Abstract
Lynch syndrome (LS), or hereditary nonpolyposis colorectal cancer, is the most common hereditary colorectal cancer (CRC) syndrome, accounting for approximately 2–5% of all newly diagnosed cases of CRC. Patients with LS have an increased lifetime risk of colorectal (52.2% in women and 68.7% in men) and endometrial cancer (15–70%), as well as certain extra-colonic cancers. Germline mutations in one of several DNA mismatch repair genes underlie LS. Molecular testing has emerged as an indispensable strategy for the diagnosis of LS. The diagnostic work-up of at-risk individuals includes a careful family history evaluation, microsatellite instability, immunohistochemistry and germline DNA analysis. A positive test result can guide clinicians in formulating the appropriate screening, surveillance and management strategies. However, because of the absence of an overt phenotype, such as a diffuse polyposis, it is not always straightforward to recognize LS clinically.
Keywords: genetic testing, immunohistochemistry, Lynch syndrome, microsatellite instability, mismatch repair genes
Colorectal cancer (CRC) is the second most common cause of cancer-related death in the USA and Europe. CRC is responsible for an estimated 52,000 deaths per year in the USA and 146,000 per year in the EU [1,2,201]. Approximately 2–5% of newly diagnosed cases of CRC can be attributed to Lynch syndrome (LS), previously referred to as hereditary nonpolyposis CRC (HNPCC). LS is an autosomal dominant disorder with incomplete but high penetrance manifested by early-onset colorectal and endometrial cancer, and an increased risk of certain extra-colonic cancers, including tumors elsewhere in the gastrointestinal tract (e.g., stomach, small bowel, biliary tract), urinary collecting system (renal pelvis, ureter) and the female reproductive system (ovaries). While initial studies of selected Lynch families estimated a 70–80% lifetime risk of colon cancer with a mean age at diagnosis in the mid-40s, more recent population-based studies have suggested a somewhat lower lifetime CRC risk (52.2% in women and 68.7% in men) and a higher median age at diagnosis of 61.2 years [3–5]. The second most common cancer in LS is endometrial cancer, and women with LS have a 15–70% and 3–14% cumulative lifetime risk of developing endometrial and ovarian cancer, respectively, with a mean age at diagnosis approximately 10 years earlier than sporadic cases [6–10]. The lifetime risk for gastric cancer in LS patients varies between populations, with a particularly high incidence in areas such as China and Korea, where there is a high endemic risk of gastric cancer, with a lifetime risk of approximately 30% [11]. In these areas, the risk for gastric cancer exceeds that for endometrial cancer. The risks for urinary tract, small bowel, brain and biliary tumors are approximately 1–12, 4–7, 1–4 and 2%, respectively [6]. Unique variants of the syndrome characterized by the presence of skin tumors (keratoacanthomas, sebaceous neoplasms) or brain tumors (glioblastomas, astrocytomas and oligodendrogliomas) are termed Muir-Torre and Turcot syndromes, respectively.
Lynch syndrome is caused by germline mutations in one of four DNA mismatch repair (MMR) genes. MMR proteins normally recognize and repair mismatched nucleotides and insertion/deletion loops caused by the slippage of DNA polymerase during replication of short repeat sequences known as microsatellites. Mutations in at least seven genes have been associated with LS: MLH1, MSH2, MSH6, PMS1, PMS2, MLH3 and EXO1, with the majority of mutations observed clinically in four genes. Germline mutations of MLH1, MSH2, MSH6 and PMS2 account for 32, 38, 14 and 15% of all known MMR mutations in LS, respectively [12]. Several missense mutations of unclear biological relevance have been identified in MLH3 and EXO1 genes [13,14].
Genotype–phenotype correlations have begun to emerge in LS (Table 1) [7–10]. MLH1 mutation carriers have a higher CRC prevalence (79 vs 69%) and younger age at cancer diagnosis when compared with MSH2 mutation carriers. However, the prevalence of extracolonic LS-associated cancers is greater among MSH2 mutation carriers compared with MLH1 mutation carriers (24 vs 9%) [7]. Compared with the incidence in the general population, carriers of a germline mutations in MSH6 have a 26-fold increased incidence of endometrial cancer, and an eightfold increased incidence of CRC, independent of sex and age [9]. Individuals with PMS2 mutations have an overall lower risk of Lynch-associated cancers, as well as an older age at cancer diagnosis [10].
Table 1.
Gene-specific cancer risks.
| Mismatch repair gene | Cumulative risk of CRC by age 70 years (%) | Cumulative risk of EC by age 70 years (%) |
|---|---|---|
| MLH1 | ||
| • Males | ~70 | |
| • Females | ~50 | ~30 |
| MSH2 | ||
| • Males | ~50 | |
| • Females | ~40 | ~45 |
| MSH6 | ||
| • Males | 22 | |
| • Females | 10 | 26 |
| PMS2 | ||
| • Males | 20 | |
| • Females | 15 | 15 |
The timely recognition of LS is essential to identify patients at high-risk who will require intensive cancer surveillance. Reductions in CRC mortality can be achieved by colonoscopic screening of individuals with this syndrome [15]. A comprehensive family history of cancer has traditionally been viewed as a simple and inexpensive way to identify hereditary forms of CRC [16]. Although family history can provide important clues to the presence of LS, this is not always reliable because of small family size, a physician's unfamiliarity with the nuances of the syndrome, lack of documentation or reduced penetrance of the MMR gene mutation in the family. However, advances in the understanding of the molecular basis of the disease have resulted in novel clinical approaches to establish the diagnosis. Approximately 90% of CRCs occurring in LS patients have a characteristic and readily detectable molecular change in the number of DNA microsatellites, making microsatellite instability (MSI) analysis of tumor samples a useful diagnostic tool to screen for LS [17]. In addition, loss of functional MMR protein that results from inactivating mutations may be assessed directly in tumor tissue through immunohistochemical (IHC) analysis, and this strategy can also pinpoint which one of the four genes is most likely to be mutated [18]. MSI and IHC analysis can therefore serve as useful screening tests for LS, and the results can guide more definitive germline mutational analysis of the appropriate DNA MMR gene. Identification of a germline mutation not only establishes the diagnosis of LS, but also provides an invaluable tool for family screening.
Establishing clinical criteria for LS
In 1895, Alfred Warthin initiated one of the most thoroughly documented cancer family histories and in 1913 he published a description of this family, designated Family ‘G’, with an unusually high prevalence of uterine and gastric cancers [19]. Decades later, Henry Lynch re-examined this family and described the clinical features of a hereditary cancer syndrome that now bears his name [20]. Several attempts have been made to define precise clinical criteria for this syndrome. In 1991, the International Collaborative Group on HNPCC at its first formal meeting in Amsterdam developed clinical criteria to provide a basis for collaborative studies and uniformity in the terminology of HNPCC (box 1) [21]. Because these criteria focused entirely on CRC diagnoses and did not fully account for all presenting features of Lynch patients, the Amsterdam Criteria II were developed to include extra-colonic Lynch-associated cancers (box 1) [22]. It is important to note that the Amsterdam criteria are relatively insensitive for the diagnosis of LS, and as many as 50% of mutation carriers do not meet the Amsterdam criteria [23]. In addition, 40–45% of families who fulfill the Amsterdam criteria fail to demonstrate MSI on tumor testing or germline mutations in any of the MMR genes [24,25–27]. Such families are unlikely to have Lynch and many represent a new colon cancer syndrome, tentatively designated ‘familial CRC type X’ [23]. These families have an increased risk of developing CRC compared with the general population, but compared with LS patients, the risks are lower, there is an older age at onset and there is no evidence of an increased risk of extra-colonic cancers.
A major concern with the Amsterdam criteria is their relative stringency, particularly with respect to the family history. Many individuals come from small families or have limited information about the medical history of relatives. In 1996, an international workshop on HNPCC hosted by the National Cancer Institute (NCI) formulated the Bethesda guidelines to capture an even wider spectrum of at-risk patients and increase sensitivity for the diagnosis of LS. Specifically, the goal was to establish broader criteria that would identify colorectal tumors that should be considered for MSI testing, thereby maximizing sensitivity, but necessarily reducing specificity for the diagnosis of LS (box 1) [28]. The Bethesda Guidelines not only incorporate broader age and family history criteria, but also include the unique histopathological features associated with Lynch tumors (tumor infiltrating lymphocytes, signet ring cells, mucinous and poorly differentiated histology). These criteria were revised and simplified in 2004 [29].
The Amsterdam criteria are associated with relatively low sensitivity (28–45%), but high specificity (up to 99%) for the diagnosis of LS in a general population, whereas the Bethesda criteria are associated with higher sensitivity (73–91%), but at the cost of lower specificity (77–82%) [12,30]. In a series of 1066 unselected patients with newly diagnosed CRC, Hampel et al. identified 23 patients with LS [26]. Only three (13%) of these 23 met the Amsterdam criteria and 18 (78%) met the Bethesda guidelines, indicating that family history alone as a screening test for Lynch will likely miss a significant proportion of cases. The difficulty of obtaining reliable family history and the overall poor sensitivity and specificity of these clinical criteria recently led the EWG (Evaluation of Genomic Applications in Practice and Prevention [EGAPP] Working Group) to remove family history from consideration as a first-line test for LS in individuals with a new diagnosis of CRC [1,12]. Nonetheless, a thorough family history remains a valuable tool to assess the risk of LS, bearing in mind that a negative or noncontributory family history cannot formally exclude the diagnosis of LS or the need for molecular testing.
Testing for MSI
The DNA MMR system recognizes and then corrects the base-pair mismatches and small insertions or deletions that can occur during DNA replication. It requires the cooperation of multiple genes from the mutS (MSH2, MSH3, MSH6) and mutL (MLH1, MLH3, PMS1 and PMS2) families. Specifically, MSH2 recognizes and binds directly to the mismatched DNA sequence, forming the MutSα heterodimeric complex with MSH6 in the presence of single base-pair mismatches or single-nucleotide insertion and deletion loops, or the MutSβ complex with MSH3 if there are larger two- to eight-nucleotide insertions or deletions [31,32]. Both MutSα or MutSβ can then recruit either MutLα or MutLβ, heterodimers of MLH1 and PMS2 or MLH1 and PMS1, respectively, that mediate crosstalk between the processes of mismatch recognition and enzymatic repair [33]. Although heterodimers of MLH1/MLH3 also form, their specific role remains to be defined.
In tumors that develop owing to defective DNA MMR, short repetitive DNA sequences known as microsatellites tend to undergo a high level of slippage that results in MSI. MSI is reported in as many as 90–95% of colorectal carcinomas, and at least 75% of endometrial carcinomas associated with LS, making MSI a sensitive marker for LS-associated tumors. Most microsatellite sequences are located in the noncoding regions of the genome, but they are also found within the coding regions of a select number of genes involved in tumor initiation and progression. Microsatellite mutations that occur in these coding sequences are directly associated with tumor pathogenesis. These genes include receptors for growth factors, such as TGFBR2 and IGF2R, cell cycle regulators (E2F4), regulators of apoptosis (BAX), and some of the MMR genes themselves (MSH3, MSH6, MLH3 and PMS2). The most commonly mutated gene in MSI tumors, seen in up to 90% of all LS cases, is the TGFBR2 gene, which harbors an (A)10 repeat that undergoes a frameshift [34]. This mutation leads to a disruption in the function of TGF-β, a tumor suppressor of critical importance in CRC. The BAX gene has a (G)8 microsatellite, which loses one or two guanines, resulting in a frameshift that inactivates the gene and disrupts the apoptosis pathway mediated by Bcl-2 in approximately 35% of all tumors with defective MMR [35].
