Abstract
Retinoschisin (RS1) is a retina-specific secreted protein encoding a conserved discoidin domain sequence. As an adhesion molecule, RS1 preserves the retinal cell architecture and promotes visual signal transduction. In young males, loss-of-function mutations in the X- linked retinoschisis gene (RS1) cause X-linked retinoschisis, a form of progressive blindness. Neither the structure of RS1 nor the nature of its anchoring and organization on the plasma membranes is fully understood. The discoidin C2 domains of coagulation factors V and VIII are known to interact with extracellular phosphatidylserine (PS). In this study we have used atomic force microscopy (AFM) to study the interactions of murine retinoschisin (Rs1) with supported anionic lipid bilayers in the presence of Ca2+. The bilayers consisting of a single lipid, PS, and mixtures of lipids with or without PS were used. Consistent with previous X-ray diffraction studies, AFM imaging showed two distinct domains in pure PS bilayers when Ca2+ was present. Upon Rs1 adsorption, these PS and PS-containing mixed bilayers underwent fast and extensive reorganization. Protein localization was ascertained by immunolabeling. AFM imaging showed the Rs1 antibody bound exclusively to the calcium-rich ordered phase of the bilayers pointing to the sequestration of Rs1 within those domains. This was further supported by the increased mechanical strength of these domains after Rs1 binding. Besides, changes in bilayer thickness suggested that Rs1 was partially embedded into the bilayer. These findings support a model whereby the Rs1 protein binds to PS in the retinal cell plasma membranes in a calcium-dependent manner.
Retinoschisin (RS1) is a cell-surface protein expressed exclusively in the retina and pineal gland (1, 2). In the retina, RS1 functions as an adhesion molecule to preserve the structural organization of the retinal cell layers, a critical factor for promoting visual signal transduction. Loss-of-function mutations in the X-linked retinoschisis gene (RS1) lead to a form of early-onset macular degeneration in young males known as X-linked retinoschisis (XLRS) (3–5). XLRS disease is characterized by schisis (splitting) of the central retina between light sensing photoreceptor outer-cell layers and light processing inner-cell layers with the appearance of cystic cavities in the macular region, which leads to impaired visual-signal processing and progressive vision loss with age. When retinal function is assessed by clinical electroretinogram (ERG), XLRS patients display markedly reduced inner retinal b-wave amplitudes relative to the photoreceptor a-wave, which implicates abnormal synaptic signaling or neuronal processing. There is no cure for XLRS; however, RS1 gene-transfer studies in preclinical mouse models of XLRS have shown therapeutic promise (6–8).
RS1 encodes two functional sites of conserved sequence motifs, the N-terminus signal sequence (aa 1–21/23) (9, 10) and a long and evolutionarily-conserved sequence motif termed the discoidin domain (DS; aa 64–219) implicated in cell adhesion and regulatory signaling processes (11, 12). The signal sequence guides protein translocation across the endoplasmic reticulum (ER) and, following secretion, RS1 associates with the outer leaflet of the plasma membrane of photoreceptor and bipolar cells within the retina (1, 13, 14). The clinical pathology and evidence in mouse models of XLRS suggest that RS1 mediates its function through DS-dependent adhesive mechanism. In addition to these two functional domains, the mature protein has an N-terminal, 39 amino-acid RS1 domain of unknown function (10). X-ray crystal structures or nuclear magnetic resonance (NMR) structures of several other proteins encoding the DS are currently known, but not that of RS1 (15, 16).
The biochemical nature of RS1 organization on the membrane is not precisely known. DS domain-containing proteins are known to bind phospholipids, carbohydrates, and other proteins (11, 12, 17). For example, coagulation factors V and VIII bind phosphatidylserine (PS), while various types of collagens have been identified as potential ligands for the two mammalian discoidin-domain receptor tyrosine kinases, DDR1 and DDR2. Proteins containing structures similar to discoidin domain (e.g., the C2 domain of protein kinase C), are also known to bind anionic lipids (16). The nature of the ligands that interact with RS1 is not precisely known. Protein-lipid overlay assay experiments point to direct binding of RS1, specifically to PS in the presence of calcium (Ca2+) (13). RS1 was also shown to associate with the complex formed between Na+/K+ ATPase and the sterile alpha and TIR motif-containing protein, SARM1(18). In the chicken retina, L-type voltage-gated calcium channel (LVGCC) has been identified as a potential RS1 ligand (19).
Recent studies have discussed the shortcomings of co-immunoprecipitation (co-IPs) and pulldown assays used to identify protein interactions and have argued for the application of biophysical techniques to directly identify and validate protein interactions (20). Here, we use atomic force microscope (AFM) to directly visualize and probe model anionic lipid bilayers and their possible interactions with murine retinoschisin (Rs1). AFM provides real-time monitoring of temporal changes in the lipid bilayers under physiological conditions before and after the introduction of the potential ligands (21–23). It has allowed the visualization of charged lipid domains as well as the lipid-protein interactions such as electrostatic binding of synapsin to anionic PS-rich domains (24) and synaptotagmin-induced phospholipid bilayer perturbation (25). Furthermore, AFM can assess the mechanical properties of bilayers undergoing changes as a result of interactions with the associated proteins. We examined the behavior of supported lipid bilayers containing PS under the influence of Rs1 in the presence and absence of calcium cations. The results show that Rs1 induces a rather fast and extensive reorganization of the PS-containing bilayer structure in the presence of Ca2+. Rs1 was at least partially embedded in the bilayer, as we observe minimal alteration of thickness of the bilayers. The changes in the bilayer were further confirmed by the decreased fluidity of the regions of the bilayer where Rs1 was adsorbed. The localization of Rs1 within the lipid was ascertained by visualizing the binding of Rs1 antibodies to the bilayer containing the adsorbed Rs1. The interactions of Rs1 with mixed bilayers were more complex but consistent with those observed with PS alone.
Materials and Methods
Construction and expression of recombinant mouse Rs1 cDNA clone
Construction of recombinant mouse Rs1 cDNA clone for protein expression was performed using Bac-to-Bac Baculovirus expression kit reagents (Invitrogen, Carlsbad, CA). The mouse Rs1 cDNA cloned into pFastBac™HT A vector in tandem with a six-histidine tag was a gift provided by Yong Zeng (NIDCD/NIH). The Rs1DNA-recombinant pFastBac™HT A was purified and used to transform DH10 Bac E. Coli cells by heat shock for the site specific transposition of the Rs1 DNA from pFastBacHTa to bacmid DNA which is contained in DH10 Bac E. coli cells. The transformed cells were plated onto LB agar containing kanamycin (50 μg/ml), gentamycin (7 μg/ml), tetracycline (10 μg/ml), Bluo-gal (100 μg/ml), and IPTG (40 μg/ml). The white colonies were selected and DNA was isolated by alkaline lysis using the S.N.A.P. MidiPrep Kit (Invitrogen). Rs1 DNA in bacmid was identified by PCR using M-13 forward and reverse priming sites. The recombinant Rs1 DNA was also confirmed by using Rs1 cDNA specific primers. The Rs1-DNA recombinant bacmid DNA was transfected into Sf9 cells using cellfectin transfection reagent (Invitrogen), and the cells were cultured for 72 h at 27°C. The clarified supernatant was collected as small scale low titer P1 viral stock. This was further amplified to produce pFastBac-Rs1 baculoviral stock with a titer of 1×108 pfu/ml. This high-titer viral stock was used to infect Sf9 cells for expression of recombinant Rs1.
