The Arabidopsis disease resistance protein RPP1 recognizes the ATR1 effector protein from Hyaloperonospora arabidopsidis. This works shows that the molecular basis of recognition is mediated by the in planta association of the LRR domain of the RPP1 protein with the ATR1 effector protein. The in planta association of specific alleles of ATR1 leads to the activation of plant immune responses.
Abstract
Activation of plant immunity relies on recognition of pathogen effectors by several classes of plant resistance proteins. To discover the underlying molecular mechanisms of effector recognition by the Arabidopsis thaliana RECOGNITION OF PERONOSPORA PARASITICA1 (RPP1) resistance protein, we adopted an Agrobacterium tumefaciens–mediated transient protein expression system in tobacco (Nicotiana tabacum), which allowed us to perform coimmunoprecipitation experiments and mutational analyses. Herein, we demonstrate that RPP1 associates with its cognate effector ARABIDOPSIS THALIANA RECOGNIZED1 (ATR1) in a recognition-specific manner and that this association is a prerequisite step in the induction of the hypersensitive cell death response of host tissue. The leucine-rich repeat (LRR) domain of RPP1 mediates the interaction with ATR1, while the Toll/Interleukin1 Receptor (TIR) domain facilitates the induction of the hypersensitive cell death response. Additionally, we demonstrate that mutations in the TIR and nucleotide binding site domains, which exhibit loss of function for the induction of the hypersensitive response, are still able to associate with the effector in planta. Thus, our data suggest molecular epistasis between signaling activity of the TIR domain and the recognition function of the LRR and allow us to propose a model for ATR1 recognition by RPP1.
INTRODUCTION
Plants have evolved a multilevel innate immune system to protect them against infection by a diverse range of pathogens, including viruses, bacteria, fungi, oomycetes, and nematodes. Despite the great evolutionary distance among phytopathogens, the outcome of the plant–pathogen interactions is controlled by the same principles: the ability of the pathogen to suppress the plant immune system to establish infection, and the ability of plants to recognize the presence of a pathogen and to induce immune responses that restrict pathogen growth.
The first line of plant defense consists of the integral plasma membrane receptors known as pattern-recognition receptors, which recognize the presence of common pathogen-associated molecular patterns (PAMPs) near the cell surface (Chisholm et al., 2006; Jones and Dangl, 2006; Trinchieri and Sher, 2007). Upon association with PAMPs, the pattern-recognition receptors activate a downstream mitogen-activated protein kinase signaling cascade that culminates in transcriptional activation and generation of the innate immune responses (Chisholm et al., 2006; Jones and Dangl, 2006). This line of defense, called PAMP-triggered immunity, is commonly suppressed by a successful pathogen in order to establish infection. To interfere with PAMP-triggered immunity, pathogens from different kingdoms of life have evolved effector proteins that are delivered into and function within the host plant cells (Desveaux et al., 2006; Kamoun, 2006; Dodds et al., 2009). The second layer of plant immunity depends on the ability of the plant to recognize these pathogen-derived effectors and trigger a robust resistance response that normally culminates in a hypersensitive cell death response (HR). While PAMPs represent conserved microbial molecules, effector molecules constitute very divergent groups of proteins and their recognition requires constant structural and evolutionary adjustment of the corresponding receptors. Effector-triggered immunity is mediated by a large group of structurally related intracellular innate immune receptors encoded by resistance (R) genes. Products of R genes either directly or indirectly recognize pathogen effectors and induce innate immunity. The major class of R proteins is characterized by the central nucleotide binding site (NBS) domain and C-terminal leucine-rich repeats (LRRs). This group can be further subdivided according to the N-terminal domain into a TIR-NBS-LRR class, which has an N-terminal domain with sequence similarity to Drosophila melanogaster Toll and human Interleukin1 Receptor (TIR), and a CC-NBS-LRR class, which has a structured coiled-coil domain (CC). Currently, the data supporting molecular functions of the TIR/CC, NBS, and LRR domains in R proteins are sparse, sometimes contradictory, and scattered across many different proteins and experimental systems. Therefore, there is a need for in planta data that would test and clearly define the roles of different domains for a given R protein.
The NBS domain of R proteins shares sequence similarity with the mammalian cell death–inducing proteins, including human apoptotic peptidase-activating factor 1 (Apaf-1), its Caenorhabditis elegans homolog CED-4, and a large group of intracellular nucleotide oligomerization domain receptors that function in mammalian innate immunity (Takken et al., 2006). In animal proteins, nucleotide binding and hydrolysis acts as a molecular switch that regulates signal transduction by conformational change, often leading to oligomerization of the protein (Kim et al., 2005). In plants, the NBS domains of the tomato (Solanum lycopersicum) R proteins I-2 and Mi-1 were shown to bind and hydrolyze ATP in vitro, supporting the function of the NBS as a nucleotide binding switch in R protein activation (Tameling et al., 2002). In addition, mutations in the conserved P-loop motif resulted in a loss of nucleotide binding in vitro (Tameling et al., 2002) and the corresponding loss of HR induction in planta (Tameling et al., 2002; Wirthmueller et al., 2007). The intact P-loop motif was also required for effector-induced oligomerization of the tobacco (Nicotiana tabacum) TIR-NBS-LRR resistance protein N (Mestre and Baulcombe, 2006). Based on these data, nucleotide binding is considered to be a molecular switch that regulates the activity of R proteins (Takken et al., 2006).
The TIR/CC N-terminal domains of R proteins were originally proposed to act in downstream signaling rather than in initial perception of the effector molecule. This view was based on the signaling function of TIRs in the Toll receptors from Drosophila, human Interleukin1 receptor, and mammalian Toll-Like Receptors (TLRs). In the case of R proteins, this model was supported by Frost et al. (2004), who showed autoactivation of the TIR domain of flax (Linum usitatissimum) L10, and by Swiderski et al. (2009), who demonstrated that the TIR domains of several R proteins, including Arabidopsis thaliana Resistance to Pseudomonas syringae 4 (RPS4) and Recognition of Peronospora parasitica 1A (RPP1A) as well as tobacco N, show different degrees of effector-independent HR when transiently expressed in N. tabacum or Nicotiana benthamiana. However, there also exists evidence of TIR involvement in determining interactions with various effectors (Ellis et al., 1999; Burch-Smith et al., 2007). The importance of the TIR domains for R protein function was highlighted in several studies that identified mutations in conserved residues that led to the loss of HR in planta (Dinesh-Kumar et al., 2000; Swiderski et al., 2009).
The LRR domains are often involved in ligand–receptor interactions in animal systems (Kobe and Deisenhofer, 1993; Alder et al., 2005). In plant innate immunity, this role of LRR domains is supported, in the case of some R proteins, by both genetic analyses and yeast two-hybrid data (Deslandes et al., 2003; Dodds et al., 2006). Evolutionary analyses suggest that many R genes and their cognate effectors are evolving under the pressure of diversifying selection (Mondragon-Palomino, 2002; Win et al., 2007). This often results in numerous duplications and rearrangements at the R gene loci as well as in high levels of polymorphism in the LRR domain (Ellis et al., 1999; Allen et al., 2004). Domain-swapping experiments confirmed the role of the LRR domain in determining specificity toward different effector gene variants (Ellis et al., 1999; Dodds et al., 2001; Rairdan and Moffett, 2006; Shen et al., 2007; Rentel et al., 2008). Direct protein–protein interaction of the LRR domain with the cognate effector was demonstrated for rice (Oryza sativa) Pi-ta and the rice blast fungus (Magnaporthe grisea) effector AvrPita. In this case, the LRR region was both necessary and sufficient for interaction in yeast two-hybrid assays and for in vitro binding (Jia et al., 2000). Despite growing evidence for the direct interactions between R proteins and their cognate effectors, there has been a lack of in planta biochemical data supporting the role of the LRR region in effector recognition.
The RECOGNITION OF PERONOSPORA PARASITICA1 (RPP1) locus, which contains a complex resistance gene cluster, was originally identified in Arabidopsis thaliana ecotype Wassilewskija (Ws; Botella et al., 1998). Several members of the RPP1 gene family have been shown to specify disease resistance against Hyaloperonospora arabidopsidis (previously known as Peronospora parasitica; Botella et al., 1998; Rehmany et al., 2005; Sohn et al., 2007), including RPP1-WsA, RPP1-WsB, RPP1-WsC, and RPP1-NdA, while RPP1-like genes from other ecotypes have been implicated in hybrid incompatibility (Bomblies et al., 2007). Currently, proteins encoded by only two RPP1 alleles have been shown to recognize the cognate effector ARABIDOPSIS THALIANA RECOGNIZED1 (ATR1) from H. arabidopsidis (Rehmany et al., 2005). One of those alleles was cloned from the Arabidopsis ecotype Ws and is denoted RPP1-WsB, whereas the other, RPP1-NdA, is from Arabidopsis ecotype Niederzenz (Nd). The R proteins RPP1-WsB and RPP1-NdA share a common TIR-NBS-LRR domain architecture and are 87% identical at the amino acid level. Although polymorphisms are present throughout their coding sequences, most of the differences occur in the LRR region and include both single amino acid polymorphisms and short insertions and deletions.
ATR1 belongs to a simple locus in H. arabidopsidis with diverse allelic variants present in different strains of the pathogen (Rehmany et al., 2005). The ATR1 protein has two regions found in most oomycete effectors thus far: the N-terminal eukaryotic signal peptide that targets the effector for secretion, followed by the putative host-targeting domain defined by the conserved RXLR motif (Rehmany et al., 2005; Birch et al., 2006). The rest of the protein lacks sequence similarity to any proteins of known function. Although this region is thought to contribute to pathogen virulence (Sohn et al., 2007), recognition by RPP1-WsB and RPP1-NdA is its only known function.
