Abstract
The replication protein A complex (RPA) plays a crucial role in DNA replication and damage response. However, it is not known whether this complex is regulated by the SUMOylation pathway. Here we show that the 70kd subunit of RPA (RPA70) associates with a Sentrin/SUMO-specific protease, SENP6, in the nucleus to maintain RPA70 in a hypo-SUMOylated state during S phase. Campothecin (CPT), an inducer of replication stress, dissociates SENP6 from RPA70 allowing RPA70 to be modified by a small ubiquitin-like modifier 2/3 (SUMO-2/3). RPA70 SUMOylation facilitates recruitment of Rad51 to the DNA damage foci to initiate DNA repair through homologous recombination (HR). Cell lines that expressed a RPA70 mutant that cannot be SUMOylated are defective in HR and have a marked increase in sensitivity to CPT. These results demonstrate that SUMOylation status of RPA70 plays a critical role in the regulation of DNA repair through homologous recombination.
The small ubiquitin-like modifier (SUMO) has been shown to regulate cellular processes by controlling the localization, function, interaction, and stability of a large number of cellular proteins (Hay, 2005; Meulmeester and Melchior, 2008; Mukhopadhyay and Dasso, 2007; Yeh, 2009). SUMOylation is catalyzed by SUMO-specific E1, E2, E3s and reversed by a family of Sentrin/SUMO-specific proteases, SENPs. In mammalian cells, six different SENPs belonging to three sub-families have been identified. The first family, consisting of SENP1 and SENP2, has broad specificity for SUMO-1, 2 and 3, and localizes to the nucleus and nuclear envelope, respectively (Gong et al., 2000; Hang and Dasso, 2002; Zhang et al., 2002). The second family consists of SENP3 and SENP5 that favors SUMO-2/3 as substrates and are localized to the nucleolus (Di Bacco et al., 2006; Gong and Yeh, 2006; Yun et al., 2008). Members of third family include SENP6 and SENP7: each has an additional loop inserted in the catalytic domain and also appears to prefer SUMO-2/3 (Mukhopadhyay and Dasso, 2007; Yeh, 2009). From an evolutionary standpoint, SENP1, SENP2, SENP3 and SENP5 are more closely related to the yeast Ulp1, whereas SENP6 and SENP7 are related to Ulp2 (Li and Hochstrasser, 1999, 2000; Mukhopadhyay and Dasso, 2007). Although de-SUMOylation has been extensively studied in vitro, its in vivo functions have only begun to be understood.
We have previously shown that deletion of the murine SENP1 gene leads to the development of severe fetal anemia as a result of erythropoietin (EPO) deficiency (Cheng et al., 2007). SENP1 regulates transcription of EPO through its ability to regulate the stability of hypoxia-inducible factor 1α (HIF1α). The in vivo functions of other SENPs are less well understood. Depletion of SENP3 by siRNA disrupts nucleolar ribosomal RNA processing, a phenotype similar to knockdown of NPM1 (Haindl et al., 2008). Knockdown of SENP5 by siRNA results in inhibition of cell proliferation and appearance of binucleate cells, suggesting that SENP5 may play a role in mitosis and/or cytokinesis (Di Bacco et al., 2006). Silencing of SENP6 causes redistribution of SUMO2 and SUMO3, into the PML bodies (Mukhopadhyay et al., 2006). Recently, an RNA interference-based screen showed that SENP6, but not other SENPs, functioned in cell proliferation (Kittler et al., 2007), but the mechanisms behind this were not reported.
Replication protein A (RPA), the main eukaryotic ssDNA binding protein complex, consists of three subunits, RPA1 (RPA70), RPA2 (RPA32), and RPA3 (RPA14). RPA70 is the major ssDNA binding subunit and is involved primarily in interactions with other DNA metabolism proteins (Fanning et al., 2006; Zou et al., 2006). TheRPA32 subunit has low affinity for ssDNA and utilizes its C-terminal α-helix domain for other protein interactions. It has been suggested that hyper-phosphorylation of RPA32 may redirect RPA from DNA replication to DNA repair (Zou et al., 2006). RPA14 does not exhibit affinity for ssDNA, but is required for stable heterotrimer formation (Iftode et al., 1999; Wold, 1997).
In response to replication stress or fork stalling, the long stretches of RPA-coated ssDNA at DNA damage sites serve as a common intermediate structure for the assembly of two independent checkpoint apparatuses, 9-1-1/Rad17-Rfc2–5 and ATR-ATRIP complexes, that initiate the replication checkpoint response (Zou and Elledge, 2003). RPA plays an important role in the repair of DSBs by homologous recombination (HR) through its ability to interact with Rad51, a recombinase(Galletto and Kowalczykowski, 2007). RPA stimulates DNA strand exchange by removing DNA secondary structure that is inhibitory to contiguous filament formation. However, RPA inhibits DNA strand exchange when it saturates ssDNA before the addition of Rad51. During HR, cofactors (mediators), such as human FANCD1/BRCA2, and Rad51 paralogs, overcome this inhibition and stimulate DNA strand exchange by recruiting Rad51 to replace RPA from ssDNA (Sugiyama and Kowalczykowski, 2002).
Here we show that RPA70 is SUMOylated on lysine residues 449 and K577, with K449 being the major site. RPA70 is associated with SENP6 during S-phase to maintain RPA70 in a hypo-SUMOylated state. However, in response to replication-mediated DSBs, SENP6 is dissociated from RPA70 causing an increase of RPA70 SUMOylation that facilitates recruitment of Rad51 to initiate HR. This identifies a specific role of SENP6 in the regulation of RPA complex and reveals that SUMOylation is important in initiating Rad51-dependent HR.
SENP6 is associated with RPA70 during the S phase
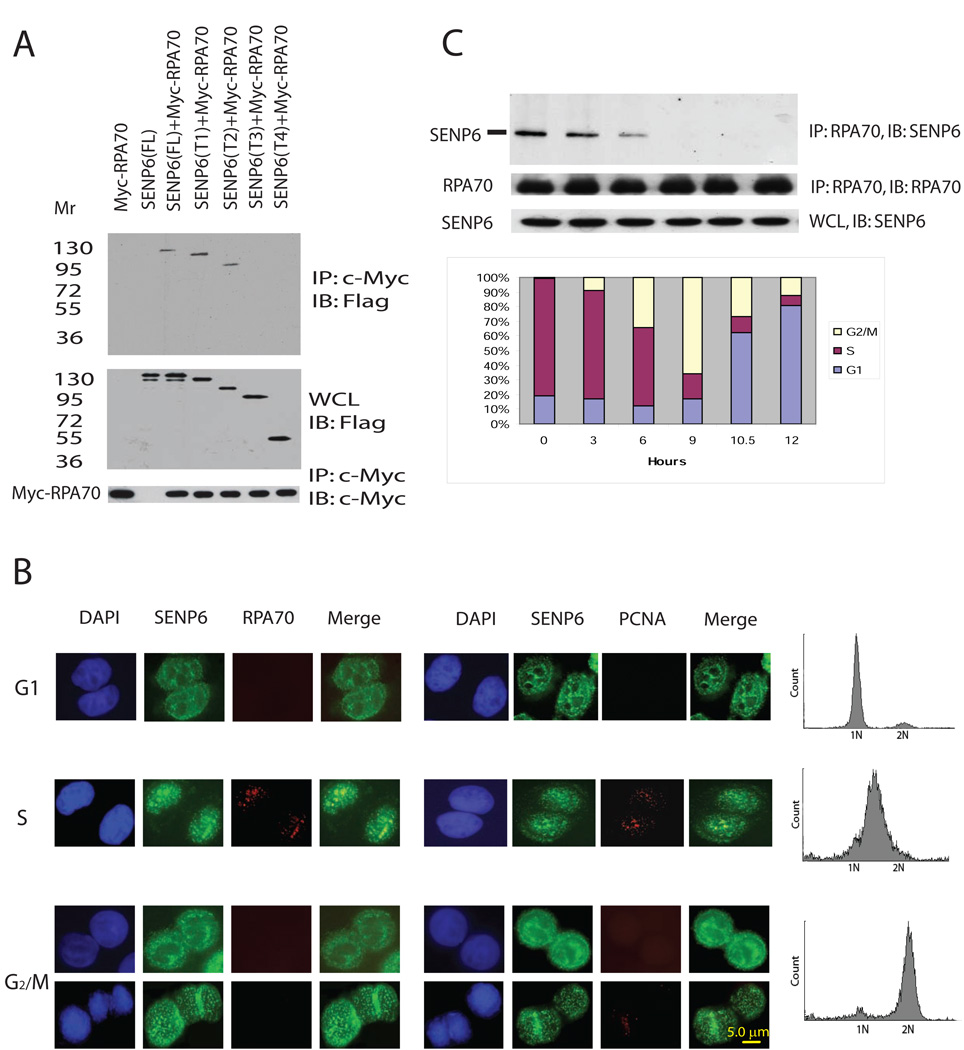
To gain further insight into the role of SENP6 in cellular function, we used a yeast two-hybrid screen to identify its potential substrates. RPA70, a key player in DNA replication and DNA damage responses (Fanning et al., 2006; Zou and Elledge, 2003), was isolated in this screen using a SENP6 catalytic-site mutant (Cys1030-Ser) as bait. Full-length SENP6 was able to co-precipitate with full length RPA70 in vivo (Figure 1A). Truncation experiments showed that residues 629–777 of SENP6 were necessary for RPA70 binding (Figure 1A and Figure S1A). It has been shown that RPA70 is associated with chromatin and is co-localized with cholorodeoxyuridine (CldU) at the replication foci/fork only during S phase in cyto-skeleton (CSK)-Triton-extracted cells, but not in G1 and G2/M (Dimitrova and Gilbert, 2000). Treatment of CSK-Triton removes soluble RPA70 and retains chromatin-associated RPA70. Using the same treatment, we found that SENP6 co-localized with RPA70 and Proliferating Cell Nuclear Antigen (PCNA), a replication foci marker, during S-phase (Figure 1B), suggesting that the association between SENP6 and RPA70 occurs at replication structures in S phase. Moreover, endogenous SENP6 co-precipitated with endogenous RPA70 during the S-phase, but not in other cell cycle phases (Figure 1C). This is not due to degradation of SENP6 at the G2/M phase because endogenous SENP6 protein levels remains constant throughout the cell cycle (Figure 1C, lower panel). This association is specific because non-specific IgG could not precipitate RPA70 and SENP6 (Figure S1B) and is independent of DNA because DNase was included in the process of immunoprecipitation. Thus, the association between SENP6 and RPA70 appears to be cell cycle-dependent.
