Abstract
Pigment epithelium-derived factor (PEDF), a potent blocker of angiogenesis in vivo, and of endothelial cell migration and tubule formation, binds with high affinity to a yet unknown protein on the surface of endothelial cells. Given that protein fingerprinting suggested a match of a ~60-kDa PEDF-binding protein in bovine retina to Bos taurus F1-ATP synthase β-subunit, and that F1F0-ATP synthase components have been identified recently as cell-surface receptors, we examined the direct binding of PEDF to F1. Size-exclusion ultrafiltration assays showed that recombinant human PEDF formed a complex with recombinant yeast F1. Real-time binding by surface plasmon resonance demonstrated that yeast F1 interacted specifically and reversibly with human PEDF. Kinetic evaluations revealed high binding affinity for PEDF, in agreement with PEDF affinities for endothelial cell-surfaces. PEDF blocked interactions between F1 and angiostatin, another antiangiogenic factor, suggesting overlapping PEDF- and angiostatin-binding sites on F1. Surfaces of endothelial cells exhibited affinity for PEDF-binding proteins of ~60-kDa. Antibodies to F1 β-subunit specifically captured PEDF-binding components in endothelial plasma membranes. Extracellular ATP synthesis activity of endothelial cells was examined in the presence of PEDF. PEDF significantly inhibited the extracellular ATP produced by endothelial cells, in agreement with direct interactions between cell-surface ATP synthase and PEDF. In addition to demonstrating that PEDF binds to cell-surface F1, these results show that PEDF is a ligand for endothelial cell-surface F1F0-ATP synthase. They suggest that PEDF-mediated inhibition of ATP synthase may be part of the biochemical mechanisms by which PEDF exerts its antiangiogenic activity.
Keywords: PEDF, F1-ATP synthase, protein-protein interactions, endothelial cells, ligand, surface plasmon resonance
Introduction
Pathologic vessel growth in the posterior segment of the eye can perturb the structure and morphology of the retina, and lead to visual loss. If this angiogenesis is prevented, retina degeneration is dramatically restricted. Therefore, endogenous angiogenic inhibitors are likely to play an important role on ocular neovascularization development. Pigment epithelium-derived factor (PEDF) is a potent antiangiogenic, neurotrophic and antitumorigenic factor [1–5]. It is an extracellular protein present in the interphotoreceptor matrix and vitreous [6, 7], believed to be responsible for the avascularity of these compartments under physiologic conditions. Moreover, the concentrations of PEDF in the eye are inversely correlated to ocular angiogenic development, and overexpression of PEDF or local PEDF protein delivery prevents ocular neovascularization, tumorigenesis and delays retinal cell death in vivo [4, 8–18]. PEDF induces endothelial cell apoptosis, inhibits the proliferation and migration of endothelial cells; and blocks formation of endothelial capillary-like networks and vessel sprouting ex vivo from chick aortic rings [1, 18, 19]. However, little is known about the molecular mechanisms by which PEDF functions to regulate endothelial cell behavior.
PEDF is a member of the serpin superfamily by structural homology, but does not have inhibitory activity vs. serine proteases [20]. Its biological activities are associated with receptor interactions at cell-surface interfaces and changes in protein expression. There is evidence for high affinity PEDF-binding sites and proteins in retinoblastoma, normal retina cells, cerebellar granule cell neurons and motor neurons, as well as in endothelial HUVEC cells [21–24]. We have recently identified a ~85-kDa PEDF-binding protein in the retina that is a phospholipase-linked membrane protein termed PEDF-R [25]. PEDF has high affinity for this protein and stimulates its phospholipase A enzymatic activity.
It is unclear if the only receptor for PEDF is PEDF-R. Studies on PEDF binding partners have also revealed a ~60-kDa PEDF-binding protein in membrane extracts from bovine retina tissues and retinoblastoma Y-79 tumor cells [21, 22] of a yet unknown identity. Preliminary investigations by peptide fingerprinting suggested a match of the bovine retina protein to Bos taurus F1-ATP synthase β-subunit (Table S1). Until recently, F1F0-ATP synthase expression was assumed to be strictly confined to mitochondria, where it generates most of the cellular ATP. Current evidence for an extra-mitochondrial expression of its components is derived from immunofluorescence, biochemistry and proteomics studies [26, 27]. F1F0-ATP synthase components have been identified as cell-surface receptors for apparently unrelated ligands during studies performed on angiogenesis, lipoprotein metabolism, innate immunity, hypertension, or regulation of food intake. One of these ligands is angiostatin which also inhibits ocular angiogenesis [28] and is antitumorigenic [29] like PEDF. It has been reported that angiostatin binds and inhibits the F1 catalytic domain of H+-ATP synthase on human umbilical vein endothelial cells (HUVECs) cell-surfaces leading to inhibition of migration and proliferation of endothelial cells [30–32]. HUVEC cells possess high ATP synthesis activity on the cell surface [33]. Extracellular ATP generation by HUVECs can be detected within 5 s after addition of ADP and inorganic phosphate, which is inhibited by mitochondrial H+-ATP synthase inhibitors (e.g., efrapeptins, resveratrol, and piceatannol) targeting the F1 catalytic domain [33]. Furthermore, these F1-targeting H+-ATP synthase inhibitors can block tube formation and proliferation of HUVECs without affecting intracellular ATP levels [33, 34]. These observations agree with the idea that the mechanisms of blocking angiogenesis might implicate binding and inhibition of the endothelial cell-surface F1F0-ATP synthase.
In the current study, we examined the potential interactions between PEDF and ATP synthase. We used highly purified recombinant yeast F1-ATPase and recombinant human PEDF in size-exclusion ultrafiltration assays and surface plasmon resonance (SPR) spectroscopy. We also assessed the binding of PEDF to endothelial cell-surface ATP synthase and examined the effect of PEDF on the extracellular ATP synthesis activity of human microvascular endothelial cells (HMVECs) and bovine retinal endothelial cells (BRECs). Our results provide evidence for high affinity interactions between PEDF and F1, as well as for a PEDF-mediated inhibition of extracellular ATP synthesis activity in endothelial cells. We discuss how these interactions provide insights into the mechanisms of action for angiogenesis inhibition.
Results
Direct binding of PEDF and F1
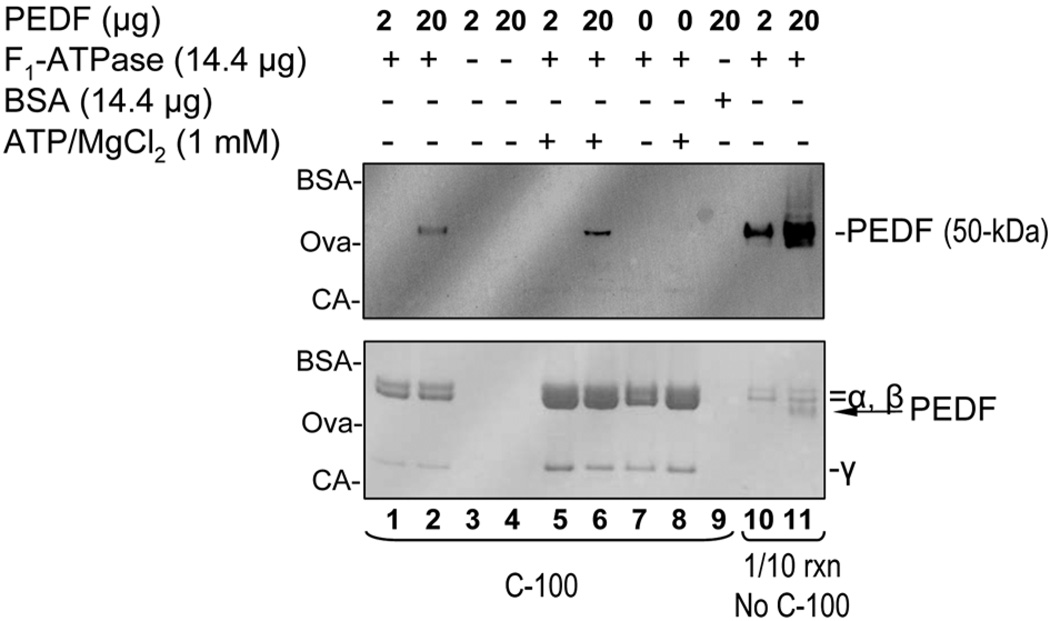
To investigate the potential interactions between PEDF and ATP synthase, mixtures of highly purified recombinant yeast F1-ATPase (~360-kDa) and human PEDF (50-kDa) were assayed first by complex formation. Solutions containing F1 (14.4 µg) and PEDF (2 or 20 µg) were mixed and incubated at room temperature for 1 hour before subjecting the mixtures to size-exclusion ultrafiltration through membranes of 100-kDa exclusion limits (C-100). Figure 1 shows PEDF immunostaining and Ponceau Red staining of bands for F1 subunits and PEDF after SDS-PAGE. One tenth of the reactions was removed before size-exclusion ultrafiltration, and analyzed in separate lanes as control of starting material (lanes 10 and 11). PEDF immunostaining was proportional to the PEDF amount added to the reactions. Ponceau Red bands for α and β subunits were detected in both reaction mixtures. In lane 10 the relative intensities of Ponceau Red bands suggested a lower PEDF:α/β subunit ratio (about or less 1:10) than in lane 11, in which the band intensities for each α/β subunits and PEDF appeared at approximately 1:1 molar ratio for these components. After ultrafiltration only the reactions with equimolar amounts of PEDF and α/β subunits showed detectable levels of PEDF (lane 2), indicating the formation of PEDF complexes with F1. Omitting F1 (lanes 3 and 4) or replacing it with bovine serum albumin (66-kDa) (lane 9) did not result in PEDF complexes. A combination of 1 mM Mg++/ATP is known to increase the stability of the F1-ATPase multimeric protein as detected by an increase in Ponceau Red staining of the F1 subunits (see bottom of figure 1 and compare lanes 1, 2 and 7 to lanes 5, 6 and 8). Binding reactions in the presence of 1 mM Mg++/ATP resulted in a proportional increase in PEDF:F1 complexes (compare lanes 2 to 6). Formation of fluorescein-conjugated PEDF complexes with F1-ATPase was also observed (see Fig. S3). These observations revealed that PEDF bound specifically to F1 complexes.