Typically, Sf9 cells were maintained on an orbital shaker in 250-ml polycarbonate Erlenmeyer flasks (Corning) with 50-ml working volume at 27°C. Sf/900 II SFM medium (Invitrogen) was used for culture maintenance and baculovirus infection. Cells were infected with viral stock at mid-logarithmic phase of growth at 4×106 cells/ml. Briefly, Sf9 cells were spun down and resuspended in fresh medium (one-fifth of final volume). An appropriate volume of recombinant Rs1 baculovirus was then added to achieve a multiplicity of infection (MOI) of 1. After 1 h in reduced volume with periodic agitation, fresh medium was added and the flasks were transferred to the orbital shaker (agitation rate, 100 rpm). 10 μM leupeptin was added 24 h post-infection to reduce proteolysis of the expressed protein in both the membrane fraction and the supernatant. The media and the cells were harvested at 72 h post-infection. The recombinant Rs1 protein in the media was detected by immunoblot analysis with rabbit polyclonal anti-Rs1 antibody (6). The media was further centrifuged at 10,000×g for 15 min and dialyzed against a buffer containing 50 mM Na2HPO4, 300 mM NaCl pH 7.4. The media containing the soluble recombinant proteins was loaded onto His Pur™-Cobalt resin (Thermo Fisher Scientific, Rockford, IL) column. The resin bound target proteins were washed with 3five-bed volumes of wash buffer (50 mM Na2HPO4, 300 mM NaCl, 10 mM imidazole pH 7.4), to elute nonspecific bound contaminants. 6X-His–Rs1 protein was eluted from the His Pur Cobalt resin with elute buffer (50 mM Na2HPO4, 250 mM NaCl and 250 mM imidazole, pH 7.4) and collected in 1.0-ml fractions. The collected fractions were examined via SDS-PAGE, Western blotting with Rs1 antibody and BCA protein assay (Pierce) to identify the 6X-His–Rs1 protein and determine concentrations. The Rs1 positive fractions were polled and reloaded on a fresh pre-washed His Pur™-Cobalt column and eluted from the His Pur-Cobalt resin with elute buffer in 1-ml linear gradients. Only the fractions that contained protein of highest purity were pooled. The His-tag was removed by TEV protease (Invitrogen) digestion according to the manufacturer’s protocols. SDS-PAGE profile of Rs1, LC-MS/MS analysis and the MS/MS spectra of the peptides corresponding to the sequence of Rs1 are provided in the Supporting Information I.
Supported Lipid Bilayer Preparation
The lipids(1-Palmitoyl-2-Oleoyl-sn-Glycero-3-[Phospho-L-Serine] (PS); (1-Palmitoyl-2-Oleoyl-sn-Glycero-3-Phosphocholine (PC); 1-Palmitoyl-2-Oleoyl-sn-Glycero-3-Phosphoethanolamine (PE), and 1,2-Dioleoyl-sn-Glycero-3-Phosphoinositol (PI) were purchased from Avanti Polar Lipids (Alabaster, AL) and cholesterol (Chol) from Sigma-Aldrich (St. Louis, MO).
For the supported lipid bilayers, PS lipid, PS-containing lipid mixtures (with PC, PE, PI and Chol), and other mixtures not containing PS (PC alone and PC with PE, PI and Chol) were examined. The lipids in chloroform stock were mixed as appropriate and the chloroform was evaporated under a stream of nitrogen gas. The lipids were rehydrated and vortexed in 20 mM HEPES, 150 mM NaCl, pH 7.5 buffer to give a total lipid concentration of 1 mg/ml. Lipid suspensions in the appropriate buffers were extruded (mini-extruder by Avanti Polar Lipids) through 100 nm polycarbonate filters (Whatman, Piscataway, NJ) to produce unilamellar vesicles. Typically, 10 μL of 0.4 mg/ml (0.5 mM in case of pure PS) lipid concentration suspensions were deposited onto freshly cleaved mica. Electrostatic forces cause the vesicles to burst and spread on the mica. After deposition, the solutions were incubated on mica for 15 min, and washed with the buffer to remove unbound and unbroken vesicles.
Atomic Force Microscopy
AFM imaging was performed under the same buffer with commercial AFM instruments (either Multimode or Bioscope with NS-V controllers, Veeco, Santa Barbara, CA). With the Multimode, a flow-through fluid cell design allowed us to inject the Rs1-protein solutions or change the buffer conditions while imaging continuously. Real-time temporal frame sequences were obtained in each experiment. Nanoindentation experiments were performed to assess the fluidity of the lipid bilayer by measuring the force required to penetrate through the bilayer to the mica substrate. Nanometer resolution mapping of the fluidity was obtained by indenting a dense array of points across the supported bilayer. In all cases, sharpened silicon nitride probes (SiNi by Budget Sensors, Bulgaria) were used with nominal stiffness of 0.06 N/m and 0.27 N/m. Both contact and non-contact AFM imaging were performed. The typical scan rate was 1 Hz, and the image resolution was ~0.6–6 nm/pixel, depending on the desired level of detail and the required image acquisition speed. The AFM images were preprocessed using the built-in instrument software and were further processed with the NIH ImageJ (ver. 1.38) software. Custom Matlab software (Mathworks, Natick, MA) was written to automatically detect and measure the bilayer break-through force in the nanoindentation experiments.