In this study, we characterize the molecular mechanisms underlying the recognition of ATR1 by RPP1. We present biochemical evidence for in planta association between RPP1-WsB and ATR1, which correlates with the ability of RPP1-WsB to recognize this effector and elicit a resistance response. Our studies reveal that this association is mediated by the LRR domain of RPP1-WsB. Moreover, we demonstrate an epistatic relationship between effector binding ability of the LRR and HR induction by the TIR and NBS domains. Finally, we exploit natural polymorphisms in ATR1 to isolate two gain-of-recognition mutants, showing that the pattern of ATR1 recognition depends on just a few amino acids that are subjected to strong diversifying selection.
RESULTS
The Phylogeny of ATR1 Correlates with Its Recognition by RPP1-WsB and RPP1-NdA
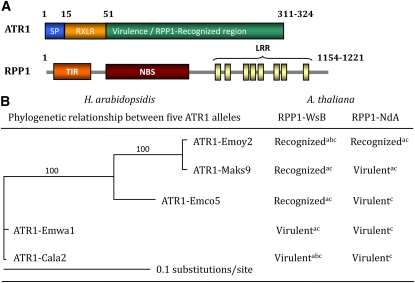
Previous studies established the gene-for-gene recognition specificities of five ATR1 alleles sequenced from natural populations of H. arabidopsidis (Figure 1; see Supplemental Figure 1 online) and two RPP1 alleles isolated from Arabidopsis ecotypes Ws and Nd (Figure 1A; see Supplemental Figure 2 online; Rehmany et al., 2005). The ATR1 sequences obtained from different H. arabidopsidis strains contained many polymorphisms and were named after the strain they were isolated from; that is, ATR1-Emoy2 comes from H. arabidopsidis strain Emoy2. It has been demonstrated that only a subset of ATR1 alleles is able to elicit a hypersensitive response (HR) in transgenic Arabidopsis plants carrying either RPP1-WsB or RPP1-NdA. Thus, transgenic plants carrying RPP1-WsB elicited HR in response to ATR1-Emoy2, ATR1-Maks9, and ATR1-Emco5 but not in response to ATR1-Cala2 or ATR1-Emwa1, and plants carrying RPP1-NdA recognized ATR1-Emoy2 but not ATR1-Maks9 (Figure 1B; Rehmany et al., 2005; Sohn et al., 2007).
Figure 1.
Domain Architecture and Allele-Specific Recognition Pattern of ATR1 and RPP1.
(A) Domain organization of the oomycete effector ATR1 and the Arabidopsis resistance protein RPP1. The ATR1 diagram shows the eukaryotic secretion sequence (SP), host translocation-targeting region (containing RXLR), and C-terminal domain, recognized by RPP1. The RPP1 diagram highlights the major domains identified by sequence similarity to known protein families: TIR, NBS, and a series of LRRs.
(B) Maximum-likelihood tree illustrating phylogenetic relationships among five ATR1 alleles (left) and the corresponding recognition pattern by RPP1 alleles RPP1-WsB and RPP1-NdA (right). Bootstrap values are shown on the tree branches.
Recognition was demonstrated in the following studies: ain Arabidopsis in cobombardment assay (Rehmany et al., 2005); bin Arabidopsis with type three secretion system delivery by Pseudomonas syringae pv tomato DC3000 (Sohn et al., 2007); cin tobacco by Agrobacterium-mediated transient coexpression (this study). The alignment used is shown in Supplemental Figure 1 online (pdf format) and Supplemental Data Set 1 online (text format).
We decided to more closely examine the relationships between the ATR1 sequence variation and recognition by RPP1. We constructed a maximum likelihood phylogenetic tree of ATR1 alleles and mapped the pattern of their recognition by RPP1. Interestingly, the three alleles recognized by RPP1-WsB clustered together, while the two nonrecognized alleles formed a separate clade (Figure 1B). Although the number of sequences within both ATR1 and RPP1 groups of genes is too small to make any concrete conclusions, such an evolutionary pattern is consistent with tight coevolution between RPP1 and ATR1 and highlights the need for a deeper sequence sampling of both genes.
Allelic sequences encoding both ATR1 and RPP1 show high levels of polymorphisms. The most divergent alleles, ATR1-Emoy2 and ATR1-Emwa1, share only 81.3% sequence identity at the amino acid level (see Supplemental Figure 1 online). Rehmany et al. (2005) examined the average pair-wise divergence between ATR1 alleles in a sliding-window analysis and found that synonymous and nonsynonymous polymorphisms are distributed unevenly throughout the coding sequence, with nonsynonymous mutations accumulating in the C-terminal region of the protein. Similarly, sequence comparison of RPP1 alleles indicated that most of the polymorphisms accumulated in the LRR portion of the protein (Botella et al., 1998; see Supplemental Figure 2 online).
High levels of polymorphisms and evidence of positive selection in the R gene and its cognate effector suggest a direct recognition of the effector protein by the corresponding resistance protein. Investigation of the molecular basis of ATR1–RPP1 interaction in a natural pathosystem presents several experimental challenges: (1) low levels of protein expression for both ATR1 and RPP1; (2) small numbers of cells undergoing a resistance response during the infection; and (3) inability to genetically manipulate H. arabidopsidis. To circumvent these problems, we adopted a transient Agrobacterium tumefaciens-mediated protein expression system in tobacco.
RPP1 Activates ATR1-Dependent HR in Tobacco
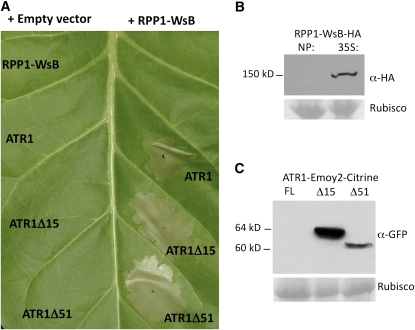
Tobacco has been widely used for Agrobacterium-mediated transient expression of proteins and for assaying the HR. However, one commonly occurring limitation of the heterologous plant expression system is autoactivation of the overexpressed resistance protein even in the absence of the effector. This autoactivation phenotype was observed in the case of Arabidopsis R proteins Resistance to Pseudomonas syringae 2 (RPS2), RPS4 (Zhang et al., 2004), and Recognition of Peronospora parasitica 13 (Rentel et al., 2008) but not RPP1-WsA (Weaver et al., 2006). We tested the expression of RPP1-WsB in tobacco, driven either by its native promoter (see Supplemental Figure 3 online) or by the strong constitutive cauliflower mosaic virus 35S promoter (35S; Figure 2A). None of the RPP1-WsB constructs exhibited autoactivation and cell death in the absence of ATR1. To find a construct amenable to biochemical analyses, we made a protein fusion between RPP1-WsB and the hemagglutinin epitope tag (HA) on both N- and C-terminal ends of the protein. While protein levels of the RPP1-WsB construct driven by its native promoter were below the detection limit, we could detect 35S:RPP1-WsB-HA by protein immunoblot (Figure 2B). The ATR1 constructs were fused with Citrine tag, a fluorescent protein variant that is detected by anti-green fluorescent protein (α-GFP) antibody. Coexpression of the RPP1-WsB-HA together with full-length ATR1-Emoy2-Citrine, ATR1δ15-Emoy2-Citrine (lacking the signal peptide sequence), or ATR1δ51-Emoy2-Citrine (lacking both the signal peptide sequence and the RXLR motif) elicited a HR ∼20 to 24 h post infiltration (Figure 2A). The full-length ATR1 protein produced a slower and less pronounced response. Protein immunoblot analysis showed that while ATR1δ15-Emoy2-Citrine and ATR1δ51-Emoy2-Citrine were expressed at very high levels, the full-length ATR1 protein levels were below the detection limit (Figure 2C), probably due to partial secretion of the protein from the plant cells and instability in the extracellular space. Based on these results, we chose to use 35S:RPP1-WsB-HA and ATR1δ51-Citrine for further biochemical analyses.
Figure 2.
Coexpression of ATR1 and RPP1 in Tobacco Triggers the HR.
(A) RPP1-WsB-HA induces HR in tobacco upon Agrobacterium-mediated coexpression with full-length ATR1-Emoy2-Citrine, ATR1δ15-Emoy2-Citrine, or ATRδ51-Emoy2-Citrine. All constructs were expressed under the control of the constitutive cauliflower mosaic virus 35S promoter. The photograph was taken 48 h post infiltration.
(B) Protein immunoblot probed with α-HA antibody showing relative levels of protein expression of RPP1-WsB-HA in tobacco driven by either its native promoter (NP) or cauliflower mosaic virus 35S promoter (35S). Ribulose-1,5-bis-phosphate carboxylase/oxygenase (Rubisco), stained on the blots with Ponceau S stain, is shown as a loading control.
(C) Protein immunoblot probed with α-GFP antibody showing relative levels of protein expression of the full-length (FL) ATR1-Emoy2-Citrine, ATR1δ15-Emoy2-Citrine, and ATRδ51-Emoy2-Citrine in tobacco at 24 h post infiltration. Rubisco is shown as a loading control.