Fig 1.
Complex formation assays between soluble recombinant human PEDF and recombinant yeast F1-ATPase. Proteins were incubated for 1 h at room temperature and then the mixtures were subjected to size-exclusion ultrafiltration using membranes with size exclusion limits of 100-kDa (lanes 1–9 indicated by C-100). The amounts of each component in each reaction mixture are indicated at the top. The total protein complexes retained by the membrane for each reaction were applied to lanes 1–9 of a 10–20% polyacrylamide gel and resolved by SDS-PAGE. One tenth of the reactions corresponding to lanes 1 and 2 before subjected to ultrafiltration were applied to lanes 10 and 11 (indicated by 1/10 rxn, No C-100) of the same gel. Proteins were transferred from the gel to a western blot, stained with Ponceau red (bottom blot) and then immunostained with anti-PEDF antibodies (top blot). The migration positions of PEDF, F1 α-β-and γ-subunits are indicated to the right, and of protein standards to the left (BSA, bovine serum albumin ~66-kDa; OVA, ovalbumin ~48-kDa; CA, carbonic anhydrase ~31-kDa).
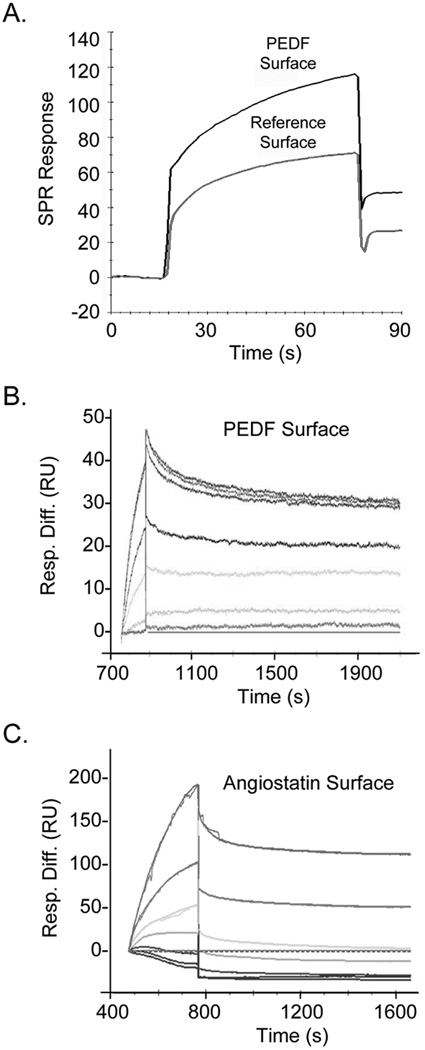
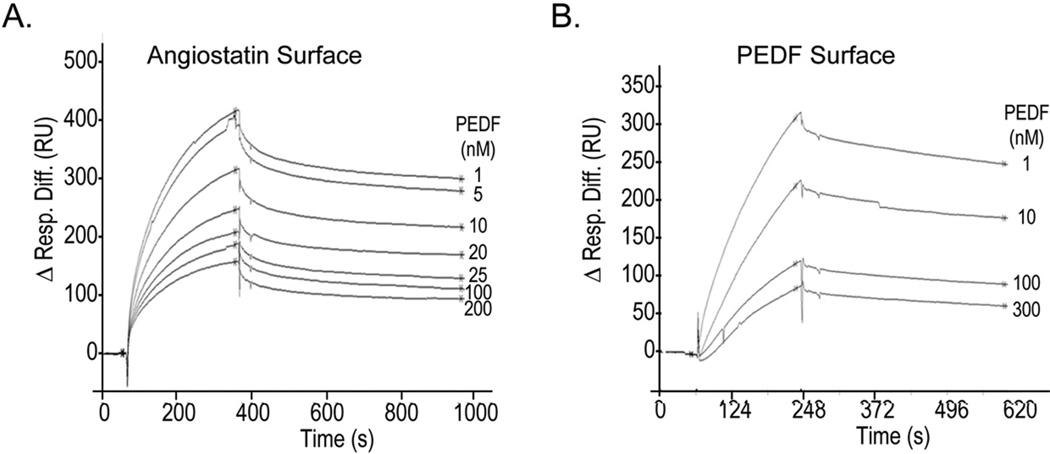
To determine the biophysical binding parameters for the PEDF:F1 interactions, real-time SPR spectroscopy was performed. Sensograms with PEDF protein immobilized on the surface of a CM5 sensor chip revealed binding response units for the yeast F1-ATPase that were above reference cells (without PEDF) (Fig. 2A). They indicated specific, reversible and concentration-response binding of F1 to PEDF (Figs. 2B). The kinetic parameters for the SPR interactions between F1-ATPase and PEDF were consistent with a 1:1 Langmuir binding, implying a one-site binding between F1 and PEDF. They revealed high binding affinities (KD = 1.51 nM) with fast association and low dissociation rates between PEDF and F1 in vitro (Fig. 2B). Similarly, the SPR interactions between F1 and angiostatin kringles 1–5 (K1–5) were assessed (Fig. 2C). Table 1 summarizes the results with several batches of F1 proteins. The yeast F1 had higher affinity for PEDF- than for angiostatin K1–5-surface sensor chips (>10-fold). Altogether, these results implied that soluble and immobilized PEDF can interact with F1.
Fig 2.
Real-time SPR binding analyses of F1 and PEDF interactions.
(A) SPR spectroscopy of recombinant yeast F1-ATPase (F1) with recombinant human PEDF immobilized on a CM5 sensor chip. Sensograms of SPR Responses (Relative Units, R.U.) of 200 nM F1 solutions injected onto surfaces with PEDF or without PEDF (reference surface) are shown.
(B and C) Sensograms were recorded with PEDF (B) or human angiostatin K1–5 (C) immobilized on CM5 sensor chips and injections of F1 solutions (100, 50, 20, 10, 5, 1, and 0 nM F1 in (B); 500, 300, 200, 100, 50, 20, and 0 nM F1 in (C)) using a BIAcore 3000 biosensor and BIAevaluation software. SPR response differences with respect to the blank surface were subtracted by the sensograms at 0 nM concentration during the BIAevaluation (Resp. Diff., y axis) and are shown as a function of time (s, x axis). The kinetic and thermodynamic values for PEDF in (B) were ka (1/Ms) = 6.89 × 103; kd (1/s) = 1.04 × 10−5; KD = 1.51 nM; and for angiostatin in (C) were ka (1/Ms) = 962; kd (1/s) = 1.88 × 10−4; KD = 195 nM.
Table 1.
