This work uses technical advances to quantify the mitochondrial DNA in single cells and provides evidence that the male gamete, rather than the female gamete, plays the most critical role in regulating maternal inheritance of mitochondria in angiosperms.
Abstract
The mechanisms that regulate mitochondrial inheritance are not yet clear, even though it is 100 years since the first description of non-Mendelian genetics. Here, we quantified the copy numbers of mitochondrial DNA (mtDNA) in the gametic cells of angiosperm species. We demonstrate that each egg cell from Arabidopsis thaliana, Antirrhinum majus, and Nicotiana tabacum possesses 59.0, 42.7, and 73.0 copies of mtDNA on average, respectively. These values are equivalent to those in Arabidopsis mesophyll cells, at 61.7 copies per cell. On the other hand, sperm or generative cells from Arabidopsis, A. majus, and N. tabacum possess minor amounts of mtDNA, at 0.083, 0.47, and 1 copy on average, respectively. We further reveal a 50-fold degradation of mtDNA during pollen development in A. majus. In contrast, markedly high levels of mtDNA are found in the male gametic cells of Cucumis melo and Pelargonium zonale (1296.3 and 256.7 copies, respectively). Our results provide direct evidence for mitochondrial genomic insufficiency in the eggs and somatic cells and indicate that a male gamete of an angiosperm may possess mtDNA at concentrations as high as 21-fold (C. melo) or as low as 0.1% (Arabidopsis) of the levels in somatic cells. These observations reveal the existence of a strong regulatory system for the male gametic mtDNA levels in angiosperms with regard to mitochondrial inheritance.
INTRODUCTION
Not long after the rediscovery of Mendel’s laws of heredity in 1900, it was reported that leaf color patterns in the angiosperm species Pelargonium zonale and Mirabilis jalapa are inherited in a manner that is contrary to established genetic laws (Baur, 1909; Correns, 1909). In sexual crosses of these plants, the filial generations always receive leaf colors from both male and female parents in Pelargonium (biparental inheritance) but from only the female parent in Mirabilis (maternal inheritance). These distinct patterns of inheritance were observed soon afterward in additional angiosperm species, such as Antirrhinum majus and a few species of Oenothera (for a historical review, see Hagemann, 2000). It was subsequently established that the transmission of plastids (normal green plastids and mutated white/yellow plastids) during sexual reproduction underlies these non-Mendelian rules (Renner, 1934). The identification of plastid DNA later in the 1960s provided a substantial basis for this nonnuclear heredity.
Several mechanisms are known to regulate plastid inheritance in angiosperms. These mechanisms appear as cellular events that block or facilitate the transmission of male or female plastids during sexual reproduction (for reviews, see Hagemann and Schröder, 1989; Kuroiwa, 1991; Mogensen, 1996). For example, an extremely polarized distribution of plastids in microspores during the first pollen mitosis results in generative cells free of plastids. This is the most prominent case supporting the maternal inheritance of plastids in angiosperms. Other evidence commonly used as indicative of maternal inheritance includes the degeneration of plastids within the generative and/or sperm cells and the exclusion of plastids from those cells. The plastids of mature sperm cells that have undergone such mechanisms are poorly inherited by the zygotes. It is clear, therefore, that these cellular events act to block the transmission of male plastids. For biparental plastid inheritance, however, male plastid transmission is facilitated by contrasting cellular events. The unequal distribution of microspore plastids does not occur in this case, and plastids distributed to the generative cells increase in number (Liu et al., 2004; Hu et al., 2008) and amplify their DNA (Nagata et al., 1999; Hu et al., 2008). Hence, mechanisms that operate in the male gametic cells regulate non-Mendelian inheritance of plastids before fertilization.
In addition to these mechanisms, plastid inheritance is also affected by cellular events within the female gametic cell. In Medicago sativa, the frequency of male plastid inheritance relies on the genotypes of the female parents (Smith, 1989). This is determined by spatial distributions of egg cell plastids (Zhu et al., 1993; Rusche et al., 1995). A strong female transmitter (a genotype exhibiting a higher frequency of maternal plastid transmission) distributes the majority of cellular plastids (more than 60%) to the apical portion of the egg cell, which then forms the embryo, and positions the minority of plastids (less than 40%) in the basal portion, which is later incorporated into the suspensor cell (and will not be inherited). In contrast, plastid polarization occurs in the opposite direction within a weak female transmitter (more than 65% in the basal portion and less than 45% in the apical portion). The inheritance of plastids (male-to-female ratio) is thus affected by the partitioning of a greater or smaller portion of female (egg) plastids to the early embryonic cells.
Mitochondria are parallel organelles that contain their own DNA genome. This genome is inherited in a strictly maternal manner in animals and, with the exception of a couple of species showing biparental or paternal inheritance, in angiosperms (Ankel-Simons and Cummins, 1996; Mogensen, 1996). Notably, the cellular events that block the maternal inheritance of male plastids do not necessarily block male mitochondria. For example, owing to the nonpolarized distribution of mitochondria in the microspore, the generative cell receives mitochondria after the first pollen mitosis. These mitochondria then remain structurally intact in the mature generative and sperm cells (for examples, see Clauhs and Grun, 1977; Yu and Russell, 1994; Sodmergen et al., 2002). This largely differs from plastids that are either excluded from or degenerated within the male gametic cells in the case of maternal inheritance. On the other hand, a small number of other cellular events, such as the discarding of male gametic cytoplasm before and during fertilization in Hordeum vulgare (Mogensen and Rusche, 1985; Mogensen, 1988; Russell et al., 1990), reduce the levels of both plastids and mitochondria in the male gametic cells.
It has been established that mitochondria contained in the male gametic cells participate in fertilization (Russell, 1980, 1983; Yu et al., 1994). The maternal mode of mitochondrial inheritance, therefore, is not straightforward. A notable cellular event that facilitates this process, however, is the degradation of mitochondrial DNA (mtDNA) within the male gametic cells. This degradation has been observed, using both fluorescence and immunoelectron microscopy, in angiosperms such as Triticum aestivum (Miyamura et al., 1987), Hordeum vulgare (Sodmergen et al., 2002), Wisteria sinensis and Robinia pseudoacacia (Hu et al., 2005), and Medicago truncatula (Matsushima et al., 2008). Interestingly, this degradation process is not observed in angiosperms showing paternal or biparental mitochondrial inheritance, in which the amount of mtDNA increases in the male gametic cells in contrast to the typical situation (Nagata et al., 1999). Therefore, it appears that the quantities of mtDNA in the cells are regulated in parallel with the modes of mitochondrial inheritance.
Maternal inheritance has been studied extensively in animals, and the elimination of male mtDNA occurs during spermatogenesis, such that a spermatozoon in the mouse maintains only ~50 copies of mtDNA (Hecht et al., 1984). This equates to a 20-fold reduction compared with somatic cells, which are known to possess 103 to 104 copies of mtDNA per cell (Cummins, 1998). On the other hand, a significant amplification of female mtDNA during oogenesis results in 5.0 × 105 copies of mtDNA in mouse oocytes (Shitara et al., 2000). When these amounts of mtDNA mix during fertilization, the ratio of male to female mtDNA in the zygote is ~1:104 (50:5.0 × 105). At this ratio of inputs, the inheritance of the male mtDNA (via the small amounts of paternal leakage that inevitably happens during maternal inheritance) occurs at a frequency of 1.0 × 10−4 (Gyllensten et al., 1991). If additional sperm mitochondria are introduced to fertilized eggs by microinjection, this results in a remarkable degree of paternal transmission (Shitara et al., 2000). Clearly, tight regulation of gametic mtDNA quantities is critical for proper maternal inheritance in animals. In addition, the ubiquitin-mediated degeneration of sperm mitochondria in the zygote is suggested to act as a trigger for removing any male contributions after fertilization (Sutovsky et al., 1999, 2000; Sutovsky, 2003; Thompson et al., 2003).
We undertook this study to examine quantitatively the cellular events that may participate in the regulation of mitochondrial inheritance in angiosperms. Our experiments were designed to clarify (1) how and at what levels the gametic mtDNA quantities are regulated in relation to mitochondrial inheritance; and (2) if mechanisms such as the polarized distribution of mitochondria occur in egg cells and affect mitochondrial inheritance in a manner similar to that of plastids. Our analyses were greatly facilitated by recent technical advances, such as the successful isolation of sperm (generative) and egg cells, the successful application of PCR to a single cell or very small quantity of cells, and the development of effective three-dimensional analysis of an egg cell. Our efforts have led to the discovery of regulatory mechanisms for mitochondrial inheritance in angiosperms that are largely different from those found in animals. We conclude that male gametic mtDNA is highly regulated (either via elimination or duplication) in angiosperms and that this regulation plays a critical role in the determination of mitochondrial inheritance.
RESULTS
Technical Advances
The gametic cells of the angiosperms are closely protected within the gametophytes. The large-scale purification of these cells for the purposes of DNA quantification is thus difficult. To resolve this issue, we attempted DNA quantification using a nested PCR protocol, a method that allows the use of a single cell or very small quantity of living cells. To ensure the accuracy of our PCR quantification method, we used an internal competitor template. We designed the reactions to amplify partial fragments of the maturase (matR) and cytochrome c oxidase I (coxI) genes encoded by the mitochondrial genome (see Supplemental Figure 1 online). First, the amplification of the target and competitor sequences in a competitive manner was confirmed (see Supplemental Figure 2 online) and the PCR conditions showed sufficient sensitivity to detect very small numbers of mtDNA copies (up to two copies; see Supplemental Figure 3 online). The amplification efficiencies for the target and competitor templates were calibrated to eliminate minor errors that may occur due to slight differences between the templates (see Supplemental Figure 4 online).
Our experimental scheme included the purification of the gamete cells and DNA quantification by PCR. Figure 1 outlines the procedure used for general cell isolation and purification and presents an illustrative example. To verify the validity of the quantification from cells, we tested the detection of mtDNA from mesophyll cells of Arabidopsis thaliana. It has been indicated previously that these cells contain ~50 copies of mtDNA per cell (Draper and Hays, 2000). Our quantification analysis yielded an equivalent value at 61.7 ± 5.5 copies per cell (Figure 2A) and thus appears to be an accurate method.
