Abstract
Obesity is a heritable trait and a major risk factor for highly prevalent common diseases such as hypertension, cardiac diseases and type 2 diabetes. Obesity is a major public health concern worldwide. Previously we showed that BMI was positively correlated with African ancestry among the African American (AA) participants in the NHLBI’s Family Blood Pressure Program (FBPP). Using Individual Ancestry (IA) estimates at 284 marker locations across the genome, we now present a Quantitative Admixture Mapping (QAM) analysis of body mass index (BMI) in the same population. We used a set of unrelated individuals from Nigeria to represent the African ancestral population and the European Americans in the FBPP as the European ancestral population. The analysis was based on a common set of 284 microsatellite markers genotyped in all three groups. We considered the quantitative trait, BMI, as the response variable in a regression analysis with the marker location specific excess European ancestry as the explanatory variable. After suitably adjusting for different covariates such as sex, age and study center, we found strong evidence for a positive association with European ancestry at chromosome locations 3q29 and 5q14 and a negative association on chromosome 15q26. These results suggest that these regions may harbor genes influencing BMI in the AA population.
Introduction
Although obesity is an individual clinical condition, it is increasingly viewed as a serious and growing worldwide public health problem. Obesity is believed to predispose to all the major killer diseases such as type 2 diabetes, cardiac disorders, hypertension, stroke, metabolic diseases and even some forms of cancer1. The prevalence of obesity has been continually rising for two decades2. A common and inexpensive surrogate to measure obesity is body mass index (BMI), defined as the ratio of weight in kilograms to squared height in meters. Another advantage to analyzing BMI is that it is a continuously measured trait, which generally provides greater power in searching for explanatory covariates than overweight or obesity defined by arbitrary cutoffs.
Though it is largely believed that excessive nutrient intake and a sedentary lifestyle of the developed world has been the major cause behind the obesity epidemic3, other factors including genetic predisposition are also deemed responsible. Genetic factors contribute significantly to obesity4,5, with heritability estimates of BMI ranging from 30 to 70%6–9. There have been numerous efforts to identify genes and chromosomal regions responsible for BMI, using genome-wide linkage and association analysis10. While a number of genes with rare mutations are known to lead to increased BMI and obesity, such as the melanocortin 4 receptor gene11, most recently convincing evidence for the role of a common polymorphism in the FTO gene with a modest effect on BMI has been presented12.
New world admixed populations provide unique opportunities for genetic admixture mapping studies 13–21. The AA population of the United States is typically represented by admixture of European and African ancestral genomes in different proportions with some spatial variation22–24. Several studies have examined the correlation between European (or African) ancestry in African Americans and BMI or obesity24–27. One study found a positive association between African ancestry and BMI25, another a positive association of European ancestry with obesity related traits26, and a third no correlation of ancestry with BMI27. In a previous analysis of the AA participants in the Family Blood Pressure Program (FBPP)28, we found a positive correlation between BMI and individual African ancestry estimated from genome-wide microsatellite markers24. In the current study, we now present results of an analysis examining the correlation of BMI with estimated ancestry proportions at each of 284 marker loci among 1344 unrelated AA subjects from the same FBPP population, in a search for potential locus-specific effects.
Materials and Methods
Subjects
The FBPP is a large multicenter genetic study of high blood pressure and related conditions in multiple racial/ethnic groups, including European Americans (EA), African Americans, Mexican Americans and Asians and Asian Americans. It includes four component networks: GenNet, GENOA, HyperGEN and SAPPHIRE. GenNet, GENOA and HyperGEN independently collected samples from EA and AA families. GenNet sampled AA and EA nuclear families in Maywood, Illinois and Tecumseh, Michigan, respectively, through identification of a young middle-aged proband with elevated blood pressure. GENOA sampled AA sibships containing sibling pairs with hypertension from Jackson, Mississippi and EA sibships with an affected proband from Rochester, Minnesota. HyperGEN recruited AA and EA hypertensive siblings and random unrelated individuals from five field centres (AA from Birmingham, Alabama and Forsyth County, North Carolina; EA from Salt Lake City, Utah, Minneapolis, Minnesota, Framingham, Massachusetts, and Forsyth County, North Carolina).
All the individuals we included in the study were unrelated AA from field centers of GenNet, GENOA and HyperGEN. Race/ethnicity information was obtained by self-description. To maximize the number of unrelated individuals in our sample, whenever possible we selected unrelated founder individuals, otherwise one randomly selected individual per family. Our final sample of 1344 individuals consisted of 280 individuals who were sampled by the GenNet network, 349 individuals sampled by the GENOA network and 715 individuals sampled by the HyperGEN network.
Genotyping
DNA was extracted from whole blood by standard methods by each of the four FBPP networks and was sent to the US National Heart, Lung, and Blood Institute(NHLBI)’s Mammalian genotyping service in Marshfield, Wisconsin, for genotyping. Screening set 8 (372 highly polymorphic microsattelite markers with an average map distance of 10 cM) was used for all four networks.
Statistical Analysis
We used the computer program Structure29 to estimate genome-wide, as well as site-specific ancestries in all African American participants. The linkage model was used, with genetic distance between markers specified according to the Marshfield map. In each analysis, the MCMC algorithm was run for 100,000 steps of burn-in followed by another 100,000 steps.