Summary of SPR kinetic parameters for the interactions between yeast F1-ATP synthase and human PEDF or human angiostatin K1–5
| SPR Surface |
F1-ATP synthase (batch No.) |
Fit method | ka (1/Ms ± SEa) |
kd (1/s ± SEa) |
KA (1/M ± AVEDEVb) |
KD (nM ± AVEDEVb) |
|---|---|---|---|---|---|---|
| PEDF | 1 | 1:1 (Langmuir) binding | 8.8E+03 ± 127 | 5.5E-05 ± 9.4E-07 | 1.6E+08 ± 2.7E+06 | 6.30 ± 0.11 |
| PEDF | 1 | 1:1 (Langmuir) binding with drifting baseline |
6.9E+03 ± 76 | 1.0E-05 ± 3.0E-07 | 6.6E+08 ± 1.9E+07 | 1.51 ± 0.04 |
| PEDF | 1 | 1:1 (Langmuir) binding | 3.6E+04 ± 323 | 1.7E-04 ± 7.8E-07 | 2.1E+08 ± 1.9E+06 | 4.82 ± 0.04 |
| PEDF | 2 | 1:1 (Langmuir) binding | 8.4E+04 ± 1030 | 1.5E-04 ± 8.4E-07 | 5.6E+08 ± 6.8E+06 | 1.79 ± 0.02 |
| PEDF | 2 | 1:1 (Langmuir) binding with drifting baseline |
8.6E+04 ± 883 | 7.2E-04 ± 4.8E-06 | 1.2E+08 ±1.2E+06 | 8.39 ± 0.09 |
| PEDF | 3 | 1:1 (Langmuir) binding with mass transfer |
4.7E+05 ± 9200 | 1.4E-03 ± 2.5E-05 | 3.2E+08 ± 6.4E+06 | 3.08 ± 0.06 |
| Angiostatin | 2 | 1:1 (Langmuir) binding | 1.3E+03 ± 29 | 2.0E-04 ± 2.2E-06 | 6.5E+06 ± 1.5E+05 | 154 ± 3.5 |
| Angiostatin | 2 | 1:1 (Langmuir) binding | 0.5E+03 ± NDc | 6.9E-05 ± NDc | 7.3E+06 ± NDc | 137 ± NDc |
| Angiostatind | 3 | 1:1 (Langmuir) binding | 0.96E+03 ± 4.5 | 1.9E-04 ± 2.9E-06 | 5.1E+06 ± 7.9E+04 | 195 ± 3.0 |
SE values were obtained from the files of the SPR kinetic analyses using the BIAevaluation software program.
AVEDEV values were calculated from ka ± SE and kd ± SE values using Excel’s Statistical functions.
ND, not available or determined.
An additional SPR bioevaluation estimated the kd value to be 2.10E-05 1/s and the KD value to be 230 nM for the interactions between F1 (batch No. 3) and angiostatin surfaces (Peter Schuck, personal communications).
Competition between PEDF and angiostatin for F1 binding
Angiostatin binds the α/β subunits of F1 [31]. To determine whether PEDF and angiostatin share a binding site(s) on F1, the SPR interactions between angiostatin and F1 were competed with PEDF. Injections of yeast F1 mixed with increasing concentrations of PEDF decreased the SPR response to angiostatin-surface sensor chips in a dose-response fashion (Fig. 3A) and with an estimated half-maximum inhibition IC50 of ~12 nM PEDF. Control injections of yeast F1 mixed with PEDF onto PEDF-surfaces also decreased the SPR response of F1 (Fig. 3B; estimated IC50 = ~17 nM PEDF), and PEDF by itself was deficient in binding either surface (data not shown). Competition of fluorescein-conjugated PEDF to F1-ATPase binding with PEDF and angiostatin was also observed by size-exclusion ultrafiltration (see Fig. S4). These results indicated that PEDF efficiently blocked the F1 interactions with angiostatin by competing for the angiostatin-binding site(s).
Fig 3.
Ligand competition of F1 binding. Ligand competition of F1 binding to angiostatin (A) or PEDF (B) surfaces was performed. F1 (100 nM) was premixed with increasing concentrations of PEDF (as indicated) and injected on each surface for 300 and 250 sec, respectively, at a flow rate of 20 ml/min. Dissociation was carried out with running buffer for 600 and 300 seconds, respectively. SPR response differences with respect to blank surfaces were aligned to 0 in the region preceding the injections (Δ Resp. Diff.) and are shown as a function of time. Half-maximal inhibition determined by non-linear regression of SPR response differences at saturation and dissociation time points as a function PEDF concentration were IC50 = 11.8 ± 0.3 nM PEDF for angiostatin surface and IC50 = 17.3 ± 2.1 nM PEDF for PEDF surface.
Binding of PEDF to endothelial cell-surface ATP synthase
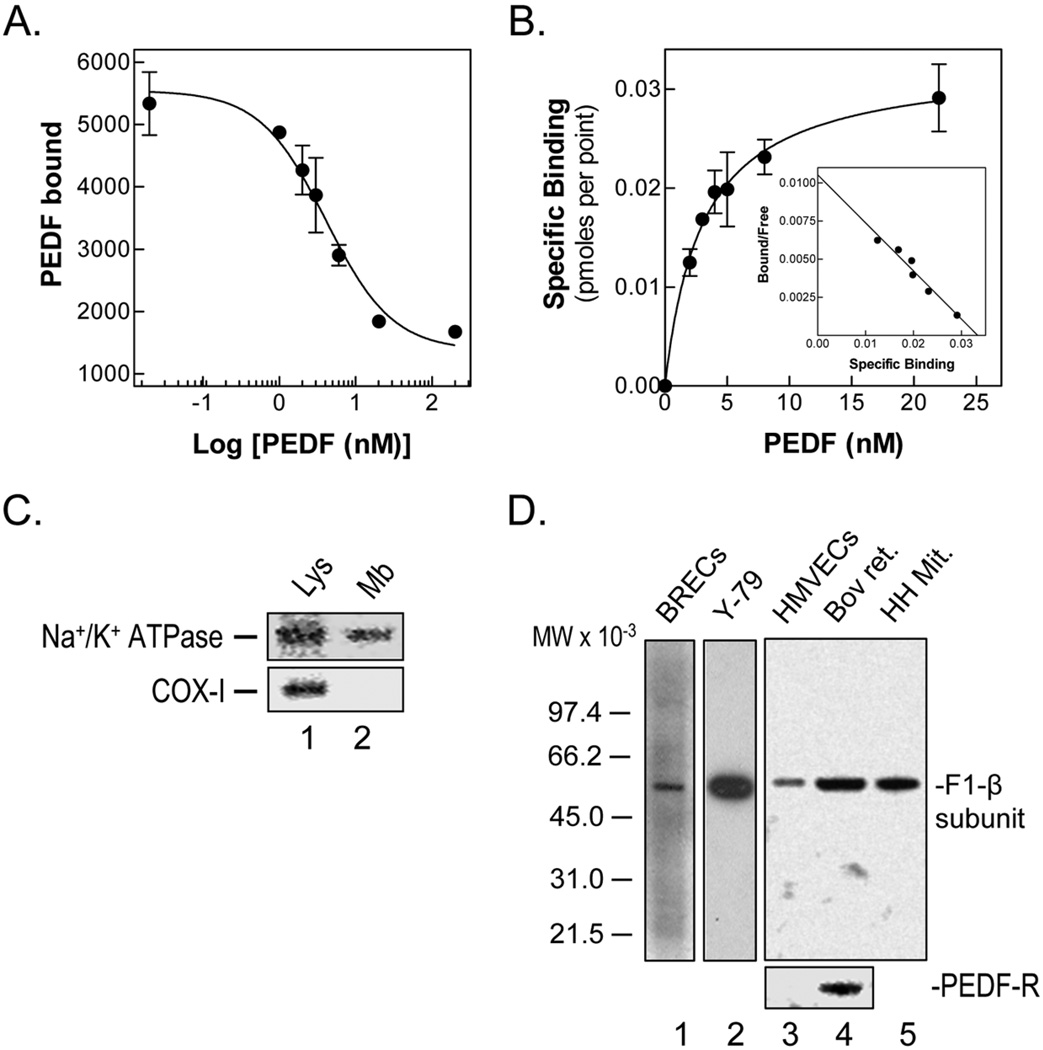
As illustrated in figures 4A–B, PEDF bound to BRECs with high affinity (KD = 3.04 – 4.97 nM) and with 39,000 – 78,000 sites per cell (two different batches of cells). Competition of the radioligand PEDF binding to cells with unlabeled ligand showed an EC50 (4.1 – 4.6 nM) similar to the KD. The physicochemical parameters of these interactions are in agreement with previously reported ones for the binding of PEDF to HUVECs (KD = 5.2 ± 2.3 nM; Bmax = 42,000 – 54,000 sites/cell; EC50 = 5.1 nM; [24]), and the affinity for purified PEDF and yeast F1 proteins (see above). These results demonstrated that the binding of PEDF to the surface of endothelial cells was specific, concentration-dependent, saturable and with high affinity, and suggested that PEDF interacts with a protein(s) at the surface of endothelial cells.
