Abstract
The E2F transcription factors play a key role in the regulation of cellular proliferation and terminal differentiation. E2F6 is the most recently identified and the least well understood member of the E2F family. It is only distantly related to the other E2Fs and lacks the sequences responsible for both transactivation and binding to the retinoblastoma protein. Consistent with this finding, E2F6 can behave as a dominant negative inhibitor of the other E2F family members. In this study, we continue to investigate the possible role(s) of E2F6 in vivo. We report the isolation of RYBP, a recently identified member of the mammalian polycomb complex, as an E2F6-interacting protein. Mapping studies indicate that RYBP binds within the known “repression domain” of E2F6. Moreover, we demonstrate that endogenous E2F6 and polycomb group proteins, including RYBP, Ring1, MEL-18, mph1, and the oncoprotein Bmi1, associate with one another. These findings suggest that the biological properties of E2F6 are mediated through its ability to recruit the polycomb transcriptional repressor complex.
Keywords: retinoblastoma protein pathway
The E2F transcription factors are a family of genes that play critical roles in the regulation of cellular proliferation and differentiation (for review, see refs. 1 and 2). They act by controlling the transcriptional state of genes whose expression is essential for cell cycle progression and DNA synthesis. Deregulated E2F activity has been shown to lead to inappropriate cell cycle entry, transformation, and apoptosis. Thus, by understanding the mechanisms by which E2F activity controls the expression of its downstream target genes, we can gain important insights into the processes of cellular proliferation, differentiation, and apoptosis.
A total of eight genes have been cloned thus far that encode components of E2F activity (for review, see ref. 1). Their protein products can be subdivided into two families, the E2Fs (1–6) and the DPs (1, 2), that form heterodimers to generate functional E2F complexes (3–6). Although the presence of a DP protein is required for activity, the functional specificity of the resulting E2F–DP heterodimer is conferred by the E2F moiety (1). The E2F family has been divided into three distinct groups on the basis of sequence homology and functional properties.
E2F1, E2F2, and E2F3 represent the first E2F subclass. These proteins, when bound to DP, have high transcriptional activity and are sufficient to drive quiescent cells into S phase (7–10). In normal cells, the activity of these complexes is controlled by their association with the retinoblastoma protein (pRB), the first tumor suppressor identified (11). Cell cycle-dependent phosphorylation of pRB causes it to dissociate from E2F–DP complexes, and this release correlates with the timing of E2F-responsive gene activation in vivo. Mouse models for E2f1 and E2f3 confirm that these E2Fs play a key role in the activation of E2F target genes and the induction of proliferation arising from mitogenic signaling or from the loss of pRB (1, 12, 13).
E2F4 and E2F5 represent the second subclass of the E2F family. Unlike the first subclass, E2F4 and E2F5 are poor transcriptional activators, and they are unable to induce quiescent cells to enter the cell cycle (10, 14, 15). Instead, these E2F proteins are believed to be important for the repression of E2F-responsive genes through their ability to bind to pRB and its related proteins p107 and p130 and to recruit histone deacetylases to the promoters of E2F-responsive genes (1, 16). Consistent with this model, analysis of E2f4 and E2f5 mutant mouse strains suggests that these proteins are not required for the regulation of cellular proliferation but play a key role in the terminal differentiation of specific cell types (17–19).
Additionally, we (6) and others (20–22) identified a sixth member of the E2F family. The central portion of the E2F6 protein shares considerable homology with the domains of E2F1 through E2F5 that mediate their heterodimerization and DNA binding properties. However, E2F6 also diverges considerably from the previous E2F subgroups within these domains. The distinction between E2F6 and the other E2Fs is underscored by the degree of sequence variation outside of these domains. The N-terminal domain of E2F6 bears no homology to those of other E2F family members and, most importantly, E2F6 terminates 42 amino acids after the dimerization domain. As a result, E2F6 lacks the sequences that mediate the transcriptional activation or pRB, p107, and p130 binding properties of the other E2F proteins. Thus, E2F6 represents a third subclass of the E2F family that is likely to display distinct biological properties.
With overexpression assays, we have shown that E2F6 can repress the transcription of known E2F-responsive genes (6). In these experiments, E2F6 appears to function as a dominant negative inhibitor through competition with other E2F family members. Consistent with these observations, other groups have reported that E2F6 can behave as an active repressor when fused to a heterologous DNA binding domain (20). However, the mechanism by which E2F6 represses transcription is not well understood.
We have used a yeast two-hybrid assay to identify E2F6-associated proteins. This analysis yielded RYBP (Ring 1 and YY1 binding protein), a known component of the mammalian polycomb complex that binds specifically to the repression domain of E2F6. By generating specific immunological reagents, we show that endogenous E2F6 associates with the Bmi1-containing polycomb complex. These data have important implications for our understanding of both E2F and polycomb complex function in vivo.
Materials and Methods
Yeast Two-Hybrid Assay.
