Abstract
Topoisomerase IIα (topoIIα) is an essential mammalian enzyme that topologically modifies DNA and is required for chromosome segregation during mitosis. Previous research suggests that inhibition of topoII decatenatory activity triggers a G2 checkpoint response, which delays mitotic entry due to insufficient decatenation of daughter chromatids. Here we examine the effects of both topoIIα and topoIIβ on decatenatory activity in cell extracts, DNA damage and decatenation G2 checkpoint function, and the frequencies of p16INK4A allele loss and gain. In diploid human fibroblast lines, depletion of topoIIα by siRNA was associated with severely reduced decatenatory activity, delayed progression from G2 into mitosis, and insensitivity to G2 arrest induced by the topoII catalytic inhibitor ICRF-193. Furthermore, interphase nuclei of topoIIα-depleted cells displayed increased frequencies of losses and gains of the tumor suppressor genetic locus p16INK4A. This study demonstrates that the topoIIα protein is required for decatenation G2 checkpoint function, and inactivation of decatenation and the decatenation G2 checkpoint leads to abnormal chromosome segregation and genomic instability.
Keywords: decatenation, G2 checkpoint, topoisomerase IIα, topoisomerase IIβ, chromosomal instability
Introduction
DNA topoisomerases are evolutionarily conserved nuclear enzymes that have major functions in the cell cycle, including DNA replication, recombination, and chromosome segregation (Larsen et al. 1996; Wang 2002). The two major families of topoisomerases are differentiated by the type of enzymatic reactions each performs. Type I topoisomerases produce protein-associated single strand breaks in DNA and relieve supercoiling tension by free rotation of the cut strand around the intact strand. Type II topoisomerases produce protein-associated DNA double-strand breaks (dsb) and are capable of passing an intact DNA duplex through the protein-associated dsb. Thus, only type II topoisomerases can separate knotted and intertwined DNA molecules. Mammalian species express two topoII isoforms: α and β. Each isoform is encoded by a separate gene located on different human chromosomes and can be distinguished by mass (Austin and Marsh, 1998; Burden and Osheroff, 1998; Capranico et al., 1992). The two isoforms share ~70% homology at the amino acid level, and exist as homodimeric enzymatic complexes with similar catalytic activities, although α/β heterodimers have been observed (Gromova et al., 1998; Biersack et al., 1996).
TopoIIβ is non-essential and constitutively expressed, whereas topoIIα is an essential gene maximally expressed in G2 and M (Goswami et al., 1996; Heck et al., 1988; Akimitsu et al., 2003). TopoII activity is required for chromosome condensation, decatenation of intertwined daughter DNA duplexes, and centromere resolution (Wang et al., 2008; Coelho et al., 2008; Maeshima et al., 2005). Catenations between chromatid arms are removed prior to mitosis, while centromeric catenations persist up to the metaphase/anaphase transition (Diaz-Martinez et al., 2006; Santamaria et al., 2007).
DNA and topoIIα form a reversible, covalent complex, often referred to as the cleavage complex (Burden and Osheroff, 1998; Cortes et al., 2003). TopoII-inhibiting drugs interfere with various stages of the catalytic cycle, and are therefore divided into two classes: poisons and catalytic inhibitors. TopoII poisons such as doxorubicin and etoposide stabilize the cleavage complex, which may block DNA replication forks or transcriptional machinery and create dsbs (Xiao et al., 2003). Upon proteolysis of the stabilized cleavage complex, the dsb is exposed; thus, these drugs are used clinically to treat cancers that typically over-express topoII (Mao et al., 2001; Jarvinen et al., 1996; Lorusso et al., 2007).
In contrast, catalytic inhibitors prevent the formation of the cleavage complex by intercalating into DNA and inhibiting topoII binding, or by stabilizing topoII in a closed-clamp conformation after the ligation step of the catalytic cycle (Sehested and Jensen, 1996; Roca et al., 1994). Therfore, topoII catalytic inhibitors are primarily used in the clinical setting as an adjunct to reduce the cardiotoxicity of topoII poisons (Lyu et al., 2007). The ability of topoII catalytic inhibitors to protect against the toxicity of topoII poisons implies that poisons and catalytic inhibitors have very different effects on topoII activity, DNA binding, and chromatin structure.
Studies using topoII catalytic inhibitors suggest that G2 cells monitor the catenation state of intertwined sister chromatids following DNA replication and actively delay progression into mitosis pending sufficient chromatid decatenation (Downes et al., 1994; Deming et al., 2001). A subset of chromatid catenations appear to be organized in the centromere, and these catenations are not separated until the onset of anaphase (Diaz-Martinez et al., 2006). Thus, the decatenation G2 checkpoint appears to monitor the sufficiency of decatenation along the chromosomal arms, not its completion. The expression of an active decatenation G2 checkpoint was supported by evidence that the G2 delay in cells with catalytic inhibition of topoII was dependent upon BRCA1 expression and could be overridden by caffeine (Deming et al., 2001). BRCA1 may be required to activate topoIIα by ubiquitylation (Lou et al., 2005). Other studies have implicated ATR, ATM, Plk1, WRN, MDC1, and Chk1 in the decatenation G2 checkpoint (Bower et al., 2010; Robinson et al., 2007; Deming et al., 2002; Franchitto et al., 2003; Luo et al., 2009). Although the presence of a decatenation G2 checkpoint that is independent of DNA damage has been supported by a variety of studies (Damelin et al., 2005; Nakagawa et al., 2004; Doherty et al., 2003), the concept is still controversial since ICRF-193 has been shown to activate DNA damage signaling in some cancer cell lines (Nakagawa et al., 2004; Park and Avraham 2006).
The work described herein utilizes normal human diploid fibroblast (NHDF) lines isolated from three different individuals and depleted of either topoIIα or topoIIβ. These data support a distinction between the decatenation and DNA damage G2 checkpoints, and suggest that topoIIα plays a role in maintaining genomic stability through chromatid decatenation and decatenation G2 checkpoint signaling.
Results
TopoIIα is required for decatenatory activity
As there are two forms of topoII in mammals, it was necessary to determine the individual contribution of each isoform to cellular decatenatory activity. NHDFs were depleted of topoIIα or topoIIβ using siRNA and assayed 48 h after electroporation. Representative western blots for three NHDF lines are shown (Fig. 1A, S1A). Nuclear extracts were prepared and assayed for in vitro decatenatory activity upon catenated circular DNA molecules isolated from trypanosome kinetoplasts (kDNA). Incubation of kDNA with NHDF nuclear extracts released free mini-circles and intermediate mobility DNA species that likely represent catenated dimers or trimers, while decatenatory activity was inhibited by the topoII catalytic inhibitor, ICRF-193 (Fig. 1B). Furthermore, decatenatory activity was severely reduced in nuclear extracts isolated from topoIIα-depleted cells relative to cells that were electroporated with non-targeting control (NTC) siRNA (Fig. 1B, S1B). TopoIIβ-depleted nuclear extracts displayed similar activity to that of the NTC extracts. Collectively, these results suggest that topoIIα accounts for the majority of decatenatory activity in actively growing NHDFs.
Figure 1. TopoIIα enzymatic activity accounts for the majority of decatenatory activity in normal human fibroblasts.
A. Western immunoblot analysis showing topoIIα depletion 48 h after electroporation in three different normal human fibroblast lines with an average reduction in protein levels of 94%. TopoIIβ depletion averaged 75%. B. The catalytic topoII inhibitor ICRF-193 decreased the in vitro decatenatory activity of NHDF nuclear extracts. siRNA depletion of topoIIα ablated the decatenatory activity of nuclear extracts, whereas topoIIβ depletion had no effect on decatenatory activity. Results are representative of three independent experiments in three different NHDF cell lines.