Fig 4.
PEDF binding to endothelial cell surfaces.
(A–B) Radioligand binding assays was with 2 nM 125I-PEDF and 0–200 nM unlabeled ligand on BRECs attached to collagen-coated plates at 4°C for 90 min. Cells were washed with binding medium and bound radioactivity was determined in cell extracts detached with 0.1 N NaOH. Binding competition with unlabeled PEDF (A), and saturation isotherm, non-linear regression of transformed binding in function of PEDF concentration (B) with a Scatchard plot, linear regression of transformed binding data (in the inset) determined by GraphPad software are shown.
(C) Western blots of HMVEC total lysate and plasma membrane extracts with antibodies to Na+/K+ ATPase, a plasma membrane marker, and to cytochrome c oxidase (COX-1), a mitochondrial membrane marker, are shown. Samples were loaded to the gel as follows: lane 1, total homogenate from HMVECs (52 µg protein); and lane 2, HMVECs membrane fraction (4 µg protein).
(D) Western blots of BRECs, Y-79, HMVECs, and bovine retina membrane extracts with antibodies to F1-β subunit. Westerns of same samples of HMVECs and bovine retina with antibodies to PEDF-R are also shown (bottom). Detergent soluble plasma membrane protein fractions were prepared and loaded to gels as follows (protein amounts): lane 1, BRECS (8 µg); lane 2, Y-79 cells (8 µg); lane 3, HMVECs (5 µg), and lane 4, bovine retina (5 µg). Lane 5 was human heart mitochondria (1 µg), a positive control for F1-ATP synthase.
To determine whether the endothelial PEDF-binding component was related to cell-surface ATP synthase, we prepared subcellular fractions of plasma membrane proteins from endothelial cells. We confirmed that they were depleted of mitochondrial membrane markers and contained plasma membrane markers (Fig. 4C). In westerns of detergent-soluble membrane protein fractions from HMVECs and BRECs we detected immunoreactive proteins to antibody to the β subunit of human heart mitochondrial F1F0-ATP synthase (anti-hF1), which comigrated with ~60-kDa proteins of yeast and bovine heart mitochondrial F1-ATPase controls (Fig. 4D). The β subunit-immunoreactive band was also detected in plasma membrane extracts from normal bovine retina and human retinoblastoma Y-79 tumor cells. However, PEDF-R was undetectable in endothelial cell membranes extracts.
SPR interactions of PEDF and endothelial cell-membrane proteins
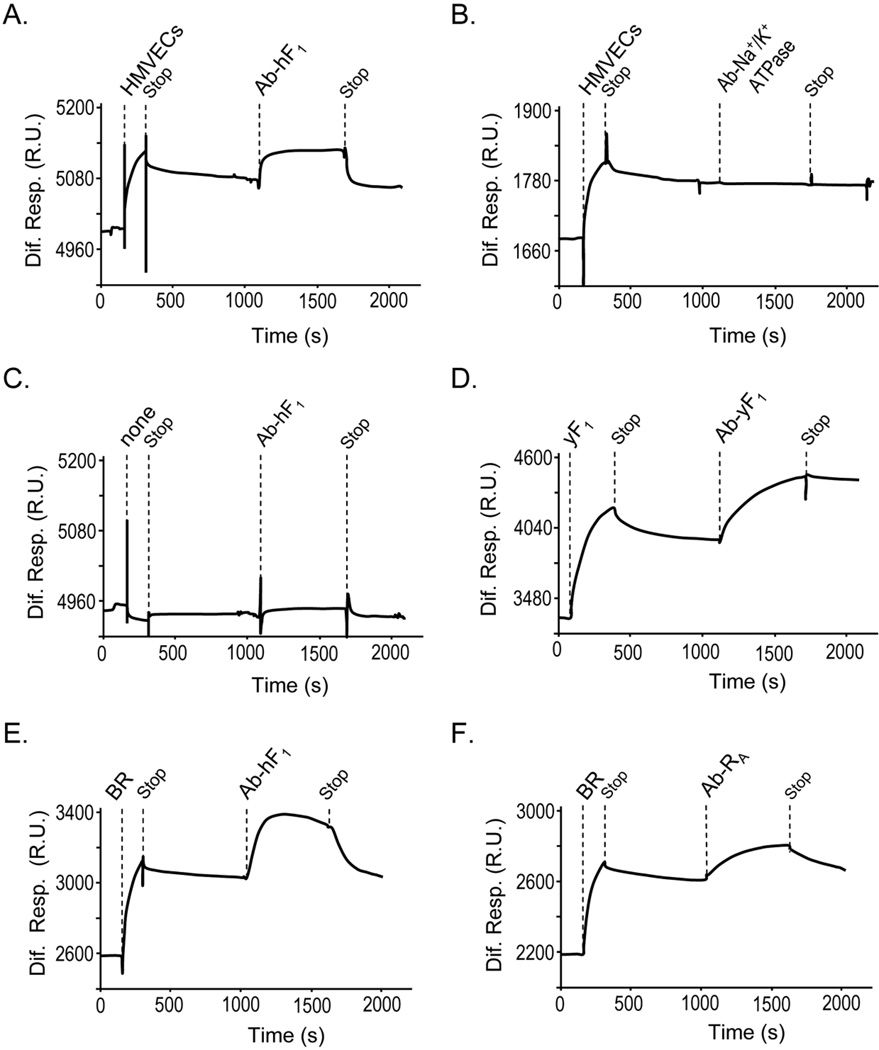
To investigate whether the endothelial cell-surface F1-ATP synthase binds to PEDF, real-time SPR spectroscopy was performed with detergent-soluble plasma membrane extracts from HMVECs on a PEDF surface sensor chip. Sensograms revealed binding response units with injections of membrane extracts that were above reference cells (without PEDF) (Fig. 5A), indicating specific binding of a component(s) in HMVEC membranes to PEDF. Upon stopping the injection of extracts, the bound components remaining on the PEDF sensor chip become available to be selectively captured with injections of specific antibodies. This was clearly demonstrated by capturing purified yeast F1 on PEDF sensor chips with polyclonal antiserum to yeast F1 (Fig. 5D). To determine whether the PEDF-binding component(s) in endothelial membranes included F1, solutions of antibodies to F1 were injected subsequently to the surface. As shown in figure 5A, injections of anti-hF1 increased the SPR response units above those of HMVECs plasma membrane extracts. In contrast, a F1-unrelated antibody that immunorecognized Na+/K+-ATPase in HMVECs plasma membrane extracts (see Fig. 4A) did not increase the SPR response (Fig. 5B), and anti-hF1 alone (control injections) did not bind to the PEDF surface (Fig. 5C). Figures 5E and 5F show that F1 β-subunit and the previously identified PEDF-R (14) from bovine retina plasma membranes bound to PEDF, respectively. PEDF-R was undetectable in endothelial cell membranes extracts by SPR capturing (personal observations, LN), in agreement with western blotting (see above). Altogether, these results clearly demonstrated that the ATP synthase F1 β-subunit in plasma membrane extracts of endothelial cells was a PEDF-binding component. They suggest that interactions of extracellular PEDF ligands with F1-β-subunit on endothelial cell surfaces may regulate ATP metabolism.
Fig 5.
PEDF-binding proteins in cell membranes from HMVECs and bovine retina. SPR spectroscopy on PEDF surfaces of detergent-soluble membrane proteins from HMVEC (A and B) and bovine retina (E and F), and no extracts (C) and control yeast F1 (yF1) (D). Antibody capturing was with antibodies to human F1-β subunit (Ab-hF1), yeast F1-β subunit (Ab-yF1) and PEDF-R (Ab-RA). Protein extracts (34 µg/ml) were injected for 300 s at a flow rate of 20 µl/min, and after 600 s of dissociation, flow rate was decreased to 5 µl/min and specific antibodies (5 µg/ml) were injected for 600 s. Sensograms relative to reference surface (without PEDF) are shown. Dotted lines to the top of each sensogram point to time of injection of proteins and antibodies, as well as stop of injection.
