Abstract
We review modeling studies concerning cytoskeletal activity of fission yeast. Recent models vary in length and time scales, describing a range of phenomena from cellular morphogenesis to polymer assembly. The components of cytoskeleton act in concert to mediate cell-scale events and interactions such as polarization. The mathematical models reduce these events and interactions to their essential ingredients, describing the cytoskeleton by its bulk properties. On a smaller scale, models describe cytoskeletal subcomponents and how bulk properties emerge.
The cell interior faces disorder from homogenizing Brownian motion. But life requires order. Cells need to control their shape, direct their motion, polarize, and divide. The cytoskeleton comprises the filamentous networks that provide scaffolding for internal order (Hayles and Nurse, 2001). Powered by ATP and GTP hydrolysis, the polymers and motor proteins of the cytoskeleton organize spontaneously into varied networks. These cytoskeletal structures span several orders of magnitude in length. The structural building blocks, nanometers across, may be a thousandth the size of the largest structures they form. Cytoskeletal dynamics span several orders of magnitude in time as well. A single polymer subunit may diffuse across the cell in seconds, a ten-thousandth of the cell’s division time.
Fission yeast is one model organism for the study of subcellular organization mediated by the cytoskeleton (La Carbona et al., 2006). Fission yeast undergoes simple and reproducible cell shape changes. Additionally, the ease of genetic manipulations and microscopic imaging make the organism ideal for quantitative studies. A growing body of theoretical work examines cytoskeletal organization in asymptotic regimes of space and time (Mogilner et al., 2006b). These theoretical models support reduction of a system to essential components by matching the emergent behavior in the model with the observed behavior from experiments. Where model behavior differs from experimental results, these models motivate further investigation. Recently, modeling contributed to understanding the role of the cytoskeleton in fission yeast cell polarization and mitosis.
POLARIZED GROWTH
Fission yeast grow along one axis. Their shape is simple: to first approximation, two hemispheres of constant radius cap a cylinder of increasing length (see Fig. 1). When the length has doubled from birth, a contractile ring halves the cell (Bathe and Chang, 2010; Pollard and Wu, 2010). Growth occurs at the tips. When growth starts, only the old end—the end not created by the previous division—grows. This monopolar growth eventually gives way to bipolar growth; this is called new end take-off (NETO) (Mitchison and Nurse, 1985). Two components of the cytoskeleton, actin filaments and microtubules, mediate growth. These cells mark their tips for growth with the help of microtubules and execute growth with the help of actin filaments (Martin, 2009; Piel and Tran, 2009; Tolić-Nørrelykke, 2010).
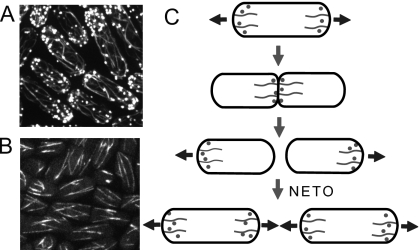
Figure 1. Images of yeast cells (Jian-Qiu Wu, Ohio State University) and yeast growth pattern.
(A) Images of the actin cytoskeleton in cells expressing GFP-CHD which binds to the sides of actin filaments. Actin cables and actin patches are seen distributed in monopolar and bipolar patterns. (B) In cells expressing GFP-atb2, microtubule bundles run across the cell. (C) Cartoon showing the redistribution of the actin cytoskeleton during the cell cycle. Prior to cytokinesis, actin accumulates at growing tips; during mitosis it accumulates in the middle; daughter cells start to grow in a monopolar manner and transition to bipolar growth at new end take-off.
Microtubules polymerize toward both tips. Stable ends anchor close to the nucleus in bundles while the dynamically unstable ends explore the interior near the cell tips (see Fig. 1) (Chang and Martin, 2009; Piel and Tran, 2009; Sawin and Tran, 2006). Although individual microtubules are short-lived, they collectively provide a directed track to the cell tips. The microtubules contribute to tip growth indirectly—motor proteins follow them to transport landmark proteins to the cell tips (Mata and Nurse, 1997).
Actin polymerizes near growing tips. Regulating proteins organize actin filaments into two major structures: cables and patches (Moseley and Goode, 2006; Pollard and Cooper, 2009). The formin For3p associates with tip markers where it nucleates and polymerizes actin cables (Martin and Chang, 2006). Cables wind from the tips through the cell body; motor proteins transport secretory vesicles and organelles along cables to the cell tips. The Arp2∕3 complex nucleates actin patches near growth sites for endocytosis (Sirotkin et al., 2005). In patches, short actin filaments form dense, highly branched networks.
These individual cytoskeletal structures are transient and disordered compared to the lifespan and order of the whole cell. Yet somehow they self-organize into a system robust enough to provide cells with a simple pattern of cell growth. These coupled growth processes, from a pool of structural components and regulator proteins, provide flexible and reliable scaffolding for the order required by living cells.
Models of polarized cell growth and NETO
The simple growth pattern of fission yeast provides an opportunity to model how cells develop order. For instance, a model of NETO may reveal basic mechanisms responsible for polarity. Several lines of evidence indicate that NETO depends on cytoskeletal dynamics (Marks et al., 1986; Martin and Chang, 2005). For example, some strains of yeast switch out of the monopolar state into the bipolar state after transient treatments with Latrunculin A (LatA), a drug that prevents actin polymerization by sequestering actin monomers (Rupes et al., 1999). Microtubules in monopolar cells are symmetrically distributed but the actin filaments and the formin nucleators concentrate at the growing old end. As the cells grow longer, they undergo NETO—transition to a state of symmetric growth, with symmetric actin and microtubule distributions. What could be the process responsible for the asymmetry in the actin distribution before NETO? And what triggers NETO?
A recent modeling study of NETO contains promising insights (Csikász-Nagy et al., 2008). The authors cast the problem in the language of nonlinear dynamics. According to their model, as cells elongate the state of asymmetric polymerization, or monopolar growth, becomes unstable. The cell assumes a stable symmetric polymerization state—bipolar growth. The model implicates length-dependent instability as the cause of NETO. This model belongs to a well-studied class of models called reaction-diffusion models (Meinhardt and Gierer, 2000; Turing, 1952).
