Bimolecular fluorescence complementation assays allow the visualization of the homotypic and heterotypic GTPase interactions in vivo. The Rsr1 homotypic interaction involves its polybasic region and depends on its GDP-GTP exchange factor. Dimerization of GTPases may be an efficient mechanism to set up cellular asymmetry.
Abstract
Cell polarization occurs along a single axis that is generally determined in response to spatial cues. In budding yeast, the Rsr1 GTPase and its regulators direct the establishment of cell polarity at the proper cortical location in response to cell type–specific cues. Here we use a combination of in vivo and in vitro approaches to understand how Rsr1 polarization is established. We find that Rsr1 associates with itself in a spatially and temporally controlled manner. The homotypic interaction and localization of Rsr1 to the mother-bud neck and to the subsequent division site are dependent on its GDP-GTP exchange factor Bud5. Analyses of rsr1 mutants suggest that Bud5 recruits Rsr1 to these sites and promotes the homodimer formation. Rsr1 also exhibits heterotypic interaction with the Cdc42 GTPase in vivo. We show that the polybasic region of Rsr1 is necessary for the efficient homotypic and heterotypic interactions, selection of a proper growth site, and polarity establishment. Our findings thus suggest that dimerization of GTPases may be an efficient mechanism to set up cellular asymmetry.
INTRODUCTION
In diverse organisms, the small-molecular-weight GTPases function as key signaling molecules in polarity development (Etienne-Manneville, 2004; Raftopoulou and Hall, 2004). One of the fundamental questions is how these GTPases are locally activated. In the budding yeast Saccharomyces cerevisiae, selection of a cortical site for growth, referred to as bud-site selection, determines the axis of cell polarization. A proper bud site is determined by the Rsr1 GTPase module, which includes the Ras-type GTPase Rsr1 (also known as Bud1), its GDP-GTP exchange factor (GEF) Bud5 and its GTPase-activating protein (GAP) Bud2 (Bender and Pringle, 1989; Chant et al., 1991; Chant and Herskowitz, 1991; Bender, 1993; Park et al., 1993). The Rsr1 GTPase module is thus thought to link spatial landmarks to polarity establishment {see Park and Bi, 2007 and references therein}.
Previous studies have identified numerous interactions between RSR1 and genes involved in polarity establishment. Overexpression of RSR1 suppresses the temperature-sensitive growth of the cdc24 and cdc42 mutants that are specifically defective in polarity establishment (Bender and Pringle, 1989; Kozminski et al., 2003). Rsr1 interacts with the Rho-type GTPase Cdc42 and its exchange factor Cdc24 in a GTP-dependent manner (Zheng et al., 1995; Park et al., 1997; Kozminski et al., 2003). RSR1 also exhibits a genetic interaction with GIC1 and GIC2 (GTPase interactive components 1 and 2), which encode two closely related proteins that interact with Cdc42. Cells lacking both Gic1 and Gic2 also exhibit temperature-sensitive defects in polarity establishment including perturbed actin cytoskeleton organization (Brown et al., 1997; Chen et al., 1997). Cells deleted for RSR1, GIC1, and GIC2 cannot undergo bud emergence (Kawasaki et al., 2003), suggesting a shared role of Rsr1 and Gic1/Gic2 in polarity establishment. Thus, Rsr1 may function not only in guiding Cdc42 or its regulators to the proper growth site, but also in regulating polarity establishment through multiple protein–protein interactions.
The Rsr1 GTPase cycle appears to be critical for its role in bud-site selection and polarity establishment. Cells deleted for BUD2 or BUD5 bud in a random manner in all cell types (Chant et al., 1991; Bender, 1993; Park et al., 1993). Similarly, the RSR1 mutant that encodes either GTP- or GDP-locked Rsr1 exhibits random bud-site selection (Ruggieri et al., 1992). Overexpression of the wild-type Rsr1, but not the GTP- or GDP-locked Rsr1, can suppress the temperature-sensitive growth of the cdc42 or gic1 gic2 mutant (Kawasaki et al., 2003; Kozminski et al., 2003). Thus, cycling of Rsr1 between the GTP- and GDP-bound states is important for its role, rather than Rsr1 functioning as a Ras-like on/off switch. However, less is understood about how and when the Rsr1 GTPase cycle is activated.
The two regulators of Rsr1, Bud2 and Bud5, are likely to play a critical role for localized activation of the Rsr1 GTPase cycle. Bud2 and Bud5 localize to the presumptive bud site in G1, but the exact localization patterns of each protein at the later stages of the cell cycle are different. Bud2 localizes to the mother-bud neck after bud emergence in the earlier stages of the cell cycle, but de-localizes during M phase (Park et al., 1999). Bud5 localizes to the bud tip after bud emergence and then to the mother-bud neck as double rings in M phase. These double rings split and become inherited by mother and daughter cells after cell division (Kang et al., 2001; Marston et al., 2001). These specific localization patterns of Bud2 and Bud5 seem to be important for proper bud-site selection, as mis-localization of these proteins caused by overexpression leads to random budding patterns (Park et al., 1999; Kang et al., 2001).
Rsr1 is distributed uniformly throughout the plasma membrane, but becomes highly concentrated at the division site and at the sites of polarized growth including the bud tips (Park et al., 2002). Although this localization pattern of Rsr1 is consistent with its role in bud-site selection and polarity establishment, Rsr1 also becomes concentrated at a single site in randomly budding cells before bud emergence. It is thus not clear which of these sites of Rsr1 localization is related to its function in bud-site selection. It is also not fully understood about how Rsr1 polarization is established. These remaining questions led us to investigate further the localization and the action of Rsr1 during yeast budding. Here we provide in vivo and in vitro evidence for Rsr1 dimerization. Our data suggest that this homotypic interaction of Rsr1 is important for its polarization and its function. Our findings thus support the idea that dimerization of GTPases is an efficient mechanism to achieve cell polarization.
MATERIALS AND METHODS
Standard methods of yeast genetics and recombinant DNA manipulation were used (Guthrie and Fink, 1991; Ausubel et al., 1999). Yeast cells were grown under standard growth conditions at 30°C unless otherwise indicated. Yeast strains used in this study are listed in Table 1. Details of plasmid constructions are described in Supplemental Methods, and plasmids used in this study are listed in Supplemental Table 1.