Effects of PEDF on the extracellular ATP synthesis activity of endothelial cells
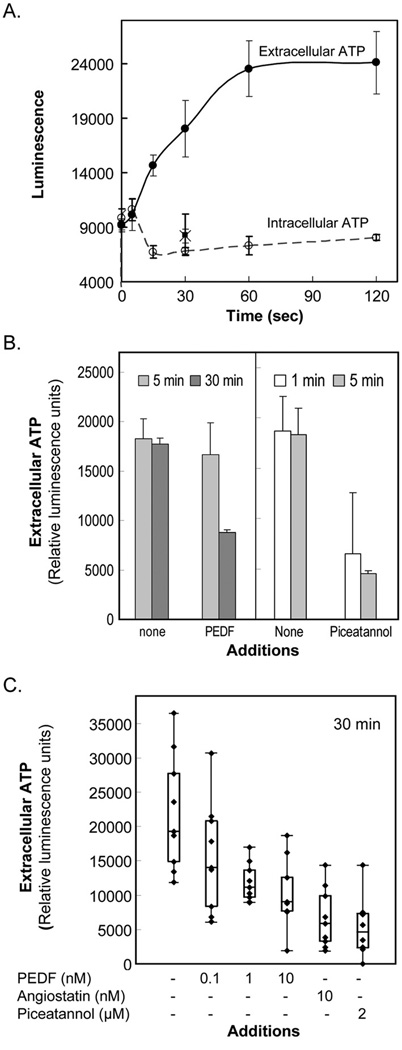
First, we determined the ATP synthesis activity of HMVECs. The cell-surface ATP synthase activity was measured by the extracellular ATP production after additions of ADP and inorganic phosphate to intact HMVEC cells. Extracellular ATP production increased linearly during the first 60 s of incubation, while their intracellular ATP levels did not change significantly with incubation time or when inorganic phosphate was not included in the reactions (Fig. 6A). These results demonstrate extracellular ATP synthase activity in these cells, as observed before for HUVECs [33].
Fig 6.
ATP production by HMVECs.
(A) Extracellular ATP production and intracellular ATP levels by HMVECs. ATP synthesis was initiated by the addition of a solution containing ADP and Pi to a culture of HMVECs. At indicated times, extracellular medium and intracellular pools were prepared, and the ATP content in those pools was determined. Each point corresponds to an average of triplicate samples for: extracellular medium (●); intracellular pools (○); and reactions without Pi (extracellular ■; intracellular ×).
(B) HMVECs were incubated in EBM2/BSA in the presence of PEDF (1 nM) or piceatannol (20 µM) for an increasing period of time (top). Extracellular ATP synthesis activity was determined upon incubations for 60 s with ADP and inorganic phosphate in the presence of indicated inhibitors (x axis).
(C) HMVECs were incubated in EBM2/BSA containing increasing PEDF concentrations, angiostatin K1–5 or piceatannol for 30 min. Extracellular ATP synthesis activity was determined as in (B). Box-and-whisker plot representations of replicates for extracellular ATP synthesis determination are shown. Each point corresponds to a measurement from one well, each condition was performed in triplicate wells, and all conditions were repeated with three batches of cells. Values inside the boxes correspond to the central 50% of measurements, their internal horizontal bars correspond to median values, and the vertical lines outside the boxes correspond to variances of measurements. Inhibitor concentrations are indicated in the x-axis. PEDF and positive controls angiostatin (10 nM) and piceatannol (2 µM) inhibited ATP synthesis.
Then, we examined the extracellular ATP synthesis activity of endothelial cells in the presence of PEDF. The cell-surface ATP synthase activity was measured in HMVECs treated with PEDF for indicated periods of time. Extracellular ATP generation was assayed within 60 s after addition of ADP and inorganic phosphate in the presence or absence of PEDF. Treatments of 30 minutes with 1 nM PEDF decreased extracellular ATP synthesis (Fig. 6B). The positive control piceatannol was also a potent inhibitor requiring even ≤5 min of preincubation time for effective blocking. Other investigators have demonstrated inhibition of extracellular ATP synthesis by pretreating HUVECs for 30 min with much higher doses of angiostatin K1–3 (50 µM; [35]) and piceatannol (1 – 20 µM [33]; 500 µM [35]) than the ones used here. As shown in figure 6C, pre-treatments with PEDF for 30 min inhibited the extracellular ATP synthesis activity in a dose-dependent fashion. The range of distribution of the measurements reflected the variability of the assay. The median values of the inhibitory activity of PEDF on extracellular ATP synthesis varied between 27%, 43% and 53% with 0.1 nM, 1 nM and 10 nM PEDF, respectively. No significant statistical difference was observed between PEDF and angiostatin at 10 nM concentration (p≤ 0.096). Moreover, the treatments with PEDF or angiostatin for up to 48 h did not decrease the intracellular levels of ATP, and, if any, both slightly increased them (see Fig. S1). These results demonstrated that extracellular PEDF additions inhibited the extracellular ATP synthesis activity of endothelial cells.
Discussion
PEDF, a potent inhibitor of neovascularization, targets endothelial cells [3]. We have shown that PEDF directly binds and inhibits endothelial cell-surface ATP synthase. These two proteins interact when they are in solution and when either one is immobilized. PEDF can bind recombinant yeast F1 in a purified version or native mammalian F1 in membrane cell extracts or in intact cells. We observed that chemically modified PEDF at primary amines (e.g., Fl-PEDF) also binds to F1-ATPase (see Figs. S3–S4). The interactions are specific, reversible and of high affinity, and are between PEDF and the F1 β-subunit. Furthermore, inhibition of extracellular ATP synthesis in intact endothelial cells demonstrates that the PEDF interaction blocks the structural determinant required for the activity of the cell-surface ATP synthase. PEDF shares these properties with angiostatin, and the observed competition for binding F1 between these two factors implies that the β subunit of F1 has an overlapping site(s) for binding both proteins. These conclusions suggest that interactions between extracellular PEDF ligands and F1 β-subunit on endothelial cell surfaces may regulate ATP metabolism. They imply that inhibition of ATP synthase may be part of the biochemical mechanisms by which PEDF exerts its antiangiogenic activity.
Previous reports have described a PEDF-binding protein of 60-kDa in plasma membranes from HUVECs [36], normal bovine retina [22] and human retinoblastoma Y-79 tumor cells [21], but had not shown its identity. The present results reveal that the ~60-kDa PEDF-binding protein is the ATP synthase F1-β subunit in endothelial cells, as well as in retina and Y-79 cells (Figs. 4D, 5E–F, Table S1; [21, 22]). Other subunits of the ATP synthase holoenzyme, such as the α and β subunits of F1 domain and the b-and d-subunits of F0 have been identified also in plasma membranes of HUVECs, several tumor cells, adipocytes and myocytes [27]. Interestingly, the entire F1F0 ATP synthase has demonstrable activity in the endothelial cell-surface, with ability to synthesize ATP and transport protons [27, 30]. Our data provide further lines of evidence for the extra-mitochondrial expression of ATP synthase in the surface of endothelial cells. The presence of F1-β subunit in retinoblastoma Y-79 cell surfaces is consistent with previously reported expression of F1F0-ATP synthase in tumor cell-surfaces [27], and suggests a role for interactions between cell-surface ATP synthase and PEDF in mediating differentiating activity in retinoblastoma cells. PEDF-affinity column chromatography of plasma membrane extracts revealed different migration patterns of PEDF-binding proteins among bovine retina, Y-79 cells and BRECs ([21, 22]; Fig. S2). All had bands corresponding to F1 α/β subunits of ~60-kDa but only bovine retina and Y-79 cells with detectable bands for PEDF-R of ~85-kDa. Peptide fingerprinting of the PEDF-binding protein of 60-kDa matched it to the F1 β-subunit (Table S1). The inability of detecting PEDF-R in endothelial cell membranes supports the idea that endothelial cell surfaces express a different set of PEDF-binding protein(s) than neural retinal cell-surfaces, which may distinctly and specifically trigger angiostatic activities upon interacting with PEDF ligand.