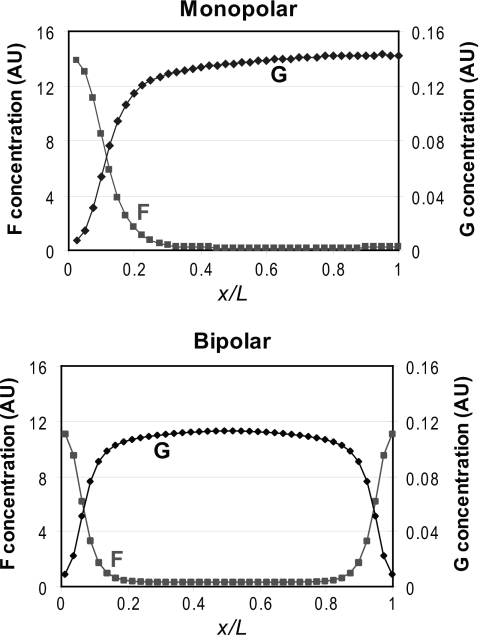
In the model of Csikász-Nagy and others, microtubules transport a continuous field of dynamic landmarks toward the cell tips symmetrically, as suggested by experiments (Martin and Chang, 2005). These markers contribute to conversion of a fast-diffusing substrate into a slowly diffusing polymer. Likely candidates for the substrate and polymer are actin monomers and actin filaments, respectively. Dynamic landmarks activate autocatalytic actin polymerization: presumably, actin filaments in cables and patches further recruit actin nucleators. This could be consistent with experiments suggesting positive feedback in the polarization system (Terenna et al., 2008). Autocatalytic polymerization amplifies local noise, polarizing the cell and breaking symmetry. Autocatalytic growth at the growing tip depletes the cytoplasmic actin-monomer pool and prevents growth at the other tip. As cells grow, the diffusion limits flow of actin monomers to the growing tip (Fig. 2), the concentration of actin monomers at the new end increases, and the new end takes off. For some lengths, stable monopolar and bipolar states coexist, consistent with switching between states after transient LatA treatment (Rupes et al., 1999)—a major success for the model.
Figure 2. Polymer (f) and substrate (g) concentration gradients across the long axis of a growing cell (distance.
x, total cell length L ). [Data generated with the model of Csikász-Nagy et al. (2008), online content with default parameters (http://www.cellcycle.bme.hu/morphopaper/).] Monopolar cell is 8.50 μm and bipolar cell is 14.0 μm. Both concentrations in arbitrary units using the same scale on both axes.
Riveline (2009) approaches the problem differently. According to his scaling arguments, asymmetric end curvature determines the post-division growth pattern. The new end has a higher radius of curvature after septation; he proposes that this inhibits growth and argues NETO occurs when turgor pressure deforms the new end, decreasing its curvature. This model predicts that length at NETO increases with cell radius—this has not yet been tested.
These models do not directly contradict each other but they do suggest different dominating factors. According to Csikász-Nagy and others, bipolar growth depends on diffusion limitation, unaccounted for by Riveline; according to Riveline, monopolar growth depends on a difference in tip curvature, unaccounted for by Csikász-Nagy and others. It remains unclear whether both mechanisms could coexist.
Modeling studies of NETO raise questions for experimentalists and theorists. Does NETO depend on the existence of a growing actin-monomer concentration gradient? So far, the actin-monomer concentration profile remains unmeasured in yeast. What new experimental evidence will emerge regarding the connections between cytoskeletal polarization and cell growth (Baumgärtner and Tolić-Nørrelykke, 2009)? What other models of bistable behavior are possible? Physical models of collective phenomena such as NETO are necessarily coarse-grained. How can such coarse-grained models be tested using advanced genetics operating at the molecular level?
Related models of polarization for budding yeast
Much recent modeling of polarization focuses on another yeast. Budding yeast draws its name from a polarity process. Wild-type cells mark their previous division site but experiments suggest that cells select a site for budding in the absence of these markings. Onsum and Rao (2009) recently reviewed modeling work on symmetry breaking in budding yeast. According to these recent Turing-type models, cells amplify a cortical Cdc42p signal and pick a spot on a homogeneous surface (Altschuler et al., 2008; Goryachev and Pokhilko, 2008; Hawkins et al., 2009; Howell et al., 2009; Marco et al., 2007; Ozbudak et al., 2005; Slaughter et al., 2009). These studies describe varied causes of autocatalytic amplification, both actin-dependent and actin-independent, and global inhibition. Actin contributes to the local amplification required by Turing models of budding-yeast symmetry breaking: Cdc42p sites nucleate actin cables that facilitate further Cdc42p delivery to those sites.
Fission yeast Cdc42p acts similarly. With other proteins, it activates actin-cable-nucleator For3p and therefore affects cytoskeletal organization (Chang and Martin, 2009; Martin et al., 2007). If a reaction-diffusion model best explains NETO, some substrate such as a signaling molecule may limit growth. Cdc42p and related proteins constitute a reasonable pool of suspects for this substrate. As in budding yeast, Cdc42p clustering and actin-dependent transport may contribute to autocatalytic assembly at the tips. Unlike in budding yeast, the actin cytoskeleton appears to mediate growth at sites defined and maintained by microtubules. Fission yeast maintains a polarized growth state throughout but budding yeast grows outward symmetrically during the first growth phase. Perhaps this lessens budding yeast’s need to maintain growth locations and therefore reduces microtubules’ importance. Future experimental and theoretical studies should examine how different cytoskeletal components adapted to achieve slightly different tasks.
Models of interphase cytoskeletal subcomponents: actin
Quantitative studies of the individual cytoskeletal subcomponents—microtubule bundles, actin cables, actin patches—explore links between molecular components and cell structure. Recent models describe these subcomponents.
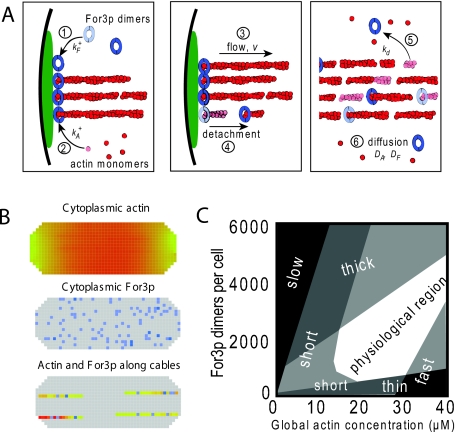
Fission-yeast formin For3p nucleates cables from cell tips. For3p molecules attach to cell tips, nucleate actin filaments, dislodge from cell tips, travel into the cell along actin cables, and travel back to the cell tip (Martin and Chang, 2006). Wang and Vavylonis (2008) modeled coupled For3p and actin cable turnover (see Fig. 3). In their model, a continuous pool of actin monomers and a discrete pool of For3p molecules diffuse through the cell. The formins bind to cortical sites at the cell tips. Once bound, formins aid local actin polymerization until dissociation. Essentially, this model augments the mechanism proposed by Martin and Chang with rate constants, diffusion coefficients, and quantified localization.