Box 1. Clinical criteria for the diagnosis of Lynch syndrome.
Amsterdam I criteria
At least three relatives with colorectal cancer
One is a first-degree relative of the other two
At least two successive generations affected
At least one colorectal cancer diagnosed at <50 years of age
Familial adenomatous polyps should be excluded
Amsterdam II criteria
At least three relatives with a hereditary nonpolyposis colorectal cancer-associated cancer†
One is a first-degree relative of the other two
At least two successive generations affected
At least one hereditary nonpolyposis colorectal cancer-associated cancer† diagnosed at <50 years of age
Familial adenomatous polyps should be excluded
Tumors should be verified by pathologic examination
Modified Amsterdam criteria
In very small families, two colon cancer cases in first-degree relatives in at least two generations, one case diagnosed at <55 years of age
In families with two first-degree relatives with colon cancer, a third relative with an unusual early-onset cancer or endometrial cancer
Bethesda guidelines
Colorectal cancer diagnosed at <50 years of age
Presence of synchronous (simultaneous) or metachronous (diagnosed at different time) colorectal cancer or other syndrome associated tumors† regardless of age
Colorectal cancer with microsatellite instability-high histology‡ diagnosed at <60 years of age
Colorectal cancer or syndrome-associated tumor† diagnosed at <50 years in at least one first-degree relative
• Colorectal cancer or syndrome-associated tumor† diagnosed at any age in two or more relatives regardless of age
The integrity of DNA MMR is routinely analyzed by extracting DNA from a paraffin-embedded tumor sample as well as control normal tissue. PCR amplification of specific microsatellite markers is performed (Table 2 & Figure 1). The PCR products can be separated electrophoretically and differences in fragment size of tumor-derived DNA versus DNA from normal tissue are scored, leading to an assessment of instability (Figure 1). Initially, the specific microsatellite loci used to test for MSI were highly variable. In an attempt to facilitate comparisons between studies, a NCI sponsored workshop in 1997 recommended a panel of five microsatellite markers, known as the Bethesda markers, that consisted of two mononucleotide (BAT25 and BAT26) and three dinucleotide (D5S346, D2S123 and D17S250) repeats [36]. Samples with instability in two or more of these markers were defined as MSI-H (for high-frequency MSI), whereas those with one unstable marker were designated MSI-L (for low-frequency MSI). Samples with no instability in any of the markers were considered to be microsatellite stable (MSS). In the event that more than five markers were tested, instability at at least 30% of analyzed markers would define MSI-H. However, this NCI five-marker microsatellite panel may underestimate the number of MSI-H tumors and overestimate the number of MSI-L tumors. Mononucleotide repeats are commonly shifted in MSI-H tumors. However, MSI-L tumors typically exhibit shifts in dinucleotide repeats, and the significance of the MSI-L phenotype for the diagnosis of LS still remains uncertain [37]. In two reports that analyzed MSI using the Bethesda markers, the sensitivities of the dinucleotide repeats were reported to be 72–89% for D2S123,50–81% for D17S250 and 50–59% for D5S346 [17,38]. Because these markers are highly polymorphic, the analysis of corresponding normal DNA is required, and this can make the MSI process relatively time-consuming and expensive.
Table 2.
List of commonly used microsatellite instability markers.
| Microsatellite marker | Gene near marker/GenBank number | Location of the repeat | Repeat motif |
|---|---|---|---|
| BAT25 | c-kit | 4q12 (intron 16) | TTTT.T.TTTT.(T)7.A(T)25 |
| BAT26 | MSH2 | 2p16.3–p21 (intron 5) | (T5)......(A)26 |
| BAT40 | HSD3B2 | 1p13.1 | TTTT.TT..(T)7................TTTT.(T)40 |
| NR21 | SLC7A8 | 14q11.2 (5′ UTR) | (T)21 |
| NR22 | HUMB5A | 11q24–25 (3′ UTR) | (T)22 |
| NR24 | ZNF2 | 2q11.2 (3′ UTR) | (T)24 |
| MONO-27 | MAP4K3 | 2q21 (intron 13) | (A)27 |
| D2S123 | MSH2 | 2p16 | (CA)13TA(CA)15(T/GA)7 |
| D5S346 | APC | 5q21/22 | (CA)26 |
| D17S250 | BRCA1 | 17q11.2–q12 | (TA)7..............................(CA)24 |
| D10S197 | GAD2 | 10p (intron 7) | CACCAGA(CA)7.A.A.(CA)12(AGAAA)2 |
| PENTA C | AL138752 | 21q22.3 | (AAAAG)3–15 |
| PENTA D | AC003656 | 9p12–13.3 | (AAAAG)2–17 |
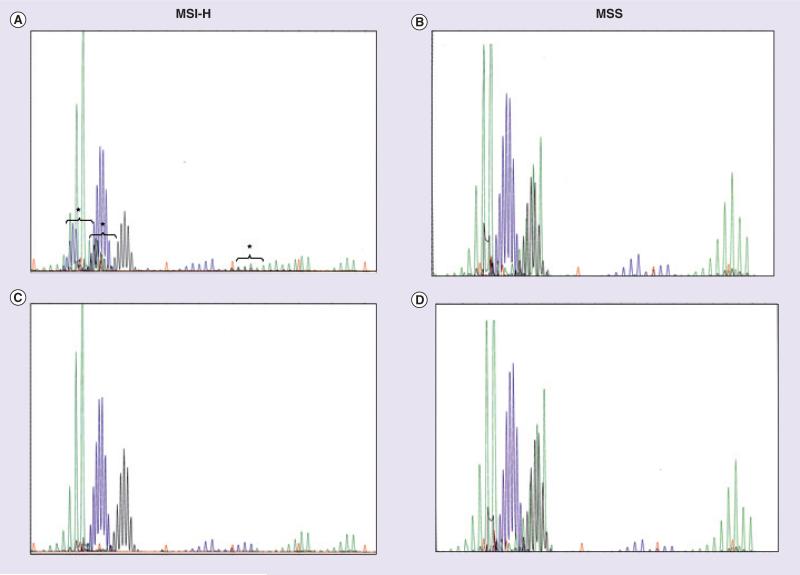
Figure 1. Microsatellite instability analysis of colorectal tumor and corresponding normal DNA.
Analysis of MSI in colorectal adenocarcinomas using the five markers of the International Workshop of Bethesda and fluorescence-based multiplex PCR. The electrophoretic patterns of an MSI-H tumor (A: tumor tissue; B: corresponding normal mucosa) and of an MSS carcinoma (C: tumor tissue; D: normal mucosa) are illustrated. New, shifted alleles present in the MSI-H tumor sample are indicated with a bracket.
MSI-H: High frequency microsatellite instability; MSS: Microsatellite stable.
Courtesy of A John Iafrate.
Suraweera et al. developed a panel of five quasi-monomorphic mononucleotide repeats (BAT25, BAT26, NR21, NR22 and NR24), which can be analyzed together in a pentaplex PCR without the need for matching normal samples. This is possible because allelic size variations in each of these repeats are observed in less than 1% of the Caucasian population, but the number is somewhat higher in the African and African–American populations (up to 10%) [39,40]. This panel performed with a sensitivity greater than 95% and a specificity greater than 98% for the determination of MSI status in a series of 64 MSI-H and 40 MSS colon primary tumors, whose MSI status was previously established by the analysis of a larger panel of dinucleotide microsatellite markers [41]. This option was included in the 2004 Revised Bethesda Guidelines for HNPCC, although the original panel of mono- and di-nucleotide markers from 1997 remained unchanged as the primary recommendation [42]. A similar panel of markers that includes five nearly monomorphic mononucleotides (BAT25, BAT26, NR21, NR24 and MONO27) for MSI determination and two polymorphic pentanucleotide markers (Penta C and Penta D) that help confirm that tumor and matching normal samples are from the same individual has also been introduced [43]. When this assay was applied to a set of 72 MSI-H and 81 control colorectal tumors that had been previously characterized, using a panel of ten microsatellite markers and IHC analysis for MLH1, MSH2, MSH6 and PMS2, Bacher et al. observed that over 97% of MSI-H samples were correctly identified and overall there was 99% concordance in MSI classification between the two methods. Of note, 43 samples classified as MSI-L by the ten microsatellite marker panel were scored as MSS with this multiplex system, confirming that this multiplex assay has a high specificity for detection of MSI-H tumors [43].
The performance of the NCI-recommended panel of markers compared with the pentaplex panel of mononucleotide repeats for the detection of MMR-deficient CRCs was also addressed by Xicola et al. [44]. The sensitivity, specificity and positive and negative predictive values to detect absence of expression of at least one MMR protein were determined using the NCI panel and the pentaplex panel in 531 and 527 colorectal tumors, respectively. Whereas specificity and negative predictive values were high for both panels, the sensitivity and positive predictive values were 76.5 and 65.0% for the NCI panel and 95.8 and 88.5% for the mononucleotide pentaplex panel. Although the two panels were not compared in the same patient populations, the analysis with both sets of markers in all MSI-L tumors and all tumors that showed a discrepancy between MSI and IHC expression of MMR protein confirmed a superior performance of the pentaplex panel of mononucleotide repeats. Furthermore, whereas a substantial number of tumors with an MSI-L phenotype was detected using the NCI panel, this phenotype was absent when the pentaplex panel of mononucleotide repeats was used.
Others have suggested that testing BAT25 and/or BAT26 alone would be sufficient to establish the MSI status of a tumor without the need to analyze normal DNA, since these markers are quasi-monomorphic in most Caucasian populations [44–46]. However, use of BAT26 alone might potentially lead to an underestimation of the true levels of MSI due to the failure to recognize those rare cases with biallelic MSH2 deletions. BAT26 lies intragenically in MSH2 and would not be amplifiable from CRCs that have a biallelic deletion of MSH2. Therefore, use of at least one other mononucleotide marker (e.g., BAT25 or NR24) has been suggested [47].
Advantages & limitations of MSI testing
There are several advantages and limitations of MSI testing that should be highlighted. One of the main advantages of MSI testing is that it provides a functional analysis of deficient MMR activity. For example, this information can be useful in scenarios where there are conflicting findings, such as a family history highly suspicious for LS, and positive MSI testing, but absence of a DNA MMR mutation. In this particular scenario, a positive MSI test would still be indicative of a diagnosis of LS and suggest that current methods of mutation detection may be inadequate or new genes involved in MMR await identification. Furthermore, some nondeleterious mutations, such as missense or in-frame insertion/deletion mutations, are reported, particularly in the MLH1 and MSH6 genes, which do not lead to a truncated protein and will not be predicted to affect protein translation, stability and antigenicity. In these cases, MSI could help determine whether there are true functional consequences of these variant mutations.