We compared the interactions of the purified F1 and PEDF proteins, and those that occur with cells. The KD values of the SPR binding of yeast F1 to immobilized human PEDF match those for the interactions between PEDF and the surface of endothelial cells (KD = 3 – 7.5 nM) ([24]; Figs. 4A–B), as well as the concentration of PEDF capable of inhibiting about 50% of the maximum extracellular ATP synthase activity in HMVECs (Fig. 6C). The estimated IC50 values of PEDF for blocking yeast F1 binding to immobilized angiostatin (~12 nM) or PEDF (~17 nM) insinuate similar affinity for PEDF when in solution and immobilized on sensor chips. This observation implies that only minimal changes in affinity occurred upon PEDF immobilization. In contrast, angiostatin K1–5 at concentrations ≤270 nM (5-fold the F1 concentration) could not compete with immobilized PEDF on sensor chips (unpublished observations, LN), in agreement with a lower affinity for the yeast F1:angiostatin interactions. In spite of the higher affinity of yeast F1 for immobilized human PEDF than for human angiostatin K1–5 by SPR (Table 1), no significant statistical difference is observed between PEDF and angiostatin in inhibiting endothelial extracellular ATP synthase activity (Figs. 5C). A previous reported value of an apparent dissociation constant (Kd(app) = 14.1 nM) for human angiostatin K1–3 binding to purified bovine heart F1-ATPase immobilized on plastic [30] suggests higher affinity for these interactions than for yeast F1 binding to angiostatin K1–5 sensor chips (KD = 130 – 237 nM; Table 1). The affinity of F1:angiostatin interactions is likely to be species-specific and the observed SPR affinity of angiostatin:F1 is lower compared to that in mammalian cells. In addition alterations of critical structural determinants in angiostatin for binding F1 might also affect the affinity of these interactions. For example, immobilization of molecules on the SPR sensor chips by conjugation of primary amines (lysine residues and amino terminal ends) to the CM5 surfaces may decrease the affinity of the angiostatin molecule for binding F1. As mentioned above, PEDF is not affected by this. Moreover, piceatannol, known to target the catalytic F1 domain at the β subunit [37], does not affect the SPR interactions of F1 to PEDF by either coinjecting it or including it in the SPR running buffer (personal observations, LN, SPB). This implies that the structural determinants required for binding PEDF and piceatannol do not overlap.
Our results have biological implications. The interactions of extracellular PEDF ligands with cell surface F1F0-ATP synthase molecules may regulate the levels of ATP and ADP, which in turn may affect the behavior of endothelial cells, e.g., PEDF may interact with ATP:P2X and ADP:P2Y receptor-mediated signaling pathways by regulating the availability of the ATP and ADP ligands, similar to angiostatin [27]. It has been shown that blocking the ATP synthase by targeting the F1 catalytic domain with angiostatin or piceatannol can trigger caspase-mediated endothelial cell apoptosis, and inhibit tube formation and proliferation necessary for antiangiogenesis [32–34]. Similarly, blocking the ATP synthase with PEDF may trigger signal transduction to mediate apoptosis in endothelial and/or tumor cells.
In summary, this is the first report demonstrating that PEDF binds endothelial cell surface F1 β-subunit, and inhibits endothelial extracellular ATP synthesis activity. The findings imply that F1F0-ATP synthase may act as a receptor for PEDF on the surface of endothelial cells, and that PEDF can inhibit this extra-mitochondrial ATP synthase, which catalyzes ATP synthesis. The interactions PEDF:ATP synthase might be a critical biochemical step for the angiostatic effects exerted by PEDF on the neovasculature.
Materials and methods
Proteins
PEDF was human recombinant PEDF as described before [38]. Recombinant yeast F1-ATPase was obtained and highly purified as described before [39]. Human angiostatin K1–5 was purchased from Calbiochem, La Jolla, CA, USA. Human angiostatin K1–3 was from Sigma, St. Louis, MO, USA. Polyclonal antibodies directed against the β-subunit of the yeast ATPase were made in rabbit using β-subunit purified from recombinant yeast F1 by SDS polyacrylamide gel electrophoresis. Mouse monoclonal antibody to human F1F0-ATP synthase β subunit, anti-F1F0-β, (Ab-hF1) (Cat# MS503), and human heart mitochondria extracts (Cat. # MS801-50) were from MitoSciences, Eugene, OR, USA.
Cells
Immortalized human microvascular endothelial cells (HMVECs) were a generous gift of Dr. Rong Shao and were cultured as described before [40]. Bovine retina endothelial cells (BRECs) were from Vec Technologies, Inc., Rensselaer, NY, USA. These cells were sensitive to the angiostatic effects of PEDF.
Size-exclusion ultrafiltration
Complex formation was analyzed by size exclusion ultrafiltration using Centricon-100 devices with membranes of 100-kDa exclusion limits, as described before [41]. This assay is based on the fact that PEDF of 50-kDa passes through the membranes and does not when forming a complex ≥100-kDa. The components retained by the membrane after centrifugation and washes of the devices were analyzed by western blotting.
Surface Plasmon Resonance (SPR) spectroscopy
The interactions between PEDF and yeast F1-ATPase were analyzed by SPR using a BIAcore 3000 instrument (BIAcore, Uppsala, Sweden) with immobilized PEDF ligands, as described previously [42]. PEDF ligand (4 ng) was immobilized on a CM5 sensor chip, by NHS/EDC activation, followed by covalent amine coupling of the protein to the surface. A reference surface without protein was prepared by the same procedure. Both surfaces were preconditioned with two injections of 50 mM NaOH, then washed with 0.5 M NaCl and re-equilibrated with binding buffer (10 mM Tris-Cl pH 7.5, 0.15 M NaCl, 0.25 M Sucrose (TBS-S)). Ten different dilutions of F1 solutions with concentrations ranging between 0.3–200 nM were injected on both surfaces. Each injection was followed by a 50 mM NaOH regeneration step. The results were analyzed using BIAevaluation software. The data were then fitted to several binding models for a kinetic analysis. The best fittings were obtained with a simple 1:1 Langmuir model for the PEDF surface binding assay. Background baseline noise was slightly adjusted using drifting baseline or accounting mass transfer limitation to produce the best fit.
Radioligand binding assays
Assays were performed using a given concentration of 125I-PEDF as radioligand and unlabeled PEDF as competitor on endothelial cells attached to the wells, as described before [21]. The radioligand binding data was analyzed by nonlinear regression with one site competition equation for ligand competition with unlabeled PEDF. The radiolabeled data was transformed to calculate bound PEDF per assay and then analyzed by non linear regression with one binding site (hyperbola) as best fitted for Saturation binding isotherm. GraphPad Prism version 4.00 for Windows (GraphPad Software, San Diego, CA, USA) was used for the data analysis and generation of plots.
Ligand competition assays
F1-ATPase (100 nM) was premixed with increasing concentrations of PEDF (1–300 nM) and injected with a kinject procedure on a CM5 BIAcore chips with immobilized PEDF (5000 RU) or angiostatin K1–5 (1400 RU). Injections were for 300 seconds followed by 600 seconds of dissociation, and regeneration of the chip surface with 50 mM NaOH. SPR differential responses were plotted as a function of PEDF concentration.
Cell-surface ATP synthesis activity
The activity of extracellular ATP synthesis by HMVECs was determined as described previously [32] with the following modifications. HMVECs were serum-starved overnight at 37°C in EBM2/EGF/hydrocortisone (Cambrex Bio Science, Walkersville, MD, USA) plus 0.2% BSA, and then for 1 hour at 37°C in EBM2/VEGF/bFGF (Cambrex Bio Science, Walkersville, MD, USA) plus 0.2% BSA, following instructions by manufacturer. Cells were preincubated with effectors for 30 min at 37°C. After 10 minutes at room temperature, cells were rinsed once with HEPES buffer (10 mM HEPES, pH 7.4, 150 mM NaCl). Then, HEPES buffer containing 1 mM MgCl2 with effectors were added to the wells. After 1 min, a solution of the same buffer with a final concentration of 100 µM ADP, 10 mM potassium phosphate (Pi), and 1 mM MgCl2 was added. The cells were then incubated at room temperature for indicated periods of time up to 120 s, and the extracellular ATP content was determined in the media using the ATP bioluminescence CellTiter-Glo assay kit (Promega, Madison, WI, USA) according to the manufacturer's instructions. The intracellular ATP content was also determined in lysed cells using Cell-Titer Glo assay kit. All measurements were performed using the Wallac Victor2 1420 multilabel counter (Wallac Oy, Turku, Finland) and the results were analyzed using an Excel spreadsheet (Microsoft, Redmond, WA, USA).