Table 1.
Yeast strains used in this study
| Straina | Relevant genotype | Source/comments | |
|---|---|---|---|
| HPY210# | a | his3-Δ200 leu2-Δ1 lys2-801 trp1Δ63 ura3-52 | Singh et al. (2008) |
| HPY211# | α | his3-Δ200 leu2-Δ1 lys2-801 trp1Δ63 ura3-52 | Singh et al. (2008) |
| HPY1200# | α | RSR1::YFPN-RSR1-TRP1 | Derived from HPY211b |
| HPY1213# | a | RSR1::YFPC-RSR1-TRP1 | Derived from HPY210b |
| HPY1214# | a/α | RSR1::YFPN-RSR1-TRP1/RSR1::YFPC-RSR1-TRP1 | HPY1200 × HPY1213 |
| DDY1300@ | a | his3Δ200 leu2-3,112 ura3-52 lys2-801am CDC42-LEU2 | Kozminski et al. (2000) |
| DDY1301@ | α | his3Δ200 leu2-3,112 ura3-52 lys2-801am CDC42-LEU2 | Kozminski et al. (2000) |
| DDY1326@ | a | cdc42-118D76A-LEU2 | Kozminski et al. (2000) |
| HPY1196@ | α | VN-CDC42-HIS3 | Derived from DDY1301c |
| HPY1197@ | a | VC-CDC42-kanMX6 | Derived from DDY1300d |
| HPY1198@ | a/α | VN-CDC42-HIS3/VC-CDC42-kanMX6 | HPY1196 × HPY1197 |
| HPY1209 | a/a | RSR1::YFPN-RSR1-TRP1/RSR1 CDC42/VC-CDC42-kanMX6 | HPY1197 × HPY1200 |
| HPY1215# | α | RSR1::YFPN-rsr1-7K260-264S-TRP1 | Derived from HPY211b |
| HPY1216# | a | RSR1::YFPC-rsr1-7K260-264S-TRP1 | Derived from HPY210b |
| HPY1217 | a/α | RSR1::YFPN-rsr1K260-264S-TRP1/RSR1 CDC42/VC-CDC42-kanMX6 | HPY1215 × HPY1197 |
| HPY1219# | a/α | RSR1::YFPN-RSR1-TRP1/RSR1::YFPC-rsr1-7K260-264S-TRP1 | HPY1200 × HPY1216 |
| HPY1220# | a/α | RSR1::YFPN-rsr1-7K260-264S-TRP1/RSR1::YFPC-rsr1-7K260-264S-TRP1 | HPY1215 × HPY1216 |
| HPY1256# | α | RSR1::YFPN-RSR1G12V-TRP1 | Derived from HPY211b |
| HPY1552# | a | RSR1::YFPC-RSR1G12V-TRP1 | Derived from HPY210b |
| HPY1565# | α | RSR1::YFPN-RSR1K16N-TRP1 | Derived from HPY211b |
| HPY1522# | a | RSR1::YFPC-RSR1K16N-TRP1 | Derived from HPY210b |
| HPY1568# | a/α | RSR1::YFPN-RSR1G12V-TRP1/RSR1::YFPC-RSR1G12V-TRP1 | HPY1256 × HPY1552 |
| HPY1608# | a/α | RSR1::YFPN-RSR1K16N-TRP1/RSR1::YFPC-RSR1K16N-TRP1 | HPY1565 × HPY1522 |
| HPY1570# | α | RSR1::YFPN-RSR1-TRP1 bud5Δ::URA3 | Derived from HPY1200e |
| HPY1571# | a | RSR1::YFPC-RSR1-TRP1 bud5Δ::URA3 | Derived from HPY1213e |
| HPY1575# | a/α | RSR1::YFPN-RSR1-TRP1/RSR1::YFPC-RSR1-TRP1 bud5Δ::URA3/bud5Δ::URA3 | HPY1570 × HPY1571 |
| HPY319# | a | bud5Δ::URA3 | Derived from HPY210e |
| HPY320# | α | bud5Δ::URA3 | Derived from HPY211e |
| HPY1822# | a | RSR1::YFPN-RSR1K16N-TRP1 bud5Δ::URA3 | Segregant from HPY319 × HPY1565 |
| HPY1823# | α | RSR1::YFPC-RSR1K16N-TRP1 bud5Δ::URA3 | Segregant from HPY320 × HPY1522 |
| HPY1824# | a/α | RSR1::YFPN-RSR1K16N-TRP1/RSR1::YFPC-RSR1K16N-TRP1 bud5Δ::URA3/bud5Δ::URA3 | HPY1822 × HPY1823 |
| YKT342# | α | gic1Δ::TRP1 gic2Δ::kanMX6 | Kawasaki et al. (2003) |
| YKT401# | a | rsr1Δ::HIS3 gic1Δ::TRP1 gic2Δ::kanMX6 [GIC1, CEN, URA3] | Kawasaki et al. (2003) |
| HPY1609# | a | rsr1Δ::HIS3::rsr1-7K260-264S-LEU2 gic1Δ::TRP1 gic2Δ::kanMX6 | Derived from YKT401b |
| HPY11* | α | ura3 leu2 trp1 his4 can1 | Park et al. (1993) |
| HPY12* | a | ura3 leu2 trp1 his4 can1 | Park et al. (1993) |
| HPY263* | α | rsr1Δ::URA3 | Park et al. (2002) |
| HPY401* | α | rsr1Δ::URA3::GFP-RSR1-TRP1 | Park et al. (2002) |
| HPY402* | α | rsr1Δ::URA3::YFP-RSR1-TRP1 | Park et al. (2002) |
| HPY423* | α | rsr1Δ::URA3::YFP-RSR1K16N-TRP1 | Park et al. (2002) |
| HPY588* | α | rsr1Δ::URA3::GFP-rsr1-8K260S,K261S-TRP1 | Park et al. (2002) |
| HPY589* | α | rsr1Δ::URA3::GFP-rsr1-9K263S,K264S-TRP1 | Park et al. (2002) |
| HPY621* | α | rsr1Δ::URA3::GFP-rsr1-7K260-264S-TRP1 | Park et al. (2002) |
| HPY318* | a | bud5Δ::URA3 | Park et al. (1993) |
| HPY593* | a | rsr1Δ::URA3::YFP-RSR1-TRP1 bud5Δ::URA3 | Segregant from HPY402 × HPY318 |
| HPY1612* | α | rsr1Δ::kanMX4::YFP-RSR1-TRP1 | Derived from HPY402f |
| HPY1625* | α | rsr1Δ::URA3::rsr1-7K260-264S-TRP1 | Derived from HPY263b |
| HPY1740* | α | rsr1Δ::URA3::rsr1-9K263S,K264S-TRP1 | Derived from HPY263b |
| HPY1741* | α | rsr1Δ::URA3::rsr1-8K260S,K261S-TRP1 | Derived from HPY263b |
| HPY1632* | α | rsr1Δ::kanMX4::YFP-RSR1-TRP1 TUB1-CFP-URA3 | Derived from HPY1612g |
| HPY1664* | α | rsr1Δ::kanMX4::YFP-RSR1-TRP1 bud2Δ::LEU2 TUB1-CFP-URA3 | Derived from HPY1632h |
