Abstract
The NAD-dependent deacetylase Sir2 was initially identified as a mediator of replicative lifespan in budding yeast and was subsequently shown to modulate longevity in worms and flies1,2. Its mammalian homologue, SIRT1, appears to have evolved complex systemic roles in cardiac function, DNA repair, and genomic stability. Recent studies suggest a functional relevance of SIRT1 in normal brain physiology and neurological disorders. However, it is unknown if SIRT1 plays a role in higher-order brain functions. We report that SIRT1 modulates synaptic plasticity and memory formation via a microRNA-mediated mechanism. Activation of SIRT1 enhances, while its loss-of-function impairs, synaptic plasticity. Surprisingly, these effects were mediated via post-transcriptional regulation of CREB expression by a brain-specific microRNA, miR-134. SIRT1 normally functions to limit expression of miR-134 via a repressor complex containing the transcription factor YY1, and unchecked miR-134 expression following SIRT1 deficiency results in the down-regulated expression of CREB and BDNF, thereby impairing synaptic plasticity. These findings demonstrate a novel role for SIRT1 in cognition and a previously unknown microRNA-based mechanism by which SIRT1 regulates these processes. Furthermore, these results describe a separate branch of SIRT1 signaling, in which SIRT1 has a direct role in regulating normal brain function in a manner that is disparate from its cell survival functions, demonstrating its value as a potential therapeutic target for the treatment of CNS disorders.
The mammalian Sir2 homolog SIRT1 is involved in a variety of complex processes relevant to aging, including the regulation of oxidative stress, metabolism control, and circadian rhythms1–4. We previously demonstrated that SIRT1 gain-of-function is neuroprotective in overactive Cdk5 and mutant SOD models of neurodegeneration, which are relevant to Alzheimer’s disease and ALS, respectively5. Moreover, SIRT1 has recently been implicated in molecular pathways regulated by cocaine6, suggesting that, in addition to its involvement in neurogenesis and neuroprotection, SIRT1 has further functions in the brain that are yet to be described.
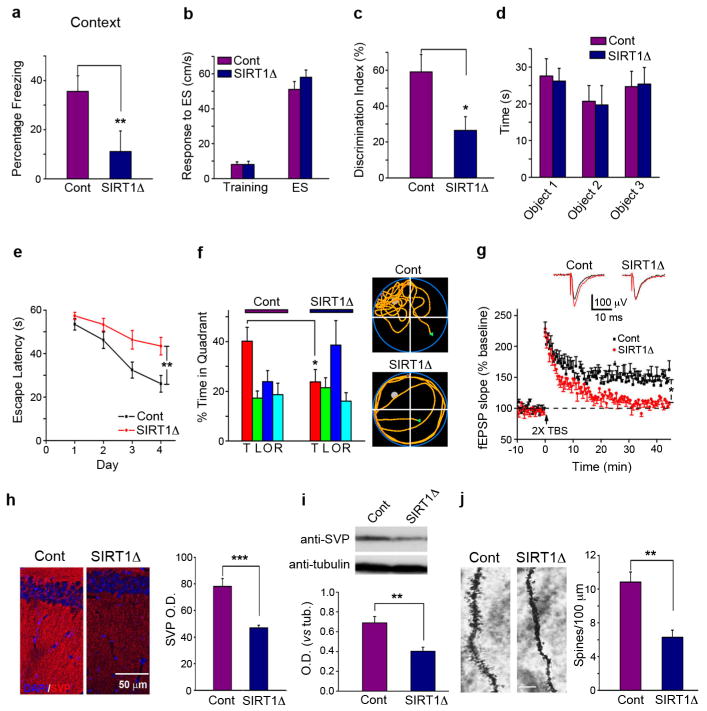
To directly evaluate the physiological role of SIRT1 in learning and memory, mutant mice lacking SIRT1 catalytic activity in a brain-specific manner (SIRT1Δ) were generated by crossing mice carrying a floxed SIRT1Δex4 allele7,8 with Nestin-Cre transgenic mice. In tests of associative memory, SIRT1Δ mice exhibited a significant decrease in freezing behavior as evaluated by both contextual (Fig. 1a) and tone-dependent (Fig. S2a) fear conditioning paradigms. Shock sensitivity and locomotor activity did not differ between SIRT1Δ mice and littermate controls (Figs. 1b, S2b). SIRT1Δ mice showed a similarly decreased memory performance in a novel object recognition task (Fig. 1c), which relies upon the hippocampus and cortex9. The time spent exploring the objects during training did not significantly differ between groups (Fig 1d). In the Morris water maze, SIRT1Δ mice displayed significantly increased escape latencies in the hidden platform paradigm (Fig. 1e), while spending less time in the target quadrant in a probe trial (Fig. 1f) compared with control mice, suggesting that SIRT1 plays a role in spatial learning. Visual function and swimming ability were not affected in the SIRT1Δ mice (Figs. S2c,d). Together, these results show that SIRT1 has an important role in several forms of memory.
