frizzell RA. Modulation of Cl- secretion by benzimidazolones. II. Coordinate regulation of apical GCl and basolateral GK. Am J Physiol Lung Cell Mol Physiol 271: L785–L795, 1996 [DOI] [PubMed] [Google Scholar]
- 11.Fanger CM, Rauer H, Neben AL, Miller MJ, Wulff H, Rosa JC, Ganellin CR, Chandy KG, Cahalan MD. Calcium-activated potassium channels sustain calcium signaling in T lymphocytes. Selective blockers and manipulated channel expression levels. J Biol Chem 276: 12249–12256, 2001 [DOI] [PubMed] [Google Scholar]
- 12.Fong P, Argent BE, Guggino WB, Gray MA. Characterization of vectorial chloride transport pathways in the human pancreatic duct adenocarcinoma cell line HPAF. Am J Physiol Cell Physiol 285: C433–C445, 2003 [DOI] [PubMed] [Google Scholar]
- 13.Fordtran JS. Speculations on the pathogenesis of diarrhea. Fed Proc 26: 1405–1414, 1967 [PubMed] [Google Scholar]
- 14.Furness JB, Robbins HL, Selmer IS, Hunne B, Chen MX, Hicks GA, Moore S, Neylon CB. Expression of intermediate conductance potassium channel immunoreactivity in neurons and epithelial cells of the rat gastrointestinal tract. Cell Tissue Res 314: 179–189, 2003 [DOI] [PubMed] [Google Scholar]
- 15.Gottardi CJ, Pietrini G, Roush DL, Caplan MJ. Sorting of ion transport proteins in polarized cells. J Cell Sci Suppl 17: 13–20, 1993 [DOI] [PubMed] [Google Scholar]
- 16.Greger R, Bleich M, Warth R. New types of K+ channels in the colon. Wien Klin Wochenschr 109: 497–498, 1997 [PubMed] [Google Scholar]
- 17.Halm ST, Liao T, Halm DR. Distinct K+ conductive pathways are required for Cl− and K+ secretion across distal colonic epithelium. Am J Physiol Cell Physiol 291: C636–C648, 2006 [DOI] [PubMed] [Google Scholar]
- 18.Hanley PJ, Musset B, Renigunta V, Limberg SH, Dalpke AH, Sus R, Heeg KM, Preisig-Muller R, Daut J. Extracellular ATP induces oscillations of intracellular Ca2+ and membrane potential and promotes transcription of IL-6 in macrophages. Proc Natl Acad Sci USA 101: 9479–9484, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishii TM, Silvia C, Hirschberg B, Bond CT, Adelman JP, Maylie J. A human intermediate conductance calcium-activated potassium channel. Proc Natl Acad Sci USA 94: 11651–11656, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jensen BS, Strobaek D, Christophersen P, Jorgensen TD, Hansen C, Silahtaroglu A, Olesen SP, Ahring PK. Characterization of the cloned human intermediate-conductance Ca2+-activated K+ channel. Am J Physiol Cell Physiol 275: C848–C856, 1998 [DOI] [PubMed] [Google Scholar]
- 21.Joiner WJ, Basavappa S, Vidyasagar S, Nehrke K, Krishnan S, Binder HJ, Boulpaep EL, Rajendran VM. Active K+ secretion through multiple KCa-type channels and regulation by IKCa channels in rat proximal colon. Am J Physiol Gastrointest Liver Physiol 285: G185–G196, 2003 [DOI] [PubMed] [Google Scholar]
- 22.Joiner WJ, Wang LY, Tang MD, Kaczmarek LK. hSK4, a member of a novel subfamily of calcium-activated potassium channels. Proc Natl Acad Sci USA 94: 11013–11018, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jung H. Topology and function of the Na+/proline transporter of Escherichia coli, a member of the Na+/solute cotransporter family. Biochim Biophys Acta 1365: 60–64, 1998 [DOI] [PubMed] [Google Scholar]
- 24.Kast C, Canfield V, Levenson R, Gros P. Membrane topology of P-glycoprotein as determined by epitope insertion: transmembrane organization of the N-terminal domain of mdr3. Biochemistry 34: 4402–4411, 1995 [DOI] [PubMed] [Google Scholar]
- 25.Kast C, Gros P. Topology mapping of the amino-terminal half of multidrug resistance-associated protein by epitope insertion and immunofluorescence. J Biol Chem 272: 26479–26487, 1997 [DOI] [PubMed] [Google Scholar]