Figure 1. SENP6 associates with RPA70 during S phase.
(A) Mapping of binding domain of SENP6 with RPA70. HEK-293 cells were co-transfected with 2 µg of c-Myc-tagged RPA70 and 2 µg of FLAG-tagged full-length or truncated SENP6, as indicated. Tagged RPA70 was precipitated by anti-c-Myc affinity matrix and immunoblotted with anti-FLAG antibodies (upper panel). Immunoblotting with anti-FLAG (middle panel) and anti-c-Myc (lower panel) antibodies were performed to ensure that equivalent amounts of truncated SENP6 were present in the lysates before immunoprecipitation or to use as the loading control for IP Western. The samples of anti-c-Myc (lower panel) came from IP. FL: full length, T1: SENP6(1–974); T2: SENP6(1–777); T3: SENP6(1–629); T4: SENP6(1–379).
(B) Co-localization of RPA70 and SENP6 in S-phase cells. HeLa cells were synchronized in mitosis following a previously described procedure (Dimitrova et al., 1999). Metaphase cells were released in the next cell cycle and aliquots of the cells were either collected 4 hr later (G1 phase) or synchronized at the G1/S-phase border through a double-thymidine block as described in Methods and subsequently released in free medium for 3 h (S-phase) and 9 hr (G2/M phase). The cells in G1 and S phases were Triton-extracted, fixed, and stained for indicated antibodies. For cells in G2/M, cells released for 9 hr from second thymidine block were trypsinized, CSK-extracted, fixed, and cytospinned onto cover slide to perform immunostaining. Representative images from different stages of cell cycle were shown. SENP6 (green), RPA70 (red), or PCNA (red). Co-localization of SENP6 with RPA70 or PCNA is visible in yellow.
(C) SENP6 associates with RPA70 during S phase.
HeLa cells were synchronized by double-thymidine block and harvested at indicated time. Endogenous SENP6 was co-precipitated with anti-RPA70 polyclonal antibodies and immunoblotted with anti-SENP6 antibodies (upper panel). Input of precipitated RPA70 was standardized with anti-RPA70 monoclonal antibodies (middle panel). Endogenous SENP6 in the WCL was confirmed by immunoblotting with anti-SENP6 monoclonal antibodies (lower panel). Inserted box shows the cell cycle profile at different time after release from double-thymidine block.
RPA70 is modified by SUMO-2/3 in vivo
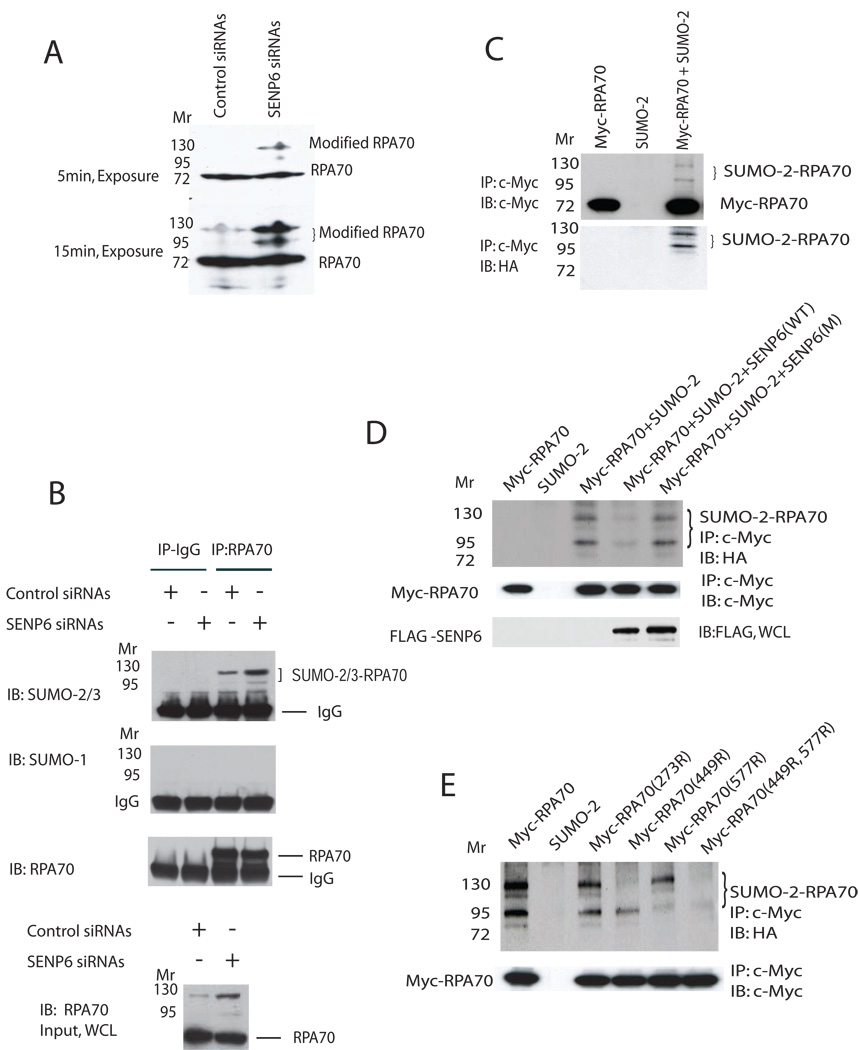
Since SENP6 is a SUMO-specific protease (Mukhopadhyay et al., 2006), we asked whether RPA70 is a substrate of SENP6. For this purpose, we knocked down endogenous SENP6 by siRNA. The knockdown was highly efficient as shown in Figure S2A. As expected, SENP6 knockdown induced the accumulation of higher molecular-weight forms of endogenous RPA70 (Figure 2A). To confirm that these higher molecular-weight bands were due to SUMOylation, endogenous RPA70 was immunoprecipitated and blotted with anti-SUMO-1 or anti-SUMO-2/3 antibodies. These higher molecular-weight bands could readily be detected with anti-SUMO-2/3 (Figure 2B), but not with anti-SUMO-1 antibody (Figure 2B) that had been used to detect endogenous SUMO-1 (Cheng et al., 2007; Hakli et al., 2005; Yamaguchi et al., 2005) or control IgG (Figure 2B). The observation that endogenous RPA70 is modified by endogenous SUMO-2/3 is consistent with the known substrate preference of SENP6 for polySUMOylated SUMO2/3 species (Mukhopadhyay et al., 2006). SUMOylation of RPA70 was further confirmed by co-transfection of Myc-tagged RPA70 and HA-tagged SUMO-2 constructs in COS-1 cells. In the presence of HA-SUMO-2 plasmids, two SUMOylated bands were generated that migrated slower than that of unmodified Myc-tagged RPA70 (Figure 2C). Over-expression of SENP6 decreased the intensity of these two slower-migrating bands, but a SENP6 catalytic mutant did not reduce these bands (Figure 2D). The ability of SENP6 to regulate RPA70 SUMOylation is biologically relevant because endogenous RPA70 SUMOylation is increased following the dissociation of RPA70 and SENP6 in the G2/M phase (Figure S2B and Figure 1C). The higher molecular weight band shown in Figure S2B was RPA70 modified by SUMO2/3; this was demonstrated by an IP-Western experiment (Figure S2C). Taken together, these results demonstrate that SENP6 regulates the SUMOylation status of RPA70 in vivo.
Figure 2. SENP6 regulates SUMOylation of RPA70.
(A) SENP6-knockdown induces RPA70 SUMOylation. Cell lysates were prepared at 72hr after transfection with indicated siRNAs, separated by SDS-PAGE, and immunoblotted with anti-RPA70 monoclonal antibodies. Two different exposures were shown.
(B) Endogenous RPA70 is modified by endogenous SUMO-2/3 in vivo. SENP6-knockdown was described in Methods. Cell lysate was aliquoted equally for performing immunoprecipitation by control IgG or RPA70 monoclonal antibody. Endogenous SUMOylated RPA70 was immunoblotted with anti-SUMO-1 or anti-SUMO-2/3 antibodies. Loading samples were standardized by blotting with anti-RPA70 monoclonal antibodies.
(C) RPA70 is modified by SUMO-2 in an over-expression system. COS-1 cells were transfected with indicated c-Myc-tagged RPA70 (2.5 ug) and HA-tagged SUMO-2 (0.5 ug) plasmids. Immunoprecipitation was performed with Myc-EZview™ Red Affinity Gel matrix 24hr after transfection and analyzed with c-Myc (upper panel) or HA antibodies (lower panel).
(D) Over-expression of SENP6 de-conjugates SUMOylated RPA70. COS-1 cells were transfected with c-Myc-tagged RPA70 (2.0 µg), HA-tagged SUMO-2 (0.5 µg), and SENP6 (0.5 µg) or SENP6 catalytic site mutant (0.5 µg) plasmids. Tagged RPA70 was precipitated and immunoblotted with anti-HA (upper panel) or anti-c-Myc antibodies (middle panel). SENP6 expression was confirmed by immunoblotting with anti-FLAG antibodies (lower panel). Over-expression of SENP6 may result in non-specific cleavage. However, the true specificity of SENP6 was shown by the siRNA knock down studies shown in Figure 2A and B.
(E) Determination of SUMOylation sites in RPA70. COS-1 cells were transfected with 2.0 µg of c-Myc-tagged RPA70 or its mutants along with HA-SUMO-2 plasmids. Immunoprecipitates of c-Myc-tagged RPA70 were immunoblotted with anti-HA (upper panel) or anti-c-Myc antibodies (lower panel).
RPA70 contains three consensus SUMOylation motifs, centered on K273, K449, and K577. We tested the ability of these sites to be SUMOylated in vivo by introducing various combinations of K273R, K449R, and K577R point mutations into RPA70. The results showed that RPA70-SUMO-2 conjugates were virtually absent in the RPA70 (K449, 577R) double-mutant, RPA70(ΔSUMO) (Figure 2E). The amount of SUMOylated conjugates in cells with the K449R mutant was significantly reduced, but in contrast, the K577R mutant only showed a small effect. This indicates that RPA70 is modified in vivo on lysine residues 449 and K577, with K449 being the major site of SUMOylation.