However, MSI testing is labor-intensive and more costly than IHC, requires expert molecular pathologic services, and while it is a hallmark for LS, it is not specific for it. Approximately 10–15% of sporadic CRCs also exhibit MSI due to somatic CpG island methylation of the promoter of the MLH1 gene, resulting in transcriptional silencing [48]. LS-associated and sporadic MSI-positive CRCs have many histopathologic features in common, such as mucinous histology, poor differentiation and the presence of lymphocytic infiltration, but differ in that sporadic MSI CRCs are not associated with a positive family history, and are more common in women at older ages. Recently, a V600E hotspot mutation in exon 15 of the BRAF gene, a member of the RAF family of kinases, has shown utility in distinguishing tumors with somatic hypermethylation of MLH1 and those arising through a germline mutation. Deng et al. found that this specific BRAF mutation occurred in 87% of sporadic tumors with hypermethylated MLH1, whereas it was not present in any MSI tumor with a germline MLH1 mutation [49]. In addition, BRAF mutations were not detected in any tumors from patients with germline MSH6 mutations and in tumors from 23 MMR-negative families, 13 of whom fulfilled the Amsterdam criteria and ten the Bethesda criteria [50]. Collectively, the detection of a BRAF mutation in a MSI CRC suggests a sporadic origin of the disease and not LS. These findings have a potential impact in the diagnostic algorithm of LS, since BRAF sequencing can define which cases with abnormal MSI and IHC results do not require further germline testing.
IHC analysis of MMR protein expression
Mutations in MMR genes usually lead to loss of expression of a detectable protein product in the nuclei of tumor cells, providing a rationale for the use of IHC techniques to detect MMR gene mutations. Adjacent normal colonic mucosa can serve as a positive control (Figure 2). Knowing how MMR proteins interact during DNA repair can help interpret IHC results and guide germline testing (Table 3). As previously reported, MMR proteins form heterodimers. MSH2 dimerizes with either MSH6 or MSH3, depending on the length of the base-pair mismatch, and then recruits heterodimers of MLH1 and PMS2, or MLH1 and PMS1 to excise the mismatched nucleotides. MSH2 and MLH1 proteins are the obligatory partners of their respective heterodimeric complexes, and mutations result in the loss of both the obligatory and secondary partner proteins by IHC. However, the converse is not true, since loss of staining of MSH6 or PMS2 alone is typically observed with germline mutations in each of these respective genes [10,51]. Other proteins, such as MSH3, MLH3 and PMS1, may compensate in the binding to the obligatory proteins.
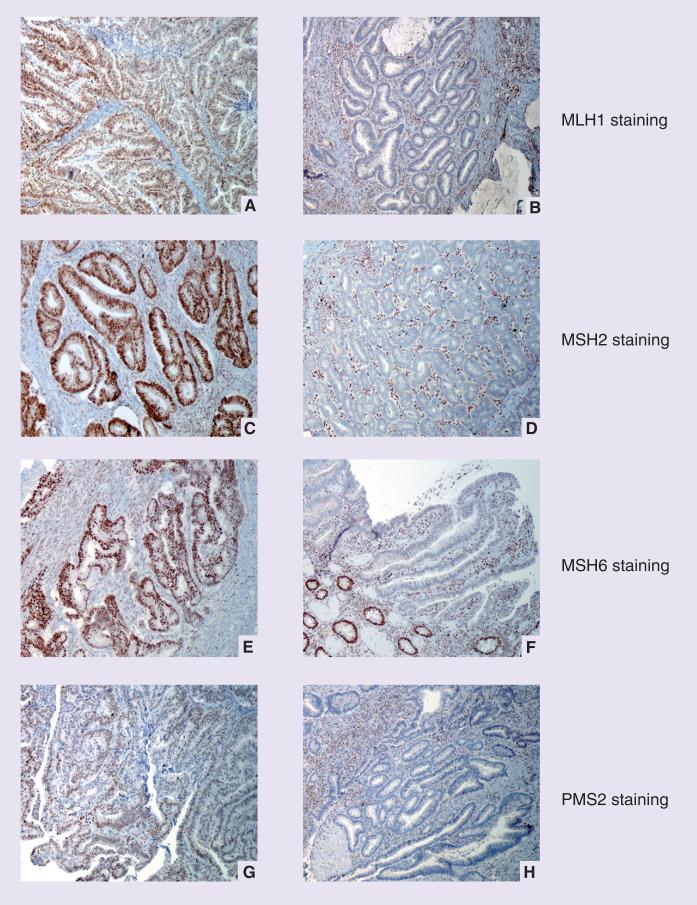
Figure 2. Immunohistochemical staining for mismatch repair proteins in colorectal carcinoma.
(A) Positive and (B) absent staining for MLH1; (C) positive and (D) absent staining for MSH2; (E) positive and (F) absent staining for MSH6; (G) positive and (H) absent staining for PMS2.
Photographs courtesy of A John Iafrate.
Table 3.
Immunohistochemistry testing results based on germline mutation status.
| Germline mutation | Protein staining | |||
|---|---|---|---|---|
| MLH1 | MSH2 | MSH6 | PMS2 | |
| MLH1 | Absent | Present | Present | Absent |
| MSH2 | Present | Absent | Absent | Present |
| MSH6 | Present | Present | Absent | Present |
| PMS2 | Present | Present | Present | Absent |
Since MSH2 and MLH1 mutations account for more than 70% of all known MMR mutations in LS, efforts to evaluate the utility of IHC in detecting the syndrome have focused primarily on MSH2 and MLH1. As recently reviewed by Shia, the sensitivity of MLH1 and MSH2 IHC in predicting germline mutations in the corresponding genes is approximately 85%. There is a lower detection rate of MLH1 mutations (74%) [52]. More than a third of the MLH1 alterations are missense mutations that may potentially result in catalytically inactive, but antigenically intact, proteins and false-positive staining by IHC [53–55]. The addition of PMS2 IHC led to the identification of 23% more patients with an MLH1 mutation in a study by de Jong et al. [56]. When 35 tumors associated with a known MLH1 germline mutation were tested by IHC, 21 tumors showed absence of both MLH1 and PMS2 staining, while eight tumors exhibited negative staining for PMS2 only. Furthermore, MSH6 immunostaining not only permits the detection of MSH6 mutations, but also supports the detection of MSH2 mutations. The initial two-antibody panel (MLH1 and MSH2) has evolved into a four-antibody panel (MLH1, MSH2, MSH6 and PMS2) with an increased sensitivity of 94% in predicting a gene mutation.
Advantages & limitations of IHC testing
The major advantage of this assay is that IHC is widely available as part of the routine services in general pathology, and therefore does not depend upon the involvement of a molecular genetics laboratory. The recent automation of immunostaining assures consistent and reproducible immunostaining procedures, allowing comparison of immunostaining patterns between different tissues and cases. Another specific advantage of IHC is that tumors with MSH6 mutations frequently lack or have low levels of MSI owing to a functional redundancy in the DNA MMR system [57,58]. Indeed, MSH2 may form heterodimers with either MSH6 to repair single-nucleotide mismatches or with MSH3 to repair small insertion and deletion mismatches larger than one nucleotide. Therefore, when MSH6 is mutated, the MSH2/MSH3 dimer is still functional, and MSI may not be apparent. However, IHC will reveal loss of MSH6 staining.
One inherent potential shortcoming is that the technique is somewhat subjective and depends upon the quality of tissue preparation, staining and interpretation of the results. Interestingly, abnormal staining patterns may be due to tissue preservation and the tumor microenvironment. For example, tissue hypoxia or oxidative stress may diminish the function of MMR proteins, even in genetically MMR-proficient tissues, leading to a focal loss or weak staining [59,60]. Secondary abnormalities in MMR genes may also lead to rare staining patterns. For example, the existence of mononucleotide repeats in the coding sequences of several MMR genes, such as MSH2, MSH6 and PMS2, may result in abnormal IHC staining when mutations in these microsatellites occur as a consequence of a germline mutation of a different MMR gene [61,62].
Is two better than one? Comparison of MSI & IHC analysis
There is no single diagnostic algorithm in LS. Variations will occur on the basis of test availability, financial considerations, availability of tumor blocks and other factors. Lindor et al. performed a comparative study to assess the strengths and weaknesses of MSI testing and IHC for MLH1 and MSH2 proteins in 1144 patients [63]. The IHC test was 92.3% sensitive and 100% specific for the detection of MSI. Furthermore, IHC as a stand-alone approach to identify mutation carriers was associated with a sensitivity of 81.8% and specificity of 93.9%, compared with 90.0% sensitivity and 93.9% specificity for MSI as a stand-alone screening test. In patients fulfilling the Bethesda guidelines, Pinol et al. found that both MSI testing and tumor immunostaining were equivalent and highly cost-effective strategies to select patients for germline mutations [64]. Sensitivities of 81.8 and 81.8%, specificities of 98 and 98.2%, and positive predictive values of 27.3 and 29%, were reported for MSI and IHC testing respectively, in patients fulfilling the Bethesda guidelines. The sensitivity of this combined approach was lower than with MSI or IHC alone in unselected patients because of the imperfect concordance between the clinical criteria and the molecular tests. However, the advantage of using the revised Bethesda guidelines in conjunction with MSI or IHC evaluation of protein expression was derived from a higher positive predictive value for the presence of a germline mutation.
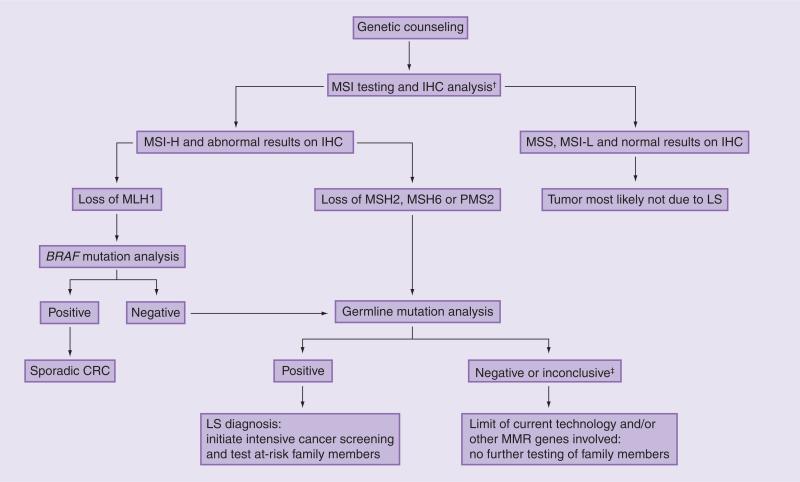
The limitations of screening on the basis of MSI or IHC alone were demonstrated by Lagerstedt Robinson et al., who used family history, MSI and IHC testing to select for MMR gene mutation analysis in 60 families who met the Amsterdam criteria, 188 non-Amsterdam families and 37 patients with early-onset CRC without a family history [65]. One of the most intriguing observations was that deleterious MMR gene mutations were found in four out of 17 MSI-negative Amsterdam families and in one of 30 MSI-negative non-Amsterdam families. However, the IHC assays correctly identified the mutant protein in three of these five cases, and these mutations would have been missed if MSI was used as a stand-alone method. Conversely, IHC alone would have missed some MSI-positive cases, particularly those due to missense MMR gene mutations, strongly suggesting that the MSI and IHC tests may serve as complementary tests that can maximize sensitivity when combined. Therefore, the most comprehensive screening approach should include both MSI and IHC testing (Figure 3). However, IHC alone may be a reasonable strategy if MSI testing is not available.