Figure 1. SIRT1 loss-of-function impairs memory and synaptic plasticity.
(a) Freezing time of SIRT1Δ mice and littermate controls (Cont) 24 hr after contextual fear conditioning training. (b) Shock sensitivities did not differ between control and SIRT1Δ mice. (c) SIRT1Δand control mice were tested for novel object discrimination 24 hrs after training. (d) Pre-test exploration times were equal for each object. (e) Escape latencies of SIRT1Δ and control mice were examined with the Morris water maze hidden platform test. (f) Left panel: the swimming time spent in each quadrant in the probe trial on day 5 (T, target; L, left; O, opposite; R, right). Right panel: representative path tracings of the probe test. (g) LTP measurements were performed in the CA1 region of acute slices from SIRT1Δ mice and controls. (h) SVP immunoreactivity in SIRT1Δ and control hippocampi. (i) Western blots from hippocampal lysates examined SVP in SIRT1Δ and control mice. (j) The density of Golgi-impregnated hippocampal neuronal dendritic spines was measured in SIRT1Δ and control mice. Scale bar: 10 μm. TBS: theta-burst stimulation, SVP: synaptophysin, OD: optical density. *p < 0.05, **p < 0.01, ***p < 0.001. All histograms represent average ± SEM.
Next, we used a long-term potentiation (LTP) paradigm to directly determine the role of SIRT1 in synaptic plasticity. LTP in hippocampal CA1 neurons, induced by two θ burst (2×TBS) stimulation of the Schaffer collaterals in control mice, was abrogated in SIRT1Δ mice, demonstrating a requirement of SIRT1 in synaptic plasticity (Fig. 1g). CA1 neurons in SIRT1Δ mice exhibited normal basal synaptic transmission (Figs. S3a,b) compared to control mice. These results demonstrate that the LTP deficits caused by SIRT1 inactivation are not due to impaired synaptic transmission.
The brains of SIRT1Δ mice had a grossly normal anatomy (data not shown). However, experiments using an antibody against synaptophysin (SVP), which labels the presynaptic terminals of functional synapses10, revealed significant decreases in SVP immunoreactivity in the hippocampal striatum radiatum of SIRT1Δ mice, as well as reduced SVP protein content in the SIRT1Δ hippocampus, compared to controls (Figs. 1h and 1i). Golgi impregnation demonstrated that the dendritic spine density of CA1 pyramidal neurons is significantly decreased in the hippocampus of SIRT1Δ mice (Fig. 1j). These results suggest that SIRT1 regulates synapse formation, synaptic plasticity, and memory formation.
