Abstract
Evolvability is the property of a biological system to quickly adapt to new requirements. Robustness seems to be the opposite. Nonetheless many biological systems display both properties—a puzzling observation, which has caused many debates over the last decades. A recently published model by Draghi et al. [Nature 463, 353–355 (2010)] elegantly circumvents complications of earlier in silico studies of molecular systems and provides an analytical solution, which is surprisingly independent from parameter choice. Depending on the mutation rate and the number of accessible phenotypes at any given genotype, evolvability and robustness can be reconciled. Further research will need to investigate if these parameter settings adequately represent the range of degrees of freedom covered by natural systems and if natural systems indeed assume a state in which both properties, robustness and evolvability, are featured.
Biological systems are widely perceived to be robust in the sense that small or occasionally even larger, disturbances will not alter the system’s behavior. Examples are known for cellular networks (Milo et al., 2002; Aldana and Cluzel, 2003), protein structures (Taverna and Goldstein, 2002), or developmental pathways (von Dassow et al., 2000). From an evolutionary perspective this observation is puzzling: how, if small variations in the genotype do not change a phenotype, can organisms adapt to changing environments? And there is plenty of evidence indeed that biological systems are also very evolvable, i.e., produce heritable phenotypic variation (Wagner, 2008) such as heat shock proteins (HSPs) facilitating variation (Queitsch et al., 2002), RNAs (Fontana and Schuster, 1998), or standing genetic variation (Wittkopp et al., 2009).
In theoretical terms, evolvability has been the focus of many approaches since long. A particularly useful metaphor, the one of fitness landscapes has been coined in the 1930 by theoretical and mathematical biologists such as Sewall Wright, John Haldane, and Ronald Fisher. Therein, a landscape depicts a scalar fitness value over a discrete genotype space and evolution becomes a walk on this landscape with populations typically concentrating on the highest mountains, representing the fittest individuals. With the advances of novel technologies over the last decades, such as affordable computers, efficient software, and novel genome sequencing technologies, many of these earlier predictions could be tested in silico and experimentally.
Among the most remarkable theoretical insights are result from computational studies on the genotype-phenotype relationship. For example, digital organisms (Wilke et al., 2001), protein lattice models (Bornberg-Bauer and Chan, 1999), or analytical considerations (Huynen, 1996; van Nimwegen et al., 1999) indicated that we may often observe the “survival of the flattest,” i.e., the genotype which has the largest number of neutral mutations or, in other words, is most robust. Many insights were obtained from studies on RNA, which is a suitable model system because RNA plays many functional roles, an RNA world may have been at the origin of life and RNA phenotypes can be reasonably approximated by calculating RNA (secondary) structures from sequence alone, much easier than for proteins. Here, the most influential notion derived was the one of neutral networks, collections of genotypes which are interconnected by a series of point mutations, and all code (leaving aside the issue of suboptimal solutions) for the same structure (Schuster et al., 1994). In silico population dynamic studies used constant population sizes and let mutants survive with a probability proportional to their fitness, arbitrarily defined by the phenotype’s similarity to a target (structure), thus enforcing adaptation to a new environmental pressure. These studied explained, at the molecular level, the interplay between evolutionary stasis, i.e., when the population drifts along neutral networks, sometimes splitting into subpopulations, and punctuated innovations (Huynen, 1996; Fontana and Schuster, 1998) when a transition to another network, representing a fitter phenotype (structure) occurs. Evolvability is generally high since almost all phenotypes (i.e., frequent structures) in the near mutational neighborhood of any genotype ni are accessible within a few mutations on virtually every major neutral network (Huynen, 1996; Fontana and Schuster, 1998). This has been metaphorically compared to “a bowl of spaghetti” (Goldstein, 2008) because every noodle (neutral network, representing one of many phenotypes K) comes (mutationally) close to any other at least somewhere in the bowl (sequence space, see Fig. 1). Proteins, on the other hand, seem to form dense blobs with a pronounced center np, featuring many neutral mutations (i.e., being robust) and the number of neutral mutationsq decreasing (robustness decreasing) with mutational distance from the center (Bornberg-Bauer and Chan, 1999) (see Fig. 2). The probability to find other phenotpyes pj in close mutational neighborhood of any given genotype bearing pi, i.e., the evolvability, is vanishingly low everywhere, but slightly higher at specific “transitions points” further away from np.
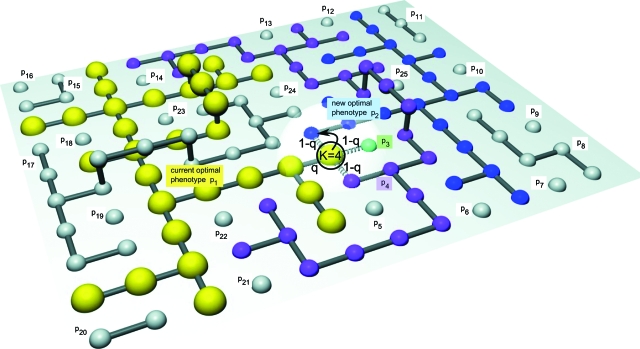
Figure 1. Sequence space of RNA genotypes and the neutral networks they form.