Figure 3. Suggested algorithm for molecular and genetic testing for Lynch syndrome.
†For purposes of feasibility, IHC alone is a reasonable strategy.
‡For inconclusive results, an intensive cancer screening may still be recommended.
CRC: Colorectal cancer; IHC: Immunohistochemistry; LS: Lynch syndrome; MMR: Mismatch repair; MSI: Microsatellite instability; MSS: Microsatellite stable.
MSI & IHC analysis in non-CRC tissues
MSI and IHC testing is not restricted to CRCs. For example, colonic adenomas are considered precursors of LS-associated colon cancers. Although the number of polyps in Lynch patients appears to be similar to the general population, the polyps are more likely to occur at a younger age, have a pre dilection for the proximal colon, be larger, display villous features or high-grade dysplasia, and most importantly, grow rapidly and progress to invasive cancer in less than 3 years. Many individuals with suspected Lynch now undergo routine colonoscopic screening with polypectomy, and in such a scenario, there is no colon cancer tissue available for testing. It is important to note that MSI and loss of MMR proteins by IHC are uncommon among adenomas in the general population. In a large series of 378 patients with adenomatous polyps, only six (1.6%) had at least one adenoma with MSI, and five of these six patients were indeed carriers of a germline MMR gene mutation [66]. We have recently shown that a combination of both MSI analysis and IHC staining for MMR proteins can detect DNA repair deficiency in Lynch-associated adenomas, and this included adenomas smaller than 5 mm [67]. The 40 adenomas removed during surveillance colonoscopy from 15 subjects with a germline MMR gene mutation were analyzed with both MSI and IHC testing. The presence of a germline mutation correlated with an abnormal MSI result in 58% of cases, an abnormal IHC result in 70% of cases, and either an abnormal MSI and IHC result in 73% of cases. Thus, in the work-up of patients suspected to have a germline MMR gene mutation, it is reasonable to analyze adenomas, and our data suggest that IHC testing alone is nearly as sensitive as a combined approach with MSI and IHC.
Endometrial cancer is the second most frequent cancer in women with LS. The frequency of MSI in LS-associated endometrial carcinoma is reported to be 75% [68,69]. de Leeuw et al. performed MSI and IHC testing for MLH1, MSH2 and MSH6 proteins in a series of LS-associated endometrial tumors from patients with known MMR gene germline mutations [70]. All endometrial tumors from MLH1 and MSH2 mutation carriers, and 36% of those from MSH6 mutation carriers demonstrated an MSI-H phenotype. IHC predicted the presence of an MSH2 or MSH6 mutation in 100 and 75% of tumors, respectively, while loss of MLH1 staining was observed in only 27% of endometrial tumors from MLH1 mutation carriers. Several studies have also demonstrated that sporadic endometrial cancers can display MSI with a frequency from 15 to 35% [69,71–81].
MSI has been demonstrated in up to 100% of ovarian cancers from LS patients [68]. Studies of unselected ovarian cancer patients have reported a range for MSI between 12 and 20%, although many had a small sample size and were performed before 1997 [77,82–86]. IHC in unselected ovarian cancers has revealed loss of MMR protein expression in 2.3–6.1% of tumors [87–89].
Detection of MMR gene mutations
Most germline mutations have been identified in the MSH2 (38%) and MLH1 (32%) genes, with MSH6 and PMS2 mutations each accounting for approximately 15% of all known LS mutations [12]. There are no consensus hotspots for mutations, and a full spectrum of nonsense, frameshift, splice and missense point mutations have been described. Prescreening techniques, such as single-strand conformation polymorphism, conformation-sensitive gel electrophoresis, denaturing gradient gel electrophoresis or denaturing high-performance liquid chromatography can be used, and such a strategy permits DNA sequencing to be targeted to specific abnormal regions. Each of these strategies takes advantage of the different physical properties of DNA fragments containing a polymorphism or mutation that distinguishes them from normal sequences. Currently, the cost and ease of large-scale DNA sequencing have obviated the need for these indirect approaches. However, certain classes of gene mutations are not detected by DNA sequencing; these include large genomic deletions, genomic rearrangements and genomic duplications. In these cases, the gold standard is Southern blotting. However, this approach is time-consuming and expensive, typically uses radionucleotides, and requires a large amount of DNA. Alternative methods such as multiplex ligation-dependent probe amplification (MLPA) have been developed. This is a relatively inexpensive, simple and reproducible PCR-based method that uses the same equipment used for DNA sequencing [90,91]. In a Finnish study, 45 mutation-negative individuals with clinical and IHC findings suggestive of LS were evaluated by MLPA, or in some cases, long-range genomic PCR. Of these individuals, 27% were found to have large genomic rearrangements caused by deletions of one or several exons of the MLH1 or more frequently, MSH2 gene [92]. Large genomic alterations account for 5–30% and 10–60% of all MLH1 and MSH2 mutations, respectively, with this wide range of frequencies owing, in part, to the fact that many studies examined small sample populations [90,93–97]. Baudhuin et al. utilized both Southern blotting and MLPA techniques in a consecutive series of 365 unrelated cases and found that although the majority of the mutations identified in MLH1 and MSH2 genes were point mutations and small insertions/deletions, large genomic alterations were present in 17.9 and 45.3% of the MLH1 and MSH2 mutation-positive carriers, respectively [98].
A diploid to haploid conversion analysis, in which maternal and paternal alleles are separated prior to mutation screening, is a more sensitive method for detecting mutations that are undetectable by routine sequencing. Somatic cell hybrids between human and rodent cells are first created. These hybrid cells lose individual human chromosomes, thereby allowing the analysis of a haploid human genome, one chromosome at a time. Casey et al. performed a blinded comparison of conventional DNA sequencing and conversion analysis to identify mutations in MLH1, MSH2 and MSH6 genes in 89 CRC patients suspected of carrying a mutation in MMR genes owing to their family history, age at diagnosis, MSI status and/or loss of MLH1, MSH2 or MSH6 protein expression of their tumors [27]. Conversion analysis increased the diagnostic yield of genetic testing by 56% compared with genomic sequencing alone. Despite this potential, this technique has not come into routine clinical use owing to the high expense and technical demands of creating somatic cell hybrids.
Recurrent & founder mutations
Most germline mutations reported in the MMR genes are unique. A mutation that arises de novo with high frequency is defined as recurrent. Mutations that occur once and are then passed on to succeeding generations are designated founder mutations and are typically limited to a certain geographic area or a certain ethnic group. One of the best-known recurrent mutations that accounts for approximately 11% of all MSH2 germline mutations is an A→T transversion in the donor splice site of intron 5 that leads to transcriptional skipping of exon 5 [99]. The recurrent nature of this mutation is explained by the fact that the adenine is the first in a stretch of 26 adenines, creating a hotspot for the slippage of DNA polymerase during replication. In Newfoundland (Canada), this mutation behaves as a founder mutation, having been introduced by an early settler some time after 1610, and it accounts for 20–25% of all LS mutations in this region [100]. A genomic deletion of exon 16 of the MLH1 gene, dating back over 1000 years, accounts for more than 50% of all LS cases in Finland [101]. The 1906 G→C mutation in the MSH2 gene has been documented as a founder mutation in Ashkenazi Jews, accounting for almost 20% of LS in Ashkenazi Jewish families [102]. The American founder mutation (AMF), a genomic deletion of exons 1–6 of MSH2, has been traced back to 1727 to a German immigrant and his wife. This mutation accounts for up to 10% of the estimated total population of LS carriers in the USA. [103]. The most important aspect of recurrent and founder mutations is that testing for these particular alterations as a first step in appropriate populations may lower the cost of a molecular diagnosis. Indeed, in populations where a high proportion of all LS are caused by a founder mutation, such as Finnish (>50%), Jewish (~20%) and Newfoundlander populations (20–25%), this testing algorithm is already in use.
Homozygous biallelic MMR mutations
Rarely, carriers of two independent MMR gene mutations have been described. These individuals exhibit a unique clinical phenotype, including hematological malignancies and/or brain tumors, CRC in childhood, and features reminiscent of neurofibromatosis type 1 (NF1), mainly café au lait spots. In 1999, there were two simultaneously published reports of offspring in LS families who developed hematological malignancies and signs of NF1 at a very early age. DNA sequence analysis and allele-specific amplification revealed homozygous MLH1 germline mutations [104,105]. Gallinger et al. first reported on children with homozygous MLH1 gene deficiency and NF1 features who developed early-onset gastrointestinal cancers in the first two decades of life [106]. In all these reports, the children were conceived from consanguineous matings between first cousins. This syndrome has been referred to as childhood cancer syndrome (CCS), Lynch III syndrome, CoLoN syndrome (colon tumors and/or leukemia/lymphoma and/or neurofibromatosis features), and very recently, constitutional mismatch repair deficiency (CMMR-D) syndrome [107–110]. As reviewed by Wimmer and Etzler, patients carrying homozygous MLH1 or MSH2 mutations have an earlier age of malignancy (mean age: 3.5 vs 9 years) and more frequently develop hematological tumors than patients with biallelic MSH6 or PMS2 mutations, who have a higher incidence of brain and typical LS-associated tumors [110].
Germline epimutations
There have been recent reports of germline methylation of the MLH1 and MSH2 gene promoter, resulting in transcriptional silencing of the affected allele in tissues derived from all three embryonic germ cell lineages. Epimutations of the MLH1 gene were first described in 2001, and affected individuals developed tumors that exhibited MSI, loss of expression of the MLH1 protein and in some cases somatic loss of the wild-type allele [111–114]. Some kindreds had a positive family history of colorectal or other Lynch-related cancers, though they generally did not satisfy the Amsterdam criteria [112,113]. One patient had a very low (<1%) proportion of spermatozoa with MLH1 promoter methylation, implying the potential for transmission to the offspring, but no intergenerational transmission has been demonstrated in his family members. Hitchins et al. recently evaluated 24 patients with early-onset MSI-positive colorectal or endometrial cancers without germline mutations in MMR genes. They found two unrelated women who had a germline MLH1 epimutation. A son of one of these patients exhibited partial methylation of MLH1, consistent with transmission of the epimutation, but methylation of MLH1 was not present in his sperm, indicating reversion of the epimutation during spermatogenesis. Although the maternal allele was inherited by several other children, there was no evidence of methylation, indicating a reversion of the epimutations to normal status, and in fact biallelic expression of MLH1 was found [115]. Although it is a very rare cause of LS, a germline epimutation of MLH1 should be suspected in individuals who have a family history of LS-associated cancers that exhibit MSI and loss of MLH1 staining in the absence of a germline mutation of MLH1. In many of these cases, the first clue may be the demonstration of MSI within the normal control tissue.