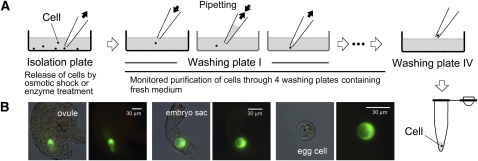
Figure 1.
Preparation of Cells for Single-Cell mtDNA Quantification.
(A) Living mesophyll, sperm (generative), and egg cells were isolated, purified in mannitol-containing medium, and prepared for PCR. All procedures were monitored with an inverted microscope. A freshly released cell was immediately selected from the isolation plate and washed by repeated pipetting on four serial washing plates containing fresh medium to remove visible contamination. Intact cells became spherical in shape during the washing procedure.
(B) An example of egg cell isolation from the Arabidopsis transgenic line (DD45:GFP) expressing green fluorescent protein in the cell. Light images of the ovule and embryo sac were merged with fluorescence images to show localizations of the egg cell.
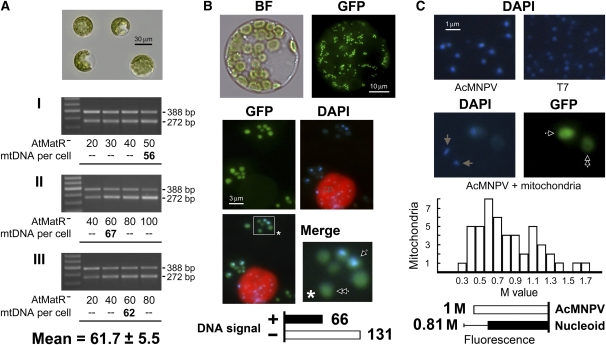
Figure 2.
Genomic Insufficiency of Mitochondria in Mesophyll Cells of Arabidopsis.
(A) Mesophyll protoplasts were isolated from young leaves of Arabidopsis (ecotype Columbia) and purified (top panel). Three competitive PCR quantifications (I, II, and III) were used to determine the mtDNA levels per cell (bottom three panels). For each quantification, four protoplasts were pretreated by freezing, heating, and proteinase K digestion, and the resulting mixture was divided equally into four PCR tubes. The mtDNA copy number per cell was calculated from the PCR samples, yielding product ratios closest to the previously determined efficiency coefficient (0.80 for Arabidopsis; see Supplemental Figure 4 online). The three quantifications were averaged to obtain the mean value.
(B) Mesophyll protoplasts isolated from young leaves of Arabidopsis (ecotype Columbia with transgenic mitochondrial GFP; 35S-mtGFP) were stained with DAPI and visualized under a bright field (BF; top left) and also at the excitation wavelengths for green fluorescent protein (GFP; top right) and DAPI (middle right). Red autofluorescence was observed from a chloroplast (cp) along with chloroplast DNA signals. It is clear that mtDNA signals were not present in a portion of the mitochondria after image merge (bottom left). The blocked area was enlarged (bottom right), and the arrow and double arrow highlight mitochondria with and without mtDNA signals, respectively. A survey of 197 mitochondria revealed that ~66% lacked mtDNA signals.
(C) Purified virus particles of AcMNPV and T7 were visualized after staining with DAPI (top left and right). Mesophyll mitochondria were costained along with AcMNPV (bottom left and right). Arrow and double arrows indicate mitochondria with and without mtDNA signals, respectively. Brown arrows indicate AcMNPV particles neighboring the mitochondria. Of the 49 mitochondria observed to have positive mtDNA signals, the mtDNA levels ranged from 0.3 to 1.7 M with an average value of 0.81 M. Error bar represents sd.
The leaf cells of Arabidopsis are known to possess ~670 mitochondria per cell (Sheahan et al., 2005). Hence, the values that we and others have obtained (50–60 copies) for mesophyll cells of this species imply a significant insufficiency of the mitochondrial genome. To verify such a possibility, we stained the mesophyll protoplasts in Arabidopsis with 4′,6-diamidino-2-phenylindole (DAPI). We inspected 197 mitochondria in total from different cells and found that 66 of these mitochondria had positive mtDNA (nucleoid) signals, while the remaining 131 mitochondria lacked this staining (Figure 2B). This indicates that mtDNA is undetectable in two-thirds (131 of 197) of the mitochondria in these cells. As controls for the positive staining, we used Autographa californica multiple nucleopolyhedrovirus (AcMNPV) and T7 bacterial phage containing 134 and 39 kb of double-stranded DNA per particle, respectively (Figure 2C). In addition, costaining of mitochondria and AcMNPV allowed a comparative detection of fluorescence between mitochondrial nucleoids and viral DNA. Through measurements from 49 mitochondria (nucleoid-containing) and equal numbers of virus particles costained with and localized in the fields neighboring the mitochondria (for an example, see images in Figure 2C), we estimated that the mitochondrial nucleoids had a relative fluorescence of 0.81 ± 0.33 M (fluorescence of an AcMNPV particle is determined as 1 M) on average (Figure 2C). This signifies that a nucleoid-containing mitochondrion may possess mtDNA of roughly 109 kb (0.81 × 134 kb) in size on average. Given that the complete genome of Arabidopsis mtDNA is 366.9 kb in size (Unseld et al., 1997), a mesophyll cell is calculated to have 66.7 copies of mtDNA (670 × 66 × 109/[197 × 366.9]). This is a value very close to that determined by our PCR quantification method. We thus conclude that the mitochondrial genome of the Arabidopsis mesophyll cell is indeed insufficient and that the method used in this study produces reliable results for the quantification of cellular DNA amounts.
mtDNA Levels in Egg Cells
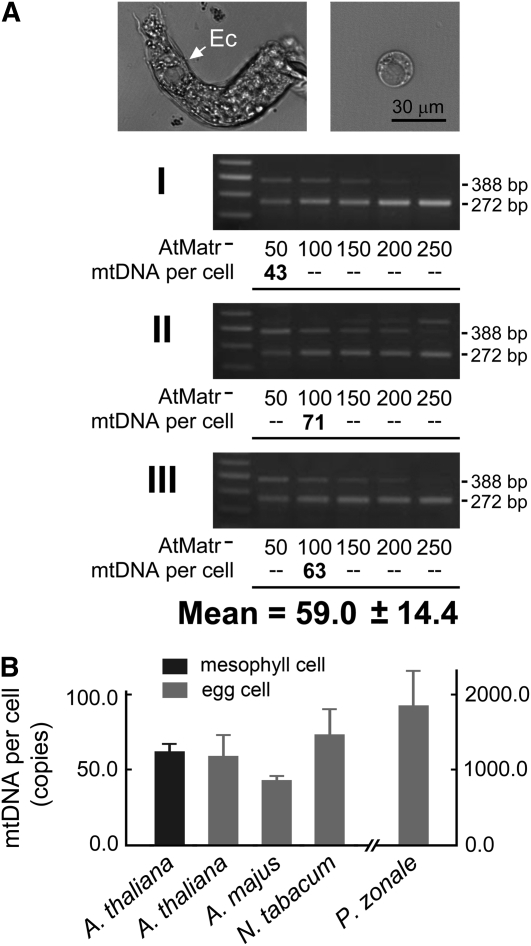
The isolation of intact egg cells involves, first, the isolation of the embryo sacs in an enzyme medium and then the discrimination and manual release of the cell under microscopic monitoring. Using this technique, we isolated egg cells from Arabidopsis, A. majus, Nicotiana tabacum, and P. zonale (see images of isolation in Figure 3A and Supplemental Figure 5 online). Three independent quantifications, using 15 egg cells in total, yielded a mean of 59.0 ± 14.4 copies of mtDNA per egg cell of Arabidopsis (Figure 3A). Similar amounts of mtDNA were detected in the egg cells of A. majus and N. tabacum (42.7 ± 3.5 and 73.0 ± 17.5 copies per cell, respectively; Figure 3B; see Supplemental Figure 5 online). Notably, these values for mtDNA quantity were equivalent to those found in somatic (mesophyll) cells. It thus appears that maternal mitochondrial inheritance in these species does not depend on mtDNA amplification in egg cells. This is a distinctly different result from those obtained in previous studies on animals (e.g., a bovine oocyte contains 100 times as much mtDNA as a bovine somatic cell, as described above).
Figure 3.
Levels of mtDNA per Egg Cell in Arabidopsis, A. majus, N. tabacum, and P. zonale.
(A) An Arabidopsis (ecotype Columbia) egg cell (Ec) was isolated from a digested embryo sac (top left) and purified (top right). Three quantifications (I, II, and III) using competitive PCR were implemented to determine the amount of mtDNA per cell (bottom three panels). For each quantification, five egg cells were pretreated by freezing, heating, and proteinase K digestion, and the resulting mixture was divided equally into five PCR tubes. The mtDNA copy number per cell was calculated for the PCR sample with a target:competitor product ratio closest to the previously determined efficiency coefficient (0.80 for Arabidopsis; see Supplemental Figure 4 online). The results from the three quantifications were averaged to obtain the mean value.
(B) Comparison of the mean mtDNA content per egg cell of Arabidopsis, A. majus, N. tabacum, and P. zonale. Quantifications of egg mtDNA in A. majus, N. tabacum, and P. zonale were performed as described in (A) for Arabidopsis (see Supplemental Figure 5 online). Error bars represent sd.
In contrast, egg cells of P. zonale were found to contain a considerably higher quantity of mtDNA (1852.7 ± 483.9 copies per cell; Figure 3B; see Supplemental Figure 5 online). This value is 30-fold greater than that of the Arabidopsis mesophyll cell (1852.7 versus 61.7), implying a possible amplification of egg mtDNA in P. zonale. It is notable also that P. zonale is a rare species among angiosperms in that it employs biparental mitochondrial inheritance. However, we could not be sure that this high quantity might relate to this biparental inheritance. More examinations of such species will be required to determine this possibility. For Cucumis melo, the only angiosperm species known to employ paternal mitochondrial inheritance (Havey et al., 1998), insurmountable technical difficulties unfortunately prevented us from isolating intact egg cells.
mtDNA Levels in Generative or Sperm Cells
The generative or sperm cells of A. majus, Arabidopsis, C. melo, N. tabacum, and P. zonale were released from mature germinating pollen tubes or pollen grains via osmotic shock. Figure 4A shows a burst Arabidopsis pollen tube, a purified Arabidopsis sperm cell, and the PCR quantification results we obtained for Arabidopsis sperm cells. In Arabidopsis (ecotype Columbia), preliminary experiments detected less than one copy of mtDNA in a sperm cell. This value is below the limit of accurate quantification. We thus performed the PCR analysis using batches of 10 cells, which yielded a finding of 7.3 ± 1.5 copies of mtDNA per 10 Arabidopsis sperm cells (Figure 4A), indicating that a sperm cell of Arabidopsis possesses as few as 0.73 mtDNA copies. However, it is known that an ~620-kb mtDNA fragment containing matR is inserted into chromosome 2 of Arabidopsis (ecotype Columbia; Stupar et al., 2001). Given that this matR sequence may be amplified along with the mitochondrial matR sequence, the actual mtDNA copy number in Arabidopsis sperm cells might be even less than 0.73.