Race-Specific Recognition of ATR1 by RPP1 in Tobacco
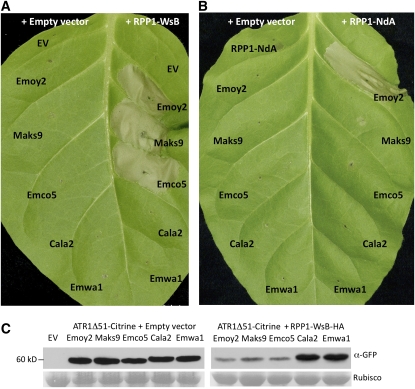
In order to further characterize whether the tobacco system reflects the previously observed ATR1/RPP1 recognition specificities, we coexpressed five ATR1 alleles, ATR1-Emoy2, ATR1-Maks9, ATR1-Emco5, ATR1-Cala2, and ATR1-Emwa1, with either RPP1-WsB or RPP1-NdA. We observed the same race-specific recognition pattern in tobacco as previously described in Arabidopsis (Rehmany et al., 2005; Sohn et al., 2007) and expanded the data for RPP1-NdA, which was previously tested only with ATR1-Emoy2 and ATR1-Maks9. RPP1-WsB induced an HR when coexpressed with ATR1-Emoy2, Maks9, and Emco5 but not with Cala2 or Emwa1 (Figure 3A); RPP1-NdA induced an HR only when coexpressed with ATR1-Emoy2 but not with any other ATR1 allele (Figure 3B). The same recognition specificities were observed for both 35S:RPP1-WsB and genomic RPP1-WsB construct (see Supplemental Figure 3 online). Protein gel blot analysis showed that, whereas all ATR1 protein variants were expressed to equally high levels in the absence of RPP1 (Figure 3C, left panel), protein levels of the recognized alleles decreased dramatically upon induction of HR (Figure 3C, right panel). These data indicate that the lack of recognition of a subset of ATR1 alleles was not due to a lack of protein expression. All ATR1 variants localized consistently to the cytoplasm and nucleus, as determined by fluorescence microscopy (see Supplemental Figure 4 online), indicating that the difference in recognition was not due to differential localization of ATR1 variants within the plant cell.
Figure 3.
Race-Specific Recognition of ATR1 Alleles by RPP1-WsB and RPP1-NdA in Tobacco.
(A) Coinfiltration of RPP1-WsB with ATR1-Emoy2, ATR1-Maks9, and ATR1-Emco5, but not ATR1-Cala2 or ATR1-Emwa1, triggers the HR in tobacco.
(B) Coinfiltration of RPP1-NdA with ATR1-Emoy2, but none of the other four alleles, triggers the HR. Photographs were taken 48 h post infiltration.
(C) Protein immunoblot probed with α-GFP antibody showing relative levels of protein abundance of five ATR1 variants coinfiltrated with empty vector (EV; left panel) or with RPP1-WsB (right panel). Samples for protein extraction were collected at 20 to 24 h post infiltration, when the HR started to appear. Rubisco is shown as a loading control.
RPP1 Coimmunoprecipitates with ATR1
To investigate whether recognition specificity results from physical interaction between the ATR1 and RPP1 proteins, we tested ATR1 variants for their ability to associate with RPP1-WsB in planta. Citrine-tagged ATR1δ51 constructs were transiently coexpressed with 35S:RPP1-WsB-HA in tobacco for 20 to 24 h. Tissue was collected when the HR began to appear. Protein extracts were incubated with α-GFP antibodies to immunoprecipitate ATR1δ51-Citrine. Resulting immunocomplexes were separated by SDS-PAGE, and RPP1-WsB was detected by immunoblot using α-HA antibody. Due to the mild conditions of the nondenaturing protein extraction buffer used for coimmunoprecipitations, the initial levels of RPP1-WsB protein were very low in all of the input samples and were usually close to the limit of detection by a protein gel blot. Therefore, we performed parallel α-HA immunoprecipitations to ensure that RPP1-WsB protein was present in all of the samples.
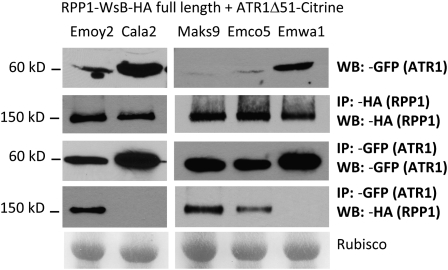
We observed that RPP1-WsB coimmunoprecipitated with the three variants of ATR1 (Emoy2, Maks9, and Emco5) that also triggered HR, but it did not associate with the nonrecognized variants ATR1-Cala2 and ATR1-Emwa1 (Figure 4). These data show a perfect correlation between the ability of RPP1 to associate with ATR1 and the activation of the immune response, suggesting that it is dependent on the ability of RPP1-WsB to associate with ATR1. Additionally, we observed a slight, but consistent, decrease in the amount of RPP1-WsB associating with ATR1-Emco5 as compared with ATR1-Maks9 and ATR1-Emoy2 (Figure 4).
Figure 4.
RPP1-WsB Associates with ATR1 Alleles in Planta in a Recognition-Specific Manner.
RPP1-WsB-HA coimmunoprecipitates with the recognized ATR1 variants ATR1-Emoy2-Citrine, ATR1-Maks9-Citrine, and ATR1-Emco5-Citrine but not with the virulent variants ATR1-Cala2-Citrine and ATR1-Emwa1-Citrine. RPP1 and ATR1 alleles were transiently expressed in tobacco leaves by Agrobacterium infiltration. Tissue samples were collected at 24 h post infiltration and analyzed by immunoprecipitation (IP) and protein immunoblot (WB). Top panel, Input probed with α-GFP for ATR1-Citrine; second panel, α-HA IP showing the presence of RPP1-WsB protein in all samples; third panel, α- GFP IP samples probed with α-GFP for immunoprecipitating ATR1; bottom panel, α-GFP IP probed with α-HA for coimmunoprecipitating RPP1. Rubisco is shown as a loading control.
Unfortunately, we were unable to detect a robust association between RPP1-NdA and ATR1-Emoy2. In one coimmunoprecipitation experiment out of three conducted, we detected a weak signal corresponding to RPP1-NdA, which was specific for an association with ATR1-Emoy2, but the level of the signal was just above the detection limit (data not shown). This could be due to a much weaker protein–protein interaction between RPP1-NdA and ATR1-Emoy2.
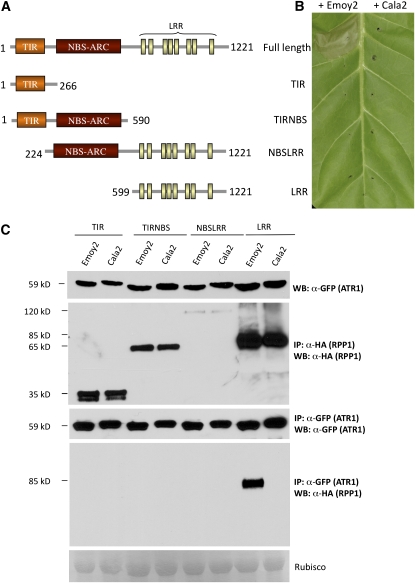
The LRR Domain of RPP1-WsB Is Necessary and Sufficient for an Association with ATR1 but Not Sufficient for Triggering the HR
Next, we decided to determine which domains of RPP1-WsB are important for an association with ATR1. We made several truncations of RPP1-WsB that carried deletions of the TIR, NBS, and LRR domains (Figure 5A). Each truncated RPP1 construct was tagged with HA and coexpressed with Citrine-tagged ATR1δ51-Emoy2 or ATR1δ51-Cala2 in tobacco for 24 h. We observed that only the full-length RPP1-WsB but none of the truncated constructs was able to elicit an ATR1-specific HR (Figure 5B). We performed coimmunoprecipitation experiments to determine whether any of the RPP1 truncations were able to associate with ATR1. The results showed that the TIR or TIR-NBS constructs without the LRR domain did not coimmunoprecipitate with ATR1, while the LRR domain alone was sufficient for interactions and maintained the same binding specificity as the full-length RPP1-WsB protein (Figure 5C). The NBS-LRR construct showed decreased protein stability; nonetheless, we could detect a weak but specific association with ATR1-Emoy when the blot was exposed for a sufficiently long time (see Supplemental Figure 5A online). Since the LRR domain alone coimmunoprecipitated with ATR1-Emoy2 but not with ATR1-Cala2, we concluded that the LRR domain is sufficient for interaction with the effector. Similar results were obtained from coimmunoprecipitation of the LRR of RPP1-WsB and ATR1-Maks9, ATR1-Emco5, and ATR1-Emwa1 (see Supplemental Figure 5B online), showing that the LRR domain is sufficient for determining RPP1-WsB specificity.
Figure 5.
The LRR Region of RPP1 Is Necessary and Sufficient for Association with ATR1 but Not for Eliciting the HR in Tobacco.
(A) Schematic organization of the RPP1-WsB truncations. Domain abbreviations are described in Figure 1A.
(B) Corresponding phenotypes of the HA-tagged RPP1-WsB deletion constructs coexpressed with ATR1δ51-Emoy2 or ATR1δ51-Cala2 in tobacco. The photograph was taken 48 h post infiltration.
(C) Coimmunoprecipitation of RPP1-WsB truncations with ATR1. The LRR domain alone, but not the TIR or TIR-NBS domain, associates with ATR1 in planta. The NBS-LRR construct is unstable in planta but shows allele-specific binding to ATR1 upon prolonged exposure (see Results). Panels are labeled as in Figure 4.
