Abstract
Histone deacetylases (HDACs) counterbalance acetylation of lysine residues, a protein modification involved in numerous biological processes. Here, Hdac1 and Hdac2 conditional knock-out alleles were used to study the function of class I Hdac1 and Hdac2 in cell cycle progression and haematopoietic differentiation. Combined deletion of Hdac1 and Hdac2, or inactivation of their deacetylase activity in primary or oncogenic-transformed fibroblasts, results in a senescence-like G1 cell cycle arrest, accompanied by up-regulation of the cyclin-dependent kinase inhibitor p21Cip. Notably, concomitant genetic inactivation of p53 or p21Cip indicates that Hdac1 and Hdac2 regulate p53–p21Cip-independent pathways critical for maintaining cell cycle progression. In vivo, we show that Hdac1 and Hdac2 are not essential for liver homeostasis. In contrast, total levels of Hdac1 and Hdac2 in the haematopoietic system are critical for erythrocyte-megakaryocyte differentiation. Dual inactivation of Hdac1 and Hdac2 results in apoptosis of megakaryocytes and thrombocytopenia. Together, these data indicate that Hdac1 and Hdac2 have overlapping functions in cell cycle regulation and haematopoiesis. In addition, this work provides insights into mechanism-based toxicities observed in patients treated with HDAC inhibitors.
Keywords: Hdac1, Hdac2, haematopoiesis, p21Cip, senescence
Introduction
Post-translational modifications (PTMs) such as phosphorylation, methylation, ubiquitination, and acetylation are crucial regulatory modules at the heart of biological processes in the cell and are tightly regulated by a multitude of enzymes that catalyse the addition or removal of PTMs (Campos and Reinberg, 2009). Lysine acetylation of histones and non-histone proteins is controlled by histone acetyl transferases and histone deacetylases (HDACs). HDACs can be classified on the basis of their homology to yeast counterparts (Yang and Seto, 2008). Class I HDACs (HDAC1, -2, -3 and -8) are highly homologous to Saccharomyces cerevisiae Rpd3. Class IIa HDACs (HDAC4, -5, -7 and -9) and Class IIb HDACs (HDAC6 and -10) consist of S. cerevisiae Hda1 homologues. HDAC11 is the sole member of the Class IV HDACs, based on homology to both class I and class II HDACs (Gregoretti et al, 2004).
Although the high sequence similarity between class I HDACs might anticipate a significant overlap in function, genetic studies in mice have revealed redundant as well as specific functions of these enzymes (Haberland et al, 2009c). Deletion of Hdac1 results in embryonic lethality as early as E9.5 of development (Lagger et al, 2002). In contrast, Hdac2 deficiency results in viable mice with reduced body weight (Trivedi et al, 2007; Zimmermann et al, 2007; Guan et al, 2009). Others have reported that Hdac2 deficiency is not compatible with life because of cardiac myopathy (Montgomery et al, 2007). The basis for these different phenotypes is not clear, but may relate to genetic background of the mice. Hdac2 also has a specific function in repression of genes involved in synaptogenesis, as evidenced by enhanced synapse formation, learning and memory in Hdac2-deficient mice (Guan et al, 2009). Hdac3 deletion results in early embryonic lethality and this enzyme has a critical function in cell cycle regulation and cardiac metabolism (Bhaskara et al, 2008; Knutson et al, 2008; Montgomery et al, 2008). Finally, Hdac8 has an important function in the differentiation of neural crest cells (Haberland et al, 2009b).
A prime function of HDACs relates to their classical function as transcriptional co-repressors through deacetylation of lysine residues in histone tails. This results in a closed chromatin structure and diminished accessibility for the basal transcription machinery. Class I HDACs are present in a variety of repressor complexes such as SIN3A, NuRD, REST and N-CoR/SMRT, which acquire their regional activities in part by interacting with sequence-specific transcription factors (Yang and Seto, 2008). The intimate link between class I HDACs and proteins involved in tumourigenesis, such as Mad/Mxi, pRB, p53 and PML–RAR fusion proteins, has established important functions for HDACs in tumourigenic processes. Correspondingly, pharmacological inhibition of HDACs, using chemical HDAC inhibitors (HDACi), results in cell cycle arrest and apoptosis of tumour cells (Minucci and Pelicci, 2006). Moreover, the use of relative selective HDACi-targeting class I HDACs has produced anti-tumourigenic effects, and genetic inactivation of class I Hdac1 and Hdac2 in transformed murine cells results in cessation of tumourigenic potential (Rasheed et al, 2008; Haberland et al, 2009a).
Despite the clinical efficacy of HDACi, treatment of patients with HDACi results in undesirable haematological side effects, such as anaemia and thrombocytopenia (Prince et al, 2009). Currently, it is unclear whether these side effects are due to the targeting of (multiple) HDACs or because of off-target effects on non-HDAC proteins. These issues prompted us to explore more precisely the function of class I Hdac1 and Hdac2 in cell proliferation and haematopoietic development.
Results
Normal cell cycle regulation and increased levels of Hdac1 in Hdac2-deficient mouse embryonic fibroblasts
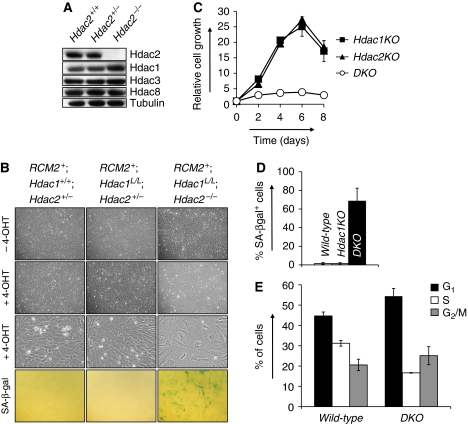
To examine the function of Hdac2 in cell cycle regulation in primary cells, we generated mouse embryonic fibroblasts (MEFs) deficient for Hdac2. Relative to wild-type controls, both Hdac2+/− and Hdac2−/− MEFs do not show any alterations in proliferation under normal culture conditions as well as under growth-restricting conditions, such as low serum, oncogene-induced senescence (OIS) and irradiation (data not shown). These results prompted us to investigate possible compensatory mechanisms of other class I Hdac members, Hdac1, -3 and -8. Western blot analysis of Hdac2-deficient MEF protein lysates revealed that Hdac1 protein levels are increased as compared with Hdac2-proficient MEFs. In contrast, protein levels of Hdac3 and Hdac8 remained unchanged (Figure 1A). Interestingly, ablation of Hdac1 results in an increase of Hdac2 protein levels (Figure 3A; Supplementary Figure 3A). These results indicate reciprocal compensatory mechanisms between Hdac1 and Hdac2 and suggest functional redundancy between these two class I Hdacs.
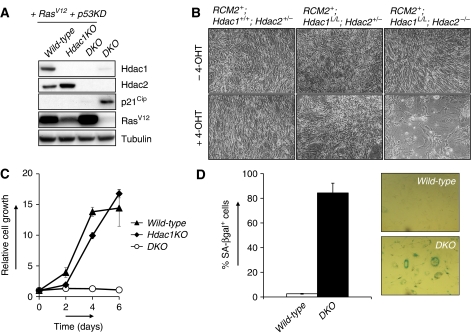
Figure 1.
Hdac1 and Hdac2 collectively control cell cycle progression. (A) Western blot analysis of Hdac2-deficient MEF protein lysates for indicated proteins. Tubulin served as a loading control. (B) Representative photographs of MEF cell cultures with indicated genotypes grown without 4-OHT or with 200 nM 4-OHT. Representative details of MEF cultures with indicated genotypes are shown in the third row. Note the presence of large, flat cells in 4-OHT-treated RCM2+;Hdac1L/L;Hdac2−/− cultures. Bottom panels show representative pictures of senescence-associated β-galactosidase-stained MEF cultures with indicated genotypes. (C) Growth curve analysis of Hdac1KO (closed squares), Hdac2KO (closed triangles) or DKO MEFs (open circles). All experiments were performed in triplicate. (D) Percentage of SA-bgalactosidase positive cells in MEF cultures with indicated genotypes. (E) Cell cycle analysis of wild-type and DKO MEFs for G1, S and G2/M cell cycle phases by BrdU-PI FACS. Values represent the average of three independent experiments.