Figure 3. Model of actin cables in fission yeast (Wang and Vavylonis, 2008).
(A) Schematic showing the basic processes of the model. (B) 3D computational lattice model (http://athena.physics.lehigh.edu/research/actin_cable_applet.html) accounting for the small number of For3p which are treated as discrete units. (C) Qualitative dynamical phase diagram describing the morphology of the actin cable system as a function of actin and For3p concentration.
Comparisons of this actin cable model to experiment suggested a set of parameter values. Due to actin-For3p coupling, actin cable thickness, polymerization speed, and length vary across parameter space [see Fig. 3C]. The actin cable network must be both robust and highly adaptable—insensitive enough to be reliable and sensitive enough to be controllable. In the model, this corresponds to broad but finite regions of stable behavior. Experiments exploring parameter space, by varying concentrations and rate constants genetically and pharmacologically (Gao and Bretscher, 2008; Nolen et al., 2009; Rizvi et al., 2009), will test the parameter-space structure suggested by this model.
According to the model of Fig. 3, actin cables remove their own nucleator and undergo retrograde flow. Cable flow depletes the actin-monomer pool at the tips, causing a concentration gradient across the cell. More complex actin-For3p interactions could introduce nonlinearity and therefore, potentially, multiple stable tip states. This may lead to another possible mechanism for NETO (Wang and Vavylonis, 2008). In such a mechanism, unlike in the model of Csikász-Nagy and others, bistability may be due to removal, rather than recruitment, of actin nucleators.
Cables are not the only actin superstructure in the cell. Fission yeast contains actin patches that contribute to endocytosis near regions of membrane remodeling [Fig. 1A]. Recent experiments quantify the kinetics of assembly of coat proteins, Arp2∕3 complex, actin, and other cofactors in patches (Galletta et al., 2008; Kaksonen et al., 2005;Sirotkin et al., 2005), opening the door for modeling studies. Motivated by such data, Liu et al. (2006, 2009) twice modeled actin-patch mechanics and assembly kinetics in budding yeast. Their model includes coat-protein assembly at the site of endocytosis, pushing forces by local actin polymerization, and phase separation of lipids and membrane proteins at the vesicle bud’s border driven by membrane curvature. These processes’ combined effects generate tension around the bud neck, causing vesicle scission for endocytosis.
Models of interphase cytoskeletal subcomponents: microtubules
To maintain their shape, fission yeast cells must define their tips. Throughout growth, proteins follow microtubules and symmetrically mark both tips. Experimental and theoretical results suggest a simple mechanism is responsible for microtubule alignment.
According to recent experiments, normal cell shape and microtubule organization reinforce each other (Minc et al., 2009; Terenna et al., 2008). The rod shape directs elongating microtubules to the ends of the cell [see Figs. 1B, 4]. Oriented to the long axis of the cell, they provide a path to the cell tips for polarity-protein deposition. These proteins contribute to further preferential growth at the cell tips, exaggerating the linear shape that directs the microtubules. This interplay provides a feedback mechanism between cell shape and microtubule alignment. This alignment process also allows microtubules to center the nucleus (Daga et al., 2006; Tolić-Nørrelykke et al., 2005; Tran et al., 2001).
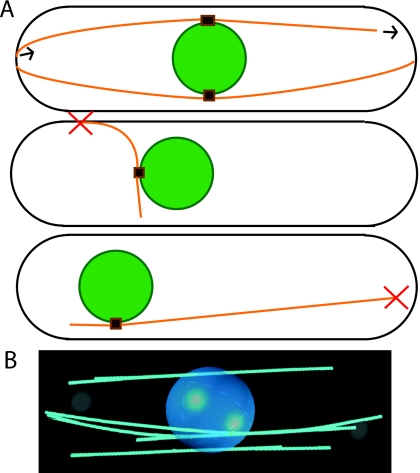
Figure 4. Illustration and results of the model of Foethke et al. (2009).
(A) Model schematic: microtubules orange, nucleus green, microtubule organizing centers (MTOCs) brown. Red X indicates likely catastrophe. The model includes four MTOCs each nucleating four microtubules. Top: normal alignment, polymerization, depolymerization. Middle: likely catastrophe due to force, preventing medially aligned microtubules. Bottom: likely catastrophe due to length, necessary to center nucleus. (B) Representative image at steady-state, using online content with default parameters (http://www.nature.com/msb).
Microtubules may self-organize by virtue of their confinement and dynamics alone but this is difficult to show experimentally: if one removes everything else conceivably affecting microtubule dynamics, what is left could not meaningfully be called a cell. Computer simulation helps here. A model extending in vitro findings shows it is possible, at least in principle, for observed organization to emerge from simple rules (Foethke et al., 2009). Indeed, simulations show a physical model can reproduce many measurable traits of interphase microtubule orientation. Foethke and others capture the essential features of microtubules in vivo, including their ability to center the nucleus, in a physical model using catastrophe rates dependent on both force and length (see Fig. 4). As a result, microtubules that happen to grow with an unfavorable orientation depolymerize. Microtubules push the nucleus opposite their direction of growth. Since shorter microtubules are more stable, more force comes from the closer tip and the microtubule system centers the nucleus, as suggested by an earlier computational model (Tran et al., 2001). These results extend easily to microtubule dynamics in differently shaped cells, such as those in recent experiments where they are confined (Minc et al., 2009; Terenna et al., 2008).
In the model mentioned above, Foethke and others assume that microtubules overlap and form stable antiparallel bundles near the nucleus. A similar model describes how they self-organize—with nucleators, crosslinkers, and motor proteins—into such bundles (Janson et al., 2007). Microtubules slide across each other but grind to a halt and form steady-state bundles when motor proteins cannot overcome friction due to crosslinkers.