Results
1. Pure PS lipid bilayer: Adsorption of Rs1 in the presence of Ca2+ leads to phase reorganization
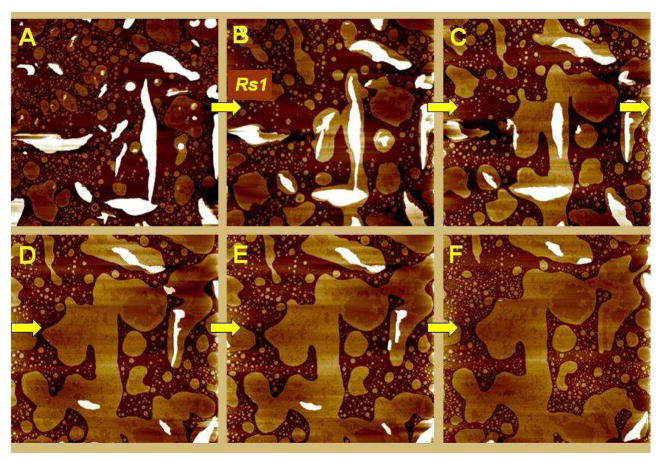
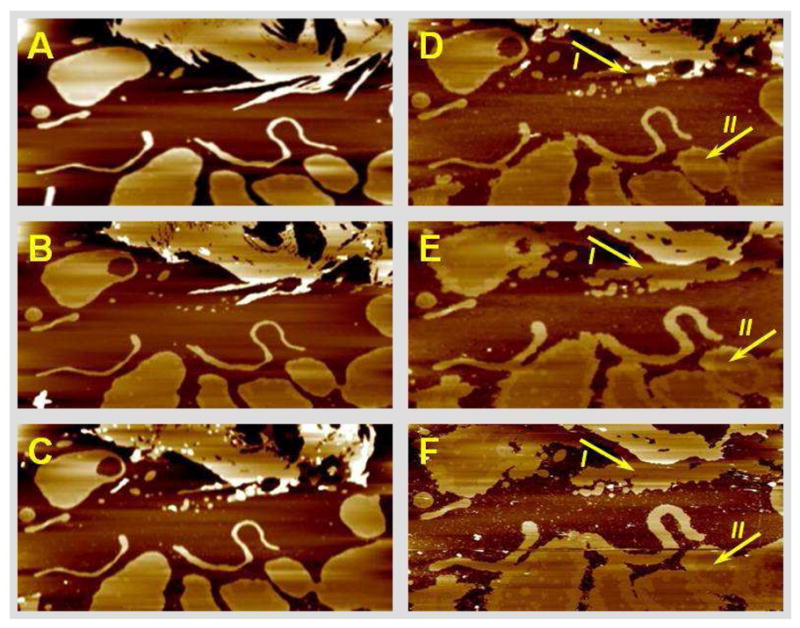
When the lipid preparation buffer included 0.5 mM Ca2+, pure PS vesicles formed patches of varying shapes and sizes in lieu of a continuous bilayer, even after long incubation times (over 1 hr). Two clearly defined domains (phases) were observed within these lipid patches. Once equilibrated on the mica, the bilayer patches were very stable, and the two domains remained robust and unchanged under scanning for many hours (Fig. 1A). As shown in Fig. 1A and the line section profile in Fig. 1C, the two phases had a height difference of 7 ± 1 Å. In addition, we frequently observed smaller lipid patches overlying larger ones (white circular patches in Fig. 1A and multilayered patches in Fig. 2A), and those stacked bilayers also exhibited phase domains similar to those on the mica substrate (basal patches). The existence of distinct phases in pure PS bilayers in the presence of calcium cations was first observed by X-ray diffraction and NMR studies (26, 27). However, this is the first time such phases are directly visualized and their different properties probed at the nanometer scale. Rapid and extensive changes were observed when Rs1 protein was introduced into PS bilayer in the presence of Ca2+. Initially there was a brief period of bilayer disruption, as evidenced by the difficulty of stable imaging right after the introduction of the protein. Stable imaging was re-established within a few minutes and the bilayer was observed undergoing extensive structural reorganization. Typically, the lower-fluid phase of the bilayer patches expanded and induced patches to confluence (Fig. 1B–D). Simultaneously, the second level bilayers overlying those directly on the mica substrate (basal bilayer) gradually dissolved, probably providing some of the material for the expansion of the patches on the mica substrate. The thicker, ordered phase domains exhibited less change during this reorganization process. The two phases kept shifting and reshaping until a new equilibrium was established. Until then, the lipids moved between the phases which was primarily seen as an expansion of the fluid phase. This movement could not have been caused by the scanning forces as the bilayer configuration eventually reached a final state, remaining unchanged even after prolonged scanning. These effects are seen best in the image animations in the Supporting Information II (Movie 1). The difference in height between the two phases did not change appreciably. The ordered phase appeared to increase by only about 1.5–2 Å which is within the instrument noise. These observations suggest that the Rs1 protein, which caused the bilayer reorganization, was at least partially embedded in the bilayer. The progression of the reorganization under the influence of Rs1 is shown in Figs. 1 and 2. In Fig. 1 it is also noticed that the small PS patches overlying those attached to the mica substrate were quickly depleted after the injection of the protein. In Fig. 2 this action of Rs1 is more striking. Fig. 2A shows a region containing multiple levels, shapes and sizes of stacked bilayers before the introduction of the protein. The protein caused an orderly deconstruction of these multilayers over time (Fig. 2B–F) along with expansion of the basal bilayer and reorganization of its phase domain structure (see also Supporting Information II – Movie 2). Additional images showing the progression of this bilayer towards equilibrium just prior to Rs1 injection are shown in the Supporting Information III (Fig. SIII-A). The findings were replicated more than ten times with similar results and the speed of the bilayer expansion correlated directly with Rs1 concentration.
Fig. 1.
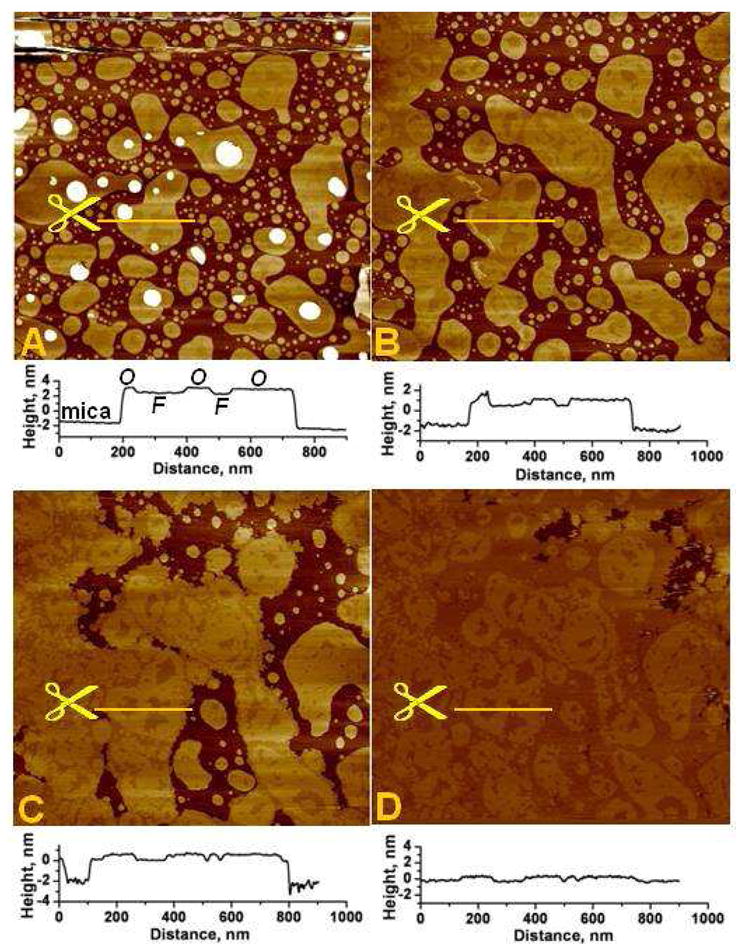
Supported PS bilayer undergoes phase reorganization upon addition of Rs1.
A. Typical PS bilayer patches prepared in 20 mM HEPES, 150 mM NaCl, 0.5 mM CaCl2 buffer. Following a brief period of equilibration, the patches and their phase configuration remained stable over several hours; shown here is the bilayer structure after about 2 hours of scanning. At the end of this period, the Rs1 solution was injected into the AFM fluid cell (four injections from initial concentration of 40 nM to the final concentration of 0.4 μM). The injection causes an imaging disturbance seen near the top part of the image which is scanned from bottom to top. White, circular, patches are “2nd level” bilayer patches overlying the bilayers attached to the mica (basal bilayers).
B – D. The same area 20 min, 2 and 2.5 hrs, respectively, after injection of Rs1; Note that the contours of the phases in image D repeat the contours of patches in image “B”. Images are 3×3 μm2.
Line plots under each image show section profiles along the lines drawn in yellow and indicated by the scissors in each corresponding image above. The sub-nm phase domain height differences are shown in these section profiles. The Ca2+-rich, ordered and the Ca2+-poor, fluid domains are respectively denoted by “O” and “F” on the line plot under image A. The total thickness of the bilayers is approximately 4nm.