Hdac1 and Hdac2 collectively regulate cell cycle progression
To directly explore possible functional redundancy between Hdac2 and its most homologous family member, Hdac1, we generated a cell culture system in which Hdac1 and Hdac2 can be deleted individually or simultaneously. Therefore, we used an Hdac1 conditional knock-out (cKO) allele in which exon 2 is flanked by loxP recombination sites (Supplementary Figure 1A). Cre-recombinase-mediated deletion of Hdac1 exon 2 produces an Hdac1-null allele (Supplementary Figure 1A) and results in embryonic lethality of Hdac1−/− mice consistent with earlier studies (Lagger et al, 2002; Montgomery et al, 2007). Intercrosses using Hdac1 cKO mice, Hdac2 cKO mice (Guan et al, 2009) and Rosa26CreERT2 (RCM2) mice (Hameyer et al, 2007) allowed us to generate a series of isogenic MEFs in which, on addition of tamoxifen (4-OHT), Hdac1 and Hdac2 could be deleted (Supplementary Figure 1B).
Ablation of Hdac2 (resulting in Hdac2KO MEFs) or Hdac1 (resulting in Hdac1KO MEFs) did not result in an overt phenotype in MEFs under normal growth conditions (Figure 1B and C). In contrast, somatic deletion of Hdac1 in germ-line Hdac2−/− cells (referred to as DKO MEFs) results in a dramatic growth arrest and induction of a large and flat, senescence-like, cell morphology (Figure 1B and C). Accordingly, up to 80% of all DKO cells stained positive for senescence-associated β-galactosidase (SA-β-gal) activity, whereas Hdac1 or Hdac2 single null cells showed wild-type-staining patterns (Figure 1B and D). To dissect the nature of the cellular proliferation arrest in DKO MEFs, BrdU-PI fluorescence-activated cell sorting (FACS) analysis was used to determine the cell cycle distribution in wild-type and DKO MEFs. As compared with wild-type controls, DKO MEFs displayed a two-fold reduction in S-phase cells and a 10% increase in the G1-phase cells (Figure 1E). Thus, Hdac1 and Hdac2 cooperate to control G1 to S transition and their absence provokes a senescence-like, G1 cell cycle arrest.
Hdac1 or Hdac2 catalytic activity is required to maintain cell cycle progression
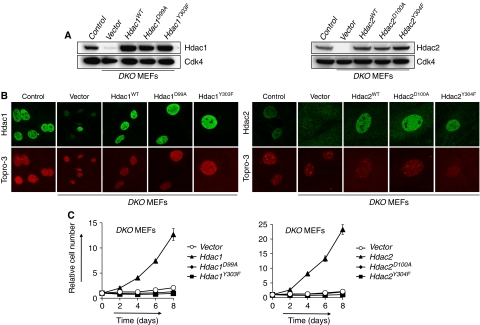
HDACs remove acetyl groups from lysine residues through a mechanism that involves deacetylase activity, which is dependent on other proteins present in HDAC multi-protein complexes (Sengupta and Seto, 2004). Inactivation of Hdac1 and Hdac2 in our experiments resulted in a complete removal of these proteins, raising the question whether the observed growth arrest in DKO MEFs is due to dissociation of HDAC protein complexes or because of the absence of HDAC activity. To address this question, we generated Hdac1 and Hdac2 catalytic inactive mutants by mutating conserved residues found to be critical for deacetylase activity in HDAC8 (Supplementary Figure 2A–D; Vannini et al, 2007). Retroviral expression in MEFs resulted in near-physiological protein levels and proper cellular localization of Hdac1D99A, Hdac1Y303F, Hdac2D100A or Hdac2Y304F mutants, similar to wild-type Hdac1 or Hdac2 (Figure 2A and B). Subsequently, expression of catalytic inactive mutants as well as wild-type Hdac1 and Hdac2 was tested for its ability to rescue a growth arrest in DKO MEFs. Although MEFs deficient for Hdac1 and Hdac2 were unable to proliferate, identical MEFs expressing exogenous wild-type Hdac1 or wild-type Hdac2 were fully rescued with respect to their proliferation capacity. In contrast, DKO MEFs expressing Hdac1D99A, Hdac1Y303F, Hdac2D100A or Hdac2Y304F catalytic inactive mutants were unable to proliferate (Figure 2C). These results further corroborate functional redundancy between Hdac1 and Hdac2, as expression of either Hdac1 or Hdac2 is sufficient to rescue DKO MEFs. Furthermore, our results establish that the deacetylase activity of Hdac1 or Hdac2 is essential for cellular proliferation.
Figure 2.
Hdac1 or Hdac2 deacetylase activity is required for cell cycle progression. (A) Western blot analysis of DKO MEFs expressing wild-type Hdac1, Hdac1D99A, Hdac1Y303F (left panel), wild-type Hdac2, Hdac2D100A or Hdac2Y304F (right panel). Lysates prepared from wild-type (control) and DKO MEFs (vector) were used as a positive and negative control, respectively. Cdk4 served as a loading control. (B) Subcellular localization of wild-type and mutant Hdac1 and Hdac2 ectopically expressed in DKO MEFs by immunofluorescence staining using antibodies for Hdac1 (left panels) or Hdac2 (right panels). Note the presence of a Hdac1-proficient nucleus in vector-treated DKO MEFs because of a non-recombined Hdac1 cKO allele. (C) Growth curve analysis of DKO MEFs expressing either wild-type or mutant Hdac1 or Hdac2.
Hdac1 and Hdac2 cooperatively regulate p21Cip expression
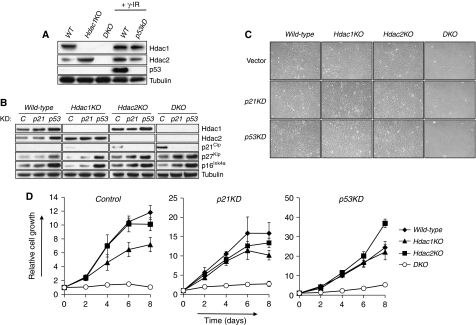
Cellular senescence is a potent proliferation arrest induced upon oncogene expression, DNA-damage or suboptimal cell culture conditions of normal, non-transformed cells. The cell cycle inhibitors p16Ink4a and p19Arf are up-regulated upon senescence-inducing conditions and activate the Retinoblastoma protein (pRb) and p53-tumour suppressors, respectively (Campisi, 2005). To see whether Hdac1 and Hdac2 prevent cellular senescence by repressing the expression of senescene-associated cell cycle inhibitors, we analysed p16Ink4a and p19Arf protein levels in wild-type, Hdac1KO, Hdac2KO and DKO MEFs. Western blot analysis revealed no up-regulation of p16Ink4a or p19Arf in the absence of Hdac1 and Hdac2 (Figure 3B; Supplementary Figure 3A). In addition, p53 protein levels were not stabilized in DKO MEFs, suggesting that Hdac1 and Hdac2 do not control p53 protein levels under normal culture conditions (Figure 3A). It also suggests that deficiency for Hdac1 and Hdac2 does not result in a p53-activating DNA-damage response. p21Cip, a cell cycle inhibitor protein that is transcriptionally regulated by Hdac1 in embryonic stem (ES) cells (Lagger et al, 2002), was modestly up-regulated in Hdac1KO MEFs and Hdac2KO MEFs as compared with control MEFs (Figure 3B). In contrast, DKO MEFs showed strong induction of p21Cip. Expression of a closely related cell cycle inhibitory protein p27Kip did not correlate with the cell cycle arrest in DKO MEFs, as it was up-regulated in Hdac2KO as well as DKO MEFs (Figure 3B). Collectively, these data point to p21Cip as a potential point of action in Hdac1/2-mediated regulation of the cell cycle.
Figure 3.