- 26.Kolski-Andreaco A, Tomita H, Shakkottai VG, Gutman GA, Cahalan MD, Gargus JJ, Chandy KG. SK3-1C, a dominant-negative suppressor of SKCa and IKCa channels. J Biol Chem 279: 6893–6904, 2004 [DOI] [PubMed] [Google Scholar]
- 27.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305: 567–580, 2001 [DOI] [PubMed] [Google Scholar]
- 28.Lauf PK, Misri S, Chimote AA, Adragna NC. Apparent intermediate K conductance channel hyposmotic activation in human lens epithelial cells. Am J Physiol Cell Physiol 294: C820–C832, 2008 [DOI] [PubMed] [Google Scholar]
- 29.Liou W, Geuze HJ, Slot JW. Improving structural integrity of cryosections for immunogold labeling. Histochem Cell Biol 106: 41–58, 1996 [DOI] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408, 2001 [DOI] [PubMed] [Google Scholar]
- 31.Logsdon NJ, Kang J, Togo JA, Christian EP, Aiyar J. A novel gene, hKCa4, encodes the calcium-activated potassium channel in human T lymphocytes. J Biol Chem 272: 32723–32726, 1997 [DOI] [PubMed] [Google Scholar]
- 32.Lomax RB, McNicholas CM, Lombes M, Sandle GI. Aldosterone-induced apical Na+ and K+ conductances are located predominantly in surface cells in rat distal colon. Am J Physiol Gastrointest Liver Physiol 266: G71–G82, 1994 [DOI] [PubMed] [Google Scholar]
- 33.Meera P, Wallner M, Song M, Toro L. Large conductance voltage- and calcium-dependent K+ channel, a distinct member of voltage-dependent ion channels with seven N-terminal transmembrane segments (S0-S6), an extracellular N terminus, and an intracellular (S9-S10) C terminus. Proc Natl Acad Sci USA 94: 14066–14071, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mense M, Rajendran V, Blostein R, Caplan MJ. Extracellular domains, transmembrane segments, and intracellular domains interact to determine the cation selectivity of Na,K- and gastric H,K-ATPase. Biochemistry 41: 9803–9812, 2002 [DOI] [PubMed] [Google Scholar]
- 35.Muth TR, Caplan MJ. Transport protein trafficking in polarized cells. Annu Rev Cell Dev Biol 19: 333–366, 2003 [DOI] [PubMed] [Google Scholar]
- 36.Neylon CB, Lang RJ, Fu Y, Bobik A, Reinhart PH. Molecular cloning and characterization of the intermediate-conductance Ca(2+)-activated K(+) channel in vascular smooth muscle: relationship between K(Ca) channel diversity and smooth muscle cell function. Circ Res 85: e33–43, 1999 [DOI] [PubMed] [Google Scholar]
- 37.Nimigean CM, Magleby KL. Functional coupling of the beta(1) subunit to the large conductance Ca(2+)-activated K(+) channel in the absence of Ca(2+). Increased Ca(2+) sensitivity from a Ca(2+)-independent mechanism. J Gen Physiol 115: 719–736, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rajendran VM, Kashgarian M, Binder HJ. Aldosterone induction of electrogenic sodium transport in the apical membrane vesicles of rat distal colon. J Biol Chem 264: 18638–18644, 1989 [PubMed] [Google Scholar]
- 39.Rajendran VM, Oesterlin M, Binder HJ. Sodium uptake across basolateral membrane of rat distal colon. Evidence for Na-H exchange and Na-anion cotransport. J Clin Invest 88: 1379–1385, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reyes R, Duprat F, Lesage F, Fink M, Salinas M, Farman N, Lazdunski M. Cloning and expression of a novel pH-sensitive two pore domain K+ channel from human kidney. J Biol Chem 273: 30863–30869, 1998 [DOI] [PubMed] [Google Scholar]
- 41.Rufo PA, Jiang L, Moe SJ, Brugnara C, Alper SL, Lencer WI. The antifungal antibiotic, clotrimazole, inhibits Cl- secretion by polarized monolayers of human colonic epithelial cells. J Clin Invest 98: 2066–2075, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sandle GI, McNicholas CM, Lomax RB. Potassium channels in colonic crypts. Lancet 343: 23–25, 1994 [DOI] [PubMed] [Google Scholar]