For the analysis of 1344 FBPP African Americans, we assumed a two-ancestral populations model. We also included 1378 unrelated non-Hispanic white participants from the FBPP as well as 127 African individuals from the Human Genome Diversity Project (HGDP)30. This latter set of individuals had been genotyped at more than three hundred STRs at the time of our analysis, and we included genotypes at 284 markers which were also genotyped in the FBPP individuals.
At each locus we calculated, for each individual, an ancestry deviation defined as the estimated ancestry at that location minus the background ancestry estimated from the genome-wide markers for that individual.
Specifically, let be the locus-specific ancestry of individual i (i = 1, 2, …, N) at marker locus l (l=1, 2, …, L) estimated from Structure. We compute the overall (genomewise) individual admixture ( ) for the individual i as the average individual admixture per locus:
We computed the ancestry difference at each locus, using the genomewide IA as baseline. Specifically for individual i, the ancestry difference for ancestral population k at marker l is defined as:
Note that we have dropped the suffix k pertaining to the population from which the ancestry coefficients are derived. The variable was then used as the primary dependent variable in a linear regression model with BMI (transformed) as the dependent variable. Age, sex and network were also included as covariates in this analysis, if significant. The standardized regression coefficient of xli, defined as , is distributed as asymptotically normal and was used to assess statistical significance.
To account for multiple testing (284 markers) we performed a permutation analysis in which we randomly reassigned the genetic ancestry estimates for the 284 marker locations to individuals, whose BMI and covariate data remained intact. This procedure preserved the correlation structure of the markers and the correlation structure of BMI and covariates, but dissociated the relationship between the markers and phenotypes. For each permuted data set, we performed the same regression analysis of BMI on excess ancestry at each marker location, as was done for the original data, and obtained the most extreme values (positive and negative) of rl (the Z-score statistics). One thousand permutations were performed. To derive P values adjusted for multiple testing, we determined the percentage of times out of 1,000 permutations that an observed value of rl was exceeded in the permuted data analysis.
Results
Sample demographics are given in Table 1. There were 878 males and 466 females. The subjects from the GenNet network were the youngest (average age 41) and subjects from GENOA the oldest (average age 60), while the HyperGen subjects were in between (average age 51). Average BMIs were generally higher in males than females, and there was little variation in BMI among networks. Average European ancestry varied modestly among the three networks, as was previously observed23.
Table 1.
Demographics of study sample.
| GenNet | GENOA | HyperGEN | ||||
|---|---|---|---|---|---|---|
| Female | Male | Female | Male | Female | Male | |
| Number of Individuals | 116 | 164 | 108 | 241 | 242 | 473 |
| Average(BMI) | 27.38 ± 6.4 | 31.71 ± 8.6 | 29.37 ± 4.6 | 31.67 ± 7.3 | 29.34 ± 6.6 | 34.28 ± 8.3 |
| Average(Age) | 40.75 ± 11.8 | 41.97 ± 12.6 | 60.85 ± 9.1 | 59.71 ± 9.8 | 50.83 ± 13.2 | 50.51 ± 12.6 |
| European Ancestry (%) | 13.87 | 13.48 | 14.76 | 14.86 | 13.06 | 12.53 |
The distribution of BMI for the 1344 unrelated individuals in this study was positively skewed. Neither the 1/BMI or the Log(BMI) transformation, generally found in the literature, provided a satisfactory normalization of the data. However, a loglog transformation of BMI did make the distribution normal (Supplementary Figure 1). The trait LLBMI, which was defined as the loglog transformation of the original BMI, was strongly affected by sex and study center but not age (or age2) when tested by analysis of variance (Table 2).
Table 2.
Analysis of variance results on Log(log(BMI)).
| df | Sum Sq | Mean Sq | F value | P | |
|---|---|---|---|---|---|
| Age | 1 | 0.0045 | 0.0045 | 1.0493 | 0.30585 |
| Sex | 1 | 0.4082 | 0.4082 | 95.0617 | <2.2 × 10−16 |
| Network | 2 | 0.1116 | 0.0558 | 12.9926 | 2.579 × 10−6 |
| Sex × Network | 2 | 0.0336 | 0.0168 | 3.9140 | 0.02019 |
| Residuals | 1337 | 5.7415 | 0.0043 |
After adjusting the LLBMI values for sex, study network and the interaction between sex and network, we regressed it on excess European Ancestry (xl) at each locus l (l= 1 to 284). The ratio (rl) of the estimated slope of the regression (bl) divided by its standard error (sl) is asymptotically normal. We looked at regions with high absolute value of rl. A positive value of the gradient bl (and hence rl) at the marker locus l implies that BMI is positively correlated with excess European ancestry (and negatively correlated with African ancestry) at that locus while a negative value of rl implies that BMI is negatively correlated with European Ancestry (and positively correlated with African ancestry) at that locus.