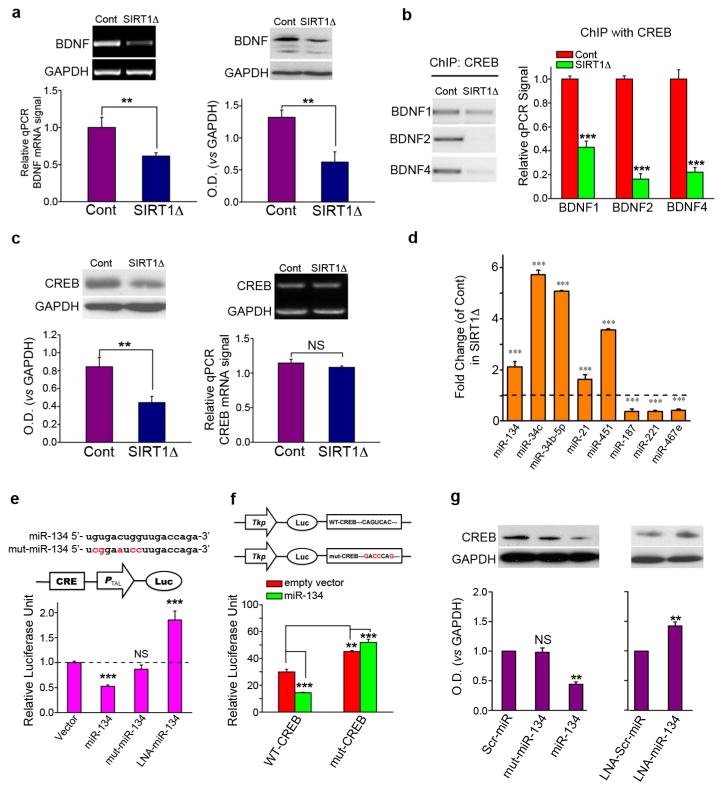
Brain-derived neurotrophic factor (BDNF) and cAMP response binding protein (CREB) are two genes that play critical roles in synaptic plasticity and modulating synapse formation11–13. Both mRNA and protein levels of BDNF were significantly decreased in SIRT1Δ hippocampi compared with controls (Fig. 2a). CREB binds to several BDNF promoters and plays a key role in the activity-dependent regulation of BDNF expression14–17. We performed chromatin immunoprecipitation (ChIP) to further examine the association of CREB and BDNF promoters. Baseline binding of CREB to BDNF promoters 1, 2, and 4 was reduced in the SIRT1Δ hippocampus (Fig. 2b). As reported previously15,17, binding of CREB to BDNF promoters 1 and 4, was increased after contextual fear conditioning training. In SIRT1Δ mice, however, this training-related increase in BDNF promoter 1 and 4 binding by CREB was abolished (Fig. S4a).
Figure 2. BDNF and CREB are downregulated, while miRNA-134 is up-regulated, in SIRT1Δ mice.
(a) BDNF in the SIRT1Δ and control mouse hippocampus. Left: mRNA levels. Right: Western blot. (b) Chromatin immunoprecipitation (ChIP) with an anti-CREB antibody was followed by qPCR for BDNF promoters 1, 2, and 4. (c) CREB in the SIRT1Δ and control mouse hippocampus. Left: Western blot. Right: mRNA levels. (d) qPCR of selected miRNAs from hippocampi of SIRT1Δ and control mice. (e) CAD cells were transfected with the plasmids or LNA indicated together with CRE-Luc. (f) Luciferase reporter constructs containing either a wildtype (WT-CREB) or a mutated (mut-CREB) CREB 3’UTR region, were cotransfected with miR-134 or control. (g) CREB protein expression in CAD cells after transfection with the indicated constructs or LNAs. Scr: scrambled, mut: mutant, LNA: locked-nucleic-acid (LNA), CRE-Luc: CREB activity reporter construct. **p < 0.01; ***p < 0.001; NS, not significant.
One possible explanation for the decreased CREB binding to BDNF promoters observed in SIRT1Δ mice is that CREB itself is downregulated. Consistent with this notion, CREB protein levels were significantly reduced in SIRT1Δ hippocampi (Fig. 2c, left). Although overall CREB levels were reduced in the SIRT1Δ mice, CREB phosphorylation was still increased by fear conditioning training (Fig. S4b). However, in contrast to BDNF, mRNA levels of CREB were not altered (Fig. 2c, right), suggesting that CREB protein levels are downregulated in SIRT1Δ brains via posttranscriptional mechanisms.
To investigate such mechanisms, we considered a well-known means of posttranscriptional regulation of gene expression, the inhibition of translation via microRNA (miRNA). miRNAs are expressed at high levels in the brain18,19, and the involvement of miRNA in numerous aspects of normal and abnormal brain function has been reported19,20. We compared the expression of miRNAs in SIRT1Δ hippocampi with littermate control hippocampi using a miRNA microarray (Exqion Inc), and found that a number of brain-enriched miRNAs differed in expression between the two groups (Fig. S5). We evaluated the expression of eight miRNAs that appeared significantly altered in the microarray analysis, including five upregulated and three downregulated miRNAs, using mature miRNA-specific quantitative PCR (qPCR). Consistent with the microarray, our qPCR analyses confirmed significant changes in expression of all eight miRNAs (Fig. 2d).