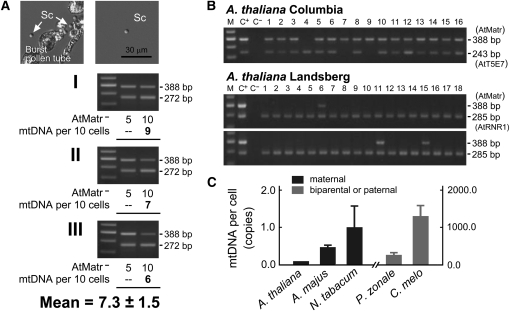
Figure 4.
Amounts of mtDNA per Sperm or Generative Cell of Arabidopsis, A. majus, N. tabacum, C. melo, and P. zonale.
(A) An Arabidopsis (ecotype Columbia) sperm cell (Sc) was isolated from a burst pollen tube (top left) and purified (top right). Three quantifications (I, II, and III) using competitive PCR were performed to determine the amount of mtDNA per batch of 10 cells (bottom three panels). For each quantification, 20 sperm cells were pretreated by freezing, heating, and proteinase K digestion, and the resulting mixture was divided equally into two PCR tubes. The mtDNA copy number per 10-cell batch was calculated for the PCR sample with a target:competitor product ratio closest to the previously determined efficiency coefficient (0.80 for Arabidopsis; see Supplemental Figure 4 online). The measurements from the three quantifications were then averaged to obtain the mean value.
(B) Detection of mtDNA from single sperm cells of Arabidopsis. Using PCR conditions similar to those described in (A), mtDNA was detected in 15 of the 16 sperm cells examined (ecotype Columbia; top panel). An internal control using a single-copy sequence found on chromosome 2 (T5E7; for structural description, see Supplemental Figure 12 online) was also detected, indicating that the quantification procedure might have amplified the known mtDNA insertion in the Columbia nuclear DNA genome. On the other hand, in cells of the ecotype Landsberg (bottom panels), mtDNA was detected in 3 of 36 sperm cells. In this case, a single-copy nuclear sequence (RNR; for structural description, see Supplemental Figure 12 online) was utilized as the internal control. A mesophyll protoplast (C+) and 0.5 μL of cell-free washing medium (C−) were used as positive and negative template controls, respectively.
(C) Mean mtDNA content per sperm or generative cell in Arabidopsis, A. majus, N. tabacum, C. melo, and P. zonale. Quantifications of sperm mtDNA in these angiosperm species were performed as described in (A) for Arabidopsis (see Supplemental Figure 6 online). Error bars represent sd.
To investigate whether our PCR procedure amplified the nuclear sequence on chromosome 2, we used PCR of single sperm cells to detect a single-copy nuclear fragment (At T5E7f) near the mtDNA insertion. This analysis yielded 14 positive detections out of 16 cells examined (Figure 4B), suggesting that nuclear matR could be amplified from most sperm cells under our PCR conditions; thus, the value of mtDNA (0.73 copies per cell) might include largely nuclear contributions. With this in mind, single-cell PCR amplifications were performed for sperm cells purified from the Landsberg ecotype of Arabidopsis. A partial fragment of ribonucleotide reductase R1 (RNR1), a single-copy nuclear gene reported previously (Chabouté et al., 1998), was amplified as a control. Strikingly, the amplification of the matR sequence from the Landsberg sperm cells was much lower (3 positive detections from 36 cells) than that from Columbia cells (15 positive detections from 16 cells; Figure 4B). From the results with Landsberg, we conclude that the average mtDNA copy number in Arabidopsis sperm cells is in fact 0.083 rather than 0.73.
Our preliminary experiments also detected low mtDNA contents in the generative cell of A. majus and the sperm cells of N. tabacum. We accordingly purified these cells and performed PCR using 15-cell batches. Three independent quantifications of A. majus generative cells (performed in duplicate for a total of 90 cells) and of N. tabacum sperm cells (performed in triplicate for a total of 135 cells) yielded mtDNA copy numbers of 7.0 ± 1.0 and 15.3 ± 8.5 copies per 15-cell batch, respectively, equivalent to 0.47 ± 0.07 and 1.0 ± 0.6 copies per cell, respectively (Figure 4C; see Supplemental Figure 6 online). These values were very consistent with the findings from our single-cell examinations using the single-copy nuclear genes Am granule-bound starch synthase 1 (GBSS1; Mérida et al., 1999) and Nt RNR2 (Chabouté et al., 1998) as internal controls (24 positive amplifications from 63 A. majus generative cells and 16 positive amplifications from 16 N. tabacum sperm cells; see Supplemental Figure 6 online).
It is clear that the sperm cells of Arabidopsis, A. majus, and N. tabacum, species known to undergo maternal mitochondrial inheritance, contribute not more than one copy of mtDNA to the zygotes. This is consistent with the maternal mode of inheritance in these plants. To understand better how male mtDNA is transmitted in cases of biparental and paternal inheritance, we performed identical quantifications in C. melo and P. zonale, respectively. A generative cell of C. melo was found to possess 1296.3 ± 310.6 copies and a sperm cell of P. zonale 256.7 ± 71.2 copies of mtDNA (Figure 4C; see Supplemental Figure 6 online). These values indicate that the sperm cells of C. melo and P. zonale may contribute male mtDNA at levels that are 4.2-fold (for P. zonale, 256.7 versus 61.7) and 21.0-fold (for C. melo, 1296.3 versus 61.7) greater than that of an Arabidopsis mesophyll cell. Given that the maternal species possess mtDNA at ~1:1000 (the Arabidopsis case, 0.083:61.7) to 16:1000 (the tobacco case, 1:61.7) of the mesophyll cell levels, it is apparent that mtDNA quantities of the male gametic cells are highly regulated in angiosperms with regard to mitochondrial inheritance.
Regulation of the mtDNA Levels in Male Gametic Cells
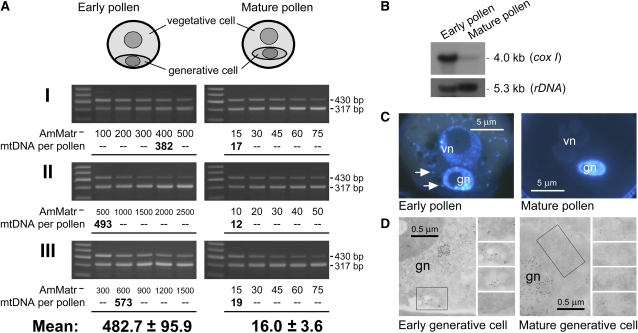
Previous studies have suggested that pollen and male gametic mtDNA may be degraded in species that undergo maternal mitochondrial inheritance (Miyamura et al., 1987; Nagata et al. 1999; Sodmergen et al., 2002; Hu et al., 2005). We performed quantitative analyses of this event. First, the total mtDNA levels within single pollen grains were quantified via our PCR technique. The results indicated 482.7 ± 95.9 copies per early binucleate pollen and 16.0 ± 3.6 copies per mature pollen of A. majus (Figure 5A). This indicates a 30-fold degradation of pollen mtDNA during pollen development. A parallel experiment, using a coxI probe for hybrid pollen total DNA on a DNA gel blot, yielded a similar result (Figure 5B). Following these analyses, pollen sections were observed under fluorescence and immunoelectron microscopy, methods routinely used to track possible pollen mtDNA degradation in the previous studies cited above. The reduction of mtDNA labeling in the pollen cells was once more detected (Figures 5C and 5D) and thus verified that microscopic examination of mtDNA quantity correlates with the results of molecular analysis. It is clear, therefore, that the level of pollen mtDNA is downregulated during pollen development (for similar results in Arabidopsis, see Supplemental Figure 8 online).
Figure 5.
Downregulation of A. majus Pollen and Generative Cell mtDNA during Pollen Development.
(A) Gently crushed pollen grains of A. majus were subjected to competitive PCR to quantify the mtDNA levels. Three quantification experiments each were performed for early and mature pollen in which five crushed pollen grains were pretreated by freezing, heating, and proteinase K digestion. The resulting mixture was then divided equally into five PCR tubes. The mtDNA copy number per early or mature pollen grain was calculated using the PCR samples with a target:competitor product ratio closest to the previously determined efficiency coefficient (0.66 for A. majus; see Supplemental Figure 4 online). The measurements from the three quantifications were then averaged to obtain the mean values.
(B) Analysis of the degradation of A. majus pollen mtDNA by DNA gel blot hybridization. Total DNA extracted from early and mature pollen grains was digested with EcoRI and probed with a coxI fragment. As a loading control, the samples were also probed with a nuclear rDNA fragment.
(C) Disappearance of mtDNA fluorescence (arrows) in DAPI-stained pollen sections. The generative cells contain intact mitochondria but no plastids (see Supplemental Figure 7 online). gn, generative nuclei; vn, vegetative nuclei.
(D) Reduction in the immunogold DNA labeling intensity in the mitochondria (boxed) of generative cells. Serial electron micrograph sections are shown to the right of each panel.