| HPY1668* | a | rsr1Δ::kanMX4::YFP-RSR1-TRP1 bud5Δ::URA3 TUB1-CFP-URA3 | Segregant from HPY1632 × HPY593 |
| HPY23@ | α | ura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2-801 ade2-101 | Park et al. (1993) |
| HPY260@ | α | gic1Δ::TRP1 | Derived from HPY23 |
| HPY1523@ | α | gic1Δ::TRP1 gic2Δ::kanMX4 | Derived from HPY260i |
| HPY16 | a | his3-Δ1 leu2 trp1-Δ63 ura3-52 prb1-1122 pep4-3 prc1-407 | Park et al. (1993) |
| Y147 | a | cdc24-4 ura3 leu2-3,112 his3 | Bender and Pringle (1989) |
a Strains marked with # are isogenic to HPY210 (Singh et al., 2008), except as indicated; strains marked with * are isogenic to HPY11 (Park et al., 1993), except as indicated; and the background of the strains marked with @ is S288C.
b The plasmid expressing each Rsr1 fusion or rsr1 mutant in pRS304 or pRS305 (see Supplemental Methods and Supplemental Table 1) was integrated into the RSR1 locus of each strain, after digestion with BssHII.
c For expression of VN-Cdc42, the PCR fragment was generated using the plasmid pFA6a-HIS3-PRPL7B-VN (Sung and Huh, 2007; a gift from W.-K. Huh), using a primer pair of oCDC42UP (5′-CGTTATTTATTATACTATTCTATTTTCCTGAGGAGATAGGGAATTCGAGCTCGTTTAAAC-3′) and oCDC42R5VN (5′-CAGCACCATCACCGACAACAACACACTTTAGCGTTTGCATAGTACCACCAGAACCCTCGATGTTGTGGCGGATC-3′). The resulting PCR fragment was directly targeted to the CDC42 locus of DDY1301, resulting VN fused to the N terminus of Cdc42.
d For expression of VC-Cdc42, the PCR fragment was generated using the plasmid pFA6a-kanMX6-PRPL7B-VC (Sung and Huh, 2007) (a gift from W.-K. Huh), using a primer pair of oCDC42UP (5′-CGTTATTTATTATACTATTCTATTTTCCTGAGGAGATAGGGAATTCGAGCTCGTTTAAAC-3′) and oCDC42R5VC (5′-CAGCACCATCACCGACAACAACACACTTTAGCGTTTGCATAGTACCACCAGAACCCTTGTACAGCTCGTCCATG-3′). The resulting PCR fragments were directly targeted to the CDC42 locus of DDY1300, resulting VC fused to the N terminus of Cdc42.
e The BUD5 gene was disrupted with pUC18-bud5Δ::URA3, as previously described (Chant et al., 1991). The bud5 deletion was confirmed by colony PCR and determining the budding pattern.
f rsr1Δ::URA3 was replaced with rsr1Δ::kanMX4 using the PCR products generated from an rsr1Δ::kanMX4 strain (purchased from Open Biosystems, Huntsville, AL) using a primer pair of oBUD1G2 (5′-GGTCATCGGTTCGATTCCGGTTGCGTCC-3′) and oBUD139 (5′-GCGTTCGTTCTTAACTACGCC-3′).
g The plasmid pKTUB1-CFP (Bailly et al., 2003; a gift from M.-N. Simon) was integrated into the URA3 locus of HPY1612 after digestion with StuI.
h The BUD2 gene was disrupted using pUC19-bud2Δ::LEU2, as previously described (Park et al., 1993). The bud2 deletion was confirmed by colony PCR and determining the budding pattern.
i The GIC2 gene was disrupted by using the PCR product generated from a gic2Δ::kanMX4 strain (purchased from Open Biosystems) using a primer pair of oGIC21 (5′-CCAGTAAAACGAGACCTGTTGATG-3′) and oGIC22 (5′-ACGAATGTATGGGATAACGCCAAG-3′).
Fluorescence Microscopy and Bimolecular Fluorescence Complementation
To visualize bud scars for determination of budding patterns, cells were grown at indicated temperatures and then stained with Calcofluor (Pringle, 1991). Image acquisition for proteins fused to yellow fluorescence protein (YFP) and cyan fluorescence protein (CFP) was carried out essentially as previously described (Kang et al., 2001) using a Nikon E800 microscope (Nikon, Tokyo, Japan) fitted with a 100× oil-immersion objective (NA = 1.30), a Uniblitz electronic shutter (Delta Photonics, Ottawa, ON, Canada), a Prior Z-axis drive (Prior Scientific, Rockland, MA), and a Hamamatsu Orca ER cooled charge-coupled device (Hamamatsu Photonics, Bridgewater, NJ). The Slidebook software (Intelligent Imaging Innovations, Denver, CO) was used to capture a series of optical sections at 0.3-μm intervals. Cells were exposed for 1 s and 200 msec for YFP-Rsr1 and Tub1-CFP, respectively. Subsequent processing (including deconvolution) and analyses of all images were performed in the same way using Slidebook software and Photoshop (Adobe Systems, San Jose, CA).