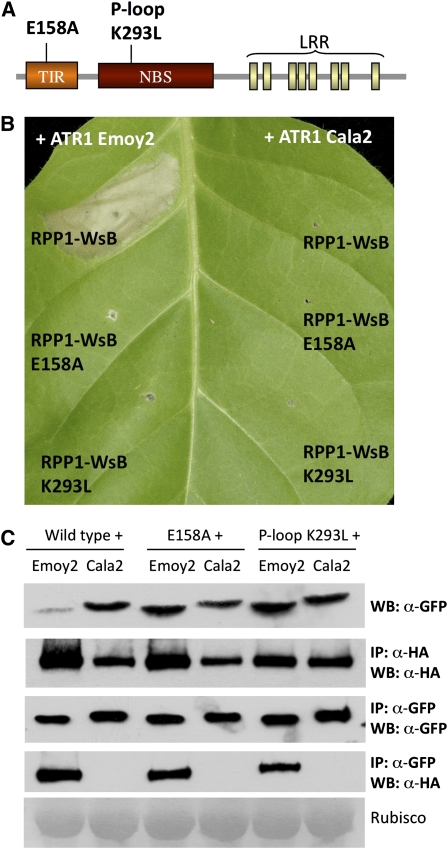
Mutants in TIR and NBS That Do Not Induce HR Are Still Able to Interact with ATR1
Previous genetic analyses of R proteins identified conserved amino acid residues in both TIR and NBS domains that are critical for the activation of HR (Dinesh-Kumar et al., 2000; Takken et al., 2006). We have made a multiple sequence alignment between the TIR domains of several TIR-NBS-LRR resistance genes, including Arabidopsis RPP1-WsB, RPP4, and RPS4, tobacco N, flax M and L, and human TLR1 (see Supplemental Figure 6A online). There are several conserved residues among those R proteins and TLR1. We used mutational analysis and identified a conserved Glu (E) at position 158 that was required for RPP1-WsB to elicit HR (Figure 6B) while not affecting protein stability. The most conserved motif within the NBS domain is the P-loop motif (also known as kinase-1a or Walker-A), which has been shown to affect ATP binding in vitro and was required for eliciting in planta HR response (Tameling et al., 2002). The P-loop has the conserved consensus sequence GxxxxGKS/T (where x is any amino acid), and mutation of the conserved Lys (K) leads to a loss-of-function phenotype (see Supplemental Figure 6B online; Takken et al., 2006). We created a K293L mutation in RPP1-WsB and found that, as with similar mutations to other R proteins, it abolished the ability of RPP1-WsB to trigger effector-mediated HR (Figure 6B).
Figure 6.
Mutations in the TIR and NBS Domains That Compromise Induction of HR Do Not Affect in Planta Association between ATR1 and RPP1.
(A) Schematic diagram of RPP1-WsB, showing positions of the E158A mutation in the TIR domain and the P-loop K293L mutation in the NBS domain.
(B) RPP1-WsB-HA E158A and RPP1-WsB-HA K293L are unable to induce HR in tobacco upon coexpression with ATR1δ51-Emoy2-Citrine. The photograph was taken at 48 h post infiltration.
(C) RPP1-WsB E158A and RPP1-WsB K293L are still able to associate with ATR1-Emoy2 but not with ATR1-Cala2. Panels are labeled as in Figure 4.
We tested the E158A and K293L mutants of RPP1-WsB for their ability to coimmunoprecipitate with ATR1. Interestingly, we discovered that both mutants retained their ability to associate with the effector (Figure 6C). These data clearly show an epistatic relationship between the recognition function of the LRR domain of RPP1-WsB and the subsequent downstream activation of signaling events mediated by the TIR and NBS domains.
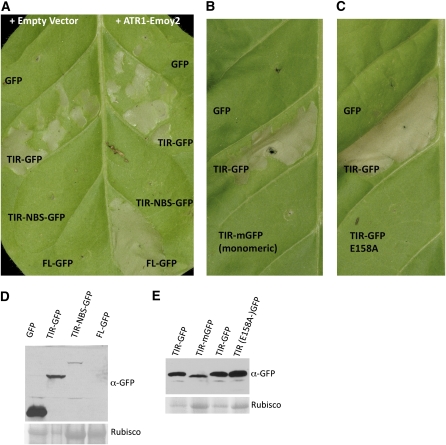
The TIR Domain of RPP1-WsB Elicits Effector-Independent HR
The TIR domains of several TIR-NBS-LRR proteins, including a RPP1-WsB paralogue, RPP1-WsA, are able to induce effector-independent HR in both tobacco and Arabidopsis (Frost et al., 2004; Swiderski et al., 2009). We were surprised that we did not observe this phenotype with our TIR-HA construct of RPP1-WsB, which was equivalent to the constructs described for RPP1-WsA. However, when we tested the same truncation variant tagged with GFP, we detected effector-independent HR that appeared around 48 h post infiltration. Thus, the TIR domain of RPP1-WsB was sufficient for induction of the HR, yet the response was slower and weaker than effector-mediated HR induced by the full-length RPP1 protein (Figure 7A). The TIR-NBS-GFP protein did not activate effector-independent HR (Figure 7A), which either could be due to decreased protein levels of this protein fusion compared with the TIR-GFP (Figure 7D) or may indicate additional negative regulation of the TIR by the NBS domain. Additionally, in the case of RPP1-WsB, HR induced by the TIR truncation was dependent upon the presence of a large fusion tag, the GFP.
Figure 7.
The TIR Domain Is Sufficient for Triggering the HR in Tobacco.
(A) The TIR truncation (amino acids 1–266) of RPP1-WsB fused to GFP is able to elicit effector-independent HR in tobacco.
(B) An A206K mutation in GFP that disrupts its ability to form dimers compromises the ability of the TIR to autoactivate.
(C) The E158A mutation in the TIR domain compromises its ability to trigger effector-independent HR.
(D) and (E) Protein immunoblots probed with α-GFP antibody showing relative expression levels of the GFP-tagged RPP1 constructs. Samples were taken at 48 h post infiltration. Rubisco is shown as a loading control. At 48 h post infiltration, HR triggers overall protein degradation (i.e., lower levels of Rubisco), while levels of TIR-GFP are unchanged.
The effect of GFP could be attributed to either stabilization of the fusion protein or its native ability to form weak dimers (Shaner et al., 2005). The dimerization property of GFP can be disrupted by a single amino acid substitution, A206K (Shaner et al., 2005), creating monomeric GFP (mGFP). We tested whether dimerization of GFP might contribute to the ability of TIR to elicit autoactive HR. We observed that the TIR fusion to mGFP was unable to elicit the response (Figure 7B). This finding suggests a requirement for TIR domain dimerization prior to triggering the HR. Protein gel blot analysis showed that the protein expression of the TIR-mGFP was slightly reduced compared with the wild-type GFP version (Figure 7E); therefore, we cannot exclude the possibility that the ability of the TIR to induce the effector-independent response depends on the relative protein stability. Additionally, we tested the E158A point mutation and observed that it abolished the ability of the TIR domain to trigger effector-independent HR (Figure 7C). Protein gel blot demonstrated that the E158A mutation did not compromise protein stability (Figure 7F); therefore, loss of function in this case is not due to reduced protein expression. Collectively, these data demonstrate that the TIR domain of RPP1 is capable of eliciting HR response independently of either the nucleotide binding ability of NBS or the effector binding ability of LRR.
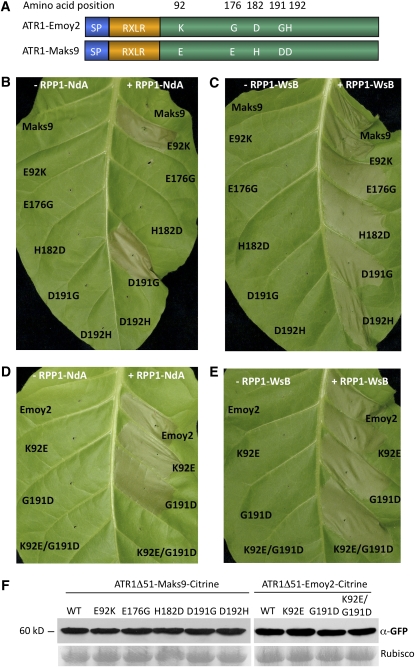
Single Amino Acid Changes in ATR1-Maks9 Result in Gain of Recognition by RPP1-NdA
ATR1 alleles Emoy2 and Maks9 differ by five amino acids, yet only ATR1-Emoy2 and not ATR1-Maks9 is recognized by RPP1-NdA (Figure 8A). We decided to evaluate the contribution of those five amino acids to the pathogen's ability to escape recognition by RPP1-NdA. We used site-directed mutagenesis to substitute each amino acid in ATR1-Maks9 to the corresponding residue in ATR1-Emoy2 and tested whether any of those single amino acid mutations could lead to recognition by RPP1-NdA. Two single substitutions, E92K and D191G, independently converted ATR1-Maks9 to a form fully recognized by RPP1-NdA, while the other three sites did not have any visible effect (Figure 8B). In the reciprocal experiment, when the K92E or G191D mutation was introduced into ATR1-Emoy2, either substitution delayed the recognition (the HR started to appear only at 48 h post infiltration) but did not reduce the intensity of HR once it started to appear. The double mutant K92E/G191D completely abolished the recognition of ATR1-Emoy2 by RPP1-NdA (Figure 8D).
Figure 8.
Two Amino Acids in ATR1-Emoy2 and ATR1-Maks9 Mediate Recognition Specificity by RPP1-NdA.