A yeast two-hybrid screen was performed with the system of Vidal et al. (23). Briefly, amino acids 9–239 of E2F6 were cloned into the pPC97 vector and the resulting plasmid was transformed into the yeast strain MaV103 by using the lithium acetate method. This strain was transformed with a library derived from activated human T cells, and the transformations were plated on synthetic complete (SC)–Leu–Trp plates. Two days later, the transformations were replica-plated onto SC–Leu–Trp–His plates supplemented with 10 mM 3-aminotriazole (3-AT; Sigma) and subsequently replica-plated again onto SC–Leu–Trp–His plates supplemented with 30 mM 3-AT. The prey plasmid was rescued and transformed into bacteria. Plasmids were retransformed into MaV103 containing pPC97-E2F6 to verify that the 3-AT resistance was conferred by the library plasmid. The inserts in the recovered prey plasmids were then sequenced.
Plasmid Construction.
Full-length or truncation mutants of human E2F6, RYBP, and mouse Ring1A were generated by PCR and subcloned into pHACMV-neo-Bam or pCMV-neo-Bam. All constructs were verified by sequencing. Full-length RYBP was additionally subcloned into pQE30 (Qiagen, Chatsworth, CA). The plasmids pCMV-E2F1, pCMV-E2F2, pCMV-E2F3, pCMV-E2F4, pQE30-E2F6, and pHA-Bmi1 have been described (3, 6, 24, 25).
Antibody Production.
Full-length His6-tagged E2F6 (amino acids 1–275) proteins were expressed in bacteria, purified over a Ni2+-nitrilotriacetic acid-agarose resin (Qiagen), and used to immunize mice. The resulting polyclonal antiserum was monitored for its ability to recognize transfected E2F6 and not E2F1 through E2F5 or polycomb group (PcG) proteins, by both Western blotting and immunoprecipitation. Mice generating E2F6-specific antibodies were killed and the spleens were removed. The recovered splenocytes were fused to the SP2/O cell line by a polyethylene glycol-mediated method, generating hybridoma cell lines. After 10 days, the tissue culture supernatants were screened for the ability to detect recombinant E2F6 by an ELISA. The positive cell lines were single-cell-cloned and tested again for E2F6 reactivity/specificity by Western blot analysis and immunoprecipitation.
mAbs to His6-tagged RYBP(1–228) peptide were generated as above. Specificity was determined against transfected RYBP and YAF-2 by Western blotting and immunoprecipitation. Anti-Ring1 (ASA8), anti-MEL-18, anti-mph1-SM, and anti-Bmi1 (Bmi1-F6) antibodies have been described (25, 26).
Transient Transfections and Immunoprecipitations.
Cells were maintained in DMEM containing 10% FCS. Transient transfections were conducted as described (6). For immunoprecipitations, C33-A cells were transfected with 10 μg of the indicated plasmid and labeled with 250 μCi of [35S]methionine Express labeling mixture (NEN; 1 Ci = 37 GBq) in methionine-free medium (GIBCO) for 5 h. Immunoprecipitation was performed exactly as described (6) with 12CA5 [anti-hemagglutinin (HA) tag], anti-E2F6 (LLF6–1), or anti-RYBP (LLRYBP-1) antibodies.
For endogenous immunoprecipitations, 108 ML-1 cells were lysed in E1A lysis buffer (24) and precleared with protein A-Sepharose beads (Amersham Pharmacia) at 4°C for 30 min. The lysates were incubated with the indicated antibodies for 1 h at 4°C, and the immunocomplexes were recovered on protein A-Sepharose beads and separated by SDS/PAGE. Western blotting was done as described (6) with the indicated antibodies.
Results
Isolation of E2F6-Interacting Proteins.
To understand better the role of E2F6, we used a yeast two-hybrid approach to identify interacting proteins. A fusion between the GAL4 DNA binding domain and amino acids 9–239 of the E2F6 protein was used as bait to screen cDNAs from an activated human T cell library fused with the Gal4 transcriptional activation domain. Four independent clones were isolated from the approximately 4 × 105 transformants on the basis of their ability to interact with E2F6 and not with pRB or DP1 fusion proteins. Sequence analysis showed that two of the four isolated clones encoded overlapping fragments of DP2, a known heterodimeric partner of the other E2F family members. Because we have shown that E2F6 and DP2 can coimmunoprecipitate (6), this provided strong validation of the screen. The remaining clones were found to contain the full-length (amino acids 1–228) or the near full-length (amino acids 12–228) coding sequence of a known zinc-finger-containing protein, RYBP. The murine homologue of RYBP was originally isolated in a yeast two-hybrid screen by virtue of its ability to bind to Ring1A (27). The human and mouse RYBP proteins are completely identical apart from three amino acid substitutions (Gln → Pro, Glu → Asp, and Thr → Ser) at residues 87, 97, and 143, respectively. RYBP is also highly related to YAF-2, a protein that was identified in a yeast two-hybrid screen with YY1 (28), suggesting that these represent a family of proteins.