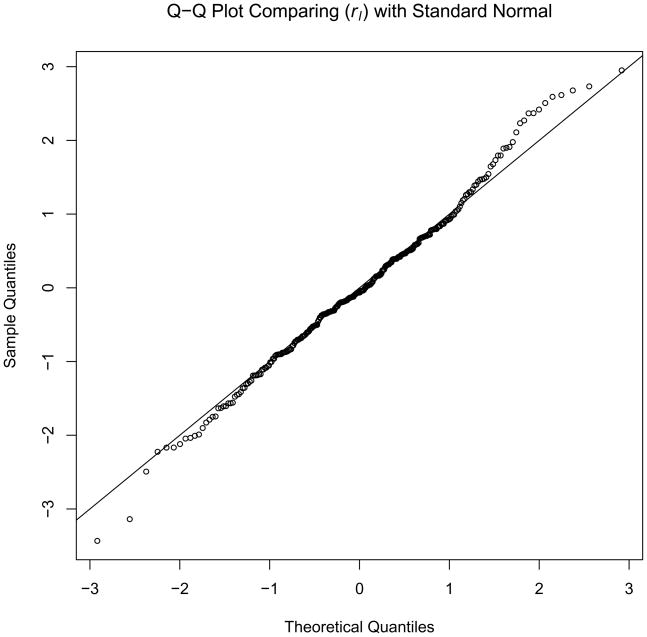
A Quantile-Quantile (Q-Q) plot of the rl values against a normal distribution reveals that the fit is good except for the tails. There is a significant bulge in the right tail and at least 3 points in the left tail of the distribution also appear to be outliers (Figure 1). Table 3 lists all markers for which the absolute value of the ratio rl was larger than 1.96 (corresponding to a two-sided unadjusted P value of .05). There are 11 points in the left tail and 13 in the right tail with absolute values larger than 1.96, compared to 7 expected in each tail by chance. The three most extreme points in the left tail of the distribution are three consecutive markers, all from chromosome region 15q25.3–26.2. The next locus lying posterior to 15q26.2 at 15q26.3 (D15S642 or GATA27A03) also has a low rl value of −2.12. The points that constitute the bulge in the right tail of the distribution are markers primarily from chromosome regions 3q28–29 and 5q14–23. There are also several points from the region 16p11.2–13.1.
Figure 1.
Q-Q plot comparing rl with a standard normal distribution.
Table 3.
Marker locations with the most significant regression on ancestry.
| DNAME | Location and Distance(cM) | Estimated (bl) | Estimated (rl) | Marker Names |
|---|---|---|---|---|
| Left Tail | ||||
| D15S816 | 15q26.2 (100.59) | −5.784 | −3.433 | GATA73F01 |
| D15S652 | 15q26.1 (90.02) | −3.572 | −3.139 | ATA24A08 |
| D15S655 | 15q25.3 (82.84) | −2.559 | −2.493 | ATA28G05 |
| Unknown | 1q32.2 (226.16) | −4.027 | −2.225 | GATA124F08 |
| D1S235 | 1q42.3 (254.64) | −1.632 | −2.168 | AFM203YG9 |
| D1S3462 | 1q42.2 (247.23) | −2.081 | −2.167 | ATA29C07 |
| D15S642 | 15q26.3 (122.14) | −3.235 | −2.120 | GATA27A03 |
| D4S1652 | 4q35.2 (208.07) | −2.236 | −2.046 | GATA5B02 |
| D1S549 | 1q41 (239.66) | −2.423 | −2.036 | GATA4H09 |
| D7S1818 | 7p12.3 (69.56) | −2.865 | −2.008 | GATA24D12 |
| D7S817 | 7p14.3 (50.29) | −2.512 | −1.990 | GATA13G11 |
| Right Tail | ||||
| D5S1480 | 5q32 (147.49) | 5.320 | 1.977 | ATA23A10 |
| D5S1501 | 5q14.1 (85.25) | 2.438 | 2.109 | GATA52A12 |
| D16S769 | 16p12.1 (50.6) | 2.756 | 2.233 | GATA71H05 |
| D16S753 | 16p11.2 (57.79) | 3.135 | 2.270 | GGAA3G05 |
| D16S748 | 16p13.13 (22.65) | 2.464 | 2.367 | ATA3A07 |
| D5S1725 | 5q14.3 (97.82) | 1.555 | 2.369 | GATA89G08 |
| D16S764 | 16p13.11 (29.97) | 2.815 | 2.418 | GATA42E11 |
| D5S1505 | 5q23.1 (129.83) | 8.552 | 2.507 | GATA62A04 |
| D5S816 | 5q31.1–31.2 (139.33) | 7.821 | 2.591 | GATA2H09 |
| D11S1984 | 11p15.5 (2.11) | 4.273 | 2.615 | GGAA17G05 |
| D3S2418 | 3q28–29 (215.84) | 4.325 | 2.678 | ATA22E01 |
| D5S2501 | 5q22.1 (116.98) | 5.953 | 2.733 | GATA68A03 |
| D3S1311 | 3q29 (224.88) | 4.282 | 2.950 | AFM254VE1 |
To check the significance of our findings we ran a permutation test in which we randomly assigned the adjusted LLBMI values with covariates to an individual, and regressed it on the locus specific excess European Ancestry (xl) (see Methods). In 1000 permutations, the minimum of 284 rl scores only once crossed −3.14 and never crossed −3.43. The maximum also crossed 2.95 only once. Hence the results associated with the markers D15S816, D15S652 and D3S1311 have empirical adjusted P values less than .002. We have done some additional analyses to show that these values are not due to ‘outlier’ effects. Specifically, we looked at the scatter plot of individual excess European ancestry and LLBMI to search for outlier points with very high (or low) individual excess European Ancestry at a locus coupled with a very high (or low) BMI value, which could distort the results. However, no such ‘outlier’ points were identified (Supplementary Figure 2).