Hdac1 and Hdac2 regulate cell cycle progression independent of p53 or p21Cip. (A) Western blot analysis of wild-type (WT), Hdac1KO and DKO MEF protein lysates for Hdac1, Hdac2 and p53. γ-Irradiated wild-type cells expressing either control or p53 shRNA were used as a positive and negative control, respectively. Tubulin served as a loading control. (B) Western blot analysis of protein lysates for Hdac1, Hdac2, p21Cip, p27Kip and p16Ink4a of MEFs with indicated genotypes infected with retroviruses expressing control shRNA (C), p21Cip shRNA (p21) or p53 shRNA (p53). Tubulin served as a loading control. (C) Representative pictures of wild-type, Hdac1KO, Hdac2KO or DKO MEFs infected with retroviruses expressing control shRNA, p21Cip shRNA or p53 shRNA. (D) Growth curve analysis of wild-type, Hdac1KO, Hdac2KO and DKO MEFs, expressing either control shRNA or shRNA directed against p21Cip and p53.
Hdac1 and Hdac2 regulate cell cycle progression independent of p53–p21Cip
To examine whether the increased expression of p21Cip is responsible for the cell cycle arrest induced by Hdac1 and Hdac2 ablation, we infected control and DKO MEFs with empty retroviruses (vector) or retroviruses expressing a short-hairpin RNA (shRNA) against p21Cip (referred to as p21KD) (Figure 3B). Despite efficient knock-down of p21Cip, cells lacking both Hdac1 and Hdac2 still entered a cell cycle arrest identical to p21Cip-proficient cells lacking Hdac1 and Hdac2 (Figure 3C and D). As p53 is a major regulator of the G1/S transition in cellular senescence and functions as a transcriptional activator of p21Cip (el-Deiry et al, 1993), we tested whether shRNA-mediated knock-down of p53 (p53KD) is required for both p21Cip induction and cell cycle arrest in DKO MEFs. Although p53 knock-down blocked p21Cip induction (Figure 3B), DKO MEFs still underwent a cell cycle arrest, indicating that p53 is required for the induction of p21Cip expression in the absence of Hdac1 and Hdac2 (Figure 3B) and independently confirm our results obtained with p21Cip knock-down (Figure 3C and D). These data indicate that the p53–p21Cip axis is dispensable for the cell cycle arrest as a result of Hdac1 and Hdac2 deficiency.
Although we obtained hardly detectable levels of p21Cip using p21Cip or p53 shRNA-mediated knock-down, it is conceivable that residual levels of p21Cip are accountable for the observed cell cycle arrest in the absence of Hdac1 and Hdac2. To address this issue, we generated Hdac2+/−p21−/− mice, which were intercrossed to obtain Hdac2−/−p21−/− MEFs (referred to as Hdac2KO;p21−/− MEFs).
To down-regulate Hdac1 levels in this cell system, we generated retroviruses expressing Hdac1 shRNA, resulting in efficient knock-down of Hdac1 (Supplementary Figure 3A). Similar to the results obtained in DKO MEFs, knock-down of Hdac1 (Hdac1KD) in Hdac2KO MEFs, resulted in a senescence-like G1 cell cycle arrest accompanied by SA-β-gal activity and increased p21Cip protein levels (Supplementary Figure 3B–D). In contrast, expression levels of p19Arf, p16Ink4a and p27Kip did not correlate with the observed phenotype (Supplementary Figure 3A).
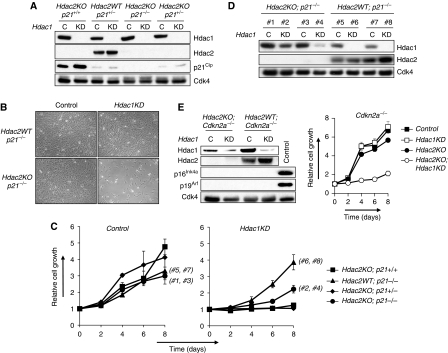
Subsequently, we infected Hdac2WT;p21+/−, Hdac2KO; p21+/+, Hdac2KO;p21+/− and Hdac2KO;p21−/− MEFs with retroviruses expressing Hdac1 shRNA or control shRNA. Hdac1KD resulted in an increased expression of p21Cip only in Hdac2KO;p21+/+ and to a lesser extent in Hdac2KO; p21+/− MEFs (Figure 4A). Regardless of p21Cip status, proliferation ceased dramatically in Hdac1KD;Hdac2KO MEFs (Figure 4B and C). During the growth curve analysis, we noted that Hdac2KO;p21−/− MEF cultures expressing Hdac1 shRNA regained proliferation capacity after 6–8 days on plating (Figure 4C). Western blot analysis of these MEF cultures, along with Hdac2WT;p21+/−MEF cultures expressing Hdac1 shRNA, revealed loss of Hdac1 knock-down specifically in Hdac2KO;p21−/− MEF cultures (Figure 4D). These results indicate a strong selection against Hdac1KD in Hdac2KO;p21−/− MEFs, supporting our results that loss of Hdac1 and Hdac2, even in the absence of p21Cip, is not compatible with proliferation. Collectively, these data strongly indicate and independently confirm that p21Cip is dispensable for establishing the cell cycle arrest in the absence of Hdac1 and Hdac2.
Figure 4.
Genetic ablation of p21Cip, p16Ink4a or p19Arf does not allow proliferation of DKO MEFs. (A) Western blot analysis of protein lysates of indicated MEFs expressing either control shRNA (C) or Hdac1 shRNA (KD) for Hdac1, Hdac2 and p21Cip. Cdk4 served as a loading control. (B) Representative pictures of MEFs with indicated genotypes infected with retroviruses expressing either control shRNA or Hdac1 shRNA. (C) Growth curve analysis of Hdac2KO;p21+/+(squares), Hdac2WT;p21−/− (triangles), Hdac2KO;p21+/− (diamonds) and Hdac2KO;p21−/− (circles), MEFs expressing either control shRNA (left panel) or Hdac1 shRNA (right panel). (D) Western blot analysis of protein lysates for Hdac1 and Hdac2 of two independent Hdac2WT;p21−/− and Hdac2KO;p21−/− MEF clones infected with either control (#1, #3, #5, #7) or Hdac1 shRNA (#2, #4, #6, #8), isolated at day 8 of the growth curve analysis as shown in (C). The clone numbers in (C) correspond with the clones and genotypes as shown in (D). Cdk4 served as a loading control. (E) Left panel: western blot analysis of Hdac2WT;Cdkn2a−/− and Hdac2KO;Cdkn2a−/− MEFs expressing either control (control) or Hdac1 shRNA (Hdac1KD) for Hdac1, Hdac2, p16Ink4a and p19Arf. Cdk4 was used as a loading control. As a control for p16Ink4a and p19Arf expression, we used late passage wild-type MEFs. Right panel: growth curve analysis of Hdac2WT;Cdkn2a−/− (squares) and Hdac2KO;Cdkn2a−/− (circles) MEFs expressing either control (filled tags) or Hdac1 shRNA (open tags).
p16Ink4a- and p19Arf-independent cell cycle arrest in Hdac1;Hdac2-deficient MEFs
It is conceivable that other cell cycle inhibitor proteins besides p21Cip are involved in the senesence-like cell cycle arrest in the absence of Hdac1 and Hdac2. p16Ink4a and p19Arf, encoded by the Cdkn2a allele, are two major cell cycle inhibitors involved in cellular senescence and activate the pRb- and p53-tumour suppressor pathways by inhibition of cyclinD/cdk4 and Mdm2, respectively. To test whether deletion of these cell cycle inhibitors allows a bypass of the observed cell cycle arrest, we generated Hdac2WT;Cdkn2a−/− and Hdac2KO;Cdkn2a−/− MEFs and subsequently down-regulated Hdac1 by expression of Hdac1 shRNA. Despite the absence of p16Ink4a and p19Arf, simultaneous inactivation of Hdac1 and Hdac2 still resulted in a cell cycle arrest, indicating that these cell cycle inhibitors are not required for inducing a cell cycle arrest in Hdac1- and Hdac2-deficient MEFs (Figure 4E).