To evaluate further the degree of regulation in relation to mitochondrial inheritance, it was necessary to quantify the mtDNA in the earliest generative cells. However, isolation of these cells is technically not possible at present because they firmly associate with the pollen intine just after the first pollen mitosis. We thus estimated the mtDNA quantity of the cell from the cytoplasmic volumes. This approach is feasible because of the nonpolarized distribution of microspore mitochondria during the first pollen mitosis (for examples, see Schröder, 1985; Sodmergen et al., 2002; also refer to the dihexyloxacarbocyanine iodide [DiOC6] images in Figure 6 and Supplemental Figure 10 online). Using sections through the center of A. majus pollen grains, we surveyed the volume of vegetative cytoplasm and found a value of 1562.0 ± 94.4 μm3 and that of the generative cytoplasm to obtain a value of 79.9 ± 6.4 μm3 (see Supplemental Figure 9 online), indicating an mtDNA compartmentalization ratio of 19.6 ± 1.7:1 between the cells. Given the total quantity of mtDNA (482.7 ± 95.9 copies) per pollen grain, an early generative cell is predicated to possess ~23.4 copies (482.7/20.6). Compared with that quantified from the mature generative cell (0.47 copies per cell), a 50-fold reduction in mtDNA was indicated to occur in the generative cell. This result indicated a more rapid mtDNA degradation in the generative cell than in the pollen grain overall, which undergoes a 30-fold reduction in mtDNA, as described above. In Arabidopsis, greater mtDNA labeling was observed in the vegetative mitochondria than in the generative mitochondria, consistent with this idea (see Supplemental Figure 8 online).
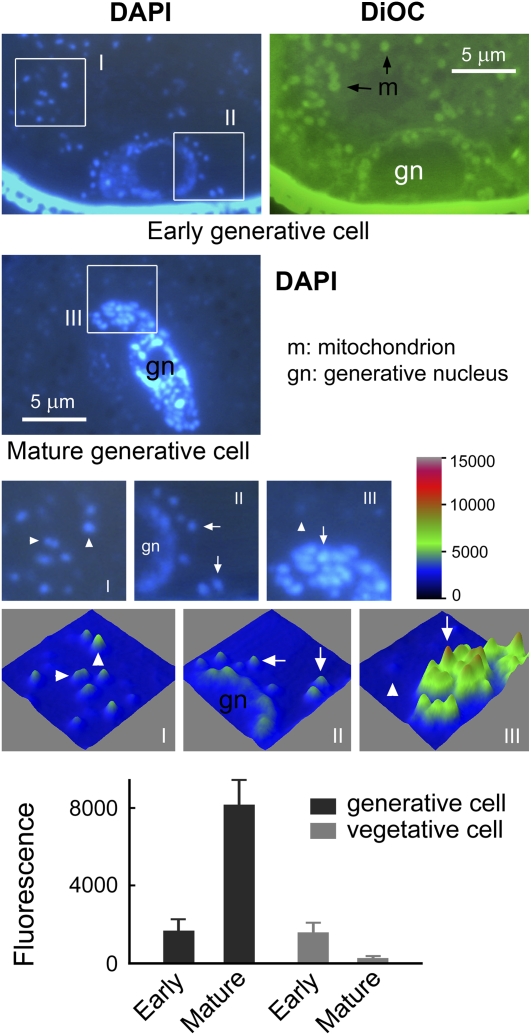
Figure 6.
Amplification of Generative mtDNA of C. melo.
C. melo pollen sections were stained with DAPI and DiOC6 (a fluorescent dye that stains mitochondria). Mitochondrial nucleoids in an early vegetative cell (I), an early generative cell (II), and mature generative and vegetative cells (III) are boxed in the top panels; these boxes are enlarged in the middle row. Digital analysis of the fluorescence images (nucleoids; n = 15) revealed a 4.9-fold increase in mtDNA fluorescence in the mature generative cell (8176.0, in arbitrary units) compared with the early generative cell (1246.2, in arbitrary units) and simultaneously a sixfold diminution of mtDNA fluorescence in the vegetative cell (1617.8 in the early cell and 269.9 in the mature cell, in arbitrary units) during pollen development. Arrows indicate mitochondrial nucleoids in the generative cell, and arrowheads indicate mitochondrial nucleoids in the vegetative cell. Error bars represent sd.
To understand better how the high quantities of male gametic mtDNA in C. melo and P. zonale were achieved, we examined sections of the corresponding cells during pollen development. It has been shown previously that mtDNA fluorescence increases in the cells of P. zonale, implying duplication of male gametic mtDNA (Nagata et al., 1999). We again observed this trend in C. melo (Figure 6). In pollen grains just after their first mitosis, fluorescence of mitochondrial nucleoids (mtDNA) was equal in intensity between the early generative and vegetative cells. This indicates that during pollen mitosis, mitochondria with equal amounts of DNA are received by the early pollen cells. Notably, however, the fluorescence of mitochondrial nucleoids in the mature generative cell was remarkably stronger. Image analysis revealed that the nucleoid fluorescence in the mature generative cell was 4.9-fold higher on average than that in the early generative cell (8176.0 ± 1246.2 per nucleoid in the mature cell and 1662.6 ± 567.8 per nucleoid in the early cell, in arbitrary units). Given that a mature C. melo generative cell contains mtDNA at a 21-fold higher amount than a mesophyll cell (1296.3 versus 61.7 copies as detected above) and that nucleoid fluorescence reflects the level of mtDNA as shown in Figure 5, it is very possible that the highly increased nucleoid fluorescence intensity may reflect a preferential amplification of mtDNA in the generative cells during pollen development. In contrast, the nucleoid fluorescence in the mature vegetative cell decreased considerably (1617.8 ± 499.1 per nucleoid in the early cell and 269.9 ± 75.5 per nucleoid in the mature cell, in arbitrary units). This decrease was phenotypically similar to plants that undergo maternal mitochondrial inheritance (see Arabidopsis and A. majus examples in this study). In C. melo, however, we did not perform PCR analysis of the mtDNA levels in vegetative cells during pollen development. The decrease in fluorescence intensity in this plant may similarly reflect a degradation of vegetative mtDNA. In P. zonale, we observed similar results (see Supplemental Figure 10 online).
Egg and Sperm Mitochondria
The spatial distribution pattern of the mitochondria within an egg cell is an early indicator of how mitochondrial inheritance is regulated in female gametes, as has been demonstrated for plastids (Zhu et al., 1993; Rusche et al., 1995). We were interested to know if the genomic insufficiency of mitochondria observed in the mesophyll cells (Figure 2) could possibly occur in the egg cell also, as our initial PCR result showed that the egg cell contained a quite similar amount of mtDNA to the mesophyll cell (61.7 ± 5.5 copies per mesophyll cell and 59.0 copies per egg cell; Figures 2 and 3). To investigate this further, we employed electron microscopy via serial ultrathin sectioning and computerized three-dimensional analysis of an unfertilized egg cell (Figure 7; see Supplemental Movie 1 online).
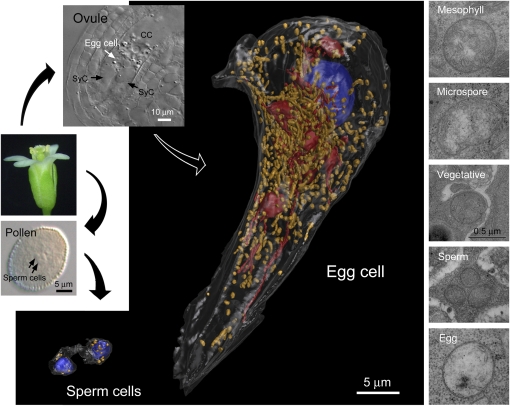
Figure 7.
Three-Dimensional Configurations of the Sperm and Egg Cells of Arabidopsis.
The cells were reconstructed from serial ultrathin sections to exhibit the number and spatial distribution of mitochondria within the cells. For the quantification results in detail, see Supplemental Table 1 online. The cell nuclei, mitochondria, and plastids are rendered in blue, yellow, and red, respectively. Note that the sperm cells contain markedly smaller mitochondria. Electron microscopic images (at right) further show that the mitochondria of the early microspore are similar in size to those of the egg and mesophyll cells, signifying that the mitochondrial volumes decrease significantly in sperm and vegetative cells during pollen development (for a statistical analysis, see Supplemental Figure 11 online). CC, central cell; SyC, synergid cell.
The cell examined was 3851.0 μm3 in volume and contained 795 mitochondria and 16 plastids (for more details of the cellular parameters, see Supplemental Table 1 online). This first indicates a very similar genomic insufficiency for egg mitochondria as compared with the mesophyll cell. It could be calculated that an Arabidopsis egg mitochondrion may contain 0.078 (59.0/759) copies of mtDNA on average, which indicates that ~13 (759/59.0) mitochondria share a complete mtDNA genome. To analyze mitochondrial and plastid distributions within the cell in relation to possible polarization of these organelles during zygote division, a partition plane is required to separate the egg cell into apical and basal portions. However, zygotic division has not been quantitatively analyzed in Arabidopsis so far. We thus predicted such a plane by referring to (1) zygote division in M. sativa, in which an apical and a basal cell form with a cytoplasm volume ratio of 1:2 (Rusche et al., 1995); and (2) the localization of the cell nucleus in an Arabidopsis zygote just before division, from which we assumed a cytoplasm volume ratio of 1:1.68 (Figure 8A). Using the partition plane of M. sativa, the apical and basal portions of the Arabidopsis egg cell were predicted to have 81 (10%) and 714 (90%) mitochondria and 2 (13%) and 14 (87%) plastids, respectively. It thus appeared that the basal portion had higher proportions of mitochondria and plastids than the apical portion. A similar result was obtained with a predicted partition plane from Arabidopsis (Figure 8B). The apical and basal portions of the cell possessed 133 (17%) and 662 (83%) mitochondria and 4 (25%) and 12 (75%) plastids, respectively. Hence, in both of these partition planes, the basal portion is rich in mitochondria and plastids (Figure 8C). We thus conclude that the distribution of mitochondria in the egg cell of Arabidopsis is not likely to exert a bias toward the maternal inheritance of this species.