Discussion
Figure 1 reveals outliers in both tails of the ancestry Z-score distribution, but otherwise a good fit to a normal distribution. Table 4 lists 24 markers out of 284 (8.5%) compared to the expected 14 (5%). If we take |rl| > 2.5 as our cutoff instead 1.96, there are 9 (3.2%) markers above that threshold while we expect only 3.5 (1.24%). If we look at the markers in Table 3 with further detail, we find the markers are mostly clustered into 6 different regions of the genome (Table 4) at 1q, 7p and 15q (excess African ancestry) and 3q, 5q nad 16p (excess European ancestry). Eight of the 9 markers for which the rl values are larger than 2.5 (shown in bold) are from the three regions, 15q25.3–q26.3, 3q28–q29 and 5q14.1–5q32.
Table 4.
Most significant chromosomal regions showing African or European ancestry association with BMI.
| Location and Distance(cM) | DNAME | Marker Name | Estimated (rl) |
|---|---|---|---|
| 1q32.2–1q42.3 | |||
| 226.16 | Unknown | GATA124F08 | −2.225 |
| 239.66 | D1S549 | GATA4H09 | −2.036 |
| 247.23 | D1S3462 | ATA29C07 | −2.167 |
| 254.64 | D1S235 | AFM203YG9 | −2.168 |
| 3q28–29 | |||
| 215.84 | D3S2418 | ATA22E01 | 2.678 |
| 224.88 | D3S1311 | AFM254VE1 | 2.950§ |
| 5q14.1–5q32 | |||
| 85.25 | D5S1501 | GATA52A12 | 2.109 |
| 97.82 | D5S1725 | GATA89G08 | 2.369 |
| 116.98 | D5S2501 | GATA68A03 | 2.733 |
| 129.83 | D5S1505 | GATA62A04 | 2.507 |
| 139.33 | D5S816 | GATA2H09 | 2.591 |
| 147.49 | D5S1480 | ATA23A10 | 1.977 |
| 7p14.3–7p12.3 | |||
| 50.29 | D7S817 | GATA13G11 | −1.990 |
| 57.79 | D7S2846 | GATA31A10 | −1.789* |
| 69.56 | D7S1818 | GATA24D12 | −2.008 |
| 15q25.3–q26.3 | |||
| 82.84 | D15S655 | ATA28G05 | −2.493 |
| 90.02 | D15S652 | ATA24A08 | −3.139§ |
| 100.59 | D15S816 | GATA73F01 | −3.433§ |
| 122.14 | D15S642 | GATA27A03 | −2.120 |
| 16p13.1–16p11.2 | |||
| 22.65 | D16S748 | ATA3A07 | 2.367 |
| 29.97 | D16S764 | GATA42E11 | 2.418 |
| 43.89 | D16S403 | AFM049XD2 | 1.899* |
| 50.6 | D16S769 | GATA71H05 | 2.233 |
| 57.79 | D16S753 | GGAA3G05 | 2.270 |
Locus does not have |rl| > 1.96 but is listed here because it lies between two points in the same region having |rl| > 1.96.
Locus is significant after the permutation test.
Among the six identified locations, the regions 7p12.3–7p14.3 and 16p11.2–16p13.1 have the lowest statistical significance. However the region on 7p is known to harbor the growth related Growth hormone-releasing hormone receptor (GHRHR) and Isolated growth hormone deficiency (IGHD) genes. Mutations in these genes have been generally associated with lower BMI31. Mouse polygenic models of obesity studying the QTL abdominal fat has found a putative human homologue in region 7p13–p1232. It may be noted here that the rl values of the region 7p are negative, implicating a positive association of BMI with excess African ancestry at that locus. In a recent linkage study among 769 subjects from 182 families in Africa, the marker D7S817 has been linked to BMI with a LOD score of 3.8333. In another linkage study consisting of 342 families, D7S1818 was associated with a LOD score of 2.2 for BMI trends from childhood to adulthood34. The marker D16S764, which has the highest rl value of 2.41 among all the markers in the 16p region, has been shown previously to be both modestly linked (LOD 2.45) and associated (p<0.0006) with BMI35. The sample in the above study was composed of 893 white sibpairs, which may be consistent with the high positive value of rl. Several markers in this region have also been shown to be linked to BMI and other obesity related phenotypes with LOD scores ranging from 1.7 to 3.21. All these studies were reported among different populations of European descent 35–38.
The region 1q32.2–1q42.3 contains the Angiotensinogen (AGT) gene. Numerous previous studies30–41 have found association between this gene and obesity related phenotypes, including BMI. Keeping in mind the negative rl values of markers in this region (African excess), the fact that linkage of this region with obesity related phenotypes was found only among black families may be of particular interest42.