Fig. 2.
Rs1 protein in the presence of Ca2+ reorganizes stacks of multiple bilayers in favor of a single, expanded, 2D bilayer. A. The top left image shows stacked bilayer patches in equilibrium before Rs1 injection. Up to five-level stacked patches are present. Rs1 was injected after image A was acquired. B–F. The same 3×3 μm2 region was imaged continuously after Rs1 injection. Notice the gradual depletion of overlying patches and the simultaneous expansion of the basal bilayer. Also noticeable is the progressive reorganization of the phase configuration on the basal bilayer.
2. PS lipid bilayer transitions with Rs1 protein require Ca2+
In the absence of calcium cations, PS formed continuous, uniform bilayers on the mica. The supported bilayer was topographically featureless and its presence was demonstrated by indenting the surface with the sharp AFM probe to observe bilayer penetration which appears as a discontinuity in the force-distance curve during probe approach. When Rs1 was introduced to pure PS bilayers, it adsorbed randomly and weakly as indicated by the ease with which the AFM probe could move or remove the adsorbed Rs1 particles (Supporting Information III, Fig. SIII-C).
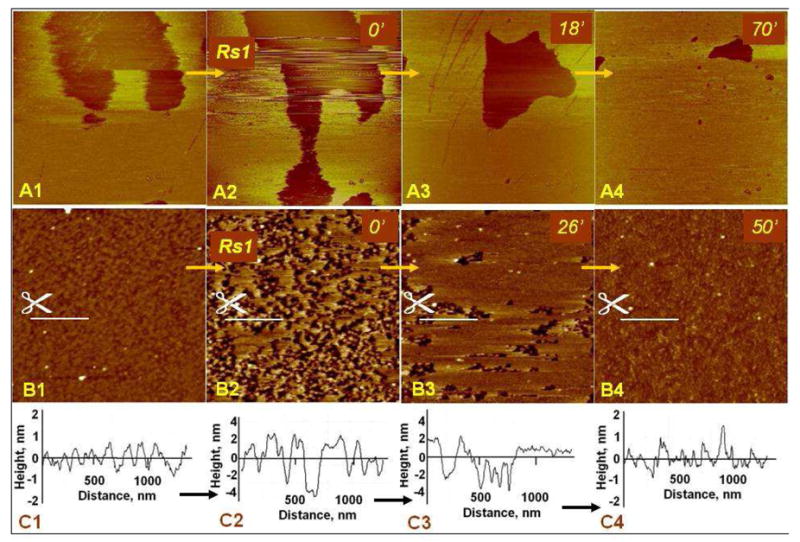
We subsequently examined the role of calcium cations in Rs1 binding starting again with a bilayer obtained from vesicles formed in 20 mM HEPES, 150 mM NaCl and 0.5 mM CaCl2. Once equilibrated on the AFM stage, these bilayers were washed several times with the same buffer minus CaCl2 to remove calcium from the bilayer. The bilayers remained stable except for a few of the fluid- phase domains that were desorbed (Fig. 3A). Some of the ordered phase domains were still present which possibly points to the strong binding of calcium to the PS lipid heads. Following the buffer wash, the equivalent of 300 nM Rs1 was introduced. The bilayers remained unperturbed during several scans and no Rs1 protein was seen on the surface of either lipid domain (Fig. 3B). Addition of another bolus of Rs1 solution similar to the first one (300 nM) did not result in any obvious surface adsorption. Only small patches of additional lipid bilayers appeared mainly as second level bilayers and sparse adsorption of protein on the mica substrate was seen (Fig. 3B, C), indicating that Rs1 remains largely in the solution. Addition of Ca2+, by 0.5 mM CaCl2 buffer exchange, caused the patches to gradually expand and the top layers dissolved as before (Fig. 3D–F). Here, addition of the calcium caused the expansion of the domains of ordered phase but, again, the fluid-phase domains also expanded as seen in the upper, right quadrant of Fig. 3F (pointed at by arrows).
Fig. 3.
Role of Ca2+ in the PS bilayer expansion under the influence of Rs1. A. Patches of PS bilayer were deposited onto mica from the buffer solution of 20 mM HEPES, 150 mM NaCl and 0.5 mM CaCl2, then washed with 20 mM HEPES-150 mM NaCl; image A was acquired after the wash. B. Same region imaged after Rs1 protein injection; C. Small changes observed following second Rs1 injection; D. CaCl2 injected to the final concentration of 0.5 mM, then additional Rs1 protein injected; E, F. Same region as in A–D imaged at two different times after CaCl2 injection. Notice the characteristic stacked bilayer depletion, the simultaneous expansion of the basal bilayer (arrows marked with I) and the confluence of separate patches into a continuous layer (arrows marked with II). The scan size: 6 × 2.8 μm2.
3. Lipid bilayers containing PS reorganize under the influence of Rs1
We explored the effect of including neutral and other anionic moieties in the lipid bilayers by forming mixtures of two lipids (PS:PC, 1:3), three lipids (PE:PS:PC, 5:3:2) and a lipid mixture mimicking in general a plasma/synaptic membrane composition (41% PC, 32% PE, 12% PS, 5% PI, 10% cholesterol) (28). Unlike pure PS lipid, these mixes tended to form continuous layers in the presence of Ca2+ (Fig. 4-A1). Phases were also present but were quite different for each of the mixtures. The two- and three-lipid mixes exhibited primarily one phase along with small patches of a second lower fluid-phase as seen in Fig. 4-A1 for the two-lipid case. Upon Rs1 injection, the bilayer was disrupted (Fig. 4-A2) followed by a gradual re-establishment of the continuous bilayer (Fig. 4-A3 and A4). The most visible interaction of Rs1 occurred with the five-component mixture in the presence of Ca2+. The bilayer exhibited a fractal-like phase structure that remained very stable while scanning for over an hour (Fig. 4-B1). Additional images showing the progression of the five-component supported bilayer towards equilibrium are shown in Supporting Information (Fig. SIII-B). Immediately upon the addition of Rs1 protein, the bilayer started undergoing structural changes. Initially, we observed fast and extensive disruption with holes appearing in a pattern similar to the phase structure of the bare bilayer (Fig. 4-B2), indicating that the protein preferentially interacted only with particular phase(s) of the lipid. The initial phase configuration had fine structure at scales too small to allow us to ascertain which part of the bilayer was exactly disrupted. The observed holes were of full thickness of the bilayer indicating selective transient desorption of a particular phase of the lipid (Fig. 4-B2). Subsequently, the holes gradually dissipated (‘healing’) (Fig. 4-B3) until the lipid bilayer reformed as a continuous sheet (Fig. 4-B4). The physical appearance of the final bilayer after Rs1 adsorption was different from the bare bilayer; while the fractal-like phase structure before Rs1 adsorption appeared as an undulation in height about a mean value (Fig. 4-C1), distinct protrusions or spikes were seen in the equilibrated structure after Rs1 adsorption (Fig. 4-C4). Quantitatively, the difference was seen as a ~50% increase in surface roughness and a significantly more skewed height distribution after Rs1 equilibration. There are probably protein molecules protruding from the bilayer, but the heights of the protrusions (1–1.5 nm) indicate that the protein is, at least partially, embedded into the bilayer. A sequence of images following Rs1 adsorption to the five-component lipid bilayer is shown as an animation in Supporting Information II (Movie 2).