Of these miRNAs, miRNA-134 was of particular interest, as it is specifically expressed in the brain and has been demonstrated to negatively regulate dendritic spine formation in vitro21. Comparative genomic analyses of the 3’UTR of the mouse CREB gene revealed three partial complementary binding sites for miR-134, with one site being well-conserved (Fig. S6a). To determine whether miR-134 directly binds to and inhibits the translation of CREB mRNA, we carried out a CREB activity luciferase reporter assay in which luciferase is driven by a minimal promoter downstream of CREB-binding elements. Co-transfection of miR-134 with CRE-Luc resulted in a significant decrease in CREB activity in cultured CAD cells, a neural cell line (Fig. 2e). As a negative control, we created a miR-134 mutant containing mutations in the miR-134 seed region (mut-miR-134). Expression of mut-miR-134 did not affect CREB activity (Fig. 2e). The expression of miR-34b-5p or miR-34c, two other miRNAs upregulated in brains of SIRT1Δ mice, had no effect on CREB activity (Fig. S6b), indicating specific regulation of CREB by miR-134. Finally, miR-134 loss-of-function using a locked-nucleic-acid (LNA)-modified oligonucleotide probe (LNA-miR-134), which achieves specific knockdown of endogenous miR-134, significantly enhanced CREB activity (Fig. 2e). These results suggest that miR-134 regulate CREB protein expression via a post-transcriptional mechanism.
To verify the direct nature of CREB inhibition by miR-134, we generated a luciferase reporter construct in which the 3’UTR of CREB, containing all three predicted miR-134 target sequences, was inserted downstream of a luciferase expression cassette (WT-CREB). As expected, co-expression of miR-134 with WT-CREB significantly attenuated reporter expression (Fig. 2f). A luciferase reporter construct linked to the 3’UTR of CREB containing multiple mutations within the three miR-134 target sequences (mut-CREB) was refractory to knockdown by miR-134 (Fig. 2f). Expression of mut-CREB alone resulted in a significant increase in luciferase activity, indicating a de-repression of the reporter construct by endogenous miR-134 (Fig. 2f). These results indicate that miR-134 attenuates CREB expression via a specific interaction with the target regions within the 3’UTR of CREB. Moreover, overexpression of miR-134, but not mut-miR-134, in cultured CAD cells resulted in reduced levels of CREB protein relative to a scrambled miR control (Scr-miR; Fig. 2g). In contrast, delivery of LNA-miR-134 resulted in increased CREB protein levels compared to LNA-scrambled-miR (LNA-scr-miR) control (Fig. 2g), suggesting that, under normal conditions, miR-134 has a limiting effect on CREB translation.
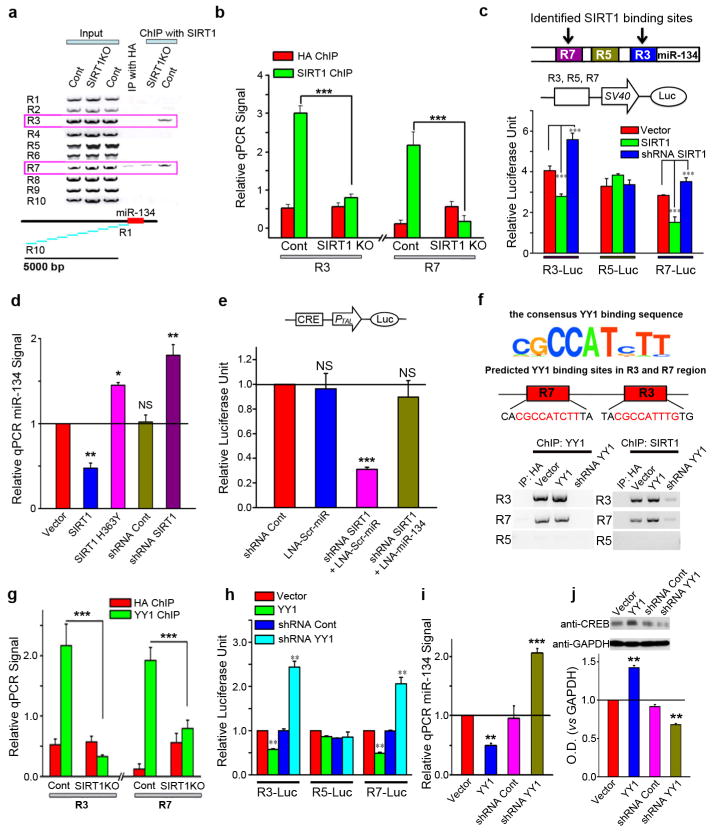
To investigate how SIRT1 modulates miR-134 transcription, we conducted ChIP with an anti-SIRT1 antibody and examined the association of SIRT1 with DNA sequences spanning ~5 Kb upstream of the Pre-miR-134 sequence. We found that two regions, R3 and R7, could be specifically amplified from SIRT1 ChIP, suggesting that SIRT1 is associated with potential regulatory elements upstream of miR-134 (Fig. 3a). The specificity of the interaction was verified by ChIP analyses from SIRT1 constitutive knockout (KO) mouse brain tissue8, which yielded a dramatically reduced pull-down of the R3 and R7 fragments (Fig. 3b). To determine the transcriptional modulating activities of these sites, we cloned R3, R7, and the control R5 fragment upstream of a minimal promoter in a pCL3-promoter luciferase reporter construct. Interestingly, co-expression of SIRT1 with reporter constructs containing either R3 or R7, but not R5, significantly reduced, while SIRT1 shRNA increased, the expression of the luciferase reporter (Fig. 3c). Collectively, these results indicate that SIRT1 inhibits miR-134 expression by directly binding to distal inhibitory elements.
Figure 3. SIRT1 regulates CREB through miR-134.
