Abstract
RNA polymerase (RNAP) is a complex molecular machine that governs gene expression and its regulation in all cellular organisms. To accomplish its function of accurately producing a full length RNA copy of a gene, RNAP performs a plethora of chemical reactions and undergoes multiple conformational changes in response to cellular conditions. At the heart of this machine is the active center - the engine, which is composed of distinct fixed and moving parts that serve as the ultimate acceptor of regulatory signals and target of inhibitory drugs. Recent advances in the structural and biochemical characterization of RNAP explain the active center at the atomic level and enable new approaches to understanding the entire transcription mechanism, its exceptional fidelity and control.
Keywords: RNA polymerase, catalysis, transcription, fidelity, elongation factors, termination, antibiotics
INTRODUCTION
DNA-dependent RNA polymerase (RNAP) executes the first step in gene expression - transcription. The enzyme synthesizes an RNA copy of a DNA template strand de novo from nucleoside triphosphate (NTP) substrates. RNAP generates force (137,33) and utilizes energy to propagate unidirectionally in two-dimensional space at a speed of 15 to 80 nt/s (105,28), but unlike a motorized vehicle, this molecular machine operates as a Brownian ratchet (7,1,14). All cellular RNAPs are multisubunit molecules with a total molecular weight of approximately half a million Daltons. Although the total number of enzyme subunits varies significantly between species, the primary sequence, ternary structure and function of the catalytically-competent core (subunit composition 2α, β', β, and ω in bacteria) is evolutionarily conserved (4,146,26,24,80).
Transcription is a cyclic process that can be divided into three major steps – promoter DNA binding/RNA chain initiation, processive RNA chain elongation, and termination. The productive phase of transcription is RNA chain elongation, in which the enzyme is processively engaged with the template DNA, moving forward in elementary steps by single nucleotide addition to the 3' terminus of the growing RNA. In this state the system is referred to as the ternary elongating complex (EC). In the transcription cycle, elongation is preceded by the abortive intitiation step, in which a static ≪open≫ promoter complex attempts to leave the transcrition start site by synthesizing short RNA oligonucleotides; one of them is successfully extended, leading to formation of the EC, the others are aborted from the complex (46,97). Elongation ends when the EC disociates in response to a specific termination signal. The resultant free enzyme may engage a new promoter, aided by intitiation factor(s), leading to formation of a closed complex, which converts to an open complex by local DNA melting. Detailed descriptions of the transcription cycle can be found in several recent reviews (115,108,38,13).
The basic enzymatic activity of RNAP is the transfer of the nucleotidyl moiety from the NTP substrate to the hydroxyl at the 3' terminus of the nascent transcript (Figure 1). The catalytic center of RNAP thus includes the binding sites for the RNA 3'-terminus (i site) and the insertion site for the incoming NTP (i+1 site, also known as A site). The formation of the phoshodiester bond is followed by translocation of the newly formed 3' terminus from the i+1 to the i site and the release of the pyrophosphate (PPi) product. This reaction can be reversed (Figure 1). PPi stimulates processive degradation of RNA with the release of the 3'-terminal NTP, a process known as pyrophosphorolysis (101). For pyrophosphorolysis to occur, the RNA 3'-terminus must reoccupy the i+1 site. Thus, during the elongation phase the EC exists in equilibrium between the i and i+1 states (14). In addition to these two basic forward and reverse enzymatic reactions, the RNAP catalytic center is capable of performing at least two types of nuclease reactions (Figure 1) (19) that are important for achieving the fidelity and processivity of transcription.
Figure 1. Catalytic activities of RNAP.
Schematic representation of enzymatic reactions performed by the EC in different states. i and i+1 represent the product and substrate binding sites of the catalytic center, respectively. Red arrows indicate the direction of the nucleophilic attack. P - phosphate group, N – nucleotide residue, solid lines represent the nascent RNA.
During the past several years, a large amount of structural and biochemical information about the molecular details of RNAP function and regulation at each step of the transcription cycle has become available. This review focuses on the intricate organization of the RNAP catalytic center, which serves as a robust, yet sensitive, molecular engine that drives and controls the rate, processivity, and fidelity of transcription.
FUNCTIONAL TOPOGRAPHY OF THE EC AT HIGH RESOLUTION
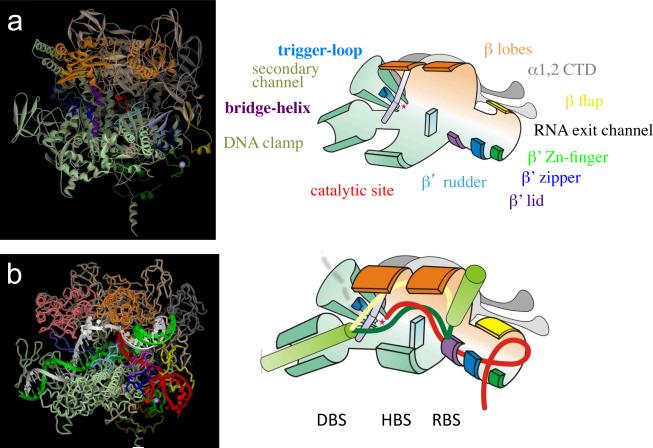
Extensive biochemical and genetic analysis in the late 1990s pinpointed the catalytic center of RNAP (144,74,83) and three ajacent nucleic acid binding sites that hold the EC together (87): the front DNA duplex binding site (DBS), the RNA-DNA hybrid binding site (HBS), and the RNA binding site (RBS) (Figure 2). Three aspartate residues within the absolutely conserved NADFDGD motif of the largest (β') subunit coordinate the catalytic Mg++, which is surrounded by several distinct domains of the β and β' sybunits (83,146) (Figure 3). The DBS interacts with ~9 bp of the DNA duplex immediately downstream of the catalytic center (89,90,63). The HBS is defined by close contacts between the β' and β subunits and the 3'-terminal 7–9 bp of the RNA:DNA hybrid located within ~12–15 bp of melted DNA, or transcription “bubble” (88,107,63,60). Finally, the RBS consists of a region of close interactions between the NH2-terminal parts of β' and a sequence of single-stranded RNA located approximately −8 to −14 nt relative to the RNA 3' terminus (90,64,63). Protein-nucleic acid interactions at all three sites contribute to the overall stability and processivity of the EC. Disturbing these contacts at any individual site sensitizes the EC to high ionic strength (e.g. 0.5 M KCl) (89,64,107,60). The recently determined high-resolution structures of yeast and bacterial ECs (34,55,139,129) reveal the precise positioning of the nucleic acid scaffold relative to the catalytic center, with all three functionally defined binding sites neatly placed in designated protein channels (Figure 2). Available structures of the EC were deduced from crystals of reconstituted complexes containing a short synthetic DNA and RNA partially annealed to the DNA template strand within a mismatched bubble (34,55,139,129).
Figure 2. Structural overview of the EC.
(a) High-resolution crystal structure of Thermus aquaticus RNAP core (146) (left panel) is shown as colored ribbons (αI, light grey; αII, grey; β, light brown; β', light green; ω, dark green; σ, magenta, as imaged by the Accelris DS-Visualizer program. Structure represents the main channel view of RNAP. Mobile elements are shown as follows: β' BH (residues I705-V835 of T.t. β'), purple; β' TL (T1234-L1256), blue; β' zipper (V26-E47), dark blue; β' Zn-finger (G51-S83), green; β' lid (R525-S538), light blue; β' rudder (N584-S602), turquoise; β lobe 1 (A29-D133, G337-S392) and β lobe 2 (R142-N330), orange; and β flap (G757-S789), yellow. The RNAP β' catalytic loop (N737-Q744) is colored red and the Mg++ ion is shown as a small red sphere. Right panel shows schematic representation of the RNAP structure as a simplified cartoon with the same color code as in the left panel with all the key structural elements indicated. The catalytic center is shown as a small red star.
(b) Structural model of the EC (left panel) based on the T.t. EC structure (129). Right panel shows the schematic representation of the structure with the same color code as in the left panel. Duplex DNA is shown as flexible green cylinders; nascent RNA, template and non-template DNA strands are shown as red, dark green and yellow worms, respectively. The main channel secures the RNA-DNA hybrid in the hybrid-binding site (HBS). β' domains clamp the downstream DNA in the DNA duplex binding site (DBS). The β' zipper, Zn-finger and lid along with α flap form the RNA exit channel. RNA-binding site (RBS) is located at the junction of HBS and the RNA exit channel and also functions as rear zip-lock that separates the nascent transcript from the hybrid (see the text for details). The front zip-lock formed by the BH and other elements of the catalytic center separates RNA (shown as a dashed red line) from the hybrid during backtracking, directing it into the secondary channel (shown as a funnel). Transcription goes from right to left.
Figure 3. Structure of the RNAP catalytic center.
Left panel shows a close-up view of the catalytic center, which is obtained using high-resolution structures of the T.t. EC in the presence of the non-hydrolizable substrate analog AMPcPP (128). Right panel is a schematics of the left panel indicating specific interactions between evolutionarily conserved amino acid residues, substrate NTP, Mg++ ions and parts of the nucleic acid scaffold, based on the structures of T.t. and yeast ECs (136,55,128,129). Residue numbers corresponding to T.t. β and β' polypeptides are shown. Residues of the Rpb1 of yeast RNAP II that differ from T.t. β' are shown in parenthesis. Three principal groups of residues are indicated: those that coordinate Mg++ ions, those that participate in binding and proper orientation of the NTP α-, β- and γ-phosphates, and those responsible for recognition of correct NTPs. The two Mg++ ions play an essential role in RNA catalysis (114, 110). The first Mg++ (Mg-A) is bound by three aspartates of the β' catalytic loop (D739, D741 and D743). The second Mg2+ (Mg-B) is less stably bound in the enzyme mostly by one aspartate of the β' catalytic loop (D739) via direct interactions, and by one aspartate of β (D686) via water-mediated interactions (128). Solid and dashed lines indicate direct and water-mediated contacts, respectively. During RNA synthesis (or pyrophosphorolysis) Mg-B is involved in the coordination of the NTP α-, β- and γ-phosphates, insuring their optimal spatial alignment for the nucleophilic attack of the activated 3'-OH group on the α-phosphate (shown by the green arrow on right panel). Additionally, the TL residues R1029 and R1239, and also β R557 and R879 bind the NTP β- and γ-phosphates via hydrogen bonding and electrostatic interactions (128). β' H1042 plays an especially important role in the binding of the α- phosphate (128) (and possibly the β-phosphate [136]) and facilitating the nucleophilic attack of the 3'-OH group. To ensure incorporation of correct nucleotides into the RNA, TL M1238 and BH β'T1088 make hydrophobic/van der Waals contacts with the NTP base and the DNA base at the i+1 position, while β' N737 and R704 establish hydrogen bonds with the 2'- and 3'-OH groups of the ribose ring of the NTP (128,50].