Catalytic inhibition of topoII with ICRF-193 inhibits mitotic entry
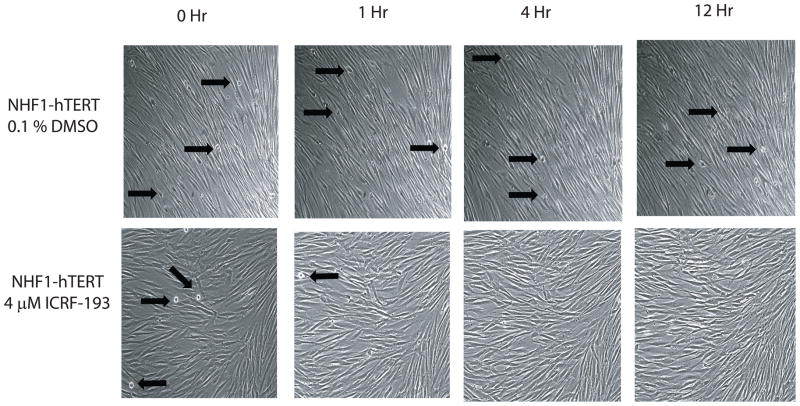
Catalytic inhibitors of topoII are thought to induce a decatenation G2 checkpoint and prevent entry into mitosis. In order to verify that the decatenation G2 checkpoint was effective in NHDFs, live-imaging bright-field microscopy was employed. Mitotic NHDFs were clearly distinguishable from interphase fibroblasts under bright-field microscopy due to their round morphology, whereas interphase fibroblasts were flat, thin, and elongated. Time-lapse images were collected every two minutes and full length movies can be viewed at our laboratory website (http://top2a.med.unc.edu/jackie/home.html). Representative images at 0, 1, 4, and 12 h after addition of DMSO or ICRF-193 are shown, with black arrows designating mitotic cells (Fig. 2). NHDFs exposed to DMSO entered and exited mitosis throughout the length of the movie, whereas catalytic inhibition of topoIIα with 4 μM ICRF-193 allowed mitotic exit, but not mitotic entry. Flow cytometric analysis of cellular DNA content further demonstrated that greater than 50% of fibroblasts were in G2 after 28 h incubation with ICRF-193 (Fig. S2). These results indicated that mitotic entry was arrested in NHDFs upon catalytic inhibition of topoII.
Figure 2. Catalytic inhibition of topoIIα prevents entry into mitosis.
NHDFs were observed via time-lapse bright-field microscopy in the presence of DMSO (vehicle control) or the catalytic topoII inhibitor ICRF-193. Control cells continued to enter and exit mitosis throughout the 24 h period of observation. Black arrows indicate several mitotic cells in each frame. After addition of ICRF-193, all initial mitotic cells exited mitosis within 2–3 hours of topoII inhibition, and no new mitotic figures were observed up to 24 h after treatment (see full length movies at http://top2a.med.unc.edu/jackie/home.html.)
ICRF-193 does not induce detectable DNA damage in NHDFs
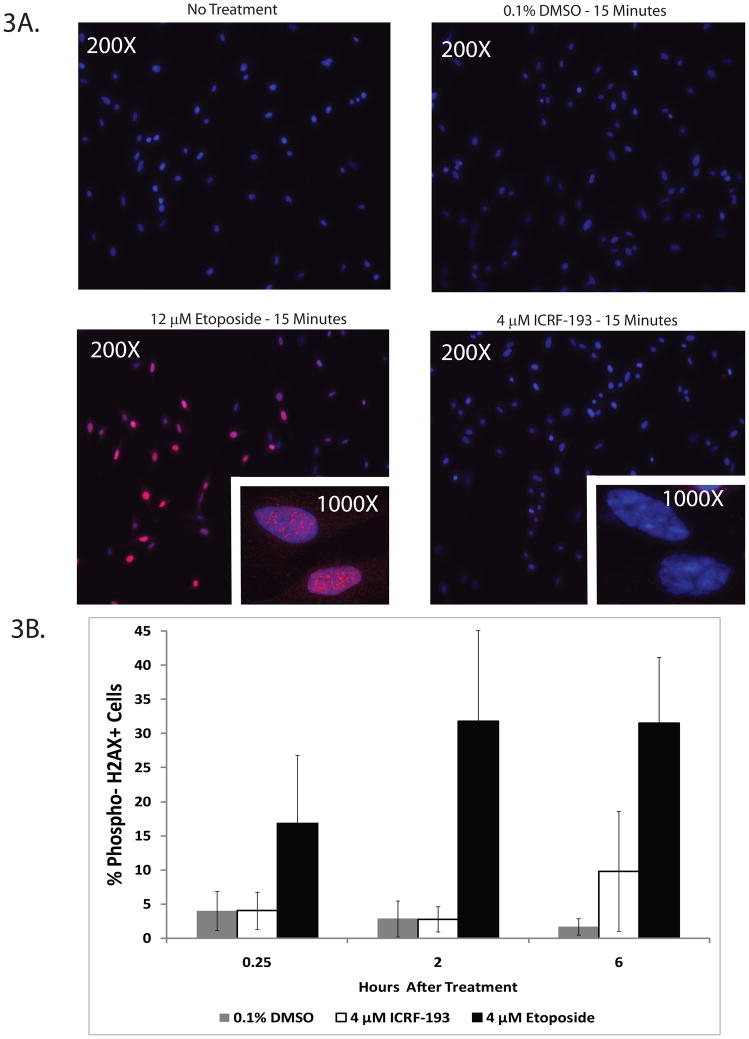
A few studies have detected DNA damage (particularly dsbs) upon inhibition of topoII with ICRF-193. In order to verify that catalytic inhibition of topoIIα with ICRF-193 did not induce a DNA damage response in NHDFs, immunofluorescence and flow cytometric studies were performed to determine the levels of a common DNA damage marker, γH2AX. In Figure 3A, representative images are shown for NHF1-hTERTs treated for 15 min with DMSO, ICRF-193, or etoposide and stained for γH2AX by immunofluorescence. Etoposide-treated NHDFs displayed many γH2AX foci after 15 min, whereas no detectable γH2AX foci were present after ICRF-193 treatment. Flow cytometric studies were performed in parallel to quantify γH2AX positive NHDFs (Fig. 3B). All NHDFs exhibited an increase in γH2AX positive cells after etoposide treatment, whereas ICRF-193 treatment induced no detectable γH2AX staining above background levels. Similar results have been observed by western immunoblot analysis in lymphoblast and fibroblast cell lines (Bower et al., 2010). The slight increase in γH2AX staining seen at the 6 h time point was determined to be statistically insignificant by a Student’s t-test, and was the result of an idiosyncratic response by the NHF3-hTERT cell line. NHF3-hTERTs also displayed higher background γH2AX staining and hypersensitivity to both ICRF-193 and topoIIα depletion (Zhou and Kaufmann, unpublished data and Fig. 7C), suggesting that these cells may display higher levels of cellular stress in culture. Overall, these results indicate that ICRF-193 does not induce dsb formation in NHDFs.
Figure 3. Catalytic inhibition of topoII with ICRF-193 does not induce phospho-H2AX foci in NHDFs.
A. NHF1-hTERTs were exposed to 0.1% DMSO, 4 μM ICRF-193, or 12 μM etoposide. Phospho-Ser 139 H2AX (γH2AX) foci were observed after a 15 min treatment with the topoII poison etoposide, which produces DNA dsbs. Catalytic inhibition of topoII with ICRF-193 did not induce detectable γH2AX foci in NHDFs. Representative images are shown at 200X and 1000X (inset). B. Quantification of the percentage of cells expressing γH2AX in three NHDF lines treated with 4 μ etoposide or 4 μM ICRF-193 by flow cytometry. γH2AX staining increased over time in the etoposide-treated cells. ICRF-193 did not produce any detectable γH2AX staining until 6 h after treatment, and the signal was variable among the different NHDF lines. NHF3-hTERT cells produced substantially more γH2AX staining than the other two fibroblast lines and exhibited higher background levels of γH2AX. Results are presented as an average of three different NHDF cell lines ± SEM. * = p < 0.05.