(A) ATR1-Emoy2 and ATR1-Maks9 protein sequences differ by five amino acid residues, depicted and numbered here.
(B) and (C) Either amino acid substitution E92K or D191G converts ATR1-Maks9 into a variant recognized by RPP1-NdA while not altering recognition by RPP1-WsB.
(D) and (E) K92E and G191D mutations in ATR1-Emoy2 collectively lead to loss of recognition by RPP1-NdA but not RPP1-WsB. Photographs were taken at 3 d post infiltration.
(F) Protein immunoblots probed with α-GFP antibody showing similar levels of protein expression for all ATR1-Maks9 and ATR1-Emoy2 variants. Rubisco is shown as a loading control. WT, Wild type.
Protein gel blot analysis showed that all of the ATR1 mutant variants produced protein amounts equal to the wild type (Figure 8F). Additionally, the mutations did not have any effect on the recognition by RPP1-WsB (Figures 8C and 8E), suggesting that additional sites can mediate interaction between ATR1 and RPP1-WsB. This genetic analysis suggests that it is unlikely that RPP1 monitors the enzymatic activity of ATR1, since several independent sites in ATR1 can activate recognition. It is interesting that the identified mutations involved charged residues, which can alter the charge of the surface area of the ATR1 molecule, influencing its ability to associate with a cognate R protein. Additional structural analyses will help us to understand the significance of those amino acid residues. This finding illustrates the narrow evolutionary line between recognition and susceptibility that puts effectors and R genes under diversifying selection; in this case, a pathogen can be only one amino acid away from being recognized by the plant.
DISCUSSION
Pathogen effector recognition by plant resistance proteins is one of the most important initial events required for a successful immune response, yet the molecular events underlying recognition specificity remain enigmatic. Our genetic and molecular analyses of the oomycete effector protein ATR1 and its cognate resistance protein RPP1 provide a mechanistic insight into the perception of an oomycete pathogen inside the plant cell. In this article, we have demonstrated that the LRR domain of the RPP1-WsB protein is able to associate with the ATR1 protein in planta. Interestingly, this association does not require functional TIR or NBS domains that function in RPP1 activation and downstream signaling, leading to activation of disease resistance responses. We also provide in planta data supporting the direct recognition of a pathogenic effector by a TIR-NBS-LRR–type resistance protein.
Evolutionary Histories of ATR1 and RPP1 Suggest Direct Recognition
Recognition mechanisms of pathogenic effector proteins by corresponding R proteins have been grouped into at least three general models. The first model proposes that recognition occurs though direct binding between the effector and cognate R protein, manifested in the “ligand and receptor” model. The direct recognition model is supported by yeast two-hybrid interaction studies between the flax R proteins L and M and their corresponding flax rust effectors AvrL and AvrM (Dodds et al., 2006; Catanzariti et al., 2010). Additional yeast two-hybrid and in vitro interactions have been demonstrated for tomato Resistance to Ralstonia solanacearum 1 and Ralstonia solanacearum Pseudomonas outer protein (Deslandes et al., 2003) and for rice Pi-ta and rice blast AvrPita (Jia et al., 2000).
The second model suggests that recognition is the result of indirect binding, in which an effector protein interacts with a host factor bound by the R protein. This mode of recognition has been supported by interactions between the Pseudomonas syringae effectors AvrPto/AvrPtoB, tomato kinase Pto, and the tomato NBS-LRR protein Pseudomonas resistance fenthion insensitivity (Prf; Mucyn et al., 2006). The Pto protein interacts directly with both the bacterial effectors and Prf, providing the bridging factor in effector recognition.
The third mode of effector recognition is also indirect, as recognition is achieved via detection of the enzymatic activity of the effector by the R protein. The most well-described examples of this type of recognition include the P. syringae effector AvrRpt2, a bacterial Cys protease that cleaves the Arabidopsis host factor RPM1 Interacting Protein 4 (RIN4). This cleavage leads to activation of the Arabidopsis CC-NBS-LRR protein RPS2 (Axtell et al., 2003; Axtell and Staskawicz, 2003). Similarly, the P. syringae effector AvrPphB, a Cys protease, cleaves the Arabidopsis protein AvrPphB susceptible 1 (Shao et al., 2003). Proteolytic activity of AvrPphB is indirectly detected by the Arabidopsis CC-NBS-LRR protein RPS5 (Ade et al., 2007), which leads to activation of resistance signaling. Additional examples of indirect enzymatic activation include Pseudomonas effectors AvrRpm1 and AvrB, which induce phosphorylation of the Arabidopsis RIN4 protein, leading to activation of the CC-NBS-LRR protein Resistance to Pseudomonas maculicola 1 (RPM1; Mackey et al., 2002).
It has been noted that the evolutionary history of the cognate effector/R gene pairs correlates with their mode of recognition. The high levels of diversifying selection in the flax rust effector alleles and corresponding flax R genes have been commonly explained by an arms race between the ligand and the receptor (Dodds et al., 2006; Jones and Dangl, 2006; Barrett et al., 2009). Indeed, this observation is consistent with the hypothesis that evolutionary selection pressure targets particular amino acid sites in the effector and the LRR portions of a resistance protein as a result of a direct interaction between the two proteins. On the other hand, effectors with enzymatic activity that induce modifications in the host targets are evolving under balancing or purifying selection (Van der Hoorn et al., 2002; Rohmer et al., 2004). Generally, it has been assumed that the R genes that have evolved to monitor host targets of the effectors are evolving more slowly and are less prone to duplications, since their function is to be stably associated with the invariant host proteins (Rohmer et al., 2004; Jones and Dangl, 2006).
The evolutionary history of ATR1 and RPP1 matches the direct recognition model. The ATR1 effector gene is evolving under very strong levels of diversifying selection (Rehmany et al., 2005). The RPP1 gene locus shows high levels of duplications, insertions/deletions, and polymorphisms, which create alleles with altered specificities, such as RPP1-WsB and RPP1-NdA. This observation is further supported by the closer examination of the interactions between the two RPP1 alleles WsB and NdA and the ATR1 alleles Emoy2 and Maks9. Since both RPP1-WsB and RPP1-NdA can recognize ATR1-Emoy2, we can conclude that both RPP1 alleles encode functional resistance gene products capable of eliciting the defense response. Similarly, since both ATR1-Emoy2 and ATR1-Maks9 are recognized by RPP1-WsB, this interaction reveals that both effector alleles are functional. However, only ATR1-Emoy2, but not ATR1-Maks9, is recognized by RPP1-NdA. Moreover, we have demonstrated that this difference in recognition can be overturned by two independent amino acid site substitutions in ATR1-Maks9. Given this evidence, we conclude that it is unlikely that RPP1 recognizes an enzymatic function of ATR1. This leaves the hypothesis of either recognition by direct binding or recognition by indirect binding, which requires additional host proteins. Using similar logic, we can conclude that if binding is indirect, there should be at least two different host targets of ATR1, guarded by RPP1-WsB and RPP1-NdA. We conclude that in light of the evolutionary history of ATR1 and RPP1, recognition through direct binding remains the most parsimonious model.
Activation of RPP1 Is Specified by in Planta Association with ATR1
The lack of published in planta data demonstrating that R proteins are able to associate with effectors inside the plant cell has led researchers to question the overall validity of the direct binding hypothesis. This lack of in planta data for R protein–effector association may be explained by difficulties using natural pathosystems in biochemical assays, including very low levels of protein expression and the low strength or transient nature of R protein–effector interactions. Here, we show that the pattern of in planta association between ATR1 and RPP1-WsB matches that of the effector recognition, suggesting that the ability of RPP1 to form a protein complex with ATR1 is required for triggering the hypersensitive cell death resistance response. At the same time, we are well aware that our in planta results do not prove direct protein–protein binding. Such a demonstration remains challenging due to our inability to purify soluble recombinant RPP1 protein or its LRR domain in the organisms commonly used for heterologous protein production, including Saccharomyces cerevisiae, Pichia pastoris, and Escherichia coli. It is possible that RPP1 may require additional plant-specific factors that provide protein stability, such as HSP90, SGT1, or RAR1, or yet unidentified factors. Thus, future research will include isolating the ATR1/RPP1 protein complex from Arabidopsis and characterizing other interacting protein complex members by mass spectrometry to elucidate the nature of the interaction between the R protein and its effector.
Roles of Different Domains of RPP1 in Effector Recognition and Signaling Responses
Although many R proteins share a common CC/TIR-NBS-LRR domain architecture, the actual biochemical role of each domain in effector recognition and downstream signaling has been subject to debate. Based on molecular genetic analyses, the LRR domain was originally proposed to mediate effector recognition; this is well supported by yeast two-hybrid studies and in vitro experiments (Deslandes et al., 2003; Dodds et al., 2006). However, recent in vivo data suggested the TIR domain of tobacco TIR-NBS-LRR protein N and the CC domain of the potato (Solanum tuberosum) CC-NBS-LRR protein Rx as the main regions involved in effector recognition (Burch-Smith et al., 2007; Rairdan et al., 2008). Despite the fact that these data contradicted previous studies of N, in which the NBS-LRR portion was shown to bind the cognate tobacco mosaic virus effector protein p50 both in vitro and in yeast (Ueda et al., 2006), a new hypothesis has emerged postulating that the N-terminal CC/TIR domain is the main determinant of effector–R protein interactions (Burch-Smith and Dinesh-Kumar, 2007; Collier and Moffett, 2009; Lukasik and Takken, 2009). Having established the in planta association between the RPP1-WsB and ATR1 proteins, we were able to identify which domain of RPP1 was responsible for effector recognition. Our deletion analysis showed that neither TIR nor TIR-NBS could associate with ATR1 in planta. On the other hand, the LRR domain was both necessary and sufficient for interaction with ATR1 in an allele-specific manner. Furthermore, we introduced single amino acid substitutions in the TIR domain and the P-loop of the NBS domain that abolished the ability of RPP1 to trigger effector-induced HR but did not disrupt the association with ATR1. This indicates that ATR1 can associate with RPP1 it its inactive state, which is consistent with the proposed model, in which R protein activation is “switched on” after perception of the effector (Takken et al., 2006; Lukasik and Takken, 2009). These data are also consistent with the step-wise model of R protein activation, in which the LRR domain associates with the effector before the activation of other domains and signaling is triggered (Collier and Moffett, 2009).