Oncogenic-transformed cells harbour a senescence-like programme suppressed by Hdac1 or Hdac2
OIS is viewed as a mechanism to protect cells from oncogenic transformation (Mooi and Peeper, 2006). Expression of oncogenic RasV12 induces the expression of the cell cycle inhibitors p16Ink4a and p19Arf, thereby activating the pRb- and p53-tumour suppressor proteins. Inactivation of p53 allows bypass of RasV12-induced senescence and as a consequence oncogenic transformation (Campisi, 2005). We wished to address whether the senescence-like arrest in the absence of Hdac1 and Hdac2 is still functional in oncogenic-transformed cells that have bypassed OIS. To this end, we oncogenically transformed control and RCM2+;Hdac1L/L;Hdac2−/− MEFs with retroviruses expressing p53 shRNA as well as oncogenic Ras (RasV12) (Figure 5A). Upon addition of 4-OHT to these cells, we obtained wild-type, Hdac1-deficient (Hdac1KO) and Hdac1/Hdac2-deficient (DKO)-transformed fibroblasts. Cells deficient for either Hdac1 or Hdac2 did not show an impairment of proliferation. Similar to our observations in primary MEFs, Hdac1 deficiency resulted in increased Hdac2 levels, suggesting a compensatory function for Hdac2 in the absence of Hdac1 (Figure 5A). Indeed, ablation of Hdac1 and Hdac2 in transformed cells resulted in a senescence-like growth arrest in short- and long-term proliferation assays (Figure 5B and C). Despite the fact that these cells have bypassed the p53-dependent RasV12-induced senescence checkpoint, we still observed SA-β-gal activity in the majority (up to 80%) of DKO cells (Figure 5D). Similar to our observations in primary MEFs, p53 knock-down in transformed fibroblasts prevented p21Cip up-regulation (Figure 5A), but not a cell cycle arrest in the absence of Hdac1 and Hdac2, suggesting that also in transformed cells Hdac1 and Hdac2 function independent of p53 and p21Cip in maintaining cellular proliferation. Together, these data show an essential and redundant function of Hdac1 and Hdac2 in suppressing a p53–p21Cip-independent pathway that is able to evoke a senescence response, even in cells that have escaped OIS.
Figure 5.
Hdac1 and Hdac2 collectively suppress a senescence-inducing pathway in transformed cells. (A) Western blot analysis of 4-OHT-treated MEFs with indicated genotypes, expressing RasV12 and p53 shRNA for Hdac1, Hdac2, p21Cip and RasV12. Tubulin served as loading control. DKO MEFs served as a control for p21Cip expression. (B) Representative pictures of oncogenic-transformed (RasV12;p53KD) MEF cultures with indicated genotypes in the absence (top panels) or presence (lower panels) of 4-OHT. (C) Growth curve analysis of wild-type (triangles), Hdac1KO (diamonds) and DKO (open circles) oncogenic-transformed MEFs. (D) Quantification and representative pictures of SA-β-galactosidase positive cells in cultures of wild-type and DKO oncogenic-transformed cells. Shown are average values of six different microscopic fields.
Ablation of Hdac1 and Hdac2 results in anaemia and thrombocytopenia
Treatment of cancer patients using HDACi is complicated by the adverse clinical impact on the haematopoietic system (Prince et al, 2009). The growth arrest conferred by simultaneous deletion of Hdac1 and Hdac2 in primary fibroblasts prompted us to explore whether the HDACi-related toxicities can be explained by selective targeting of (multiple) HDACs or to off-target effects. Deletion of Hdac1 and/or Hdac2 in the haematological compartment of the mouse will enable us to address these questions. To this end, we generated mice harbouring the interferon-inducible MxCre transgene (Kuhn et al, 1995) and cKO alleles for Hdac1 and Hdac2. Administration of polyinosine-polycytidylic acid (pI;pC) induces an interferon response, thereby activating MxCre expression predominantly in the haematopoietic system and liver. pI;pC-induced MxCre expression resulted in successful deletion of Hdac1 and Hdac2 in bone marrow (Supplementary Figure 4) and liver (Supplementary Figure 5). Ablation of Hdac1, or simultaneous deletion of Hdac1 and Hdac2 in the liver, did not result in histological abnormalities, indicating that Hdac1 and Hdac2 are not critical in the maintenance of hepatocytes (Supplementary Figure 5). In contrast, although pI;pC-treated MxCre+ or MxCre+; Hdac1L/L (referred to as MxCre+;Hdac1KO) mice appeared normal, similar treated MxCre+;Hdac1L/L;Hdac2L/L (referred to as MxCre+;DKO) mice became rapidly moribund at approximately 9 days after pI;pC injections displaying anaemic features and internal bleedings (Figure 6A). Indeed, peripheral blood analysis showed a five-fold reduction in red blood cells and thrombocyte numbers were 16-fold reduced in MxCre+;DKO mice as compared with MxCre+ mice. Although thrombocyte numbers in MxCre+;Hdac1KO mice were decreased, this reduction is not significant (P>0.05) (Figure 6B). Histological examination of bone-marrow sections as well as total bone-marrow cell counts revealed a reduction in total cell numbers in MxCre+;DKO mice compared with MxCre+ mice (Figure 6B and C). Strikingly, as compared with MxCre+ and MxCre+;Hdac1KO mice, MxCre+;DKO bone marrow contained 20-fold less megakaryocytes, which are responsible for thrombocyte production, providing a rational for the strong reduction in circulating thrombocytes (Figure 6C and D). Accordingly, we observed the presence of many apoptotic megakaryocytes, as identified by histological morphology, pyknotic nuclei in histology slides and immunohistochemistry for activated caspase-3 (Figure 6C and E). These results show that, similar to our observations in primary and transformed fibroblasts, Hdac1 and Hdac2 have overlapping functions in the development of the erythrocyte-megakaryocyte lineage.
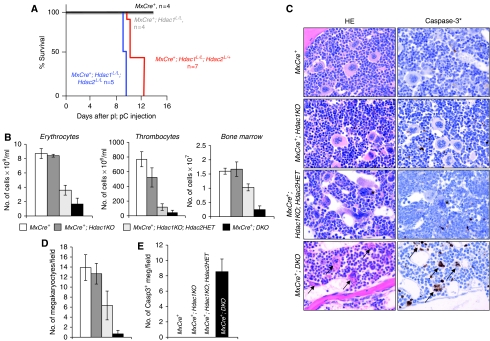
Figure 6.
Hdac1 and Hdac2 have overlapping functions in haematopoiesis. (A) Kaplan–Meier curves of pI;pC-treated MxCre+, MxCre+;Hdac1L/L, MxCre+Hdac1L/L;Hdac2L/+ and MxCre+Hdac1L/L;Hdac2L/L mice. (B) Total bone marrow (per femur), erythrocyte and thrombocyte numbers in peripheral blood of mice with indicated genotypes. (C) Bone marrow histology of pI;pC-treated MxCre+, MxCre+;Hdac1KO, MxCre+;Hdac1KO;Hdac2HET and MxCre+;DKO mice; left panels show haematoxylin–eosin-stained paraffin tissue sections, right panels show immunohistochemistry on paraffin tissue sections using antibodies against activated caspase-3. Note the presence of mitotic figures (asterix) in megakaryocytes of MxCre+;Hdac1KO;Hdac2HET mice. Magnification is × 200. (D) Average megakaryocyte number in bone marrow in six different microscopic fields of three independent mice with indicated genotype. (E) Quantification of activated caspase-3 positive cells in bone marrow of indicated pI;pC-treated genotypes. Shown are average counts in three different microscopic fields in three independent mice per genotype.