The 6 consecutive markers on chromosome 5 with high positive rl values span a large region from 78 Mb to 144 Mb (5q14.1–5q32). In a study involving 321 sibpairs the second marker in this region, D5S1725 (with an rl value of 2.37) was previously found to be linked to body fat and fat mass with LOD scores of 2.56 and 2.25 respectively43. It is worth mentioning that these findings are from a study in Western Africa. However in two different linkage studies among white families, markers in this region, including D5S1505, have been found to be linked with more direct BMI related traits34,44. Chen et al34 analyzed 342 sibships and found D5S1453 at 5q21.3 to be linked with trends in BMI from childhood to adulthood with a LOD score of 2 and D5S1505 to be linked with long term burden in BMI with a LOD score of 2.2. The French study44 was based on 447 subjects in 109 pedigrees chosen through a proband with BMI>27. They found a linkage peak with LOD score 2.68 at marker D5S1463, located at 5q14.3. Rice et al45 studied abdominal fat and high BMI, involving 453 subjects in 99 white families, and found two peaks in this region. Of these two locations, marker D5S658 at 5q31.3 had a LOD score of 2.06 and D5S1480 had a LOD score of 2.1. In another study involving 88 families, BMI was shown to be associated with polymorphisms in the gene Nuclear Receptor subfamily 3, group C, member 1 (glucocorticoid receptor) (NR3C1) at 5q31 with a p-value of 0.00946.
The chromosome 3 markers D3S2418 and D3S1311, spanning a 5 Mb region, show the strongest excess European ancestry with BMI in our study. Associations of markers in the candidate gene Apolipoprotein D (3q26.2-qter) with BMI was reported by Vijayraghaban et al47. Marker D3S1311, which was statistically significant after the permutation test, also lies in a promising linkage region (LOD score 2.5) in the study of Rice et al45. The region 15q25.3–15q26.3 harbors the 2 markers (D15S652 and D15S816) with the most significant rl values in our study. It is also the site for the gene neuromedin B (NMB) (15q22-qter) which is a candidate for type 1 diabetes, obesity and hunger disorder48. Polymorphisms in NMB have been shown to be associated with BMI and other obesity related phenotypes46. In a recent study, Bouchard et al49 fine mapped a 20-megabase region around a quantitative trait locus on chromosome 15q26 for abdominal subcutaneous fat (ASF) in an extended sample of 707 subjectsfrom 202 families from the Quebec Family Study. Chagnon et al50 studied 336 sibpairs and 609 relative pairs and found D15S652 linked with fat free mass with a LOD score of 3.56. Another marker in this region, D15S657 at 15q26.2, also had a LOD score of 2. Using a subset of the data that we have analyzed here, Lewis et al51 have reported linkage (LOD score 3) of body fat (%) with marker D15S655 among males in the HyperGEN network.
To our knowledge, this is the largest Quantitative admixture mapping effort in terms of sample size and marker locus involvement. We took care to eliminate possible errors in the locus-wise ancestry estimates, because any systematic bias could affect the final results. Statistical variation in locus-wise ancestry estimates was kept to a minimum by running the MCMC for a long period. To further check the robustness of our results, we also examined results of analyses based on different random selections of unrelated individuals from the AA families and obtained very similar results. Overall our findings are encouraging and provide regions for follow up analyses of genes influencing BMI in these and other African American families.
Supplementary Material
Figure S1. Histograms and normal Q-Q plots of BMI, Log(BMI) and Log(log(BMI)).
Figure S2. Scatter plots of excess European ancestry at three loci versus Log(log(BMI)).
References
- 1.U.S. Dept. of Health and Human Services, National Institutes of Health. Clinical Guidelines on the Identification, Evaluation, and Treatment of Overweight and Obesity in Adults: The Evidence Report. NHLBI document 98–4083 2000 [Google Scholar]
- 2.Centers for Disease Control and Prevention. US Obesity Trends. 1984–2002. [Google Scholar]
- 3.Bleich S, Cutler D, Murray C, Adams A. National Bureau of Economic Research Working Paper No. 12954. 2007. Why is the Developed World Obese? [Google Scholar]
- 4.Friedman JM. Modern science versus the stigma of obesity. Nature Medicine. 2004;10:563–569. doi: 10.1038/nm0604-563. [DOI] [PubMed] [Google Scholar]