Knocking down SENP6 induces replication defects that result in DNA breaks
To study the relevance of RPA70 and SENP6 association in S phase, we examined the cell cycle profile of SENP6-knockdown cells. Knocking down SENP6 in Hela cells caused a delay in the S phase and accumulation of cells in the S and G2/M phases after release from the second thymidine block (Figure S3A). This cell cycle defect can be repaired by a siRNA-resistant SENP6, but not by the catalytic inactive SENP6 (Figure S3B). Furthermore, DNA synthesis was markedly reduced, as the number of cells incorporating BrdU dropped from 36.2% (control cells) to 6.4% (SENP6-knockdown cells) (Figure S3C). Furthermore, SENP6-knockdown induced co-localization of RPA70 and SUMO-2/3 at many punctate foci (Figure S3D). Moreover, many of the SUMO-2/3 foci also co-localized with γ-H2AX foci, a marker for DSBs (Figure S3E) and with Rad51 (Figure S3F). There was also an increase in sister chromatid exchange (Figure S3G). These results suggest that depletion of SENP6 caused replication-mediated DSB and initiation of DNA repair pathways.
Campothecin and ionized radiation induces separation of SENP6 from RPA70 and RPA70 SUMOylation
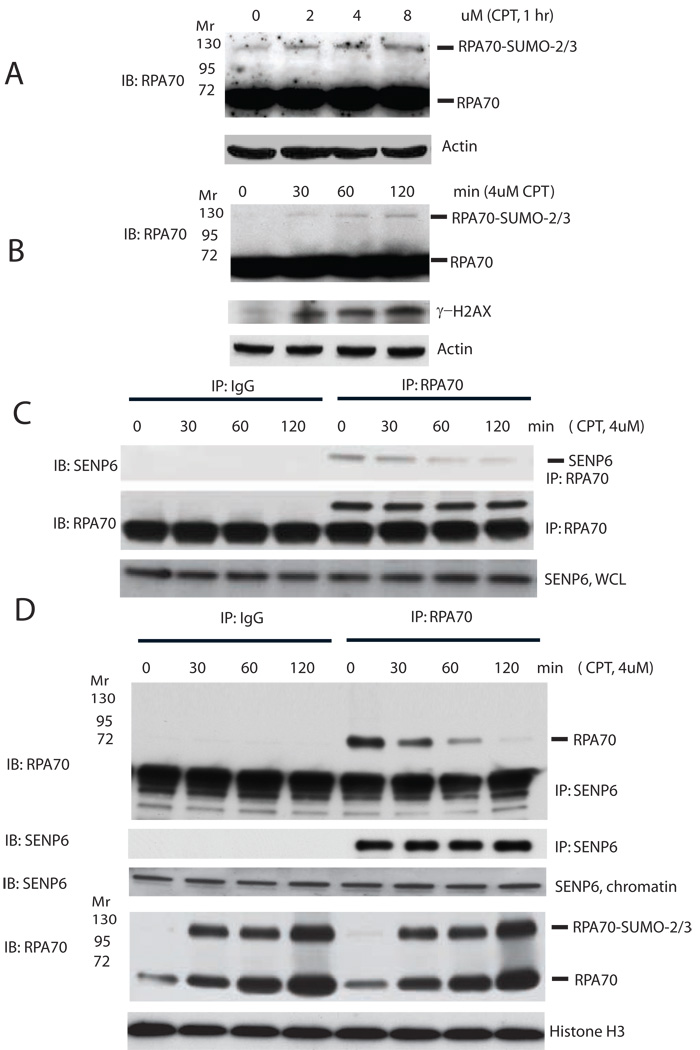
To extend these observations to other replication-mediated DSB, we tested the effect of campothecin (CPT) on SENP6 and RPA70 association. CPT specifically traps DNA topoisomerase I (Topo I)-DNA cleavage complex to form single strand breaks (SSBs) resulting in the conversion to replication-mediated DSBs at DNA replication forks in S phase (Kuzminov, 2001; Saleh-Gohari et al., 2005). We found CPT to be a potent inducer of RPA70 SUMOylation in a dose- and time-dependent manner (Figure 3A, B) in total cell lysates. Temporal RPA70 SUMOylation correlated with the separation of SENP6 from RPA70 and an increase of γ-H2AX following CPT treatment. We showed in synchronized Hela cells, CPT treatment caused separation of SENP6 from RPA70 (Figure S4A). Therefore, replication-mediated DSBs triggered RPA70 SUMOylation following its dissociation with SENP6 (Figure 3 B , C). Because a replication-mediated DSB is preferentially repaired by the HR pathway to re-establish the DNA replication fork, these results also suggest that RPA70 SUMOylation may play a role in HR. To confirm that the regulation of RPA70 SUMOylation occurs on the chromatin, the chromatin fraction was prepared from CPT-treated cells. Consistent with the observations that replication stress induces accumulation of RPA on chromatin (Sleeth et al., 2007), we found that both RPA70 and SUMOylated RPA70 accumulated in the chromatin fraction following CPT treatment (Figure 3D). Moreover, there is a relative increase of SUMOylated RPA70 over un-modified RPA70 in the chromatin fraction concomitant with the dissociation of SENP6 from RPA70 (Figure 3D). Taken together, these results suggest that CPT treatment causes separation of SENP6 from RPA70, leading to SUMOylation of RPA70 at the level of the chromatin.
Figure 3. CPT induces separation of SENP6 from RPA70 and SUMOylation of RPA70.
(A, B) RPA70 SUMOylation in response to CPT. Asynchronous HeLa cells were treated with different concentration of CPT (A) or collected at indicated time points after CPT treatment (B). The whole cell lysates were analyzed by immunoblotting with anti-RPA70, anti-γ-H2AX or anti-actin antibodies.
(C) CPT induces dissociation of RPA70 from SENP6. Endogenous RPA70 was immunoprecipitated by rabbit anti-RPA70 polyclonal antibodies or control IgG and immunoblotted with anti-SENP6 (upper panel) or anti-RPA70 (middle panel) monoclonal antibodies. WCL was blotted with anti-SENP6 antibodies (lower panel).
(D) CPT regulates SENP6/RPA70 association and RPA70 SUMOylation on the chromatin. Chromatin fractions of HeLa cells treated with CPT for different times were prepared as described in Methods. Chromatin-associated SENP6 and RPA70 were released by micrococal nuclease and were immunoprecipitated with anti-SENP6 antibody or control IgG and immunoblotted with anti-RPA70 (upper panel) or anti-SENP6 antibody (second panel). Chromatin fraction was also immunoblotted with anti-SENP6 (third panel), anti-RPA70 (fourth panel), or anti-Histone H3 (lowest panel).
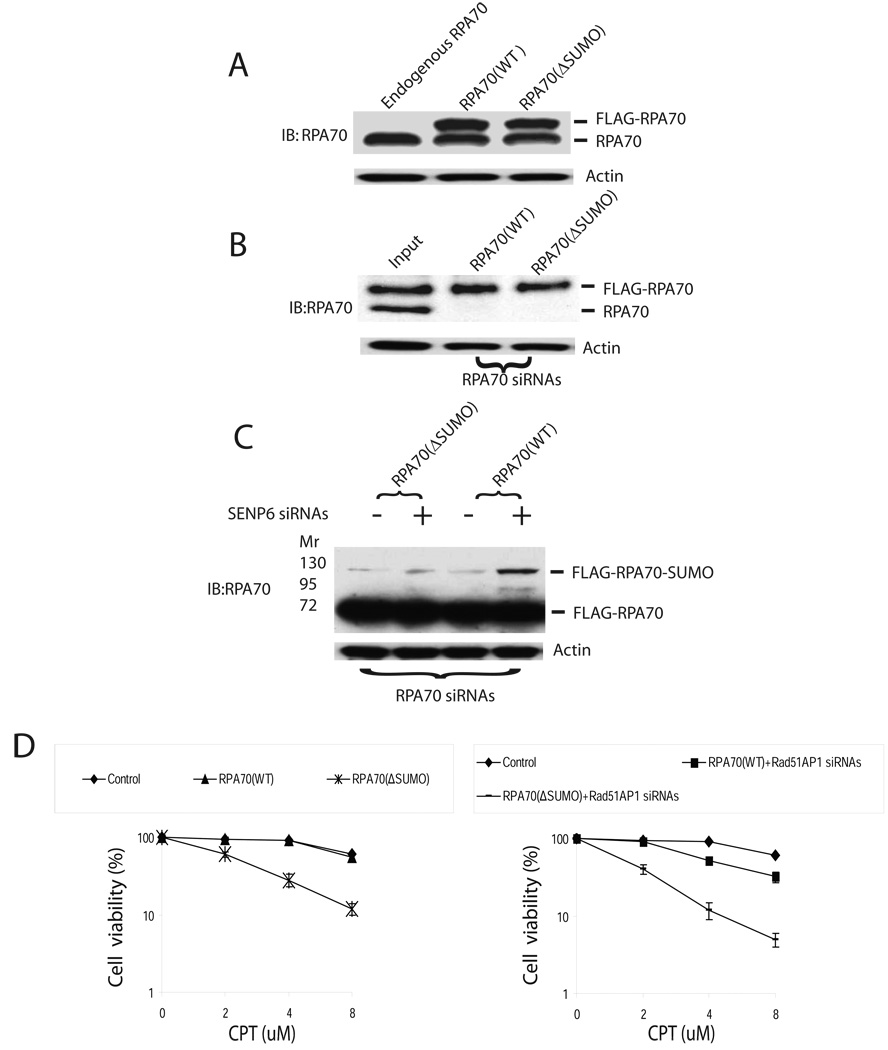
To further evaluate the function of RPA70 SUMOylation in vivo, we established HeLa cell lines that expressed siRNA-resistant, FLAG-tagged wild-type RPA70 (WT) or RPA70 that are refractory to SUMOylation (ΔSUMO). In these cell lines, the expression level of FLAG-tagged RPA70 or ΔSUMO was similar to that of endogenous RPA70 (Figure 4A). Knocking down endogenous RPA70 led to identical expression of FLAG-tagged RPA70 (WT) or RPA70 (ΔSUMO) in HeLa cells (Figure 4B). Furthermore, there was a robust SUMOylation of the FLAG-tagged RPA70 (WT), but not RPA70 (ΔSUMO), following knocking down of both endogenous RPA70 and SENP6 (Figure 4C). The RPA70 (ΔSUMO) cells were very sensitive to CPT compared to RPA70 (WT) cells (Figure 4D). This is most likely due to the inability to SUMOylate RPA70 in the RPA70 (ΔSUMO) cells (Figure S4B). As a control, knocking down Rad51AP1, a structure-specific DNA binding protein that stimulates joint molecule formation during RAD51-mediated HR (Modesti et al., 2007), also caused a marked increase in CPT sensitivity in RPA70 (ΔSUMO) cells compared to RPA70 (WT) cells (Figure 4D, Figure S4C).