Fewer cases of germline MSH2 epimutations have been reported. A unique set of Dutch and Chinese families developed early-onset colorectal or endometrial cancers, all with MSI and MSH2 protein loss, but without germline mutations in the MSH2 gene [116,117]. Deletions that disrupt the 3′ end of the adjacent TACSTD1 gene were identified, and this led to inactivation of the downstream MSH2 gene through the induction of methylation of the MSH2 promoter [117].
Polymorphisms with variable penetrance
Recent genome-wide association studies (GWAS) have identified several polymorphisms in DNA MMR genes that are associated with increased CRC risk. Because these polymorphisms may result in minor reductions in DNA repair capacity, it has been hypothesized that these polymorphisms may increase the risk of developing CRC [118]. Using GWAS, Lipkin et al. identified a new MLH1 variant, 415 G→C, which results in the amino acid substitution D132H that attenuates MLH1 function. This variant confers a clinically significant susceptibility to CRC and accounts for 1.3% of all CRC cases in Israel [119]. The MLH1 -93 G→A polymorphism is located in the gene promoter, potentially reducing MLH1 transcription and thereby reducing overall DNA repair function. This variant allele, either in the homozygous or heterozygous state, is associated with a higher risk of developing MSI tumors among patients from Ontario and Newfoundland [120]. Finally, the MSH6 variant 116 G→A is associated with an increased risk of colon cancer among men but not women [118].
Genetic test results
When LS is suspected, genetic testing is ideally initiated in a proband with an early-onset Lynch-associated cancer. A positive test confirms the diagnosis, but the absence of a mutation is a ‘true negative’ result only when a mutation has been previously identified in another family member. A particularly difficult result to explain is the so-called ‘variant of uncertain significance (VUS)’, which is usually a single-nucleotide substitution that results in a missense mutation. In contrast to mutations that result in a prematurely truncated protein, a VUS results in a single amino acid substitution and the resulting change in protein function is not known. Most such variants are benign polymorphisms,but determining the functional and clinical significance of a specific VUS with certainty is difficult. Since DNA variants are found in many CRC families, it is important to register the variants found worldwide in an accessible database, such as the LS mutational database maintained by the International Society for Gastrointestinal Hereditary Tumors [202].
A positive test result may result in anxiety, depression and concerns about discrimination in insurance coverage or the work-place. In addition to statewide efforts, a landmark passage of the federal genetic information nondiscrimination act (GINA) in the USA provides legal protection for patients who choose to have testing [121]. Furthermore, some carriers may feel guilty and isolated from their families, and this can occur with either a positive or negative test result. If a mutation is identified, it is then feasible for family members at-risk to undergo mutation-specific testing, which is less costly than the initial full gene testing.
The knowledge of a positive test result may then influence both cancer screening and surgical care of MMR gene mutation carriers. Colonoscopy starting at an early age and at frequent intervals has been shown to reduce mortality from CRC [15]. When a diagnosis of CRC is made in a MMR gene mutation carrier, subtotal colectomy with ileorectal anastomosis is recommended owing to the 16% risk for developing a second primary CRC within 10 years [122]. Currently, no modifications in chemotherapeutic management are recommended in CRC patients with LS. In women with a MMR gene mutation, endometrial cancer surveillance, including transvaginal ultrasound in combination with endometrial aspiration biopsy, should be considered every 1–2 years, beginning at age 30–35 years. Risk-reducing hysterectomy and bilateral salpingo-oophorectomy should also be presented as an option when childbearing is completed. In families with ureter/renal pelvis, gastric or small bowel cancer, annual ultrasound and urinalysis with cytological examination, and periodic upper endoscopy are also recommended for mutation carriers [6,123].
Despite major advances in diagnostic capabilities, there will be families in whom no testing is feasible (i.e., no tumor tissue or affected relative available for testing), or in which testing is negative, but in which the clinical suspicion for LS is still strong. No single test or combination of tests is completely sensitive or specific for diagnosing or excluding the syndrome. Clinical judgment should not be overridden by laboratory results if the clinical presentation for LS is compelling.
Conclusion
The work-up of patients with an early-onset CRC and/or family history of cancer is an elaborate and time-consuming process that can be intimidating for many primary-care providers and specialists. Molecular testing has emerged as an indispensable strategy for the diagnosis of LS. Currently, our algorithm for testing includes both MSI and IHC, which is the most comprehensive screening approach. However IHC alone is a reasonable choice, particularly when molecular pathologic services are not available. Germline sequencing of the genes of interest can then be pursued in those patients with a positive MSI or IHC test. Because of the absence of an overt polyposis phenotype, LS can be the most challenging hereditary CRC syndrome to recognize, and there are many patients with LS that remain undiagnosed. It is crucial for clinicians to be fully aware of the LS because of the implications for the prognosis, screening and management. An estimated 12,000 individuals could be diagnosed with LS on a yearly basis in the USA, and molecular testing strategies can make this possible.
Expert commentary
There have been considerable advances in the understanding of the molecular genetics of hereditary CRC syndromes. As a consequence, unique molecular diagnostic algorithms have emerged in LS. The most comprehensive approach should include the assessment of both MSI and IHC for DNA MMR proteins. However, if MSI testing is not available, IHC alone is a reasonable strategy. Germline sequencing of the genes of interest, now widely available, can then be pursued in those patients with a positive MSI or IHC test. Early recognition of LS can reduce mortality from cancer through targeted surveillance measures. Distinctive genotype–phenotype correlations have also begun to emerge. As the number of genes associated with LS increases, it is apparent that the clinical phenotype can differ based upon the specific gene mutated, although these differences have not yet led to formal gene-specific screening and management recommendations.
Five-year view
Microsatellite instability and IHC analysis are excellent screening tests for LS. As molecular pathologic services continue to expand, this should facilitate access to both MSI and IHC tests. Currently, these tests are offered to individuals who fulfill certain high-risk criteria, but unbiased testing in all cases of CRC may emerge as the most effective way to identify all cases of LS. Large-scale studies are warranted to establish the accuracy and cost–effectiveness of routine screening of all patients with newly diagnosed CRC for LS. Insight into specific genotype–phenotype correlations may improve the clinical management of LS by refining cancer surveillance programs based upon the particular gene mutated. Finally, new strategies for chemoprevention are needed.
Key issues.
Lynch syndrome is the most common hereditary colorectal carcinoma syndrome, accounting for approximately 2–5% of all newly diagnosed cases of colorectal carcinoma.
Patients with Lynch syndrome have an increased risk of colorectal and endometrial cancer as well as certain extra-colonic cancers, including tumors of the stomach, small intestine, urinary collecting system and ovary.
Germline mutations in one of four DNA mismatch repair genes (MLH1, MSH2, MSH6 and PMS2) are linked to the disease.
Microsatellite instability (MSI) is present in 90–95% of colorectal cancers and 75% of endometrial cancers in Lynch syndrome, and MSI testing is therefore a useful diagnostic tool to screen for this disease.
Mismatch repair (MMR) gene defects can be assessed in tumor tissue through immunohistochemical (IHC) analysis, and absence of staining of a specific MMR protein can pinpoint the specific gene most likely to be mutated.
MSI and IHC testing can define which patients should further pursue germline DNA mutational analysis. Once a specific mutation is identified, the diagnosis of Lynch syndrome is confirmed, and carriers of a mutation can be differentiated from noncarriers within a family.
The early recognition of this syndrome is essential to identify patients who will require intensive cancer surveillance and other risk-reducing approaches, including prophylactic surgery.
The correlation between abnormal MSI/IHC tests and genetic testing for MMR gene mutations is excellent, but imperfect. When laboratory results are inconclusive, the clinical presentation should guide management.
Acknowledgments
Maria S Pino was supported by the MGH ECOR Fund for Medical Discovery; Daniel C Chung was supported in part by National Institutes of Health CA92594 and P50 CA127003 and also the Kate J and Dorothy L Clapp Fund.
No writing assistance was utilized in the production of this manuscript.
Footnotes
Syndrome-associated tumors include colorectal, endometrial, ovarian, stomach, small bowel, ureter or renal pelvis, biliary tract, pancreas, brain and sebaceous gland.
Presence of tumor infiltrating lymphocytes, Crohn's disease-like lymphocytic reaction, mucinous/signet-ring differentiation or medullary growth pattern. Data from [21,22,28].