Bimolecular fluorescence complementation (BiFC) assays were performed essentially as previously described (Singh et al., 2008), except that YFPN (N-terminal fragment of YFP) and YFPC (C-terminal fragment of YFP) fusions were coexpressed in diploid cells, where indicated. The diploid strains were generated by mating two haploid strains, each of which expresses an YFPN or YFPC fusion. YFPN or YFPC was fused to the N terminus of Rsr1 and its mutant forms (see Table 1 and Supplemental Methods). Similarly, the N- and C-terminal fragments, VN and VC, of Venus (a variant of YFP) were fused to the N terminus of Cdc42 (see Table 1). To monitor BiFC signals, a single optical section was captured using the YFP filter and by exposing cells to UV for 6 or 8 s as indicated.
Protein Purification and Chemical Cross-Linking
Glutathione S-transferase (GST)-Rsr1 was purified from Escherichia coli as previously described (Park et al., 1997). For chemical cross-linking, GST-Rsr1 was preloaded with GTPγS, a nonhydrolyzable GTP analogue, or GDP, and then the GST moiety was cleaved with thrombin as previously described (Kozminski et al., 2003). The protein concentration of Rsr1 was adjusted to 10 μg/ml after removing the GST moiety and incubated with 0.1 mM ethylene glycol-bis(succinic acid N-hydroxysuccinimide ester) (EGS; Sigma-Aldrich, St. Louis, MO), an irreversible cross-linker, at room temperature for 15 min. The reaction was stopped by addition of Tris (100 mM final concentration) and by cooling the reaction mixture on ice for 10 min. The protein mixtures were then boiled for 5 min and subjected to SDS-PAGE, after centrifugation at 10,000 × g for 15 min to remove any aggregates. Rsr1 was detected by immunoblotting using polyclonal antibodies against Rsr1 (a kind gift from A. Bender, Indiana University, Bloomington, IN).
Size-Exclusion Chromatography
The GST moiety of GST-Rsr1, purified from E. coli, was removed after digestion with thrombin as previously described (Kozminski et al., 2003), before size-exclusion chromatography. Gel filtration columns, Sephacryl S-200 HR (Amersham Pharmacia Biotech, Piscataway, NJ; bed volume: 1.6 ml) were equilibrated with column buffer (20 mM Tris, pH 7.5, 1 mM DTT, 100 mM NaCl) at room temperature. Rsr1 was preloaded with [3H]GTP or [3H]GDP as previously described (Kozminski et al., 2003) and then applied to the column at room temperature at the indicated concentrations. Fractions were collected at a flow rate of one drop/30 s (four drops for the first two fractions and one drop for each fraction thereafter) and then the radioactivity was quantified by scintillation counting.
N-methylanthraniloyl-GTP binding
GTP binding of Rsr1 and Rsr1K260–264S (Rsr1KS) was tested using N-methylanthraniloyl (mant)-GTP, essentially as previously described (Rojas et al., 2003) with slight modifications as follows. Affinity-purified GST-Rsr1 (∼2 μM), GST-Rsr1KS (∼3 μM), and GST (∼3 μM) proteins were immobilized on glutathione-agarose beads and then loaded with mantGTP (Molecular Probes, Eugene, OR) using 20-fold molar excess of mantGTP in nucleotide exchange buffer (50 mM Tri-HCl, pH 8.0, 20 mM EDTA, 5 mM DTT). After washing off unbound nucleotides with buffer (50 mM Tri-HCl, pH 7.5, 100 mM NaCl, 10 mM MgCl2, 5 mM DTT), the mantGTP-loaded proteins were eluted from the beads using the elution buffer (50 mM Tri-HCl, pH 8.0, 10 mM reduced glutathione, 1 mM DTT) and then subjected to fluorescence measurement using Cary Eclipse fluorescence spectrophotometer (Varian, Palo Alto, CA). MantGTP was excited at 360 nm, and emission spectra were collected from 400 to 600 nm using the SCAN program, according to the instrument's manual.
In Vitro Binding Assays
Self-association of Rsr1 was determined by in vitro binding assays as previously described (Kozminski et al., 2003) with slight modification. All steps were performed at 4°C or on ice until proteins were eluted for immunoblotting. First, hemagglutinin (HA)-tagged Rsr1 was immunoprecipitated from yeast extract prepared from cells (HPY16) carrying pRS425-HA-RSR1 (pHP582), as previously described (Kozminski et al., 2003). HA-Rsr1 (on agarose beads) was then incubated with GST-Rsr1 or GST-Rsr1KS, which was purified from E. coli and preloaded with GTPγS or GDP for 1 h. After washing with buffer (10 mM Tri-HCl, pH 7.5, 5 mM MgCl2, 2 mM DTT, 0.1% Triton, 10% glycerol), HA-Rsr1 and its associated proteins were eluted with Laemmli buffer and then subjected to SDS-PAGE and immunoblotting using polyclonal antibodies against GST (Santa Cruz Biotechnology, Santa Cruz, CA) to detect GST-Rsr1 and GST-Rsr1-7KS and monoclonal anti-HA antibody (HA.11 from Covance Research Products, Denver, PA) to detect HA-Rsr1.