- 5.Lyon HN, Hirschhorn JN. Genetics of common forms of obesity: a brief overview. Am J Clin Nutr. 2005;82:215S–217S. doi: 10.1093/ajcn/82.1.215S. [DOI] [PubMed] [Google Scholar]
- 6.Bell CG, Walley AJ, Froguel P. The genetics of human obesity. Nat Rev Genet. 2005;6:221–234. doi: 10.1038/nrg1556. [DOI] [PubMed] [Google Scholar]
- 7.Farooqi IS, O’Rahilly S. Genetic factors in human obesity. Obes Rev. 2007;1:37–40. doi: 10.1111/j.1467-789X.2007.00315.x. [DOI] [PubMed] [Google Scholar]
- 8.Hebebrand J, Friedel S, Schauble N, Geller F, Hinney A. Perspectives: molecular genetic research in human obesity. Obes Rev. 2003;4:139–146. doi: 10.1046/j.1467-789x.2003.00106.x. [DOI] [PubMed] [Google Scholar]
- 9.Schousboe K, Willemsen G, Kyvik KO, Mortensen J, Boomsma DI, Cornes BK, Davis CJ, Fagnani C, Hjelmborg J, Kaprio J, De Lange M, Luciano M, Martin NG, Pedersen N, Pietiläinen KH, Rissanen A, Saarni S, Sørensen TI, Van Baal GC, Harris JR. Sex Differences in Heritability of BMI: A Comparative Study of Results from Twin Studies in Eight Countries. Twin Res. 2003;6:409–421. doi: 10.1375/136905203770326411. [DOI] [PubMed] [Google Scholar]
- 10.Rankinen T, Zuberi A, Chagnon YC, Weisnagel SJ, Argyropoulos G, Walts B, Perusse L, Bouchard C. The human obesity gene map: the 2005 update. Obesity. 2006;14:529–644. doi: 10.1038/oby.2006.71. [DOI] [PubMed] [Google Scholar]
- 11.Vaisse C, Clément K, Durand E, Hercberg S, Guy-Grand B, Froguel P. Melanocortin-4 receptor mutations are a frequent and heterogeneous cause of morbid obesity. J Clin Invest. 2000;106:253–262. doi: 10.1172/JCI9238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW, Shields B, Harries LW, Barrett JC, Ellard S, Groves CJ, Knight B, Patch AM, Ness AR, Ebrahim S, Lawlor DA, Ring SM, Ben-Shlomo Y, Jarvelin MR, Sovio U, Bennett AJ, Melzer D, Ferrucci L, Loos RJ, Barroso I, Wareham NJ, Karpe F, Owen KR, Cardon LR, Walker M, Hitman GA, Palmer CN, Doney AS, Morris AD, Smith GD, Hattersley AT, McCarthy MI. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Risch N. Mapping genes for complex diseases using association studies in recently admixed populations. Am J Hum Genet. 1992;51(suppl):13. [Google Scholar]
- 14.Stephens JC, Briscoe D, O’Brien SJ. Mapping by admixture linkage disequilibrium in human populations: limits and guidelines. Am J Hum Genet. 1994;55:809–824. [PMC free article] [PubMed] [Google Scholar]
- 15.Chakraborty R, Weiss KM. Admixture as a tool for finding linked genes and detecting that difference from allelic association between loci. Proc Natl Acad Sci U S A. 1988;85:9119–23. doi: 10.1073/pnas.85.23.9119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith MW, Lautenberger JA, Shin HD, Chretien JP, Shrestha S, Gilbert DA, O’Brien SJ. Markers for mapping by admixture linkage disequilibrium in African American and Hispanic populations. Am J Hum Genet. 2001;69:1080–1094. doi: 10.1086/323922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith MW, Smith MW, Patterson N, Lautenberger JA, Truelove AL, McDonald GJ, Waliszewska A, Kessing BD, Malasky MJ, Scafe C, Le E, De Jager PL, Mignault AA, Yi Z, De The G, Essex M, Sankale JL, Moore JH, Poku K, Phair JP, Goedert JJ, Vlahov D, Williams SM, Tishkoff SA, Winkler CA, De La Vega FM, Woodage T, Sninsky JJ, Hafler DA, Altshuler D, Gilbert DA, O’Brien SJ, Reich D. A high-density admixture map for disease gene discovery in African Americans. Am J Hum Genet. 2004;74:1001–1013. doi: 10.1086/420856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoggart CJ, Shriver MD, Kittles RA, Clayton DG, McKeigue PM. Design and analysis of admixture mapping studies. Am J Hum Genet. 2004;74:965–978. doi: 10.1086/420855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKeigue PM. Mapping genes that underlie ethnic differences in disease risk: methods for detecting linkage in admixed populations, by conditioning on parental admixture. Am J Hum Genet. 1998;63:241–251. doi: 10.1086/301908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patterson N, Hattangadi N, Lane B, Lohmueller KE, Hafler DA, Oksenberg JR, Hauser SL, Smith MW, O’Brien SJ, Altshuler D, Daly MJ, Reich D. Methods for high-density admixture mapping of disease genes. Am J Hum Genet. 