(a) From SIRT1 ChIP, two genomic regions, R3 and R7, corresponding to base pairs 1418~830 and 3427~2901 upstream of miR-134, respectively, were amplified from the wild type, but not the SIRT1 total knockout (SIRT1KO), brain tissue. (b) miR-134 promoter regions R3 and R7 were quantified from SIRT1 ChIP with qPCR. (c) Reporter constructs containing R3, R5, or R7 regions upstream of a minimal promoter in a luciferase reporter were co-transfected with SIRT1, SIRT1 shRNA, or empty vector. (d) qPCR for miR-134 in CAD cells after transfection with the indicated plasmids. (e) CRE-luciferase reporter assay in CAD cells. (f) ChIP was performed from CAD cells with anti-YY1 and anti-SIRT1 antibodies. (g) Anti-YY1 ChIP followed by qPCR for regions R3 and R7. (h) Luciferase reporter constructs containing R3, R5, or R7 regions were co-transfected with the indicated plasmids. (i) miR-134 levels were measured in CAD cells with qPCR after transfection with the indicated plasmids. (j) Western blotting measured CREB protein levels in CAD cells after transfections with the indicated plasmids. shRNA Cont: scrambled shRNA control. *p < 0.05; **p < 0.01; ***p < 0.001; NS: not significant. All histograms represent average ± SEM.
To verify the contribution of miR-134 to the modulation of CREB by SIRT1, we created SIRT1 loss-of-function in CAD cells by either shRNA-mediated knockdown (shRNA-SIRT1) or overexpression of a catalytically-inactive mutant of SIRT1 (H363Y)22, both of which elevated the level of miR-134 in these cells (Fig. 3d). The loss of SIRT1 markedly inhibited CREB activity, but this effect was reversed by the administration of LNA-miR-134 (Fig. 3e). These results provide strong evidence that SIRT1 loss-of-function attenuates CREB activity via a miR-134-mediated posttranscriptional mechanism. We searched the R3 and R7 regions for transcription factor consensus binding motifs and discovered Yin Yang 1 (YY1) binding sites within both the R3 and R7 fragments (Fig. 3f, top), but not in any of the other fragments upstream of the miR-134 coding region. YY1 is a ubiquitous and highly-conserved transcription factor that can activate or repress gene expression, depending upon the cellular context23. To determine if YY1 binds to miR-134 regulatory sequences, we performed ChIP experiments using anti-YY1 and anti-SIRT1 antibodies after both overexpression and knockdown of YY1 in CAD cells. The anti-YY1 antibody immunoprecipitated the R3 and R7, but not the R5, fragments, suggesting an association of YY1 with DNA elements within these fragments (Fig. 3f). The knockdown of YY1 also reduced YY1 binding to the R3 and R7 miR-134 promoter regions in cultured CAD cells (Fig. 3f, lower left). Moreover, the binding of SIRT1 to R3 and R7 was impaired after YY1 knockdown (Fig. 3f, lower right), suggesting that SIRT1 cooperates with YY1 in binding to the upstream regulatory elements of miR-134. Consistent with this idea, in SIRT1 constitutive KO mice, YY1 binding to R3 and R7 of miR-134 was also reduced (Fig. 3g). To determine the functional consequence of YY1 binding to R3 and R7, we used the pCL3-promoter luciferase reporter constructs containing R3, R5, or R7 as described in Fig 3c. We found that YY1 overexpression repressed, whereas YY1 knockdown potentiated, the luciferase activity driven by R3 and R7 (Fig. 3h). Conversely, YY1 abundance had no influence on the luciferase activity driven by R5. These results suggest that the binding of YY1 to R3 and R7 represses transcription. We then investigated the influence of YY1 upon miR-134 and CREB protein abundance in CAD cells, finding that overexpression of YY1 reduced miR-134 expression, while miR-134 abundance was increased following the shRNA-mediated knockdown of YY1 (Fig. 3i). Furthermore, CREB levels were positively regulated by YY1 expression (Fig. 3j). Collectively, these observations support the concept that SIRT1 is recruited to YY1 DNA binding elements and that the two proteins collaborate to suppress miR-134 expression.