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
References
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1•.Recommendations from the EGAPP Working Group genetic testing strategies in newly diagnosed individuals with colorectal cancer aimed at reducing morbidity and mortality from Lynch syndrome in relatives. Genet. Med. 2009;11(1):35–41. doi: 10.1097/GIM.0b013e31818fa2ff. [Comprehensive and critical evaluation of genetic testing strategies in newly diagnosed individuals with colorectal cancer] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Teutsch SM, Bradley LA, Palomaki GE, et al. The evaluation of genomic applications in practice and prevention (EGAPP) initiative: methods of the EGAPP working group. Genet. Med. 2009;11(1):3–14. doi: 10.1097/GIM.0b013e318184137c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vasen HF, Wijnen JT, Menko FH, et al. Cancer risk in families with hereditary nonpolyposis colorectal cancer diagnosed by mutation analysis. Gastroenterology. 1996;110(4):1020–1027. doi: 10.1053/gast.1996.v110.pm8612988. [DOI] [PubMed] [Google Scholar]
- 4.Watson P, Lynch HT. Cancer risk in mismatch repair gene mutation carriers. Fam. Cancer. 2001;1(1):57–60. doi: 10.1023/a:1011590617833. [DOI] [PubMed] [Google Scholar]
- 5.Hampel H, Stephens JA, Pukkala E, et al. Cancer risk in hereditary nonpolyposis colorectal cancer syndrome: later age of onset. Gastroenterology. 2005;129(2):415–421. doi: 10.1016/j.gastro.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 6.Vasen HF, Möslein G, Alonso A, et al. Guidelines for the clinical management of Lynch syndrome (hereditary non-polyposis cancer). J. Med. Genet. 2007;44(6):353–362. doi: 10.1136/jmg.2007.048991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kastrinos F, Stoffel EM, Balmaña J, Steyerberg EW, Mercado R, Syngal S. Phenotype comparison of MLH1 and MSH2 mutation carriers in a cohort of 1,914 individuals undergoing clinical genetic testing in the United States. Cancer Epidemiol. Biomarkers Prev. 2008;17(8):2044–2051. doi: 10.1158/1055-9965.EPI-08-0301. [DOI] [PubMed] [Google Scholar]
- 8.Stoffel E, Mukherjee B, Raymond VM, et al. Calculation of risk of colorectal and endometrial cancer among patients with Lynch syndrome. Gastroenterology. 2009;137(5):1621–1627. doi: 10.1053/j.gastro.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9•.Baglietto L, Lindor NM, Dowty JG, et al. Risks of Lynch syndrome cancers for MSH6 mutation carriers. J. Natl Cancer Inst. 2010;102(3):193–201. doi: 10.1093/jnci/djp473. [Comprehensive evaluation of the absolute and relative cancer risks for MSH6 mutation carriers] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10•.Senter L, Clendenning M, Sotamaa K, et al. The clinical phenotype of Lynch syndrome due to germ-line PMS2 mutations. Gastroenterology. 2008;135(2):419–428. doi: 10.1053/j.gastro.2008.04.026. [The first report that PMS2 mutations significantly contribute to Lynch syndrome] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park YJ, Shin KH, Park JG. Risk of gastric cancer in hereditary nonpolyposis colorectal cancer in Korea. Clin. Cancer Res. 2000;6(8):2994–2998. [PubMed] [Google Scholar]
- 12.Palomaki GE, McClain MR, Melillo S, Hampel HL, Thibodeau SN. EGAPP supplementary evidence review: DNA testing strategies aimed at reducing morbidity and mortality from Lynch syndrome. Genet. Med. 2009;11(1):42–65. doi: 10.1097/GIM.0b013e31818fa2db. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu HX, Zhou XL, Liu T, et al. The role of hMLH3 in familial colorectal cancer. Cancer Res. 2003;63(8):1894–1899. [PubMed] [Google Scholar]
- 14.Wu Y, Berends MJ, Post JG, et al. Germline mutations of EXO1 gene in patients with hereditary nonpolyposis colorectal cancer (HNPCC) and atypical HNPCC forms. Gastroenterology. 2001;120(7):1580–1587. doi: 10.1053/gast.2001.25117. [DOI] [PubMed] [Google Scholar]
- 15.Järvinen HJ, Aarnio M, Mustonen H, et al. Controlled 15-year trial on screening for colorectal cancer in families with hereditary nonpolyposis colorectal cancer. Gastroenterology. 2000;118(5):829–834. doi: 10.1016/s0016-5085(00)70168-5. [DOI] [PubMed] [Google Scholar]
- 16.Lynch HT. Cancer and the family history trail. NY State J. Med. 1991;91:145–147. [PubMed] [Google Scholar]
- 17.Loukola A, Eklin K, Laiho P, et al. Microsatellite marker analysis in screening for hereditary nonpolyposis colorectal cancer (HNPCC). Cancer Res. 2001;61:4545–4549. [PubMed] [Google Scholar]
- 18.Müller W, Burgart LJ, Krause-Paulus R, et al. The reliability of immunohistochemistry as a prescreening method for the diagnosis of hereditary nonpolyposis colorectal cancer (HNPCC) – results of an international collaborative study. Fam. Cancer. 2001;1:87–93. doi: 10.1023/a:1013840907881. [DOI] [PubMed] [Google Scholar]
- 19.Warthin A. Hereditary with reference to carcinoma. Arch. Intern. Med. 1913;12:546–555. [Google Scholar]
- 20.Lynch HT, Shaw MW, Magnuson CW, Larsen AL, Krush AJ. Hereditary factors in cancer. Study of two large midwestern kindreds. Arch. Intern. Med. 1966;117:206–212. [PubMed] [Google Scholar]
- 21.Vasen HF, Mecklin JP, Khan PM, Lynch HT. The international collaborative group on hereditary non-polyposis colorectal cancer (ICG-HNPCC). Dis. Colon Rectum. 1991;34(5):424–425. doi: 10.1007/BF02053699. [DOI] [PubMed] [Google Scholar]
- 22.Vasen HF, Watson P, Mecklin JP, Lynch HT. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the international collaborative group on HNPCC. Gastroenterology. 1999;116(6):1453–1456. doi: 10.1016/s0016-5085(99)70510-x. [DOI] [PubMed] [Google Scholar]
- 23••.Lindor NM, Rabe K, Petersen GM, et al. Lower cancer incidence in Amsterdam-I criteria families without mismatch repair deficiency: Familial colorectal cancer type X. JAMA. 2005;293(16):1979–1985. doi: 10.1001/jama.293.16.1979. [First description of a new colon cancer syndrome tentatively designated as ‘familial colorectal cancer type X syndrome’] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Llor X, Pons E, Xicola RM, et al. Differential features of colorectal cancers fulfilling Amsterdam criteria without involvement of the mutator pathway. Clin. Cancer Res. 2005;11(20):7304–7310. doi: 10.1158/1078-0432.CCR-05-0965. [DOI] [PubMed] [Google Scholar]
- 25.Barnetson RA, Tenesa A, Farrington SM, et al. Identification and survival of carriers of mutations in DNA mismatch-repair genes in colon cancer. N. Engl. J. Med. 2006;354:2751–2763. doi: 10.1056/NEJMoa053493. [DOI] [PubMed] [Google Scholar]
- 26.Hampel H, Frankel WL, Martin E, et al. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer). N. Engl. J. Med. 2005;352(18):1851–1860. doi: 10.1056/NEJMoa043146. [DOI] [PubMed] [Google Scholar]
- 27.Casey G, Lindor NM, Papadopoulos N, et al. Conversion analysis for mutation detection in MLH1 and MSH2 in patients with colorectal cancer. JAMA. 2005;293(7):799–809. doi: 10.1001/jama.293.7.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28••.Rodriguez-Bigas MA, Boland CR, Hamilton SR, et al. A National Cancer Institute workshop on hereditary nonpolyposis colorectal cancer syndrome: meeting highlights and Bethesda guidelines. J. Natl Cancer Inst. 1997;89(23):1758–1762. doi: 10.1093/jnci/89.23.1758. [Highlights of the National Cancer Institute workshop on hereditary nonpolyposis colorectal cancer during which the Bethesda guidelines were established] [DOI] [PubMed] [Google Scholar]
- 29.Umar A, Boland CR, Terdiman JP, et al. Revised Bethesda guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J. Natl Cancer Inst. 2004;96(4):261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonis PA, Trikalinos TA, Chung M, et al. Hereditary nonpolyposis colorectal cancer: diagnostic strategies and their implications. Evid. Rep. Technol. Assess. (Full Rep.) 2007;(150):1–180. [PMC free article] [PubMed] [Google Scholar]
- 31.Prolla TA, Pang Q, Alani E, Kolodner RD, Liskay RM. MLH1, PMS1, and MSH2 interactions during the initiation of DNA mismatch repair in yeast. Science. 1994;265:1091–1093. doi: 10.1126/science.8066446. [DOI] [PubMed] [Google Scholar]
- 32.Fishel R, Ewel A, Lee S, Lescoe MK, Griffith J. Binding of mismatched microsatellite DNA sequences by the human MSH2 protein. Science. 1994;266:1403–1405. doi: 10.1126/science.7973733. [DOI] [PubMed] [Google Scholar]
- 33.Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLα in human mismatch repair. Cell. 2006;126(2):297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 34.Markowitz S, Wang J, Myeroff L, et al. Inactivation of the type II TGF-β receptor in colon cancer cells with microsatellite instability. Science. 1995;268(5215):1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 35.Rampino N, Yamamoto H, Ionov Y, et al. Somatic frameshift mutations in the Bax gene in colon cancers of the microsatellite mutator phenotype. Science. 1997;275(5302):967–969. doi: 10.1126/science.275.5302.967. [DOI] [PubMed] [Google Scholar]
- 36.Boland CR, Thibodeau SN, Hamilton SR, et al. A National Cancer Institute workshop on microsatellite instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58(22):5248–5257. [PubMed] [Google Scholar]
- 37.Perucho M, Boland CR, et al. A National Cancer Institute workshop on microsatellite instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. Correspondence re. [PubMed] [Google Scholar]; Cancer Res. 1999;59(1):249–256. [PubMed] [Google Scholar]
- 38.Nash GM, Gimbel M, Shia J, et al. Automated, multiplex assay for high-frequency microsatellite instability in colorectal cancer. J. Clin. Oncol. 2003;21(16):3105–3112. doi: 10.1200/JCO.2003.11.133. [DOI] [PubMed] [Google Scholar]
- 39.Samowitz WS, Slattery ML, Potter JD, Leppert MF. BAT-26 and BAT-40 instability in colorectal adenomas and carcinomas and germline polymorphisms. Am. J. Pathol. 1999;154(6):1637–1641. doi: 10.1016/S0002-9440(10)65418-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pyatt R, Chadwick RB, Johnson CK, et al. Polymorphic variation at the BAT-25 and BAT-26 loci in individuals of African origin. Implications for microsatellite instability testing. Am. J. Pathol. 1999;155(2):349–353. doi: 10.1016/S0002-9440(10)65131-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suraweera N, Duval A, Reperant M, et al. Evaluation of tumor microsatellite instability using five quasimonomorphic mononucleotide repeats and pentaplex PCR. Gastroenterology. 2002;123(6):1804–1811. doi: 10.1053/gast.2002.37070. [DOI] [PubMed] [Google Scholar]