2004;74:979–1000. doi: 10.1086/420871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parra EJ, Kittles RA, Argyropoulos G, Pfaff CL, Hiester K, Bonilla C, Sylvester N, Parrish-Gause D, Garvey WT, Jin L, McKeigue PM, Kamboh MI, Ferrell RE, Pollitzer WS, Shriver MD. Ancestral proportions and admixture dynamics in geographically defined African Americans living in South Carolina. Am J Phys Anthropol. 2001;114:18–29. doi: 10.1002/1096-8644(200101)114:1<18::AID-AJPA1002>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 22.Parra EJ, Marcini A, Akey J, Martinson J, Batzer MA, Cooper R, Forrester T, Allison DB, Deka R, Ferrell RE, Shriver MD. Estimating African American admixture proportions by use of population-specific alleles. Am J Hum Genet. 1998;63:1839–1851. doi: 10.1086/302148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang H, Jorgenson E, Gadde M, Kardia SL, Rao DC, Zhu X, Schork NJ, Hanis CL, Risch N. Racial admixture and its impact on BMI and blood pressure in African and Mexican Americans. Hum Genet. 2006;119:624–33. doi: 10.1007/s00439-006-0175-4. [DOI] [PubMed] [Google Scholar]
- 24.Fernandez JR, Shriver MD, Beasley TM, Rafla-Demetrious N, Parra E, Albu J, Nicklas B, Ryan AS, McKeigue PM, Hoggart CL, Weinsier RL, Allison DB. Association of African genetic admixture with resting metabolic rate and obesity among women. Obes Res. 2003;11:904–11. doi: 10.1038/oby.2003.124. [DOI] [PubMed] [Google Scholar]
- 25.Schaffer JR, Kammerer CM, Reich D, McDonald G, Patterson N, Goodpaster B, Bauer DC, Li J, Newman AB, Cauley JA, Harris TB, Tylavsky F, Ferrell RE, Zmuda JM. Genetic markers for ancestry are correlated with body composition traits in older African Americans. Osteoporos Int. 2007;18:733–41. doi: 10.1007/s00198-006-0316-6. [DOI] [PubMed] [Google Scholar]
- 26.Reiner AP, Carlson CS, Ziv E, Iribarren C, Jaquish CE, Nickerson DA. Genetic ancestry, population sub-structure, and cardiovascular disease-related traits among African-American participants in the CARDIA Study. Hum Genet. 2007;121:565–75. doi: 10.1007/s00439-007-0350-2. [DOI] [PubMed] [Google Scholar]
- 27.FBPP Investigators. Multi-center genetic study of hypertension: The Family Blood Pressure Program (FBPP) Hypertension. 2002;39:3–9. doi: 10.1161/hy1201.100415. [DOI] [PubMed] [Google Scholar]
- 28.Zhu X, Luke A, Cooper RS, Quertermous T, Hanis C, Mosley T, Gu CC, Tang H, Rao DC, Risch N, Weder A. Admixture mapping for hypertension loci with genome-scan markers. Nat Genet. 2005;37:177–81. doi: 10.1038/ng1510. [DOI] [PubMed] [Google Scholar]
- 29.Pereira RMC, Aguiar-Oliveira MH, Sagazio A, Oliveira CRP, Oliveira FT, Campos VC, Farias CT, Vicente TAR, Gois MB, Oliveira JLM, Marques-Santos C, Rocha IES, Barreto-Filho JAS, Salvatori R. Heterozygosity for a Mutation in the Growth Hormone-Releasing Hormone Receptor Gene Does Not Influence Adult Stature, But Affects Body Composition. J Clin Endocrinol Metab. 2007;92:2353–2357. doi: 10.1210/jc.2007-0092. [DOI] [PubMed] [Google Scholar]
- 30.Brockmann GA, Haley CS, Renne U, Knott SA, Schwerin M. Quantitative trait loci affecting body weight and fatness from a mouse line selected for extreme high growth. Genetics. 1998;150:369–381. doi: 10.1093/genetics/150.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adeyemo A, Luke A, Cooper R, Wu X, Tayo B, Zhu X, Rotimi C, Bouzekri N, Ward R. A genome-wide scan for body mass index among Nigerian families. Obes Res. 2003;11:266–273. doi: 10.1038/oby.2003.40. [DOI] [PubMed] [Google Scholar]
- 32.Chen W, Li S, Cook NR, Rosner BA, Srinivasan SR, Boerwinkle E, Berenson GS. An autosomal genome scan for loci influencing longitudinal burden of body mass index from childhood to young adulthood in white sibships: The Bogalusa Heart Study. Int J Obes Relat Metab Disord. 2004;28:462–469. doi: 10.1038/sj.ijo.0802610. [DOI] [PubMed] [Google Scholar]
- 33.Gorlova O, Amos C, Wang N, Shete S, Turner S, Boerwinkle E. Genetic linkage and imprinting effects on body mass index in children and young adults. Eur J Hum Genet. 2003;11:425–432. doi: 10.1038/sj.ejhg.5200979. [DOI] [PubMed] [Google Scholar]
- 34.Hsueh WC, Mitchell BD, Schneider JL, St Jean PL, Pollin TI, Ehm MG, Wagner MJ, Burns DK, Sakul H, Bell CJ, Shuldiner AR. Genome-wide scan of obesity in the Old Order Amish. J Clin Endocrinol Metab. 2001;86:1199–1205. doi: 10.1210/jcem.86.3.7358. [DOI] [PubMed] [Google Scholar]