Genotypes (spheres) encoding the same phenotype (p1 to p25) are connected via neutral mutations (with probability q). Non-neutral mutations (with probability 1−q) are only explicitly shown (dashed connections) for one highlighted genotype (black circle). The number of phenotypes available to this genotype is K=4, these are drawn in color (p1 to p4). The yellow neutral network (p1) is under selection and has high fitness (large spheres), all other genotypes are lethal. After an environmental shift the blue phenotype (p2) might become beneficial so that the highlighted genotype could make a transition with only one mutation (black arrow).
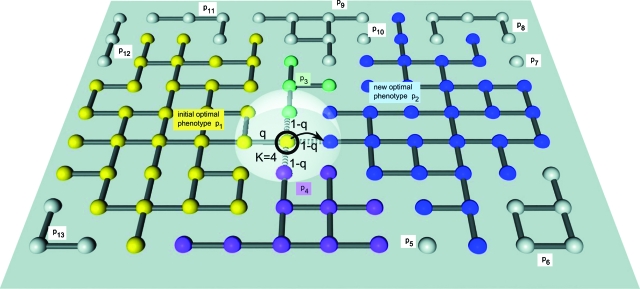
Figure 2. Sequence space and neutral networks of proteins.
Similar to Fig. 1, but here neutral networks form condensed topologies with many neutral neighbors, i.e., a high probability q at the center and a lower q toward the rims. Again, one example genotype is highlighted at a potential transition point.
A NEW MODEL RECONCILING EVOLVABILITY AND ROBUSTNESS
So far, these insights have been derived from simulations explicitly representing neutral networks or at least modeling a genotype-phenotype relationship. A particularly intriguing and elegant approach has now been put forward by Draghi et al., (2010), combining an effort of researchers from University of Pennsylvania and Yale and offering a framework, which unifies many of the afore mentioned model features. The model proposed by the authors depends on a small set of parameters μ,q,K,P,N. A population of N individuals evolves. In this population, a mutation occurs with probability μ, and if it occurs, the mutation is neutral with probability q. If it is not neutral, it picks randomly one of K phenotypes. The total number of phenotypes that can be reached from some genotype through a mutation is P and typically P is much bigger than the number K of “accessible” phenotypes, i.e., phenotypes that can be reached from a fixed genotype. The authors then work with a time-continuous approximation of a discrete Markov process. They compare their analytical results with statistical experiments; these data fit together in an impressive way. One important result is that the mean adaptation time, i.e., the mean waiting time before the arrival of a beneficial mutation, is minimal for a moderately high robustness value q provided that the ratio K∕P is small.
Intuitively, the model emphasizes the role of two factors: the existence of a moderate degree of neutrality and the rapid accessibility (in terms of nonlethal mutations) of a small fraction of all possible phenotypes. In other words, the genotype-phenotype relation must be structured in a way that at any given genotype a substantial—but not arbitrarily large—fraction of all phenotypes can be reached but at the same time information about the current phenotype is not completely lost (Eigen et al., 1989). The model does thus not represent networks explicitly but nonetheless has implicitly framed insights from network analysis in an analytical approach.
There are, of course, various properties of the fitness landscape, which cannot be easily captured by such a model. One would expect that the phenotypic neighborhoods of nearby genotypes are not independent of each other, which may be particularly true for modeling proteins (see the “blob” versus “spaghetti” analogy discussed above). More importantly, one would expect that the growth rate of the neighborhood of a genotype would influence the behavior of a system. By the growth rate, we mean the number of genotypes that can be reached in a fixed number of neutral mutations from a fixed starting point.
Moreover, this growth rate will very likely be different at different genotypes, i.e., the space of genotypes will in reality be rather anisotropic.
Most surprisingly however, the model seems to be independent from the choice of several parameters. For example the authors argue that the resampling of neighborhoods, i.e., the assumption that at every genotype the number of accessible phenotypes is equally (randomly) distributed and thus independent from the accessible phenotypes at neighboring genotypes, does not affect the qualitative behavior.
FURTHER IMPLICATIONS AND RAMIFICATIONS
So has the model then resolved all afore mentioned issues? Undoubtedly, it has improved our understanding of a complex fitness landscape, in particular in understanding the ratio between neutrality (corresponding to robustness in the model) and evolvability (number of immediate accessible phenotypes around every given instance). An “acid test” will require a couple of more observations to be brought in line: first of all, the anisotropy issue needs to be reconsidered for the model to account for proteins. Second, robustness in evolutionary theory refers to a population principle involving complex interactions, which may be represented by standing genetic variation. The precise architecture of these interactions is unclear as of yet and the target of many experimental approaches. Finally, recent results on lattice proteins have suggested that suboptimal structures may provide a very strong selection forces, influencing the dynamics of evolving populations to phenotypic transition points (Wroe et al., 2007)—a feature, which would however make the model substantially more complex.