DNA-binding site (DBS)
Downstream DNA traverses the deep cleft formed by β' (Rpb1 in yeast RNAP II) (Figure 2). The DNA remains in an intact B-form conformation up to position +2 in T.t. RNAP or +4 in yeast pol II (counting from the +1 NTP binding site) where a ~90° kink occurs (34,55,129). Such a sharp kink has been observed previously by atomic force microscopy of the intact ECs (96), suggesting that the crystal packaging did not alter the nucleic acid geometry within the complex. DNA distortion in front of the catalytic center is likely to be facilitated by polar residues of the Rpb1 switch 1 and 2 domains that together with the β (Rpb2) fork-loop 2 induce strand separation (55,129). The fork-loop 2 sterically blocks the advance of the DNA helix into the active center and prevents reassociation of DNA strands to maintain the downstream edge of the bubble (136,129). The ~9 bp DNA forms direct contacts within the protein cleft, thus providing structural justification for the DBS. Most of these contacts are long-distance electrostatic and van der Waals interactions, which should not impede the threading of DNA through the clamp. Therefore DNA stabilization in the DBS is achieved primarily via topological linking, without strong affinity to any specific DNA bases, - a feature that explains the remarkable processivity of the EC. The major polar and van der Waals contacts with the downstream DNA occur within the +2 and +5 registers. They involve phosphates interactions with the β' subunit three helix bundle (101–131 aa in T.t.) (129) and minor groove contacts with the carboxy-terminal clamp domain of Rpb1 in the yeast RNAP II (34,55,139). Consistently, alterations in the DNA duplex at the +3 to +5 positions have the strongest effect on the EC stability and termination (89,31) compared to other downstream DNA sites. Thus, taken together, the functional and structural data precisely map the DBS in the EC.
Hybrid-binding site (HBS)
The 8–9 bp RNA/DNA hybrid is enclosed in the main channel formed by the largest subunits, which defines the HBS. It is situated between the catalytic Mg++ and the bridge-helix (BH) of β' (Rbp1) at the downstream end and the lid domain of β' at its upstream end (63,34,55,139,129) (Figure 2). The size of the hybrid (~8 bp), determined by the RNA-DNA crosslinking in the native EC (88), is consistent with that observed in EC structures obtained using artificial scaffolds. The phosphate backbone of the hybrid makes multiple polar and van der Waals contacts with conserved protein residues, including those of β that are involved in binding the antibiotic rifampicin (19). The majority of these contacts, however, are weak due to repulsion between the hydrophobic side chains and charged phosphate groups. This should diminish friction and facilitate back-and-forth sliding of the main channel along the hybrid during forward translocation and backtracking. When the EC backtracks, the catalytic center disengages from the RNA 3' terminus and the enzyme slides backward, unzipping the hybrid and displacing the 3' portion of the nascent RNA (61,62,88). In this regard the catalytic center acts like a zip-lock in front of the hybrid. A key structural determinant of the front zip-lock is the β' BH that extends toward the downstream face of the hybrid, acting like a wall that separates the RNA from the DNA template strand and directs its 3' extruded portion into the adjacent secondary channel (Figure 2). A rear zip-lock at the upstream edge of the hybrid (−9 position) consists primarily of the β' lid loop (Figure 2), which serves as a wedge to facilitate RNA displacement by sterically blocking the formation of the overextended hybrid (34,138,55,84,122,129). Rpb1 residue Phe252, at the tip of the wedge, splits the RNA-DNA hybrid at position −10, forming a stacking interaction with the DNA base (138). Bubble rewinding, however, also contributes to RNA displacement (83,122). The lid structure is flexible, allowing the hybrid size to fluctuate between 7 to 10 bp, as has been observed biochemically (88,64). The lid may also stabilize it by forming stacking-like contacts with the last base pairs of the hybrid. The other domain that is involved in hybrid stabilization is the β' rudder, which interacts with the upstream edge of the hybrid (67,138,129). It contacts the DNA primarily at positions −9, −10 and −11 and likely prevents reassociation of these bases with the RNA. Another element implicated in the stabilization of the hybrid is fork-loop-I of β (Rbp2) (138,129). It interacts with the RNA phosphates at positions −5, −6 and −7 and may prevent hybrid unwinding beyond positions −7/−8. The three proteins loops, lid, rudder and fork-loop-I, interact not only with nucleic acids but also with each other and other protein elements to form a complex and a dynamic network that constitutes the rear zip-lock. Acting together, the front and rear zip-locks maintain the hybrid at a relatively constant size during elongation and backtracking and also assist in maintaining the boundaries of the transcription bubble.
RNA-binding site (RBS)
In both bacterial and yeast EC, the nascent transcript is separated from the hybrid at positions −8 to −10 (88,64,55,138,129) (Figure 2). At this position the transcript makes direct contacts with evolutionary conserved protein residues (55). These contacts constitute the RBS, which contributes to the stability of the EC (64,90). A lack of these contacts would explain the negative effect of an overextended hybrid on the stability of the EC (59) and the destabilizing effect of antisense oligonucleotides that anneal to the −8/−10 RNA bases (141). Moreover, the presence of RNA bases at the −8/ −10 positions, regardless of whether or not they are complementary to the DNA template strand, is essential for the resistance of the EC to high ionic strength conditions (64).
The lid and β “saddle” form a narrow pore to the position to which the ssRNA is extended (34,55,129). The pore leads to a wider channel formed by the flexible zipper and Zn-finger domains of β' on one side, and the β flap on the other side (Figure 2) (63,129). Only a few direct contacts and water-mediated hydrogen bonds are made between RNA and the protein in the channel (129). Consistently, biochemical data show that the 5' portion of the transcript beyond −10 does not contribute to the stability of the EC (64,107). Thus, the RBS is defined as a region of tight protein-RNA contacts 8–10 nt upstream of the catalytic center.
As discussed below, all three nucleic acid binding sites, DBS, HBS and RBS, communicate allosterically with the catalytic center, thereby influencing catalysis in response to changes in the conformational status of nucleic acids.
STRUCTURAL BASIS OF CATALYSIS: COUPLING CHEMISTRY WITH MOVEMENT
The nucleotide addition cycle (NAC) involves three essential steps: substrate entry and binding to the EC (NTP binding step), nucleotide incorporation into the nascent transcript by phosphodiester bond formation with the displacement of pyrophosphate (PPi) (chemistry step), and translocation of the nucleic acids scaffold in relation to the RNAP mainframe to free the NTP insertion site, also known as the i+1 or A site (translocation step) (Figures 3 and 4). Whereas the chemistry step is well understood from structural analyses, the exact mechanisms of the two other steps remain to be determined.
Chemistry
The catalitic center of RNAPs, like that of all nucleic acid polymerasing enzymes, contains two metal ions that are a few Å apart and responsible for phosphodiester bond formation (115,24,127,110) (Figure 3). Mg ion A (Mg-A) is bound by three aspartate residues in the β' (Rpb1) catalytic loop D739, D741, and D743 in T.t. It is responsible primarily for the activation of the RNA 3'-terminal OH group and for coordination of the α-phosphate of the incoming NTP. The second Mg ion (Mg-B) is located 5–6 Å from Mg++A. It is held less stably in the enzyme, primarily by direct interactions with β' D739, and by one aspartate residue of β (D686) via water-mediated interactions (128). Also, acidic residues β' D741 and β E685 contribute to the stabilization of Mg-B through water-mediated contacts (136,128). During each NTP addition cycle, Mg++B is likely to be delivered to the catalytic center bound to the incoming NTP substrate (116). During RNA synthesis (or the reverse reaction of pyrophosphorolysis) Mg-B is responsible for coordinating the α-, β- and γ-phosphates of the NTP substrate to keep them in proper spatial alignment for the SN-2 type nucleophilic attack of the activated 3'-OH group on the α-phosphate (Figure 1 and 3).
NTP Binding
Several high-resolution structures of RNAP bound to a substrate or its analogs defined the insertion site (i+1 or A-site) and revealed at least two additional NTP-binding sites that overlap with the insertion site (Figure 4). The existence of multiple NTP-binding sites suggests that nucleotide loading is a complex process with intermediate states that serve as checkpoints to distinguish between a correct and incorrect substrate prior to its incorporation into the nascent transcript.
Figure 4. Nucleotide addition cycle (NAC) and the dynamics of the catalytic center.
(a) Schematic diagram depicting conformational changes in the TL and BH during NTP incorporation and subsequent translocation (14,116). DNA template and RNA strands are shown in blue and red, respectively. The BH is shown in grey and the TL is in blue. Magenta circles indicate catalytic Mg-A (I) and Mg-B (II).
In the initial, post-translocated (active) EC, the TL is in the unfolded, “open” conformation (136,128). The binding of the correct NTP (orange) leads to refolding of the TL into an extended α-helical conformation (see panel b), which, in turn stabilizes NTP binding through interactions with the TL. Since NTP binding in the i+1 site stabilizes the EC in the post-translocated state, the substrate serves as a stationary pawl in the ratchet mechanism of RNAP translocation (7). After incorporation of the NTP into RNA and release of pyrophosphate, the TL/BH unit may oscillate between the unfolded (open) conformation and intermediate state in which the TL wedges into the BH (see panel c), forcing the latter to “push” against the RNA-DNA hybrid, thereby inducing translocation of the EC to an active, post-translocated state. According to this model, the TL/BH unit serves as a second, reciprocal pawl, facilitating RNAP translocation in the absence of a substrate (34,7,14). α-amanitin (yellow) and steptolydigin antibiotics have been proposed to interfere with NAC by either interrupting TL-dependent loading of a substrate (50,120) or inhibiting TL/BH-dependent RNAP translocation (14). For further details, refer to the text.
(b) TL refolding in the T.t. EC (128). BH (gray cartoon) is packed against the TL refolded as trigger-helices (blue cartoon) as a result of NTP binding (i+1, insertion site, orange sticks). i site RNA is shown as red sticks. Trigger-helixes residues interacting with NTP (Met1238, Arg1239, Phe1241, and His 1242) are shown as blue sticks.
(c) Distortion of the yeast RNAPII BH (grey cartoon) by the wedging TL (cartoon blue) in the presence of α-amanitin (sticks yellow) (14). i site RNA is shown as red sticks, wedging residue of the TL, Leu1081 is shown as teal sticks, interacting with α-amanitin His1085 is shown as magenta sticks (14,50).
Binding of the correct NTP in the insertion site was first demonstrated by examining crystals of a reconstituted yeast RNAP II EC that contained 3'-deoxy terminated RNA (139). The NTP was observed paired with the i+1 coding base of the DNA and stabilized in the catalytically-competent configuration by two metal ions. However, a slightly different configuration of the i+1-paired substrate was observed when a correct, but non-hydrolysable NTP analog (NMPcPP; with the methylene group between α and β phosphates) was used (55). Since the substrate was not in the catalytically-competent configuration (despite being paired with the i+1 DNA base) the authors suggested that it occupied a distinct, but largely overlapping pre-insertion site. The pre-insertion site is different from the previously postulated entry site (E-site), in which the incorrect (non-complementary) NTP was localized in the EC crystals of RNAP II (139). In the E-site, the incorrect NTP was flipped relative to the correct NTP so that its base faced away from the DNA template, whereas the triphosphate moiety remained engaged with the active site metals. The E-site was proposed to serve as a checkpoint to monitor correct base pairing in the A-site by rotating the NTP (139). An NTP-binding site analogous to, but not identical to, the E-site has been proposed based on characterization of the 3'-5' exonuclease reaction stimulated by mismatched NTP (110).