A major advantage of our chosen yeast two-hybrid system is the ability to assess the relative strength of protein–protein interactions by measuring the growth of clones in the presence of 3-aminotriazole (23). With this assay, we demonstrated that E2F6 had a higher affinity for RYBP than it did for its known interactor, DP2. This finding strongly suggested that RYBP would be a genuine E2F6-interacting protein.
Interaction Between RYBP and E2F6.
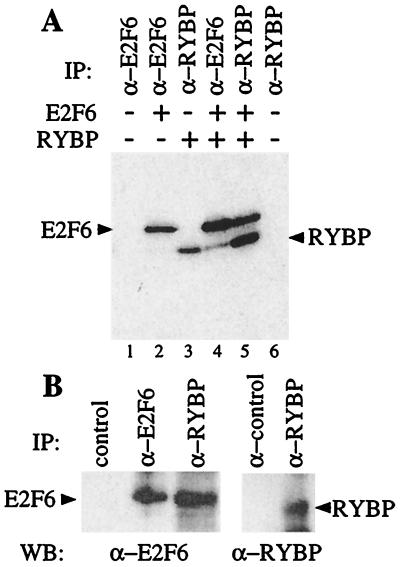
To confirm that E2F6 and RYBP can interact in mammalian cells, we expressed the full-length RYBP protein in C33-A cells by transient transfection in the presence and absence of a full-length E2F6 and then labeled the cells with [35S]methionine. The resultant complexes were recovered and analyzed by SDS/PAGE and mAbs that we had generated to specifically recognize E2F6 (LLF6–1) or RYBP (LLRYBP-1). We were consistently able to recover RYBP in the anti-E2F6 immunoprecipitate, albeit at low levels (Fig. 1A, lane 4). Moreover, the anti-RYBP antibody was able to coimmunoprecipitate E2F6 (Fig. 1A, lane 5). Thus, E2F6 and RYBP can form a complex that is poorly recognized by the anti-E2F6 antibody but is efficiently recovered by the anti-RYBP mAb.
Figure 1.
RYBP is an E2F6-interacting protein. (A) C-33A cells were transiently transfected with expression vectors encoding E2F6 and/or RYBP, labeled with [35S]methionine and immunoprecipitated with the specific mAbs LLF6–1 and LLRYBP-1. (B) ML-1 cell lysates were immunoprecipitated with control (12CA5), anti-E2F6 (LLF6–1), or anti-RYBP (LLRYBP-1) antibodies; resolved by SDS/PAGE; and immunoblotted with additional anti-E2F-6 (LLF6–2) or anti-RYBP (LLRYBP-2) antibodies. WB, Western blot; IP, immunoprecipitate.
Given these observations, we looked for an association between the endogenous E2F6 and RYBP. To address this issue, lysates of ML-1 cells were immunoprecipitated with control, anti-E2F6 (LLF6–1), or anti-RYBP (LLRYBP-1) antibodies; resolved by SDS/PAGE; and then immunoblotted with additional anti-E2F6 (LLF6–2) or an anti-RYBP (LLRYBP-2) mAbs. Consistent with the reduced amount of RYBP recovered in E2F6 immunoprecipitations of transfected cells, we were unable to detect RYBP in E2F6 immunoprecipitations of ML-1 lysates. However, we did recover a significant proportion of E2F6 in the RYBP immunoprecipitate (Fig. 1B). We therefore conclude that endogenous E2F6 and RYBP can associate with one another.
Mapping RYBP and E2F6 Interaction Sites.
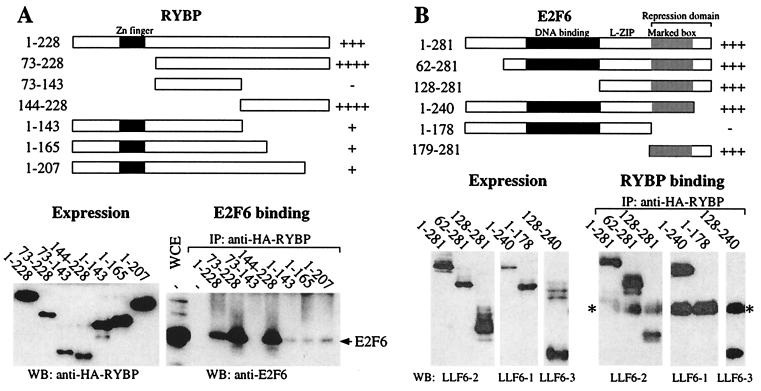
Because the manner in which these two proteins associate will influence their functions in vivo, we used a deletion mutant strategy to map the sites of interaction (Fig. 2). For these experiments, we generated a panel of RYBP and E2F6 deletion mutants and tested their ability to interact with their full-length partner by using transient transfection and coimmunoprecipitation assays. Because anti-E2F6 antibodies appear to destabilize the E2F6–RYBP complex, association was determined by immunoprecipitation through RYBP [of either full-length or HA-tagged deletion mutants] and Western blotting for E2F6 (either full-length or deletion mutants). In each case, expression of the RYBP and E2F6 deletion mutants (named according to the residues they retain) was confirmed by Western blotting of the whole cell lysate.
Figure 2.