- 42.Murphy KM, Zhang S, Geiger T, et al. Comparison of the microsatellite instability analysis system and the Bethesda panel for the determination of microsatellite instability in colorectal cancers. J. Mol. Diagn. 2006;8(3):305–311. doi: 10.2353/jmoldx.2006.050092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bacher JW, Flanagan LA, Smalley RL, et al. Development of a fluorescent multiplex assay for detection of MSI-high tumors. Dis. Markers. 2004;20(4–5):237–250. doi: 10.1155/2004/136734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xicola RM, Llor X, Pons E, et al. Performance of different microsatellite marker panels for detection of mismatch repair-deficient colorectal tumors. J. Natl Cancer Inst. 2007;99(3):244–252. doi: 10.1093/jnci/djk033. [DOI] [PubMed] [Google Scholar]
- 45.Malesci A, Laghi L, Bianchi P, et al. Reduced likelihood of metastases in patients with microsatellite-unstable colorectal cancer. Clin. Cancer Res. 2007;13(13):3831–3839. doi: 10.1158/1078-0432.CCR-07-0366. [DOI] [PubMed] [Google Scholar]
- 46.Hatch SB, Lightfoot HM, Garwacki CP, et al. Microsatellite instability testing in colorectal carcinoma: choice of markers affects sensitivity of detection of mismatch repair-deficient tumors. Clin. Cancer Res. 2005;11(6):2180–2187. doi: 10.1158/1078-0432.CCR-04-0234. [DOI] [PubMed] [Google Scholar]
- 47•.Laghi L, Bianchi P, Malesci A. Differences and evolution of the methods for the assessment of microsatellite instability. Oncogene. 2008;27(49):6313–6321. doi: 10.1038/onc.2008.217. [Comprehensive review on the evolution and differences of the methods used for the assessment of microsatellite instability status] [DOI] [PubMed] [Google Scholar]
- 48.Kondo Y, Issa JP. Epigenetic changes in colorectal cancer. Cancer Metastasis Rev. 2004;23(1–2):29–39. doi: 10.1023/a:1025806911782. [DOI] [PubMed] [Google Scholar]
- 49.Deng G, Bell I, Crawley S, et al. BRAF mutation is frequently present in sporadic colorectal cancer with methylated hMLH1, but not in hereditary nonpolyposis colorectal cancer. Clin. Cancer Res. 2004;10(1):191–195. doi: 10.1158/1078-0432.ccr-1118-3. [DOI] [PubMed] [Google Scholar]
- 50.Domingo E, Niessen RC, Oliveira C, et al. BRAF-V600E is not involved in the colorectal tumorigenesis of HNPCC in patients with functional MLH1 and MSH2 genes. Oncogene. 2005;24(24):3995–3998. doi: 10.1038/sj.onc.1208569. [DOI] [PubMed] [Google Scholar]
- 51.Hendriks YM, Wagner A, Morreau H, et al. Cancer risk in hereditary nonpolyposis colorectal cancer due to MSH6 mutations: impact on counseling and surveillance. Gastroenterology. 2004;127(1):17–25. doi: 10.1053/j.gastro.2004.03.068. [DOI] [PubMed] [Google Scholar]
- 52.Shia J. Immunohistochemistry versus microsatellite instability testing for screening colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome: part I. The utility of immunohistochemistry. J. Mol. Diagn. 2008;10(4):293–300. doi: 10.2353/jmoldx.2008.080031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raevaara TE, Vaccaro C, Abdel-Rahman WM, et al. Pathogenicity of the hereditary colorectal cancer mutation hMLH1 del616 linked to shortage of the functional protein. Gastroenterology. 2003;125:501–509. doi: 10.1016/s0016-5085(03)00905-3. [DOI] [PubMed] [Google Scholar]
- 54.Salahshor S, Koelble K, Rubio C, et al. Microsatellite instability and hMLH1 and hMSH2 expression analysis in familial and sporadic colorectal cancer. Lab. Invest. 2001;81:535–541. doi: 10.1038/labinvest.3780262. [DOI] [PubMed] [Google Scholar]
- 55.Wahlberg SS, Schmeits J, Thomas G, et al. Evaluation of microsatellite instability and immunohistochemistry for the prediction of germ-line MSH2 and MLH1 mutations in hereditary nonpolyposis colon cancer families. Cancer Res. 2002;62:3485–3492. [PubMed] [Google Scholar]
- 56.De Jong AE, Van Puijenbroek M, Hendriks Y, et al. Microsatellite instability, immunohistochemistry, and additional PMS2 staining in suspected hereditary nonpolyposis colorectal cancer. Clin. Cancer Res. 2004;10(3):972–980. doi: 10.1158/1078-0432.ccr-0956-3. [DOI] [PubMed] [Google Scholar]
- 57.Acharya S, Wilson T, Gradia S, et al. hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc. Natl Acad. Sci. USA. 1996;93(24):13629–13634. doi: 10.1073/pnas.93.24.13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marsischky GT, Filosi N, Kane MF, Kolodner R. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 1999;10:407–420. doi: 10.1101/gad.10.4.407. [DOI] [PubMed] [Google Scholar]
- 59.Chang CL, Marra G, Chauhan DP, et al. Oxidative stress inactivates the human DNA mismatch repair system. Am. J. Physiol. Cell Physiol. 2002;283(1):C148–154. doi: 10.1152/ajpcell.00422.2001. [DOI] [PubMed] [Google Scholar]
- 60.Bindra RS, Crosby ME, Glazer PM. Regulation of DNA repair in hypoxic cancer cells. Cancer Metastasis Rev. 2007;26:249–260. doi: 10.1007/s10555-007-9061-3. [DOI] [PubMed] [Google Scholar]
- 61.Watson N, Grieu F, Morris M, et al. Heterogeneous staining for mismatch repair proteins during population-based prescreening for hereditary nonpolyposis colorectal cancer. J. Mol. Diagn. 2007;9:472–478. doi: 10.2353/jmoldx.2007.060162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hampel H, Frankel WL, Martin E, et al. Feasibility of screening for Lynch syndrome among patients with colorectal cancer. J. Clin. Oncol. 2008;26:5783–5788. doi: 10.1200/JCO.2008.17.5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lindor NM, Burgart LJ, Leontovich O, et al. Immunohistochemistry versus microsatellite instability testing in phenotyping colorectal tumors. J. Clin. Oncol. 2002;20(4):1043–1048. doi: 10.1200/JCO.2002.20.4.1043. [DOI] [PubMed] [Google Scholar]
- 64.Pinol V, Castells A, Andreu M, et al. Accuracy of revised Bethesda guidelines, microsatellite instability, and immunohistochemistry for the identification of patients with hereditary nonpolyposis colorectal cancer. JAMA. 2005;293(16):1986–1994. doi: 10.1001/jama.293.16.1986. [DOI] [PubMed] [Google Scholar]
- 65.Lagerstedt Robinson K, Liu T, Vandrovcova J, et al. Lynch syndrome (hereditary nonpolyposis colorectal cancer) diagnostics. J. Natl Cancer Inst. 2007;99(4):291–299. doi: 10.1093/jnci/djk051. [DOI] [PubMed] [Google Scholar]
- 66.Halvarsson B, Lindblom A, Johansson L, Lagerstedt K, Nilbert M. Loss of mismatch repair protein immunostaining in colorectal adenomas from patients with hereditary nonpolyposis colorectal cancer. Mod. Pathol. 2005;18:1095–1101. doi: 10.1038/modpathol.3800392. [DOI] [PubMed] [Google Scholar]
- 67.Pino MS, Mino-Kenudson M, Wildemore BM, et al. Deficient DNA mismatch repair is common in Lynch syndrome-associated colorectal adenomas. J. Mol. Diagn. 2009;11(3):238–247. doi: 10.2353/jmoldx.2009.080142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ichikawa Y, Lemon S, Wang S, et al. Microsatellite instability and expression of MLH1 and MSH2 in normal and malignant endometrial and ovarian epithelium in hereditary nonpolyposis colorectal cancer family members. Cancer Genet. Cytogenet. 1999;112(1):2–8. doi: 10.1016/s0165-4608(98)00252-0. [DOI] [PubMed] [Google Scholar]
- 69.Risinger JI, Berchuck A, Kohler MF, Watson P, Lynch HT, Boyd J. Genetic instability of microsatellites in endometrial carcinoma. Cancer Res. 1993;53(21):5100–5103. [PubMed] [Google Scholar]
- 70.De Leeuw WJ, Dierssen J, Vasen HF, et al. Prediction of a mismatch repair gene defect by microsatellite instability and immunohistochemical analysis in endometrial tumours from HNPCC patients. J. Pathol. 2000;192:328–335. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH701>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 71.Lim PC, Tester D, Cliby W, et al. Absence of mutations in DNA mismatch repair genes in sporadic endometrial tumors with microsatellite instability. Clin.Cancer Res. 1996;2:1907–1911. [PubMed] [Google Scholar]
- 72.Helland A, Børresen-Dale AL, Peltomäki P, et al. Microsatellite instability in cervical and endometrial carcinomas. Int. J. Cancer. 1997;70:499–501. doi: 10.1002/(sici)1097-0215(19970304)70:5<499::aid-ijc1>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 73.Kobayashi K, Sagae S, Kudo R, Saito H, Koi S, Nakamura Y. Microsatellite instability in endometrial carcinomas: frequent replication errors in tumors of early onset and/or of poorly differentiated type. Genes Chromosomes Cancer. 1995;14:128–132. doi: 10.1002/gcc.2870140207. [DOI] [PubMed] [Google Scholar]
- 74.Kihana T, Fujioka T, Hamada K, et al. Association of replication error positive phenotype with lymphocyte infiltration in endometrial cancers. Jpn. J. Cancer Res. 1998;89:895–902. doi: 10.1111/j.1349-7006.1998.tb00646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Catasus L, Machin P, Matias-Guiu X, Prat J. Microsatellite instability in endometrial carcinomas: clinicopathologic correlations in a series of 42 cases. Hum. Pathol. 1998;29:1160–1164. doi: 10.1016/s0046-8177(98)90430-0. [DOI] [PubMed] [Google Scholar]
- 76.Sakamoto T, Murase T, Urushibata H, et al. Microsatellite instability and somatic mutations in endometrial carcinomas. Gynecol. Oncol. 1998;71:53–58. doi: 10.1006/gyno.1998.5154. [DOI] [PubMed] [Google Scholar]
- 77.Krajinovic M, Richer C, Gorska-Flipot I, et al. Genomic loci susceptible to replication errors in cancer cells. Br. J. Cancer. 1998;78:981–985. doi: 10.1038/bjc.1998.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Parc YR, Halling KC, Burgart LJ, et al. Microsatellite instability and hMLH1/hMSH2 expression in young endometrial carcinoma patients: associations with family history and histopathology. Int. J. Cancer. 2000;86:60–66. doi: 10.1002/(sici)1097-0215(20000401)86:1<60::aid-ijc9>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 79.Gurin CC, Federici MG, Kang L, Boyd J. Causes and consequences of microsatellite instability in endometrial carcinoma. Cancer Res. 1999;59:462–466. [PubMed] [Google Scholar]
- 80.Tibiletti M, Furlan D, Taborelli M, et al. Microsatellite instability in endometrial cancer: relation to histological subtypes. Gynecol. Oncol. 1999;73:247–252. doi: 10.1006/gyno.1999.5351. [DOI] [PubMed] [Google Scholar]
- 81.Berends MJ, Wu Y, Sijmons RH, et al. Toward new strategies to select young endometrial cancer patients for mismatch repair gene mutation analysis. J. Clin. Oncol. 2003;21:4364–4370. doi: 10.1200/JCO.2003.04.094. [DOI] [PubMed] [Google Scholar]
- 82.Geisler JP, Goodheart MJ, Sood AK, et al. Mismatch repair gene expression defects contribute to microsatellite instability in ovarian carcinoma. Cancer. 2003;98:2199–2206. doi: 10.1002/cncr.11770. [DOI] [PubMed] [Google Scholar]