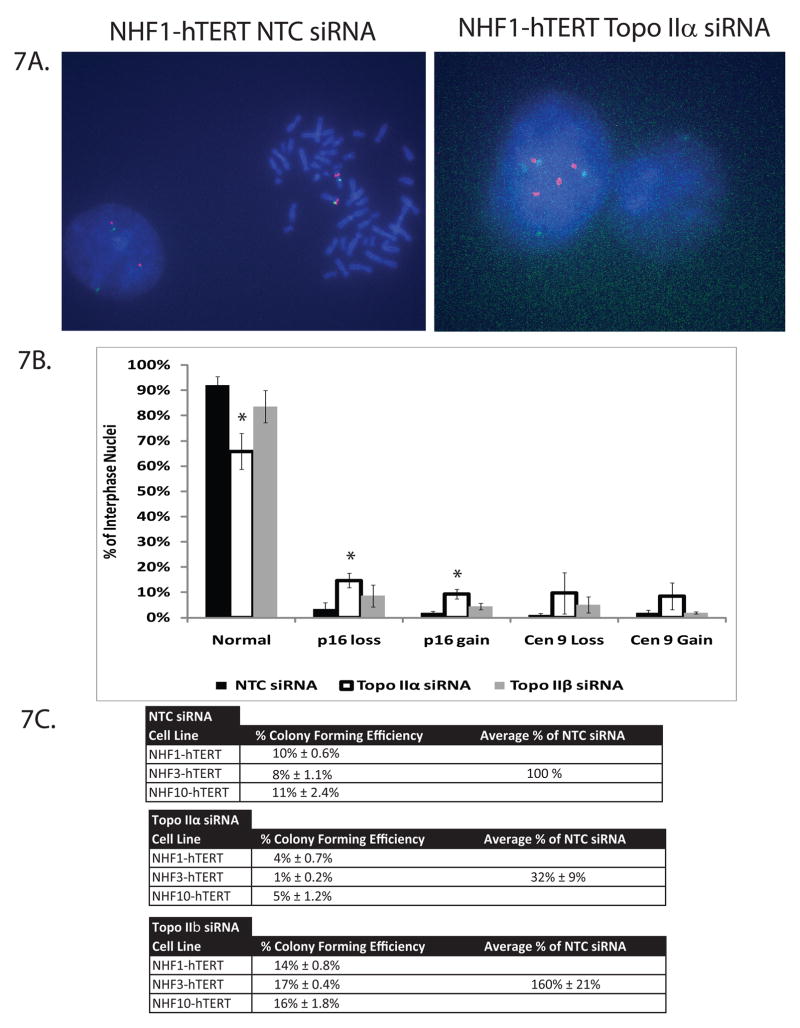
Figure 7. TopoIIα-depleted NHDFs display allelic gains and losses of the tumor suppressor p16INK4A.
A. Examples of NHDF metaphase spreads and interphase nuclei after NTC and topoIIα depletion. The example of topoIIα-depletion depicts a bridged nucleus in which four copies of the p16INK4A allele and centromere 9 appear to undergo improper segregation to the spindle poles. B. Quantification of the number of p16INK4A and centromere 9 alleles in interphase nuclei from NTC-, topoIIα-, and topoIIβ-depleted fibroblasts. Results are an average of three independent blind experiments, one for each NHDF line. At least 100 interphase nuclei per siRNA-depletion were analyzed for each line. * = p < 0.05. C. NHDF lines were treated with NTC, topoIIα, or topoIIβ siRNA and assayed for clonogenic survival. Results are presented as an average of three different NHDF cell lines, assayed in triplicate, and expressed as a percentage of NTC siRNA. Individual colony forming efficiencies are also displayed.
TopoIIα depletion ablates the decatenation G2 checkpoint response, but has no effect on the DNA damage G2 checkpoint response
ICRF-193 inhibits topoII function by holding the enzyme as a closed clamp that is tethered to DNA but incapable of DNA strand passage, thus rendering cells unable to remove DNA catenations (Xiao et al., 2003; Roca et al., 1994; Jensen et al., 2000). Previous studies have suggested that G2 cells sense the presence of entangled and/or catenated chromatids and actively delay mitosis until decatenation is sufficiently advanced to permit mitotic cell division (Downes et al., 1994). Therefore, conditions that slow the rate of chromatid decatenation should reduce the rate of mitotic entry by causing a G2 delay, leading to an increase in the G2 cell fraction.
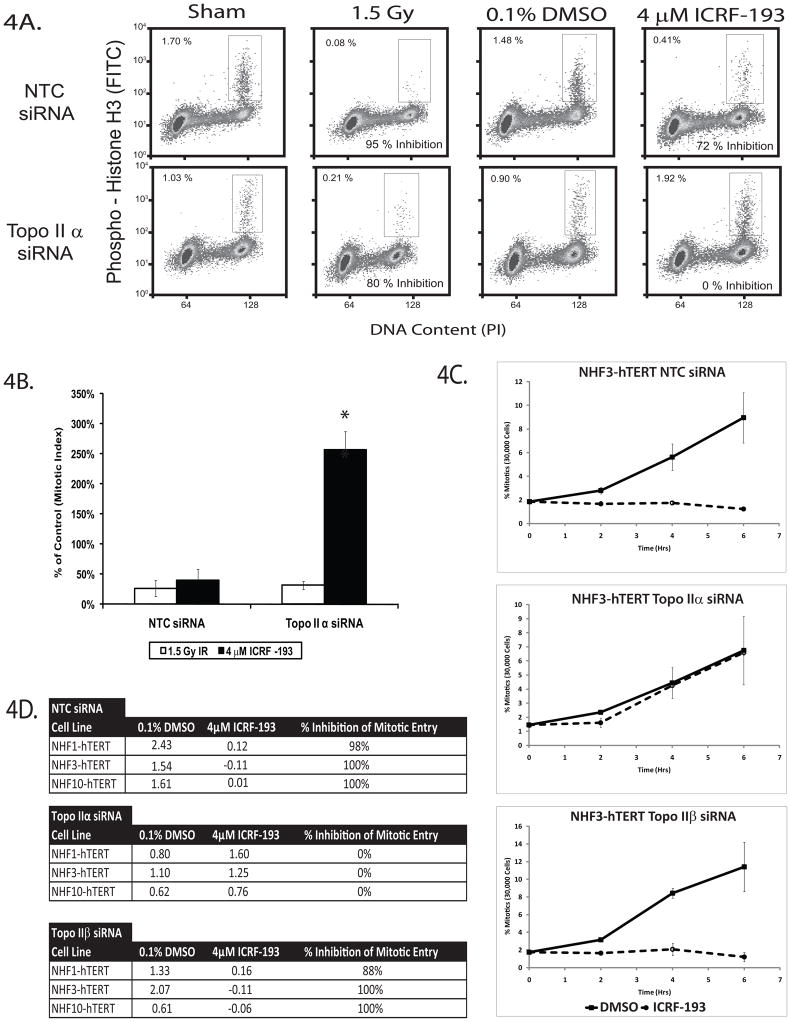
To test this hypothesis, fibroblasts were electroporated with NTC or topoIIα siRNA and analyzed for their ability to arrest in G2 and exit mitosis upon DNA damage (IR) or catalytic inhibition (ICRF-193) of topoII. Representative flow cytometry plots of NHF1-hTERTs are shown in Figure 4A. In both NTC and topoIIα-depleted fibroblasts, IR produced a 75–80% average reduction in mitotic index 2 h after treatment (Fig. 4B). This reduction of the mitotic index has previously been thought to reflect both an arrest of G2 cells and the exit of mitotic cells into interphase after irradiation. The NTC-depleted fibroblasts retained a functional decatenation G2 checkpoint; they displayed a mean 60% reduction in mitotic index upon ICRF-193 treatment. In contrast, topoIIα-depleted fibroblasts displayed a mean 257% increase in mitotic index upon treatment with ICRF-193. These results suggested that while topoIIα depletion did not significantly alter the DNA damage G2 checkpoint response, it ablated the decatenation G2 checkpoint response.
Figure 4. TopoIIα is required for the decatenation G2 checkpoint, but not for the DNA damage G2 checkpoint response.