Mapping the E2F6–RYBP interaction domains. (A) C33-A cells were transiently transfected with expression vectors encoding full-length E2F6 and the HA-tagged RYBP mutants as indicated. Expression of the HA-RYBP mutants was confirmed by Western blotting a fraction of the cell lysate. The remainder was immunoprecipitated with an anti-HA antibody (12CA5) and then immunoblotted with an anti-E2F6 antibody (LLF6–2) to assess the ability of these mutants to bind to E2F6. A summary of the interaction data is shown at the top. (B) The panel of E2F6 mutants including the location of DNA binding, leucine zipper (L-ZIP), marked box, and repression domains. The mutants were transiently transfected into C33-A cells with full-length HA-RYBP. The expression of these proteins was confirmed by Western blotting a fraction of the cell lysate with the particular anti-E2F6 mAb (LLF6–1, LLF6–2, or LLF6–3) that best recognizes each mutant. The remainder of the lysate was immunoprecipitated with an anti-RYBP antibody (LLRYBP-1) and then immunoblotted with the same anti-E2F6 antibody that was used to confirm its expression. The Ig light chain is denoted by an asterisk. An interaction summary is depicted at the top. WB, Western blot; IP, immunoprecipitate; WCE, whole cell extract.
First, we mapped the E2F6 binding site on RYBP (Fig. 2A). Because zinc-finger motifs can mediate protein–protein interactions, we began our analysis by deleting the N-terminal 72 amino acids of RYBP. This truncated RYBP protein (residues 73–228) and the full-length RYBP bound to E2F6 with similar affinity (Fig. 2A). Because there are no recognizable motifs within the remaining portion (residues 73–228), we generated mutants corresponding to the N-terminal (residues 73–143) and C-terminal (residues 144–228) portions of this fragment. Although HA-RYBP(73–143) was unable to associate with E2F6, HA-RYBP(144–228) bound as well as the full-length protein. Unfortunately, additional 5′ or 3′ deletions of this coding sequence did not yield detectable protein products (data not shown). We therefore generated a panel of C-terminal deletions within the context of the full-length RYBP. Although we were now able to generate stable proteins, the absence of the C-terminal 85, 63, or 21 amino acids greatly impaired the interaction between RYBP and E2F6. Thus, the E2F6 binding site is contained within amino acids 144–228 of RYBP, and residues at the very C terminus of this domain appear to be critical for the interaction of these two proteins.
We next mapped sequences in E2F6 that were required for RYBP binding (Fig. 2B). Because the N- and C-terminal domains of E2F6 are completely distinct from those of the other E2F family members, we began our analysis by deleting each of these regions. Given that the C-terminal sequence was absent from the yeast two-hybrid bait, we initially focused our attention on the N-terminal 62 amino acids. Surprisingly, deletion of these residues had no detectable effect on the interaction between E2F6 and RYBP. Additional N-terminal deletion showed that the DNA binding domain (residues 62–128) was also fully dispensable for interaction. Given this finding, we examined the consequences of C-terminal deletions. Consistent with the yeast two-hybrid data, the unique C-terminal sequences (residues 241–281) were not required for RYBP binding. However, additional deletion of the marked-box domain (residues 179–281) abolished the interaction between E2F6 and RYBP. Thus, this analysis indicates that the RYBP binding domain is contained within residues 129–240 and depends on residues between positions 179 and 240. Consistent with this conclusion, we were able to show that E2F6(129–240) is sufficient to bind RYBP in this coimmunoprecipitation assay. Moreover, although we were unable to express E2F6(179–240) in mammalian cells, this domain was sufficient to interact with RYBP in the yeast-two hybrid assay (data not shown). Thus, the RYBP binding domain of E2F6 maps to the dimerization domain, specifically to the marked-box domain.
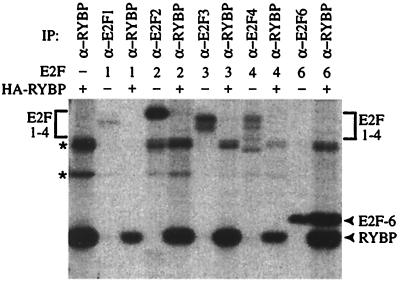
The marked-box domain is highly conserved among all members of the E2F family. This raised the possibility that RYBP might interact with one or more of the other E2F proteins. To test this hypothesis, C33-A cells were transfected with expression plasmids encoding E2F1, E2F2, E2F3, E2F4, or E2F6 in the presence or absence of HA-RYBP. The transfected cells were labeled with [35S]methionine and then immunoprecipitated with anti-E2F antibodies (to confirm expression of the E2F proteins) or an anti-HA antibody (to detect RYBP and associated proteins). Although E2F6 was readily detected in the RYBP immunoprecipitation, we did not recover E2F1 through E2F4 (Fig. 3). Similarly, RYBP was not detected in immunoprecipitates with antibodies for E2F1 through E2F4 (data not shown). We therefore conclude that RYBP interacts specifically with E2F6 and not other members of the E2F family through its marked-box domain.
Figure 3.