The catalytic center of all cellular RNAPs has two moving domains: the BH and the trigger-loop (TL), which play a crucial role in NTP binding and translocation. The TL is highly flexible and adopts various conformations, several of which have been captured in RNAP crystal structures (146,24,128,34,55,136,128,50,14). In the `closed' or `folded' conformation the TL, together with the BH, participates in the selection and binding of the correct NTP at the i+1 site (136,128,50,14). This is achieved through a network of interactions encompassing all moieties of the NTP, i.e. the base, sugar, and phosphates (Figures 3 and 4) (136,128,50,14). These contacts monitor the chemical nature of the NTP and ensure that the NTP is correctly paired with the template DNA base at position i+1 (Figure 4). The recent structure of T.t. EC complexed with the NMPcPP provides strong support for the existence of the preinsertion site and argues that NTP loading occurs in two steps. First, the open configuration of the TL allows the NTP to enter the inactive (preinsertion) site. If the NTP matches the coding base in the i+1 site, refolding of the trigger loop occurs to sequester the substrate in the i+1 site and fix it in the catalytically active state (128). The sequestration of the substrate by the TL may provide a structural explanation for the brief delay in EC inactivation by the Mg++ chelator EDTA in contrast to the instantaneous inactivation by HCl and the characteristic biphasic kinetics of the formation of the EDTA-resistant intermediate (85,35,36). It also explains the partial inability of the EC to exchange substrates, as demonstrated in a pulse-chase experiment in which the complex was pretreated with a radioactive NTP for a short period of time before being exposed to a large excess of the same non-radiolabeled NTP (57). Apparently, the access of EDTA or a substrate to the active site is temporarily blocked by the closed conformation of the trigger loop that is stabilized by the Mg++-NTP paired with the i+1 base. Based on the fast kinetics of EDTA quenching, the forward rates of NTP binding and sequestration (reverse isomerization of the TL) by RNAP II were estimated to be ~2.5 μM−1s−1 and ~1200 s−1, respectively (57). Mutations in the TL can change its isomerization rate dramatically (e.g. to 15 s−1 in the case of E1103G mutation in Rpb1 [57]), resulting in severely altered elongation rates and fidelity (7,57,50).
NTP Entry
The active site is buried deep inside the enzyme with all the major channels connecting it to the surrounding solution occupied by DNA and RNA. The only obvious path for NTP entry is through a ~20Å long secondary channel that is shaped like a funnel (Figure 2). At the junction with the catalytic center the secondary channel is only ~12Å in diameter, allowing the passage of only one NTP at a time (9). Moreover, the negative electrostatic potential that predominates in the secondary pore imposes a further constraint on NTP diffusion. Nevertheless, several indirect experimental observations suggest that NTPs enter via the secondary channel. The peptide antibiotic Microcin J25 (MccJ25) binds within the secondary channel and at least partially competes with NTP binding, suggesting that its inhibitory effect on transcription elongation is due to occlusion of the secondary channel (2,81). Alternatively, however, MccJ25 could negatively affect elongation by interfering with the movement of the TL and bridge-helix that constitute a part of the secondary channel. Moreover, MccJ25 could block PPi escape through the secondary channel, which would also result in reduction of the elongation rate. Another observation supporting NTP entry via the secondary channel is the binding of NTP to the E-site seen in crystals of the RNAP II EC (139,136). The E-site is located proximal to the secondary channel at its junction with the NTP insertion site. Furthermore, computer simulation indicates that the NTP binding in the E-site may compensate for the reduced diffusion rate imposed by the secondary pore so as to sustain an elongation rate at 10–20 nt/s (9). This predicted rate would be within the range of 15–80 nt/s established for cellular RNAPs.
An alternative NTP entry model has been proposed based on fast kinetic studies and computer modeling (85,35,36). It postulates that substrates enter through the main channel after pairing with the DNA template strand immediately ahead of the i+1 site. Several suggestive observations favor this scenario (16). The presence of NTPs complementary to the i+2 and i+3 bases seems to stimulate the rate of i+1 NTP loading and may reduce misincorporation. Also, the release of the NTP that had previously been sequestered in the i+1 site (reverse isomerization) in the presence of the RNAP II inhibitor α-amanitin is stimulated by i+2 and i+3 complementary substrates. It is speculated that exposure to i+2 and i+3 NTPs during an α-amanitin translocation blockade causes strain in the EC resulting in ejection of the i+1 NTP (35). At least some of the effects of “downstream” NTPs on elongation and misincorporation could be explained by the template misalignment detected in bacterial and T7 RNAPs (52), which would not contradict the hypothesis of NTP entry via the secondary channel. These data suggest that template misalignment due to occasional flipping of a template base out of the hybrid, rather than misincorporation in the i+1 site per se, is a preferred mechanism to explain substitution errors by cellular RNAPs (52).
The main channel NTP entry model requires that the i+2 and i+3 template bases be available for pairing with NTP substrates. Although in some EC crystal structures the bubble is indeed open at positions i+2 and i+3 (34,139), it is closed in others (55,129). Biochemical data argue that only one base pair is melted in front of the catalytic site (51). An unequivocal distinction between the main and secondary channel NTP entry models awaits at least some direct experimental evidence supporting or contradicting either of them.
Translocation
At any given position during RNA chain elongation, the EC oscillates between an elongation-competent post-translocated (i+1) state and inactive pre-translocated (i) and backtracked states (87,88,62,14). In the active conformation, the i+1 site is open to bind the next NTP, whereas during backtracking it is occupied by RNA (Figure 1). The energy of NTP hydrolysis is not used directly for RNAP translocation. Instead, a large body of evidence shows that the enzyme moves as a Brownian ratchet (7,1,14), driven forward by the energy gained from binding the correct substrate in the active site, which stabilizes the enzyme in the active conformation and suppresses backtracking. In other words, NTP acts as a stationary pawl in the EC ratchet mechanism (7). A similar ratchet mechanism was originally proposed for T7 RNAP (39). Several structural models have been proposed to describe the details of the translocation mechanism. A common feature of these models is a coordinated binding of NTP coupled with a conformational change of the BH/TL unit, resulting in a stepwise forward motion of RNAP with efficient incorporation of a correct substrate (116).
The initial translocation model was based on a comparative analysis of the first structures of bacterial and yeast RNAP. It proposed an elementary transcription cycle in which bending of the BH at the 3' face of the RNA:DNA hybrid induces translocation of the nucleic acid by 1 nt within the RNAP mainframe, while a subsequent relaxation of the BH opens the substrate binding site for the next complementary NTP (34). A bent BH was observed in the first bacterial RNAP structure (146), and a straight BH was observed in several subsequent bacterial and RNAP II structures (24,55,136,128). Consistently, protein-RNA crosslinking experiments on the E. coli EC revealed the ability of the BH to change its conformation within a single enzyme (29,7). It was proposed, therefore, that BH acts as a reciprocating pawl, pushing RNAP forward in relation to the nucleic acid scaffold, while the incoming substrate acts as a second, stationary pawl, preventing RNAP from slipping backward (Figure 4). The two pawls compete for the same substrate-binding site. The adjacent TL controls BH motion, thereby maintaining the proper balance between EC productive and inactive states (7) (Figure 4).
A modified version of the translocation model was proposed in which the TL, rather than the BH, plays the central role in RNAP movement (124,128). The binding of a correct NTP at the i+1 site restricts the lateral movement of RNAP and leads to the refolding of a TL into an extended α-helical conformation, which, in turn, further stabilizes NTP binding and results in formation of a `trapped' EC (128,136). In the `trapped' conformation, the TL obstructs the secondary channel. Thus, the NTP-induced TL refolding would favor forward translocation of RNAP simply by preventing backtracking. According to this proposal, the TL itself serves as a second reciprocal pawl, coupling RNA synthesis and translocation. This model, however, cannot fully explain the effect of TL mutations on RNAP translocation in the absence of substrates. For example, the G1136S mutation in the TL of E. coli RNAP favors the active state of the EC, whereas I1134V mutation favors the backtracking state (7). According to the structural predictions (128,136), TL refolding is not supposed to happen without correct substrates. Moreover, NTP-assisted translocation may, in principle, proceed independently of the TL, by the displacement of the 3' RNA nucleotide from Mg-A by the incoming NTP bound in the pre-insertion site (120). This may shift the EC equilibrium toward the post-translocated state.
The actual translocation mechanism is likely to combine features of both models, with the BH and TL serving as a “pushing” and anti-backtracking device, respectively, acting in concert. The recent biochemical and structural data obtained with the RNAP II-α-amanitin EC favor this mechanism (14). The refined structure of the RNAP II EC shows that the binding of α-amanitin shifts the central part of the BH by 2.5Å toward the 3' face of the RNA:DNA hybrid (Figure 4). Such a shift occludes the i+1 site and could result in the pushing of the hybrid in relation to the protein mainframe. The TL apparently stabilizes the shifted conformation of the BH by forming a wedge next to it (14) (Figure 4). These observations support a two-pawl ratchet mechanism of elongation in which the concerted movement of the BH and the TL facilitate translocation (7). It is worth noting that the “bent” conformation of the BH originally reported (146) is substantially distinct from two other conformations (straight and shifted) observed in more recent bacterial and yeast RNAP II structures and, thus, may not reflect any of the actual states of the BH. Nevertheless, the concept of a BH-based ratchet mechanism appears to be valid in light of recent structural and biochemical studies (34,7,14).
STRUCTURAL BASIS OF TRANSCRIPTION FIDELITY
Transcription is a remarkably accurate process with an error rate estimated to be less than 10−5 for bacterial and eukaryotic RNAPs (99,10,25). Since the free energy difference between the formation of correct vs. mismatched base pairs provides an accuracy of the order of 10−3 (71) the RNAP must enhance fidelity by at least two orders of magnitude. Available experimental data indicate that, similar to DNA replication, the high transcription fidelity is achieved by a combination of active NTP selection and proofreading by RNAP. Recent structural and biochemical analyses of misincorporation suggest detailed mechanisms for these processes.
NTP Selection
Recent genetic, biochemical and strcutural data implicated the TL as a central element responsible for NTP selection. Genetic screens of E.coli and yeast identified several TL mutations that severly impaired RNAP fidelity (7,57,50). β' G1136S mutation rendered the E.coli EC less discriminitive with regard to mismatched NTPs or matched 2'-dNTP, whereas the I1134V mutant enzyme was more accurate than wild type (7). Similiraly, genetically selected TL mutations H1085Y and E1103G compromised the fidelity of yeast RNAP II in vitro and in vivo (57). How does the TL function to select the correct substrate? As mentioned above, in the absense of a substarte the TL exists predominantly in an open conformation that is located ~30Å away from the NTP insertion site. However, it refolds from the open to closed state in response to binding of the correct NTP in the i+1 site (136,128). By closing around the active center the trigger loop transiently captures (or sequesters) the correct NTP, thus increasing the probability of its incorporation (128,50,57). In its folded conformation the TL contacts the NTP base and phosphate groups directly, forming a network of interactions that enables the substrate to bind by an induced fit mechnaism (Figures 3 and 4). Some TL contacts are not involved directly in substrate binding, but act via a network of interactions to stabilize the proper NTP in the i+1 site. For example, Q1078 of the TL helix interacts with N479 in RNAP II, which, in turn, forms a hydrogen bond with the 3'-OH group of the NTP. This interaction increases, by ~10 fold, the discrimination between the correct rNTP and one lacking a 3'-OH group (136). Precise alignment of interacting moieties not only fixes the correct substrate in the active center, but also properly positions it for catalysis (128,50). An incorrect NTP causes a misalignment to occur that would reduce the rate of NTP addition. Correct NTP selection may occur as a result of the preferential refolding of the TL back into the open conformation with release of the mismatched substrate and/or by affecting the chemistry step directly by the misalignment with catalytic residues. The former scenario is consistent with observations of an error-prone TL mutant, which displays prolonged NTP sequensteration for both correct and incorrect NTPs, and, thus, increases the probability of misincorporation (57). On the other hand, α-amanitin also renders RNAP II error-prone by directly binding to the TL and fixing it in the ≪half-opened≫ intermediate state (50,14,116). The induced fit mechnanism of NTP preselection is inhibited by the drug, resulting in increased rates of misincorporation.