Fig. 4.
The effect of Rs1 on mixed bilayers containing PS in the presence of calcium. A. Mix of PS:PC 1:3 in 20 mM HEPES, 150 mM NaCl, 0.5 mM CaCl2. A1 - Defects are seen in the bilayer before protein injection; A2 - Protein injection causes a fast bilayer disruption; A3 and A4 - The disruption was followed by gradual re-establishment of continuity (healing). B. Lipid mix mimicking synaptic membrane composition (41% PC, 32% PE, 12% PS, 5% PI, 10% cholesterol) in 20 mM HEPES, 150 mM NaCl, 0.5 mM CaCl2. B1 - A complex pattern of phases is seen in the equilibrated, multi-component mixed bilayer before Rs1 injection; B2 - Immediately after injection of Rs1 the bilayers were disrupted and full thickness holes appeared; B3 and B4 - The continuity of the bilayers was gradually restored via continuing Rs1 action. C. Line plots show the section profiles of the bilayers along the lines drawn in B. Notice the complex phase structure before protein injection (C1) and the changes in roughness after Rs1 injection. Primed numbers indicate time, in minutes, after Rs1 injection. All images are 3×3 μm2.
In the absence of Ca2+ the above five-component lipid mix also formed a continuous bilayer and exhibited complex phase structure. The addition of Rs1, however, caused only minor and temporary changes apparently because of very weak interaction. Images showing the progression of the lipid bilayer towards equilibrium before and after Rs1 protein introduction are shown in Supporting Information III (Fig. SIII-D).
We also performed a separate set of negative-control experiments to examine the possible interactions between Rs1 and bilayers not containing PS. Rs1 did not interact with pure PC bilayers, and only at very high concentrations of Rs1 did we observe random deposition of the protein onto the bilayers. This points to non-specific association of Rs1 with the neutrally charged PC. PC vesicle solution deposition resulted in continuous substrate coverage that was not disturbed by the introduction of the protein. A mixture of PC, PE, PI, and cholesterol (all synaptic membrane components minus PS) resulted in discontinuous bilayers with parts of the substrate exposed. The protein preferentially adsorbed to the exposed substrate rather than on top of such a bilayer (data not shown). Note that the presence of another anionic lipid, PI, did not favor Rs1 association, either in the presence or absence of Ca2+. Collectively, these results demonstrate that Rs1 specifically interacts with PS, the common element in all the lipid mixes that exhibited bilayer reorganization with the concurrent rearrangement of Rs1 within the lipid bilayer.
4. Rs1 binding and embedding enhances the local robustness of the lipid bilayer phase where it resides
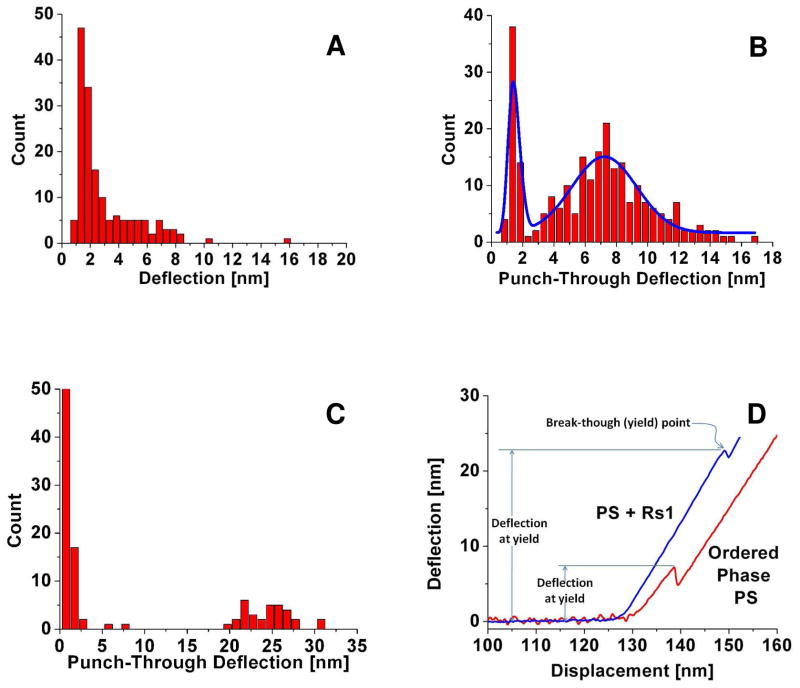
We measured bilayer yield strength by AFM nano-indentation tests on the supported bilayers. Using regular sharp probes we measured the punch-through forces required to penetrate the supported PS bilayers, before and after introducing Rs1 protein (Fig. 5). In the absence of Ca2+ and before Rs1 injection the punch-through deflections were tightly concentrated around 2 nm with an extended tail of higher deflections reaching approximately 8 nm (Fig. 5A). Addition of Rs1 protein did not affect these properties which is consistent with Rs1 not binding to PS in the absence of Ca2+. In the presence of calcium, two well-defined phases were observed. The punch-through deflection showed two distinct peaks (Fig. 5B), one corresponding to the peak observed in the absence of calcium, and a second, broader peak indicating a much less fluid bilayer with a punch-through deflection 5–6 times higher than the lower peak. Adding Rs1 protein changed this further. The lower peak, presumably that of the calcium-poor, fluid phase, remained unchanged, but the second peak shifted to a much higher deflection value than that of the ordered phase before protein addition (Fig. 5C). This indicates that adsorption of Rs1 protein onto the calcium-rich, ordered phase resulted in a significantly more robust bilayer structure. Conversely, the absence of Rs1 protein would result in increased membrane fluidity. In the retina, this would possibly disrupt the integrity of cell-to-cell adhesion machinery.
Fig. 5.
The effect of Rs1 on membrane fluidity. PS bilayer indentations were performed using the AFM cantilever and the cantilever deflection at which the bilayer ruptured were measured. The effect of Ca2+ and of Rs1 on PS bilayer fluidity is reflected in the membrane yield strength. A. PS bilayer in 20 mM HEPES-150 mM NaCl. Although “phases” appear in PS even without Ca2+, punch-through forces differ little in the two domains. B. PS bilayer in 20 mM HEPES, 150 mM NaCl, 0.5 mM CaCl2. In the presence of Ca2+, the punch through deflection shows two separate peaks with different yield strengths. C. PS bilayer in 20 mM HEPES, 150 mM NaCl, 0.5 mM CaCl2 after injection of Rs1. Protein binding greatly enhances membrane robustness of the ordered phase as opposed to the fluid phase that remains unaffected. D. A pair of force curves obtained by indenting PS ordered domain in the presence of calcium (red) and the same phase after Rs1 adsorption (blue). The discontinuities in the deflection curve due to the bilayer break-through (yield) and the cantilever deflections at which the bilayers are punctured are indicated.