RYBP interacts specifically with E2F6. C33-A cells were transiently transfected with expression vectors for E2F1, E2F2, E2F3, E2F4, or E2F6 in the presence and absence of HA-RYBP and immunoprecipitated with the indicated antibodies. Nonspecific bands are indicated by asterisks. IP, immunoprecipitate.
E2F6 and the Polycomb Complex.
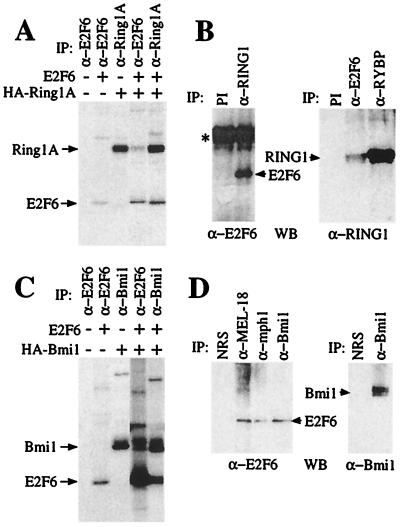
RYBP was originally identified by virtue of its association with Ring1A, a component of the mammalian polycomb complex that has been shown to participate in transcriptional repression (27). Given that E2F6 associates with RYBP in vivo, we speculated that it might also associate with other PcG proteins (Fig. 4). First, we sought to establish if E2F6 and Ring1A were associated. C-33A cells were transfected with expression vectors encoding E2F6 and a HA-tagged version of Ring1A (HA-Ring1A). The cells were then labeled with [35S]methionine and immunoprecipitated with antibodies specific for E2F6 or the HA tag. Consistent with our hypothesis, Ring1A was recovered in E2F6 immunoprecipitates and E2F6 was coimmunoprecipitated with Ring1A (Fig. 4A). Given this finding, we also tested for an interaction between the endogenous E2F6 and Ring1 proteins. Whole-cell lysates from human ML-1 cells were incubated with an anti-Ring1A polyclonal antibody (ASA8), and the resulting immunocomplexes were resolved by SDS/PAGE and then immunoblotted with an anti-E2F6 mAb (LLF6–2). No E2F6 protein was recovered by the preimmune serum but we detected a significant level of E2F6 in the Ring1 immunoprecipitates (Fig. 4B). Importantly, we were also able recover Ring1 protein in anti-E2F6 immunoprecipitates (Fig. 4B). This strongly suggests that there is a physical association between endogenous E2F6 and Ring1 that can be recovered through the immunoprecipitation of either of these proteins.
Figure 4.
E2F6 is a component of the mammalian polycomb complex. (A) C-33A cells were transiently transfected with expression vectors encoding E2F6 and/or HA-Ring1A, labeled with [35S]methionine, and immunoprecipitated with the indicated antibodies. (B) ML-1 cell lysates were immunoprecipitated with either control (preimmune serum) or anti-Ring1 antibodies (ASA8), resolved by SDS/PAGE, and immunoblotted with an anti-E2F-6 (LLF6–2) antibody (Left). Additionally, ML-1 lysates were immunoprecipitated with control (12CA5), anti-E2F6 (LLF6–1), or anti-RYBP (LLRYBP-1) antibodies; resolved by SDS/PAGE; and immunoblotted with the anti-Ring1 (ASA8) antibody (Right). The Ig heavy chain band is denoted by an asterisk. (C) C33-A cells were transfected as in A with either E2F6 or HA-Bmi1 and immunoprecipitated with the indicated antibodies. The data shown are from the same exposure of a single gel. We consistently immunoprecipitated a higher level of E2F6 protein in the presence rather than the absence of Bmi1, which reflects a comparable increase in the E2F6 protein levels (judged by Western blotting of whole-cell lysates) that occurs when it is coexpressed with Bmi1. (D) ML-1 cells were immunoprecipitated as in B with normal rabbit serum, anti-MEL-18, anti-mph1, or anti-Bmi1antibodies and immunoblotted with anti-E2F6 (LLF6–2) or anti-Bmi1 antibodies. WB, Western blot; IP, immunoprecipitate; NRS, normal rabbit serum.
One of the best characterized members of the mammalian PcG proteins is Bmi1, an oncogene whose tumorigenic properties at least partially depend on its ability to repress the expression of the p16INK4a and p19ARF tumor suppressors (29, 30). We therefore investigated whether E2F6 also associates with Bmi1. As described above, E2F6 and/or HA-Bmi1 were expressed in cells by transient transfection and association was assessed by immunoprecipitation. These experiments showed that E2F6 did associate with cotransfected Bmi1 (Fig. 4C). Finally, we also examined the ability of the endogenous E2F6 to associate with Bmi1 and two additional PcG proteins, MEL-18 and mph1. In each case, Western blotting confirmed that E2F6 was present in Bmi1, MEL-18, and mph1 immunoprecipitates but not in those derived from a variety of control antibodies (Fig. 4D and data not shown). Thus, our data indicate that endogenous E2F6 and Bmi1 proteins associate with one another.