In addition to the TL, some secondary channel residues have been implicated in nucleotide discrimination (45), consistent with the hypothesis that NTPs enter through the secondary channel. In contrast, the main-channel model of NTP entry postulates that, prior to the induced-fit step of NTP selection in the active site, NTP substrates are prescreened in the main channel by pairing with the i+2 and i+3 DNA bases, which are separated from the active site by the BH (85,145). Although the model is attractive because RNAP could achieve high fidelity with a relatively low-affinity substrate recognition and low-energy cost, it still awaits direct experimental proof.
Proofreading
In vitro and in vivo evidence indicate that RNAP is capable of detecting and removing incorrect nucleotides that have been mistakenly incorporated into the RNA chain. There are at least two distinct mechanisms of transcriptional error correction. One relies on the transcript cleavage activity of RNAP, which is strongly stimulated by the bacteria elongation factors GreA and GreB (13,92) and eukaryotic TFIIS (45). The second mechanism exploits the intrinsic exonuclease activity of RNAP (135,110,144).
The first demonstration that a cleavage factor (GreA) can enhance RNAP fidelity arose from a pre-steady state kinetic analysis of NTP misincorporation by E. coli RNAP (32). A similar demonstration was later obtained with TFIIS (48,121). A mechanism for this phenomenon became apparent when the concept of RNAP backtracking was introduced (62,88). In the backtracked EC, the extruded 3' terminal RNA portion becomes a substrate for the cleavage reaction, which is stimulated by cleavage factors. The factors gain access to the catalytic center via the secondary channel and donate the catalytic Mg++ to activate the cleavage reaction (70,54,91,111). Backtracking depends on the stability of the hybrid and the nature of the 3' terminal residue (88,110). The weaker the hybrid, the higher the probability of backtracking (88). Therefore, any mismatch would readily promote backtracking, which, in turn, would result in cleavage of the aberrant 3' terminal fragment. The structure of the T.t EC indicates that protein contacts with the hybrid may act as `shape sensors' that monitor the hybrid state to provide feedback in response to mismatches and/or dNMP incorporations, thereby contributing to transcription fidelity (129). For backtracking to occur, the TL must be in the open conformation, not to occlude the secondary channel. It is possible that protein domains that simultaneously interact with the hybrid and TL (e.g. fork-loop 1) stimulate opening of the TL in response to aberrations within the hybrid.
Transcriptional proofreading can also be achieved in vitro with the aid of intrinsic endonuclease activity, which is chemically distinct from the transcript cleavage reaction (Figure 1) (135,110,144). It has been proposed that the 3' mismatched nucleotide rotates into the pre-insertion or E-site from where it provides active groups and coordination bonds to the RNAP active center. As a result, hydrolysis of the penultimate phosphodiester bond occurs with the release of a dinucleotide monophosphate (144). This mechanism is analogous to the Mg++-dependent 3'-5' exonuclease mechanism in which a non-complementary NTP stimulates hydrolysis of the ultimate phosphodiester bond with release of the 3' terminal NMP (110,139).
The physiological role of exonuclease-mediated proofreading remains to be demonstrated. The in vitro rates of this reaction seem to be too slow to contribute significantly to transcriptional fidelity in vivo. On the other hand, at least one cleavage factor (TFIIS) and the Rpb9 subunit of RNAP II, which apparently stimulates transcript cleavage, have been shown to increase fidelity in vivo (86,65). Therefore, cleavage-mediated proofreading is a physiologically relevant mechanism of transcriptional error correction.
REGULATION OF CATALYSIS
The catalytic center of RNAP is the unltimate target for the factors and signals that regulate the eznyme activity. Control of the catalytic center can be achived by direct contact with its elements, or allosterically via other elements of RNAP that are connected to its moving and stationary parts. Factors that are capable of controlling RNAP include small molecules, protein factors, and signals encoded in nascent RNA.
Natural Small Molecule Inhibitors of Catalysis
Rifamycins (Rifs)
Ansamycin rifampicin and other Rifs, such as rifabutin and rifapentine, are the first antibiotics to be isolated that target RNAP (42). Rifs, at nanomolar concentrations, block formation of the second or third phosphodiester bond (77). Genetic, biochemical and structural evidence show that rifamycins bind to the β subunit pocket in the hybrid-binding site (HBS) and occlude the path of the elongating transcript at a length of 2–3 nt (82,104,19). In addition to the steric mechanism of inhibition, an allosteric model of the action of rifamycins has recently been proposed (5). According to this model, rifamycins also reduce the affinity of the catalytic Mg++A in the active site, which is ~19Å away from the rifamycins binding site. The structural basis for the allosteric mechanism remains unknown, although the available data suggest that it may involve the β and σ subunit domains that interact with DNA (5). The allosteric model can explain the absence of Mg++ in the RNAP-rifamycins structures, the ability of Mg++ to partially counteract rifampicin inhibition (53), and the ability of rifampicin to inhibit transcript cleavage and RNA synthesis in the RNAP-RNA binary complexes (3). The allosteric model of rifamycin action is intriguing, but requires a quantitative assessment of its relative contribution to the steric model. It would also be interesting to investigate whether the macrolide antibiotic sorangicin, which binds to the same β subunit pocket as does rifampicin, thereby blocking formation of the second and third phosphodiester bonds (18), may act via an allosteric mechanism as well.
Streptolydigin (Stl)
The tetramic acid antibiotic streptolydigin (Stl) is a less potent inhibitor of RNAP than are Rifs, with a Ki in the range of 10–100 μM (106,78). Stl binds about 20Å away from the active center and interacts with both the TL and BH (43,103,120,126). The mechanism of Stl inhibition, however, is not clearly understood. Stl does not compete with the substrate, but rather interferes with the moving parts of the catalytic center. One model proposed that Stl directly freezes the BH in its straight conformation, thereby preventing its oscillation, which may be required for translocation (126). This model also postulates that the straight BH conformation favors the active (post-translocated) state of the EC. However, the straight BH was observed in RNAP and EC structures without any drugs (55,136,127,129), indicating that Stl is unlikely to directly stabilize this conformation of the BH. Moreover, exonuclease III footprinting demonstrates no bias towards the post-translocated state of the EC in the presence of Stl (G. Bar-Nahum, V. Epshtein, E. Nudler, unpublished observations). Instead, Stl inhibits extensive backtracking, suggesting that it occludes the secondary channel either directly or indirectly. The alternative model of Stl action suggests that it stabilizes the inactive, substrate-bound pre-insertion state, presumably by interfering with closure of the active site by the TL (120).
Tagetitoxin (Tgt)
Tgt is a phytotoxin produced by Pseudomonas syringe pv. tagetis that naturally inhibits bacterial-type RNAP in chloroplasts of infected plants (76). Similar to Stl, Tgt suppresses all the activities of RNAP in vitro at low micromolar concentrations without directly competing with NTP binding (113,130). Structural data show that Tgt binds at the base of the secondary channel in the vicinity of the active center and may interfere with its moving parts, the BH and TL, thereby freezing the EC in the inactive state (130). Interestingly, the phosphate moiety of Tgt together with β' and β acidic residues coordinates the third Mg++ ion in the catalytic site, which is different from Mg-A and Mg-B (130). Because Tgt-Mg++ is coordinated by the same catalytic residue (β' Asp460 in T.t.) that is involved in the coordination of Mg-B, it could compromise Mg-B binding or disturb the correct geometry of the Mg-A and Mg-B, thereby inhibiting all catalytic reactions.
α-amanitin (Amt)
Amt is a bicyclic octapeptide toxin from the fungus Amanita phalloides that binds tightly to and inhibits the activity of RNAP II at very low concentrations (37). The Amt binding site is adjacent to the BH, suggesting that the drug interferes with its movement and, hence, RNAP translocation (17). The real time kinetic data also support this idea (35). The most recent structures of the RNAP II EC in complex with Amt provide further details of the mechanism of inhibition (14,50). Both EC structures demonstrate interactions between Amt and RNAP II that are somewhat different from those observed in the RNAP II-Amt binary complex, including additional direct and indirect contacts with the TL and BH domains of the catalytic center. Most significantly, the structures demonstrate that Amt fixes the TL/BH unit in a particular conformation that apparently favors the post-translocation state of the enzyme in relation of the RNA/DNA hybrid. In the absence of amanitin the enzyme oscillates between the pre- and post-translocated states, which could be observed directly within the crystals using 5-bromouracil analogs as the diffraction reference points (14). These structural data provide direct evidence supporting the previously proposed ratchet mechanism of elongation for bacterial RNAP (described above), in witch the oscillating BH acts as a pawl (7).
(p)ppGpp
ppGpp (guanosine 5'-diphosphate, 3'-diphosphate) is a small molecule alarmone that induces a set of responses to starvation known as the “stringent response” (72,95). The major effect ppGpp on transcription is the inhibition of initiation at ribosomal and tRNA gene promoters (72,95,23). This effect can be partially reconstituted in vitro (132), indicating that ppGpp acts directly on RNAP. ppGpp can also activate transcription from amino acid biosynthetic promoters in vivo and in vitro (8). It also decreases the elongation rate and promotes pausing (56,131). Studies of ppGpp cross-links to RNAP and RNAP structural studies provided somewhat conflicting information about a ppGpp-binding site. One crosslinking study involving the azido derivative of ppGpp indicates that it binds to the C-terminal part of the β subunit (22). A subsequent study utilizing a different crosslinkable analog, 6-thio-ppGpp, mapped its binding site primarily in the N-terminus of β' (125). However, ppGpp bound to RNAP leads to protection of both the N-terminal part of β' and a C-terminal part of the β-subunit (125). Taken together, these protein chemistry results suggest that together, the N terminus of the β' subunit and the C terminus of β subunit constitute a modular ppGpp-binding site, which is far from the catalytic center. In contrast, analysis of RNAP-ppGpp cocrystals positioned ppGpp at the base of the secondary channel near the catalytic center (6). Two modes of ppGpp binding were detected based on the proximity of the 5' and 3' diphosphates of ppGpp to the catalytic residues. The authors proposed that in the 5' orientation ppGpp could increase the affinity for the catalytic Mg++ ions and facilitate substrate binding and catalysis, which may explain the stimulating effect of ppGpp on transcription from certain promoters. However, the 3' orientation would lead to the opposite effect. Moreover, in this orientation ppGpp could compete directly with substrate molecules (6). A competition between ppGpp and NTPs has been observed in vitro (49). Interestingly, ppGpp bound in the 3' orientation might form base pairs with cytosines in the non-template DNA strand, potentially disrupting the protein-DNA interactions that stabilize the open promoter complex. The latter hypothesis could, in principle, explain the formation of inactive, stable, closed complexes induced by ppGpp (73). The recent genetic studies, however, raise doubts about the physiological relevance of the ppGpp-binding site revealed by the X-ray structures (132). Analysis of ten E.coli RNAPs with substitutions of the residues that supposedly form key contacts with ppGpp in T.t. cocrystals revealed no significant defect in responses to ppGpp in vivo (132). Therefore, the actual ppGpp binding site(s) and the exact mechanism of ppGpp-mediated control of RNAP remain to be clarified.
Protein Modulators of Catalysis
Factors acting through the secondary channel
Unlike the main and RNA exit channels that are occupied by nucleic acids, the secondary channel provides a direct and unobstructed passage from solution to the RNAP catalytic center (Figure 2). Several factors have been described that bind RNAP in the secondary channel to directly affect catalysis, including GreA/GreB and TFIIS transcript cleavage factors, the pH-dependent inhibitor of transcription Gfh1, and the ppGpp cofactor, DksA.