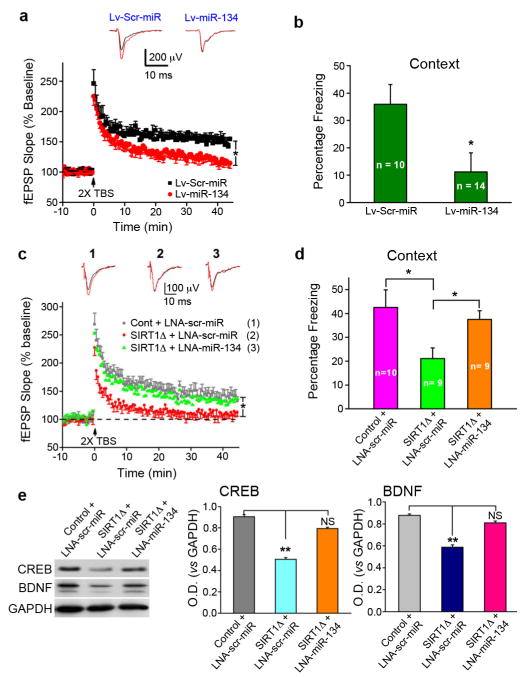
We next examined the role of miR-134 in synaptic plasticity and memory formation. miR-134 was overexpressed in area CA1 of the hippocampus via lentiviral-mediated delivery (Fig. S7a). As observed in SIRT1Δ mice, overexpression of miR-134 abrogated LTP in CA1 neurons (Fig. 4a), while basal synaptic transmission was not impaired (Fig. S7b). miR-134 overexpression in area CA1 also resulted in a significant impairment in long-term memory formation in the contextual fear-conditioning paradigm (Fig. 4b), but not in the tone-dependent fear conditioning paradigm (Fig. S7c), a task reliant upon amygdalar function24. miR-134 overexpression did not affect shock sensitivity (Fig. S7d). Thus, miR-134 overexpression in the hippocampus closely mimics the effects of SIRT1 loss. These data demonstrate that the upregulation of miR-134 plays a role in the synaptic plasticity impairment observed in SIRT1Δ mice.
Figure 4. MiR-134 knockdown rescues the LTP and memory impairments caused by SIRT1 deficiency.
(a) LTP was measured in acute hippocampal slices of mice six weeks after injection with Lv-miR-134 or Lv-Scr-miR. (b) Lv-miR-134 and Lv-Scr-miR-injected mice were tested with a contextual fear conditioning task. (c) Following hippocampal injections with LNA-miR-134 or LNA-scr-miR, LTP was measured in SIRT1Δ and control mouse acute hippocampal slices. (d) Freezing behavior in the contextual fear conditioning task was examined in SIRT1Δ and control mice after hippocampal injections of LNA-miR-134 or LNA-scr-miR. (e). Western blot for CREB and BDNF in brain lysate after in vivo miR-134 knockdown. Control scrambled miR lentivirus: Lv-Scr-miR. **p < 0.01, NS: not significant.