Although the LRR portion of RPP1 is sufficient for association with ATR1, it is not sufficient to induce the hypersensitive cell death immune response. Mutagenesis of the TIR and NBS domains of RPP1 revealed that they control the ability of RPP1 to activate the response after association with ATR1. Involvement of the TIR and NBS domains in signaling has been previously reported for the TIR-NBS-LRR proteins L10, RPS4, RPP1A, and N (Frost et al., 2004; Zhang et al., 2004; Weaver et al., 2006; Swiderski et al., 2009). The minimal region that was sufficient to induce the HR in those proteins included the TIR domain and the first 20 amino acids of the NBS domain (which does not include the P-loop motif; Frost et al., 2004; Swiderski et al., 2009). In this study, we report that the analogous region of RPP1-WsB (amino acids 1–266) was also able to elicit effector-independent HR when fused to the large epitope tag, GFP. Intrigued by the fact that fusion to GFP was required for the induction of the cell death, we examined whether the dimerizing property of GFP could contribute to the signaling of the TIR domain. Indeed, when we constructed a monomeric GFP and fused it to the TIR domain, the latter lost its ability to activate effector-independent cell death. These data suggest a hypothesis whereby the activation of the TIR domain is regulated by oligomerization of RPP1 upon perception of ATR1. Effector-induced oligomerization of R proteins has been proposed previously and has been supported by oligomerization of N, which was dependent on the presence of its cognate effector and the intact P-loop motif (Mestre and Baulcombe, 2006).
We do not attempt to propose that our data explain how all of the R proteins would be activated, nor that functions of the structurally similar domains would be exactly the same in different R proteins. Different modes of effector perception predict the existence of different modes of R protein activation. Indeed, if a protein is negatively regulated by another host factor, as in the case of Arabidopsis RPS2 and RIN4 (Axtell and Staskawicz, 2003), the initial mechanism of its activation might be different from a protein that is autoinhibited due to intramolecular interactions, as demonstrated for the potato R protein Rx (Moffett et al., 2002; Rairdan and Moffett, 2006). Herein, we provide the missing biochemical in planta data that support a particular simple model, in which effector recognition is mediated through the LRR domain while the TIR domain is involved in signaling.
RPP1 Activation Model
Given these data, we propose the following series of molecular events that lead to the activation of RPP1-WsB in response to ATR1. First, the pathogen infects the host and delivers ATR1 protein across the haustorial extracellular matrix into the plant cell. Recognition of ATR1 ultimately depends on which variant of ATR1 is delivered and which variant of RPP1 is present in the host. Our mutational analyses of ATR1 demonstrated that the pathogen could be just one amino acid away from being recognized. The LRR domain of RPP1 monitors for the presence of ATR1 inside the plant cell. Upon successful association between the LRR and the ATR1 variant, RPP1-WsB undergoes a conformational change, possibly oligomerization coupled to ATP binding by the NBS domain. This activates the TIR domain, which initiates a signal transduction cascade that leads to the HR and expression of plant disease resistance.
Several key questions will need to be resolved before we can claim full understanding of the RPP1–ATR1 interaction: (1) the composition of the protein complex they form; (2) the structural basis of effector recognition by the LRR domain; (3) the mode of TIR domain activation by LRR and NBS domains; and (4) the signaling cascade leading to the induction of the HR and plant disease resistance. The elucidation of these molecular mechanisms would be of great benefit to both plant and animal immunity and will ultimately allow researchers to rationally design broad-spectrum R genes for applications in agriculture.
METHODS
Multiple Sequence Alignment
Alignment of the amino acid sequences of Arabidopsis thaliana was done using the MUSCLE algorithm (Edgar, 2004) and visualized with belvu (Sonnhammer and Hollich, 2005). For phylogenetic analyses, we used an alignment between coding DNA sequences, in which codons were aligned corresponding to the amino acid sequence alignment using the pal2nal algorithm (Suyama et al., 2006).
Phylogenetic Analysis
Phylogenetic analysis was performed using the Phylip 3.66 software package (Felsenstein, 2005). The unrooted maximum likelihood tree was constructed from the nucleotide sequence alignment discussed above using the dnaml algorithm (Felsenstein and Churchill, 1996) with default parameters (see Supplemental Table 2 online). Bootstrapping was performed for 1000 replicates using the seqboot algorithm with the default parameters and the consense algorithm with the user-tree option (see Supplemental Table 2 online). The tree was visualized using the TreeView program (Page, 1996).
Strains and Growth Conditions
Escherichia coli DH5a and Agrobacterium tumefaciens GV3101 were grown in Luria–Bertani medium supplemented with the appropriate antibiotics at 37°C and 28°C, respectively. Bacterial DNA transformation was conducted using chemically competent E. coli (Invitrogen) and through freeze/thaw transformation of CaCl2-competent Agrobacterium (Wise et al., 2006). Tobacco (Nicotiana tabacum var Turk) plants were grown in a controlled growth chamber at 24°C with a 16-h-light/8-h-dark photoperiod before infiltrations and switched to 24 h of light after infiltrations.
Constructs
The sequences for all of the primers used in this study are shown in Supplemental Table 1 online in 5′ to 3′ orientation, restriction sites are indicated in boldface, the sequence encoding the HA tag is underlined, and the pENTR/D-TOPO targeting sequence CACC is shown in the forward primers. All point mutations were introduced by site-directed mutagenesis using the Quick-Change SDM kit (Stratagene), and primers are specified in Supplemental Table 1 online.
The open reading frame of RPP1-WsB was amplified via PCR using an Arabidopsis Ws-0 cDNA template. Cloning of the full-length RPP1-WsB gene was done in two fragments, taking advantage of the unique NdeI restriction site within the gene. The reverse primer incorporated an in-frame HA epitope tag 5′ to the stop codon in order to create a C-terminal fusion protein with RPP1-WsB. The PCR products were directly subcloned in pENTR/D-TOPO vector (Invitrogen). Two RPP1-WsB fragments were combined in pENTR/D-/TOPO with a NotI-NdeI digest. Deletion variants of RPP1 were amplified with the following primers: RPP1-WsB F/TIR R; RPP1-WsB F/NBS R; RPP1-WsB NBS F/Spe1-HA R; RPP1-WsB LRR F/Spe1-HA R. A SpeI site was incorporated in the reverse primers, which allowed creation of a C-terminal fusion with the HA or GFP epitope tags by restriction digest/ligation. For in planta analyses, all resulting RPP1-WsB constructs were introduced into pEarleyGate destination binary vector pEG201 (35S promoter, N-terminal HA tag fusion; Earley et al., 2006) using LR clonase (Invitrogen). Sequence information for RPP1-NdA was acquired from Gordon (2002). Unfortunately, we could not isolate any functional cDNA for RPP1-NdA. The full-length genomic sequence, including the native promoter and native terminator, was subcloned in three fragments using the following primers, RPP1-NdA fragment 1 F/R, fragment 2 F/R, fragment 3 F/R, and joined together by restriction digest/ligation. An XbaI restriction site was introduced at the 3′ of the RPP1-NdA open reading frame by site-directed mutagenesis. The sequence encoding for a 3xHA epitope tag was PCR-amplified from the pBJ36-3xVenus-3xHA vector (provided by Jeff Long, Salk Institute), column-purified, digested with SpeI/XbaI, and ligated into XbaI of RPP1-NdA. Resulting construct was introduced into pEG301 (no promoter) using LR clonase (Invitrogen).
All ATR1 variants were amplified from genomic DNA templates extracted from Hyaloperonospora arabidopsidis spores of the appropriate pathovar (provided by John McDowell, Thomas Eulgem, and Jonathan Jones). The ATR1 allelic variants and deletions were amplified by PCR with the primers indicated in Supplemental Table 1 online and subcloned in pENTR/D-TOPO vector (Invitrogen). A SpeI site was included in the reverse primers before the stop codon to facilitate epitope tagging. The Citrine gene was cloned on the 3′ end of all ATR1 alleles using SpeI digestion. The ATR1 constructs were introduced in the pEG103 (35S promoter, C-terminal GFP fusion) and pEG202 (35S promoter, N-terminal Flag tag fusion) binary destination vectors via LR recombination.
Agrobacterium-Mediated Transient Expression
Agrobacterium was grown in Luria–Bertani broth cultures (supplemented with 50 μg/mL gentamycin and 25 μg/mL kanamycin) overnight at 28°C with constant shaking. The cultures were spun down at a tabletop centrifuge at 10,000 rpm for 2 min. The resulting pellet was resuspended in 1 mL of induction medium (10 mM MgCl, 10 mM MES, and 150 μM acetosyringone, adjusted to pH 5.6 with KOH). Bacterial concentrations were measured and adjusted with induction medium to OD600 = 0.9. Resulting cultures were preinduced for 2 to 3 h at 28°C. For coinfiltrations, cultures carrying individual constructs were induced separately and mixed in a 1:1 ratio just before infiltration. Young tobacco leaves were inoculated with Agrobacterium cultures using a blunt syringe.