Despite the absence of any homology between bacterial DksA, Gre, and eukaryotic TFIIS, these factors share a structural similarity in their extended N-terminal coiled-coiled domains (NTDs) (112,94,54,55). All three factors bind RNAP and deliver their NTDs through the secondary channel to the catalytic center. DksA augments the effects of ppGpp on transcription during the stringent response, possibly by coordinating its Mg++ ion and stabilizing the RNAP–ppGpp complex (93,94). GreA/GreB and TFIIS participate directly in the catalysis of RNA hydrolysis by donating two acidic residues at the tip of NTD to coordinate Mg-B in the catalytic center (70,91,111,54,55). One of the Gre acidic residues (D41) also coordinates the attacking water molecule to stabilize the pentavalent transition state of the phosphate. Gre and TFIIS are the first known examples of transcription factors that modify RNAP activity by participating directly in the catalytic processes. Notably, the structural modeling suggests that the occupation of the secondary channel by Gre-NTD or TFIIS-NTD allows diffusion of the substrates and extrusion of backtracked RNA (11). In addition to coordinating the metal-ion, Gre-NTD may stimulate cleavage and facilitating backtracking due to its interaction with the TL/BH and with RNA (through its NTD basic patch residues) (70). The cleavage factors are important for overcoming promoter-proximal arrests and other elongation blocks that occur as a result of RNAP backtracking (88,140,75,11,66), for overcoming transcriptional roadblocks such as site-specific DNA-binding proteins and nucleosomes (30,58), and for transcriptional fidelity (32,48,121,65).
Gfh1 is present in some thermophilic bacteria. Although this factor shares substantial sequence similarity with GreA/GreB, it lacks any transcript cleavage activity (44,11). Instead, Gfh1 competes with GreA for binding to RNAP and serves as a general inhibitor of RNAP catalytic functions (69,118). Localized protein footprinting shows that the tip of Gfh1–NTD reaches the catalytic center of RNAP, implying that Gfh1 also acts through the RNAP secondary channel (69). Indeed, the structure of Tth Gfh1 shows an overall similarity to the structure of GreA with a notable exception of a flipped C-terminal domain (CTD) (68,118). Interestingly, the Gfh1-CTD switches its conformation depending on the pH. At higher pH (>7), it assumes an inactive `flipped' conformation, which prevents the factor from binding to RNAP. At lower pH, the Gfh1-CTD adopts an active `Gre-like' conformation, which enables Gfh1 to bind to and inhibit RNAP (69). Thus Gfh1 represents a pH-sensor that directly modulates the catalytic activity of RNAP.
Factors that act allosteriaclly
Since the secondary channel is the only direct unosbtructed route toward the catalytic center, factors that bind other parts of RNAP to control its catalytic activity must rely on allosteric mechanisms.
Bacterial Nus factors play a critical role in controlling the overall rate of elongation and efficiency of termination and antitermination (131,23,11,20,98). In vitro, NusG and NusA accelerate and decelerate elongation, respectively, by altering pausing at various sites (102,15). The eukaryotic homologue of NusG, DSIF, increases the rate of RNAP II elongation (134). In vitro analysis of E.coli TL mutants suggests that NusG and NusA control elongation by interfering with the TL/BH ratchet module (7). Thus, NusG modifies RNAP the same way as does the TL G1136S mutation (which accelerates elongation), by stabilizing the EC in the post-translocated (active) state. This “fast” mutant loses its responsiveness to NusG during elongation or translocation, suggesting that NusG is unable to promote TL/BH oscillation in this mutant enzyme. In contrast, NusA decreases the elongation rate and suppresses forward translocation (7). However, the TL mutant, I1134V, which slows elongation, becomes resistant to NusA-mediated pausing and translocation, suggesting that NusA is unable to exert any further effect on the TL/BH oscillation in this mutant. It remains to be determined exactly how Nus factors transmit their signals to the catalytic center. NusA has been proposed to act allosterically via the flexible β flap domain (123). NusG and its close paralog, the antitermination factors RfaH, which also accelerates elongation and stabilizes the post-translocated state of the EC (117), may directly or indirectly utilize the TL for this purpose. Indeed, the entire TL domain may be a universal adaptor for various factors, since it is large, unstructured, and surface-exposed in most RNAPs. Parts of the TL can be targeted by antibodies that modulate catalytic activity of the EC (142,100,31).
RNA Modulators of Catalysis
Signals in the nascent RNA control the rate and pocessivity of transcription by inducing pausing and termination. Unlike terminators, pauses do not directly cause the EC to dissociate, although they do have a crucial role in the termination process (79,40,41). There are two classes of pauses: RNA hairpin-dependent and backtracking-type pauses. The hairpin-dependent pause signal is complex and involves sequences adjacent to the pause hairpin, which forms 10–12 nt from the RNA 3' end to halt the EC (21). Backtracking-type pauses do not require any specific RNA structure and/or sequence. These pauses are determined primarily by the free energy of the base pairs of the RNA-DNA hybrid, of the DNA duplexes flanking the transcription bubble, and of those that form co-transcriptionally in the RNA immediately upstream of the RNA exit channel (88,119). Recent biochemical data indicate that there is a common `trapped' intermediate for both types of pauses from which the EC either backtracks or remains in the trapped conformation, stabilized by the RNA pause hairpin (7,123). The trapping is likely to occur as a result of jamming of the BH/TL module, which incapacitates RNAP, preventing it from bringing the correct substrate to the i+1 site and prohibiting forward translocation (7,136,128,14,50,116). How exactly the pause hairpin transmits the signal to the catalytic center, however, is unclear. It has been proposed that the hairpin acts allosterically by moving the flap domain of β, thereby changing the network of protein-nucleic acid interactions that maintain alignment of the RNA 3' OH in the catalytic center (123).
A similar trapped state can be detected during intrinsic termination just prior to EC dissociation (40,31) (Figure 5). Intrinsic termination signals appear in RNA as a strong RNA hairpin 7–8 nt upstream of the RNA 3' terminus, followed by a stretch of several U residues. The pathway to termination begins with a backtracking-type pause caused by a weak A:U hybrid, which gives the hairpin enough time to initiate folding in the RNA exit channel (40). The fully folded hairpin can then protrude to the main channel to unwind most of the hybrid and irreversibly trap the EC by clashing with the TL (31) (Figure 5). A direct contact between the TL and hairpin has two consequences. First, since the TL is part of the catalytic site, its hairpin-induced folding into the closed or folded state can inhibit substrate binding (7,136,128), resulting in an irreversible trapping of RNAP in a pre-translocated position (trapped complex). This idea is supported by the crosslinking of the 3'-end of the RNA to the TL in the trapped complex and by the fact that point mutations in the TL change the efficiency of termination by affecting the rate at which RNAP enters the trapped state (31). In addition, the forced motion of the TL, which constitutes the sidewall of the DNA-binding clamp, may induce the movement of adjacent structural elements (such as β fork-2) (136,128,27) resulting in the simultaneous opening of the clamp. In turn, opening of the clamp would remove all major interactions between RNAP and nucleic acids, thus inducing simultaneous release of the transcript and DNA. Since the structural organization and principles by which RNA and DNA are retained in the EC are highly conserved among all cellular RNAPs (63,34,55,136,129), the basic steps leading to termination are likely to be universal. Prokaryotic and eukaryotic termination factors and intrinsic signals are likely to utilize the same strategy to rapidly disrupt the extremely stable and processive ECs. Thus, similar to intrinsic terminators, bacterial termination factor Rho (20) first rapidly inactivates (traps) the EC allosterically and then invades the RNAP main channel to disrupt the trapped complex (V. Epshtein, D. Dutta, and E. Nudler, unpublished).
Figure 5. Structural model of the trapped intermediate during termination.
RNAP structure with the explanatory scheme represents the last stage of the termination process (40,31). Key structural elements are color coded (see Figure 2b): gray (green) – non-template (template) strand of DNA; red – nascent RNA; yellow - β flap; violet – β' lid; blue – β' zipper; green – Zn-finger; rose and gold - β lobes 1 and 2; aqua - β rudder; dark blue - TL; teal – DNA clamp and other parts of β'. Star - the catalytic center. The termination RNA hairpin stem grows to its final size of 7–8 bp invading the primary channel, unwinding the upstream portion of the hybrid and initiating collapse of the upstream edge of the transcription bubble. The head of the hairpin bends around the downstream edge of the transcription bubble, clashing with the TL. This may force the TL to move towards the active site and permanently fold into a “closed” conformation. Additionally, the hairpin action results in disruption of interactions in the RBS and HBS and distortion of the hybrid, which could cause complete and irreversible enzyme inactivation. At the same time, the TL movement induces DNA-binding-clamp (DBS) opening, resulting in a simultaneous release of both the nascent RNA and the DNA template.
CONCLUSIONS
A series of high-resolution structures of RNAP supported by extensive biochemical studies provide an elaborate view of RNAP as a complex molecular machine built around its catalytic center. The latter is revealed as a highly dynamic and flexible structure that serves as both the molecular engine and ultimate regulatory sensor. Many fundamental aspects of catalytic center function have become known. We now understand the general structural principles of the nucleotide addition cycle, substrate selection and transcriptional fidelity. We also understand the mechanism by which some natural, small molecular drugs and transcription factors interfere with this process. However, many important details concerning the mechanics of regulation and refinement of RNAP function remain to be elucidated. These include the precise molecular mechanism of translocation, intrinsic and factor-dependent termination, and factor-dependent regulation of these processes. The prospects are exciting, as structures of elongating RNAP in various conformational states complexed with regulatory factors are likely to be solved in the next few years. At the same time, biochemistry and protein chemistry, coupled with fast kinetic analysis and mathematical modeling, should clarify the structural intricacies of the elongation and termination processes.
SUMMARY POINTS.
Structural and biochemical studies of bacterial and yeast RNAPs reveal the precise organization of the EC: location of nucleic acids relative to the catalytic center.
The multifunctional catalytic center is revealed as a complex dynamic structure, serving as the ultimate effector of regulatory signals and inhibitory drugs.
Recent structural and biochemical studies explain the basic principles of substrate (NTP) binding, selection and transcription fidelity.
RNAP translocation is proven to occur by a Brownian ratchet mechanism.
Structural and biochemical studies of RNAP complexes with natural small molecular inhibitors and regulatory factors reveal both direct and allosteric mechanisms of modulation of the catalytic center.
The active role of the catalytic center in transcription termination and pausing is revealed.
FUTURE ISSUES.
What are the fine details of the structural mechanism of RNAP translocation?
How do various elongation and termination factors that control the rate and processivity of transcription transmit their signals to the catalytic center?
How does the catalytic center sense the intrinsic RNA and DNA regulatory signals that control the rate and processivity of transcription?
Will rational drug design be useful to target the RNAP catalytic center for application as antimicrobial therapy and gene regulation?
ACKNOWLEDGMENTS
I apologize to the many investigators whose specific results could not be cited owing to space limitations but whose work framed the questions and ideas that are discussed in this review. This work is supported by grants from the NIH.
MINI-GLOSSARY
- Nucleotide addition cycle (NAC)
the elementary enzymatic cycle of RNAP that involves three basic steps: substrate binding to the EC (NTP binding), nucleotide incorporation into the nascent transcript by phosphodiester bond formation (chemistry), and translocation of the nucleic acids scaffold in relation to the RNAP mainframe (translocation).
- BH and TL
the mobile elements of the active center, working in concert to ensure proper placement of the correct NTP at the active site and incorporation into RNA, as well as RNAP translocation, pausing, and termination.
- Rifamycins, steptolydigin, tagetitoxin, sorangicin, microcin J25
antibiotics working as natural small molecule inhibitors of bacterial RNAP. The high-resolution structures of their complexes with RNAP have been solved.