Figure 4. RPA70(ΔSUMO) has increased sensitivity to CPT treatment.
(A) Establishment of stable siRNA-resistant FLAG-tagged RPA70 cell cells. This was accomplished by stable expression of FLAG-tagged, siRNA-resistant FLAG-tagged RPA70(WT) or RPA70(ΔSUMO) by infecting HeLa cells with PQCXIP vectors. The expression levels of FLAG-tagged RPA70(WT) or RPA70(ΔSUMO) was similar to endogenous RPA70.
(B) Endogenous RPA70 was knocked down with transfection of 50nM siRNAs and cell lysates were analyzed by RPA70 monoclonal antibodies. Loading samples were standardized by immunoblotting of actin.
(C) Endogenous RPA70 and SENP6 was knocked down in RPA70(WT) or RPA70(ΔSUMO) cell lines. Cells were harvested by trypsinization 72 hr after transfection and analyzed by RPA70 monoclonal antibodies. Loading samples were standardized by immunoblotting of actin. As shown, SENP6 knockdown induces SUMOylation of FLAG-tagged RPA70 only in the RPA70(WT), but not in the RPA70(ΔSUMO) cell lines. There is a slight increase in the upper band in the RPA70(ΔSUMO) in the SENP6 knockdown cells as compared to control. This could be due to the presence of minor SUMOylation site or experimental variations. (D) RPA70(ΔSUMO) was more sensitive to CPT than wild type RPA70(WT). In left panel, endogenous RPA70 was knockdown as described in (B) in HeLa cells that carried siRNA-resistant RPA70 (WT) or RPA70 (ΔSUMO). Knockdown of Rad51AP1 was used as a positive control (right panel). HeLa cells that carried siRNA-resistant RPA70 (WT) or RPA70 (ΔSUMO) were simultaneously transfected with 50nM siRNAs against RPA70 and Rad51AP1 to knock down endogenous RPA70 and Rad51AP1. These cells were trypsinized and 1000 cells were seeded onto 60 mm diameter dishes. After a 12hr attachment period, cells were treated with increasing doses of CPT for 12hr. Cells were subsequently washed twice with PBS and incubated in fresh media for 7–10 days, after which the colonies were fixed, stained and counted. The efficiency of Rad51AP1 knockdown was shown in Figure S4C. Error bars indicate s.d. for the results of triplicate assays.
To further examine the role of RPA70 SUMOylation in the DSBs response, we tested whether ionizing radiation (IR), which also generates replication-mediated DSBs through SSBs (Jeggo and Lobrich, 2006; Liu et al., 2000; Wu and Liu, 1997), could induce similar changes in RPA SUMOylation. Indeed, IR also induced RPA70 SUMOylation in a dose- and time-dependent manner, triggered dissociation of RPA70 and SENP6 in a manner similar to CPT (Figure S5A, B, C), and the sensitivity of RPA70 (ΔSUMO) cells to IR was also increased (Figure S5D). Thus, DSBs during DNA replication could cause dissociation of SENP6 and RPA70, resulting in RPA70 SUMOylation.
SUMOylated RPA70 recruits Rad51 and regulates HR
Because RPA is known to play a role in Rad51-dependent HR (Galletto and Kowalczykowski, 2007), SUMOylation of RPA70 may enhance recruitment of Rad51 to initiate HR. To examine this possibility, RPA70 was SUMOylated in vivo using a Ubc9 fusion-dependent SUMOylation strategy (Jakobs et al., 2007). RPA70-Ubc9 fusion significantly increased RPA70 SUMOylation (Figure S6A). These SUMOylated forms of RPA70 could be de-conjugated in vitro by immunoprecipitated SENP6 proteins (Figure S6A). SUMOylation of RPA70 did not affect the assembly of RPA complex (Figure S6B, C). Furthermore, SUMOylation also did not affect RPA70’s ability to bind to ssDNA (Figure S6D).
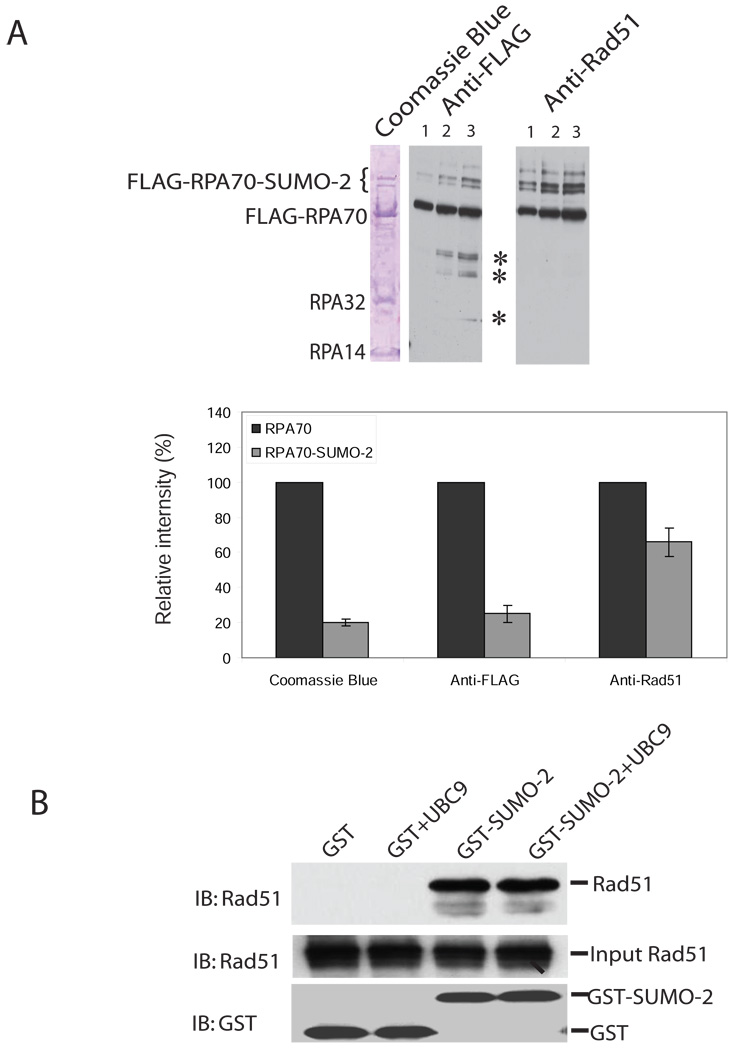
We next studied the role of SUMO-2 in the interaction between Rad51 and RPA70. SUMOylation may affect Rad51 binding to RPA70 by SUMOylation alone or by recruiting a factor that serves to bridge RPA70 and Rad51. To distinguish between these two possibilities, a Far Western analysis was performed. FLAG-immunoprecipitate was first subjected to SDS/PAGE, and then transferred to a membrane. Proteins bound to the membrane were re-natured, incubated with recombinant Rad51, and then probed with anti-Rad51 antibodies. As shown in Figure 5A, incubation of Rad51 enhanced the signal intensity of slowly migrating bands more than 2.6 times compared to that of un-modified RPA70 (Figure 5A). This suggests that RPA70 SUMOylation directly enhanced the recruitment of Rad51. Consistent with a previous report (Golub et al., 1998), in vitro Far-western blotting demonstrated that RPA70, either un-modified or SUMOylated, could interact with Rad51, but not with RPA32 or RPA14. The increase in the binding of SUMOylated RPA70 to Rad51 is mediated by SUMO because SUMO-2 directly interacted with Rad51 (Figure 5B).
Figure 5. SUMOylation enhances the interaction of RPA70 and Rad51.
(A) SUMOylation of RPA70 enhances its association with Rad51. COS-1 cells were co-transfected with FLAG-tagged RPA70-UBC9 or FLAG-tagged RPA70(ΔSUMO)-UBC9 and HA-tagged SUMO-2 plasmids. FLAG-tagged protein was purified by FLAG-EZview™ Red Affinity Gel. C-terminal fused UBC9 was removed by thrombin (see Methods). The precipitates were separated by SDS-PAGE stained with coomassie blue (left lane). The same samples were serially diluted ((1/4(lane 1):1/2(lane 2):1(lane3)) and applied for western blot by probing with HRP-conjugated anti-FLAG antibody (middle lane) or for far-western blot (right panel) that FLAG-immunoprecipitation was first subjected to SDS/PAGE, and then transferred to a membrane. The membrane-bound proteins were re-natured, incubated with recombinant Rad51, and then probed with anti-Rad51 antibodies. Two lower bands are RPA32 and RPA14. *, indicates non-specific bands. Relative intensity of the FLAG-RPA70 or FLAG-RPA70-SUMO2 regions of the coomassie blue gel (left panel) was determined by densitometry. FLAG-RPA70 was set as 100%. Error bar indicates s.d. for the results of three densitometry measurements. For the middle and right panels, the average relative intensity was determined by three measurements of the different serial dilutions. Error bars indicate s.d. for three relative intensity measurements.
(B) SUMO-2 interacts with Rad51. Recombinant Rad51 (500ng) was incubated with GST (500ng) and GST-SUMO-2 (500ng) in 0.5ml binding buffer for 2 hours at 4°C. 10 µl Glutathione-Sepharose™-4B beads were used for pull-down experiments. The precipitates were immunoblotted with anti-Rad51 or anti-GST antibodies. Ubc9 has previously been shown to be involved in binding of SUMO-2 to Rad51. However, we and others showed that SUMO-2 can directly bind to Rad51 in the absence of UBC9 (Ouyang et al., 2009).