5. Immunolabeling localizes Rs1 embedded into the ordered domains of PS bilayers
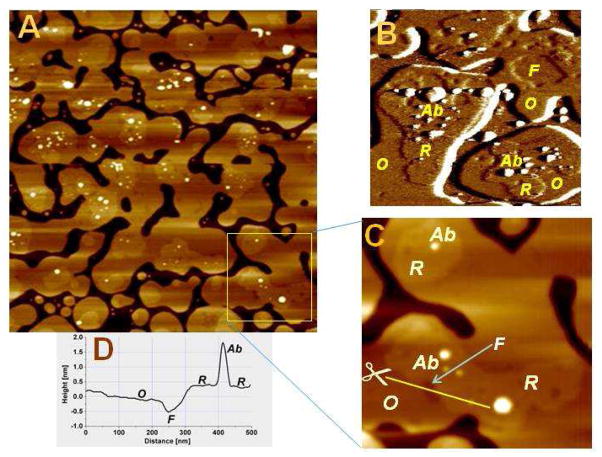
We observed that the addition of Rs1 protein caused lipid bilayer reorganization. However, the Rs1 protein could not be seen on the bilayer surface apparently because it incorporates into the ordered lipid structure with exact fit of size and conformation. While Rs1 was apparently embedded in the lipid bilayer and remained mostly invisible in the regular AFM images, we did observe labile elevated patches on top of the lipid bilayer at suitable imaging conditions (combinations of tip geometry and appropriately adjusted interaction forces). However, these patches disappeared upon prolonged imaging. We used anti-Rs1 antibody to ascertain the presence of Rs1 protein in those patches. We first injected the protein onto the PS bilayer and allowed time for the usual expansion of the fluid phase and the resulting confluence of the lipid patches (Fig. 6) until PS-Rs1 equilibrium was established. Imaging revealed additional 3Å thick, smooth patches lying on top of the ordered phase of PS. We subsequently injected Rs1 antibody while continuously scanning. Shortly after the antibody was introduced several protrusions appeared exclusively on the smooth, tallest patches hypothesized to be formed by the protein partially embedded into the ordered PS phase. Judging from their sizes, these protrusions are individual antibodies and small antibody clusters (Fig. 6B). These observations suggest that the Rs1 protein initially binds to the calcium-rich regions and then rapidly rearranges itself within the lipid bilayer leading to the reorganization of the bilayer.
Fig. 6.
Immunolocalization of Rs1 on PS bilayer. A. Rs1 was injected over a PS bilayer formed in 20 mM HEPES, 150 mM NaCl and 0.5 mM CaCl2. After Rs1-PS equilibration, Rs1-antibody was introduced. The image shows the bilayer about 30 min after antibody injection. The cross-section shows multiple height steps: “F” fluid phase (Ca2+-poor); “O” ordered Phase (Ca2+-rich); “R” Rs1 protein; “Ab” Rs1 -antibody. Note the antibody (single molecules and clusters) is present only on the elevated patches and nowhere else on the bilayer surface. These elevated patches are, therefore, assigned to Rs1. Image is 5×5 μm2. B. “Deflection” image of a 1.1×1.1 μm2 region clearly showing the antibody binding only to the most elevated patches assigned to Rs1. All the different components are indicated as noted in “a”. C. Magnified 1.1×1.1 μm2 region from image “a” showing the different regions. D. Section profile along the line drawn in “c”. Note that the Rs1 patch (“R”) is only 0.3–0.4 nm taller than the ordered PS phase (“O”) and that, in turn, is taller than the fluid phase (“F”) by about 0.7 nm.
6. The interaction between the DS domain-containing FVIII and PS
Based on the sequence alignment of RS1 with the C2 discoidin domain of coagulation factors, FV, and FVIII, Wu and Molday (29) proposed a structural model for the RS1 discoidin domain. The C2 discoidin domain of FVIII is known to interact with PS and this interaction does not require calcium (30). Hence, to compare the nature of the interaction of FVIII and Rs1 with supported PS bilayers, we imaged PS bilayers before and after FVIII introduction. FVIII indeed bound to PS bilayers and formed patches. Although such binding does not require calcium, we did observe FVIII patches on top of the ordered PS phase (Fig. 7). The taller (~1 nm) patches formed by FVIII resembled those formed by Rs1. However, changes neither in the shape of the lipid bilayer patches nor phase configuration were observed after FVIII deposition. Prolonged imaging for several hours showed no expansion of the lipid bilayer. In contrast to the Rs1 patches, the FVIII patches on top of the bilayer appeared very stable. The discoidin domain being the common element between Rs1 and FVIII structures, we infer that the Rs1 binding to PS is mediated through the discoidin domain. In summary, these findings demonstrate the direct and specific effect of Rs1 on anionic lipid bilayers containing PS in the presence of Ca2+.
Fig. 7.
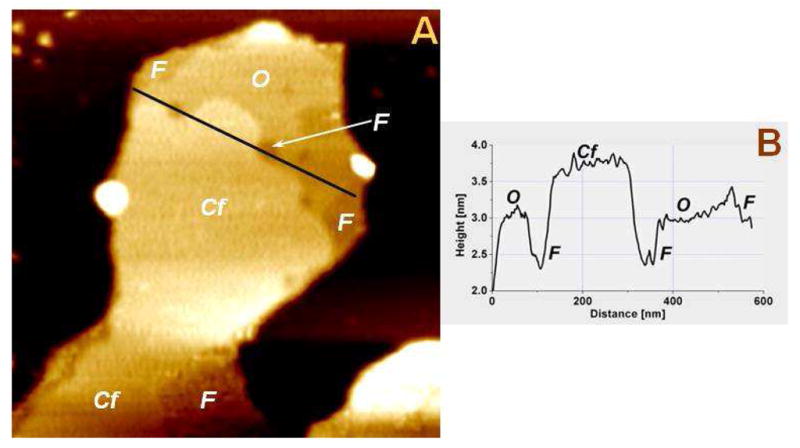
Association of FVIII with PS in the presence of calcium cations. FVIII formed smooth patches on top of the ordered PS phase. No lipid phase reorganization or any other changes were observed on the bilayer. A. A 1×1 μm2 patch of PS bilayer formed in 20 mM HEPES, 150 mM NaCl, and 0.5 mM CaCl2, after injection of FVIII and equilibration. B. Line section profile along the black line in “a”. “F” - fluid phase (Ca2+-poor); “O” – ordered Phase (Ca2+-rich); “Cf” – FVIII protein. Recombinant FVIII (Kogenate(r) FS Antihemophilic Factor) was obtained from Bayer Health Care, Tarrytown, NY.
Discussion
Using AFM, we imaged Rs1 in association with the lipid bilayers. The binding of Rs1 to the bilayers required an anionic phospholipid, phosphatidylserine and Ca2+. The results of this study are consistent with biochemical observations, which demonstrated that (a) Ca2+ increases the affinity of Rs1 binding to the retinal cell membranes and (b) Rs1 exhibits selective affinity for binding to anionic phospholipid, phosphatidylserine (13). Together with the biochemical findings, the AFM imaging provides a strong evidence of direct interaction between Rs1 and the PS.