- 35.Geller F, Dempfle A, Gorg T. Genome scan for body mass index and height in the Framingham Heart Study. BMC Genet. 2003;4(Suppl 1):S91. doi: 10.1186/1471-2156-4-S1-S91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meyre D, Lecoeur C, Delplanque J, Francke S, Vatin V, Durand E, Weill J, Dina C, Froguel P. A genome-wide scan for childhood obesity-associated traits in French families shows significant linkage on chromosome 6q22.31–q23.2. Diabetes. 2004;53:803–811. doi: 10.2337/diabetes.53.3.803. [DOI] [PubMed] [Google Scholar]
- 37.Chaves FJ, Giner V, Corella D, Pascual J, Marin P, Armengod ME, Redon J. Body weight changes and the A-6G polymorphism of the angiotensinogen gene. Int J Obes Relat Metab Disord. 2002;26:1173–1178. doi: 10.1038/sj.ijo.0802064. [DOI] [PubMed] [Google Scholar]
- 38.Hegele RA, Brunt JH, Connelly PW. Genetic variation on chromosome 1 associated with variation in body fat distribution in men. Circulation. 1995;92:1089–1093. doi: 10.1161/01.cir.92.5.1089. [DOI] [PubMed] [Google Scholar]
- 39.Prat-Larquemin L, Oppert JM, Clement K, Prat-Larquemin L, Oppert JM, Clement K, Hainault I, Basdevant A, Guy-Grand B, Quignard-Boulangé A. Adipose angiotensinogen secretion, blood pressure, and AGT M235T polymorphism in obese patients. Obes Res. 2004;12:556–561. doi: 10.1038/oby.2004.63. [DOI] [PubMed] [Google Scholar]
- 40.Collaku A, Rankinen T, Rice T, Collaku A, Rankinen T, Rice T, Leon AS, Rao DC, Skinner JS, Wilmore JH, Bouchard C. A genome-wide linkage scan for dietary energy and nutrient intakes: the health, risk factors, exercise training, and genetics (HERITAGE) family study. Am J Clin Nutr. 2004;79:881–886. doi: 10.1093/ajcn/79.5.881. [DOI] [PubMed] [Google Scholar]
- 41.Chen G, Adeyemo AA, Johnson T, Zhou J, Amoah A, Owusu S, Acheampong J, Agyenim-Boateng K, Eghan BA, Oli J, Okafor G, Abbiyesuku F, Dunston GM, Chen Y, Collins F, Rotimi C. A genome-wide scan for quantitative trait loci linked to obesity phenotypes among West Africans. Int J Obes (Lond) 2005;29:255–259. doi: 10.1038/sj.ijo.0802873. [DOI] [PubMed] [Google Scholar]
- 42.Bell C, Benzinou M, Siddiq A, Lecoeur C, Dina C, Lemainque A, Clément K, Basdevant A, Guy-Grand B, Mein CA, Meyre D, Froguel P. Genome-wide linkage analysis for severe obesity in French Caucasians finds significant susceptibility locus on chromosome 19q. Diabetes. 2004;53:1857–1865. doi: 10.2337/diabetes.53.7.1857. [DOI] [PubMed] [Google Scholar]
- 43.Rice T, Chagnon YC, Perusse L, Borecki IB, Ukkola O, Rankinen T, Gagnon J, Leon AS, Skinner JS, Wilmore JH, Bouchard C, Rao DC. A genomewide linkage scan for abdominal subcutaneous and visceral fat in black and white families: The HERITAGE Family Study. Diabetes. 2002;51:848–855. doi: 10.2337/diabetes.51.3.848. [DOI] [PubMed] [Google Scholar]
- 44.Clement K, Philippi A, Jury C, Pividal R, Hager J, Demenais F, Basdevant A, Guy-Grand B, Froguel P. Candidate gene approach of familial morbid-obesity: linkage analysis of the glucocorticoid receptor gene. Int J Obes Relat Metab Disord. 1996;20:507–512. [PubMed] [Google Scholar]
- 45.Vijayaraghavan S, Hitman GA, Kopelman PG. Apolipoprotein D polymorphism: a genetic marker of obesity and hyperinsulinemia. J Clin Endocrinol Metab. 1994;79:568–570. doi: 10.1210/jcem.79.2.7913935. [DOI] [PubMed] [Google Scholar]
- 46.Bouchard L, Drapeau V, Provencher V, Lemieux S, Chagnon Y, Rice T, Rao DC, Vohl MC, Tremblay A, Bouchard C, Pérusse L. Neuromedin beta: a strong candidate gene linking eating behaviors and susceptibility to obesity. Am J Clin Nutr. 2004;80:1478–1486. doi: 10.1093/ajcn/80.6.1478. [DOI] [PubMed] [Google Scholar]
- 47.Bouchard L, Bouchard C, Chagnon YC, Perusse L. Evidence of Linkage and Association with Body Fatness and Abdominal Fat on Chromosome 15q26. Obesity. 2007;15:2061–2070. doi: 10.1038/oby.2007.245. [DOI] [PubMed] [Google Scholar]
- 48.Chagnon YC, Borecki IB, Perusse L, Roy S, Lacaille M, Chagnon M, Ho-Kim MA, Rice T, Province MA, Rao DC, Bouchard C. Genome-wide search for genes related to the fat-free body mass in the Quebec family study. Metabolism. 2000;49:203–207. doi: 10.1016/s0026-0495(00)91299-x. [DOI] [PubMed] [Google Scholar]
- 49.Lewis CE, North KE, Arnett D, Borecki IB, Coon H, Ellison RC, Hunt SC, Oberman A, Rich SS, Province MA, Miller MB. Sex-specific findings from a genome-wide linkage analysis of human fatness in non-Hispanic whites and African Americans: the HyperGEN study. Int J Obes (Lond) 2005;29:639–649. doi: 10.1038/sj.ijo.0802916. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Histograms and normal Q-Q plots of BMI, Log(BMI) and Log(log(BMI)).
Figure S2. Scatter plots of excess European ancestry at three loci versus Log(log(BMI)).