A. Dual color flow cytometry using a mitosis-specific antibody directed towards phospho-Ser10 histone H3 and propidium iodide to analyze DNA content was employed to examine the G2 checkpoint response to catalytic inhibition of topoIIα. NHDFs depleted of topoIIα emptied mitosis 2 h after treatment with 1.5 Gy of IR. In contrast, topoIIα-depleted fibroblasts failed to empty mitosis 2 h after ICRF-193 treatment, and instead appeared to accumulate in mitosis. Results are representative of three independent experiments with NHF1-hTERTs. B. Quantification of flow cytometry results illustrated in Figure 4A. Results are the average of three independent experiments with NHF1-hTERTs. * = p < 0.0025. C. Depletion of topoIIα allows NHDFs to continuously enter mitosis in the presence of ICRF-193. The rate of mitotic entry of topoIIα-depleted NHDFs was approximately half the rate of those cells electroporated with NTC siRNA on average. TopoIIβ-depleted fibroblasts displayed similar mitotic entry rates and a similar response to ICRF-193 as the NTC-treated fibroblasts. The results depicted in the illustration are the average of three independent experiments with NHF3-hTERTs, and were robustly conserved in all three NHDFs (shown in Fig. S3). D. Rates of mitotic entry for NTC-, topoIIα-, and topoIIβ-depleted NHDFs. Mitotic entry rates were calculated as linear regression slopes from Fig. 4C. The ratios of the slopes (4 μM ICRF-193:0.1% DMSO) were used to determine the percentage of mitotic entry inhibition (1-ratio × 100%). Maximum and minimum values of inhibition were set at 100% and 0%, respectively. Results are an average of three independent experiments for each NHDF line. * = p < 0.005.
One explanation for the increase in topoIIα-depleted mitotic cells after ICRF-193 treatment was that NHDFs were delaying mitotic exit due to the persistence of centromeric DNA catenations (Wang et al., 2008; Coelho et al., 2008; Diaz-Martinez et al., 2006; Santamaria et al., 2007). This effect might be enhanced upon further inhibition of residual topoIIα and topoIIβ with ICRF-193, thus prolonging the amount of time spent in mitosis. If the amount of time NHDFs spent in mitosis was altered by a decrease in topoIIα activity, it was necessary to monitor the G2/M transition by examining the mitotic entry rate (MER) of NHDFs in order to discriminate between the G2/M and spindle assembly checkpoint mechanisms. NHDFs were collected over time in colcemid, a microtubule polymerization inhibitor that arrests cells with an intact spindle assembly checkpoint at metaphase. This assay allows for a strict examination of the G2 to M transition, and calculation of the MER by linear regression provides a quantitative measure of decatenation G2 checkpoint function.
NTC-, topoIIα-, and topoIIβ-depleted fibroblast lines displayed an increase in the percentage of mitotic cells over time in the presence of DMSO and colcemid (Fig. 4C, S3). In contrast, when topoII activity was inhibited with ICRF-193 in NTC- or topoIIβ-depleted fibroblasts, MERs were approximately zero. Surprisingly, the topoIIα-depleted fibroblasts continued to enter mitosis, regardless of the presence of ICRF-193. Comparison of MERs indicated that topoIIα-depleted cells entered mitosis at a rate that was approximately half that of NTC-treated fibroblasts (Fig. 4D). Mitotic entry in the topoIIα-depleted NHF1-hTERTs was independently verified by time-lapse bright-field microscopy and full length videos can be viewed at the laboratory’s website (http://top2a.med.unc.edu/jackie/home.html). Quantification of G2 fractions in this assay also revealed that a significant G2 accumulation occurred upon topoIIα depletion or catalytic inhibition (Fig. S4).
Several attempts to correct the phenotype of NHDFs by over-expressing a siRNA-resistant form of topoIIα were unsuccessful due to the toxicity of topoIIα over-expression. Therefore, a second siRNA directed towards topoIIα was tested in the mitotic entry assay. The results confirmed that the observed phenotype was not due to off-target effects of either siRNA (Fig. S5). Taken together, these data suggest that a catalytically inactive topoIIα protein is required to arrest cells in G2 during incubation with ICRF-193 and prevent mitotic entry in the presence of entangled/catenated chromosomes. Furthermore, these results indicate that the decatenation G2 checkpoint does not sense DNA catenations per se, but rather senses the catalytically inactive form of topoIIα.
TopoIIα depletion does not affect the ability of NHDFs to synthesize DNA
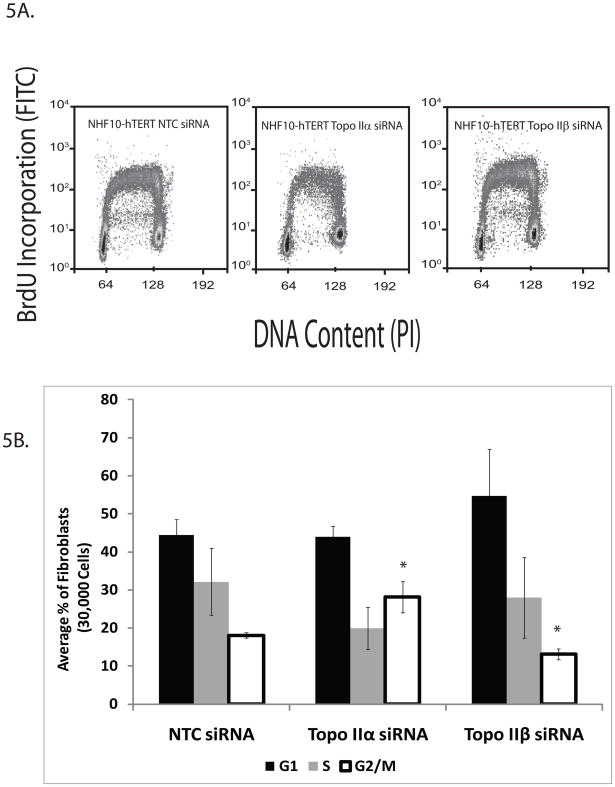
Since topoIIα protein expression is cell cycle-dependent and a previous report suggested that type II topoisomerases are required for the completion of S phase in budding yeast (Baxter and Diffley, 2008), it was important to determine whether topoIIα played a role in DNA synthesis and/or the completion of S phase in NHDFs. If DNA replication or completion of S phase were impaired, this could slow down the MERs. Flow cytometry was utilized to determine the capacity of topoIIα-depleted cells to incorporate the thymidine analog BrdU during S phase and representative flow cytometry profiles are shown (Fig. 5A). While topoIIα-depleted cells displayed normal levels of BrdU incorporation, there were slightly fewer fibroblasts present in S phase when compared to NTC-treated fibroblasts (Fig. 5B). This difference was determined to be insignificant by Student’s t-test. However, the percentage of G2 cells increased in topoIIα-depleted fibroblasts, suggesting that cells were indeed progressing through S phase and into G2. Similar results were obtained upon ICRF-193 treatment (Fig. S6) and by using synchronized fibroblasts (not shown), suggesting that topoIIα depletion or inhibition does not interfere with DNA replication or completion of S phase in NHDFs. In addition, recently published data from our laboratory has suggested that neither ATR nor Chk1 depletion affected the MERs or decatenation G2 checkpoint function of NHDFs (Bower et al., 2010). Similarly, no Chk1 activation was observed in response to ICRF-193 treatment. These data further support the conclusion that topoII catalytic inhibition or depletion does not induce a replicative stress response in NHDFs.
Figure 5. Depletion of topoII α or topoIIβ does not affect DNA replication or completion of S phase in NHDFs.
A. Representative flow cytometry images of NHF10-hTERTs analyze for the ability to incorporate BrdU and plotted against DNA content. BrdU-incorporating cells between 2N and 4N DNA content were designated as S phase cells. B. Total percentages of fibroblasts in G0/G1, S, or G2/M compartments in the presence or absence of topoIIα or topoIIβ. There was no statistically significant change in the percentage of cells found in G1 among the three groups of NHDFs. TopoIIα- -depleted NHDFs exhibited an increase in the percentage of G2/M fibroblasts compared to the NTC treated NHDFs. However, neither topoIIα- nor topoIIβ-depletion affected BrdU incorporation or initiated replication fork collapse due to replication stress (cells containing S phase DNA content but no longer incorporating BrdU). Results are presented as the average of three independent experiments, each using a different NHDF lines. * = p< 0.05.