DIVIDING THE CELL
The simple growth pattern of fission yeast suggests a robust cytoskeletal system. It also rapidly adapts for division. Cells dramatically disassemble both microtubule and actin interphase cytoskeletal systems during mitosis and each performs a major task in cell division. Microtubules mediate nuclear division, actin filaments mediate cytoplasmic division.
To finish division, an equatorial ring contracts and separates the daughter cells. This ring, the contractile ring [Fig. 6A], is a narrow bundle composed of actin filaments, myosin motors, and other proteins. A broad band of cortical nodes, made up of myosin and other proteins, tightens and becomes the contractile ring, apparently by self-organization (Pollard and Wu, 2010).
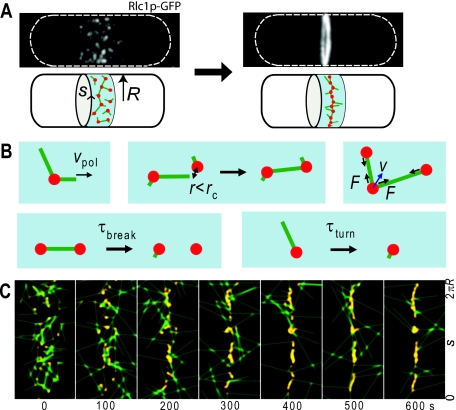
Figure 6. Search, capture, pull, and release model of contractile ring assembly.
(A) Contraction of a broad band of myosin nodes into a narrow ring in fission yeast. (B) Processes in the model. (C) Simulated images of condensing broad band of nodes as a function of time (seconds). The y-axis in each simulated image is the arc length around the cylindrical body of a cell of radius R. The x-axis shows 4.5 μm along the long axis of the cell. Nodes are shown in red, actin filaments in green. Using parameter values measured in experiments, nodes formed an equatorial ring within a time consistent with experiment
Assembly of the contractile ring
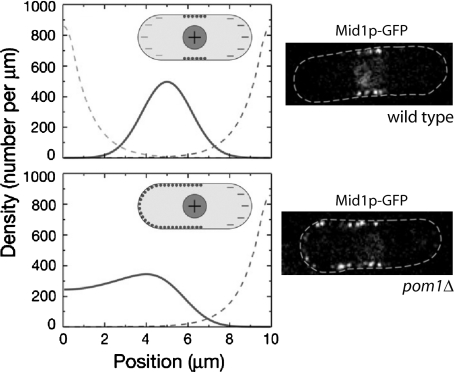
Successful contractile ring assembly depends on the initial node distribution. One recent experimental and theoretical study provides quantitative insight into the positioning of node scaffolding component Mid1p (Padte et al., 2006). In their one-dimensional reaction-diffusion model (Fig. 5), an active form of Mid1p associates with the inner plasma membrane and the inactive form cannot. Activation occurs in the nucleus, deactivation occurs in the cytoplasm. Padte and others report that localized activation of Mid1p by the nucleus is insufficient to position Mid1p within a band in the center of the cell—active Mid1p diffuses across the cell before binding to the cortex. To match observed behavior, they introduce deactivators at the cells tips. This leads to a sharper Mid1p profile centered in the middle. Additionally, they identify some of the inhibitory interactions involving Pom1p. Some inhibitors may remain unidentified—cells lacking Pom1p still exclude Mid1p from one cell tip.
Figure 5. Results of model of positioning of Mid1p in nodes in the middle of the cell and experimental images (Padte et al., 2006).
The nodes are precursor components of the contractile ring. The model involves activation of Mid1p in the nucleus, deactivation near cell tips (dashed line), and binding of active Mid1p to the membrane. Top graph shows simulated distribution of membrane-bound Mid1p (solid line) as a function of position along the cell. Bottom graph shows membrane-bound Mid1p in cells lacking one of the polar inhibitors, Pom1p. Micrographs show the corresponding experimental images. Images reproduced with permission.
At least two recent experimental studies reveal further coupling of node distribution to the cell cycle (Martin and Berthelot-Grosjean, 2009; Moseley et al., 2009). Both studies argue that physical elongation causes Pom1p depletion near the cell’s equator. Cdr2p signaling, inhibited by Pom1p in short cells, triggers cell-cycle progression in long cells. The proposed mechanism provides a physical sensor within a signaling pathway. These studies further illustrate that cytoskeleton assembly couples with cytoplasmic gradients, local activation, and deactivation mechanisms to regulate internal organization in fission yeast.
Cortical nodes attach firmly to the membrane, effectively restricting their movement to two dimensions. On this surface, the nodes condense, becoming a ring. Myosin motors in the nodes exert the force responsible for this condensation. The motors act on a dynamic meshwork of actin filaments nucleated by formin Cdc12p, another node protein (Coffman et al., 2009). Once actin filaments polymerize within the medial band, nodes move in bursts at velocities of ∼30 nm∕s, starting, stopping, and changing direction. They condense into the contractile ring in 10 min (Vavylonis et al., 2008).
On the basis of these observations, Vavylonis et al. (2008) proposed a stochastic mechanism for self-assembly. According to their model, nodes search, capture, pull, and release (see Fig. 6). Nodes search the cortical surface by nucleating actin filaments, which elongate in random directions along the cortex. Nodes capture each other by binding to filaments with myosin-II. Nodes pull each other because, once bound, myosin-II exerts a tensile force and reduces separation. Finally, nodes release as myosin-II dissociates and actin filaments disassemble. The Monte Carlo simulations of this model—search, capture, pull, and release—reproduce the start-stop motion of nodes and generate contractile rings within a range of parameter values consistent with experiments. Varying parameter values lead to disconnected clumps, consistent with observations of Cdc12p-defective cells (Hachet and Simanis, 2008). A recent theoretical work further quantifies requirements for clump formation (Ojkic and Vavylonis, submitted).
This mechanism further illustrates how random growth of filaments can establish transient connections between distant parts within the cell. The success of this unassisted random search and capture process suggests a reason for inherent randomness—it may confer resistance to rupture or relaxation during deformation, endowing assembly with a measure of robustness.
According to another proposed mechanism, actin filaments for the contractile ring may originate from a single spot as a leading actin cable (Bathe and Chang, 2010; Mishra and Oliferenko, 2008; Roberts-Galbraith and Gould, 2008). No quantitative models yet describe this mechanism.