RESULTS
Rsr1 Associates with Itself and Cdc42 In Vivo
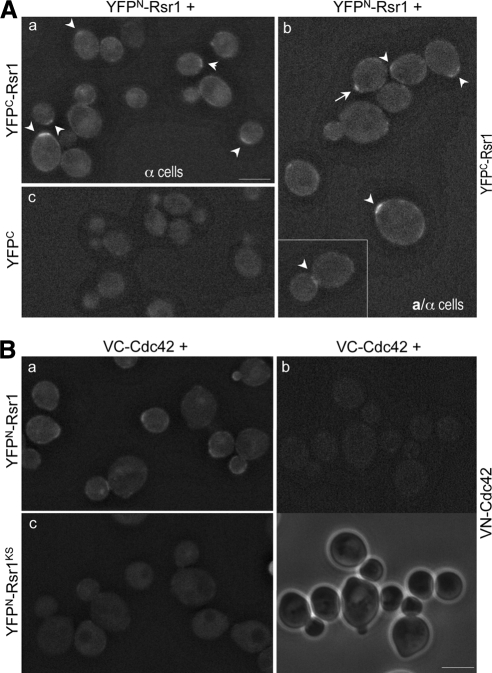
Because we previously found that Rsr1 interacts with another GTPase Cdc42 (Kozminski et al., 2003), we wondered whether Rsr1's function also involves its homotypic interaction. To determine whether and where the homotypic interaction of Rsr1 occurs in vivo, we used a BiFC assay. This technique allows visualization of protein–protein associations in live cells by monitoring YFP fluorescence, which appears when truncated YFP fragments (YFPN and YFPC) are brought together by association of two proteins fused to them (Hu et al., 2002). When YFPN-Rsr1 and YFPC-Rsr1 were coexpressed in haploid α cells and diploid a/α cells, the YFP fluorescence was detectable (Figure 1A, a and b), indicating that Rsr1 associates with itself. The YFP signal appeared as a band at one pole in haploid unbudded cells (50.6%; n = 164) and in diploid unbudded cells (47.5%; n = 183). The YFP signal was sometimes observed at the mother-bud neck in cells with large buds (as shown in the inset, Figure 1Ab). Occasionally a weaker fluorescence signal was also detected at the periphery of cells (∼15%). However, the YFP signal was rarely detected at the tips of small- and medium-sized buds. As a control, when YFPN-Rsr1 was coexpressed with YFPC (which carries only the first amino acid 30 residues of Rsr1), little fluorescence was detectable (Figure 1Ac).
Figure 1.
Rsr1 associates with itself and Cdc42 in vivo. (A) BiFC assays in haploid α cells (HPY1200), which express YFPN-Rsr1 from the RSR1 locus and carry YCpYFPC-RSR1 (a) or YCpYFPC-rsr1Δ (c); and in diploid a/α cells coexpressing YFPN-Rsr1 and YFPC-Rsr1 (HPY1214; b). Arrowheads mark cells with clear BiFC signals. Pixel intensity of the cell marked with an arrow is shown in comparison to other cells in Figure 5D. Images were captured with the YFP filter for 6-s exposure and deconvolved. Bar, 5 μm. (B) BiFC assays in diploid a/α cells coexpressing YFPN-Rsr1 and VC-Cdc42 (HPY1209) (a); VN-Cdc42 and VC-Cdc42 (HPY1198) (b); and YFPN-Rsr1KS and VC-Cdc42 (HPY1217) (c). Phase images are also shown below for the same cells of HPY1198. Images were captured with the YFP filter for 8-s exposure and deconvolved. Bar, 5 μm.
Because Rsr1 has been shown to interact with Cdc42 by in vitro studies (Kozminski et al., 2003), we examined by BiFC assays whether such heterotypic GTPase association occurs in vivo. Coexpression of VC-Cdc42 and YFPN-Rsr1 exhibited fluorescence at the sites of polarized growth including incipient bud sites, tips of growing buds and mother-bud neck (Figure 1Ba). This result thus indicates that Rsr1 associates with Cdc42 in vivo. We wondered whether these homotypic and heterotypic interactions of Rsr1 simply resulted from its enrichment at the sites of polarized growth. However, the Rsr1-Rsr1 bimolecular fluorescent complex was not observed at the vacuolar membrane or at the site of bud emergence where YFP-Rsr1 became enriched (see Figure 3 and Supplemental Figure S3; Park et al., 2002). In addition, a strain coexpressing VN-Cdc42 and VC-Cdc42 exhibited little fluorescence despite similar clustering of Cdc42 at the polarized growth sites (Figure 1Bb), suggesting that Cdc42 does not associate with itself. Taken together, the BiFC results indicate that Rsr1 interacts with itself and Cdc42 in vivo.
Figure 3.
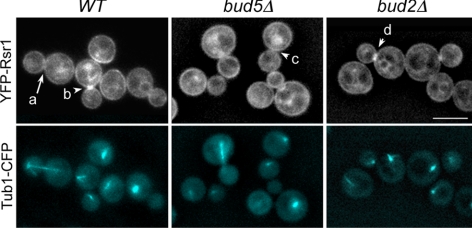
Localization of YFP-Rsr1 to the mother-bud neck occurs after anaphase and is dependent on Bud5. Localization of YFP-Rsr1 and Tub1-CFP in haploid wild-type (HPY1632), bud5Δ (HPY1668), and bud2Δ (HPY1664) cells. YFP-Rsr1 and Tub1-CFP were expressed from the RSR1 and URA3 loci on the chromosomes, respectively. A cell marked with an arrow (a) undergoes nuclear division; and cells marked with arrowheads (b–d) have completed nuclear division. Pixel intensities of these cells are shown in Supplemental Figure S1. A series of Z-sections were captured with the YFP and CFP filters for YFP-Rsr1 and Tub1-CFP, respectively. Images were deconvolved, and a single Z section is shown. Bar, 5 μm.
Homotypic Association of Rsr1 Is Likely to Depend on Its GEF Bud5 In Vivo
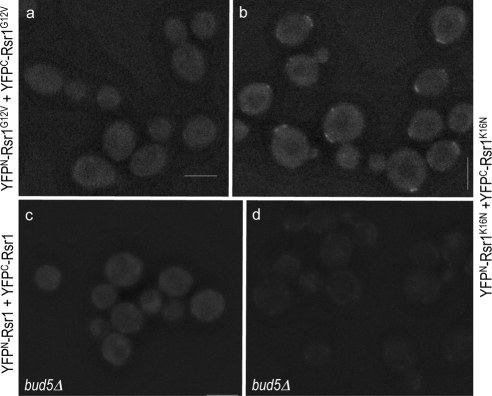
The appearance of the Rsr1 BiFC signal at a specific site raised the question of whether its self-association might depend on its guanine nucleotide-bound state or its regulator in vivo. To address the question, we performed BiFC assays using the strains expressing YFPN and YFPC fusions of Rsr1G12V and Rsr1K16N, which are expected to be in the GTP- and GDP-locked states in vivo, respectively (Ruggieri et al., 1992). Little YFP signal was detected in cells coexpressing YFPN and YFPC fusions of Rsr1G12V (1.6% of unbudded cells; n = 188; Figure 2a). In contrast, the YFP signal appeared at one pole of the cells coexpressing YFPN-Rsr1K16N and YFPC-Rsr1K16N (84% of unbudded cells; n = 183) and remained even after bud emergence (Figure 2B), indicating that self-association of Rsr1K16N is more persistent than that of wild type (see Figure 1A, a and b). The BiFC signals were sometimes observed at more than one spot in unbudded a/α cells, which were likely to reflect the signals from the previous and new division sites. Because these rsr1 mutations do not affect the steady-state level of the protein (Park et al., 1997), the difference of the fluorescence is unlikely to be caused by the different level of each mutant protein.