Plasma membrane extracts
Endothelial cell membrane extracts were prepared as described before [25]. Briefly, cells were grown to confluence, starved in serum free medium for 16 hours, harvested, homogenized and subjected to differential centrifugation. Separation of final supernatant (cytosolic fraction) and particulate material (membrane fraction) from transfected cells was with 150,000 × g centrifugation. Detergent-soluble plasma membrane fractions were subjected to SPR spectroscopy or to Western blotting.
Western blotting
Western transfers were performed as described before [7]. Immunoreactions were with mouse monoclonal anti-PEDF (Chemicon, Temecula, CA, USA) diluted 1:1000; rabbit polyclonal antibody against PEDF-R (Notari et al., 2006) diluted 1:5000; mouse anti-hF1 (MitoScience, Eugene, OR, USA) diluted 1:5,000; mouse anti cytochrome c oxidase (COX-I) complex IV (Santa Cruz Biotechnology, Santa Cruz, CA, USA) diluted 1:2,000, or rabbit anti-human Na+/K+-ATPase (Santa Cruz Biotechnology, CA, USA) diluted 1:5,000 in 5% BSA in TBS-Tween (50 mM Tris-HCl pH 7.5, 150 mM NaCl, and 0.1% Tween-20). Secondary antibodies were rabbit anti-mouse biotinylated IgG-POD (Vector Laboratories, Burlingame, CA, USA) diluted at 1:1,000 for Western vs. anti-PEDF antibodies, peroxidase labeled goat anti-mouse IgG (H+L) (KPL, Gaithersburg, MD, USA) diluted 1:100,000 for westerns with anti-COX-1 or peroxidase labeled goat anti-rabbit IgG (H+L) (KPL) diluted 1:100,000 for westerns with anti-Na+/K+ ATPase. Immunoreactive bands were detected by chemiluminescence (Super Signal® West Dura Extended Duration Substrate; Pierce Biotechnology, Rockford, IL, USA) and signal was acquired using a Typhoon 9410 laser based scanner (Amersham, Piscataway, NJ, USA). For PEDF analyses, westerns were performed as described before [7].
Supplementary Material
Acknowledgements
This research was supported in part by the Intramural Research Program of the NIH, NEI and NIH Grant: R01-GM066223. We thank Tomihiko Higuti for starting collaboration between N. Arakaki and our group, Peter Schuck for interesting discussions on SPR kinetic evaluations and confirming SPR evaluations with our data, Ignacio Rodriguez and Vicente Notario for critically reading our manuscript.
Abbreviations
- PEDF
pigment epithelium-derived factor
- F1
F1-ATP synthase protein complex
- HMVECs
human microvascular endothelial cells immortalized with telomerase
- BRECs
bovine retina endothelial cells
- HUVECs
human umbilical vascular endothelial cells
- SPR
surface plasmon resonance
- PEDF-R
PEDF receptor
References
- 1.Amaral J, Becerra SP. Pigment epithelium-derived factor and angiogenesis: Therapeutic implications. In: Penn JS, editor. Retinal and Choroidal Angiogenesis. Dordrecht, The Netherlands: Springer; 2008. pp. 311–337. [Google Scholar]
- 2.Barnstable CJ, Tombran-Tink J. Neuroprotective and antiangiogenic actions of PEDF in the eye: molecular targets and therapeutic potential. Prog Retin Eye Res. 2004;23:561–577. doi: 10.1016/j.preteyeres.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Bouck N. PEDF: anti-angiogenic guardian of ocular function. Trends Mol Med. 2002;8:330–334. doi: 10.1016/s1471-4914(02)02362-6. [DOI] [PubMed] [Google Scholar]
- 4.Dawson DW, Volpert OV, Gillis P, Crawford SE, Xu H, Benedict W, Bouck NP. Pigment epithelium-derived factor: a potent inhibitor of angiogenesis. Science. 1999;285:245–248. doi: 10.1126/science.285.5425.245. [DOI] [PubMed] [Google Scholar]
- 5.Ek ET, Dass CR, Choong PF. PEDF: a potential molecular therapeutic target with multiple anti-cancer activities. Trends Mol Med. 2006;12:497–502. doi: 10.1016/j.molmed.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 6.Wu YQ, Becerra SP. Proteolytic activity directed toward pigment epithelium-derived factor in vitreous of bovine eyes. Implications of proteolytic processing. Invest Ophthalmol Vis Sci. 1996;37:1984–1993. [PubMed] [Google Scholar]
- 7.Wu YQ, Notario V, Chader GJ, Becerra SP. Identification of pigment epithelium-derived factor in the interphotoreceptor matrix of bovine eyes. Protein Expr Purif. 1995;6:447–456. doi: 10.1006/prep.1995.1060. [DOI] [PubMed] [Google Scholar]
- 8.Cayouette M, Smith SB, Becerra SP, Gravel C. Pigment epithelium-derived factor delays the death of photoreceptors in mouse models of inherited retinal degenerations. Neurobiol Dis. 1999;6:523–532. doi: 10.1006/nbdi.1999.0263. [DOI] [PubMed] [Google Scholar]
- 9.Crawford SE, Stellmach V, Ranalli M, Huang X, Huang L, Volpert O, De Vries GH, Abramson LP, Bouck N. Pigment epithelium-derived factor (PEDF) in neuroblastoma: a multifunctional mediator of Schwann cell antitumor activity. J Cell Sci. 2001;114:4421–4428. doi: 10.1242/jcs.114.24.4421. [DOI] [PubMed] [Google Scholar]
- 10.Doll JA, Stellmach VM, Bouck NP, Bergh AR, Lee C, Abramson LP, Cornwell ML, Pins MR, Borensztajn J, Crawford SE. Pigment epithelium-derived factor regulates the vasculature and mass of the prostate and pancreas. Nat Med. 2003;9:774–780. doi: 10.1038/nm870. [DOI] [PubMed] [Google Scholar]
- 11.Mori K, Duh E, Gehlbach P, Ando A, Takahashi K, Pearlman J, Mori K, Yang HS, Zack DJ, Ettyreddy D, Brough DE, Wei LL, Campochiaro PA. Pigment epithelium-derived factor inhibits retinal and choroidal neovascularization. J Cell Physiol. 2001;188:253–263. doi: 10.1002/jcp.1114. [DOI] [PubMed] [Google Scholar]
- 12.Mori K, Gehlbach P, Yamamoto S, Duh E, Zack DJ, Li Q, Berns KI, Raisler BJ, Hauswirth WW, Campochiaro PA. AAV-mediated gene transfer of pigment epithelium-derived factor inhibits choroidal neovascularization. Invest Ophthalmol Vis Sci. 2002;43:1994–2000. [PubMed] [Google Scholar]
- 13.Renno RZ, Youssri AI, Michaud N, Gragoudas ES, Miller JW. Expression of pigment epithelium-derived factor in experimental choroidal neovascularization. Invest Ophthalmol Vis Sci. 2002;43:1574–1580. [PubMed] [Google Scholar]
- 14.Spranger J, Osterhoff M, Reimann M, Mohlig M, Ristow M, Francis MK, Cristofalo V, Hammes HP, Smith G, Boulton M, Pfeiffer AF. Loss of the antiangiogenic pigment epithelium-derived factor in patients with angiogenic eye disease. Diabetes. 2001;50:2641–2645. doi: 10.2337/diabetes.50.12.2641. [DOI] [PubMed] [Google Scholar]
- 15.Stellmach V, Crawford SE, Zhou W, Bouck N. Prevention of ischemia-induced retinopathy by the natural ocular antiangiogenic agent pigment epithelium-derived factor. Proc Natl Acad Sci U S A. 2001;98:2593–2597. doi: 10.1073/pnas.031252398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takita H, Yoneya S, Gehlbach PL, Duh EJ, Wei LL, Mori K. Retinal neuroprotection against ischemic injury mediated by intraocular gene transfer of pigment epithelium-derived factor. Invest Ophthalmol Vis Sci. 2003;44:4497–4504. doi: 10.1167/iovs.03-0052. [DOI] [PubMed] [Google Scholar]
- 17.Wang L, Schmitz V, Perez-Mediavilla A, Izal I, Prieto J, Qian C. Suppression of angiogenesis and tumor growth by adenoviral-mediated gene transfer of pigment epithelium-derived factor. Mol Ther. 2003;8:72–79. doi: 10.1016/s1525-0016(03)00128-x. [DOI] [PubMed] [Google Scholar]