Several studies abstract contractile ring formation, postulating a mechanism shared among several organisms. One hypothesis couples myosin-induced cortical filament flow to filament alignment (Salbreux et al., 2009). According to this hydrodynamic approach, an active gel of filaments self-organizes in response to an inhomogeneous myosin distribution. In an earlier work, another group also employs the continuum modeling to explain ring formation with active gel theory (Zumdieck et al., 2005). This differs from the model of Salbreux and others because a ring is one among many patterns—such as a state with four rings and another with two oscillating rings—emergent from instability of a homogeneous state rather than reactive to myosin inhomogeneities.
In addition to cytoskeletal effects, membrane modifications assist motor proteins during ring formation and contraction. The cell’s repertoire of membrane-associated proteins sense and affect curvature, anchor proteins to the membrane, and mark membrane regions (Frost et al., 2009). In particular, fission yeast Cdc15p contains BAR domains, making it sensitive to local curvature (Aspenström et al., 2006). Nodes contain this protein, which may link them to sterol-rich domains in the middle of the cell (Takeda et al., 2004). One group supposes that interplay between contraction and curvature may cause ring formation in many cell types (Shlomovitz and Gov, 2008). According to their model, a contractile network develops a ring due to instability of uniformly distributed membrane-bound protein clusters, such as of Cdc15p.
The general models discussed above have the advantage of providing physical insight and offer qualitative frameworks for describing ring formation. Quantifying physical properties—measuring rate constants, flow velocities, interaction strengths—may eliminate or support these models. At present, they depend on many unmeasured quantities and describe ring formation phenomenologically.
Modeling ring constriction
Following formation, the contractile ring constricts around the cell’s equator and a septum forms. Several models describe actin filaments and myosin motors in contractile steady-states (Carlsson, 2006; Kruse and Jülicher, 2003;Larripa and Mogilner, 2006; Liverpool and Marchetti, 2006). But contractile rings exchange proteins with the cytoplasm during constriction. One model describes ring constriction in nematode embryos (Zumdieck et al., 2007) and, according to the authors, may apply to fission yeast. Their model includes dynamic exchange between ring and cytoplasm. This description identifies possible sources of the contractile stress needed to cleave the cell, including depolymerization of filaments cross-linked by end-tracking motor proteins. This model may inform a future model specific to fission yeast but details—magnitudes of forces, identities of proteins, inclusion of nodes—will necessarily differ.
Modeling the mitotic spindle
As the ring constricts, the spindle apparatus ensures both compartments contain the proper genetic material. One study describes how microtubule cross-linkers and sliding motors regulate spindle elongation during mitosis (Fu et al., 2009). They present a computational model to describe their experimental results. In the model, microtubule bundling protein Ase1p, after dephosphorylation, recruits dephosphorylated kinesin motors to the spindle. These motors control the overlap distance of antiparallel microtubules and, accordingly, the spindle length. This mechanism ties spindle length to the cell cycle.
Most modeling studies of the spindle address other organisms. Reviewing efforts to understand mitosis, Mogilner et al. (2006a) stress the role of modeling in understanding how dividing cells form, maintain, and position their mitotic spindle. And they describe the progress, highlighting models that led to experiments that further led to refined models.
One such loop starts with a theoretical study of the budding yeast spindle during metaphase (Sprague et al., 2003). The authors model how microtubules emanating from the spindle pole body position kinetochores—the protein complex that attaches them to chromosomes. Their models generate images of simulated kinetochores and spindle pole bodies, which they compare statistically to corresponding images of live budding yeast. This rules out several models. Their models, which included dynamic instability of microtubules, failed to match their data with rescue and catastrophe frequencies independent of distance from the spindle. With this dependence included, simulated microtubules behaved as observed. This model did not include effects of kinetochore tension on microtubule dynamics. But more data emerged (Pearson et al., 2004). This led Gardner and others to refine the earlier model and conclude that a model with tension-dependent regulation better fits experimental data (Gardner et al., 2005). Continuing this line of inquiry, Gardner et al. (2008) modeled how kinesin motor proteins may allow cells to raise the catastrophe rate of longer microtubules. Another group employed image analysis to quantify the relationship between microtubules and kinetochores in budding yeast (Dorn et al., 2005).
The models reviewed by Mogilner et al. (2006a) consider the spindle apparatus as it functions in many organisms. But animal cells, budding yeast, and fission yeast solve this problem differently. The solution evolution found for fission yeast may differ due to cell geometry, chromosome count, and the closed nature of mitosis, among other factors. Efficient organelle positioning by microtubules depends on cell shape (Tolić-Nørrelykke, 2008). Pushing forces predominate in short and symmetric cell interiors such as the fission-yeast nucleus. Pulling forces are more complex but more capable in asymmetric or large interiors. Future quantitative models may support this distinction.
Meiosis
The fission-yeast spindle pole body oscillates during meiosis. Selecting this system since oscillations often signal collective behavior, Vogel et al. (2009) propose a minimal model to capture the cause of spindle pole body oscillations. They focus on the collective behavior of dynein motor proteins. According to their study, dynein may self-organize and pull dynamic microtubules toward the tips of the cell. In the model, a large-scale behavior, oscillations across the cell, arises from simple interaction at the molecular level.
CONCLUSIONS AND OUTLOOK
Healthy fission yeast depends on many systems and mechanisms. Modeling them individually provides progress toward a piecemeal mathematical description of the whole, to be refined according to experimental findings. Although the current description may seem more patchwork than quilt, recent models exhibit some overlap. Models of polarized growth assume cells define their tips and models of microtubule organization explain how cells could define their tips. Accumulating accurate quantitative models of cytoskeletal components reveals new questions about how they fit together and the extent to which this modular approach can provide an integrated description of a living cell.
To apply these models to more complex organisms, they will almost certainly require modifications. But the need for modifications will inform us. Cytoskeletal elements and regulators are often strongly conserved. In some cases, asking why the models must be adjusted despite this may lead to insight about the organism—may reveal why the organism required the adjusted mechanism. To confidently address these complex questions, we must first demonstrate the accuracy and predictive power of modeling simpler biological systems. The studies described in the review provide first steps toward this demonstration. Fission yeast provides, at least, a testing ground for modeling methods—a foundation for modeling more complex organisms.
ACKNOWLEDGMENTS
TD was supported as a GAANN fellow through a grant from the U.S. Department of Education to the Department of Physics of Lehigh University. DV was supported by NIH Grant No. R21GM083928 and by the Human Frontiers Science Program Grant No. RGP0061∕2009-C.