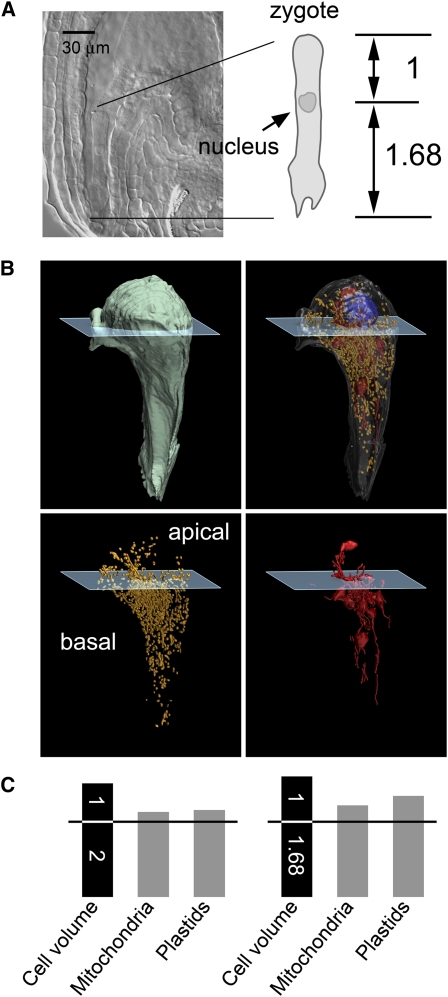
Figure 8.
Distribution of Mitochondria and Plastids in the Egg Cell of Arabidopsis.
(A) Soon after fertilization, the egg cell of Arabidopsis elongates rapidly to form a tubular zygote, and the first zygote division takes place within the tubular cell (Mansfield and Briarty, 1990). To predict the division plane, we traced a zygotic nucleus during deformation of fertilized egg cells and observed via a clear sample that the nucleus assigns the tubular cell into apical and basal portions at a 1:1.68 length ratio from the apical end. As the cell was almost tubular, we used this length ratio as a ratio of the volume between the apical and basal portions.
(B) A computer-generated partition plane separating the egg cell into apical and basal portions with a 1:1.68 ratio of cytoplasm volumes. Mitochondria and plastids were counted from both these portions (see Supplemental Table 1 online). The cell nuclei, mitochondria, and plastids are rendered in blue, yellow, and red, respectively.
(C) Distribution of mitochondria and plastids between the apical and basal portions of the egg cell. Relative to the volume of the cell portions (shown by the numbers in the black bars), both the mitochondria and plastids showed a clear tendency toward partitioning into the basal portion, regardless of the partition plane (represented by the horizontal line) used in our analysis (for details, see Supplemental Table 1 online).
Interestingly, the sperm cells of Arabidopsis possess an equivalent mtDNA quantity (0.083 copies per cell; Figure 4) to that of an egg mitochondrion (0.078 copies per mitochondrion, as described above). We thus speculated that the sperm cells may possess as few as one mitochondrion per cell, if the degradation of sperm mtDNA is not taken into account. Using three-dimensional analysis, two paired sperm cells containing 9 and 15 intact mitochondria, respectively, were observed (Figure 7; see Supplemental Movie 2 online). This again revealed the degradation of sperm mtDNA and suggested that degradation is not likely to be accompanied by the destruction of mitochondrial structures.
Another cytological result that may be important for our increased understanding of mitochondrial inheritance is that the sperm and vegetative mitochondria of Arabidopsis were found to be much smaller than those in the egg cells, as revealed by electron microscopy (Figure 7). From our examination of ultrathin sections, average mitochondrial volumes of 12.8 × 10−2, 11.8 × 10−2, and 11.4 × 10−2 μm3 were estimated in the mesophyll cell, early microspore, and egg cell, respectively. Because the subsequent statistical analysis did not reveal significant deviations among these values (see Supplemental Figure 11 online), it was concluded that these volumes are “normal.” In contrast, in the mature vegetative and sperm cells, the average mitochondrial volumes were 3.2 × 10−2 and 1.1 × 10−2 μm3, respectively, or one-quarter and one-tenth the size of the normal mitochondria, respectively. It is apparent, therefore, that the degradation of mtDNA and loss of mitochondrial volume are parallel events in pollen cells.
DISCUSSION
Genomic Insufficiency of Mitochondria
The concept of mtDNA copy number is used both here and in other studies to denote the amount of mtDNA that is equivalent to certain full sets of mitochondrial genes (Laser et al., 1997; Draper and Hays, 2000). Since all of the mitochondrial genes are necessary for proper mitochondrial function, it is a widely held view that each mitochondrion must possess at least one full copy of mtDNA. However, previous reports and the results of this study strongly suggest that this may not always be the case, at least in plants. The first evidence for this was found in cells of Cucurbitaceae, containing 110 to 140 copies of mtDNA and 360 to 1100 mitochondria per cell (Bendich and Gauriloff, 1984). In Arabidopsis, it has been reported that leaf cells contain ~670 mitochondria (Sheahan et al., 2005), whereas the copy number of mtDNA per cell is ~50 (Draper and Hays, 2000). These values obtained from separate studies imply that the amount of mtDNA is largely less than one copy per mitochondrion. We further verified this finding in our current experiments. The mean level of mtDNA we detected was 61.7 ± 5.5 copies per cell (Figure 2A), very close to the value reported previously. More interestingly, we observed an absence of fluorescent nucleoids within nearly 67% of mitochondria (Figure 2B). This may be the first visualization of genomic insufficiency in plant mitochondria. Our results also showed that the amount of mtDNA contained in the fluorescent nucleoids of the remaining 33% of the mitochondria was ~109 kb (Figure 2C). Compared with the full genome of Arabidopsis mtDNA at 366.9 kb in size (Unseld et al., 1997), it appears that even the mitochondria with mtDNA possess insufficient genomes.
In plants, a complete mitochondrial genome may be carried on several mtDNA molecules, owing to the substantial length and heteromorphy of plant mtDNA. In N. tabacum, for example, the mitochondrial genome has been measured at 430.6 kb of mtDNA (Sugiyama et al., 2005). It is indicated, however, by direct observations that most of the mtDNA molecules (~60%) of N. tabacum are 21 to 33 kb in length (Satoh et al., 1993). Clearly, the full mitochondrial genome is necessarily composed of heterogeneous and complementary molecules that are shorter than 430.6 kb in this species. This molecular feature of plant mtDNA also enables the subpackaging of a complete genome into several mitochondria. Nucleoids with mtDNA of ~109 kb, as have been detected in Arabidopsis mesophyll cells, can thus be reasonably predicted to contain a partial mitochondrial genome. In addition, as the mtDNA value we quantified directly from cells (61.7 ± 5.5 copies per cell; Figure 2A) matches the value that could be predicted from our cellular analysis (66.7 copies per cell: 670 mitochondria per cell on average × 33.5% containing nucleoids × 109 kb of mtDNA per nucleoid/366.9 kb of DNA per mitochondrial genome), we speculate that most mesophyll mitochondria (the 66.5% without fluorescent nucleoids) may possess no, or very small amounts of, mtDNA. It has been further demonstrated that plant mitochondria undergo massive and frequent fusions and fissions that are not accompanied by notable changes in the mitochondrial number (Arimura et al., 2004). This may underlie material exchange among mitochondria and explain how ostensibly insufficient mitochondrial genomic information is shared within plant cells.
It is notable that the genomic insufficiency of mitochondria is also observed in the egg cells of Arabidopsis. Our results indicate that 59.0 ± 14.4 copies of mtDNA (Figure 3A) and 795 mitochondria (Figure 7; see Supplemental Table 1 online) exist per egg cell. This indicates a similarity between angiosperm eggs and somatic cells in terms of maintaining similar numbers of mitochondria and similar amounts of mtDNA (~670 mitochondria and 61.7 ± 5.5 copies of mtDNA per mesophyll cell; Figure 2; Sheahan et al., 2005). Egg cells of A. majus and N. tabacum possess mtDNA at an equivalent level (Figure 3B; see Supplemental Figure 5 online). This indicates that the egg cells of angiosperms (except for P. zonale) may maintain mtDNA at amounts that are equivalent to those of somatic cells. This largely differs from animal cells; in a bovine oocyte, for example, the mtDNA is amplified to 2.6 × 105 copies, which is ~100-fold higher than the levels in a somatic cell (Michaels et al., 1982). Given that the relative inputs of male and female mtDNA into the zygotes is critical for proper maternal inheritance (Shitara et al. 2000), the remarkable amplification of egg mtDNA is undoubtedly a key cellular event that regulates this process in animal cells. Our current results indicate that such an event does not occur in the egg cells of A. majus, Arabidopsis, or N. tabacum, suggesting that maternal mitochondrial inheritance in these species is not regulated by this mechanism in the egg cell.
The lack of a regulatory function for the egg cell in maternal mitochondrial inheritance is also manifested by the distribution of mitochondria within this cell. In M. sativa, the apportioning of more plastids to the apical portion of the egg cell heightens the ratio of maternal plastid inheritance (Zhu et al., 1993; Rusche et al., 1995). We initially expected to observe an apical-biased distribution of mitochondria and plastids in the Arabidopsis egg cell, in line with observations in M. sativa and the maternal mode of organelle inheritance in Arabidopsis. Our present results, however, show that higher proportions of both plastids and mitochondria are distributed to the basal region (Figures 8B and 8C; see Supplemental Table 1 online). This appears to be inconsistent with the regulatory functions observed in the egg cell of M. sativa. We thus speculate that (1) for plastid inheritance, regulation by the egg cell may differ between maternal (Arabidopsis) and biparental (M. sativa) species; and (2) the mechanism that regulates plastid inheritance in the egg cell may simply not function for mitochondria. Because only one egg cell was examined in this study, the data could not be analyzed statistically. Notably, a similar nonpolarized distribution of mitochondria has been observed previously in egg cells of M. sativa (Zhu et al., 1993), although the mitochondrial distribution was not specifically analyzed in this earlier study.