However, considering the robustness of the presented model, i.e., the insensitivity toward parameter changes, suggests that it has at least captured a wide range of phenomena and will serve as a blueprint for further experiments and investigations on model behavior with respect to molecular fitness landscapes.
ACKNOWLEDGMENT:
We thank Tobias Sikosek for help with preparation of the figures. EBB acknowledges the support from DFG under Grant No. BO-2544-4∕1.
REFERENCES
- Aldana, M, and Cluzel, P (2003). “A natural class of robust networks.” Proc. Natl. Acad. Sci. U.S.A. 100, 8710–8714. 10.1073/pnas.1536783100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornberg-Bauer, E, and Chan, H S (1999). “Modeling evolutionary landscapes: mutational stability, topology, and superfunnels in sequence space.” Proc. Natl. Acad. Sci. U.S.A. 96, 10689–10694. 10.1073/pnas.96.19.10689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draghi, J A, Parsons, T L, Wagner, G P, and Plotkin, J B (2010). “Mutational robustness can facilitate adaptation.” Nature (London) 463, 353–355. 10.1038/nature08694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigen, M, McCaskill, J, and Schuster, P (1989). “The molecular quasi-species.” Adv. Chem. Phys. 75, 149–263. 10.1002/9780470141243.ch4 [DOI] [Google Scholar]
- Fontana, W, and Schuster, P (1998). “Continuity in evolution: on the nature of transitions.” Science 280, 1451–1455. 10.1126/science.280.5368.1451 [DOI] [PubMed] [Google Scholar]
- Goldstein, R A (2008). “The structure of protein evolution and the evolution of protein structure.” Curr. Opin. Struct. Biol. 18, 170–177. [DOI] [PubMed] [Google Scholar]
- Huynen, M A (1996). “Exploring phenotype space through neutral evolution.” J. Mol. Evol. 43, 165–169. 10.1007/BF02338823 [DOI] [PubMed] [Google Scholar]
- Milo, R, Shen-Orr, S, Itzkovitz, S, Kashtan, N, Chklovskii, D, and Alon, U (2002). “Network motifs: simple building blocks of complex networks.” Science 298, 824–827. 10.1126/science.298.5594.824 [DOI] [PubMed] [Google Scholar]
- Queitsch, C, Sangster, T A, and Lindquist, S (2002). “Hsp90 as a capacitor of phenotypic variation.” Nature (London) 417, 618–624. 10.1038/nature749 [DOI] [PubMed] [Google Scholar]
- Schuster, P, Fontana, W, Stadler, P F, and Hofacker, I L (1994). “From sequences to shapes and back: a case study in RNA secondary structures.” Proc. R. Soc. London, Ser. B 255, 279–284. 10.1098/rspb.1994.0040 [DOI] [PubMed] [Google Scholar]
- Taverna, D M, and Goldstein, R A (2002). “Why are proteins so robust to site mutations?” J. Mol. Biol. 315, 479–484. 10.1006/jmbi.2001.5226 [DOI] [PubMed] [Google Scholar]
- van Nimwegen, E, Crutchfield, J P, and Huynen, M (1999). “Neutral evolution of mutational robustness.” Proc. Natl. Acad. Sci. U.S.A. 96, 9716–9720. 10.1073/pnas.96.17.9716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Dassow, G, Meir, E, Munro, E M, and Odell, G M (2000). “The segment polarity network is a robust developmental module.” Nature (London) 406, 188–192. 10.1038/35018085 [DOI] [PubMed] [Google Scholar]
- Wagner, A (2008). “Robustness and evolvability: a paradox resolved.” Proc. R. Soc. London, Ser. B 275, 91–100. 10.1098/rspb.2007.1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke, C O, Wang, J L, Ofria, C, Lenski, R E, and Adami, C (2001). “Evolution of digital organisms at high mutation rates leads to survival of the flattest.” Nature (London) 412, 331–333. 10.1038/35085569 [DOI] [PubMed] [Google Scholar]
- Wittkopp, P J, Stewart, E E, Arnold, L L, Neidert, A H, Haerum, B K, Thompson, E M, Akhras, S, Smith-Winberry, G, and Shefner, L (2009). “Intraspecific polymorphism to interspecific divergence: genetics of pigmentation in Drosophila.” Science 326, 540–544. 10.1126/science.1176980 [DOI] [PubMed] [Google Scholar]
- Wroe, R, Chan, H S, and Bornberg-Bauer, E (2007). “A structural model of latent evolutionary potentials underlying neutral networks in proteins.” HFSP J. 1, 79–87. 10.2976/1.2739116 [DOI] [PMC free article] [PubMed] [Google Scholar]