References
- Altschuler, S J, Angenent, S B, Wang, Y, and Wu, L F (2008). “On the spontaneous emergence of cell polarity.” Nature (London) 454, 886–889. 10.1038/nature07119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspenström, P, Fransson, Å, and Richnau, N (2006). “Pombe cdc15 homology proteins: regulators of membrane dynamics and the actin cytoskeleton.” Trends Biochem. Sci. 31, 670–679. 10.1016/j.tibs.2006.10.001 [DOI] [PubMed] [Google Scholar]
- Bathe, M, and Chang, F (2010). “Cytokinesis and the contractile ring in fission yeast: towards a systems-level understanding.” Trends Microbiol. 18, 38–45. 10.1016/j.tim.2009.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgärtner, S, and Tolić-Nørrelykke, I M (2009). “Growth pattern of single fission yeast cells is bilinear and depends on temperature and dna synthesis.” Biophys. J. 96, 4336–4347. 10.1016/j.bpj.2009.02.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson, A E (2006). “Contractile stress generation by actomyosin gels.” Phys. Rev. E 74, 051912. 10.1103/PhysRevE.74.051912 [DOI] [PubMed] [Google Scholar]
- Chang, F, and Martin, S G (2009). “Shaping fission yeast with microtubules.” Cold Spring Harbor Perspect. Biol. 1, a001347. 10.1101/cshperspect.a001347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman, V C, Nile, A H, Lee, I J, Liu, H, and Wu, J Q (2009). “Roles of formin nodes and myosin motor activity in Mid1p-dependent contractile-ring assembly during fission yeast cytokinesis.” Mol. Biol. Cell 20, 5195–5210. 10.1091/mbc.E09-05-0428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csikász-Nagy, A, Gyorffy, B, Alt, W, Tyson, J, and Novák, B (2008). “Spatial controls for growth zone formation during the fission yeast cell cycle.” Yeast 25, 59–69. 10.1002/yea.1571 [DOI] [PubMed] [Google Scholar]
- Daga, R R, Yonetani, A, and Chang, F (2006). “Asymmetric microtubule pushing forces in nuclear centering.” Curr. Biol. 16, 1544–1550. 10.1016/j.cub.2006.06.026 [DOI] [PubMed] [Google Scholar]
- Dorn, J F, Jaqaman, K, Rines, D R, Jelson, G S, Sorger, P K, and Danuser, G (2005). “Yeast kinetochore microtubule dynamics analyzed by high-resolution three-dimensional microscopy.” Biophys. J. 89, 2835–2854. 10.1529/biophysj.104.058461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foethke, D, Makushok, T, Brunner, D, and Nédélec, F (2009). “Force- and length-dependent catastrophe activities explain interphase microtubule organization in fission yeast.” Mol. Syst. Biol. 5, 241. 10.1038/msb.2008.76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost, A, Unger, V M, and De Camilli, P (2009). “The bar domain superfamily: membrane-molding macromolecules.” Cell 137, 191–196. 10.1016/j.cell.2009.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, C, Ward, J J, Loïodice, I, Velve-Casquillas, G, Nédélec, F J, and Tran, P T (2009). “Phospho-regulated interaction between kinesin-6 klp9p and microtubule bundler ase1p promotes spindle elongation.” Dev. Cell 17, 257–267. 10.1016/j.devcel.2009.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletta, B J, Chuang, D Y, and Cooper, J A (2008). “Distinct roles for Arp2∕3 regulators in actin assembly and endocytosis.” PLoS Biol. 6, e1. 10.1371/journal.pbio.0060001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, L, and Bretscher, A (2008). “Analysis of unregulated formin activity reveals how yeast can balance f-actin assembly between different microfilament-based organizations.” Mol. Biol. Cell 19, 1474–1484. 10.1091/mbc.E07-05-0520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner, M K et al. (2008). “Chromosome congression by kinesin-5 motor-mediated disassembly of longer kinetochore microtubules.” Cell 135, 894–906. 10.1016/j.cell.2008.09.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner, M K, Pearson, C G, Sprague, B L, Zarzar, T R, Bloom, K, Salmon, E D, and Odde, D J (2005). “Tension-dependent regulation of microtubule dynamics at kinetochores can explain metaphase congression in yeast.” Mol. Biol. Cell 16, 3764–3775. 10.1091/mbc.E05-04-0275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goryachev, A B, and Pokhilko, A V (2008). “Dynamics of Cdc42 network embodies a Turing-type mechanism of yeast cell polarity.” FEBS Lett. 582, 1437–1443. 10.1016/j.febslet.2008.03.029 [DOI] [PubMed] [Google Scholar]
- Hachet, O, and Simanis, V (2008). “Mid1p∕anillin and the septation initiation network orchestrate contractile ring assembly for cytokinesis.” Genes Dev. 22, 3205–3216. 10.1101/gad.1697208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins, R J, Bénichou, O, Piel, M, and Voituriez, R (2009). “Rebuilding cytoskeleton roads: active-transport-induced polarization of cells.” Phys. Rev. E 80, 040903. 10.1103/PhysRevE.80.040903 [DOI] [PubMed] [Google Scholar]
- Hayles, J, and Nurse, P (2001). “A journey into space.” Nat. Rev. Mol. Cell Biol. 2, 647–656. 10.1038/35089520 [DOI] [PubMed] [Google Scholar]
- Howell, A S, Savage, N S, Johnson, S A, Bose, I, Wagner, A W, Zyla, T R, Nijhout, H F, Reed, M C, Goryachev, A B, and Lew, D J (2009). “Singularity in polarization: rewiring yeast cells to make two buds.” Cell 139, 731–743. 10.1016/j.cell.2009.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janson, M E, Loughlin, R, Loïodice, I, Fu, C, Brunner, D, Nédélec, F J, and Tran, P T (2007). “Crosslinkers and motors organize dynamic microtubules to form stable bipolar arrays in fission yeast.” Cell 128, 357–368. 10.1016/j.cell.2006.12.030 [DOI] [PubMed] [Google Scholar]
- Kaksonen, M, Toret, C P, and Drubin, D G (2005). “A modular design for the clathrin- and actin-mediated endocytosis machinery.” Cell 123, 305–320. 10.1016/j.cell.2005.09.024 [DOI] [PubMed] [Google Scholar]