TopoIIα-depleted fibroblasts display abnormal metaphase chromosome structure
Inhibition of topoII with ICRF-193 produces characteristic abnormalities in cells that evade the decatenation G2 checkpoint. Metaphase preparations of ICRF-193-treated cells often display under-condensed and entangled chromosomes (Deming et al., 2002). Since the results in Figures 1B and 4C indicated that NHDFs enter mitosis in the absence of significant topoIIα activity, cytogenetic studies were performed to examine chromosome morphology. TopoIIα-depletion significantly increased the frequencies of metaphase preparations with under-condensed or entangled chromosomes, whereas topoIIβ-depleted fibroblasts displayed no significant increases in chromosomal aberrations (Fig. 6A, 6B). Thus, depletion of topoIIα produced similar effects on chromosome structure as catalytic inhibition of topoII with ICRF-193.
Figure 6. NHDFs entering mitosis without topoIIα activity have abnormal metaphase chromosome morphology and exit mitosis with abnormal nuclear morphology.
A. Examples of NHDF metaphase spreads after NTC siRNA treatment and topoIIα depletion. Two phenotypes were observed in the topoIIα-depleted fibroblasts: under-condensation and entanglement. B. Quantification of metaphase spreads from NTC-, topoIIα-, and topoIIβ-depleted fibroblasts. Results are an average of three independent experiments, each using a different NHDF lines. At least 50 metaphase spreads per siRNA-depletion were analyzed in a blinded manner for each line. * = p< 0.0025. C. Examples of NHDF interphase nuclei and the types of abnormalities scored. D. Quantification of the abnormal interphase nuclei in cells treated with NTC, topoIIα, or topoIIβ siRNA. Approximately 33% and 14% of interphase nuclei were abnormal after topoIIα or topoIIβ depletion, respectively. Results are the average of three independent experiments in the NHF1-hTERT line. Greater than one thousand interphase nuclei per treatment were analyzed. * = p < 0.01.
Depletion of topoIIα increases the frequency of abnormal interphase nuclei and promotes losses and gains of the p16INK4A tumor suppressor allele
Although the cytogenetic experiments in Fig. 6A/B established that NHDFs would enter mitosis with entangled chromosomes, it was unclear whether these cells would exit mitosis, re-enter interphase, and potentially duplicate a set of damaged or aneuploid chromosomes during subsequent rounds of DNA replication and cell division. Since entangled chromosomes may not segregate properly at anaphase, failures of segregation (non-disjunction) can generate anaphase bridges, lagging chromosomes, and/or chromosomal breakage. After restitution of interphase nuclear structure, thin bridges connecting two nuclei may be apparent or one nucleus may have two lobes. Laggard or broken chromosomes are often manifested as micronuclei. The frequencies of interphase nuclei with structural alterations such as bridges, blebs and micronuclei were therefore examined (Fig. 6C). While NTC-treated fibroblasts exhibited a low frequency of nuclear abnormalities (<5%), the frequencies of abnormalities were increased to ~33% or ~14% after depletion of topoIIα or topoIIβ, respectively (Fig. 6D). These results suggested that NHDFs can exit mitosis with chromosomal mis-segregation and/or non-disjunction errors.
Abnormal chromosome segregation can lead to the loss and gain of genetic material and promote gross chromosomal rearrangements, thereby generating chromosomal instability (reviewed in Holland and Cleveland, 2009). To determine whether decreased topoIIα levels and/or activity could generate genomic instability in NHDFs, fluorescence in situ hybridization (FISH) probes directed towards the tumor suppressor p16INK4A genetic locus (red) and the centromere of chromosome 9 (green) were hybridized to cytogenetic preparations (Fig. 7A). NHDFs with 2–4 copies of each locus were considered to retain a normal allelic copy number to account for DNA replication in log phase cells. At least 100 interphase cells were analyzed from each NHDF line. The topoIIβ-depleted fibroblasts failed to exhibit any significant allelic copy number changes when compared to NTC-treated fibroblasts (Fig. 7B). However, the topoIIα-depleted NHDFs displayed increased frequencies of gains and deletions of the p16INK4A allele. These results indicate that topoIIα is required for proper chromatid segregation during mitosis, and that decreased topoIIα levels and/or activity may generate or enhance the losses/gains of the tumor suppressor p16INK4A genetic locus. Collectively, these results suggest that topoIIα is required to maintain a barrier against tumor initiation and/or progression.
Due to the large increase in the number of abnormal interphase nuclei and the changes in the copy number of the p16INK4A genetic locus after topoIIα depletion, topoIIα-depleted NHDFs were examined for their ability to undergo clonal expansion. A clonogenic survival assay was performed in NHDFs after transient topoIIα depletion (Fig. 7C). These results confirm that topoIIα is an essential gene with transient depletion of the protein reducing colony formation by an average of 68%. The colonies that escaped clonal inactivation after depletion of topoIIα may have arisen from cells that recovered topoIIα expression before lethality was manifested.
Discussion
The decatenation G2 checkpoint was originally proposed to delay entry into mitosis when chromosomes were insufficiently decatenated by topoII. Previous evidence for this checkpoint was largely derived from pharmacologic analyses using topoII catalytic inhibitors, such as ICRF-193 (Downes et al., 1994; Deming et al., 2001). This study provides biological evidence for the existence of a decatenation G2 checkpoint that is molecularly distinct from the DNA damage G2 checkpoint in three genetically diverse NHDFs (Fig. 3, 4). The previous studies in which ICRF-193 triggered a DNA damage response in some cancer lines may have used cell lines with a lax decatenation G2 checkpoint response, such as some HeLa cell lines (Bower and Kaufmann, unpublished data). HeLa cells would therefore enter mitosis in the presence of catenated chromosomes, acquire dsbs upon mitotic exit, and thus trigger a γH2AX and DNA damage response.
TopoIIα-depleted mitotic cells displayed high frequencies of entangled and under-condensed chromosomes (Figure 6A/B) similar to biological effects recognized in cells treated with ICRF-193 that escape the decatenation G2 checkpoint. Abnormal nuclear structures that arise upon failure of chromatid decatenation and subsequent mal-segregation were also observed in topoIIα-depleted fibroblasts (Figure 6C/D), suggesting that the decatenation G2 checkpoint is required to prevent the breakage of sister chromatids during the metaphase/anaphase transition. Furthermore, ICRF-193 treatment failed to prevent mitotic entry in topoIIα-depleted cells, indicating that the topoIIα protein plays an important role during the G2/M transition. NHDFs that enter mitosis in the absence of topoIIα, and thus escape decatenation G2 checkpoint activation, displayed losses and gains of the p16INK4A tumor suppressor gene. Collectively, these observations suggest that attenuation of the decatenation G2 checkpoint is one mechanism by which genomic instability may be generated, leading to tumor initiation and/or progression.
Although topoIIα and topoIIβ have similar in vitro biochemical activities, the results herein imply that the physiological roles of these proteins are very different. This is not unexpected, given that previous studies have suggested an independent role for topoIIβ in repairing DNA crosslinks and dsb-induced transcription factor recruitment (Emmons et al., 2006; Ju et al., 2006). Furthermore, the data above suggest that topoIIβ activity is sufficient for S phase progression in NHDFs, but insufficient for decatenation G2 checkpoint activation (Fig. 4 and 5).
Our results are comparable to other studies in that topoIIα appears to be required for condensation and segregation of daughter chromosomes, with little contribution by topoIIβ to this process. Targeted disruption of the TOP2A gene in the HT1080 fibrosarcoma line, siRNA depletion of topoIIα in HeLa cells, and DT40 conditional topoIIα-depleted avian B cells produced similar results (Carpenter et al., 2004; Johnson et al., 2009; Li et al., 2008). Recent reports also suggest that topoIIα may interact with MDC1 to enforce the decatenation G2 checkpoint (Luo et al., 2009), and that topoIIα can be SUMOylated to assist in centromere localization and proper chromosome segregation (Diaz-Martinez et al., 2006; Dawlaty et al., 2008). However, none of these studies have addressed the role of the decatenation G2 checkpoint in genomic stability.