To assess, in vivo, whether miR-134 upregulation underlies the impairment of LTP observed in SIRT1Δ mice, we examined the effect of miR-134 knockdown using injections of the LNA-miR-134 probe into hippocampal CA1 region (Figs. S8a,b). We found that knockdown of miR-134 restored LTP in acute hippocampal slices from SIRT1Δ hippocampi (Fig. 4c), whereas the LNA probe containing a scrambled sequence (LNA-scr-miR) failed to ameliorate the LTP defects in SIRT1Δ mice. Moreover, injections of LNA-miR-134, but not LNA-scr-miR, into SIRT1Δ mouse hippocampus markedly rescued contextual (Fig. 4d), but not tone-dependent (Fig. S8c), fear memory formation. All groups exhibited normal shock sensitivity (Fig. S8d). Examination of hippocampal lysates harvested from control and SIRT1Δ mice injected with LNA-scr-miR revealed that both CREB and BDNF protein levels were reduced in SIRT1Δ mice (Fig 4e). However, LNA-miR-134 treatment restored CREB and BDNF protein to levels comparable to control mice (Fig. 4e).
Our data indicate that the impairments in memory and synaptic plasticity observed in SIRT1Δ mice are, at least, partly mediated via an up-regulation of miR-134 and consequent translational inhibition of miR-134 target genes. SIRT1 normally functions in cooperation with YY1, and potentially additional proteins, to restrict the expression of miR-134 and that, upon SIRT1 loss-of-function, higher levels of miR-134 negatively regulate synaptic plasticity via the translational block of key plasticity proteins such as CREB (Fig. S1), which subsequently mediates the various synaptic plasticity impairments observed following SIRT1 loss-of-function.
We previously demonstrated that SIRT1 promotes neuronal survival in age-dependent neurodegenerative disorders5. We have now found that SIRT1 activity also promotes plasticity and memory in a direct manner through a mechanism distinct from its established neuroprotective activity. This result demonstrates a multi-faceted role of SIRT1 in the brain, further highlighting its potential as a target for the treatment of neurodegeneration and conditions with impaired cognition, with implications for a wider range of CNS disorders.
Methods Summary
For detailed methods, please see the supplemental materials. The SIRT1 KO and SIRT1Δex4/Nestin-Cre mice were provided by the laboratory of Leonard Guarente. The plasmids and LNA-miRNA (Ambion) were transfected into mouse CAD cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. RNA extraction, purification, and quantitative PCR were performed according to the manufacturer's protocols. Tissue and cell lysis, protein concentrations, and western blot analyses were prepared as described previously5. Immunoblot data were quantified by measuring the band intensity with NIH imaging software and UN-SCAN-it gel digitizing software (Silk Scientific). Immunostaining was performed as described previously3 with LSMeta10 software and a confocal microscope (Zeiss). All behavioral testing was performed as described previously5 and elsewhere. The data were analyzed by unpaired Student’s t-test. Two-way ANOVA was used to compare differences between groups at several time points.
Supplementary Material
Acknowledgments
We thank Drs. Rudolf Jaenisch and Maribel Rios for providing BDNF KO frozen brains, Drs. Leonard Guarente and Dena Cohen for providing SIRT1Δ and SIRT1 knockout mice, Dr. Yang Shi for providing the plasmids expressing YY1 and YY1 shRNA, Dr. Alison Mungenast for manuscript editing, Dr. Karun Singh and Matt Dobbin for critical reading of the manuscript, M Dobbin for help with Supplemental Figure 5, Dr. Xuecai Ge for help with model illustration, and Nadine Joseph for help with mouse colony maintenance. This work is partially supported by an NIH grant, PO1 AG027916, to L.H.-T. W-Y. W is supported by a Postdoctoral Fellowship from the Simons Foundation; J. Gräff is supported by a Prospective Researchers Fellowship from the Swiss National Science Foundation; J.G. is supported by the Howard Hughes Medical Institute. L.H.-T. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Author Contributions
L.-H.T. designed, directed, and coordinated the project. J.G. designed and performed electrophysiological recordings, behavior tests, biochemical assays, and morphological analyses; W.-Y.W. contributed to the design and generation of microRNA constructs, and performed viral injections, behavior tests, and biochemical analyses; Y.-W.M., J.G. and L.P. performed luciferase assays and biochemical analyses; J. Gräff and G.M. performed behavior tests; S.C.S. contributed to viral injection; D.K. contributed to SIRT1 plasmid construction. The manuscript was written by J.G., D.K., S.C.S., W.-Y.W., and L.-H.T. and commented upon by all the authors.