- 43.Sangan P, Kolla SS, Rajendran VM, Kashgarian M, Binder HJ. Colonic H-K-ATPase β-subunit: identification in apical membranes and regulation by dietary K depletion. Am J Physiol Cell Physiol 276: C350–C360, 1999 [DOI] [PubMed] [Google Scholar]
- 44.Sangan P, Thevananther S, Sangan S, Rajendran VM, Binder HJ. Colonic H-K-ATPase α- and β-subunits express ouabain-insensitive H-K-ATPase. Am J Physiol Cell Physiol 278: C182–C189, 2000 [DOI] [PubMed] [Google Scholar]
- 45.Sharma NR, Davis MJ. Mechanism of substance P-induced hyperpolarization of porcine coronary artery endothelial cells. Am J Physiol Heart Circ Physiol 266: H156–H164, 1994 [DOI] [PubMed] [Google Scholar]
- 46.Shepherd MC, Duffy SM, Harris T, Cruse G, Schuliga M, Brightling CE, Neylon CB, Bradding P, Stewart AG. KCa3.1. Ca2+ activated K+ channels regulate human airway smooth muscle proliferation. Am J Respir Cell Mol Biol 37: 525–531, 2007 [DOI] [PubMed] [Google Scholar]
- 47.Singh S, Syme CA, Singh AK, Devor DC, Bridges RJ. Benzimidazolone activators of chloride secretion: potential therapeutics for cystic fibrosis and chronic obstructive pulmonary disease. J Pharmacol Exp Ther 296: 600–611, 2001 [PubMed] [Google Scholar]
- 48.Stevenson BR, Siliciano JD, Mooseker MS, Goodenough DA. Identification of ZO-1: a high molecular weight polypeptide associated with the tight junction (zonula occludens) in a variety of epithelia. J Cell Biol 103: 755–766, 1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stieger B, Marxer A, Hauri HP. Isolation of brush-border membranes from rat and rabbit colonocytes: is alkaline phosphatase a marker enzyme? J Membr Biol 91: 19–31, 1986 [DOI] [PubMed] [Google Scholar]
- 50.Tao R, Lau CP, Tse HF, Li GR. Regulation of cell proliferation by intermediate-conductance Ca2+-activated potassium and volume-sensitive chloride channels in mouse mesenchymal stem cells. Am J Physiol Cell Physiol 295: C1409–C1416, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Toyama K, Wulff H, Chandy KG, Azam P, Raman G, Saito T, Fujiwara Y, Mattson DL, Das S, Melvin JE, Pratt PF, Hatoum OA, Gutterman DD, Harder DR, Miura H. The intermediate-conductance calcium-activated potassium channel KCa3.1 contributes to atherogenesis in mice and humans. J Clin Invest 118: 3025–3037, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vandorpe DH, Shmukler BE, Jiang L, Lim B, Maylie J, Adelman JP, de Franceschi L, Cappellini MD, Brugnara C, Alper SL. cDNA cloning and functional characterization of the mouse Ca2+-gated K+ channel, mIK1. Roles in regulatory volume decrease and erythroid differentiation. J Biol Chem 273: 21542–21553, 1998 [DOI] [PubMed] [Google Scholar]
- 53.Wang J, Morishima S, Okada Y. IK channels are involved in the regulatory volume decrease in human epithelial cells. Am J Physiol Cell Physiol 284: C77–C84, 2003 [DOI] [PubMed] [Google Scholar]
- 54.Warth R, Hamm K, Bleich M, Kunzelmann K, von Hahn T, Schreiber R, Ullrich E, Mengel M, Trautmann N, Kindle P, Schwab A, Greger R. Molecular and functional characterization of the small Ca(2+)-regulated K+ channel (rSK4) of colonic crypts. Pflügers Arch 438: 437–444, 1999 [DOI] [PubMed] [Google Scholar]
- 55.Wulff H, Miller MJ, Hansel W, Grissmer S, Cahalan MD, Chandy KG. Design of a potent and selective inhibitor of the intermediate-conductance Ca2+-activated K+ channel, IKCa1: a potential immunosuppressant. Proc Natl Acad Sci USA 97: 8151–8156, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]