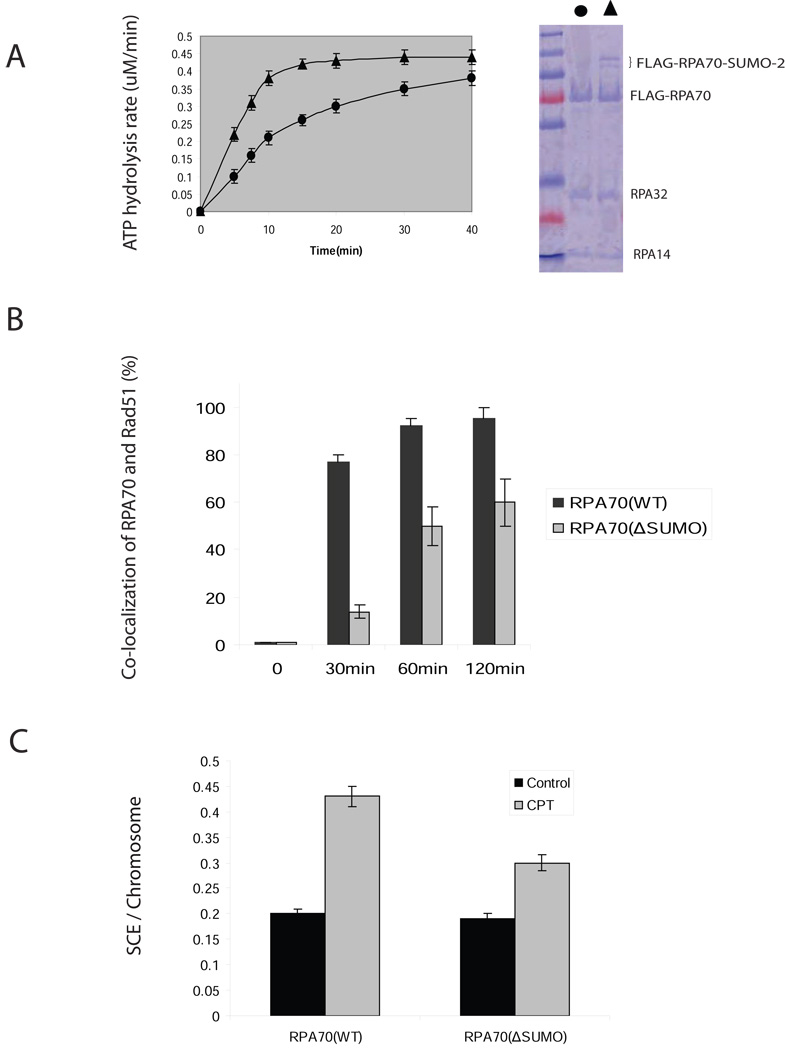
To better understand the role of RPA70 SUMOylation in Rad51 presynaptic complex formation, we examined the effects of RPA70(WT) or RPA70(ΔSUMO) on the time course of filament formation. ATP hydrolysis that accompanied the formation of a Rad51 protein-ssDNA complex was monitored (Sugiyama and Kowalczykowski, 2002). In the absence of ssDNA, we did not observe changes in ATP hydrolysis rate between RPA70 immunoprecipitates with or without SUMOylation (data not shown). However, in the presence of ssDNA, the RPA70-SUMO immunoprecipitates significantly stimulated ATP hydrolysis rate of Rad51 (Figure 6A). The increase in ATP hydrolysis in RPA70(WT) over RPA70(ΔSUMO) suggests that SUMOylation of RPA70 accelerates its replacement by Rad51 from ssDNA.
Figure 6. SUMOylation facilitates Rad51 to displace RPA in vitro and in vivo.
(A) SUMOylation facilitates Rad51 to displace RPA from ssDNA as measured by a time courses of ATPase activity. Input of FLAG-RPA70 (ΔSUMO) (circle) or a mixture of FLAG-RPA70 and FLAG-RPA70-SUMO-2 (triangle) is shown in right panel. FLAG-tagged recombinant proteins were prepared by immunoprecipitation with FLAG-EZview™ Red Affinity Gel matrix from the lysate of COS-1 cells that were co-transfected with FLAG-RPA70-UBC9 or FLAG-RPA70(ΔSUMO)-UBC9 and HA-SUMO-2 cDNAs, eluted with 3XFLAG peptide, and C-terminal fused-UBC9 was removed by thrombin. The reactions were started by addition of Rad51 protein (2.5 µM) to a preformed complex of 1 µM FLAG-RPA70 (ΔSUMO) or a mixture of FLAG-RPA70 and FLAG-RPA70-SUMO-2 with poly (dT) (1.5 µM). The rate of ATP hydrolysis was calculated from the rate of change in absorbance at 340 nm (see Methods). Error bars indicate s.d. for the results of triplicate assays.
(B) SUMOylation mutant is defective in the recruitment of Rad51 to RPA70 foci following CPT treatment. HeLa cells that carried siRNA-resistant RPA70 (WT) or RPA70(∆SUMO) were transfected with siRNAs against RPA70 to knock down endogenous RPA70. Cells treated by 4 µM CTP for 30min were extracted with CSK buffer at indicated time and stained with rabbit anti-Rad51 and mouse anti-RAP70 antibodies. Only nuclei showing more than 5 clearly co-localized-foci were considered as positive. More than 300 cells per sample were analyzed at each independent experiment. Error bars indicate s.d. for the results of triplicate assays.
(C) SUMOylation mutant causes decrease of SCE events in response to CPT. Cells were treated as described in Methods. 30 metaphases were analyzed. Error bars indicate s.d. for the results of triplicate assays.
Regulation of recruitment of Rad51 to the RPA foci was also tested in vivo in RPA70(WT) and RPA70(ΔSUMO) cells. RPA70 foci were induced by CPT treatment in a time-dependent manner (Figure 6B). Co-localization of RPA70 foci with Rad51 was reduced in RPA70(ΔSUMO) cells at all time points compared to the RPA70(WT) cells after CPT treatment. This was due to the delay of formation of Rad51 foci because we did not observe clear Rad51 foci in RPA70(ΔSUMO) cells at different time points after treatment of CPT. Representative pictures of RPA70 foci and Rad51 co-localization were shown in Figure S7A. These in vivo results indicate that the RPA70 SUMOylation is important for Rad51 foci formation.
To demonstrate the role of RPA70 SUMOylation in HR, we measured HR events in RPA70(WT) or RPA70(ΔSUMO) cells using a sister chromatid exchange (SCE) assay that reflects the reciprocal DNA interchange between sister chromatids during replication. Consistent with defects in Rad51 recruitment, we found the level of spontaneous SCEs was essentially the same in both RPA70(WT) and RPA70(ΔSUMO) cells. However, treatment of CPT induced a 30% (P<0.001) decrease in RPA70(ΔSUMO) cells compared to that in RPA70(WT) cells (Figure 6C). Taken together, SUMOylation of RPA70 is important for efficient initiation of HR through recruitment of Rad51 in response to replication stress. The impact of RPA70(ΔSUMO) on HR was further assessed through a Dr-GFP reporter (Pierce et al., 1999). The assay works through gene conversion repair of a double strand break caused by I-SceI digestion and DR-GFP plasmids repaired by homologous recombination expressing GFP (Pierce et al., 1999). We observed that GFP intensity in RPA70(ΔSUMO) cells was reduced 2.4 folds when compared with that in RPA70(WT) cells (Figure S7B), suggesting that deficiency of RPA70 SUMOylation impairs HR. Since SceI-induced DSB is independent of replication stress, the results also indicate that RPA70 SUMOylation plays a role in non-replication-mediated DSB repair through HR.
Discussion
In this manuscript, we show that SENP6 regulates the SUMOylation status of RPA70. SENP6 is associated with RPA70 in the S phase in the nucleus. Separation of SENP6 from RPA70 during S phase induces replication stress that initiates DNA repair pathways. CPT and IR, which can directly generate replication-mediated DSBs through SSBs (Jeggo and Lobrich, 2006; Liu et al., 2000), induces rapid dissociation of SENP6 from RPA70. When a replication fork encounters SSB on the template strand, the SSB can be converted to a DSB causing collapse of the fork (Branzei and Foiani, 2005; McGlynn and Lloyd, 2002). In contrast to CPT and IR, UV and hydroxyurea, which mainly causes replication fork stalling, did not perturb the association between SENP6 and RPA70 (data not shown). Therefore, dissociation of SENP6 from RPA70 could be induced by collapse of the replication forks. Separated from SENP6, RPA70 is quickly modified by SUMO2/3 through the action of unknown SUMO E3 ligases. Recent publications suggest that PIAS1 and/or PIAS4 could be potential candidates (Galanty et al., 2009; Morris et al., 2009). Two lysine residues were identified as SUMO acceptor sites. Lys 449 was modified by a poly SUMO2/3 chain, whereas Lys577 was modified by a single SUMO moiety. The major SUMOylated RPA70 band that accumuated in the G2/M phase or with SENP6 knockdown had a molecular weight of 130 kDa, most likely corresponding to the Lys449 modified specie. Although RPA70 was modified by a poly-SUMO chain, it was not a target for a SUMO-targeted ubiquitin ligase, such as RNF4 (Heideker et al., 2009; Sun et al., 2007; Tatham et al., 2008), because SUMO-RPA70 was not affected by proteasome inhibitors (data not shown).
The process of HR includes three key steps: strand invasion, branch migration and resolution of HR-intermediates (Hiom, 2001). Although it is clear that the key players in the initiation of HR and strand invasion, such as RPA and Rad51, are conserved from bacteriophages to humans, different organisms appear to repair DNA with different sets of mediator proteins. Although yeast Rad52 is an essential factor, but its murine ortholog, MmRad52, is not required for cell viability. MmRad52−/− ES cells are not hypersensitive to agents that induce DSBs and null mutant mice are viable and display no abnormalities in fertility and development of the immune system. Therefore, the presence of genes functionally related to MmRAD52 can partially compensate for the absence of MmRad52 protein (Rijkers et al., 1998; San Filippo et al., 2008). It is not known which mammalian protein is the functional equivalent of the yeast Rad52 protein. The potential candidates are human Rad51 paralogs or BRCA2 (San Filippo et al., 2008). Although we showed that SUMOylation of RPA70 plays a critical role in Rad51 recruitment and initiation of HR, other HR-related factors and their post-translation modifications, such as phosphorylation, could also participate in initiating HR. We showed that RPA70 (ΔSUMO) delayed, but did not abolish the formation of Rad51 foci. It has been reported that the association of Rad51with BRCA2 was delayed 60–75min following the replication stress (Yu et al., 2003). Therefore, it is possible that BRCA2 could initiate HR in a later stage in response to CPT in RPA70(ΔSUMO) cells. In addition, RPA32 phosphorylation has been shown to associate with Rad52 and Rad51 preferentially (Wu et al., 2005). However, we did not observe a difference in RPA32 phosphorylation in RPA70 (wt) and RPA70 (ΔSUMO) cells after treatment of CPT (data not shown). Thus, the relationship between RPA32 phosphorylation and RPA70 SUMOylation remains unclear.