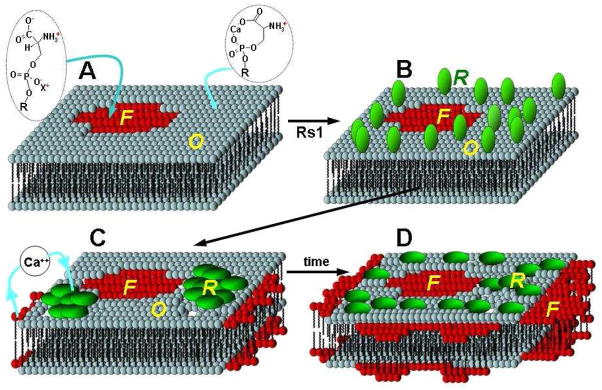
The AFM study provides biophysical evidence that Rs1 causes major reorganization of PS-containing supported bilayers in the presence of calcium cations. This reorganization can be dissected into different steps and possible mechanisms discussed. First, it is interesting that calcium cations induce phase separation in a single component bilayer. Calcium induced two distinct phases on PS by selectively interacting with certain regions and not with the others. The mechanism that underlies this is unknown. The two domains equilibrate into robust structures that remained unchanged even after prolonged imaging. The fraction of the ordered, calcium-rich, phase correlates with the relative concentrations of PS and Ca2+. It is known that calcium binds to PS by bridging the carboxyl and phosphate moieties on the lipid head, altering its configuration and leaving a net positive charge on the amino group (31). As a result, PS bilayers consist of negatively and positively charged patches. Rs1 at pH 7.0 carries a net negative charge of approximately −3 (pI = 5.6) as calculated from its primary structure (http://expasy.org/tools/pi_tool.html). This would drive the electrostatic binding of the negatively charged protein to the positively charged ordered phase of the PS patches. Such binding would bring the putative hydrophobic patches on the protein surface in close proximity to the hydrophobic core of the bilayer. The subsequent embedding of the protein into the bilayer core leads to lipid bilayer reorganization. This reorganization appears to displace and reposition lipid material, with the protein probably displacing the bound calcium from the PS causing a change in the configuration of the PS headgroup and its transition into the fluid phase. This would account for the observed expansion of the lower fluid phase that was consistently seen upon introduction of Rs1 to pure PS bilayers in the presence of calcium. A schematic representation of the hypothetical steps of the PS-Rs1 interaction in the presence of Ca2+ is depicted in Fig. 8.
Fig. 8.
Schematic illustration of the hypothesized interaction of Rs1 with PS in the presence of Ca2+. A. Ca2+ controlled PS phases before Rs1 introduction; The PS bilayer exists in two phases, a fluid phase devoid of calcium (“F”) and a calcium-rich ordered phase (“O”). B. Rs1 adsorbs randomly onto the ordered lipid phase. Electrostatic interactions lead the protein to preferentially adsorb to the ordered lipid domain. Adsorption is followed by protein insertion toward the hydrophobic lipid core causing lipid disruption and apparent expansion of the fluid phase. C. At this point the protein assembles into labile patches. D. These patches tend to diffuse toward a more homogeneous distribution over time. Anchoring of the protein to the lipid enhances the membrane robustness significantly. This is consistent with the role of Rs1 as an adhesion molecule that helps maintain the architectural integrity of the retina. “F” fluid phase (Ca2+-poor); “O” ordered Phase (Ca2+-rich); “R” Rs1 protein;
The previously described structure of the PS bilayer in the presence of Ca2+ (27) also partially explains the enhanced mechanical robustness measured with the nano-indentation tests. In addition to the tighter packing of the carbon chains in the ordered phase, the latter phase will attach electrostatically to the negatively charged mica more strongly than in the fluid-phase. When Rs1 is introduced to the Ca2+-modified PS bilayer, the protein-lipid and protein-protein interactions, such as oligomerization before or after binding, would further act to reinforce and enhance the bilayer robustness as seen in the punch-through force histograms (Fig. 5).
The nature of Rs1 interactions with ligands other than PS remains unclear. Membrane proteins often preferentially reside and function optimally within lipid domains enriched in certain types of lipids. The function of Na+/K+ ATPase, for example, has been shown to be regulated by acidic lipids, meaning that the ATPase resides in lipid domains rich in anionic lipids. It is likely that a lipid binding protein, such as Rs1, might be acting as a stabilizer of membrane microdomains into which membrane proteins, such as Na+/K+ ATPase, SARM, and LVGCC assemble (30, 32). The associations of Rs1 with these complexes combined with the findings in this study are consistent with a hypothesis that Rs1 has dual functionality, as a cell-adhesion molecule and also as a regulatory accessory of several membrane protein assemblies. The crystal structure of RS1 is not known, but a homology model of the RS1 discoidin domain, based on the 3-D structure of the C2 domain of coagulation factor V, suggests that RS1 has a lipid-binding potential (33). The model reveals a probable hydrophobic patch exposed on the surface of the protein, which is rich in aromatic amino acid residues, including Tyr-89, Trp-92 and Phe-108. The patch would naturally be directed to the hydrophobic core of a bilayer. The mechanism by which calcium would regulate this binding remains to be investigated.
Supplementary Material
Acknowledgments
We thank Sean Finnegan (NEI) for editorial assistance with manuscript preparation.
This work was supported by the Intramural Programs of the National Eye Institute, National Institute of Biomedical Imaging and Bioengineering and the National Institute on Deafness and Other Communication Disorders, at the National Institutes of Health.
Abbreviations
- RS1
human protein (NP_00032.1)
- RS1
human gene (ID: 6247, MIM 312700)
- Rs1
murine protein (NP_035432)
- Rs1
murine gene (ID: 20147)
Footnotes
Supporting information with additional data on the protein purification procedures, movies showing the evolution of the bilayers under the influence of Rs1 and composite images showing progression toward equilibration and additional control experiments is available free of charge at the ACS website http://pubs.acs.org.