Figure 2.
Bud5 is necessary for efficient homotypic interaction of Rsr1. BiFC assays were done in a/α wild-type cells coexpressing YFPN-Rsr1G12V and YFPC-Rsr1G12V (HPY1568; a); and YFPN-Rsr1K16N and YFPC-Rsr1K16N (HPY1608; b). Similarly, BiFC assays were done in a/α bud5Δ/bud5Δ cells coexpressing YFPN-Rsr1 and YFPC-Rsr1 (HPY1575; c); and YFPN-Rsr1K16N and YFPC-Rsr1K16N (HPY1824; d). Images were captured and processed as in Figure 1A. Bars, 5 μm.
Because Rsr1K16N is expected to be constitutively in the GDP-bound state in vivo, the BiFC signal from YFPN-Rsr1K16N and YFPC-Rsr1K16N is likely to be caused by self-association of Rsr1-GDP, which might be initially recruited to the division site by its GEF Bud5. To test whether self-association of Rsr1 depends on Bud5, we examined the interaction between YFPN-Rsr1 and YFPC-Rsr1 in a bud5 deletion (bud5Δ) mutant. The BiFC signal was observed in much fewer bud5Δ cells (7.6% of unbudded cells; n = 160) compared with the wild-type cells, although it was not completely absent (Figure 2c). Similarly, coexpression of YFPN-Rsr1K16N and YFPC-Rsr1K16N in bud5Δ cells did not exhibit clear YFP signal, although faint signals were seen as dots or diffuse near the periphery of some cells (compare d with b in Figure 2). Taken together, these results suggest that Bud5 is necessary for efficient homotypic association of Rsr1 in vivo.
YFP-Rsr1 Is Enriched at the Division Site after Nuclear Separation But Poorly in the Absence of Bud5
YFP-Rsr1 (as well as GFP-Rsr1) localizes to the plasma membrane and becomes enriched at the sites of polarized growth and at the division sites of the newly born G1 cells (Park et al., 2002). Because the Rsr1-Rsr1 BiFC complex was observed at the division site and the complex formation was dependent on Bud5, we asked whether localization of Rsr1 to the mother-bud neck (which becomes the subsequent division site) also depends on Bud5. We examined localization of YFP-Rsr1 in the cells which express α-tubulin (Tub1)-CFP (Figure 3) and analyzed the pixel intensity across the long axis of each cell as shown for a few representative cells in Supplemental Figure S1. These analyses indicated that Rsr1 concentrated at the mother-bud neck after nuclear separation but before cytokinesis. YFP-Rsr1 was hardly observed at the mother-bud neck in cells before or at anaphase, as indicated with the mitotic spindles shown with Tub1-CFP (compare large-budded cells with long microtubules and with short microtubules, marked with an arrow and an arrowhead, respectively, in Figure 3, a and b). Almost all postanaphase cells with short microtubules exhibited YFP-Rsr1 localized at the division site (88%; n = 120), whereas <4% of cells with long mitotic spindles (n = 50) showed enrichment of YFP-Rsr1 at the division site. After subsequent cell division, newly born G1 cells exhibited enrichment of YFP-Rsr1 at the division site. In contrast, YFP-Rsr1 localized poorly to the mother-bud neck in postanaphase bud5Δ cells (17%; n = 60; Figure 3, c), although it still localized to the plasma membrane and was also concentrated at the periphery of the growing buds. Localization of YFP-Rsr1 in postanaphase bud2Δ cells was similar to that in the wild-type cells (70%; n = 60; Figure 3d). Taken together, these data suggest that Bud5 is necessary for efficient localization of Rsr1 to the mother-bud neck and to the subsequent division site.
Rsr1 Forms a Homodimer in Vitro
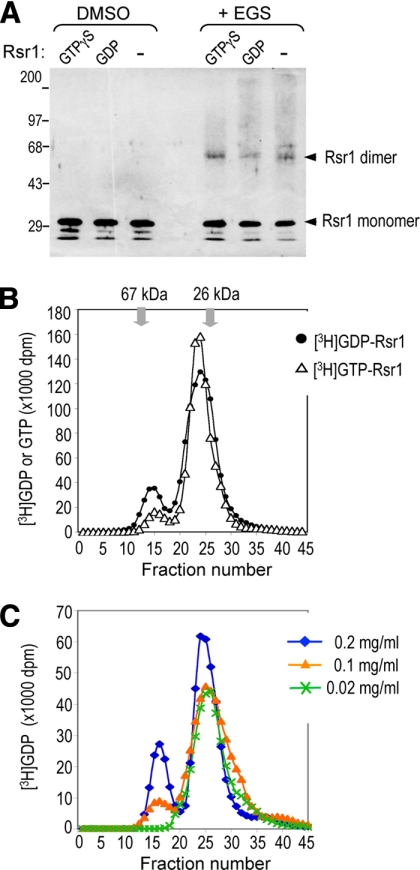
Because mammalian Ras and Rho GTPases can form dimers and oligomers (Zhang and Zheng, 1998; Inouye et al., 2000), we wondered whether homotypic association of Rsr1 detected by BiFC indicates its direct interaction. We thus tested whether Rsr1, purified from bacteria, can form oligomers in vitro by SDS-PAGE after chemical cross-linking (see Materials and Methods). When Rsr1 was treated with an irreversible cross-linker, a slower migrating band of the predicted dimer size (∼62 kDa) appeared with Rsr1 preloaded with GTPγS or GDP (and even with Rsr1 not preloaded with any nucleotides), but not with the mock-treated control (Figure 4A).