Discussion
E2F6 is the most recently identified member of the E2F family and its sequence is only distantly related to those of the other E2Fs (6, 20–22). In particular, E2F6 lacks the sequences required for pRB binding or transactivation and it can act as a dominant negative inhibitor of the other E2F family members (6). By fusing E2F6 sequences to a heterologous DNA binding domain, Gaubatz et al. (20) mapped its repression function to a C-terminal portion of E2F6 that encompasses the marked-box domain. With a yeast two-hybrid screen, we have now discovered that E2F6 binds to RYBP, a recently isolated member of the mammalian PcG complex. Consistent with this observation, we demonstrate a physical association between endogenous E2F6 and numerous PcG proteins, including RYBP, Ring1, MEL-18, mph1, and Bmi1. Because our yeast screen was far from saturating, it will be interesting to establish whether we can find other PcG proteins that bind to E2F6 in this assay. Importantly, RYBP binding is a specific property of E2F6, and not other members of the E2F family, and it maps to the marked-box domain. Thus, these data are consistent with E2F6 acting as a component of the mammalian Bmi1-containing polycomb complex in vivo, and they suggest that E2F6's ability to repress the transcription of E2F-responsive genes depends on its ability to recruit this known transcriptional repressor.
Our observations also raise the possibility that E2F6 will play a key role, beyond E2F regulation, in mediating the changes in transcriptional regulation that are essential for normal developmental patterning. The PcG was originally identified in Drosophila as one of two groups of genes whose mutation causes homeotic transformations and alterations in homeotic gene expression patterns (31). Further analysis showed that the PcG proteins normally act to maintain homeobox (hox) genes in a silenced state, whereas the trithorax complex (trxG) is critical for allowing the transcriptional activation of these genes (31). The PcG proteins are believed to function through the formation of large multisubunit complexes that arise via mutual interactions among conserved protein motifs (32, 33). Promoter mapping studies have identified DNA fragments, called polycomb response elements (PREs), that appear sufficient to mediate the silencing effect of the PcG complex, but these are several hundred base pairs long (34). Consequently, the exact mechanism by which PcG proteins interact with the PRE is not well understood but it is believed to involve pleiohomeotic, a partial homologue of the mammalian zinc-finger protein YY1 (35).
Many mammalian PcG proteins have also been identified, primarily through their homology with the Drosophila PcG proteins or as a result of yeast two-hybrid screens (36–39). Mutant mouse models confirm that a number of these mammalian PcG proteins, including Bmi1, M33, MEL-18, and Ring1A, play an important role in controlling the developmental regulation of hox gene expression and, therefore, the formation of the axial skeleton (40–43). Like their Drosophila counterparts, these proteins appear to act as multimeric complexes (25). In many cases, however, there are multiple mammalian homologues of each Drosophila protein and the phenotypes of the mutant mouse strains suggests that these have overlapping functions in vivo (44). Several distinct PcG components have been implicated in DNA binding, including MEL-18 and YY1, but these factors cannot account for the known DNA binding properties of PcG complexes (35, 45). It has therefore been proposed that additional DNA binding factors must exist in the polycomb complex (35).
In addition to its developmental role, Bmi1 has been shown to play a key role in the regulation of senescence and tumorigenicity. Indeed, Bmi1 was originally identified as a common insertion site in Moloney murine leukemia virus-induced B cell lymphomas in EμMyc transgenic mice and only subsequently was shown to be a mammalian PcG protein (46, 47). The analysis of Bmi1 mutant mice has yielded considerable insight into the role of Bmi1 in both normal development and tumorigenicity (40). The loss of Bmi1 results in the derepression of the p16INK4A and p19ARF tumor suppressor genes that are expressed from the INK4 locus and thereby inhibits cellular proliferation and induces premature senescence (29). Consistent with this observation, mouse crosses indicate that the developmental and tumorigenic properties of Bmi1 both at least partially depend on its ability to regulate the expression of the INK4 locus (29, 30). Although it is widely inferred that Bmi1 mediates the direct transcriptional repression of the INK4 locus via its participation in the polycomb complex, this has not yet been demonstrated. Moreover, it is currently unclear how the Bmi1-containing polycomb complex is recruited to this locus.
The present study shows that E2F6 is a component of the mammalian polycomb complex and, therefore, suggests that it will participate in the developmental regulation of gene transcription in a distinct manner from the other E2F family members. Previous studies have established that p19ARF is an E2F-responsive gene (48). This raises the possibility that E2F6 could contribute to the DNA binding specificity of the polycomb complex during the regulation of normal development and tumorigenicity. Generation and analysis of E2f6 mutant mouse strains will be required to test this hypothesis.
Acknowledgments
We thank M. van Lohuizen and A. Otte for Ring1A, MEL-18, mph1, and Bmi1 reagents and M. Vidal for assistance with the yeast two-hybrid assay. We are grateful to members of the Lees laboratory for helpful discussions and critical reading of this manuscript. This work was supported by a grant from the National Institutes of Health (to J.A.L.).