Male Gametes as Key Players in Mitochondrial Inheritance
As described above, the relative input ratio of male to female mtDNA into the zygote is critical for maternal inheritance. In the mouse, this ratio has been measured at ~1:104 (i.e., ~50 copies per sperm cell; Hecht et al., 1984) and 5.0 × 105 copies per egg cell (Shitara et al., 2000). In this study, the results have revealed an extremely low mtDNA content in an Arabidopsis sperm cell, at merely 0.083 copies on average (Figure 4), more than 700-fold less than the level in a somatic cell (61.7 copies per mesophyll cell; Figure 2). This indicates a far more pronounced degradation of male gametic mtDNA in angiosperms compared with animal cells (e.g., a 20-fold decrease is observed in the mouse as described above). Similarly, the male gametic cells of A. majus and N. tabacum contained mtDNA at 0.47 and 1.0 copies per cell, respectively (Figure 4C; see Supplemental Figure 6 online), much less than the levels found in an animal sperm cell. Using the generative cells of A. majus, we here demonstrated a 50-fold degradation of mtDNA during pollen development (Figure 5). Moreover, because the possible effects of nuclear artifacts are not corrected for in the results with A. majus and N. tabacum, the true amounts of male gametic mtDNA in these plants may well be lower than the values indicated. Given that the egg cells of A. majus, Arabidopsis, and N. tabacum contain copies of mtDNA that are equivalent in number to a somatic cell, it is clear that in these angiosperm species, the ratio of mtDNA inputs required for maternal inheritance is achieved in the main from the male gametes. In addition, our results indicate that a sperm cell may contribute less than a full mtDNA genome to the zygote. This provides further evidence that the male gamete, rather than the female gamete, plays the most critical role in regulating maternal inheritance in angiosperms.
The angiosperm species C. melo and P. zonale provide a rare resource for the study of paternal and biparental mitochondrial inheritance, which are unknown in animals. Our results here show that a key role of the male gametes is also manifested in the regulation of paternal and biparental mitochondrial inheritance in these plants. The male gametic cells of C. melo and P. zonale possess mtDNA at 1296.3 ± 310.6 and 256.7 ± 71.2 copies per cell (Figure 4C; see Supplemental Figure 6 online), ~4 to 20 times more than Arabidopsis mesophyll cells. These examples provide evidence that it is not the small size of the sperm cells that blocks paternal transmission and, therefore, that the difference in volume between the male and female gametic cells is not a determinant of non-Mendelian genetics. At the same time, these results indicate that the male gametic cells act as the key regulators of biparental and paternal mitochondrial inheritance.
Possible Genetic Approaches to the Study of Mitochondrial Inheritance
The aim of this study was to determine the key point at which mitochondrial inheritance is regulated during plant gametic development. We expect to be able to predict important cellular events that may participate in this regulatory mechanism in the not too distant future, thereby gaining insight that will allow further genetic investigations. Our findings here have localized the regulation of non-Mendelian genetics to the male gamete. We thus conclude that the mechanisms that regulate the amount of male gametic mtDNA are also important regulatory mechanisms for mitochondrial inheritance in angiosperms. This suggests a clearer target for further genetic studies. For example, the rapid upregulation and downregulation of mtDNA that occurs after the first pollen mitosis (Figures 5 and 6; Nagata et al., 1999) implies that mitochondrial DNase and DNA polymerase function within the developing generative cell. A focus on the possible involvement of these enzymes in the regulation of mitochondrial inheritance is thus warranted in the near future.
For a deeper insight into these regulatory mechanisms, we note that after the massive degradation of mtDNA in the male gametic cells, a minor amount of mtDNA does remain in the mature cell (i.e., 0.083, 0.47, and 1.0 copy per cell in A. majus, Arabidopsis, and N. tabacum, respectively; Figure 4C; see Supplemental Figure 6 online). This may explain the minor degree of paternal leakage during the maternal inheritance process (Gyllensten et al., 1991) and indicates an incomplete degradation of mtDNA in the male gametic cells in vivo. Similarly incomplete degradation of mtDNA was also observed in the vegetative cell (Figure 5) in vitro, indicating the possibility that non-DNase factors participate in the regulatory mechanism. This idea is supported by evidence from both mammals and yeast. The mammalian mitochondrial transcription factor (Tfam) restricts mtDNA levels; a homozygous knockout of Tfam results in a lethal phenotype with the embryos lacking mtDNA, and the cellular mtDNA copy number depends on the Tfam level (Larsson et al., 1998). Abf2p, a mitochondrial protein from yeast, has also been reported to restrict mtDNA levels (Newman et al., 1996). Notably, both of these proteins appear to bind mtDNA and stabilize it against degradation by DNase (Newman et al., 1996; Alam et al., 2003; Kucej et al., 2008). It has thus been speculated that Tfam and Abf2p might package mtDNA into nucleoids that thereby protect the mtDNA from degradation (Kang et al., 2007; Sia et al., 2009).
In angiosperms, however, although the dynamics of the mtDNA levels have been described, little is known about the regulation of these levels. We speculate that the mtDNA copy number in plant cells may be similarly regulated by factors with Tfam- and/or Abf2p-like functions and thus recommend further research into mtDNA binding proteins in plant cells. It is notable in this regard that animal sperm mtDNA codegrades with a testis-specific Tfam (Rantanen and Larsson, 2000; Rantanen et al., 2001). Similarly, plant mitochondrial inheritance (i.e., the mtDNA levels) may be regulated by nucleoid proteins within the male gametic cells. The significant loss of the sperm and vegetative mitochondrial volumes during pollen development (Figure 7; see Supplemental Figure 11 online) that occurs in concert with mtDNA degradation probably coincides with a rapid loss of mitochondrial proteins. Previous studies showing the lack of mitochondrial division within pollen cells (Mogensen and Rusche, 1985; Yu and Russell, 1994) may support this degradation. Given also that pollen germination and tube growth require active oxidative phosphorylation, any proteins lost from the mitochondria must be only indirectly involved in energy conversion. Hence, a fuller understanding of the molecular regulation of mitochondrial inheritance may require further study of mitochondrial, and particularly nucleoid, proteins that are degraded in the male gametic mitochondria.
METHODS
Plant Materials
Plant materials used in this study were extracted from Antirrhinum majus (line Tahiti), Arabidopsis thaliana (ecotypes Columbia and Landsberg), Nicotiana tabacum (var Wisconsin 38), Cucumis melo (var Reticulatus), and Pelargonium zonale (line Xidu 20) angiosperm species cultivated in greenhouses at Peking University. Transgenic Arabidopsis lines with green fluorescent protein expressed in the egg cell (DD45:GFP) and somatic mitochondria (35S-mtGFP) were kindly provided by Gary Drews of the University of Utah and Wataru Sakamoto of Okayamo University, respectively. Purified samples of Autographa californica MNPV and T7 bacterial phage were kindly provided by Jian-Guo Chen of Peking University and the Virus Resources Department, Wuhan Institute of Virology (Chinese Academy of Sciences), respectively.
Isolation of Gametic Cells and Leaf Protoplasts
Gametic cells were isolated from fresh ovules and pollen tubes (or pollen grains) based on the method of Tian and Russell (1997). Briefly, to obtain mature egg cells, unfertilized ovules were gathered onto a culture plate (3.0 mm diameter) containing 3 mL of an enzyme solution (1% [w/v] cellulase RS [Yakult], 0.1% [w/v] pectolyase Y-23 [Yakult], 13% mannitol, pH adjusted to 5.8 with MES) and incubated for 2 to 2.5 h at room temperature with occasional gentle rotation. The solution was then replaced with fresh 13% mannitol, and the ovules were dissected with a glass needle with an inverted microscope to obtain egg cells. P. zonale ovules were predissected into two parts before enzyme treatment, and the halves containing the egg cells were incubated in the enzyme solution.
Generative and sperm cells were released from pollen tubes or pollen grains by osmotic shock. Pollen tubes were germinated in Suc-containing medium containing the basic components for pollen germination, including boric acid and calcium. For A. majus, pollen grains were germinated in 15% Suc containing 0.05% H3BO3 and 5 mM CaCl2 for 4 h at 28°C. The pollen tubes were then shocked with 6.5% mannitol solution to release the generative cells. For Arabidopsis, germination was performed according to the method of Boavida and McCormick (2007), with the exception that the medium contained 15% Suc. For N. tabacum, generative cells were induced to divide within pollen tubes using a semi-in vivo procedure (Tian and Russell, 1997), after which the sperm cells were released. For C. melo and P. zonale, the generative and sperm cells were released directly from the mature pollen grains. The grains were then collected into 30% Suc and transferred to 13% mannitol for release of the cells.
Isolation of leaf protoplasts of Arabidopsis used the same enzyme solution used for the isolation of egg cells. Fresh leaves were cut into thin strips 0.5 mm wide and immersed immediately into the enzyme solution. Mesophyll protoplasts were collected from the solution after 1 h of incubation at room temperature.
Quantitative PCR
The quantitative PCR method used in this study is based on a previous protocol (Gilliland et al., 1990). Briefly, after careful washing, as shown in Figure 1, gametic cells were transferred to PCR tubes containing 5 μL of double-distilled water (ddH2O) supplemented with 1 ng/mL denatured salmon sperm DNA. After two rapid freeze/thaw cycles, the PCR tubes were immediately heated at 95°C for 5 min. To release the DNA templates, 5 μL of proteinase K solution (200 μg/mL Proteinase K [Merck] in 1× Taq polymerase buffer) was added to the PCR tubes, which were incubated overnight at 56°C and stored at –80°C. Before PCR amplification, the tubes were incubated at 95°C for 5 min to permanently inactivate the Proteinase K. The quantification of mtDNA from pollen of A. majus and Arabidopsis used the same procedure for sample treatment. In this case, however, pollen grains placed on a silicon plate were gently crushed in ddH2O with a glass needle. After transfer of the ddH2O containing the crushed pollen grains to the PCR tubes, two washes using 5 μL of ddH2O in total were performed to elute the pollen cell cytoplasm that may have attached to the silicon plate and the glass needle, and these portions of ddH2O were combined in the PCR tubes.
The mtDNA was quantified using two rounds of competitive PCR with mitochondrial matR and coxI as targets. The construction, cloning, and validation of the competitor templates are described in Supplemental Figures 1 to 3 online. The primers used for PCR amplification are shown in Supplemental Figure 1 online and listed in Supplemental Table 2 online. The first-round PCR was performed in 25-μL reaction mixtures containing 1× Taq polymerase buffer, 0.2 mM deoxynucleotide triphosphates, 0.4 μM each of first-round primer, 1 unit of Taq polymerase, competitor template plasmid, and cell samples prepared as described above. The PCR conditions consisted of an initial denaturation at 95°C for 5 min; 20 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 40 s; and a final elongation at 72°C for 5 min. One microliter of the first-round PCR product mixture was used as the template for second-round PCR, which was performed according to the first-round PCR procedure except that second-round primers were used and 27 rather than 20 cycles were used. For PCR of the mtDNA from single sperm cells, a similar procedure was used, except that the first-round PCR was performed for 25 cycles and no competitor template was added. As internal controls, single-copy nuclear fragments were amplified with specific primers (see Supplemental Figure 12 and Supplemental Table 2 online).