In yeast, SUMOylation of PCNA has been shown to interact with Srs2 helicase to prevent recombinant events in lesion bypass (Papouli et al., 2005; Pfander et al., 2005). SUMOylated PCNA recruits Srs2 to prevent HR (Bergink and Jentsch, 2009). PCNA SUMOylation also facilitates a Rad51-dependent template switch (Branzei et al., 2008), which is similar to the initiation step of HR. However, SUMOylation of mammalian PCNA has not been observed. Although it has been suggested that the yeast homolog of RPA70, rfa1, is SUMOylated, its SUMOylation site(s) and function have yet to be reported (Burgess et al., 2007). Importantly, the predicted SUMOylation sites in mammalian RPA70 are not conserved in rfa1. These observations as well as others (Shrivastav et al., 2008) suggest that yeast and mammalian cells may regulate HR differently.
In mammalian cells, the SMC5/6 complex facilitates telomere HR and elongation by promoting formation of alternative lengthening of telomeres-associated PML bodies through SUMOylation of telomere-binding proteins (Potts and Yu, 2005). After our manuscript was submitted, Galanty et al showed that SUMO E3 ligases promote responses to DNA double-strand break (Galanty et al., 2009; Morris et al., 2009). Morris et al also demonstrated that SUMOylation of BRCA1 plays a critical role in DSBs repair (Galanty et al., 2009; Morris et al., 2009). Moreover, BLM SUMOylation relieves its inhibitory effects on HR and promotes RAD51 function (Ouyang et al., 2009). Therefore, SUMOylation plays a role in various aspects of HR-mediated DNA repair.
The association between SENP6 and RPA70 during the S phase appears to be biologically important. However, the mechanism whereby SENP6 is recruited to the RPA foci is still under investigation. In a yeast two hybrid assay, we identifeid DBF4, one of the components of the replication initiation complex, as a SENP6 binding protein. It is possible that SENP6 is initially recruited to the replication foci by DBF4, then transfers to RPA70. Knocking down SENP6 by siRNA caused slowing down of the S phase, hyper-SUMOylation of RPA70, and activation of DSBs. This is dependent on the catalytic activity of SENP6 becasue the cell cycle defect can be rescued by siRNA resistant wild type SENP6, but not the catalytic inactive mutant. However, RPA70(ΔSUMO) cells could not fully rescue the cell cycle defect caused by SENP6 knockdown (data not shown), suggesting that SENP6 may be required to maintain multiple proteins in a hypo-SUMOylated state during normal cell cycle progression. Works are in progress to identify other SENP6 substrates that may play a role in DNA replication. A recent report showing that SENP6 regulates SUMOylation of CENP-1 in G2/M also supports the idea of multiple SENP6 substrates in cell cycle progression (Mukhopadhyay et al., 2010).
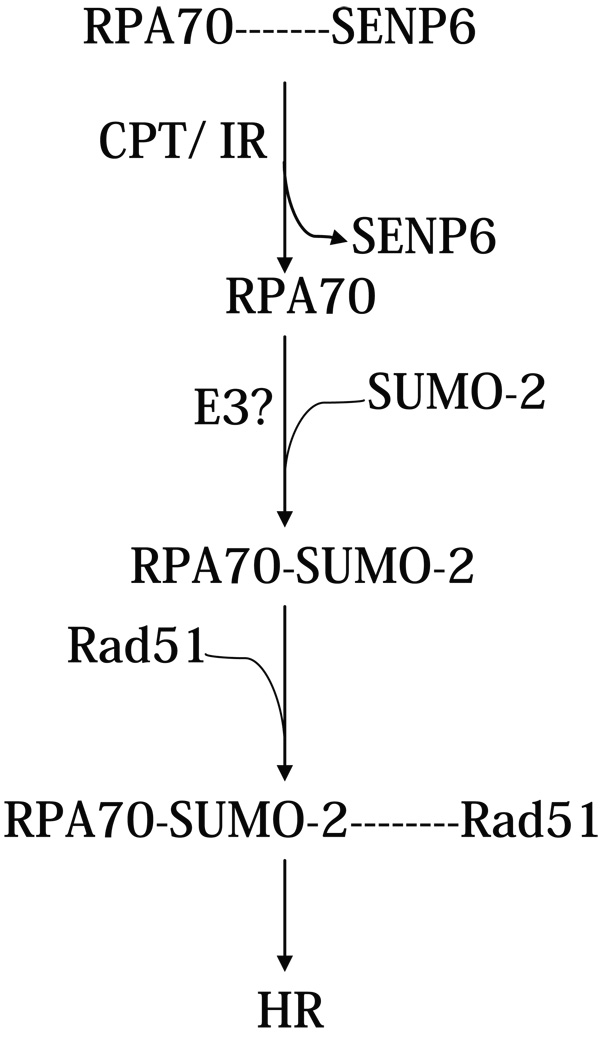
In conclusion, the SUMOylation status of RPA70 is regulated by a unique SUMO-specific protease, SENP6. Association of SENP6 with RPA70 during normal DNA replication maintains RPA70 in a hypo-SUMOylated state. However, when DSBs are generated, SENP6 is dissociated from RPA70 to allow RPA70 to be SUMOylated to initiate HR (Figure 7). Thus, the SUMOylation status of RPA70 plays a critical role in DNA repair through HR.
Figure 7. Role of RPA70 SUMOylation in the regulation of HR.
Association of SENP6 with RPA70 during normal S phase maintains RPA70 in a hypo-SUMOylated state. This is shown by a dashed line to indicate that SENP6 and RPA70 association is not stoichiometric. With CPT treatment or exposure to IR, SENP6 becomes dissociated from RPA70. SUMOylation of RPA70 then leads to enhanced recruitment of Rad51 to initiate HR.
METHODS
Plasmids, siRNAs, recombinant proteins, and antibodies
The full-length cDNAs of SENP6, RPA70-UBC9, and RPA70 were cloned into a 3X FLAG (Invitrogen, Carlsbad, CA) and 3X Myc expression (Stratagene, La Jolla, CA) plasmids respectively by using standard techniques. Site-specific mutation was introduced by using a QuickChange® Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA). All siRNA oligos were purchased from Dharmacon (Lafayette, CO). Dharmacon siGENOME™ Control was used as the control oligo. We identified two SENP6 siRNA sequences (SENP6 siRNA1: GGACAAATCTGCTCAGTGT and SENP6 siRNA2: GCACAGATACCAGTAGTAA) that both efficiently reduced SENP6 mRNA and protein levels and inhibited cell growth. SENP6 siRNA1 was used to perform all the knockdown experiments throughout this study. RPA70 siRNAs were based on an oligonucleotide sequence, AACACTCTATCCTCTTTCATG. Rad51AP1 siRNAs pool was purchased from Santa Cruz (Santa Cruz, CA).
Recombinant GST-SUMO-2 was purchased from LAE Bio (Rockville, MD). Human Rad51 cDNA was cloned into pET28 (Novagene Inc., Madison, WI). BL21(DE3) strain of E. coli was transformed with pET-RAD51, induced with isopropyl-1-thio-β-D-galactoside, and harvested for protein purification. Recombinant Rad51were purified by a combination of metal-chelate affinity and Mono Q ion exchange column chromatography.
Mouse monoclonal antibodies against influenza hemagglutinin (HA) were purchased from Covance (Richmond, CA). The coding region of SENP6, EKPKYEPNPHYHEN, was used as animmunogen to generate rabbit anti-SENP6 polyclonal antibodies, which was further purified for immunoblot and immunofluorescence staining. Mouse anti-SENP6 monoclonal antibodies were purchased from Abnova Corporation (Taipei, Taiwan). Rabbit anti-Rad51 polyclonal and mouse anti-RPA70 monoclonal antibodies were purchased from Santa Cruz (Santa Cruz, CA). Rabbit anti-RPA70 anti-Rad51AP1, and anti γ-H2AX polyclonal antibodies were from Bethyl Lab (Montgomery, TX). Monoclonal anti-SUMO-1 abtibodies (anti-GMP-1) from Zymed (Carlsbad, CA). Mouse anti-SUMO-2/3 monoclonal antibodies were kindly provided by Dr. Michael Matunis.
siRNAs transfection
The cells were split in six-well plates (5×104/ml) and grown in antibiotic-free DMEM overnight. The next day, both siRNAs and the Dharmafect reagent (Dharmacon, Lafayette, CO) were diluted with serum-free DMEM and incubated for 10 min. These diluted reagents were then mixed, incubated for 20 min at RT to form a siRNA- Dharmafect complex, and added drop-wise to the cells. After 4hr incubation at 37°C, these cells were used in experiments at indicated time. We found that 50–100 nM siRNAs could efficiently knock down endogenous SENP6, RPA70, or Rad51AP1.
Generation of stable cell lines
siRNA-resistant, FLAG-tagged RPA70(WT) and RPA70(ΔSUMO) cDNAs were digested with BamH1 and NotI, and subcloned into pQCXIP (Clontech, Mountain View, CA). Five silent mutations were introduced and indicated by bold/italic letters in RPA70 siRNA-targeted region: AACT A GT C GA T GA G AG C GGT G. These plasmids were transfected into Platinum A package cells (Cell Biolabs, San Diego, CA) by using Fugene6 transfection reagent (Roche, Indianapolis, IN). 2 days after transfection, the supernatant was collected and filtered through 0.45µm filter. 1 ml medium was removed from a 3-cm plate with the cells around 40–50% confluence, and 1ml filtered supernatant was added with polybrene (final polybrene conc. is 8 µg/ml). Stable cells were selected by puromycin (2 µg/ml) after a 24hr infection.
Immunofluorescence
Control and SENP6-knockdown cells were either fixed directly or washed with cold-skeleton buffer (CSK:10mM HEPES/KOH, pH7.4, 300mM sucrose, 100mM NaCl, 3mM MgCl2), further extracted for 2min on ice with 0.5% Triton X-100 in CSK buffer, and then fixed with 4% paraformaldehyde. Coverslips were blocked with 5% goat serum and 0.3% Tween 20 in PBS, probed with primary antibodies, washed with PBS, and then incubated with Alexa 488– and Alexa 594–labeled secondary antibodies (Molecular Probes, Carlsbad, CA) at a dilution of 1:2,000. Immunostained coverslips were mounted with Permount® (Sigma, St. Louis, MO) and sealed with nail polish. Examination was carried out immediately using appropriate excitation wavelength, depending on fluorochrome for best results or store flat at 4°C in the dark. For immunostaining of G2/M cells, G2/M HeLa cells were collected at 9 hr after release form second thymidine block. Cells were collected by centrifugation (4°C), and resuspended in CSK buffer. After 2 min, cells were pelleted, gently resuspended in fixative at 4°C for 15 min, washed with PBS and cytospined onto cover slides to process for immunofluorescence as described above.