Figure 4.
Rsr1 forms a dimer in vitro. (A) Purified Rsr1 (10 μg/ml) was preloaded with GTPγS or GDP, or in nucleotide-empty state (−), recovered after cleaving off the GST moiety, treated with 0.1 mM EGS or mock-treated (DMSO), and then subjected to SDS-PAGE. Rsr1 was detected with polyclonal antibodies against Rsr1. (B) Rsr1 forms a stable homodimer in solution. Purified Rsr1 (50 μl; 0.05 mg/ml) was applied to a size exclusion chromatography column (Sephacryl-200HR; 1.6 ml bed volume) after preloading with [3H]GDP (•) or [3H]GTP (▵). The molecular-weight standards, BSA (67 kDa) and GST (26 kDa), eluted in fractions 13–14 and 26–27, respectively. Fractions 14–16 and 24–25 contain most of the Rsr1 dimers and monomers, respectively. The amount of [3H]GDP or [3H]GTP bound to Rsr1 was measured by scintillation counting (dpm), and plotted after subtracting the background dpm. (C) Dimerization of Rsr1 is concentration-dependent. Rsr1 (5 μl of each indicated concentration) preloaded with [3H]GDP was applied to a size exclusion chromatography column as in Figure 4B. An equal molar concentration of [3H]GDP was used for GDP loading of Rsr1, which was kept at each concentration.
To gain further insight into the oligomerization process, we next analyzed Rsr1 protein by size exclusion chromatography after preloading with [3H]GTP or [3H]GDP. Both nucleotide-bound states of Rsr1 spontaneously formed dimers in solution (Figure 4B, fraction 14–16). The homodimer formation was slightly affected by the guanine nucleotide-bound state of Rsr1—[3H]GDP-Rsr1 being more efficient than [3H]GTP-Rsr1—based on the gel filtration results. When the fractions containing most of the Rsr1 dimers were passed through the column again after dilution, Rsr1 ran in the monomer fractions (data not shown). In addition, when Rsr1 was analyzed at various protein concentrations using a similar size exclusion chromatography column, Rsr1 dimerization was more favorable at a higher protein concentration (Figure 4C). Even at the highest protein concentration tested (0.2 mg/ml), Rsr1 appeared as a uniform dimer but not in a higher oligomeric state. Taken together, these data indicate that Rsr1 forms a homodimer in vitro and that Rsr1 dimerization is reversible and concentration-dependent.
An intact Polybasic Region of Rsr1 Is Required for Efficient Homotypic and Heterotypic Interactions
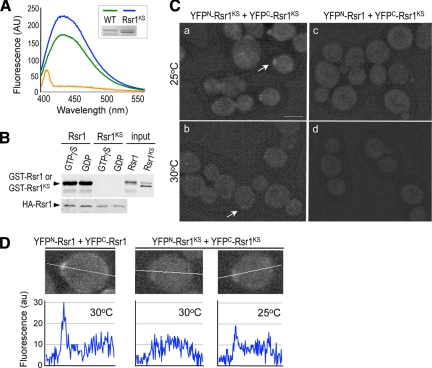
To address the functional significance of the homotypic interaction of Rsr1, we attempted to identify an Rsr1 mutant that is specifically defective in self-association. A number of Ras and Rho GTPases contain a polybasic region (PBR) near the C terminus, which mediates diverse functions of these small GTPases (Williams, 2003). Because the PBR is shown to be involved in oligomerization of the Rac1 GTPase (Zhang et al., 2001), we used Rsr1-7K260–264S (denoted by Rsr1KS), in which five Lys residues (a.a. 260–264) in the PBR were mutated to Ser (Park et al., 2002; see Figure 6A). We first monitored whether GTP or GDP binding of Rsr1 was affected by this mutation using the fluorescent GTP or GDP analog, mant-GTP or mant-GDP. The Rsr1KS mutant protein appeared to be stable and was not defective in binding to mant-GTP, compared with wild type (Figure 5A; Supplemental Figure S2).
Figure 6.
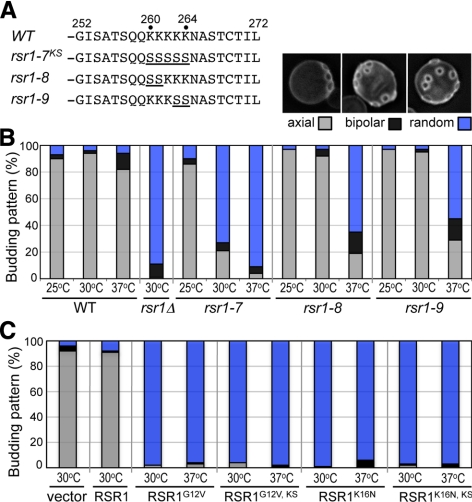
The intact PBR of Rsr1 is necessary for bud-site selection. (A) The carboxy-terminal region of Rsr1 and the mutations in the PBR are shown. Each rsr1 mutant carries substitutions of Lys to Ser (underlined). (B) Budding patterns of haploid wild-type (HPY11), rsr1Δ (HPY263), rsr1-7 (HPY1625), rsr1-8 (HPY1741), and rsr1-9 (HPY1740) cells, grown at the indicated temperature, were determined after Calcofluor staining. Each rsr1 mutant was expressed from its native promoter at the RSR1 locus. About 300–400 cells were counted for each strain in three independent sets of experiments. The average percentage of each budding pattern is shown (SD <2%); axial (gray bar), bipolar (black bar), and random (blue bar). Calcofluor staining of a representative cell budding in each pattern is shown above the graph. (C) Budding pattern of wild-type cells (HPY11) carrying each RSR1 plasmid (on YEplac195) or empty vector was determined as in Figure 6B. RSR1G12V, KS and RSR1K16N, KS represent the RSR1 plasmids carrying the K260-264S mutation (rsr1-7KS) as well as each dominant mutation. Three independent transformants of each plasmid were grown overnight in SD-URA media at 30 or 37°C, and their budding patterns were determined after Calcofluor staining. About 300 cells of each sample were counted twice and the mean (%) is shown; axial (gray bar), bipolar (black bar), and random (blue bar) pattern.