Extraction and Analysis of Pollen DNA
For DNA gel blot hybridization analysis, pollen genomic DNA was extracted using a standard cetyltrimethylammonium bromide procedure. To obtain large amounts of A. majus pollen, anthers containing early pollen (undergoing first mitosis) and mature pollen were collected in 30% Suc and squeezed to release the pollen grains. The anther debris was removed by filtration through a 50-μm nylon mesh. After three rounds of washing in fresh Suc solution with brief centrifugation at 100g, pure pollen grains were obtained. The DNA was then digested, separated, and transferred to a nylon membrane according to standard procedures. The membrane was hybridized with Arabidopsis mitochondrial coxI and A. majus 18S nuclear rDNA fragments. These fragments were amplified by PCR using the primers listed in Supplemental Table 2 online and labeled with [α-32P]dCTP using a random priming DNA labeling kit (TaKaRa).
Cytology
Gametic cells were examined using standard fluorescence microscopy and immunoelectron microscopy procedures. Samples were fixed with 3% (v/v) glutaraldehyde and 1% (w/v) paraformaldehyde and embedded in Technovit (Kulzer) or LR White (Sigma–Aldrich) resin. For electron microscopy, the fixed samples were postfixed with 2% (w/v) osmium tetroxide and embedded in Spurr’s resin. Sections for fluorescence microscopy and immunoelectron microscopy (500- and 75-nm thickness, respectively) were produced using a microtome (Leica).
Sections for fluorescence microscopy were stained with DAPI and DiOC6 based on the methods of Nagata et al. (1999), and sections for electron microscopy were stained with 1% uranyl acetate and lead citrate before observation. Immunoelectron microscopy for the detection of cellular DNA was based on the method of Johnson and Rosenbaum (1990). Sections were incubated first with a mouse anti-DNA antibody (Sigma–Aldrich) and then with goat anti-mouse IgM conjugated to 10-nm colloidal gold (British BioCell International). Sections for immunoelectron microscopy were briefly incubated with 1% uranyl acetate. Negative control sections were pretreated with DNase before processing as above.
For the three-dimensional analysis of gametic cells, pollen grains and ovules of Arabidopsis were fixed and embedded for electron microscopy. Complete serial sections of the sperm and egg cells were produced and inspected. Cell membranes, plastids, mitochondria, and cell nuclei were examined using ultrastructural images manually calibrated for electronic deformation. Three-dimensional models of the cells were reconstructed from this information using our newly written computer programs (available upon request).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: mitochondrial maturase of A. majus, AY453102; mitochondrial maturase of Arabidopsis, Y08501; mitochondrial maturase of N. tabacum, BA000042; mitochondrial maturase of C. melo, FJ791301; cytochrome c oxidase I of P. zonale, DQ317047; ribonucleotide reductase R1 of Arabidopsis (At RNR), AC007019; T5E7 fragment of Arabidopsis, AC006225; T18C6W fragment of Arabidopsis, AC007729; granule-bound starch synthase I of A. majus (Am GBSS), AJ006293 and AJ006294; and ribonucleotide reductase R2 of N. tabacum (Nt RNR), AJ276622 and X92443.
Supplemental Data
The following materials are available in the online version of this article:
Supplemental Figure 1. Construction of Competitor Templates.
Supplemental Figure 2. Competitive Amplification of Target and Competitor DNA.
Supplemental Figure 3. Sensitivity of the Quantification Method.
Supplemental Figure 4. Target and Competitor DNA Amplification Efficiencies.
Supplemental Figure 5. Quantification of mtDNA in the Egg Cells of A. majus, N. tabacum, and P. zonale.
Supplemental Figure 6. Quantification of mtDNA in Sperm (Generative) Cells of A. majus, N. tabacum, C. melo, and P. zonale.
Supplemental Figure 7. The Generative Cells of A. majus Contain Mitochondria but Not Plastids.
Supplemental Figure 8. Degradation of mtDNA in the Pollen Cells of Arabidopsis.
Supplemental Figure 9. Cytoplasmic Volume Estimates in Early Pollen Cells of A. majus.
Supplemental Figure 10. Amplification of Male Gametic mtDNA in P. zonale.
Supplemental Figure 11. Loss of Mitochondrial Volume in Arabidopsis Pollen.
Supplemental Figure 12. Structure and Usage of Nuclear Genes as Indicators for Single-Copy Detection.
Supplemental Table 1. Quantitative Analysis of Arabidopsis Egg and Sperm Cells.
Supplemental Table 2. Primers Used in This Study.
Supplemental Movie 1. Three-Dimensional Configuration of an Arabidopsis Egg Cell.
Supplemental Movie 2. Three-Dimensional Configuration of an Arabidopsis Sperm Cell Pair.
Supplementary Material
Acknowledgments
We thank Gary Drews, Wataru Sakamoto, and Jian-Guo Chen for generously providing transgenic Arabidopsis and the AcMNPV virus (DD45:GFP from G. Drews, 35S-mtGFP from W. Sakamoto, and AcMNPV from J.-G. Chen). This work was supported by the National Basic Research Program of China (Program 973, Grant 2007CB108700) and the National Natural Science Foundation of China (Creative Research Group Program, Grant 30421004; Key Program, Grant 30430040).
References
- Alam T.I., Kanki T., Muta T., Ukaji K., Abe Y., Nakayama H., Takio K., Hamasaki N., Kang D. (2003). Human mitochondrial DNA is packaged with TFAM. Nucleic Acids Res. 31: 1640–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ankel-Simons F., Cummins J.M. (1996). Misconceptions about mitochondria and mammalian fertilization: Implications for theories on human evolution. Proc. Natl. Acad. Sci. USA 93: 13859–13863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimura S., Yamamoto J., Aida G.P., Nakazono M., Tsutsumi N. (2004). Frequent fusion and fission of plant mitochondria with unequal nucleoid distribution. Proc. Natl. Acad. Sci. USA 101: 7805–7808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur E. (1909). Das Wesen und die Erblichkeitsverhältnisse der “Varietates albomarginatae hort.” von Pelargonium zonale Z. Indukt. Abstammungs-Vererbungsl. 1: 330–351 [Google Scholar]
- Bendich A.J., Gauriloff L.P. (1984). Morphometric analysis of Cucurbit mitochondria: The relationship between chondriome volume and DNA content. Protoplasma 119: 1–7 [Google Scholar]
- Boavida L.C., McCormick S. (2007). Temperature as a determinant factor for increased and reproducible in vitro pollen germination in Arabidopsis thaliana. Plant J. 52: 570–582 [DOI] [PubMed] [Google Scholar]
- Chabouté M.E., Combettes B., Clément B., Gigot C., Philipps G. (1998). Molecular characterization of tobacco ribonucleotide reductase RNR1 and RNR2 cDNAs and cell cycle-regulated expression in synchronized plant cells. Plant Mol. Biol. 38: 797–806 [DOI] [PubMed] [Google Scholar]
- Clauhs R.P., Grun P. (1977). Changes in plastid and mitochondrion content during maturation of generative cells of Solanum (Solanaceae). Am. J. Bot. 64: 377–383 [Google Scholar]
- Correns C. (1909). Vererbungsversuche mit blass(gelb)grünen und buntblättrigen Sippen bei Mirabilis jalapa, Urtica pilulifera und Lunaria annua. Z. Indukt. Abstammungs-Vererbungsl. 1: 291–329 [Google Scholar]
- Cummins J. (1998). Mitochondrial DNA in mammalian reproduction. Rev. Reprod. 3: 172–182 [DOI] [PubMed] [Google Scholar]
- Draper C.K., Hays J.B. (2000). Replication of chloroplast, mitochondrial and nuclear DNA during growth of unirradiated and UVB-irradiated Arabidopsis leaves. Plant J. 23: 255–265 [DOI] [PubMed] [Google Scholar]
- Gilliland G., Perrin S., Blanchard K., Bunn H.F. (1990). Analysis of cytokine mRNA and DNA: Detection and quantitation by competitive polymerase chain reaction. Proc. Natl. Acad. Sci. USA 87: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyllensten U., Wharton D., Josefsson A., Wilson A.C. (1991). Paternal inheritance of mitochondrial DNA in mice. Nature 352: 255–257 [DOI] [PubMed] [Google Scholar]
- Hagemann R. (2000). Erwin Baur or Carl Correns: Who really created the theory of plastid inheritance? J. Hered. 91: 435–440 [DOI] [PubMed] [Google Scholar]
- Hagemann R., Schröder M.B. (1989). The cytological basis of the plastid inheritance in angiosperms. Protoplasma 152: 57–64 [Google Scholar]
- Havey M.J., McCreight J.D., Rhodes B., Taurick G. (1998). Differential transmission of the Cucumis organellar genomes. Theor. Appl. Genet. 97: 122–128 [Google Scholar]
- Hecht N.B., Liem H., Kleene K.C., Distel R.J., Ho S.M. (1984). Maternal inheritance of the mouse mitochondrial genome is not mediated by a loss or gross alteration of the paternal mitochondrial DNA or by methylation of the oocyte mitochondrial DNA. Dev. Biol. 102: 452–461 [DOI] [PubMed] [Google Scholar]
- Hu Y., Zhang Q., Sodmergen (2005). Potential cytoplasmic inheritance in Wisteria sinensis and Robinia pseudoacacia (Leguminosae). Plant Cell Physiol. 46: 1029–1035 [DOI] [PubMed] [Google Scholar]
- Hu Y.C., Zhang Q., Rao G.Y., Sodmergen (2008). Occurrence of plastids in the sperm cells of Caprifoliaceae: Biparental plastid inheritance in angiosperms is unilaterally derived from maternal inheritance. Plant Cell Physiol. 49: 958–968 [DOI] [PubMed] [Google Scholar]