Immunoprecipitation
Cells were transfected with indicated plasmids. 24hr after transfection, cells were lysed with immmunoprecipition buffer (10 mM phosphate buffer/10 mM Tris/150 mM NaCl/1% Triton-X100, 20mM NEM, pH 7.5), treated with TURBO DNase (Ambion, Foster City, CA), and immunoprecipitated with the indicated antibodies or EZview™ Red Affinity Gel matrix (Sigma, St. Louis, MO). Immunoprecipitated proteins were resolved by SDS/PAGE, and analyzed by immunoblotting. This protocol was also used to purify a mixture of FLAG-RPA70 and FLAG-RPA70-SUMO-2 in a larger scale. Briefly, 3XFLAG-RPA70-UBC9 expression plasmid was constructed by fusing human UBC9 cDNA to the 3’ end of RPA70 cDNA through a nucleotide sequence (poly-Gly, 5mer). FLAG-tagged recombinant proteins were immunoprecipitated with FLAG-EZview™ Red Affinity Gel matrix from the lysate of COS-1 cells that were co-transfected with FLAG-RPA70-UBC9 and HA-SUMO-2 cDNAs, and eluted with 3XFLAG peptides (100ug/ml). C-terminal fused-UBC9 was removed by thrombin (Sigma, St. Louis, MO) according to the manufacturer’s instruction.
Chromatin preparation and micrococcal nuclease digestion
Chromatin of Hela cells was prepared as described in previous publications with modification (Eide et al., 2003; Gilbert and Allan, 2001). Briefly, cell cultures were harvested in PBS containing 0.25 mg/ml trypsin and 1 mM EDTA and were washed in PBS. The cell pellet was resuspended in a small volume of ice-cold buffer A (85 mM KCl/10 mM TrisHCl• (pH 7.6)/5.5% (wt/vol) sucrose/0.5 mM spermidine/0.2 mM EDTA/0.25 mM PMSF). To this an equal volume of NBA plus 0.1% (vol/vol) Nonidet P-40 was added, and the cells were incubated on ice for 3 min. Nuclei were collected by centrifugation (360 × g for 3.5 min at 4°C) and washed in buffer B (NBA without EDTA). Sedimented nuclei were resuspended in buffer C (10 mM Pipes, pH 6.8, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, and a protease inhibitor mixture) containing 1% Triton X-100, and extracted at 4 °C for 15 min. The extract was centrifuged at 2,000 × g for 5 min. The pellet was digested in buffer D (Buffer C without sucrose) supplemented with 0.5% Triton X-100, 5 mM CaCl2 and 40 units/500ug DNA micrococcal nuclease at room temperature for 40 min. The lysate was sedimented at 10,000 × g, and the supernatant constituted the chromatin fraction.
Far-western blotting
The protocol for far-western blotting was described earlier (Golub et al., 1998). Briefly, proteins were electrophoresed through a 10% SDS-polyacrylamide gel and transferred to PVDF membrane. The membrane was immersed in 8 M urea and 1% mercaptoethanol in buffer FW (20 mM Tris-HCl, pH 7.5, 60 mM NaCl, 10 mM MgCl2, 0.1 mM EDTA, 5% glycerol, 0.02% NP-40). Proteins adsorbed to the membrane were re-natured by incubation in 10 sequential 2-fold dilutions of urea in buffer FW. After blocking in buffer FW containing 5% non-fat dry milk, the membrane was incubated for 1 h at room temperature with 0.5 mg HsRad51 protein/ml in buffer FW containing 2% BSA. The bands which retained HsRad51 were detected using anti-HsRad51 antibodies.
In vitro protein binding
1µg of recombinant Rad51 and 3 µg of GST or GST fusion protein were mixed in 500 µl of binding buffer (50 mM Tris–HCl, pH 7.9, 200 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1 mM dithiothreitol). After 4 hr incubation at 4°C, 20 µl of glutathione–sepharose 4B was added, the mixture was incubated at 4°C for 1hr and centrifuged. The pellet was washed four times and proteins were eluted by SDS-PAGA loading buffer, and applied to immunoblot.
ATPase Assay
1 µM FLAG-RPA70 (ΔSUMO) or a mixture of FLAG-RPA70 and FLAG-RPA70-SUMO-2 were pre-incubated for 30 min with the reaction buffer (1.5 µM poly-dT (20mer), 2.5 mM ATP, 10 units/ml pyruvate kinase, 10 units/ml lactate dehydrogenase, 0.3 mM phosphoenolpyruvate, 256 µM NADH, 50 µg/ml bovine serum albumin, 1 mM dithiothreitol, 5 mM magnesium acetate, 50 mM KCl, and 30 mM Tris acetate (pH 7.5)), and then added 2.5 µM Rad51 protein. Hydrolysis of ATP is coupled to the oxidation of NADH, which results in a decrease in absorbance at 340 nm. The rate of ATP hydrolysis was calculated from the rate of change in absorbance using the following formula: rate of A340 decrease (s−1) × 9880 = rate of ATP hydrolysis (µM/min).
Clonogenic cell survival assay
Wild type, siRNA-resistant, FLAG-tagged RPA70(WT) and RPA70(ΔSUMO) cells were trypsinized, counted, and seeded into 60-mm cloning dishes at densities of 200–20,000 cells/dish after 24hr post-transfection of RPA70 siRNAs or in combining of Rad51AP1 siRNAs, and then irradiated or incubated with CPT, Colonies were allowed to grow in a humidified 5% CO2 atmosphere at 37°C for 7–12 days, after which cells were fixed, stained, counted.
Sister chromatid exchange assay
Briefly, 4hr after transfection with RPA70 siRNAs, HeLa cells (~30% confluence) were grown for two cell cycles in the presence of 20 µM BrdU and CPT was added at 2.5nM for 8 hr before the end of the second cell cycle. The cells were then incubated with 0.2µg/ml colcemid for 2hr, harvested with trypsin, swelled for 20min in 60 mM KCl and fixed for 30min in methanol:acetic Acid (3:1). The fixed cells were dropped onto moist 45°C pre-warmed glass slides and allowed to air dry. The slides were aged overnight and stained with 100 µg/ml Hoechst 33258 in ddH2O for 20 min at 25°C, and then bleached with a 120-watt plant light at a distance of 20 cm for 3hr. Next, the slides were stained with 10% Giemsa stain for 15min, air-dried and mounted with Permount® (Sigma, St. Louis, MO).
Yeast two-hybrid screening
Yeast two-hybrid screening was performed with the Pretransformed Human Testis Matchmaker™ cDNA Library (Clontech, Mountain View, CA, Cat. No. 638832,) according to the manufacturer’s instructions. In brief, pGBK-T7-SENP6 (M) was transformed into the yeast strain AH109. Transformants containing bait plasmid were mated with the Pre-transformed Human Testis cDNA Library. Candidates from the two-hybrid interaction were initially selected on SD medium (His, Leu, and Trp) and confirmed on SD medium (Ade, His, Leu, and Trp) containing X-gal. Plasmid DNA was isolated from the positive clones and sequenced according to the Yeastmaker™ Yeast Plasmid Isolation Kit instructions (Clontech).
Homologous Recombination (HR) Repair Assay of DR-GFP
An HR repair assay was carried out as described previously (Pierce et al., 1999). The plasmids were kindly provided by M. Jasin. Briefly, the efficiency of HR was assessed using an I-SceI expression plasmid (pCBASce) and an I-SceI repair reporter plasmid (DR-GFP) composed of two differentially mutated GFP genes, one of which contained a unique I-SceI restriction site. The assay works through gene conversion repair of a double strand break caused by I-SceI digestion. DR-GFP plasmids repaired by homologous recombination express GFP. The relevant cells were transfected with either 1ug of DR-GFP plus 2ug of pCBASce or 1ug of DR-GFP plus 2ug of control plasmids. 48 h after transfection, the cells were harvested, and the number of GFP-expressing cells was assessed by flow cytometry.
Densitometry
Auto-radiographs of the blots were scanned with Fluorchem™ 8900 (Biomedical Solution Inc.) and analyzed by AlphaEaseFC (Biomedical Solution Inc.).