- 83.Sood AK, Holmes R, Hendrix MJ, Buller RE. Application of the National Cancer Institute international criteria for determination of microsatellite instability in ovarian cancer. Cancer Res. 2001;61:4371–4374. [PubMed] [Google Scholar]
- 84.King BL, Carcangiu ML, Carter D. Microsatellite instability in ovarian neoplasms. Br. J. Cancer. 1995;72:376–382. doi: 10.1038/bjc.1995.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fujita M, Enomoto T, Yoshino K, et al. Microsatellite instability and alterations in the hMSH2 gene in human ovarian cancer. Int. J. Cancer. 1995;64:361–366. doi: 10.1002/ijc.2910640602. [DOI] [PubMed] [Google Scholar]
- 86.Buller RE, Shahin MS, Holmes RW, Hatterman M, Kirby PA, Sood AK. p53 mutations and microsatellite instability in ovarian cancer: yin and yang. Am. J. Obstet. Gynecol. 2001;184:891–902. doi: 10.1067/mob.2001.113856. [DOI] [PubMed] [Google Scholar]
- 87.Malander S, Rambech E, Kristoffersson U, et al. The contribution of the hereditary nonpolyposis colorectal cancer syndrome to the development of ovarian cancer. Gynecol. Oncol. 2006;101:238–243. doi: 10.1016/j.ygyno.2005.10.029. [DOI] [PubMed] [Google Scholar]
- 88.Domanska K, Malander S, Masback A, Nilbert M. Ovarian cancer at young age: the contribution of mismatch-repair defects in a population-based series of epithelial ovarian cancer before age 40. Int. J. Gynecol. Cancer. 2007;17:789–793. doi: 10.1111/j.1525-1438.2007.00875.x. [DOI] [PubMed] [Google Scholar]
- 89.Rosen DG, Cai KQ, Luthra R, Liu J. Immunohistochemical staining of hMLH1 and hMSH2 reflects microsatellite instability status in ovarian carcinoma. Mod. Pathol. 2006;19:1414–1420. doi: 10.1038/modpathol.3800672. [DOI] [PubMed] [Google Scholar]
- 90.Gille JJ, Hogervorst FB, Pals G, et al. Genomic deletions of MSH2 and MLH1 in colorectal cancer families detected by a novel mutation detection approach. Br. J. Cancer. 2002;87:892–897. doi: 10.1038/sj.bjc.6600565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30(12):e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gylling A, Ridanpää M, Vierimaa O, et al. Large genomic rearrangements and germline epimutations in Lynch syndrome. Int. J. Cancer. 2009;124(10):2333–2340. doi: 10.1002/ijc.24230. [DOI] [PubMed] [Google Scholar]
- 93.Wijnen J, Van Der Klift H, Vasen H, et al. MSH2 genomic deletions are a frequent cause of HNPCC. Nat. Genet. 1998;20:326–328. doi: 10.1038/3795. [DOI] [PubMed] [Google Scholar]
- 94.Charbonnier F, Olschwang S, Wang Q, et al. MSH2 in contrast to MLH1 and MSH6 is frequently inactivated by exonic and promoter rearrangements in hereditary nonpolyposis colorectal cancer. Cancer Res. 2002;62:848–853. [PubMed] [Google Scholar]
- 95.Taylor CF, Charlton RS, Burn J, Sheridan E, Taylor GR. Genomic deletions in MSH2 or MLH1 are a frequent cause of hereditary non-polyposis colorectal cancer: identification of novel and recurrent deletions by MLPA. Hum. Mutat. 2003;22:428–433. doi: 10.1002/humu.10291. [DOI] [PubMed] [Google Scholar]
- 96.Wagner A, Barrows A, Wijnen JT, et al. Molecular analysis of hereditary nonpolyposis colorectal cancer in the United States: high mutation detection rate among clinically selected families and characterization of an American founder genomic deletion of the MSH2 gene. Am. J. Hum. Genet. 2003;72:1088–1100. doi: 10.1086/373963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang Y, Friedl W, Lamberti C, et al. Hereditary nonpolyposis colorectal cancer: frequent occurrence of large genomic deletions in MSH2 and MLH1 genes. Int. J. Cancer. 2003;103:636–641. doi: 10.1002/ijc.10869. [DOI] [PubMed] [Google Scholar]
- 98.Baudhuin LM, Ferber MJ, Winters JL, et al. Characterization of hMLH1 and hMSH2 gene dosage alterations in Lynch syndrome patients. Gastroenterology. 2005;129:846–854. doi: 10.1053/j.gastro.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 99.Desai DC, Lockman JC, Chadwick RB, et al. Recurrent germline mutation in MSH2 arises frequently de novo. J. Med. Genet. 2000;37(9):646–652. doi: 10.1136/jmg.37.9.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Green J, O'Driscoll M, Barnes A, et al. Impact of gender and parent of origin on the phenotypic expression of hereditary nonpolyposis colorectal cancer in a large newfoundland kindred with a common MSH2 mutation. Dis. Colon Rectum. 2002;45(9):1223–1232. doi: 10.1007/s10350-004-6397-4. [DOI] [PubMed] [Google Scholar]
- 101.Nyström-Lahti M, Kristo P, Nicolaides NC, et al. Founding mutations and Alu-mediated recombination in hereditary colon cancer. Nat. Med. 2005;1:1203–1206. doi: 10.1038/nm1195-1203. [DOI] [PubMed] [Google Scholar]
- 102.Foulkes WD, Thiffault I, Gruber SB, et al. The founder mutation MSH2*1906G-->C is an important cause of hereditary nonpolyposis colorectal cancer in the Ashkenazi Jewish population. Am. J. Hum. Genet. 2002;71(6):1395–1412. doi: 10.1086/345075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Clendenning M, Baze ME, Sun S, et al. Origins and prevalence of the American founder mutation of MSH2. Cancer Res. 2008;68(7):2145–2153. doi: 10.1158/0008-5472.CAN-07-6599. [DOI] [PubMed] [Google Scholar]
- 104.Ricciardone MD, Ozcelik T, Cevher B, et al. Human MLH1 deficiency predisposes to hematological maligancy and neurofibromatosis type 1. Cancer Res. 1999;59(2):290–293. [PubMed] [Google Scholar]
- 105.Wang Q, Lasset C, Desseigne F, et al. Neurofibromatosis and early onset of cancers in hMLH1-deficient children. Cancer Res. 1999;59(2):294–297. [PubMed] [Google Scholar]
- 106.Gallinger S, Aronson M, Shayan K, et al. Gastrointestinal cancers and neurofibromatosis type 1 features in children with a germline homozygous MLH1 mutation. Gastroenterology. 2004;126(2):576–585. doi: 10.1053/j.gastro.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 107.Krüger S, Kinzel M, Walldorf C, et al. Homozygous PMS2 germline mutations in two families with early-onset haematological malignancy, brain tumours, HNPCC-associated tumours, and signs of neurofibromatosis type 1. Eur. J. Hum. Genet. 2008;16(1):62–72. doi: 10.1038/sj.ejhg.5201923. [DOI] [PubMed] [Google Scholar]
- 108.Felton KE, Gilchrist DM, Andrew SE. Constitutive deficiency in DNA mismatch repair: is it time for Lynch III? Clin. Genet. 2007;71(6):499–500. doi: 10.1111/j.1399-0004.2007.00801.x. [DOI] [PubMed] [Google Scholar]
- 109.Bandipalliam P. Syndrome of early onset colon cancers, hematologic malignancies and features of neurofibromatosis in HNPCC families with homozygous mismatch repair gene mutations. Fam. Cancer. 2005;4(4):323–333. doi: 10.1007/s10689-005-8351-6. [DOI] [PubMed] [Google Scholar]
- 110•.Wimmer K, Etzler J. Constitutional mismatch repair-deficiency syndrome: Have we so far seen only the tip of an iceberg? Hum. Genet. 2008;124(2):105–122. doi: 10.1007/s00439-008-0542-4. [Overview of the current knowledge on the phenotypes of patients with biallelic inheritance of mutations in mismatch repair genes] [DOI] [PubMed] [Google Scholar]
- 111.Gazzoli I, Loda M, Garber J, Syngal S, Kolodner RD. A hereditary nonpolyposis colorectal carcinoma case associated with hypermethylation of the MLH1 gene in normal tissue and loss of heterozygosity of the unmethylated allele in the resulting microsatellite instability-high tumor. Cancer Res. 2002;62:3925–3928. [PubMed] [Google Scholar]
- 112.Miyakura Y, Sugano K, Akasu T, et al. Extensive but hemiallelic methylation of the hMLH1 promoter region in early-onset sporadic colon cancers with microsatellite instability. Clin. Gastroenterol. Hepatol. 2004;2(2):147–156. doi: 10.1016/s1542-3565(03)00314-8. [DOI] [PubMed] [Google Scholar]
- 113.Suter CM, Martin DI, Ward RL. Germline epimutation of MLH1 in individuals with multiple cancers. Nat. Genet. 2004;36:497–501. doi: 10.1038/ng1342. [DOI] [PubMed] [Google Scholar]
- 114.Hitchins M, Williams R, Cheong K, et al. MLH1 germline epimutations as a factor in hereditary nonpolyposis colorectal cancer. Gastroenterology. 2005;129(5):1392–1399. doi: 10.1053/j.gastro.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 115••.Hitchins MP, Wong JJ, Suthers G, et al. Inheritance of a cancer-associated MLH1 germ-line epimutation. N. Engl. J. Med. 2007;356(7):697–705. doi: 10.1056/NEJMoa064522. [First evidence of transgenerational epigenetic inheritance of Lynch syndrome mutations] [DOI] [PubMed] [Google Scholar]
- 116.Chan TL, Yuen ST, Kong CK, et al. Heritable germline epimutation of MSH2 in a family with hereditary nonpolyposis colorectal cancer. Nat. Genet. 2006;38(10):1178–1183. doi: 10.1038/ng1866. [DOI] [PubMed] [Google Scholar]
- 117.Ligtenberg MJ, Kuiper RP, Chan TL, et al. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3′ exons of TACSTD1. Nat. Genet. 2009;41(1):112–117. doi: 10.1038/ng.283. [DOI] [PubMed] [Google Scholar]
- 118.Campbell PT, Curtin K, Ulrich CM, et al. Mismatch repair polymorphisms and risk of colon cancer, tumour microsatellite instability and interactions with lifestyle factors. Gut. 2009;58(5):661–667. doi: 10.1136/gut.2007.144220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lipkin SM, Rozek LS, Rennert G, et al. The MLH1 D132H variant is associated with susceptibility to sporadic colorectal cancer. Nat. Genet. 2004;36(7):694–699. doi: 10.1038/ng1374. [DOI] [PubMed] [Google Scholar]
- 120.Raptis S, Mrkonjic M, Green RC, et al. MLH1-93G>A promoter polymorphism and the risk of microsatellite-unstable colorectal cancer. J. Natl Cancer Inst. 2007;99(6):463–474. doi: 10.1093/jnci/djk095. [DOI] [PubMed] [Google Scholar]
- 121.Hudson KL, Holohan MK, Collins FS. Keeping pace with the times – the genetic information nondiscrimination act of 2008. N. Engl. J. Med. 2008;358(25):2661–2663. doi: 10.1056/NEJMp0803964. [DOI] [PubMed] [Google Scholar]
- 122.Aaltonen LA, Salovaara R, Kristo P, et al. Incidence of hereditary nonpolyposis colorectal cancer and the feasibility of molecular screening for the disease. N. Engl. J. Med. 1998;338(21):1481–1487. doi: 10.1056/NEJM199805213382101. [DOI] [PubMed] [Google Scholar]
- 123.Lindor NM, Petersen GM, Hadley DW, et al. Recommendations for the care of individuals with an inherited predisposition to Lynch syndrome: a systematic review. JAMA. 2006;296(12):1507–1517. doi: 10.1001/jama.296.12.1507. [DOI] [PubMed] [Google Scholar]
Websites
- 201. European Commision: implementation of the council recommendation of 2 December 2003 on cancer screening (2003/878/ec) http://eur-lex.europa.eu/LexUriServ/LexUriServ.do?uri=OJ:L:2003:327:0034:0038:EN:PDF.
- 202. InSIGHT Database www.insight-group.org.