Total levels of Hdac1 and Hdac2 are critical for erythrocyte-megakaryocyte development
Although MxCre+;Hdac1KO mice do not display a phenotype in liver or bone marrow, we noted that reduction of Hdac2 levels in the absence of Hdac1 in the bone marrow resulted in an intermediate haematological phenotype as compared with MxCre+;Hdac1KO and MxCre+;DKO mice. pI;pC treatment of MxCre+;Hdac1L/LHdac2L/+ mice (referred to as MxCre+;Hdac1KO;Hdac2HET) resulted in efficient deletion of Hdac1, but retained expression of Hdac2 in liver and bone marrow (Supplementary Figures 4C and 5C). Surprisingly, MxCre+;Hdac1L/LHdac2L/+ mice, like MxCre+;Hdac1L/LHdac2L/L mice, became lethargic on pI;pC injection and displayed anaemic features (Figure 6A). Indeed, MxCre+; Hdac1KO;Hdac2HET displayed a two-fold reduction in peripheral erythrocytes and six-fold reduction in peripheral thrombocytes, as compared with MxCre+ mice. Histological analysis of bone marrow and whole bone-marrow cell counts revealed a 30% reduction in total bone-marrow numbers in MxCre+;Hdac1KO;Hdac2HET mice as compared with MxCre+ bone marrow (Figure 6B). Most strikingly, MxCre+;Hdac1KO;Hdac2HET bone marrow contained reduced amounts of megakaryocytes (Figure 6C). This reduction cannot be explained by apoptosis of Hdac1KO;Hdac2HET megakaryocytes, as these cells are active caspase-3 negative (Figure 6C). Interestingly, these megakaryocytes show an aberrant nuclear morphology and an increased number of megakaryocytes displayed mitotic figures (Figure 6D). In addition, although normal megakaryocytes reside in the bone marrow, Hdac1KO;Hdac2HET megakaryocytes were frequently found intra-vascular or extravagating into bone-marrow blood vessels (Figure 6C). Indeed, we observed significant amounts of Hdac1KO;Hdac2HET megakaryocytes in the liver (Supplementary Figure 6A, B), providing a rational for the reduction of megakaryocytes residing in the bone marrow (Figure 6D). These observations suggest that reduced Hdac2 levels in the absence of Hdac1 result in dys-functional megakaryocytes leading to a severe decrease in thrombocyte counts (Figure 6B).
In summary, although Hdac1 and Hdac2 do not have a critical function in liver homeostasis, these class I Hdacs have redundant functions in erythrocyte-megakaryocyte development. Whereas inactivation of Hdac1 (Figure 6) or Hdac2 (Guan et al, 2009, data not shown) does not result in a haematopoietic phenotype, simultaneous inactivation of Hdac1 and Hdac2 results in severe anaemia and thrombocytopenia. These data suggest that total levels of Hdac1 and Hdac2 are critical for normal development, function and survival of megakaryocytes. Moreover, these results indicate that undesired haematological ‘side' effects of HDACi in the clinic, such as anaemia and thrombocytopenia, at least in part, relate to on-target actions on HDAC1 and HDAC2.
Discussion
Here, we show that Hdac1 and Hdac2 collectively regulate cell cycle progression in primary and transformed fibroblasts by suppressing a senescence-inducing signal transduction pathway. Simultaneous loss of Hdac1 and Hdac2 or specific inactivation of their deacetylase activity results in a senescence-like arrest at the G1-phase of the cell cycle. Interestingly, it was earlier shown that primary human and mouse cells enter a senescent state on treatment with HDACi (Ogryzko et al, 1996; Munro et al, 2004; Place et al, 2005), suggesting a function for HDACs in replicative senescence. Our results indicate that Hdac1 and Hdac2 are primary targets of the relative non-specific HDACi in the induction of senescence. In contrast to deletion of Hdac1 and Hdac2, ablation of mSin3A or mSin3B, two core components of the mSin3/HDAC protein complex, does not result in senescence, but rather in a G2/M cell cycle arrest and apoptosis (Dannenberg et al, 2005), or escape from cellular senescence (David et al, 2008; Grandinetti et al, 2009). Together, these results suggest that Hdac1- and Hdac2-containing complexes, other than the mSin3/HDAC complex, are involved in G1 cell cycle control by Hdac1 and Hdac2. Interestingly, loss of NuRD complex components, such as MTA3 and HDAC1, is associated with premature and normal ageing in human cells (Pegoraro et al, 2009), suggesting a function for an NuRD/Hdac1 and possibly Hdac2 in controlling cell cycle progression during ageing and senescence. Senescent human cells exhibit low levels of HDAC1 and harbour a specific form of HDAC2 (Wagner et al, 2001; Pegoraro et al, 2009), giving further support for a function of these enzymes in promoting proliferation and preventing cellular senescence.
To get insight into the mechanism underlying the senescence-like G1 arrest in cells lacking both Hdac1 and Hdac2, we tested several candidate genes involved in senescence and cell cycle control. Although DKO MEFs show high levels of the cell cycle inhibitor p21Cip, genetic inactivation of p21Cip using p21Cip-specific shRNAs or knock-out alleles showed that p21Cip is not required for the cell cycle arrest observed in DKO cells. Numerous studies have shown that treatment of primary and tumour cell lines with HDACi results in transcriptional up-regulation of p21Cip, which is generally viewed as a critical target for the anti-tumour activity of HDACi (Marks et al, 2004). Although several studies addressed the question whether pRB and/or p53 pathway components are required for an HDACi-induced cell cycle arrest, the results of these studies are not conclusive and seem to depend on cell-type, HDACi concentrations, and the molecular nature of HDACi (Archer et al, 1998; Munro et al, 2004; Matheu et al, 2005). Here, we show, using genetic experiments, that p21Cip and p53 are not required for the cell cycle arrest in DKO MEFs. Inactivation of p16Ink4a and p19Arf, governing the pRb and p53 pathways, also does not allow escape from a senescent-like arrest in DKO MEFs. This suggests that Hdac1 and Hdac2 regulate the pRb and p53 pathways at a more downstream level, or, alternatively, suppress pRb- and p53-independent pathways. In an effort to identify such pathways, we analysed Hdac1/2-deficient MEF gene-expression profiles. This analysis confirmed transcriptional up-regulation of p21Cip, but did not reveal candidate pathways explaining the cause of the cell cycle arrest (data not shown). Interestingly, others have shown that inactivation of p21Cip in Hdac1-deficient ES cells reverts the slow-growth phenotype of these cells (Zupkovitz et al, 2010). Although p21Cip deficiency could not rescue the lethality of Hdac1-deficient embryos, these results show that p21Cip at least in some cell types contributes to the phenotype caused by Hdac1 deficiency. Recently, others have shown that, similar to our results, loss of Hdac1 and Hdac2 results in a G1 cell cycle arrest accompanied by up-regulation of p21Cip (Yamaguchi et al, 2010). In addition, this study showed that Hdac1/2 deficiency results in elevated levels of the cell cycle inhibitor p57Kip. shRNA-mediated down-regulation of p21Cip and p57Kip allowed, to some extent, cell cycle progression in the absence of Hdac1 and Hdac2. Gene-expression profiles of our DKO MEFs did not reveal up-regulation of p57Kip, which could be related to a difference in the sensitivity of the micro-array platform used (Illumina versus Affymetrix) or because of the genetic background of the MEFs. In any case, these results suggest that besides p21Cip and p57Kip, Hdac1 and Hdac2 probably suppress other pathways involved in cell cycle regulation.
Furthermore, our results suggest that the molecular pathways activating a senescence-like arrest are still functional in oncogenic-transformed fibroblasts. Ablation of Hdac1 and Hdac2 in RasV12/p53KD-transformed cells, which have escaped OIS, still enter a senescent state. Recently, Haberland et al (2009a) showed that ablation of Hdac1 and Hdac2 in SV40 Large T-transformed fibroblasts results in a G2/M cell cycle arrest. As SV40 Large T inactivates the pRb- and p53-tumour suppressor pathways, these results are consistent with our observations that Hdac1 and Hdac2 regulate cell cycle progression independent of p16Ink4a, p19Arf and p53. Given that the majority of tumours exhibit genetic alterations resulting in the inactivation of the pRB and p53 tumour suppressor pathways, specific pharmacological inhibition of HDAC1 and HDAC2 activity might be a suitable therapeutic approach in the treatment of tumours harbouring mutations in these pathways.