Abbreviations
- RYBP
Ring 1 and YY1 binding protein
- PcG
polycomb group
- pRB
retinoblastoma protein
- HA
hemagglutinin
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.041597698.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.041597698
References
- 1.Dyson N. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 2.Helin K. Curr Opin Genet Dev. 1998;8:28–35. doi: 10.1016/s0959-437x(98)80058-0. [DOI] [PubMed] [Google Scholar]
- 3.Helin K, Wu C L, Fattaey A R, Lees J A, Dynlacht B D, Ngwu C, Harlow E. Genes Dev. 1993;7:1850–1861. doi: 10.1101/gad.7.10.1850. [DOI] [PubMed] [Google Scholar]
- 4.Krek W, Livingston D M, Shirodkar S. Science. 1993;262:1557–1560. doi: 10.1126/science.8248803. [DOI] [PubMed] [Google Scholar]
- 5.Wu C L, Zukerberg L R, Ngwu C, Harlow E, Lees J A. Mol Cell Biol. 1995;15:2536–2546. doi: 10.1128/mcb.15.5.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trimarchi J M, Fairchild B, Verona R, Moberg K, Andon N, Lees J A. Proc Natl Acad Sci USA. 1998;95:2850–2855. doi: 10.1073/pnas.95.6.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kowalik T F, DeGregori J, Schwarz J K, Nevins J R. J Virol. 1995;69:2491–2500. doi: 10.1128/jvi.69.4.2491-2500.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeGregori J, Leone G, Miron A, Jakoi L, Nevins J R. Proc Natl Acad Sci USA. 1997;94:7245–7250. doi: 10.1073/pnas.94.14.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lukas J, Herzinger T, Hansen K, Moroni M C, Resnitzky D, Helin K, Reed S I, Bartek J. Genes Dev. 1997;11:1479–1492. doi: 10.1101/gad.11.11.1479. [DOI] [PubMed] [Google Scholar]
- 10.Verona R, Moberg K, Estes S, Starz M, Vernon J P, Lees J A. Mol Cell Biol. 1997;17:7268–7282. doi: 10.1128/mcb.17.12.7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hiebert S W, Chellappan S P, Horowitz J M, Nevins J R. Genes Dev. 1992;6:177–185. doi: 10.1101/gad.6.2.177. [DOI] [PubMed] [Google Scholar]
- 12.Humbert P O, Verona R, Trimarchi J M, Rogers C, Dandapani S, Lees J A. Genes Dev. 2000;14:690–703. [PMC free article] [PubMed] [Google Scholar]
- 13.Ziebold, U., Reza, T., Caron, A. & Lees, J. A. (2001) Genes Dev., in press. [DOI] [PMC free article] [PubMed]
- 14.Lukas J, Petersen B O, Holm K, Bartek J, Helin K. Mol Cell Biol. 1996;16:1047–1057. doi: 10.1128/mcb.16.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muller H, Moroni M C, Vigo E, Petersen B O, Bartek J, Helin K. Mol Cell Biol. 1997;17:5508–5520. doi: 10.1128/mcb.17.9.5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brehm A, Kouzarides T. Trends Biochem Sci. 1999;24:142–145. doi: 10.1016/s0968-0004(99)01368-7. [DOI] [PubMed] [Google Scholar]
- 17.Lindeman G J, Dagnino L, Gaubatz S, Xu Y, Bronson R T, Warren H B, Livingston D M. Genes Dev. 1998;12:1092–1098. doi: 10.1101/gad.12.8.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Humbert P O, Rogers C, Ganiatsas S, Landsberg R L, Trimarchi J M, Dandapani S, Brugnara C, Erdman S, Schrenzel M, Bronson R T, et al. Mol Cell. 2000;6:281–291. doi: 10.1016/s1097-2765(00)00029-0. [DOI] [PubMed] [Google Scholar]
- 19.Gaubatz S, Lindeman G J, Ishida S, Jakoi L, Nevins J R, Livingston D M, Rempel R E. Mol Cell. 2000;6:729–735. doi: 10.1016/s1097-2765(00)00071-x. [DOI] [PubMed] [Google Scholar]
- 20.Gaubatz S, Wood J G, Livingston D M. Proc Natl Acad Sci USA. 1998;95:9190–9195. doi: 10.1073/pnas.95.16.9190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morkel M, Wenkel J, Bannister A J, Kouzarides T, Hagemeier C. Nature (London) 1997;390:567–568. doi: 10.1038/37507. [DOI] [PubMed] [Google Scholar]
- 22.Cartwright P, Muller H, Wagener C, Holm K, Helin K. Oncogene. 1998;17:611–623. doi: 10.1038/sj.onc.1201975. [DOI] [PubMed] [Google Scholar]