Since many dietary components are known topoII catalytic inhibitors/poisons (Bandele and Osheroff, 2007; Bandele and Osheroff, 2008; Barjesteh van Waalwijk van Doorn-Khosrovani et al., 2007), it is possible that successive rounds of bypassing the decatenation G2 checkpoint may be an important consideration for the study of tumor initiation and progression. As TOP2A allelic deletions have been observed in breast cancer patients, exposure to these dietary compounds may increase genomic instability in these cancers and accelerate tumor progression (Beser et al., 2007). A further understanding of the signaling mechanisms involved in the decatenation G2 checkpoint could potentially lead to the identification of early-stage biomarkers and new therapeutic targets for a subset of cancers.
Materials and Methods
Experimental Design and Cell Culture
NHDFs were described previously (Simpson et al., 2005). Each fibroblast line was isolated from a different individual and underwent <100 population doublings. Experiments were performed in all three NHDF lines to assess biological reproducibility and inter-individual genetic variation. NHDFs were grown in DMEM/10% FBS/2 mM L-Glutamine and maintained at 5% CO2 and 37°C. Periodic tests for mycoplasma contamination using a commercial kit (Gen-Probe, San Diego, CA) were negative.
In vitro Decatenation Assay
107 fibroblasts were fractionated with a ProtoJET Nuclear Extraction Kit (Fermentas, Inc., Glen Burnie, MD). Nuclear extracts were assayed for decatenatory activity using the TopoGEN Eukaryotic TopoII Assay kit (TopoGEN, Inc., Port Orange, FL). kDNA was electrophoresed in a 1% agarose gel to separate topoisomers, stained with SYBR Gold (Invitrogen, Carlsbad, CA), analyzed using a Typhoon 9400 (GE Healthcare, Piscataway, NJ), and quantified by ImageQuant TL v2005 Software (GE Healthcare).
Time Lapse Microscopy
NHDFs were treated with 0.1% DMSO or 4 μM ICRF-193 (Sigma, St. Louis, MO) at time zero. Bright-field time-lapse images were captured every two minutes at the Michael Hooker Microscopy Facility (MHMF) and the Microscopy Services Laboratory Core Facility at UNC-CH.
Immunofluorescence and γH2AX Flow Cytometry
NHDFs plated on coverslips were treated for 0, 0.25, 2, or 6 h with 4–12 μM Etoposide, 4 μM ICRF-193, or 0.1% DMSO. Coverslips were removed and fixed with 95% ethanol:5% acetic acid. The remaining cells were analyzed by flow cytometry. Samples were stained with a Phospho-Ser139 H2AX antibody (Millipore, Billerica, MA) and an anti-mouse Texas Red secondary antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Nuclei were counter-stained with DAPI. Images were obtained on a Leica DMIRB inverted fluorescence microscope at the MHMF. Flow cytometry samples were measured on a Dako CyAN ADP instrument at the Flow Cytometry Core Facility at UNC-CH and plots were analyzed using Summit 4.3 software. Percentages of γH2AX positive cells were plotted against time and linear regression slopes were calculated for each treatment group. Statistical analysis was performed using a Student’s t-test.
Ionizing Radiation
A Gamma Cell 40 Cs137 irradiation source was used for IR treatment. Mitotic index was determined by the flow cytometry protocol described below two hours after irradiation.
Electroporation of Fibroblasts
The NTC (5′-UGGUUUACAU GUCGACUAA-3′), topoIIα (5′-CUGCCUGUUUAGUCGCUUUC A-3′), and topoIIβ (5′-GAAGUUGUCUGUUGAGAGAUU-3′) siRNA were obtained from Dharmacon (Dharmacon, Inc., Lafayette, CO). A second topoIIα siRNA was obtained from Invitrogen (5′-GAGAUGUCACUAAUGAUGACCAUUA-3′). NHDFs were electroporated using an NHDF Nucleofector Kit (Lonza, Basel, Switzerland), and assayed for biological effects 48 h post-electroporation. Protein depletion for each individual experiment was confirmed by western immunoblot at time of assay.
Western Immunoblot
Standard immunoblotting techniques were used to detect expression of topoIIα and topoIIβ. Briefly, equal amounts of total cellular protein were electrophoresed on 7% SDS-PAGE gels and transferred to nitrocellulose membranes and blocked in 4% BSA/TBST. TopoIIα and topoII β antibodies were obtained from BD Biosciences (San Jose, CA). Secondary anti-mouse Cy3/Cy5 antibodies were obtained from GE Healthcare and fluorescence was measured on a Typhoon 9400. Quantification of protein expression was performed using ImageQuant TL.
Flow Cytometric Mitotic Entry Assay
Electroporated fibroblasts were plated at a density of 106 cells (day 0), and fed with fresh medium on day 1. On day 2, cells were treated with 100 ng/mL colcemid and either 0.1% DMSO or 4 μM ICRF-193 for 2, 4, and 6 h. Fibroblasts were fixed with 95% ethanol:5% acetic acid. A phospho-Ser10 histone H3 primary antibody (Millipore) and a FITC-labeled secondary (Santa Cruz Biotechnology) were used to identify mitotic cells. Propidium iodide was used to measure DNA content. Summit 4.3 plots were used to quantify the percentage of NHDFs with 4N DNA content and phospho-Ser10 histone H3. The percentage of mitotic cells for each sample was plotted against time and the resulting slope of the line was used to measure the MER (% entering mitosis/hour).
Flow Cytometry BrdU Incorporation
Fibroblasts were incubated with 10 μM BrdU for 2 hours and fixed with 70% ethanol. An anti-BrdU-FITC antibody (BD Biosciences) was used to identify cells with actively replicating DNA. Propidium iodide was used to measure DNA content. Summit 4.3 plots were used to quantify the percentage of fibroblasts in G0/G1, S, and G2/M.
Cytogenetic Studies
Fibroblasts were incubated with 100 ng/mL colcemid for 1 h, incubated in hypotonic KCl (75 mM), and fixed with 3:1 methanol:acetic acid. Nuclei were dropped onto microscope slides and Geimsa-stained for visualization (Kaufmann et al., 1997). Metaphases were examined using an Olympus BH2 microscope with a SPOT RT camera. 50+ metaphases were scored for each treatment group/cell line. All analyses were performed blinded to sample identity.
Nuclear Abnormality Assessment
NHF1-hTERT cells were electroporated with siRNA as described above and allowed to adhere to coverslips. After 48 h, the coverslips were fixed with 3:1 methanol:acetic acid and stained with DAPI. Coverslips were mounted onto slides and 1000+ interphase nuclei were analyzed in a blind manner for structural abnormalities (blebs, bridges, and micronuclei) at the MHMF.
Fluorescence in situ Hybridization (FISH)
A second set of cytogenetic metaphase preparations were artificially aged in 2X SSC/0.1% NP-40 for 30 min at 37°C, successively dehydrated in 70%, 85%, and 100% ethanol, and air dried. A Vysis LSI p16 (9p21) Spectrum Orange/CEP9 Spectrum Green probe (Abbott Molecular, Des Plaines, IL) was hybridized to metaphase preparations overnight using a Vysis HyBrite (Abbott Diagnostics, Abbott Park, IL) according to the probe manufacturer’s instructions. Gene copy numbers were assessed in a blind manner for both the p16INK4A locus and the CEP9 locus for at least 100 interphase nuclei per sample.
Clonogenic Survival Assay
Single cell suspensions of fibroblasts were electroporated with NTC, topoIIα-, or topoIIβ-directed siRNA as described. One thousand cells were plated in triplicate. Colonies were fed every 2–3 days with fresh medium and stained 14 days after electroporation with 0.05% crystal violet/40% methanol. Colonies with >50 cells were counted (Zhou et al., 2006).