Whereas primary and transformed fibroblasts strongly depend on either Hdac1 or Hdac2 to maintain their proliferative potential, we show that these class I Hdacs are not essential for the maintenance of post-mitotic hepatocytes. The difference in phenotypes might be the result of a cell-type-specific context such as the presence or absence of HDAC-recruiting sequence-specific transcription factors. Alternatively, these phenotypes might also be related to the proliferation state of the cell. Recent observations suggest that the proliferation state of cardiac cells (cycling versus post-mitotic) determines whether Hdac1/2 deficiency results in apoptosis (Haberland et al, 2009a). Similar observations have been made in resting versus cycling B-lymphocytes (Yamaguchi et al, 2010). These observations predict that Hdac1 and/or Hdac2 are essential for proliferating hepatocytes, for example when forced to proliferate on a partial hepatectomy.
Conditional inactivation of Hdac1 and Hdac2 in bone marrow results in severe anaemia and thrombocytopenia indicative for overlapping functions of Hdac1 and Hdac2 in the differentiation of the megakaryocyte-erythrocyte cell lineage. Interestingly, the nucleosome and remodelling complex (NuRD), harbouring Hdac1 and Hdac2, was found to be a critical protein complex involved in megakaryocyte-erythrocyte differentiation by recruiting GATA-1 and FOG-1, two essential transcription factors for the differentiation of megakaryocytes and erythrocytes (Hong et al, 2005; Rodriguez et al, 2005; Miccio et al, 2009; Gao et al, 2010). In addition, it was found that Hdac1 and Hdac2 are components of an NuRD and SIN3 complex involved in erythrocyte differentiation (Brand et al, 2004). In concordance with observations described here, knock-down of HDAC1 in early human haematopoietic progenitors results in a decrease in erythroid differentiation (Wada et al, 2009).
Our results suggest that the redundancy between Hdac1 and Hdac2 in haematopoiesis may be related to the total levels of Hdac1 and Hdac2. Ablation of Hdac1 or Hdac2 results in compensatory up-regulation of Hdac2 or Hdac1, respectively. The levels of Hdac1 or Hdac2 are sufficient to allow cell proliferation or haematopoiesis. A reduction in Hdac2 combined with Hdac1 deficiency results in total Hdac1/2 levels, which are not compatible with normal haematopoiesis as evidenced by the anaemia and thrombocytopenia in MxCre+;Hdac1KO;Hdac2HET mice. Interestingly, preliminary analysis of germ-line Hdac1HET;Hdac2KO mice suggests that these mice are viable and do not develop anaemia or thrombocytopenia (data not shown). This suggests that despite the observed functional redundancy between Hdac1 and Hdac2, these proteins are not equally redundant.
Moreover, these results establish that HDACi-treatment-related haematological toxicities in the clinic, such as anaemia and thrombocytopenia, are caused, at least in part, by on-target effects on HDAC1 and HDAC2. This knowledge may guide the design of new HDACi or the development of new HDACi-treatment regimen. Elucidation of the molecular pathways contributing to senescence-like cell cycle arrest on inhibition of either Hdac1 or Hdac2 will improve our understanding of the function of these class I HDACs in cellular proliferation, differentiation and will allow the identification of cancer patients' response to pharmacological inhibition of HDAC1 and HDAC2.
Materials and methods
Mouse strains and generation of Hdac1 cKO mice
To generate a cKO allele for Hdac1, a targeting construct was generated by flanking Hdac1 exon 2 by loxP sites. Upon targeting in ES cells, successful germ-line transmission was obtained by breeding chimeric mice to C57Bl6 mice. Mice carrying an Hdac1 cKO allele (Hdac1cKO) were intercrossed with germ-line Cre-transgenic mice to generate an Hdac1-null allele. Hdac1 cKO mice were crossed onto mice carrying an Hdac2 (conditional) knock-out allele (Guan et al, 2009). Conditional deletion in MEFs was obtained by breeding Hdac1 and Hdac2 cKO mice onto Rosa26CreERT2 mice (Hameyer et al, 2007). To delete Hdac1 and Hdac2 in the haematopoietic system, we crossed the interferon-inducible MxCre allele (Kuhn et al, 1995) onto an Hdac1L/L;Hdac2L/L background. Cre-recombinase was induced by intraperitoneal injection of 300 mg pI:pC (Sigma) in 150 ml of phosphate-buffered saline (PBS), five times, every other day. Mice were killed 4–10 days after the last injection. p21−/− and Cdkn2a−/− mice have been described before (Deng et al, 1995; Krimpenfort et al, 2001).
Cell culture, retroviral infection, retroviral constructs and growth curve analysis
MEFs were isolated from E13.5 embryos and cultured in DMEM supplemented with 10% FBS, glutamate, penicillin and streptomycin. MEFs harbouring the Rosa26CreERT2 allele were treated with 200 nM tamoxifen (4-OHT, Sigma) to activate Cre-recombinase. Retroviruses were generated by CaPO4 co-transfection of pCL helper plasmid (Naviaux et al, 1996) and retroviral plasmid into 293T cells. After 48 h of transfection, viral supernatants were filtered (22 μm). Fresh viral supernatants were supplemented with 4 μg/ml polybrene and added to target cells for 6 h. Fresh medium was added to the infected cells overnight followed by a second infection with fresh viral supernatant. Infected cells were selected using puromycin (2 μg/ml) for at least 2 days. To delete Hdac1, infected cells were also treated with 200 nM 4-OHT until the end of the experiment. The p53 and p21Cip shRNA constructs are described (Dirac and Bernards, 2003; Foijer et al, 2005). Hdac1 shRNA or control constructs were generated by cloning 19-mer Hdac1 shRNA sequences (5′-GGCAAGTACTATGCTGTGA-3′) or control sequences (5′-GTCTGTTACTACTACGACG-3′) into pRetroSuper (Brummelkamp et al, 2002). Growth curve analyses were performed as described (Dannenberg et al, 2000, 2005).
BrdU-PI FACS analysis
BrdU-PI FACS analysis was performed identical as described (Dannenberg et al, 2005). MEF cultures were analysed 8–10 days after treatment with 200 nM 4-OHT.
SA-β-gal staining
Staining for SA-β-gal was performed after washing cells with PBS and fixing them in 2% formaldehyde, 0.2% glutaraldehyde in PBS for 5 min. Subsequently, cells were washed with PBS and incubated for 24–36 h at 37°C in a staining solution containing 5 mM K3(CN)6, 5 mM K4Fe(CN)6, 2 mM MgCl2, 150 mM NaCl, 1 mg/ml X-gal (in DMF) and 40 mM citric acid/Na2HPO4 (pH 6.0). Phase contrast pictures were taken from stained cultures stored in 4% formaldehyde and number of SA-β-gal positive cells were quantified.
Generation of Hdac1 and Hdac2 catalytic inactive mutants
Hdac1 and Hdac2 mouse cDNA were cloned from MEF cDNA and recombined into pDONR223 plasmids (Rual et al, 2004). Hdac1D99A, Hdac1Y303F, Hdac2D100A and Hdac2Y304F mutants were generated using pDONR223-Hdac1 and pDONR223-Hdac2 plasmids in combination with the QuickChange Lightning Site-Directed Mutagenesis kit (Stratagene) according to the manufacturers' protocol. After mutagenesis, the Hdac1 and Hdac2 inserts were transferred from the pDONR223 donor vector into the pQCXIP-DEST retroviral destination vector (kind gift of Dr Kenneth Scott) using Gateway LR Clonase recombination (Invitrogen).
Hdac enzymatic-activity assay
Wild-type and mutant Hdac1 or Hdac2 were expressed in 293T cells by transfecting 12.5 μg of retroviral vectors. Hdac1 or Hdac2 was immunoprecipitated from 1 mg of total cell lysate with 10 μg Hdac1 or Hdac2 antibody using the Pierce Crosslink Immunoprecipitation kit (Pierce Biotechnology) according to the manufacturers' protocol. As a negative control, we used 10 μg IgG antibody to determine non-specific-bound Hdac activity. Immunoprecipitates were subsequently assayed for Hdac activity using the HDAC Fluorimetric Activity Assay kit (Enzo life Sciences), according to the manufacturers' instructions. Hdac activity values were corrected for non-specific (IgG)-bound Hdac activity and divided by wild-type Hdac1 or Hdac2 values to determine the relative Hdac activity.