References
- 1.Molday LL, Hicks D, Sauer CG, Weber BH, Molday RS. Expression of X-linked retinoschisis protein RS1 in photoreceptor and bipolar cells. Invest Ophthalmol Vis Sci. 2001;42:816–825. [PubMed] [Google Scholar]
- 2.Takada Y, Fariss RN, Muller M, Bush RA, Rushing EJ, Sieving PA. Retinoschisin expression and localization in rodent and human pineal and consequences of mouse RS1 gene knockout. Mol Vis. 2006;12:1108–1116. [PubMed] [Google Scholar]
- 3.Sikkink SK, Biswas S, Parry NR, Stanga PE, Trump D. X-linked retinoschisis: an update. J Med Genet. 2007;44:225–232. doi: 10.1136/jmg.2006.047340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tantri A, Vrabec TR, Cu-Unjieng A, Frost A, Annesley WH, Jr, Donoso LA. X-linked retinoschisis: a clinical and molecular genetic review. Surv Ophthalmol. 2004;49:214–230. doi: 10.1016/j.survophthal.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Vijayasarathy C, Ziccardi L, Zeng Y, Smaoui N, Caruso RC, Sieving PA. Null retinoschisin-protein expression from an RS1 c354del1-ins18 mutation causing progressive and severe XLRS in a cross-sectional family study. Invest Ophthalmol Vis Sci. 2009;50:5375–5383. doi: 10.1167/iovs.09-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeng Y, Takada Y, Kjellstrom S, Hiriyanna K, Tanikawa A, Wawrousek E, Smaoui N, Caruso R, Bush RA, Sieving PA. RS-1 gene delivery to an adult Rs1h knockout mouse model restores ERG b-wave with reversal of the electronegative waveform of X-Linked retinoschisis. Invest Ophthalmol Vis Sci. 2004;45:3279–3285. doi: 10.1167/iovs.04-0576. [DOI] [PubMed] [Google Scholar]
- 7.Min SH, Molday LL, Seeliger MW, Dinculescu A, Timmers AM, Janssen A, Tonagel F, Tanimoto N, Weber BH, Molday RS, Hauswirth WW. Prolonged recovery of retinal structure/function after gene therapy in an Rs1h-deficient mouse model of x-linked juvenile retinoschisis. Mol Ther. 2005;12:644–651. doi: 10.1016/j.ymthe.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 8.Park TK, Wu Z, Kjellstrom S, Zeng Y, Bush RA, Sieving PA, Colosi P. Intravitreal delivery of AAV8 retinoschisin results in cell type-specific gene expression and retinal rescue in the Rs1-KO mouse. Gene Ther. 2009;16:916–926. doi: 10.1038/gt.2009.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vijayasarathy C, Gawinowicz MA, Zeng Y, Takada Y, Bush RA, Sieving PA. Identification and characterization of two mature isoforms of retinoschisin in murine retina. Biochem Biophys Res Commun. 2006;349:99–105. doi: 10.1016/j.bbrc.2006.07.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Molday RS. Focus on molecules: retinoschisin (RS1) Exp Eye Res. 2007;84:227–228. doi: 10.1016/j.exer.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 11.Vogel WF, Abdulhussein R, Ford CE. Sensing extracellular matrix: An update on discoidin domain receptor function. Cellular Signalling. 2006;18:1108–1116. doi: 10.1016/j.cellsig.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 12.Kiedzierska A, Smietana K, Czepczynska H, Otlewski J. Structural similarities and functional diversity of eukaryotic discoidin-like domains. Biochim Biophys Acta. 2007;1774:1069–1078. doi: 10.1016/j.bbapap.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 13.Vijayasarathy C, Takada Y, Zeng Y, Bush RA, Sieving PA. Retinoschisin is a peripheral membrane protein with affinity for anionic phospholipids and affected by divalent cations. Invest Ophthalmol Vis Sci. 2007;48:991–1000. doi: 10.1167/iovs.06-0915. [DOI] [PubMed] [Google Scholar]
- 14.Vijayasarathy C, Takada Y, Zeng Y, Bush RA, Sieving PA. Organization and molecular interactions of retinoschisin in photoreceptors. Adv Exp Med Biol. 2008;613:291–297. doi: 10.1007/978-0-387-74904-4_34. [DOI] [PubMed] [Google Scholar]
- 15.Aragao KS, Satre M, Imberty A, Varrot A. Structure determination of Discoidin II from Dictyostelium discoideum and carbohydrate binding properties of the lectin domain. Proteins. 2008;73:43–52. doi: 10.1002/prot.22038. [DOI] [PubMed] [Google Scholar]
- 16.Fuentes-Prior P, Fujikawa K, Pratt KP. New insights into binding interfaces of coagulation factors V and VIII and their homologues lessons from high resolution crystal structures. Curr Protein Pept Sci. 2002;3:313–339. doi: 10.2174/1389203023380639. [DOI] [PubMed] [Google Scholar]
- 17.Baumgartner S, Hofmann K, Chiquet-Ehrismann R, Bucher P. The discoidin domain family revisited: new members from prokaryotes and a homology-based fold prediction. Protein Sci. 1998;7:1626–1631. doi: 10.1002/pro.5560070717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Molday LL, Wu WW, Molday RS. Retinoschisin (RS1), the protein encoded by the X-linked retinoschisis gene, is anchored to the surface of retinal photoreceptor and bipolar cells through its interactions with a Na/K ATPase-SARM1 complex. J Biol Chem. 2007;282:32792–32801. doi: 10.1074/jbc.M706321200. [DOI] [PubMed] [Google Scholar]
- 19.Shi L, Jian K, Ko ML, Trump D, Ko GY. Retinoschisin, a new binding partner for L-type voltage-gated calcium channels in the retina. J Biol Chem. 2009;284:3966–3975. doi: 10.1074/jbc.M806333200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mackay JP, Sunde M, Lowry JA, Crossley M, Matthews JM. Protein interactions: is seeing believing? Trends Biochem Sci. 2007;32:530–531. doi: 10.1016/j.tibs.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 21.Mingeot-Leclercq MP, Deleu M, Brasseur R, Dufrene Y. Atomic force microscopy of supported lipid bilayres. Nature Protocols. 2008;3:1654–1659. doi: 10.1038/nprot.2008.149. [DOI] [PubMed] [Google Scholar]
- 22.Giocondi MC, Seantier B, Dosset P, Milhiet PE, Le Grimellec C. Characterizing the interactions between GPI-anchored alkaline phosphatases and membrane domains by AFM. Pflugers Arch - Eur J Physiol. 2008;456:179–188. doi: 10.1007/s00424-007-0409-x. [DOI] [PubMed] [Google Scholar]
- 23.Picas L, Montero MT, Morros A, Cabañas ME, Seantier B, Milhiet PE, Hernández-Borrell J. Calcium-induced formation of subdomains on phosphatidylethanolamine-phosphatidylglycerol bilayers: a combined DSC, 31P NMR and AFM Study. J Phys Chem B. 2009;113:4648–4655. doi: 10.1021/jp8102468. [DOI] [PubMed] [Google Scholar]
- 24.Murray J, Cuccia L, Ianoul A, Cheetham JJ, Johnston LJ. Imaging the selective binding of synapsin to anionic membrane domains. Chembiochem. 2004;5:1489–1494. doi: 10.1002/cbic.200400097. [DOI] [PubMed] [Google Scholar]
- 25.Shahin V, Datta D, Hui E, Henderson RM, Chapman ER, Edwardson JM. Synaptotagmin perturbs the structure of phospholipid bilayers. Biochemistry. 2008;47:2143–2152. doi: 10.1021/bi701879g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feigenson GW. On the nature of calcium ion binding between phosphatidylserine lamellae. Biochemistry. 1986;25:5819–5825. doi: 10.1021/bi00367a071. [DOI] [PubMed] [Google Scholar]
- 27.Roux M, Bloom M. Calcium binding by phosphatidylserine headgroups. Deuterium NMR study. Biophys J. 1991;60:38–44. doi: 10.1016/S0006-3495(91)82028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li L, Shin OH, Rhee JS, Arac D, Rah JC, Rizo J, Sudhof T, Rosenmund C. Phosphatidylinositol phosphates as co-activators of Ca2+ binding to C2 domains of synaptotagmin 1. J Biol Chem. 2006;281:15845–15852. doi: 10.1074/jbc.M600888200. [DOI] [PubMed] [Google Scholar]
- 29.Wu WW, Molday RS. Defective discoidin domain structure, subunit assembly, and endoplasmic reticulum processing of retinoschisin are primary mechanisms responsible for X-linked retinoschisis. J Biol Chem. 2003;278:28139–28146. doi: 10.1074/jbc.M302464200. [DOI] [PubMed] [Google Scholar]
- 30.Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- 31.Christie WW. The Lipid Library. The American Oil Chemists’ Society; 2010. Phosphatidylserine and related lipids: Structure, occurance, biochemistry and analysis. [Google Scholar]
- 32.Bandorowicz-Pikula J. Lipid-binding proteins as stabilizers of membrane microdomains--possible physiological significance. Acta Biochim Pol. 2000;47:553–564. [PubMed] [Google Scholar]
- 33.Fraternali F, Cavallo L, Musco G. Effects of pathological mutations on the stability of a conserved amino acid triad in retinoschisin. FEBS Letters. 2003;544:21–26. doi: 10.1016/s0014-5793(03)00433-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.