- α-amanitin
a powerful fungal toxin that inhibits eukaryotic RNAP II. The high-resolution structures of α-amanitin complexed with the EC have been solved.
- ppGpp
guanosine 5'-diphosphate, 3'-diphosphate - a signaling molecule that induces a set of responses to starvation known as the “stringent response” in bacteria.
- GreA/GreB and TFIIS
transcription elongation factors that stimulate intrinsic transcript cleavage activity of bacterial RNAP and eukaryotic RNAP II, respectively.
- NusA and NusG
transcription factors that control the rate of elongation and the efficiency of termination and antitermination in bacteria.
- Intrinsic terminator
a signal in the nascent RNA composed of a strong hairpin followed by a strecth of several U residiues that causes rapid inactivation and dissociation of the EC in bacteria.
ACRONYMS
- Amt
α-amanitin
- EC
elongation complex
- BH
bridge-helix
- CTD
carboxy terminal domain
- DBS
DNA duplex binding site
- HBS
RNA:DNA hybrid binding site
- i
the product (3' terminal RNA residue) binding site in the RNAP catalytic center
- i+1
the substrate binding site in the RNAP catalytic center
- MccJ25
peptide antibiotic Microcin J25
- NAC
nucleotide addition cycle
- NTD
amino terminal domain
- NTP
ribonucleoside triphosphate
- ppGpp
guanosine tetraphosphate
- PPi
pyrophosphate
- RBA
single stranded RNA binding site
- Rif
rifampicin
- RNAP
RNA polymerase
- Stl
streptolydidgin
- Tgt
tagetitoxin
- TL
trigger-loop
Footnotes
DISCLOSURE STATEMENT The author is not aware of any conditions that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Abbondanzieri E, et al. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005;438:460–465. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adelman K, et al. Molecular mechanism of transcription inhibition by peptide antibiotic Microcin J25. Mol. Cell. 2004;14:753–762. doi: 10.1016/j.molcel.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 3.Altmann CR, Solow-Cordero DE, Chamberlin MJ. RNA cleavage and chain elongation by Escherichia coli DNA-dependent RNA polymerase in a binary enzyme. RNA complex. Proc Natl Acad Sci U S A. 1994;91:3784–8. doi: 10.1073/pnas.91.9.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Archambault J, Friesen J. Genetics of RNA polymerases I, II and III. Microbiol Rev. 1993;57:703–724. doi: 10.1128/mr.57.3.703-724.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Artsimovitch I, et al. Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins. Cell. 2005;122:351–63. doi: 10.1016/j.cell.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 6.Artsimovitch I, et al. Structural basis for transcription regulation by alarmone ppGpp. Cell. 2004;117:299–310. doi: 10.1016/s0092-8674(04)00401-5. [DOI] [PubMed] [Google Scholar]
- 7.Bar-Nahum, et al. A ratchet mechanism of transcription elongation and its control. Cell. 2005;120:183–193. doi: 10.1016/j.cell.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 8.Barker M, Gaal T, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. II. Models for positive control based on properties of RNAP mutants and competition for RNAP. J Mol Biol. 2001;305:689–702. doi: 10.1006/jmbi.2000.4328. [DOI] [PubMed] [Google Scholar]
- 9.Batada N, Westover K, Bushnell D, Levitt M, Kornberg RD. Diffusion of nucleoside triphosphates and role of the entry site to the RNA polymerase II active center. Proc Natl Acad Sci U S A. 2004;101(50):17361–4. doi: 10.1073/pnas.0408168101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blank A, Gallant J, Burgess R, Loeb L. An RNA polymerase mutant with reduced accuracy of chain elongation. Biochemistry. 1986;25:5920–8. doi: 10.1021/bi00368a013. [DOI] [PubMed] [Google Scholar]
- 11.Borukhov S, Lee J, Laptenko O. Bacterial transcription elongation factors: new insights into molecular mechanism of action. Mol Microbiol. 2005;55:1315–24. doi: 10.1111/j.1365-2958.2004.04481.x. [DOI] [PubMed] [Google Scholar]
- 12.Borukhov S, Nudler E. RNA polymerase: the vehicle of transcription. Trends Microbiol. 2008;16:126–34. doi: 10.1016/j.tim.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 13.Borukhov S, Sagitov V, Goldfarb A. Transcript cleavage factors from E. coli. Cell. 1993;72:459–466. doi: 10.1016/0092-8674(93)90121-6. [DOI] [PubMed] [Google Scholar]
- 14.Brueckner F, Cramer P. Structural basis of transcription inhibition by alpha-amanitin and implications for RNA polymerase II translocation. Nat Struct Mol Biol. 2008;15:811–818. doi: 10.1038/nsmb.1458. [DOI] [PubMed] [Google Scholar]
- 15.Burova E, Hung S, Sagitov V, Stitt B, Gottesman M. Escherichia coli NusG protein stimulates transcription elongation rates in vivo and in vitro. J Bacteriol. 1995;177:1388–1392. doi: 10.1128/jb.177.5.1388-1392.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burton Z, Feig M, Gong X, Zhang C, Nedialkov Y, Xiong Y. NTP-driven translocation and regulation of downstream template opening by multi-subunit RNA polymerases. Biochem Cell Biol. 2005;83:486–496. doi: 10.1139/o05-059. [DOI] [PubMed] [Google Scholar]
- 17.Bushnell D, Cramer P, Kornberg R. Structural basis of transcription: alpha-amanitin-RNA polymerase II cocrystal at 2.8 A resolution. Proc Natl Acad Sci U S A. 2002;99:1218–1222. doi: 10.1073/pnas.251664698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campbell E, et al. Structural, functional, and genetic analysis of sorangicin inhibition of bacterial RNA polymerase. EMBO J. 2005;24:674–682. doi: 10.1038/sj.emboj.7600499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campbell E, et al. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell. 2001;104:901–12. doi: 10.1016/s0092-8674(01)00286-0. [DOI] [PubMed] [Google Scholar]
- 20.Cardinale CJ, et al. Termination factor Rho and its cofactors NusA and NusG silence foreign DNA in E. coli. Science. 2008;320:935–938. doi: 10.1126/science.1152763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan C, Wang D, Landick R. Multiple interactions stabilize a single paused transcription intermediate in which hairpin to 3' end spacing distinguishes pause and termination pathways. J Mol Biol. 1997;268:54–68. doi: 10.1006/jmbi.1997.0935. [DOI] [PubMed] [Google Scholar]
- 22.Chatterji D, Fujita N, Ishihama A. The mediator for stringent control, ppGpp, binds to the beta-subunit of Escherichia coli RNA polymerase. Genes Cells. 1998;3:279–287. doi: 10.1046/j.1365-2443.1998.00190.x. [DOI] [PubMed] [Google Scholar]
- 23.Condon C, Squires C, Squires C. Control of rRNA transcription in Escherichia coli. Microbiol Rev. 1995;59:623–645. doi: 10.1128/mr.59.4.623-645.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cramer P, Bushnell D, Kornberg R. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 25.de Mercoyrol L, Corda Y, Job C, Job D. Accuracy of wheat-germ RNA polymerase II. General enzymatic properties and effect of template conformational transition from right-handed B-DNA to left-handed Z-DNA. Eur J Biochem. 1992;206:49–58. doi: 10.1111/j.1432-1033.1992.tb16900.x. [DOI] [PubMed] [Google Scholar]
- 26.Ebright R. RNA polymerase: structural similarities between bacterial RNA polymerase and eukaryotic RNA polymerase II. J Mol Biol. 2000;304:687–698. doi: 10.1006/jmbi.2000.4309. [DOI] [PubMed] [Google Scholar]
- 27.Ederth J, Mooney R, Isaksson L, Landick R. Functional interplay between the jaw domain of bacterial RNA polymerase and allele-specific residues in the product RNA-binding pocket. J Mol Biol. 2006;356:1163–1179. doi: 10.1016/j.jmb.2005.11.080. [DOI] [PubMed] [Google Scholar]
- 28.Epshtein V, Nudler E. Cooperation between RNA polymerase molecules in transcription elongation. Science. 2003;300:801–805. doi: 10.1126/science.1083219. [DOI] [PubMed] [Google Scholar]
- 29.Epshtein V, et al. Swing-gate model of nucleotide entry into the RNA polymerase active center. Mol Cell. 2002;10:623–634. doi: 10.1016/s1097-2765(02)00640-8. [DOI] [PubMed] [Google Scholar]
- 30.Epshtein V, Toulmé F, Rahmouni A, Borukhov S, Nudler E. Transcription through the roadblocks: the role of RNA polymerase cooperation. EMBO J. 2003;22:4719–4727. doi: 10.1093/emboj/cdg452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Epshtein V, Cardinale C, Ruckenstein A, Borukhov S, Nudler E. An allosteric path to transcription termination. Mol. Cell. 2007;28:991–1001. doi: 10.1016/j.molcel.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 32.Erie D, Hajiseyedjavadi O, Young MC, von Hippel P. Multiple RNA polymerase conformations and GreA: control of the fidelity of transcription. Science. 1993;262:867–873. doi: 10.1126/science.8235608. [DOI] [PubMed] [Google Scholar]
- 33.Galburt E, et al. Backtracking determines the force sensitivity of RNAP II in a factor-dependent manner. Nature. 2007;446:820–823. doi: 10.1038/nature05701. [DOI] [PubMed] [Google Scholar]
- 34.Gnatt A, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 35.Gong X, Nedialkov Y, Burton Z. Alpha-amanitin blocks translocation by human RNA polymerase II. J Biol Chem. 2004;279:27422–27427. doi: 10.1074/jbc.M402163200. [DOI] [PubMed] [Google Scholar]
- 36.Gong X, Zhang C, Feig M, Burton Z. Dynamic error correction and regulation of downstream bubble opening by human RNA polymerase II. Mol Cell. 2005;18:461–470. doi: 10.1016/j.molcel.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 37.Greenleaf A. Amanitin-resistant RNA polymerase II mutations are in the enzyme's largest subunit. J Biol Chem. 1983;258:13403–13406. [PubMed] [Google Scholar]
- 38.Greive S, von Hippel P. Thinking quantitatively about transcriptional regulation. Nat Rev Mol Cell Biol. 2005;6:221–232. doi: 10.1038/nrm1588. [DOI] [PubMed] [Google Scholar]
- 39.Guajardo R, Sousa R. A model for the mechanism of polymerase translocation. J. Mol. Biol. 1997;265:8–19. doi: 10.1006/jmbi.1996.0707. [DOI] [PubMed] [Google Scholar]
- 40.Gusarov I, Nudler E. The mechanism of intrinsic transcription termination. Mol. Cell. 1999;3:495–504. doi: 10.1016/s1097-2765(00)80477-3. [DOI] [PubMed] [Google Scholar]
- 41.Gusarov I, Nudler E. Control of intrinsic transcription termination by N and NusA: the basic mechanisms. Cell. 2001;107:437–449. doi: 10.1016/s0092-8674(01)00582-7. [DOI] [PubMed] [Google Scholar]
- 42.Hartmann G, Honikel K, Knüsel F, Nüesch J. The specific inhibition of the DNA-directed RNA synthesis by rifamycin. Biochim Biophys Acta. 1967;145:843–844. doi: 10.1016/0005-2787(67)90147-5. [DOI] [PubMed] [Google Scholar]
- 43.Heisler L, Suzuki H, Landick R, Gross C. Four contiguous amino acids define the target for streptolydigin resistance in the beta subunit of Escherichia coli RNA polymerase. J Biol Chem. 1993;268:25369–25375. [PubMed] [Google Scholar]
- 44.Hogan B, Hartsch T, Erie D. Transcript cleavage by Thermus thermophilus RNA polymerase. Effects of GreA and anti-GreA factors. J Biol Chem. 2002;277:967–975. doi: 10.1074/jbc.M108737200. [DOI] [PubMed] [Google Scholar]