Western blot analysis
Protein levels were determined by western blot analyses using routine protocols. Antibodies against Hdac2 (SC-7899), Hdac8 (E5), Cdk4 (C-22), p21Cip (C-19), p16Ink4a (M156) were obtained from Santa Cruz Biotechnology; Hdac1 antibody (IMG-337) from Imgenex; p27Kip (3698) and Hdac3 (3949) antibodies from Cell Signaling; p19Arf antibody (Ab80) from Abcam and p53 (IMX25) antibody from Monosan. Peroxidase-conjugated goat anti-rabbit and goat anti-mouse secondary antibodies were from Dako.
Immunofluorescence staining on cells
For immunodetection of proteins in MEFs, we cultured MEFs on 0.8 cm2 glass bottom chamber wells (Nunc) at a density of 2 × 105 cells per well in DMEM containing 200 nM 4-OHT (Sigma). In case of retroviral infection, MEFs were selected using 2 μg/ml puromycin for at least 2 days. MEFs were fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100 and blocked with 1% bovine serum albumin in PBS. Cells were stained with antibodies directed against Hdac1 (IMG-337, Imgenex) and Hdac2 (SC-7899, Santa Cruz Biotechnology) and analysed by confocal microscopy. The DNA was counterstained with TO-PRO-3 (Invitrogen). Alexa fluorisothicyanaat (FITC)-488 nm (Invitrogen) was used as secondary antibody.
Immunohistochemistry on tissue sections
For immunodetection of proteins in tissue sections of mice, we dissected mice and fixed tissues in ethanol–acetic acid–formol saline for 24 h. Tissues were subsequently embedded in paraffin. Tissue sections were dried on glass slides overnight at 37°C. Subsequently, tissue slides were incubated for 2 h at 56°C and de-paraffinized. Antigen retrieval was obtained by heating slides for 1 min at 900 W and 15 min at 250 W in 0.01 M citrate buffer, pH 6. Endogenous peroxidase activity was blocked for 10 min with 3% H2O2 in distilled water. Slides were washed in distilled water for 5 min, followed by a 5 min wash step in PBS (pH 7.4). Pre-incubation was performed for 30 min with 5% normal goat serum (Sanquin) in 1% PBS/BSA followed by an overnight incubation with primary antibodies at 4°C. Hdac1 (Imgenex) and Hdac2 (Santa Cruz Biotechnology) antibodies were used in a 1:500 dilution in 1% PBS/BSA, whereas the caspase-3 antibody (Cell Signalling Technology) was used in a 1:100 dilution. Subsequently, slides were rinsed three times for 5 min in PBS on which slides were incubated for half an hour with secondary antibody and HRP complex (PowerVision poly-HRP-Anti-Rabbit IgG, ImmunoLogic). Slides were rinsed in PBS and incubated with DAB substrate chromogen system (Dako). DAB-reaction was stopped by rinsing slides in running tap water followed by a 2 min rinse in distilled water. Counterstain was performed with haematoxylin (Merck) and subsequently with water for 5 min. Finally, slides were dehydrated and mounted with a xylene-based mount solution (Vectashield).
Whole blood analysis
Mouse peripheral blood was obtained by cardiac puncture and collected in EDTA-coated vials. A total of 15 μl of whole blood was analysed using the Beckman Coulter ACT 10 Haematology Blood Analyser.
Supplementary Material
Acknowledgments
We thank Thomas Rosahl and Merck & Co. Genetically Engineered Models Center of Excellence for providing the unpublished Hdac1 cKO mice designed and produced in collaboration with Xavier Warot, Marie-Christine Birling and Guillaume Pavlovic at the Institut Clinique Souris, Strasbourg, France. We thank Victoria Richon, Chantale Guy, Judy Fleming and Andreas Bloecher at Merck and Co. for discussions on this project. We thank Paul Krimpenfort and Anton Berns for generously providing p21−/− and Cdkn2a cKO mice. We thank Ton Schrauwers, Corine van Langen, Auke Zwerver, Cor Spaan and Dienke Jonkers for animal care. We thank Hein te Riele and Floris Foijer for providing p21Cip shRNA constructs, Hilda de Vries for p53 reagents and Rene Bernards for p53 shRNA constructs. We are grateful to Martin van der Valk for histology review. We thank Kenneth Scott for providing the pQXCIP-DEST retroviral vector. We thank Hein te Riele and Paul Krimpenfort for critical reading of the paper. This work was supported by grants from the NWO to J-HD (NWO-VIDI 864.07.008) and HJ (NWO-VIDI 917.56.328) and the Dutch Cancer Society (KWF-2007-3978).
Footnotes
The authors declare that they have no conflict of interest.
References
- Archer SY, Meng S, Shei A, Hodin RA (1998) p21(WAF1) is required for butyrate-mediated growth inhibition of human colon cancer cells. Proc Natl Acad Sci USA 95: 6791–6796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskara S, Chyla BJ, Amann JM, Knutson SK, Cortez D, Sun ZW, Hiebert SW (2008) Deletion of histone deacetylase 3 reveals critical roles in S phase progression and DNA damage control. Mol Cell 30: 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand M, Ranish JA, Kummer NT, Hamilton J, Igarashi K, Francastel C, Chi TH, Crabtree GR, Aebersold R, Groudine M (2004) Dynamic changes in transcription factor complexes during erythroid differentiation revealed by quantitative proteomics. Nat Struct Mol Biol 11: 73–80 [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R (2002) Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell 2: 243–247 [DOI] [PubMed] [Google Scholar]
- Campisi J (2005) Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell 120: 513–522 [DOI] [PubMed] [Google Scholar]
- Campos EI, Reinberg D (2009) Histones: annotating chromatin. Annu Rev Genet 43: 559–599 [DOI] [PubMed] [Google Scholar]
- Dannenberg JH, David G, Zhong S, van der Torre J, Wong WH, Depinho RA (2005) mSin3A corepressor regulates diverse transcriptional networks governing normal and neoplastic growth and survival. Genes Dev 19: 1581–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannenberg JH, van Rossum A, Schuijff L, te Riele H (2000) Ablation of the retinoblastoma gene family deregulates G(1) control causing immortalization and increased cell turnover under growth-restricting conditions. Genes Dev 14: 3051–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David G, Grandinetti KB, Finnerty PM, Simpson N, Chu GC, Depinho RA (2008) Specific requirement of the chromatin modifier mSin3B in cell cycle exit and cellular differentiation. Proc Natl Acad Sci USA 105: 4168–4172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C, Zhang P, Harper JW, Elledge SJ, Leder P (1995) Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell 82: 675–684 [DOI] [PubMed] [Google Scholar]
- Dirac AM, Bernards R (2003) Reversal of senescence in mouse fibroblasts through lentiviral suppression of p53. J Biol Chem 278: 11731–11734 [DOI] [PubMed] [Google Scholar]
- el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B (1993) WAF1, a potential mediator of p53 tumor suppression. Cell 75: 817–825 [DOI] [PubMed] [Google Scholar]
- Foijer F, Wolthuis RM, Doodeman V, Medema RH, te Riele H (2005) Mitogen requirement for cell cycle progression in the absence of pocket protein activity. Cancer Cell 8: 455–466 [DOI] [PubMed] [Google Scholar]
- Gao Z, Huang Z, Olivey HE, Gurbuxani S, Crispino JD, Svensson EC (2010) FOG-1-mediated recruitment of NuRD is required for cell lineage re-enforcement during haematopoiesis. EMBO J 29: 457–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandinetti KB, Jelinic P, DiMauro T, Pellegrino J, Fernandez Rodriguez R, Finnerty PM, Ruoff R, Bardeesy N, Logan SK, David G (2009) Sin3B expression is required for cellular senescence and is up-regulated upon oncogenic stress. Cancer Res 69: 6430–6437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoretti IV, Lee YM, Goodson HV (2004) Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol 338: 17–31 [DOI] [PubMed] [Google Scholar]
- Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, Nieland TJ, Zhou Y, Wang X, Mazitschek R, Bradner JE, DePinho RA, Jaenisch R, Tsai LH (2009) HDAC2 negatively regulates memory formation and synaptic plasticity. Nature 459: 55–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland M, Johnson A, Mokalled MH, Montgomery RL, Olson EN (2009a) Genetic dissection of histone deacetylase requirement in tumor cells. Proc Natl Acad Sci USA 106: 7751–7755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland M, Mokalled MH, Montgomery RL, Olson EN (2009b) Epigenetic control of skull morphogenesis by histone deacetylase 8. Genes Dev 23: 1625–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland M, Montgomery RL, Olson EN (2009c) The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet 10: 32–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hameyer D, Loonstra A, Eshkind L, Schmitt S, Antunes C, Groen A, Bindels E, Jonkers J, Krimpenfort P, Meuwissen R, Rijswijk L, Bex A, Berns A, Bockamp E (2007) Toxicity of ligand-dependent Cre recombinases and generation of a conditional Cre deleter mouse allowing mosaic recombination in peripheral tissues. Physiol Genomics 31: 32–41 [DOI] [PubMed] [Google Scholar]
- Hong W, Nakazawa M, Chen YY, Kori R, Vakoc CR, Rakowski C, Blobel GA (2005) FOG-1 recruits the NuRD repressor complex to mediate transcriptional repression by GATA-1. EMBO J 24: 2367–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson SK, Chyla BJ, Amann JM, Bhaskara S, Huppert SS, Hiebert SW (2008) Liver-specific deletion of histone deacetylase 3 disrupts metabolic transcriptional networks. EMBO J 27: 1017–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimpenfort P, Quon KC, Mooi WJ, Loonstra A, Berns A (2001) Loss of p16Ink4a confers susceptibility to metastatic melanoma in mice. Nature 413: 83–86 [DOI] [PubMed] [Google Scholar]
- Kuhn R, Schwenk F, Aguet M, Rajewsky K (1995) Inducible gene targeting in mice. Science 269: 1427–1429 [DOI] [PubMed] [Google Scholar]
- Lagger G, O'Carroll D, Rembold M, Khier H, Tischler J, Weitzer G, Schuettengruber B, Hauser C, Brunmeir R, Jenuwein T, Seiser C (2002) Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J 21: 2672–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks PA, Richon VM, Miller T, Kelly WK (2004) Histone deacetylase inhibitors. Adv Cancer Res 91: 137–168 [DOI] [PubMed] [Google Scholar]
- Matheu A, Klatt P, Serrano M (2005) Regulation of the INK4a/ARF locus by histone deacetylase inhibitors. J Biol Chem 280: 42433–42441 [DOI] [PubMed] [Google Scholar]
- Miccio A, Wang Y, Hong W, Gregory GD, Wang H, Yu X, Choi JK, Shelat S, Tong W, Poncz M, Blobel GA (2009) NuRD mediates activating and repressive functions of GATA-1 and FOG-1 during blood development. EMBO J 29: 442–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minucci S, Pelicci PG (2006) Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer 6: 38–51 [DOI] [PubMed] [Google Scholar]
- Montgomery RL, Davis CA, Potthoff MJ, Haberland M, Fielitz J, Qi X, Hill JA, Richardson JA, Olson EN (2007) Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev 21: 1790–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery RL, Potthoff MJ, Haberland M, Qi X, Matsuzaki S, Humphries KM, Richardson JA, Bassel-Duby R, Olson EN (2008) Maintenance of cardiac energy metabolism by histone deacetylase 3 in mice. J Clin Invest 118: 3588–3597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooi WJ, Peeper DS (2006) Oncogene-induced cell senescence—halting on the road to cancer. N Engl J Med 355: 1037–1046 [DOI] [PubMed] [Google Scholar]
- Munro J, Barr NI, Ireland H, Morrison V, Parkinson EK (2004) Histone deacetylase inhibitors induce a senescence-like state in human cells by a p16-dependent mechanism that is independent of a mitotic clock. Exp Cell Res 295: 525–538 [DOI] [PubMed] [Google Scholar]
- Naviaux RK, Costanzi E, Haas M, Verma IM (1996) The pCL vector system: rapid production of helper-free, high-titer, recombinant retroviruses. J Virol 70: 5701–5705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogryzko VV, Hirai TH, Russanova VR, Barbie DA, Howard BH (1996) Human fibroblast commitment to a senescence-like state in response to histone deacetylase inhibitors is cell cycle dependent. Mol Cell Biol 16: 5210–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegoraro G, Kubben N, Wickert U, Gohler H, Hoffmann K, Misteli T (2009) Ageing-related chromatin defects through loss of the NURD complex. Nat Cell Biol 11: 1261–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Place RF, Noonan EJ, Giardina C (2005) HDACs and the senescent phenotype of WI-38 cells. BMC Cell Biol 6: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince HM, Bishton MJ, Harrison SJ (2009) Clinical studies of histone deacetylase inhibitors. Clin Cancer Res 15: 3958–3969 [DOI] [PubMed] [Google Scholar]
- Rasheed W, Bishton M, Johnstone RW, Prince HM (2008) Histone deacetylase inhibitors in lymphoma and solid malignancies. Expert Rev Anticancer Ther 8: 413–432 [DOI] [PubMed] [Google Scholar]
- Rodriguez P, Bonte E, Krijgsveld J, Kolodziej KE, Guyot B, Heck AJ, Vyas P, de Boer E, Grosveld F, Strouboulis J (2005) GATA-1 forms distinct activating and repressive complexes in erythroid cells. EMBO J 24: 2354–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual JF, Hirozane-Kishikawa T, Hao T, Bertin N, Li S, Dricot A, Li N, Rosenberg J, Lamesch P, Vidalain PO, Clingingsmith TR, Hartley JL, Esposito D, Cheo D, Moore T, Simmons B, Sequerra R, Bosak S, Doucette-Stamm L, Le Peuch C et al. (2004) Human ORFeome version 1.1: a platform for reverse proteomics. Genome Res 14 (10B): 2128–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta N, Seto E (2004) Regulation of histone deacetylase activities. J Cell Biochem 93: 57–67 [DOI] [PubMed] [Google Scholar]
- Trivedi CM, Luo Y, Yin Z, Zhang M, Zhu W, Wang T, Floss T, Goettlicher M, Noppinger PR, Wurst W, Ferrari VA, Abrams CS, Gruber PJ, Epstein JA (2007) Hdac2 regulates the cardiac hypertrophic response by modulating Gsk3 beta activity. Nat Med 13: 324–331 [DOI] [PubMed] [Google Scholar]
- Vannini A, Volpari C, Gallinari P, Jones P, Mattu M, Carfi A, De Francesco R, Steinkuhler C, Di Marco S (2007) Substrate binding to histone deacetylases as shown by the crystal structure of the HDAC8-substrate complex. EMBO Rep 8: 879–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Kikuchi J, Nishimura N, Shimizu R, Kitamura T, Furukawa Y (2009) Expression levels of histone deacetylases determine the cell fate of hematopoietic progenitors. J Biol Chem 284: 30673–30683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M, Brosch G, Zwerschke W, Seto E, Loidl P, Jansen-Durr P (2001) Histone deacetylases in replicative senescence: evidence for a senescence-specific form of HDAC-2. FEBS Lett 499: 101–106 [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Cubizolles F, Zhang Y, Reichert N, Kohler H, Seiser C, Matthias P (2010) Histone deacetylases 1 and 2 act in concert to promote the G1-to-S progression. Genes Dev 24: 455–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Seto E (2008) The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol 9: 206–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann S, Kiefer F, Prudenziati M, Spiller C, Hansen J, Floss T, Wurst W, Minucci S, Gottlicher M (2007) Reduced body size and decreased intestinal tumor rates in HDAC2-mutant mice. Cancer Res 67: 9047–9054 [DOI] [PubMed] [Google Scholar]
- Zupkovitz G, Grausenburger R, Brunmeir R, Senese S, Tischler J, Jurkin J, Rembold M, Meunier D, Egger G, Lagger S, Chiocca S, Propst F, Weitzer G, Seiser C (2010) The cyclin-dependent kinase inhibitor p21 is a crucial target for histone deacetylase 1 as a regulator of cellular proliferation. Mol Cell Biol 30: 1171–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.