- Kruse, K, and Jülicher, F (2003). “Self-organization and mechanical properties of active filament bundles.” Phys. Rev. E 67, 051913. 10.1103/PhysRevE.67.051913 [DOI] [PubMed] [Google Scholar]
- La Carbona, S L, Goff, C L, and Goff, X L (2006). “Fission yeast cytoskeletons and cell polarity factors: connecting at the cortex.” Biol. Cell 98, 619–631. 10.1042/BC20060048 [DOI] [PubMed] [Google Scholar]
- Larripa, K, and Mogilner, A (2006). “Transport of a 1d viscoelastic actin-myosin strip of gel as a model of a crawling cell.” Physica A 372, 113–123. 10.1016/j.physa.2006.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J, Kaksonen, M, Drubin, D G, and Oster, G (2006). “Endocytic vesicle scission by lipid phase boundary forces.” Proc. Natl. Acad. Sci. U.S.A. 103, 10277–10282. 10.1073/pnas.0601045103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J, Sun, Y, Drubin, D G, and Oster, G F (2009). “The mechanochemistry of endocytosis.” PLoS Biol. 7, e1000204. 10.1371/journal.pbio.1000204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liverpool, T B, and Marchetti, M C (2006). “Rheology of active filament solutions.” Phys. Rev. Lett. 97, 268101. 10.1103/PhysRevLett.97.268101 [DOI] [PubMed] [Google Scholar]
- Marco, E, Wedlich-Soldner, R, Li, R, Altschuler, S J, and Wu, L F (2007). “Endocytosis optimizes the dynamic localization of membrane proteins that regulate cortical polarity.” Cell 129, 411–422. 10.1016/j.cell.2007.02.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks, J, Hagan, I, and Hyams, J (1986). “Growth polarity and cytokinesis in fission yeast: the role of the cytoskeleton.” J. Cell Sci. Suppl. 5, 229–241. [DOI] [PubMed] [Google Scholar]
- Martin, S G (2009). “Microtubule-dependent cell morphogenesis in the fission yeast.” Trends Cell Biol. 19, 447–454. 10.1016/j.tcb.2009.06.003 [DOI] [PubMed] [Google Scholar]
- Martin, S G, and Berthelot-Grosjean, M (2009). “Polar gradients of the dyrk-family kinase pom1 couple cell length with the cell cycle.” Nature (London) 459, 852–856. 10.1038/nature08054 [DOI] [PubMed] [Google Scholar]
- Martin, S G, and Chang, F (2005). “New end take off: regulating cell polarity during the fission yeast cell cycle.” Cell Cycle 4, 1046–1049. [PubMed] [Google Scholar]
- Martin, S G, and Chang, F (2006). “Dynamics of the formin for3p in actin cable assembly.” Curr. Biol. 16, 1161–1170. 10.1016/j.cub.2006.04.040 [DOI] [PubMed] [Google Scholar]
- Martin, S G, Rincón, S A, Basu, R, Pérez, P, and Chang, F (2007). “Regulation of the formin for3p by cdc42p and bud6p.” Mol. Biol. Cell 18, 4155–4167. 10.1091/mbc.E07-02-0094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata, J, and Nurse, P (1997). “Tea1 and the microtubular cytoskeleton are important for generating global spatial order within the fission yeast cells.” Cell 89, 939–949. 10.1016/S0092-8674(00)80279-2 [DOI] [PubMed] [Google Scholar]
- Meinhardt, H, and Gierer, A (2000). “Pattern formation by local self-activation and lateral inhibition.” BioEssays 22, 753–760. [DOI] [PubMed] [Google Scholar]
- Minc, N, Bratman, S V, Basu, R, and Chang, F (2009). “Establishing new sites of polarization by microtubules.” Curr. Biol. 19, 83–94. 10.1016/j.cub.2008.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra, M, and Oliferenko, S (2008). “Cytokinesis: catch and drag.” Curr. Biol. 18, R247–R250. 10.1016/j.cub.2008.01.029 [DOI] [PubMed] [Google Scholar]
- Mitchison, J M, and Nurse, P (1985). “Growth in cell length in the fission yeast Schizosaccharomyces pombe.” J. Cell Sci. 75, 357–376. [DOI] [PubMed] [Google Scholar]
- Mogilner, A, Wollman, R, Civelekoglu-Scholey, G, and Scholey, J (2006a). “Modeling mitosis.” Trends Cell Biol. 16, 88–96. 10.1016/j.tcb.2005.12.007 [DOI] [PubMed] [Google Scholar]
- Mogilner, A, Wollman, R, and Marshall, W F (2006b). “Quantitative modeling in cell biology: what is it good for?” Dev. Cell 11, 279–287. 10.1016/j.devcel.2006.08.004 [DOI] [PubMed] [Google Scholar]
- Moseley, J B, and Goode, B L (2006). “The yeast actin cytoskeleton: from cellular function to biochemical mechanism.” Microbiol. Mol. Biol. Rev. 70, 605–645. 10.1128/MMBR.00013-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley, J B, Mayeux, A, Paoletti, A, and Nurse, P (2009). “A spatial gradient coordinates cell size and mitotic entry in fission yeast.” Nature (London) 459, 857–860. 10.1038/nature08074 [DOI] [PubMed] [Google Scholar]
- Nolen, B J, Tomasevic, N, Russell, A, Pierce, D W, Jia, Z, McCormick, C D, Hartman, J, Sakowicz, R, and Pollard, T D (2009). “Characterization of two classes of small molecule inhibitors of Arp2∕3 complex.” Nature (London) 460, 1031–1034. 10.1038/nature08231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onsum, M D, and Rao, C V (2009). “Calling heads from tails: the role of mathematical modeling in understanding cell polarization.” Curr. Opin. Cell Biol. 21, 74–81. 10.1016/j.ceb.2009.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozbudak, E M, Becskei, A, and van Oudenaarden, A (2005). “A system of counteracting feedback loops regulates Cdc42p activity during spontaneous cell polarization.” Dev. Cell 9, 565–571. 10.1016/j.devcel.2005.08.014 [DOI] [PubMed] [Google Scholar]
- Padte, N N, Martin, S G, Howard, M, and Chang, F (2006). “The cell-end factor pom1p inhibits mid1p in specification of the cell division plane in fission yeast.” Curr. Biol. 16, 2480–2487. 10.1016/j.cub.2006.11.024 [DOI] [PubMed] [Google Scholar]
- Pearson, C G, Yeh, E, Gardner, M, Odde, D, Salmon, E D, and Bloom, K (2004). “Stable kinetochore-microtubule attachment constrains centromere positioning in metaphase.” Curr. Biol. 14, 1962–1967. 10.1016/j.cub.2004.09.086 [DOI] [PubMed] [Google Scholar]