- 45.Holmes S, Santangelo T, Cunningham C, Roberts J, Erie D. Kinetic investigation of Escherichia coli RNA polymerase mutants that influence nucleotide discrimination and transcription fidelity. J Biol Chem. 2006;281:18677–18683. doi: 10.1074/jbc.M600543200. [DOI] [PubMed] [Google Scholar]
- 46.Hsu L, Vo N, Kane C, Chamberlin M. In vitro studies of transcript initiation by Escherichia coli RNA polymerase. 1. RNA chain initiation, abortive initiation, and promoter escape at three bacteriophage promoters. Biochemistry. 2003;42:3777–3786. doi: 10.1021/bi026954e. [DOI] [PubMed] [Google Scholar]
- 47.Izban M, Luse D. The RNA polymerase II ternary complex cleaves the nascent transcript in a 3'----5' direction in the presence of elongation factor SII. Genes Dev. 1992;6:1342–1356. doi: 10.1101/gad.6.7.1342. [DOI] [PubMed] [Google Scholar]
- 48.Jeon C, Agarwal K. Fidelity of RNA polymerase II transcription controlled by elongation factor TFIIS. Proc Natl Acad Sci U S A. 1996;93(24):13677–82. doi: 10.1073/pnas.93.24.13677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jöres L, Wagner R. Essential steps in the ppGpp-dependent regulation of bacterial ribosomal RNA promoters can be explained by substrate competition. J Biol Chem. 2003;278:16834–16843. doi: 10.1074/jbc.M300196200. [DOI] [PubMed] [Google Scholar]
- 50.Kaplan C, Larsson K, Kornberg R. The RNA polymerase II trigger loop functions in substrate selection and is directly targeted by alpha-amanitin. Mol Cell. 2008;30:547–556. doi: 10.1016/j.molcel.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kashkina E, et al. Multisubunit RNA polymerases melt only a single DNA base pair downstream of the active site. J Biol Chem. 2007;282:21578–21582. doi: 10.1074/jbc.C700098200. [DOI] [PubMed] [Google Scholar]
- 52.Kashkina E, et al. Template misalignment in multisubunit RNA polymerases and transcription fidelity. Mol Cell. 2006;24:257–266. doi: 10.1016/j.molcel.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 53.Kerrich-Santo R, Hartmann G. Influence of temperature on the action of rifampicin on RNA polymerase in presence of DNA. Eur J Biochem. 1974;43:521–532. doi: 10.1111/j.1432-1033.1974.tb03439.x. [DOI] [PubMed] [Google Scholar]
- 54.Kettenberger H, Armache K, Cramer P. Architecture of the RNA polymerase II-TFIIS complex and implications for mRNA cleavage. Cell. 2003;114:347–357. doi: 10.1016/s0092-8674(03)00598-1. [DOI] [PubMed] [Google Scholar]
- 55.Kettenberger H, Armache K, Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol Cell. 2004;16:955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 56.Kingston R, Nierman W, Chamberlin M. A direct effect of guanosine tetraphosphate on pausing of Escherichia coli RNA polymerase during RNA chain elongation. J Biol Chem. 1981;256:2787–2797. [PubMed] [Google Scholar]
- 57.Kireeva M, et al. Transient reversal of RNA polymerase II active site closing controls fidelity of transcription elongation. Mol Cell. 2008;30:557–566. doi: 10.1016/j.molcel.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kireeva M, et al. Nature of the nucleosomal barrier to RNA polymerase II. Mol Cell. 2005;18:97–108. doi: 10.1016/j.molcel.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 59.Kireeva M, Komissarova N, Kashlev M. Overextended RNA:DNA hybrid as a negative regulator of RNA polymerase II processivity. J Mol Biol. 2000;299:325–335. doi: 10.1006/jmbi.2000.3755. [DOI] [PubMed] [Google Scholar]
- 60.Kireeva M, et al. The 8-nucleotide-long RNA:DNA hybrid is a primary stability determinant of the RNA polymerase II elongation complex. J Biol Chem. 2000;275:6530–6536. doi: 10.1074/jbc.275.9.6530. [DOI] [PubMed] [Google Scholar]
- 61.Komissarova N, Kashlev M. Transcriptional arrest: Escherichia coli RNA polymerase translocates backward, leaving the 3' end of the RNA intact and extruded. Proc Natl Acad Sci U S A. 1997;94(5):1755–60. doi: 10.1073/pnas.94.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Komissarova N, Kashlev M. RNA polymerase switches between inactivated and activated states by translocating back and forth along the DNA and the RNA. J Biol Chem. 1997;272:15329–15338. doi: 10.1074/jbc.272.24.15329. [DOI] [PubMed] [Google Scholar]
- 63.Korzheva N, et al. A structural model of transcription elongation. Science. 2000;289:619–625. doi: 10.1126/science.289.5479.619. [DOI] [PubMed] [Google Scholar]
- 64.Korzheva N, Mustaev A, Nudler E, Nikiforov V, Goldfarb A. Mechanistic model of the elongation complex of Escherichia coli RNA polymerase. Cold Spring Harb Symp Quant Biol. 1998;63:337–45. doi: 10.1101/sqb.1998.63.337. [DOI] [PubMed] [Google Scholar]
- 65.Koyama H, Ito T, Nakanishi T, Sekimizu K. Stimulation of RNA polymerase II transcript cleavage activity contributes to maintain transcriptional fidelity in yeast. Genes Cells. 2007;12:547–559. doi: 10.1111/j.1365-2443.2007.01072.x. [DOI] [PubMed] [Google Scholar]
- 66.Kulish D, Struhl K. TFIIS enhances transcriptional elongation through an artificial arrest site in vivo. Mol Cell Biol. 2001;21:4162–4168. doi: 10.1128/MCB.21.13.4162-4168.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kuznedelov K, Korzheva N, Mustaev A, Severinov K. Structure-based analysis of RNA polymerase function: the largest subunit's rudder contributes critically to elongation complex stability and is not involved in the maintenance of RNA-DNA hybrid length. EMBO J. 2002;21:1369–1378. doi: 10.1093/emboj/21.6.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lamour V, Hogan B, Erie D, Darst S. Crystal structure of Thermus aquaticus Gfh1, a Gre-factor paralog that inhibits rather than stimulates transcript cleavage. J Mol Biol. 2006;356:179–188. doi: 10.1016/j.jmb.2005.10.083. [DOI] [PubMed] [Google Scholar]
- 69.Laptenko O, et al. pH-dependent conformational switch activates the inhibitor of transcription elongation. EMBO J. 2006;25(10):2131–41. doi: 10.1038/sj.emboj.7601094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Laptenko O, Lee J, Lomakin I, Borukhov S. Transcript cleavage factors GreA and GreB act as transient catalytic components of RNA polymerase. EMBO J. 2003;22:6322–6334. doi: 10.1093/emboj/cdg610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Loeb L, Kunkel T. Fidelity of DNA synthesis. Annu Rev Biochem. 1982;51:429–57. doi: 10.1146/annurev.bi.51.070182.002241. [DOI] [PubMed] [Google Scholar]
- 72.Magnusson L, Farewell A, Nystrom T. ppGpp: a global regulator in Escherichia coli. Trends Microbiol. 2005;13:236–242. doi: 10.1016/j.tim.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 73.Maitra A, Shulgina I, Hernandez V. Conversion of active promoter-RNA polymerase complexes into inactive promoter bound complexes in E. coli by the transcription effector, ppGpp. Mol Cell. 2005;17:817–829. doi: 10.1016/j.molcel.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 74.Markovtsov V, Mustaev A, Goldfarb A. Protein-RNA interactions in the active center of transcription elongation complex. Proc Natl Acad Sci U S A. 1996;93(8):3221–6. doi: 10.1073/pnas.93.8.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Marr M, Roberts J. Function of transcription cleavage factors GreA and GreB at a regulatory pause site. Mol Cell. 2000;6:1275–1285. doi: 10.1016/s1097-2765(00)00126-x. [DOI] [PubMed] [Google Scholar]
- 76.Mathews D, Durbin R. Tagetitoxin inhibits RNA synthesis directed by RNA polymerases from chloroplasts and Escherichia coli. J Biol Chem. 1990;265:493–498. [PubMed] [Google Scholar]
- 77.McClure W, Cech C. On the mechanism of rifampicin inhibition of RNA synthesis. J Biol Chem. 1978;253:8949–8956. [PubMed] [Google Scholar]
- 78.McClure W. On the mechanism of streptolydigin inhibition of Escherichia coli RNA polymerase. J. Biol. Chem. 1980;255:1610–1616. [PubMed] [Google Scholar]
- 79.McDowell J, et al. Determination of intrinsic transcription termination efficiency by RNA polymerase elongation rate. Science. 1994;266:822–825. doi: 10.1126/science.7526463. [DOI] [PubMed] [Google Scholar]
- 80.Minakhin L, et al. Bacterial RNA polymerase subunit w and eukaryotic RNA polymerase subunit RPB6 are sequence, structural, and functional homologs and promote RNA polymerase assembly. Proc Natl Acad Sci USA. 2001;98:892–897. doi: 10.1073/pnas.98.3.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mukhopadhyay J, et al. Antibacterial peptide microcin J25 inhibits transcription by binding within and obstructing the RNA polymerase secondary channel. Mol. Cell. 2004;14:739–751. doi: 10.1016/j.molcel.2004.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mustaev A, et al. Topology of the RNA polymerase active center probed by chimeric rifampicin-nucleotide compounds. Proc Natl Acad Sci U S A. 1994;91:12036–12340. doi: 10.1073/pnas.91.25.12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mustaev A, et al. Modular organization of the catalytic center of RNA polymerase. Proc Natl Acad Sci U S A. 1997;94:6641–6645. doi: 10.1073/pnas.94.13.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Naryshkina T, Kuznedelov K, Severinov K. The role of the largest RNA polymerase subunit lid element in preventing the formation of extended RNA-DNA hybrid. J Mol Biol. 2006;361:634–643. doi: 10.1016/j.jmb.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 85.Nedialkov Y, et al. NTP-driven translocation by human RNA polymerase II. J Biol Chem. 2003;278:18303–18312. doi: 10.1074/jbc.M301103200. [DOI] [PubMed] [Google Scholar]
- 86.Nesser N, Peterson D, Hawley D. RNA polymerase II subunit Rpb9 is important for transcriptional fidelity in vivo. Proc Natl Acad Sci U S A. 2006;103:3268–3273. doi: 10.1073/pnas.0511330103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nudler E. Transcription elongation: structural basis and mechanisms. J Mol Biol. 1999;288:1–12. doi: 10.1006/jmbi.1999.2641. [DOI] [PubMed] [Google Scholar]
- 88.Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. The RNA/DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell. 1997;89:33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- 89.Nudler E, Avetissova E, Markovtsov V, Goldfarb A. Transcription processivity: protein DNA interactions holding together the elongation complex. Science. 1996;273:211–217. doi: 10.1126/science.273.5272.211. [DOI] [PubMed] [Google Scholar]
- 90.Nudler E, Gusarov I, Avetissova E, Kozlov M, Goldfarb A. Topology of transcription elongation complex in Escherichia coli. Science. 1998;281:424–428. doi: 10.1126/science.281.5375.424. [DOI] [PubMed] [Google Scholar]
- 91.Opalka N, et al. Structure and function of the transcription elongation factor GreB bound to bacterial RNA polymerase. Cell. 2003;114:335–345. doi: 10.1016/s0092-8674(03)00600-7. [DOI] [PubMed] [Google Scholar]