- Johnson K.A., Rosenbaum J.L. (1990). The basal bodies of Chlamydomonas reinhardtii do not contain immunologically detectable DNA. Cell 62: 615–619 [DOI] [PubMed] [Google Scholar]
- Kang D., Kim S.H., Hamasaki N. (2007). Mitochondrial transcription factor A (TFAM): Roles in maintenance of mtDNA and cellular functions. Mitochondrion 7: 39–44 [DOI] [PubMed] [Google Scholar]
- Kucej M., Kucejova B., Subramanian R., Chen X.J., Butow R.A. (2008). Mitochondrial nucleoids undergo remodeling in response to metabolic cues. J. Cell Sci. 121: 1861–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroiwa T. (1991). The replication, differentiation, and inheritance of plastids with emphasis on the concept of organelle nuclei. Int. Rev. Cytol. 128: 1–62 [Google Scholar]
- Larsson N.G., Wang J., Wilhelmsson H., Oldfors A., Rustin P., Lewandoski M., Barsh G.S., Clayton D.A. (1998). Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat. Genet. 18: 231–236 [DOI] [PubMed] [Google Scholar]
- Laser B., Mohr S., Odenbach W., Oettler G., Kück U. (1997). Parental and novel copies of the mitochondrial orf25 gene in the hybrid crop-plant triticale: Predominant transcriptional expression of the maternal gene copy. Curr. Genet. 32: 337–347 [DOI] [PubMed] [Google Scholar]
- Liu Y., Zhang Q., Hu Y.F., Sodmergen (2004). Heterogeneous pollen in Chlorophytum comosum, a species with a unique mode of plastid inheritance intermediate between the maternal and biparental modes. Plant Physiol. 135: 193–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield S.G., Briarty L.G. (1990). Endosperm cellularization in Arabidopsis thaliana. Arabidopsis Inf. Serv. 27: 65–72 [Google Scholar]
- Matsushima R., Hu Y.C., Toyoda K., Sodmergen, Sakamoto W. (2008). The model plant Medicago truncatula exhibits biparental plastid inheritance. Plant Cell Physiol. 49: 81–91 [DOI] [PubMed] [Google Scholar]
- Mérida A., Rodríguez-Galán J.M., Vincent C., Romero J.M. (1999). Expression of the granule-bound starch synthase I (Waxy) gene from snapdragon is developmentally and circadian clock regulated. Plant Physiol. 120: 401–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels G.S., Hauswirth W.W., Laipis P.J. (1982). Mitochondrial DNA copy number in bovine oocytes and somatic cells. Dev. Biol. 94: 246–251 [DOI] [PubMed] [Google Scholar]
- Miyamura S., Kuroiwa T., Nagata T. (1987). Disappearance of plastid and mitochondrial nucleoids during the formation of generative cells of higher plants revealed by fluorescence microscopy. Protoplasma 141: 149–159 [Google Scholar]
- Mogensen H.L. (1988). Exclusion of male mitochondria and plastids during syngamy in barley as a basis for maternal inheritance. Proc. Natl. Acad. Sci. USA 85: 2594–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen H.L. (1996). The hows and whys of cytoplasmic inheritance in seed plants. Am. J. Bot. 83: 383–404 [Google Scholar]
- Mogensen H.L., Rusche M.L. (1985). Quantitative ultrastructural analysis of barley sperm. I. Occurrence and mechanism of cytoplasm and organelle reduction and the question of sperm dimorphism. Protoplasma 128: 1–14 [Google Scholar]
- Nagata N., Saito C., Sakai A., Kuroiwa H., Kuroiwa T. (1999). The selective increase or decrease of organellar DNA in generative cells just after pollen mitosis one controls cytoplasmic inheritance. Planta 209: 53–65 [DOI] [PubMed] [Google Scholar]
- Newman S.M., Zelenaya-Troitskaya O., Perlman P.S., Butow R.A. (1996). Analysis of mitochondrial DNA nucleoids in wild-type and a mutant strain of Saccharomyces cerevisiae that lacks the mitochondrial HMG box protein Abf2p. Nucleic Acids Res. 24: 386–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rantanen A., Jansson M., Oldfors A., Larsson N.G. (2001). Downregulation of Tfam and mtDNA copy number during mammalian spermatogenesis. Mamm. Genome 12: 787–792 [DOI] [PubMed] [Google Scholar]
- Rantanen A., Larsson N.G. (2000). Regulation of mitochondrial DNA copy number during spermatogenesis. Hum. Reprod. 15 (suppl. 2): 86–91 [DOI] [PubMed] [Google Scholar]
- Renner O. (1934). Die pflanzlichen Plastiden als selbständige Elemente der genetischen konstitution. Ber. Verhandl. Sächs Akad. Wiss Leipzig Math-Phys. Kl. 86: 241–266 [Google Scholar]
- Rusche M.L., Mogensen H.L., Zhu T., Smith S.E. (1995). The zygote and proembryo of alfalfa: Quantitative, three-dimensional analysis and implications for biparental plastid inheritance. Protoplasma 189: 88–100 [Google Scholar]
- Russell S.D. (1980). Participation of male cytoplasm during gamete fusion in an angiosperm, Plumbago zeylanica. Science 210: 200–201 [DOI] [PubMed] [Google Scholar]
- Russell S.D. (1983). Fertilization in Plumbago zeylanica: Gametic fusion and fate of the male cytoplasm. Am. J. Bot. 70: 416–434 [Google Scholar]
- Russell S.D., Rougier M., Dumas C. (1990). Organization of the early post-fertilization megagametophyte of Populus deltoides: Ultrastructure and implications for male cytoplasmic transmission. Protoplasma 155: 153–165 [Google Scholar]
- Satoh M., Nemoto Y., Kawano S., Nagata T., Hirokawa H., Kuroiwa T. (1993). Organization of heterogeneous mitochondrial DNA molecules in mitochondrial nuclei of cultured tobacco cells. Protoplasma 175: 112–120 [Google Scholar]
- Schröder M.B. (1985). Ultrastructural studies on plastids of generative and vegetative cells in Liliaceae. 3. Plastid distribution during the pollen development in Gasteria verrucosa (Mill.) Duval. Protoplasma 124: 123–129 [Google Scholar]
- Sheahan M.B., McCurdy D.W., Rose R.J. (2005). Mitochondria as a connected population: Ensuring continuity of the mitochondrial genome during plant cell dedifferentiation through massive mitochondrial fusion. Plant J. 44: 744–755 [DOI] [PubMed] [Google Scholar]
- Shitara H., Kaneda H., Sato A., Inoue K., Ogura A., Yonekawa H., Hayashi J.I. (2000). Selective and continuous elimination of mitochondria microinjected into mouse eggs from spermatids, but not from liver cells, occurs throughout embryogenesis. Genetics 156: 1277–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia R.A., Carrol S., Kalifa L., Hochmuth C., Sia E.A. (2009). Loss of the mitochondrial nucleoid protein, Abf2p, destabilizes repetitive DNA in the yeast mitochondrial genome. Genetics 181: 331–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.E. (1989). Influence of parental genotype on plastid inheritance in Medicago sativa. J. Hered. 80: 214–217 [DOI] [PubMed] [Google Scholar]
- Sodmergen, Zhang Q., Zhang Y., Sakamoto W., Kuroiwa T. (2002). Reduction in amounts of mitochondrial DNA in the sperm cells as a mechanism for maternal inheritance in Hordeum vulgare. Planta 216: 235–244 [DOI] [PubMed] [Google Scholar]
- Stupar R.M., Lilly J.W., Town C.D., Cheng Z., Kaul S., Buell C.R., Jiang J. (2001). Complex mtDNA constitutes an approximate 620-kb insertion on Arabidopsis thaliana chromosome 2: Implication of potential sequencing errors caused by large-unit repeats. Proc. Natl. Acad. Sci. USA 98: 5099–5103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama Y., Watase Y., Nagase M., Makita N., Yagura S., Hirai A., Sugiura M. (2005). The complete nucleotide sequence and multipartite organization of the tobacco mitochondrial genome: Comparative analysis of mitochondrial genomes in higher plants. Mol. Genet. Genomics 272: 603–615 [DOI] [PubMed] [Google Scholar]
- Sutovsky P. (2003). Ubiquitin-dependent proteolysis in mammalian spermatogenesis, fertilization, and sperm quality control: Killing three birds with one stone. Microsc. Res. Tech. 61: 88–102 [DOI] [PubMed] [Google Scholar]
- Sutovsky P., Moreno R.D., Ramalho-Santos J., Dominko T., Simerly C., Schatten G. (1999). Ubiquitin tag for sperm mitochondria. Nature 402: 371–372 [DOI] [PubMed] [Google Scholar]
- Sutovsky P., Moreno R.D., Ramalho-Santos J., Dominko T., Simerly C., Schatten G. (2000). Ubiquitinated sperm mitochondria, selective proteolysis, and the regulation of mitochondrial inheritance in mammalian embryos. Biol. Reprod. 63: 582–590 [DOI] [PubMed] [Google Scholar]
- Thompson W.E., Ramalho-Santos J., Sutovsky P. (2003). Ubiquitination of prohibitin in mammalian sperm mitochondria: Possible roles in the regulation of mitochondrial inheritance and sperm quality control. Biol. Reprod. 69: 254–260 [DOI] [PubMed] [Google Scholar]
- Tian H.Q., Russell S.D. (1997). Micromanipulation of male and female gametes of Nicotiana tabacum. I. Isolation of gametes. Plant Cell Rep. 16: 555–560 [DOI] [PubMed] [Google Scholar]
- Unseld M., Marienfeld J.R., Brandt P., Brennicke A. (1997). The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat. Genet. 15: 57–61 [DOI] [PubMed] [Google Scholar]
- Yu H.S., Huang B.Q., Russell S.D. (1994). Transmission of male cytoplasm during fertilization in. Nicotiana tabacum. Sex. Plant Reprod. 7: 313–323 [Google Scholar]
- Yu H.S., Russell S.D. (1994). Populations of plastids and mitochondria during male reproductive cell maturation in Nicotiana tabacum L.: A cytological basis for occasional biparental inheritance. Planta 193: 115–122 [Google Scholar]
- Zhu T., Mogensen H.L., Smith S.E. (1993). Quantitative, three-dimensional analysis of alfalfa egg cells in two genotypes: Implications for biparental plastid inheritance. Planta 190: 143–150 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.