- 18.Amaral J, Becerra SP. Effects of human recombinant PEDF protein and PEDF-derived peptide 34-mer on choroidal neovascularization. Invest Ophthalmol Vis Sci. doi: 10.1167/iovs.09-4455. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Notari L, Miller A, Martinez A, Amaral J, Ju M, Robinson G, Smith LE, Becerra SP. Pigment epithelium-derived factor is a substrate for matrix metalloproteinase type 2 and type 9: implications for downregulation in hypoxia. Invest Ophthalmol Vis Sci. 2005;46:2736–2747. doi: 10.1167/iovs.04-1489. [DOI] [PubMed] [Google Scholar]
- 20.Becerra SP. Focus on Molecules: Pigment epithelium-derived factor (PEDF) Exp Eye Res. 2006;82:739–740. doi: 10.1016/j.exer.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 21.Alberdi E, Aymerich MS, Becerra SP. Binding of pigment epithelium-derived factor (PEDF) to retinoblastoma cells and cerebellar granule neurons. Evidence for a PEDF receptor. J Biol Chem. 1999;274:31605–31612. doi: 10.1074/jbc.274.44.31605. [DOI] [PubMed] [Google Scholar]
- 22.Aymerich MS, Alberdi EM, Martinez A, Becerra SP. Evidence for pigment epithelium-derived factor receptors in the neural retina. Invest Ophthalmol Vis Sci. 2001;42:3287–3293. [PubMed] [Google Scholar]
- 23.Bilak MM, Becerra SP, Vincent AM, Moss BH, Aymerich MS, Kuncl RW. Identification of the neuroprotective molecular region of pigment epithelium-derived factor and its binding sites on motor neurons. J Neurosc. 2002;22:9378–9386. doi: 10.1523/JNEUROSCI.22-21-09378.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Filleur S, Volz K, Nelius T, Mirochnik Y, Huang H, Zaichuk TA, Aymerich MS, Becerra SP, Yap R, Veliceasa D, Shroff EH, Volpert OV. Two functional epitopes of pigment epithelial-derived factor block angiogenesis and induce differentiation in prostate cancer. Cancer Res. 2005;65:5144–5152. doi: 10.1158/0008-5472.CAN-04-3744. [DOI] [PubMed] [Google Scholar]
- 25.Notari L, Baladron V, Aroca-Aguilar JD, Balko N, Heredia R, Meyer C, Notario PM, Saravanamuthu S, Nueda ML, Sanchez-Sanchez F, Escribano J, Laborda J, Becerra SP. Identification of a lipase-linked cell membrane receptor for pigment epithelium-derived factor. J Biol Chem. 2006;281:38022–38037. doi: 10.1074/jbc.M600353200. [DOI] [PubMed] [Google Scholar]
- 26.Champagne E, Martinez LO, Collet X, Barbaras R. Ecto-F1Fo ATP synthase/F1 ATPase: metabolic and immunological functions. Curr Opin Lipidol. 2006;17:279–284. doi: 10.1097/01.mol.0000226120.27931.76. [DOI] [PubMed] [Google Scholar]
- 27.Chi SL, Pizzo SV. Angiostatin is directly cytotoxic to tumor cells at low extracellular pH: a mechanism dependent on cell surface-associated ATP synthase. Cancer Res. 2006;66:875–882. doi: 10.1158/0008-5472.CAN-05-2806. [DOI] [PubMed] [Google Scholar]
- 28.Raisler BJ, Berns KI, Grant MB, Beliaev D, Hauswirth WW. Adeno-associated virus type-2 expression of pigmented epithelium-derived factor or Kringles 1–3 of angiostatin reduce retinal neovascularization. Proc Natl Acad Sci U S A. 2002;99:8909–8914. doi: 10.1073/pnas.122247299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wahl ML, Kenan DJ, Gonzalez-Gronow M, Pizzo SV. Angiostatin's molecular mechanism: aspects of specificity and regulation elucidated. J Cell Biochem. 2005;96:242–261. doi: 10.1002/jcb.20480. [DOI] [PubMed] [Google Scholar]
- 30.Moser TL, Kenan DJ, Ashley TA, Roy JA, Goodman MD, Misra UK, Cheek DJ, Pizzo SV. Endothelial cell surface F1-F0 ATP synthase is active in ATP synthesis and is inhibited by angiostatin. Proc Natl Acad Sci U S A. 2001;98:6656–6661. doi: 10.1073/pnas.131067798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moser TL, Stack MS, Asplin I, Enghild JJ, Hojrup P, Everitt L, Hubchak S, Schnaper HW, Pizzo SV. Angiostatin binds ATP synthase on the surface of human endothelial cells. Proc Natl Acad Sci U S A. 1999;96:2811–2816. doi: 10.1073/pnas.96.6.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veitonmaki N, Cao R, Wu LH, Moser TL, Li B, Pizzo SV, Zhivotovsky B, Cao Y. Endothelial cell surface ATP synthase-triggered caspase-apoptotic pathway is essential for k1–5-induced antiangiogenesis. Cancer Res. 2004;64:3679–3686. doi: 10.1158/0008-5472.CAN-03-1754. [DOI] [PubMed] [Google Scholar]
- 33.Arakaki N, Nagao T, Niki R, Toyofuku A, Tanaka H, Kuramoto Y, Emoto Y, Shibata H, Magota K, Higuti T. Possible role of cell surface H+ -ATP synthase in the extracellular ATP synthesis and proliferation of human umbilical vein endothelial cells. Mol Cancer Res. 2003;1:931–939. [PubMed] [Google Scholar]
- 34.Kimura Y, Baba K, Okuda H. Inhibitory effects of active substances isolated from Cassia garrettiana heartwood on tumor growth and lung metastasis in Lewis lung carcinoma-bearing mice (Part 2) Anticancer Res. 2000;20:2923–2930. [PubMed] [Google Scholar]
- 35.Burwick NR, Wahl ML, Fang J, Zhong Z, Moser TL, Li B, Capaldi RA, Kenan DJ, Pizzo SV. An Inhibitor of the F1 subunit of ATP synthase (IF1) modulates the activity of angiostatin on the endothelial cell surface. J Biol Chem. 2005;280:1740–1745. doi: 10.1074/jbc.M405947200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamagishi S, Inagaki Y, Nakamura K, Abe R, Shimizu T, Yoshimura A, Imaizumi T. Pigment epithelium-derived factor inhibits TNF-alpha-induced interleukin-6 expression in endothelial cells by suppressing NADPH oxidase-mediated reactive oxygen species generation. J Mol Cell Cardiol. 2004;37:497–506. doi: 10.1016/j.yjmcc.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 37.Gledhill JR, Montgomery MG, Leslie AG, Walker JE. Mechanism of inhibition of bovine F1-ATPase by resveratrol and related polyphenols. Proc Natl Acad Sci U S A. 2007;104:13632–13637. doi: 10.1073/pnas.0706290104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stratikos E, Alberdi E, Gettins PG, Becerra SP. Recombinant human pigment epithelium-derived factor (PEDF): characterization of PEDF overexpressed and secreted by eukaryotic cells. Protein Sci. 1996;5:2575–2582. doi: 10.1002/pro.5560051220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mueller DM, Puri N, Kabaleeswaran V, Terry C, Leslie AG, Walker JE. Ni-chelate-affinity purification and crystallization of the yeast mitochondrial F1-ATPase. Protein Expr Purif. 2004;37:479–485. doi: 10.1016/j.pep.2004.06.035. [DOI] [PubMed] [Google Scholar]
- 40.Shao R, Guo X. Human microvascular endothelial cells immortalized with human telomerase catalytic protein: a model for the study of in vitro angiogenesis. Biochem Biophys Res Commun. 2004;321:788–794. doi: 10.1016/j.bbrc.2004.07.033. [DOI] [PubMed] [Google Scholar]
- 41.Alberdi E, Hyde CC, Becerra SP. Pigment epithelium-derived factor (PEDF) binds to glycosaminoglycans: analysis of the binding site. Biochemistry. 1998;37:10643–10652. doi: 10.1021/bi9802317. [DOI] [PubMed] [Google Scholar]
- 42.Meyer C, Notari L, Becerra SP. Mapping the type I collagen-binding site on pigment epithelium-derived factor. Implications for its antiangiogenic activity. J Biol Chem. 2002;277:45400–45407. doi: 10.1074/jbc.M208339200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.