Supplementary Material
Acknowledgments
This work was supported by an NIH-NCI Ruth L. Kirschstein National Research Service Award Grant # 1F32CA134155-01 (J.J. Bower) and PHS grants CA81343 and P30-ES10126 (W.K. Kaufmann).
Abbreviations used
- topoII
topoisomerase II
- dsb
double strand break
- NHDF
normal human diploid fibroblast
- NTC
non-targeting control
- MER
mitotic entry rate
Footnotes
Conflict of Interest: There are no conflicts of interest to report.
References
- Akimitsu N, Adachi N, Hirai H, Hossain MS, Hamamoto H, Kobayashi M, et al. Enforced cytokinesis without complete nuclear division in embryonic cells depleting the activity of DNA topoisomerase IIalpha. Genes Cells. 2003;8:393–402. doi: 10.1046/j.1365-2443.2003.00643.x. [DOI] [PubMed] [Google Scholar]
- Austin CA, Marsh KL. Eukaryotic DNA topoisomerase II beta. Bioessays. 1998;20:215–26. doi: 10.1002/(SICI)1521-1878(199803)20:3<215::AID-BIES5>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Bandele OJ, Osheroff N. Bioflavanoids as poisons of human topoisomerase IIα and IIβ. Biochemistry. 2007;46:6097–108. doi: 10.1021/bi7000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandele OJ, Osheroff N. (−)-epigallocatechin gallate, a major constituent of green tea, poisons human type II topoisomerases. Chem Res Toxicol. 2008;21:936–43. doi: 10.1021/tx700434v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barjesteh van Waalwijk van Doorn-Khosrovani S, Janssen J, Maas LM, Godschalk RWL, Nijhuis JG, van Schooten FJ. Dietary flavonoids induce MLL translocations in primary human CD34+ cells. Carcinogenesis. 2007;28:1703–9. doi: 10.1093/carcin/bgm102. [DOI] [PubMed] [Google Scholar]
- Barrett JF, Gootz TD, McGuirk PR, Farrell CA, Sokolowski SA. Use of in vitro topoisomerase II assays for studying quinolone antibacterial agents. Antimicrob Agents Ch. 1989;33:1697–1703. doi: 10.1128/aac.33.10.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter J, Diffley JFX. Topoisomerase II inactivation prevents the completion of DNA replication in budding yeast. Mol Cell. 2008;30:790–802. doi: 10.1016/j.molcel.2008.04.019. [DOI] [PubMed] [Google Scholar]
- Beser AR, Tuzlali S, Guzey D, Dolec Guler S, Hacihanefioglu S, Dalay N. Her-2, TOP2A, and chromosome 17 alterations in breast cancer. Pathol Oncol Res. 2007;13:180–5. doi: 10.1007/BF02893497. [DOI] [PubMed] [Google Scholar]
- Biersack H, Jensen S, Gromova I, Nielsen IS, Westergaard O, Andersen AH. Active heterodimers are formed from human DNA topoisomerase IIα and IIβ isoforms. Proc Natl Acad Sci USA. 1996;93:8288–93. doi: 10.1073/pnas.93.16.8288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower JJ, Zhou Y, Zhou T, Simpson DA, Arlander SJ, Paules RS, et al. Revised genetic requirements for the decatenation G2 checkpoint: the role of ATM. Cell Cycle. 2010;9:1617–28. doi: 10.4161/cc.9.8.11470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burden DA, Osheroff N. Mechanism of action of eukaryotic topoisomerase II and drugs targeted to the enzyme. Biochim Biophys Acta. 1998;1400:139–54. doi: 10.1016/s0167-4781(98)00132-8. [DOI] [PubMed] [Google Scholar]
- Capranico G, Tinelli S, Austin CA, Fisher ML, Zunino F. Different patterns of gene expression of topoisomerase II isoforms in differentiated tissues during murine development. Biochim Biophys Acta. 1992;1132:43–8. doi: 10.1016/0167-4781(92)90050-a. [DOI] [PubMed] [Google Scholar]
- Carpenter AJ, Porter AC. Construction, characterization, and complementation of a conditional-lethal DNA topoisomerase II alpha mutant human cell line. Mol Biol Cell. 2004;15:5700–11. doi: 10.1091/mbc.E04-08-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho PA, Queiroz-Machado J, Carmo AM, Moutinho-Pereira S, Maiato H, Sunkel CE. Dual role of topoisomerase II in centromere resolution and aurora B activity. PLoS Biol. 2008;6:1758–77. doi: 10.1371/journal.pbio.0060207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes F, Pastor N, Mateos S, Dominguez I. Roles of DNA topoisomerases in chromosome segregation and mitosis. Mutat Res. 2003;543:59–66. doi: 10.1016/s1383-5742(02)00070-4. [DOI] [PubMed] [Google Scholar]
- Damelin M, Sun YE, Sodja VB, Bestor TH. Decatenation checkpoint deficiency in stem and progenitor cells. Cancer Cell. 2005;8:479–84. doi: 10.1016/j.ccr.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Dawlaty MM, Malureanu L, Jeganathan KB, Kao E, Sustmann C, Tahk S, et al. Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIα. Cell. 2008;133:103–15. doi: 10.1016/j.cell.2008.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deming PB, Cistulli CA, Zhao H, Graves PR, Piwnica-Worms H, Paules RS, et al. The human decatenation checkpoint. Proc Natl Acad Sci USA. 2001;98:12044–9. doi: 10.1073/pnas.221430898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deming PB, Flores KG, Downes CS, Paules RS, Kaufmann WK. ATR enforces the topoisomerase II-dependent G2 checkpoint through inhibition of Plk1 kinase. J Biol Chem. 2002;277:36832–8. doi: 10.1074/jbc.M206109200. [DOI] [PubMed] [Google Scholar]
- Diaz-Martinez LA, Gimenez-Abian JF, Azuma Y, Guacci V, Gimenez-Martin G, Lanier LM, et al. PIASgamma is required for faithful chromosome segregation in human cells. PLoS ONE. 2006;1:e53. doi: 10.1371/journal.pone.0000053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty SC, McKeown SR, McKelvey-Martin V, Downes CS, Atala A, Yoo JJ, et al. Cell cycle checkpoint function in bladder cancer. J Natl Cancer Inst. 2003;95:1859–68. doi: 10.1093/jnci/djg120. [DOI] [PubMed] [Google Scholar]
- Downes CS, Clarke DJ, Mullinger AM, Gimenez-Abian JF, Creighton AM, Johnson RT. A topoisomerase II-dependent G2 cycle checkpoint in mammalian cells/[published erratum appears in Nature 1994 Dec 15;372(6507):710] Nature. 1994;372:467–70. doi: 10.1038/372467a0. [DOI] [PubMed] [Google Scholar]
- Emmons M, Boulware D, Sullivan DM, Hazlehurst LA. Topoisomerase II beta levels are a determinant of melphalan-induced DNA crosslinks and sensitivity to cell death. Biochem Pharmacol. 2006;72:11–8. doi: 10.1016/j.bcp.2006.03.017. [DOI] [PubMed] [Google Scholar]
- Franchitto A, Oshima J, Pichierri P. The G2-phase decatenation checkpoint is defective in Werner syndrome cells. Cancer Res. 2003;63:3289–95. [PubMed] [Google Scholar]
- Goswami PC, Roti Roti JL, Hunt CR. The cell cycle-coupled expression of topoisomerase IIα during S phase is regulated by mRNA stability and is disrupted by heat shock or ionizing radiation. Mol Cell Biol. 1996;16:1500–8. doi: 10.1128/mcb.16.4.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromova I, Biersack H, Jensen S, Nielsen OF, Westergaard O, Andersen AH. Characterization of DNA topoisomerase II α/β heterodimers in HeLa cells. Biochemistry. 1998;37:16645–52. doi: 10.1021/bi981391l. [DOI] [PubMed] [Google Scholar]