Supplementary Material
ACKNOWLEDGEMENTS
We thank S. Mitra, J. Jin, L. Li, M. Matunis, X. Huang, M. Jasin, and S. Zhang for reagents and/or discussion, and Y. Long, M. Cao, J. Ren for technical support. This work was supported in part by National Institute of Health Grants to ETHY (CA 239520), PBC (2R56GM06581, IR21AI076747-01), and a Welch Grant to PBC (AU-1569). ETHY is the McNair Scholar of the Texas Heart Institute/St. Luke’s Episcopal Hospital.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bergink S, Jentsch S. Principles of ubiquitin and SUMO modifications in DNA repair. Nature. 2009;458:461–467. doi: 10.1038/nature07963. [DOI] [PubMed] [Google Scholar]
- Branzei D, Foiani M. The DNA damage response during DNA replication. Curr Opin Cell Biol. 2005;17:568–575. doi: 10.1016/j.ceb.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Branzei D, Vanoli F, Foiani M. SUMOylation regulates Rad18-mediated template switch. Nature. 2008;456:915–920. doi: 10.1038/nature07587. [DOI] [PubMed] [Google Scholar]
- Burgess RC, Rahman S, Lisby M, Rothstein R, Zhao X. The Slx5-Slx8 complex affects sumoylation of DNA repair proteins and negatively regulates recombination. Mol Cell Biol. 2007;27:6153–6162. doi: 10.1128/MCB.00787-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Kang X, Zhang S, Yeh ET. SUMO-Specific Protease 1 Is Essential for Stabilization of HIF1alpha during Hypoxia. Cell. 2007;131:584–595. doi: 10.1016/j.cell.2007.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bacco A, Ouyang J, Lee HY, Catic A, Ploegh H, Gill G. The SUMO-specific protease SENP5 is required for cell division. Mol Cell Biol. 2006;26:4489–4498. doi: 10.1128/MCB.02301-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova DS, Gilbert DM. Stability and nuclear distribution of mammalian replication protein A heterotrimeric complex. Exp Cell Res. 2000;254:321–327. doi: 10.1006/excr.1999.4770. [DOI] [PubMed] [Google Scholar]
- Dimitrova DS, Todorov IT, Melendy T, Gilbert DM. Mcm2, but not RPA, is a component of the mammalian early G1-phase prereplication complex. J Cell Biol. 1999;146:709–722. doi: 10.1083/jcb.146.4.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide T, Tasken KA, Carlson C, Williams G, Jahnsen T, Tasken K, Collas P. Protein kinase A-anchoring protein AKAP95 interacts with MCM2, a regulator of DNA replication. J Biol Chem. 2003;278:26750–26756. doi: 10.1074/jbc.M300765200. [DOI] [PubMed] [Google Scholar]
- Fanning E, Klimovich V, Nager AR. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34:4126–4137. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanty Y, Belotserkovskaya R, Coates J, Polo S, Miller KM, Jackson SP. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature. 2009;462:935–939. doi: 10.1038/nature08657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletto R, Kowalczykowski SC. RecA. Curr Biol. 2007;17:R395–R397. doi: 10.1016/j.cub.2007.03.009. [DOI] [PubMed] [Google Scholar]
- Gilbert N, Allan J. Distinctive higher-order chromatin structure at mammalian centromeres. Proc Natl Acad Sci U S A. 2001;98:11949–11954. doi: 10.1073/pnas.211322798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub EI, Gupta RC, Haaf T, Wold MS, Radding CM. Interaction of human rad51 recombination protein with single-stranded DNA binding protein, RPA. Nucleic Acids Res. 1998;26:5388–5393. doi: 10.1093/nar/26.23.5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong L, Millas S, Maul GG, Yeh ET. Differential regulation of sentrinized proteins by a novel sentrin-specific protease. J Biol Chem. 2000;275:3355–3359. doi: 10.1074/jbc.275.5.3355. [DOI] [PubMed] [Google Scholar]
- Gong L, Yeh ET. Characterization of a family of nucleolar SUMO-specific proteases with preference for SUMO-2 or SUMO-3. J Biol Chem. 2006;281:15869–15877. doi: 10.1074/jbc.M511658200. [DOI] [PubMed] [Google Scholar]
- Haindl M, Harasim T, Eick D, Muller S. The nucleolar SUMO-specific protease SENP3 reverses SUMO modification of nucleophosmin and is required for rRNA processing. EMBO Rep. 2008;9:273–279. doi: 10.1038/embor.2008.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakli M, Karvonen U, Janne OA, Palvimo JJ. SUMO-1 promotes association of SNURF (RNF4) with PML nuclear bodies. Exp Cell Res. 2005;304:224–233. doi: 10.1016/j.yexcr.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Hang J, Dasso M. Association of the human SUMO-1 protease SENP2 with the nuclear pore. J Biol Chem. 2002;277:19961–19966. doi: 10.1074/jbc.M201799200. [DOI] [PubMed] [Google Scholar]
- Hay RT. SUMO: a history of modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Heideker J, Perry JJ, Boddy MN. Genome stability roles of SUMO-targeted ubiquitin ligases. DNA Repair (Amst) 2009;8:517–524. doi: 10.1016/j.dnarep.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiom K. Recombination: homologous recombination branches out. Curr Biol. 2001;11:R278–R280. doi: 10.1016/s0960-9822(01)00138-5. [DOI] [PubMed] [Google Scholar]
- Iftode C, Daniely Y, Borowiec JA. Replication protein A (RPA): the eukaryotic SSB. Crit Rev Biochem Mol Biol. 1999;34:141–180. doi: 10.1080/10409239991209255. [DOI] [PubMed] [Google Scholar]
- Jakobs A, Koehnke J, Himstedt F, Funk M, Korn B, Gaestel M, Niedenthal R. Ubc9 fusion-directed SUMOylation (UFDS): a method to analyze function of protein SUMOylation. Nat Methods. 2007;4:245–250. doi: 10.1038/nmeth1006. [DOI] [PubMed] [Google Scholar]
- Jeggo P, Lobrich M. Radiation-induced DNA damage responses. Radiat Prot Dosimetry. 2006;122:124–127. doi: 10.1093/rpd/ncl495. [DOI] [PubMed] [Google Scholar]
- Kittler R, Pelletier L, Heninger AK, Slabicki M, Theis M, Miroslaw L, Poser I, Lawo S, Grabner H, Kozak K, et al. Genome-scale RNAi profiling of cell division in human tissue culture cells. Nat Cell Biol. 2007;9:1401–1412. doi: 10.1038/ncb1659. [DOI] [PubMed] [Google Scholar]
- Kuzminov A. Single-strand interruptions in replicating chromosomes cause double-strand breaks. Proc Natl Acad Sci U S A. 2001;98:8241–8246. doi: 10.1073/pnas.131009198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SJ, Hochstrasser M. A new protease required for cell-cycle progression in yeast. Nature. 1999;398:246–251. doi: 10.1038/18457. [DOI] [PubMed] [Google Scholar]
- Li SJ, Hochstrasser M. The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol Cell Biol. 2000;20:2367–2377. doi: 10.1128/mcb.20.7.2367-2377.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LF, Desai SD, Li TK, Mao Y, Sun M, Sim SP. Mechanism of action of camptothecin. Ann N Y Acad Sci. 2000;922:1–10. doi: 10.1111/j.1749-6632.2000.tb07020.x. [DOI] [PubMed] [Google Scholar]
- McGlynn P, Lloyd RG. Recombinational repair and restart of damaged replication forks. Nat Rev Mol Cell Biol. 2002;3:859–870. doi: 10.1038/nrm951. [DOI] [PubMed] [Google Scholar]
- Meulmeester E, Melchior F. Cell biology: SUMO. Nature. 2008;452:709–711. doi: 10.1038/452709a. [DOI] [PubMed] [Google Scholar]
- Modesti M, Budzowska M, Baldeyron C, Demmers JA, Ghirlando R, Kanaar R. RAD51AP1 is a structure-specific DNA binding protein that stimulates joint molecule formation during RAD51-mediated homologous recombination. Mol Cell. 2007;28:468–481. doi: 10.1016/j.molcel.2007.08.025. [DOI] [PubMed] [Google Scholar]
- Morris JR, Boutell C, Keppler M, Densham R, Weekes D, Alamshah A, Butler L, Galanty Y, Pangon L, Kiuchi T, et al. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature. 2009;462:886–890. doi: 10.1038/nature08593. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D, Arnaoutov A, Dasso M. The SUMO protease SENP6 is essential for inner kinetochore assembly. J Cell Biol. 2010;188:681–692. doi: 10.1083/jcb.200909008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D, Ayaydin F, Kolli N, Tan SH, Anan T, Kametaka A, Azuma Y, Wilkinson KD, Dasso M. SUSP1 antagonizes formation of highly SUMO2/3-conjugated species. J Cell Biol. 2006;174:939–949. doi: 10.1083/jcb.200510103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D, Dasso M. Modification in reverse: the SUMO proteases. Trends Biochem Sci. 2007;32:286–295. doi: 10.1016/j.tibs.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Ouyang KJ, Woo LL, Zhu J, Huo D, Matunis MJ, Ellis NA. SUMO modification regulates BLM and RAD51 interaction at damaged replication forks. PLoS Biol. 2009;7:e1000252. doi: 10.1371/journal.pbio.1000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papouli E, Chen S, Davies AA, Huttner D, Krejci L, Sung P, Ulrich HD. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol Cell. 2005;19:123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts PR, Yu H. Human MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol Cell Biol. 2005;25:7021–7032. doi: 10.1128/MCB.25.16.7021-7032.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijkers T, Van Den Ouweland J, Morolli B, Rolink AG, Baarends WM, Van Sloun PP, Lohman PH, Pastink A. Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol Cell Biol. 1998;18:6423–6429. doi: 10.1128/mcb.18.11.6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh-Gohari N, Bryant HE, Schultz N, Parker KM, Cassel TN, Helleday T. Spontaneous homologous recombination is induced by collapsed replication forks that are caused by endogenous DNA single-strand breaks. Mol Cell Biol. 2005;25:7158–7169. doi: 10.1128/MCB.25.16.7158-7169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- Sleeth KM, Sorensen CS, Issaeva N, Dziegielewski J, Bartek J, Helleday T. RPA mediates recombination repair during replication stress and is displaced from DNA by checkpoint signalling in human cells. J Mol Biol. 2007;373:38–47. doi: 10.1016/j.jmb.2007.07.068. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Kowalczykowski SC. Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J Biol Chem. 2002;277:31663–31672. doi: 10.1074/jbc.M203494200. [DOI] [PubMed] [Google Scholar]
- Sun H, Leverson JD, Hunter T. Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 2007;26:4102–4112. doi: 10.1038/sj.emboj.7601839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, Palvimo JJ, Hay RT. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol. 2008;10:538–546. doi: 10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
- Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- Wu J, Liu LF. Processing of topoisomerase I cleavable complexes into DNA damage by transcription. Nucleic Acids Res. 1997;25:4181–4186. doi: 10.1093/nar/25.21.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Yang Z, Liu Y, Zou Y. Preferential localization of hyperphosphorylated replication protein A to double-strand break repair and checkpoint complexes upon DNA damage. Biochem J. 2005;391:473–480. doi: 10.1042/BJ20050379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Sharma P, Athanasiou M, Kumar A, Yamada S, Kuehn MR. Mutation of SENP1/SuPr-2 reveals an essential role for desumoylation in mouse development. Mol Cell Biol. 2005;25:5171–5182. doi: 10.1128/MCB.25.12.5171-5182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh ET. SUMOylation and De-SUMOylation: Wrestling with Life's Processes. J Biol Chem. 2009;284:8223–8227. doi: 10.1074/jbc.R800050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu DS, Sonoda E, Takeda S, Huang CL, Pellegrini L, Blundell TL, Venkitaraman AR. Dynamic control of Rad51 recombinase by self-association and interaction with BRCA2. Mol Cell. 2003;12:1029–1041. doi: 10.1016/s1097-2765(03)00394-0. [DOI] [PubMed] [Google Scholar]
- Yun C, Wang Y, Mukhopadhyay D, Backlund P, Kolli N, Yergey A, Wilkinson KD, Dasso M. Nucleolar protein B23/nucleophosmin regulates the vertebrate SUMO pathway through SENP3 and SENP5 proteases. J Cell Biol. 2008;183:589–595. doi: 10.1083/jcb.200807185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Saitoh H, Matunis MJ. Enzymes of the SUMO modification pathway localize to filaments of the nuclear pore complex. Mol Cell Biol. 2002;22:6498–6508. doi: 10.1128/MCB.22.18.6498-6508.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- Zou Y, Liu Y, Wu X, Shell SM. Functions of human replication protein A (RPA): from DNA replication to DNA damage and stress responses. J Cell Physiol. 2006;208:267–273. doi: 10.1002/jcp.20622. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.