Transient Protein Expression in Tobacco
To detect transient protein expression in tobacco, two leaf discs (1.5 cm diameter) were collected 24 to 48 h post infiltration. The samples were frozen in liquid nitrogen and ground with a prechilled plastic pestle. Protein was extracted with 150 μL of the Laemmli buffer (0.24 M Tris-Cl, pH 6.8, 6% SDS, 30% glycerol, 16% β-mercaptoethanol, 0.006% bromophenol blue, and 5 M urea). Samples were boiled for 5 min and centrifuged at maximum speed for 10 min in a tabletop centrifuge at room temperature; supernatants were transferred into fresh tubes before analysis by SDS-PAGE and immunoblotting as described below.
Coimmunoprecipitations
All constructs were transiently expressed in tobacco using Agrobacterium-mediated transformation with strain GV3101. Leaf tissue was collected 24 h post infiltration, when the first visible signs of HR started to appear, weighed, and snap-frozen in liquid nitrogen. The weight of the tissue undergoing HR was estimated based on its surface area. We have routinely used ∼1 g of tissue for each coimmunoprecipitation experiment, which corresponds to about one youngest fully expanded leaf of 3- to 4-week-old tobacco. Each tissue sample was ground with mortar and pestle to a homogeneous powder in liquid nitrogen, transferred into a precooled mortar containing 2 mL of the protein extraction buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% Triton X-100, 0.2% Nonidet P-40, 6 mM β-mercaptoethanol, 0.3 μM aprotinin, 10 μM leupeptin, 1 μM pepstatin A, and a Complete protease inhibitor cocktail [Roche]), and homogenized with a fresh precooled pestle until the sample was completely thawed. Resulting samples were transferred to 1.5-mL Eppendorf tubes and centrifuged for 20 min, 14,000 rpm, and 4°C. The supernatant was split into two fresh Eppendorf tubes and used as input for the α-GFP and α-HA immunoprecipitations. All the steps in the immunoprecipitations were performed at 4°C.
In order to immunoprecipitate the target protein, either 2 μL of α-GFP (rabbit, polyclonal; Abcam) or 20 μL of α-HA (mouse, clone 16B12; Roche) was added to 1 mL of protein extract. The antibody–lysate mixture was incubated with gentle tumbling for 1.5 h at 4°C. Next, 50 μL of Protein G beads (Protein G Sepharose for Fast Flow; GE Healthcare), prewashed three times in the protein extraction buffer, was added to each sample and incubated with gentle tumbling for 4 h at 4°C. Then, the beads were spun down for 2 min at 1000g, washed three times with 1 mL of the protein extraction buffer, and supplemented with fresh protease inhibitors and fresh β-mercaptoethanol. The protein was eluted by boiling for 5 min in 50 μL of the Laemmli buffer. The samples, 5 μL per lane for detecting immunoprecipitated protein and 25 μL per lane for detecting coimmunoprecipitating protein, were separated on 10% SDS-PAGE gels or commercial NuPAGE SDS gradient 4% to 12% gels (Invitrogen), transferred to nitrocellulose membrane (Fisher), and analyzed by immunoblotting using mouse α-GFP (clone B34; Covance) and goat anti-mouse-horseradish peroxidase (Bio-Rad) or rat α-HA-horseradish peroxidase (clone 3F10; Roche). All coimmunoprecipitation experiments were performed at least three times from different leaf tissue samples and gave robust and repeatable results.
Accession Numbers
Sequence data that were used in this paper can be found in the National Center for Biotechnology Information databases under the following accession numbers: ATR1-Emoy2 (gi61660946), ATR1-Maks9 (gi61660952), ATR1-Emco5 (gi61660954), ATR1-Cala2 (gi61660958), ATR1-Emwa1 (gi61660960), RPP1-WsB (gi3860164). The genomic sequence of RPP1-NdA was cloned based on Gordon (2002) and deposited in the GenBank database under accession number HM209027.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Multiple Sequence Alignment of the ATR1 Protein Sequences Used in This Study.
Supplemental Figure 2. Multiple Sequence Alignment of the Protein Sequences Coding for RPP1-WsB and RPP1-NdA.
Supplemental Figure 3. Race-Specific Recognition of ATR1 Alleles by the Genomic RPP1-WsB Construct Expressed under the Control of Its Native Promoter.
Supplemental Figure 4. Localization of Different ATR1 Alleles in Tobacco.
Supplemental Figure 5. Additional Coimmunoprecipitation Experiments Showing Interactions between NBS-LRR and LRR Domains of RPP1-WsB and Different ATR1 Variants.
Supplemental Figure 6. Multiple Sequence Alignment of the TIR Domain and P-Loop Motif in Different R Proteins and the TIR Domain of Human TLR1.
Supplemental Table 1. List of Primers Used in This Study for Gene Amplification and Site-Directed Mutagenesis.
Supplemental Table 2. Set of Parameters Used in dnaml, seqboot, and consense Algorithms for Constructing the Phylogenetic Tree of the ATR1 Alleles.
Supplemental Data Set 1. Text File of the Alignment Used for the Phylogenetic Analysis Shown in Figure 1.
Acknowledgments
We are grateful to Jim Beynon (University of Warwick) for providing sequence information and plant material that facilitated making RPP1-NdA constructs; to John McDowell (Virginia Tech), Thomas Eulgem (University of California Riverside), and Jonathan Jones (Sainsbury Laboratory) for providing H. arabidopsidis inocula and DNA samples; and to Kimmen Sjölander (University of California Berkeley) for providing access to the belvu program used in visualizing multiple sequence alignments. We also thank Daniil Prigozhin (University of California Berkeley), Sandra Goritschnig (University of California Berkeley), Peter Dodds (Commonwealth Scientific and Industrial Research Organisation), Lauriebeth Leonelli (University of California Berkeley), and Baomin Zhang (University of California Berkeley) for helpful discussion of the presented material and critical reading of the manuscript. This work was supported by the National Science Foundation (grant NSF 2010 0726229).
References
- Ade J., DeYoung B.J., Golstein C., Innes R.W. (2007). Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc. Natl. Acad. Sci. USA 104: 2531–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alder M.N., Rogozin I.B., Iyer L.M., Glazko G.V., Cooper M.D., Pancer Z. (2005). Diversity and function of adaptive immune receptors in a jawless vertebrate. Science 310: 1970–1973 [DOI] [PubMed] [Google Scholar]
- Allen R.L., Bittner-Eddy P.D., Grenville-Briggs L.J., Meitz J.C., Rehmany A.P., Rose L.E., Beynon J.L. (2004). Host-parasite coevolutionary conflict between Arabidopsis and downy mildew. Science 306: 1957–1960 [DOI] [PubMed] [Google Scholar]
- Axtell M.J., Chisholm S.T., Dahlbeck D., Staskawicz B.J. (2003). Genetic and molecular evidence that the Pseudomonas syringae type III effector protein AvrRpt2 is a cysteine protease. Mol. Microbiol. 49: 1537–1546 [DOI] [PubMed] [Google Scholar]
- Axtell M.J., Staskawicz B.J. (2003). Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112: 369–377 [DOI] [PubMed] [Google Scholar]
- Barrett L., Thrall P., Dodds P., Van Der Merwe M., Linde C., Lawrence G., Burdon J. (2009). Diversity and evolution of effector loci in natural populations of the plant pathogen Melampsora lini. Mol. Biol. Evol. 26: 2499–2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch P., Rehmany A., Pritchard L., Kamoun S., Beynon J. (2006). Trafficking arms: Oomycete effectors enter host plant cells. Trends Microbiol. 14: 8–11 [DOI] [PubMed] [Google Scholar]
- Bomblies K., Lempe J., Epple P., Warthmann N., Lanz C., Dangl J., Weigel D. (2007). Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol. 5: e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botella M.A., Parker J.E., Frost L.N., Bittner-Eddy P.D., Beynon J.L., Daniels M.J., Holub E.B., Jones J.D. (1998). Three genes of the Arabidopsis RPP1 complex resistance locus recognize distinct Peronospora parasitica avirulence determinants. Plant Cell 10: 1847–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch-Smith T., Dinesh-Kumar S. (2007). The functions of plant TIR domains. Sci. STKE 401: pe46. [DOI] [PubMed] [Google Scholar]
- Burch-Smith T., Schiff M., Caplan J., Tsao J., Czymmek K., Dinesh-Kumar S. (2007). A novel role for the TIR domain in association with pathogen-derived elicitors. PLoS Biol. 5: e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catanzariti A.M., Dodds P.N., Ve T., Kobe B., Ellis J.G., Staskawicz B.J. (2010). The AvrM effector from flax rust has a structured C-terminal domain and interacts directly with the M resistance protein. Mol. Plant Microbe Interact. 23: 49–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm S., Coaker G., Day B., Staskawicz B. (2006). Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 124: 803–814 [DOI] [PubMed] [Google Scholar]
- Collier S.M., Moffett P. (2009). NB-LRRs work a “bait and switch” on pathogens. Trends Plant Sci. 14: 521–529 [DOI] [PubMed] [Google Scholar]
- Deslandes L., Olivier J., Peeters N., Feng D.X., Khounlotham M., Boucher C., Somssich I., Genin S., Marco Y. (2003). Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl. Acad. Sci. USA 100: 8024–8029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desveaux D., Singer A., Dangl J. (2006). Type III effector proteins: Doppelgangers of bacterial virulence. Curr. Opin. Plant Biol. 9: 376–382 [DOI] [PubMed] [Google Scholar]