- Piel, M, and Tran, P T (2009). “Cell shape and cell division in fission yeast.” Curr. Biol. 19, R823–R827. 10.1016/j.cub.2009.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard, T D, and Cooper, J A (2009). “Actin, a central player in cell shape and movement.” Science 326, 1208–1212. 10.1126/science.1175862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard, T D, and Wu, J Q (2010). “Understanding cytokinesis: lessons from fission yeast.” Nat. Rev. Mol. Cell Biol. 11, 149–155. 10.1038/nrm2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riveline, D (2009). “Explaining lengths and shapes of yeast by scaling arguments.” PLoS ONE 4, e6205. 10.1371/journal.pone.0006205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi, S A, Neidt, E M, Cui, J, Feiger, Z, Skau, C T, Gardel, M L, Kozmin, S A, and Kovar, D R (2009). “Identification and characterization of a small molecule inhibitor of formin-mediated actin assembly.” Chem. Biol. 16, 1158–1168. 10.1016/j.chembiol.2009.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts-Galbraith, R H, and Gould, K L (2008). “Stepping into the ring: the SIN takes on contractile ring assembly.” Genes Dev. 22, 3082–3088. 10.1101/gad.1748908 [DOI] [PubMed] [Google Scholar]
- Rupes, I, Jia, Z, and Young, P G (1999). “Ssp1 promotes actin depolymerization and is involved in stress response and new end take-off control in fission yeast.” Mol. Biol. Cell 10, 1495–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salbreux, G, Prost, J, and Joanny, J F (2009). “Hydrodynamics of cellular cortical flows and the formation of contractile rings.” Phys. Rev. Lett. 103, 058102. 10.1103/PhysRevLett.103.058102 [DOI] [PubMed] [Google Scholar]
- Sawin, K E, and Tran, P T (2006). “Cytoplasmic microtubule organization in fission yeast.” Yeast 23, 1001–1014. 10.1002/yea.1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomovitz, R, and Gov, N S (2008). “Physical model of contractile ring initiation in dividing cells.” Biophys. J. 94, 1155–1168. 10.1529/biophysj.107.111351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirotkin, V, Beltzner, C C, Marchand, J B, and Pollard, T D (2005). “Interactions of wasp, myosin-i, and verprolin with Arp2∕3 complex during actin patch assembly in fission yeast.” J. Cell Biol. 170, 637–648. 10.1083/jcb.200502053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaughter, B D, Das, A, Schwartz, J W, Rubinstein, B, and Li, R (2009). “Dual modes of cdc42 recycling fine-tune polarized morphogenesis.” Dev. Cell 17, 823–835. 10.1016/j.devcel.2009.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague, B L, Pearson, C G, Maddox, P S, Bloom, K S, Salmon, E, and Odde, D J (2003). “Mechanisms of microtubule-based kinetochore positioning in the yeast metaphase spindle.” Biophys. J. 84, 3529–3546. 10.1016/S0006-3495(03)75087-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda, T, Kawate, T, and Chang, F (2004). “Organization of a sterol-rich membrane domain by cdc15p during cytokinesis in fission yeast.” Nat. Cell Biol. 6, 1142–1144. 10.1038/ncb1189 [DOI] [PubMed] [Google Scholar]
- Terenna, C R, Makushok, T, Velve-Casquillas, G, Baigl, D, Chen, Y, Bornens, M, Paoletti, A, Piel, M, and Tran, P T (2008). “Physical mechanisms redirecting cell polarity and cell shape in fission yeast.” Curr. Biol. 18, 1748–1753. 10.1016/j.cub.2008.09.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolić-Nørrelykke, I M (2008). “Push-me-pull-you: how microtubules organize the cell interior.” Eur. Biophys. J. 37, 1271–8. 10.1007/s00249-008-0321-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolić-Nørrelykke, I M (2010). “Force and length regulation in the microtubule cytoskeleton: lessons from fission yeast.” Curr. Opin. Cell Biol. 22, 21–28. 10.1016/j.ceb.2009.12.011 [DOI] [PubMed] [Google Scholar]
- Tolić-Nørrelykke, I M, Sacconi, L, Stringari, C, Raabe, I, and Pavone, F S (2005). “Nuclear and division-plane positioning revealed by optical micromanipulation.” Curr. Biol. 15, 1212–1216. 10.1016/j.cub.2005.05.052 [DOI] [PubMed] [Google Scholar]
- Tran, P, Marsh, L, Doye, V, Inoue, S, and Chang, F (2001). “A mechanism for nuclear positioning in fission yeast based on microtubule pushing.” J. Cell Biol. 153, 397–412. 10.1083/jcb.153.2.397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turing, A M (1952). “The chemical basis of morphogenesis.” Philos. Trans. R. Soc. London, Ser. B 237, 37–72. 10.1098/rstb.1952.0012 [DOI] [Google Scholar]
- Vavylonis, D, Wu, J Q, Hao, S, O’Shaughnessy, B, and Pollard, T D (2008). “Assembly mechanism of the contractile ring for cytokinesis by fission yeast.” Science 319, 97–100. 10.1126/science.1151086 [DOI] [PubMed] [Google Scholar]
- Vogel, S K, Pavin, N, Maghelli, N, Jülicher, F, and Tolić-Nørrelykke, I M (2009). “Self-organization of dynein motors generates meiotic nuclear oscillations.” PLoS Biol. 7, e1000087. 10.1371/journal.pbio.1000087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H, and Vavylonis, D (2008). “Model of for3p-mediated actin cable assembly in fission yeast.” PLoS ONE 3, e4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumdieck, A, Kruse, K, Bringmann, H, Hyman, A A, and Jülicher, F (2007). “Stress generation and filament turnover during actin ring constriction.” PLoS ONE 2, e696. 10.1371/journal.pone.0000696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumdieck, A, Lagomarsino, M C, Tanase, C, Kruse, K, Mulder, B, Dogterom, M, and Jülicher, F (2005). “Continuum description of the cytoskeleton: ring formation in the cell cortex.” Phys. Rev. Lett. 95, 258103. 10.1103/PhysRevLett.95.258103 [DOI] [PubMed] [Google Scholar]