- 23.Vidal M, Brachmann R K, Fattaey A, Harlow E, Boeke J D. Proc Natl Acad Sci USA. 1996;93:10315–10320. doi: 10.1073/pnas.93.19.10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moberg K, Starz M A, Lees J A. Mol Cell Biol. 1996;16:1436–1449. doi: 10.1128/mcb.16.4.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alkema M J, Bronk M, Verhoeven E, Otte A, van't Veer L J, Berns A, van Lohuizen M. Genes Dev. 1997;11:226–240. doi: 10.1101/gad.11.2.226. [DOI] [PubMed] [Google Scholar]
- 26.Saurin A J, Shiels C, Williamson J, Satijn D P, Otte A P, Sheer D, Freemont P S. J Cell Biol. 1998;142:887–898. doi: 10.1083/jcb.142.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcia E, Marcos-Gutierrez C, del Mar Lorente M, Moreno J C, Vidal M. EMBO J. 1999;18:3404–3418. doi: 10.1093/emboj/18.12.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalenik J L, Chen D, Bradley M E, Chen S J, Lee T C. Nucleic Acids Res. 1997;25:843–849. doi: 10.1093/nar/25.4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacobs J J, Scheijen B, Voncken J W, Kieboom K, Berns A, van Lohuizen M. Genes Dev. 1999;13:2678–2690. doi: 10.1101/gad.13.20.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacobs J J, Kieboom K, Marino S, DePinho R A, van Lohuizen M. Nature (London) 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 31.Kennison J A. Annu Rev Genet. 1995;29:289–303. doi: 10.1146/annurev.ge.29.120195.001445. [DOI] [PubMed] [Google Scholar]
- 32.Kyba M, Brock H W. Mol Cell Biol. 1998;18:2712–2720. doi: 10.1128/mcb.18.5.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shao Z, Raible F, Mollaaghababa R, Guyon J R, Wu C T, Bender W, Kingston R E. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- 34.Strutt H, Paro R. Mol Cell Biol. 1997;17:6773–6783. doi: 10.1128/mcb.17.12.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown J L, Mucci D, Whiteley M, Dirksen M L, Kassis J A. Mol Cell. 1998;1:1057–1064. doi: 10.1016/s1097-2765(00)80106-9. [DOI] [PubMed] [Google Scholar]
- 36.Gould A. Curr Opin Genet Dev. 1997;7:488–494. doi: 10.1016/s0959-437x(97)80075-5. [DOI] [PubMed] [Google Scholar]
- 37.Schumacher A, Magnuson T. Trends Genet. 1997;13:167–170. [PubMed] [Google Scholar]
- 38.Satijn D P, Otte A P. Mol Cell Biol. 1999;19:57–68. doi: 10.1128/mcb.19.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bardos J I, Saurin A J, Tissot C, Duprez E, Freemont P S. J Biol Chem. 2000;275:28785–28792. doi: 10.1074/jbc.M001835200. [DOI] [PubMed] [Google Scholar]
- 40.van der Lugt N M, Domen J, Linders K, van Roon M, Robanus-Maandag E, te Riele H, van der Valk M, Deschamps J, Sofroniew M, van Lohuizen M, et al. Genes Dev. 1994;8:757–769. doi: 10.1101/gad.8.7.757. [DOI] [PubMed] [Google Scholar]
- 41.Core N, Bel S, Gaunt S J, Aurrand-Lions M, Pearce J, Fisher A, Djabali M. Development (Cambridge, UK) 1997;124:721–729. doi: 10.1242/dev.124.3.721. [DOI] [PubMed] [Google Scholar]
- 42.Akasaka T, Kanno M, Balling R, Mieza M A, Taniguchi M, Koseki H. Development (Cambridge, UK) 1996;122:1513–1522. doi: 10.1242/dev.122.5.1513. [DOI] [PubMed] [Google Scholar]
- 43.del Mar Lorente M, Marcos-Gutierrez C, Perez C, Schoorlemmer J, Ramirez A, Magin T, Vidal M. Development (Cambridge, UK) 2000;127:5093–5100. doi: 10.1242/dev.127.23.5093. [DOI] [PubMed] [Google Scholar]
- 44.Bel S, Core N, Djabali M, Kieboom K, Van der Lugt N, Alkema M J, Van Lohuizen M. Development (Cambridge, UK) 1998;125:3543–3551. doi: 10.1242/dev.125.18.3543. [DOI] [PubMed] [Google Scholar]
- 45.Kanno M, Hasegawa M, Ishida A, Isono K, Taniguchi M. EMBO J. 1995;14:5672–5678. doi: 10.1002/j.1460-2075.1995.tb00254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haupt Y, Alexander W S, Barri G, Klinken S P, Adams J M. Cell. 1991;65:753–763. doi: 10.1016/0092-8674(91)90383-a. [DOI] [PubMed] [Google Scholar]
- 47.van Lohuizen M, Verbeek S, Scheijen B, Wientjens E, van der Gulden H, Berns A. Cell. 1991;65:737–752. doi: 10.1016/0092-8674(91)90382-9. [DOI] [PubMed] [Google Scholar]
- 48.Bates S, Phillips A C, Clark P A, Stott F, Peters G, Ludwig R L, Vousden K H. Nature (London) 1998;395:124–125. doi: 10.1038/25867. [DOI] [PubMed] [Google Scholar]