Author Information
Reprints and permissions information is available at www.nature.com/reprints. Correspondence and requests for materials should be sent to L.-H.T. (lhtsai@mit.edu)
Literature citations
- 1.Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakahata Y, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim D, et al. SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer's disease and amyotrophic lateral sclerosis. EMBO J. 2007;26:3169–3179. doi: 10.1038/sj.emboj.7601758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Renthal W, et al. Genome-wide analysis of chromatin regulation by cocaine reveals a role for sirtuins. Neuron. 2009;62:335–348. doi: 10.1016/j.neuron.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen DE, Supinski AM, Bonkowski MS, Donmez G, Guarente LP. Neuronal SIRT1 regulates endocrine and behavioral responses to calorie restriction. Genes Dev. 2009;23:2812–2817. doi: 10.1101/gad.1839209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng HL, et al. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci U S A. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broadbent NJ, Squire LR, Clark RE. Spatial memory, recognition memory, and the hippocampus. Proc Natl Acad Sci U S A. 2004;101:14515–14520. doi: 10.1073/pnas.0406344101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calhoun ME, et al. Comparative evaluation of synaptophysin-based methods for quantification of synapses. J Neurocytol. 1996;25:821–828. doi: 10.1007/BF02284844. [DOI] [PubMed] [Google Scholar]
- 11.Kang H, Schuman EM. Long-lasting neurotrophin-induced enhancement of synaptic transmission in the adult hippocampus. Science. 1995;267:1658–1662. doi: 10.1126/science.7886457. [DOI] [PubMed] [Google Scholar]
- 12.Frank DA, Greenberg ME. CREB: a mediator of long-term memory from mollusks to mammals. Cell. 1994;79:5–8. doi: 10.1016/0092-8674(94)90394-8. [DOI] [PubMed] [Google Scholar]
- 13.Flavell SW, Greenberg ME. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu Rev Neurosci. 2008;31:563–590. doi: 10.1146/annurev.neuro.31.060407.125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tao X, Finkbeiner S, Arnold DB, Shaywitz AJ, Greenberg ME. Ca2+ influx regulates BDNF transcription by a CREB family transcription factor-dependent mechanism. Neuron. 1998;20:709–726. doi: 10.1016/s0896-6273(00)81010-7. [DOI] [PubMed] [Google Scholar]
- 15.Hong EJ, McCord AE, Greenberg ME. A biological function for the neuronal activity-dependent component of Bdnf transcription in the development of cortical inhibition. Neuron. 2008;60:610–624. doi: 10.1016/j.neuron.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Timmusk T, et al. Multiple promoters direct tissue-specific expression of the rat BDNF gene. Neuron. 1993;10:475–489. doi: 10.1016/0896-6273(93)90335-o. [DOI] [PubMed] [Google Scholar]
- 17.Chiaruttini C, Sonego M, Baj G, Simonato M, Tongiorgi E. BDNF mRNA splice variants display activity-dependent targeting to distinct hippocampal laminae. Mol Cell Neurosci. 2008;37:11–19. doi: 10.1016/j.mcn.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 18.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fiore R, Siegel G, Schratt G. MicroRNA function in neuronal development, plasticity and disease. Biochim Biophys Acta. 2008;1779:471–478. doi: 10.1016/j.bbagrm.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 20.Bushati N, Cohen SM. MicroRNAs in neurodegeneration. Curr Opin Neurobiol. 2008;18:292–296. doi: 10.1016/j.conb.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 21.Schratt GM, et al. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 22.van der Veer E, et al. Extension of human cell lifespan by nicotinamide phosphoribosyltransferase. J Biol Chem. 2007;282:10841–10845. doi: 10.1074/jbc.C700018200. [DOI] [PubMed] [Google Scholar]
- 23.Shi Y, Lee JS, Galvin KM. Everything you have ever wanted to know about Yin Yang 1. Biochim Biophys Acta. 1997;1332:F49–66. doi: 10.1016/s0304-419x(96)00044-3. [DOI] [PubMed] [Google Scholar]
- 24.Fanselow MS, Gale GD. The amygdala, fear, and memory. Ann N Y Acad Sci. 2003;985:125–134. doi: 10.1111/j.1749-6632.2003.tb07077.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.