- Dinesh-Kumar S.P., Tham W.H., Baker B.J. (2000). Structure-function analysis of the tobacco mosaic virus resistance gene N. Proc. Natl. Acad. Sci. USA 97: 14789–14794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds P., Lawrence G.J., Catanzariti A., Teh T., Wang C., Ayliffe M.A., Kobe B., Ellis J. (2006). Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl. Acad. Sci. USA 103: 8888–8893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds P.N., Lawrence G.J., Ellis J.G. (2001). Six amino acid changes confined to the leucine-rich repeat β-strand/β-turn motif determine the difference between the P and P2 rust resistance specificities in flax. Plant Cell 13: 163–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds P.N., Rafiqi M., Gan P.H., Hardham A.R., Jones D.A., Ellis J.G. (2009). Effectors of biotrophic fungi and oomycetes: Pathogenicity factors and triggers of host resistance. New Phytol. 183: 993–1000 [DOI] [PubMed] [Google Scholar]
- Earley K., Haag J., Pontes O., Opper K., Juehne T., Song K., Pikaard C. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J.G., Lawrence G.J., Luck J.E., Dodds P.N. (1999). Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell 11: 495–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. (2005). PHYLIP (Phylogeny Inference Package) version 3.6. http://evolution.genetics.washington.edu/phylip.html [Google Scholar]
- Felsenstein J., Churchill G.A. (1996). A hidden Markov model approach to variation among sites in rate of evolution. Mol. Biol. Evol. 13: 93–104 [DOI] [PubMed] [Google Scholar]
- Frost D., Way H., Howles P., Luck J., Manners J., Hardham A., Finnegan J., Ellis J. (2004). Tobacco transgenic for the flax rust resistance gene L expresses allele-specific activation of defense responses. Mol. Plant Microbe Interact. 17: 224–232 [DOI] [PubMed] [Google Scholar]
- Gordon A. (2002). Analysis of the RPP1 Resistance Gene Cluster in Arabidopsis Accession Niederzenz (Nd-1). PhD dissertation (Birmingham, UK: University of Birmingham; ). [Google Scholar]
- Jia Y., McAdams S.A., Bryan G.T., Hershey H.P., Valent B. (2000). Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19: 4004–4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J., Dangl J. (2006). The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kamoun S. (2006). A catalogue of the effector secretome of plant pathogenic oomycetes. Annu. Rev. Phytopathol. 44: 41–60 [DOI] [PubMed] [Google Scholar]
- Kim H.E., Du F., Fang M., Wang X. (2005). Formation of apoptosome is initiated by cytochrome c–induced dATP hydrolysis and subsequent nucleotide exchange on Apaf-1. Proc. Natl. Acad. Sci. USA 102: 17545–17550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobe B., Deisenhofer J. (1993). Crystal structure of porcine ribonuclease inhibitor, a protein with leucine-rich repeats. Nature 366: 751–756 [DOI] [PubMed] [Google Scholar]
- Lukasik E., Takken F.L. (2009). STANDing strong, resistance proteins instigators of plant defence. Curr. Opin. Plant Biol. 12: 427–436 [DOI] [PubMed] [Google Scholar]
- Mackey D., Holt B.F., III, Wiig A., Dangl J.L. (2002). RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108: 743–754 [DOI] [PubMed] [Google Scholar]
- Mestre P., Baulcombe D.C. (2006). Elicitor-mediated oligomerization of the tobacco N disease resistance protein. Plant Cell 18: 491–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffett P., Farnham G., Peart J., Baulcombe D.C. (2002). Interaction between domains of a plant NBS−LRR protein in disease resistance-related cell death. EMBO J. 17: 4511–4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondragon-Palomino M. (2002). Patterns of positive selection in the complete NBS-LRR gene family of Arabidopsis thaliana. Genome Res. 12: 1305–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucyn T., Clemente A., Andriotis V., Balmuth A., Oldroyd G., Staskawicz B., Rathjen J. (2006). The tomato NBARC-LRR protein Prf interacts with Pto kinase in vivo to regulate specific plant immunity. Plant Cell 18: 2792–2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page R.D. (1996). TreeView: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12: 357–358 [DOI] [PubMed] [Google Scholar]
- Rairdan G., Moffett P. (2006). Distinct domains in the ARC region of the potato resistance protein Rx mediate LRR binding and inhibition of activation. Plant Cell 18: 2082–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rairdan G.J., Collier S.M., Sacco M.A., Baldwin T.T., Boettrich T., Moffett P. (2008). The coiled-coil and nucleotide binding domains of the potato Rx disease resistance protein function in pathogen recognition and signaling. Plant Cell 20: 739–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmany A., Gordon A., Rose L.E., Allen R.L., Armstrong M.R., Whisson S.C., Kamoun S., Tyler B.M., Birch P.R.J., Beynon J.L. (2005). Differential recognition of highly divergent downy mildew avirulence gene alleles by RPP1 resistance genes from two Arabidopsis lines. Plant Cell 17: 1839–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentel M.C., Leonelli L., Dahlbeck D., Zhao B., Staskawicz B.J. (2008). Recognition of the Hyaloperonospora parasitica effector ATR13 triggers resistance against oomycete, bacterial, and viral pathogens. Proc. Natl. Acad. Sci. USA 105: 1091–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmer L., Guttman D.S., Dangl J.L. (2004). Diverse evolutionary mechanisms shape the type III effector virulence factor repertoire in the plant pathogen Pseudomonas syringae. Genetics 167: 1341–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N.C., Steinbach P.A., Tsien R.Y. (2005). A guide to choosing fluorescent proteins. Nat. Methods 2: 905–909 [DOI] [PubMed] [Google Scholar]
- Shao F., Golstein C., Ade J., Stoutemyer M., Dixon J.E., Innes R.W. (2003). Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science 301: 1230–1233 [DOI] [PubMed] [Google Scholar]
- Shen Q., Saijo Y., Mauch S., Biskup C., Bieri S., Keller B., Seki H., Ulker B., Somssich I., Schulze-Lefert P. (2007). Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315: 1098–1103 [DOI] [PubMed] [Google Scholar]
- Sohn K.H., Lei R., Nemri A., Jones J.D. (2007). The downy mildew effector proteins ATR1 and ATR13 promote disease susceptibility in Arabidopsis thaliana. Plant Cell 19: 4077–4090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer E.L., Hollich V. (2005). Scoredist: A simple and robust protein sequence distance estimator. BMC Bioinformatics 6: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M., Torrents D., Bork P. (2006). PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34: 609–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiderski M., Birker D., Jones J. (2009). The TIR domain of TIR-NB-LRR resistance proteins is a signaling domain involved in cell death induction. Mol. Plant Microbe Interact. 22: 157–165 [DOI] [PubMed] [Google Scholar]
- Takken F., Albrecht M., Tameling W. (2006). Resistance proteins: Molecular switches of plant defence. Curr. Opin. Plant Biol. 9: 383–390 [DOI] [PubMed] [Google Scholar]
- Tameling W.I., Elzinga S.D., Darmin P.S., Vossen J.H., Takken F.L., Haring M.A., Cornelissen B.J. (2002). The tomato R gene products I-2 and MI-1 are functional ATP binding proteins with ATPase activity. Plant Cell 14: 2929–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinchieri G., Sher A. (2007). Cooperation of Toll-like receptor signals in innate immune defence. Nat. Rev. Immunol. 7: 179–190 [DOI] [PubMed] [Google Scholar]
- Ueda H., Yamaguchi Y., Sano H. (2006). Direct interaction between the tobacco mosaic virus helicase domain and the ATP-bound resistance protein, N factor during the hypersensitive response in tobacco plants. Plant Mol. Biol. 61: 31–45 [DOI] [PubMed] [Google Scholar]
- Van der Hoorn R.A., De Wit P.J., Joosten M.H. (2002). Balancing selection favors guarding resistance proteins. Trends Plant Sci. 7: 67–71 [DOI] [PubMed] [Google Scholar]
- Weaver M.L., Swiderski M., Li Y., Jones J. (2006). The Arabidopsis thaliana TIR-NB-LRR R-protein, RPP1A: Protein localization and constitutive activation of defence by truncated alleles in tobacco and Arabidopsis. Plant J. 47: 829–840 [DOI] [PubMed] [Google Scholar]
- Win J., Morgan W., Bos J., Krasileva K.V., Cano L.M., Chaparro-Garcia A., Ammar R., Staskawicz B.J., Kamoun S. (2007). Adaptive evolution has targeted the C-terminal domain of the RXLR effectors of plant pathogenic oomycetes. Plant Cell 19: 2349–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirthmueller L., Zhang Y., Jones J., Parker J. (2007). Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr. Biol. 17: 2023–2029 [DOI] [PubMed] [Google Scholar]
- Wise A.A., Liu Z., Binns A.N. (2006). Three methods for the introduction of foreign DNA into Agrobacterium. Agrobacterium Protocols, 2nd ed, Wang K., ed (Totowa, NJ: Humana Press Inc; ), pp. 43–54 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Dorey S., Swiderski M., Jones J.D. (2004). Expression of RPS4 in tobacco induces an AvrRps4-independent HR that requires EDS1, SGT1 and HSP90. Plant J. 40: 213–224 [DOI] [PubMed] [Google Scholar]