- Heck MMS, Hittelman WN, Earnshaw WC. Differential expression of DNA topoisomerases I and II during the eukaryotic cell cycle. Proc Natl Acad Sci USA. 1988;85:1086–90. doi: 10.1073/pnas.85.4.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy, and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10:478–87. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvinen TAH, Kononen J, Pelto-Huikko M, Isola J. Expression of topoisomerase IIα is associated with rapid cell proliferation, aneuploidy, and c-erbB2 overexpression in breast cancer. Am J Pathol. 1996;148:2073–82. [PMC free article] [PubMed] [Google Scholar]
- Jensen LH, Nitiss KC, Rose A, Dong J, Zhou J, Hu T, et al. A novel mechanism of cell killing by anti-topoisomerase II bisdioxopiperazines. J Biol Chem. 2000;275:2137–46. doi: 10.1074/jbc.275.3.2137. [DOI] [PubMed] [Google Scholar]
- Johnson M, Phua HH, Bennett SC, Spence JM, Farr CJ. Studying vertebrate topoisomerase 2 function using a conditional knockdown system in DT40 cells. Nuc Acid Res. 2009;37:e98. doi: 10.1093/nar/gkp480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, et al. A topoisomerase IIβ-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- Kaufmann WK, Schwartz JL, Hurt JC, Byrd LL, Galloway DA, Levedakou E, et al. Inactivation of G2 checkpoint function and chromosomal destabilization are linked in human fibroblasts expressing human papillomavirus type 16 E6. Cell Growth Differ. 1997;8:1105–14. [PubMed] [Google Scholar]
- Larsen AK, Skladanowski A, Bojanowski K. The roles of DNA topoisomerase II during the cell cycle. Prog Cell Cycle Res. 1996;2:229–39. doi: 10.1007/978-1-4615-5873-6_22. [DOI] [PubMed] [Google Scholar]
- Li H, Wang Y, Liu X. Plk1-dependent phosphorylation regulates functions of DNA topoisomerase II in cell cycle progression. J Biol Chem. 2008;283:6209–21. doi: 10.1074/jbc.M709007200. [DOI] [PubMed] [Google Scholar]
- Liu LF. DNA topoisomerase poisons as antitumor drugs. Annu Rev Biochem. 1989;58:351–75. doi: 10.1146/annurev.bi.58.070189.002031. [DOI] [PubMed] [Google Scholar]
- Loehberg CR, Thompson T, Kastan MB, Maclean KH, Edwards DG, Kitrell FS, et al. Ataxia telangiectasia-mutated and p53 are potential mediators of chloroquine-induced resistance to mammary carcinogenesis. Cancer Res. 2007;67:12026–33. doi: 10.1158/0008-5472.CAN-07-3058. [DOI] [PubMed] [Google Scholar]
- Lorusso V, Manzione L, Silvestris N. Role of liposomal anthracyclines in breast cancer. Ann Oncol. 2007;18:vi70–3. doi: 10.1093/annonc/mdm229. [DOI] [PubMed] [Google Scholar]
- Lou Z, Minter-Dykhouse K, Chen J. BRCA1 participates in DNA decatenation. Nat Struct Mol Biol. 2005;12:589–93. doi: 10.1038/nsmb953. [DOI] [PubMed] [Google Scholar]
- Luo K, Yuan J, Chen J, Lou Z. Topoisomerase IIα controls the decatenation checkpoint. Nat Cell Biol. 2009;11:204–210. doi: 10.1038/ncb1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu YL, Kerrigan JE, Lin CP, Azarova AM, Tsai YC, Ban Y, et al. Topoisomerase IIβ Mediated DNA Double-Strand Breaks: Implications in Doxorubicin Cardiotoxicity and Prevention by Dexrazoxane. Cancer Res. 2007;67:8839–46. doi: 10.1158/0008-5472.CAN-07-1649. [DOI] [PubMed] [Google Scholar]
- Maeshima K, Eltsov M, Laemmli UK. Chromosome structure: improved immunolabeling for electron microscopy. Chromosoma. 2005;114:365–75. doi: 10.1007/s00412-005-0023-7. [DOI] [PubMed] [Google Scholar]
- Mao Y, Desai SD, Ting CY, Hwang J, Liu LF. 26 S proteasome-mediated degradation of topoisomerase II cleavable complexes. J Biol Chem. 2001;276:40652–8. doi: 10.1074/jbc.M104009200. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Hayashita Y, Maeno K, Masuda A, Sugito N, Osada H, et al. Identification of decatenation G2 checkpoint impairment independently of DNA damage G2 checkpoint in human lung cancer cell lines. Cancer Res. 2004;64:4826–32. doi: 10.1158/0008-5472.CAN-04-0871. [DOI] [PubMed] [Google Scholar]
- Park I, Avraham HK. Cell cycle-dependent DNA damage signaling induced by ICRF-193 involves ATM, ATR, CHK2, and BRCA1. Exp Cell Res. 2006;312:1996–2008. doi: 10.1016/j.yexcr.2006.02.029. [DOI] [PubMed] [Google Scholar]
- Robinson HM, Bratlie-Thoresen S, Brown R, Gillespie DA. Chk1 is required for G2/M checkpoint response induced by the catalytic topoisomerase II inhibitor ICRF-193. Cell Cycle. 2007;6:1265–7. doi: 10.4161/cc.6.10.4225. [DOI] [PubMed] [Google Scholar]
- Roca J, Ishida R, Berger JM, Andoh T, Wang JC. Antitumor bisdioxopiperazines inhibit yeast DNA topoisomerase II by trapping the enzyme in the form of a closed protein clamp. Proc Natl Acad Sci U S A. 1994;91:1781–5. doi: 10.1073/pnas.91.5.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaria A, Neef R, Eberspacher U, Eis K, Husemann M, Mumberg D, et al. Use of the novel Plk1 inhibitor ZK-Thiazolidinone to elucidate functions of Plk1 in early and late stages of mitosis. Mol Biol Cell. 2007;18:4024–36. doi: 10.1091/mbc.E07-05-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehested M, Jensen PB. Mapping of DNA topoisomerase II poisons (etoposide, clerocidin) and catalytic inhibitors (aclarubicin, ICRF-189) to four distinct steps in the topoisomerase II catalytic cycle. Biochem Pharm. 1996;51:879–86. doi: 10.1016/0006-2952(95)02241-4. [DOI] [PubMed] [Google Scholar]
- Simpson DA, Livanos E, Heffernan TP, Kaufmann WK. Telomerase exression is sufficient for chromosomal integrity in cells lacking p53-dependent G1 checkpoint function. J Carcinog. 2005;4:18. doi: 10.1186/1477-3163-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart DJ, Halicka HD, Traganos F, Darzynkiewicz Z, Williams G. Ciprofloxacin-induced G2 arrest in TK6 lymphoblastoid cells is not dependent on DNA double-strand break formation. Cancer Biol Ther. 2008;7:113–9. doi: 10.4161/cbt.7.1.5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder RD, Arnone MR. Putative identification of functional interactions between DNA intercalating agents and topoisomerase II using the V79 in vitro micronucleus assay. Mut Res. 2002;503:21–35. doi: 10.1016/s0027-5107(02)00028-3. [DOI] [PubMed] [Google Scholar]
- Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–40. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- Wang LHC, Schwarzbraun T, Speicher MR, Nigg EA. Persistence of DNA threads in human anaphase cells suggests late completion of sister chromatid decatenation. Chromosoma. 2008;117:123–35. doi: 10.1007/s00412-007-0131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Mao Y, Desai SD, Zhou N, Ting CY, Hwang J, et al. The topoisomerase II beta circular clamp arrests transcription and signals a 26S proteasome pathway. Proc Natl Acad Sci U S A. 2003;100:3239–44. doi: 10.1073/pnas.0736401100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Chou JW, Simpson DA, Zhou Y, Mullen TE, Medeiro M, et al. Profiles of global gene expression in ionizing-radiation-damaged human diploid fibroblasts reveal synchronization behind the G1 checkpoint in a G0-like state of quiescence. Environ Health Persp. 2006;114:553–9. doi: 10.1289/ehp.8026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.