- 92.Orlova M, Newlands J, Das A, Goldfarb A, Borukhov S. Intrinsic transcript cleavage activity of RNA polymerase. Proc Natl Acad Sci U S A. 1995;92:4596–4600. doi: 10.1073/pnas.92.10.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Paul B, et al. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell. 2004;118:311–322. doi: 10.1016/j.cell.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 94.Perederina A, et al. Regulation through the secondary channel–structural framework for ppGpp-DksA synergism during transcription. Cell. 2004;118:297–309. doi: 10.1016/j.cell.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 95.Potrykus K, Cashel M. (p)ppGpp: Still Magical? Annu Rev Microbiol. 2008 May 2; doi: 10.1146/annurev.micro.62.081307.162903. 2008. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 96.Rees W, Keller R, Vesenka J, Yang G, Bustamante C. Evidence of DNA bending in transcription complexes imaged by scanning force microscopy. Science. 1993;260:1646–1649. doi: 10.1126/science.8503010. [DOI] [PubMed] [Google Scholar]
- 97.Revyakin A, Liu C, Ebright RH, Strick TR. Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science. 2006;314:1139–1143. doi: 10.1126/science.1131398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Roberts J, Shankar S, Filter J. RNA polymerase elongation factors. Annu. Rev. Microbiol. 2008;62:211–33. doi: 10.1146/annurev.micro.61.080706.093422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rosenberger R, Hilton J. The frequency of transcriptional and translational errors at nonsense codons in the lacZ gene of Escherichia coli. Mol Gen Genet. 1983;191:207–212. doi: 10.1007/BF00334815. [DOI] [PubMed] [Google Scholar]
- 100.Rouby J, et al. Characterization of monoclonal antibodies against Escherichia coli core RNA polymerase. Biochem J. 2002;361:347–354. doi: 10.1042/0264-6021:3610347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rozovskaia T, Chenchik A, Bibilashvili R. Reaction of pyrophosphorolysis catalyzed by Escherichia coli RNA polymerase. Mol Biol (Mosk) 1981;15:636–652. [PubMed] [Google Scholar]
- 102.Schmidt M, Chamberlin M. Amplification and isolation of Escherichia coli nusA protein and studies of its effects on in vitro RNA chain elongation. Biochemistry. 1984;23:197–203. doi: 10.1021/bi00297a004. [DOI] [PubMed] [Google Scholar]
- 103.Severinov K, et al. Streptolydigin-resistant mutants in an evolutionarily conserved region of the beta' subunit of Escherichia coli RNA polymerase. J Biol Chem. 1995;270:23926–23929. doi: 10.1074/jbc.270.41.23926. [DOI] [PubMed] [Google Scholar]
- 104.Severinov K, et al. The beta subunit Rif-cluster I is only angstroms away from the active center of Escherichia coli RNA polymerase. J Biol Chem. 1995;270:29428–29432. doi: 10.1074/jbc.270.49.29428. [DOI] [PubMed] [Google Scholar]
- 105.Shilatifard A, Conaway R, Conaway J. The RNA polymerase II elongation complex. Annu Rev Biochem. 2003;72:693–715. doi: 10.1146/annurev.biochem.72.121801.161551. [DOI] [PubMed] [Google Scholar]
- 106.Siddhikol C, Erbstoeszer J, Weisblum B. Mode of action of streptolydigin. J. Bacteriol. 1969;99:151–155. doi: 10.1128/jb.99.1.151-155.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sidorenkov I, Komissarova N, Kashlev M. Crucial role of the RNA:DNA hybrid in the processivity of transcription. Mol Cell. 1998;2:55–64. doi: 10.1016/s1097-2765(00)80113-6. [DOI] [PubMed] [Google Scholar]
- 108.Sims RJ, 3rd, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 109.Sosunov V, et al. The involvement of the aspartate triad of the active center in all catalytic activities of multisubunit RNA polymerase. Nucleic Acids Res. 2005;33:4202–4211. doi: 10.1093/nar/gki688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sosunov V, et al. Unified two-metal mechanism of RNA synthesis and degradation by RNA polymerase. EMBO J. 2003;22:2234–2244. doi: 10.1093/emboj/cdg193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sosunova E, et al. Donation of catalytic residues to RNA polymerase active center by transcription factor Gre. Proc Natl Acad Sci U S A. 2003;100:15469–15474. doi: 10.1073/pnas.2536698100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Stebbins C, et al. Crystal structure of the GreA transcript cleavage factor from Escherichia coli. Nature. 1995;373:636–640. doi: 10.1038/373636a0. [DOI] [PubMed] [Google Scholar]
- 113.Steinberg T, Burgess R. Tagetitoxin inhibition of RNA polymerase III transcription results from enhanced pausing at discrete sites and is template-dependent. J Biol Chem. 1992;267:20204–20211. [PubMed] [Google Scholar]
- 114.Steitz T. DNA and RNA polymerases: structural diversity and common mechanisms. Harvey Lect. 1997–1998;93:75–93. [PubMed] [Google Scholar]
- 115.Svejstrup J. The RNA polymerase II transcription cycle: cycling through chromatin. Biochim Biophys Acta. 2004;1677:64–73. doi: 10.1016/j.bbaexp.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 116.Svetlov V, Nudler E. Jamming the ratchet of transcription. Nat Struct Mol Biol. 2008;15:777–779. doi: 10.1038/nsmb0808-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Svetlov V, Belogurov G, Shabrova E, Vassylyev D, Artsimovitch I. Allosteric control of the RNA polymerase by the elongation factor RfaH. Nucleic Acids Res. 2007;35:5694–5705. doi: 10.1093/nar/gkm600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Symersky J, et al. Regulation through the RNA polymerase secondary channel. Structural and functional variability of the coiled-coil transcription factors. J Biol Chem. 2006;281:1309–1312. doi: 10.1074/jbc.C500405200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tadigotla V, et al. Thermodynamics and kinetics based identification of transcriptional pauses. Proc Natl Acad Sci USA. 2006;103:4439–4444. doi: 10.1073/pnas.0600508103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Temiakov D, et al. Structural basis of transcription inhibition by antibiotic streptolydigin. Mol Cell. 2005;19:655–666. doi: 10.1016/j.molcel.2005.07.020. [DOI] [PubMed] [Google Scholar]
- 121.Thomas M, Platas A, Hawley D. Transcriptional fidelity and proofreading by RNA polymerase II. Cell. 1998;93:627–637. doi: 10.1016/s0092-8674(00)81191-5. [DOI] [PubMed] [Google Scholar]
- 122.Toulokhonov I, Landick R. The role of the lid element in transcription by E. coli RNA polymerase. J Mol Biol. 2006;361(4):644–58. doi: 10.1016/j.jmb.2006.06.071. [DOI] [PubMed] [Google Scholar]
- 123.Toulokhonov I, Landick R. The flap domain is required for pause RNA hairpin inhibition of catalysis by RNA polymerase and can modulate intrinsic termination. Mol Cell. 2003;12:1125–1136. doi: 10.1016/s1097-2765(03)00439-8. [DOI] [PubMed] [Google Scholar]
- 124.Toulokhonov I, Zhang J, Palangat M, Landick R. A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol Cell. 2007;27:406–419. doi: 10.1016/j.molcel.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 125.Toulokhonov I, Shulgina I, Hernandez V. Binding of the transcription effector ppGpp to Escherichia coli RNA polymerase is allosteric, modular, and occurs near the N terminus of the beta'-subunit. J Biol Chem. 2001;276:1220–1225. doi: 10.1074/jbc.M007184200. [DOI] [PubMed] [Google Scholar]
- 126.Tuske S, et al. Inhibition of bacterial RNA polymerase by streptolydigin: stabilization of a straight-bridge-helix active-center conformation. Cell. 2005;122:541–552. doi: 10.1016/j.cell.2005.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Vassylyev D, et al. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 128.Vassylyev D, et al. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007;448:163–168. doi: 10.1038/nature05931. [DOI] [PubMed] [Google Scholar]
- 129.Vassylyev D, et al. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 130.Vassylyev D. Structural basis for transcription inhibition by tagetitoxin. Nat. Struct. Mol. Biol. 2005;12:1086–1093. doi: 10.1038/nsmb1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Vogel U, Jensen K. Effects of ppGpp on rate of transcription elongation in isoleucine-starved Escherichia coli. J Biol Chem. 1994;269:16236–16241. [PubMed] [Google Scholar]
- 132.Vrentas C, et al. Still looking for the magic spot: the crystallographically defined binding site for ppGpp on RNA polymerase is unlikely to be responsible for rRNA transcription regulation. J Mol Biol. 2008;377:551–564. doi: 10.1016/j.jmb.2008.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Vrentas C, Gaal T, Ross W, Ebright R, Gourse R. Response of RNA polymerase to ppGpp: requirement for the omega subunit and relief of this requirement by DksA. Genes Dev. 2005;19:2378–2387. doi: 10.1101/gad.1340305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wada T, et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wang D, Hawley DK. Identification of a 3'-->5' exonuclease activity associated with human RNA polymerase II. Proc Natl Acad Sci U S A. 1993;90:843–847. doi: 10.1073/pnas.90.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wang D, Bushnell D, Westover K, Kaplan C, Kornberg R. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang M, et al. Force and velocity measured for single molecules of RNA polymerase. Science. 1998;282:902–907. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- 138.Westover K, Bushnell D, Kornberg R. Structural basis of transcription: separation of RNA from DNA by RNA polymerase II. Science. 2004;303:1014–1016. doi: 10.1126/science.1090839. [DOI] [PubMed] [Google Scholar]
- 139.Westover K, Bushnell D, Kornberg R. Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center. Cell. 2004;119:481–489. doi: 10.1016/j.cell.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 140.Wind M, Reines D. Transcription elongation factor SII. Bioessays. 2000;22:327–336. doi: 10.1002/(SICI)1521-1878(200004)22:4<327::AID-BIES3>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yarnell W, Roberts J. Mechanism of intrinsic transcription termination and antitermination. Science. 1999;284:611–615. doi: 10.1126/science.284.5414.611. [DOI] [PubMed] [Google Scholar]
- 142.Zakharova N, Bass I, Arsenieva E, Nikiforov V, Severinov K. Mutations in and monoclonal antibody binding to evolutionary hypervariable region of Escherichia coli RNA polymerase beta' subunit inhibit transcript cleavage and transcript elongation. J Biol Chem. 1998;273:24912–24920. doi: 10.1074/jbc.273.38.24912. [DOI] [PubMed] [Google Scholar]
- 143.Zaychikov E, et al. Mapping of catalytic residues in the RNA polymerase active center. Science. 1996;273:107–109. doi: 10.1126/science.273.5271.107. [DOI] [PubMed] [Google Scholar]
- 144.Zenkin N, Yuzenkova Y, Severinov K. Transcript-assisted transcriptional proofreading. Science. 2006;313:518–520. doi: 10.1126/science.1127422. [DOI] [PubMed] [Google Scholar]
- 145.Zhang C, Burton Z. Transcription factors IIF and IIS and nucleoside triphosphate substrates as dynamic probes of the human RNA polymerase II mechanism. J Mol Biol. 2004;342:1085–1099. doi: 10.1016/j.jmb.2004.07.070. [DOI] [PubMed] [Google Scholar]
- 146.Zhang G. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 Å resolution. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]