Figure 5.
Homotypic interaction of Rsr1 requires an intact polybasic region near the C terminus. (A) Rsr1K260–264S (Rsr1KS) binds mantGTP as efficiently as wild type. Purified GST-Rsr1 (∼2 μM; green line) and GST-Rsr1KS (∼3 μM; blue line) were used to test mantGTP binding. The amount of wild-type and Rsr1KS mutant proteins used for mantGTP binding was estimated by Coomassie blue staining (as shown in inset) with the BSA standards. Rsr1KS runs slightly faster than wild type in a protein gel, and a slower migrating band presumably corresponds to nascent protein with unmodified CaaX box. GST (∼3 μM; orange line) was used to detect background fluorescence. MantGTP was excited at 360 nm, and emission spectra were collected from 400 to 600 nm. (B) HA-Rsr1 interacts with GST-Rsr1 but not with GST-Rsr1KS in vitro. HA-Rsr1, immunoprecipitated from yeast extract, was incubated with purified GST-Rsr1 or GST-Rsr1KS, preloaded with GTPγS or GDP. GST-Rsr1 or GST-Rsr1KS was detected with polyclonal antibodies against GST (top panel); and HA-Rsr1 was detected with a mAb against HA epitope (bottom panel). Purified GST-Rsr1 and GST-Rsr1KS added in the binding reaction are shown (input). (C) BiFC assays were performed in diploid a/α cells coexpressing YFPN-Rsr1KS and YFPC-Rsr1KS (HPY1220) grown at 25°C (a) or 30°C (b); and YFPN-Rsr1 and YFPC-Rsr1KS (HPY1219) at 25°C (c) or 30°C (d). Images were captured and processed as in Figure 1A. Size bar, 5 μm. Note: Although cells were grown at the indicated temperatures, these cells were kept at the same, room temperature during microscopic observation. This might have contributed to some faint BiFC signal observed in the cells grown at 30°C (b). (D) Pixel intensity is shown along the line for the cells marked with arrows in Figure 1Ab and Figure 5C, a and b.
We then tested in two different ways whether the rsr1-7 mutation affects homotypic interaction of Rsr1. First, we carried out in vitro–binding assays using purified GST-Rsr1 or GST-Rsr1KS (preloaded with GTPγS or GDP) and HA-Rsr1 immunoprecipitated from yeast extract (see Materials and Methods). Consistent with our BiFC results, GST-Rsr1 associated with HA-Rsr1 (Figure 5B). In contrast, little GST-Rsr1KS was recovered with HA-Rsr1, indicating that the PBD mutation disrupts the self-association of Rsr1 (Figure 5B).
Next, to determine how the PBR mutation affects the homotypic interaction of Rsr1 in vivo, BiFC assays were carried out in a strain expressing the YFPN and YFPC fusions of Rsr1KS from its chromosomal locus. Surprisingly, the formation of the Rsr1KS bimolecular fluorescent complex was dependent on growth temperature. YFP fluorescence was barely detectable when cells coexpressing YFPN-Rsr1KS and YFPC-Rsr1KS were grown at 30°C: Cells even with faint YFP signals were <10% of unbudded cells (n = 298; Figure 5Cb). In contrast, ∼36% of unbudded cells (n = 337) exhibited the concentrated BiFC signal at 25°C (Figure 5C, a), although the signal was weaker than that in wild-type cells. Pixel intensity of many unbudded cells was compared, and representative cells marked with arrows (in Figure 5C and Figure 1Ab) are shown in Figure 5D. In the strain coexpressing YFPN-Rsr1 and YFPC-Rsr1KS, almost no BiFC signals were detectable at 25°C (< 2%; n = 160) and 30°C (1.7%; n = 60; Figure 5C, c and d), consistent with the in vitro–binding results (see Figure 5B). Because the fluorescence intensity of YFP-Rsr1 fusions and the level of the proteins were about the same when cells were grown at 25 and 30°C (Supplemental Figures S2 and S3), the stronger BiFC signal suggests more efficient homotypic interaction of Rsr1KS at 25°C than at 30°C. Taken together, these results suggest that the intact PBR of Rsr1 is necessary for its homotypic interaction, and that Rsr1KS is partially functional in vivo for the homotypic interaction.
We took a similar approach to test whether the PBR is also important for the heterotypic association of Rsr1 with Cdc42. When YFPN-Rsr1KS and VC-Cdc42 were coexpressed from their chromosomal loci, the bimolecular fluorescent complex formation greatly diminished at 30°C, compared with the case of YFPN-Rsr1 and VC-Cdc42 complex (Figure 1Bc), suggesting that the PBR of Rsr1 is also important for the heterotypic interaction.
The PBR of Rsr1 Is Necessary for Bud-Site Selection
Because in vitro interaction and in vivo BiFC assays suggested that Rsr1-7KS did not efficiently self-associate, we asked how the rsr1 mutations in the PBR affect Rsr1's function in bud-site selection and polarity establishment. First, we examined the budding pattern of the rsr1-7 mutant. The rsr1-7 mutant exhibited mostly a random budding pattern at 30 and 37°C, similar to rsr1Δ cells, whereas it exhibited an axial budding pattern at 25°C (Figure 6B). Thus the rsr1-7 mutant is defective in bud-site selection in a temperature-dependent manner.
We wondered whether the rsr1-7 mutation resulted in a gross structural alteration, leading to a completely nonfunctional protein, despite its ability to bind GTP/GDP. We thus carried out additional tests to confirm that rsr1-7 encodes a functional protein that maintains its ability to interact with its GEF Bud5 and a downstream effector Cdc24. To test whether the Rsr1-7KS protein was defective in interaction with Bud5 or Cdc24, we combined the rsr1-7 mutation with the dominant mutations RSR1K16N or RSR1G12V. Expression of RSR1K16N in a wild-type strain leads to random bud-site selection, presumably because the GDP-locked Rsr1 binds Bud5 but cannot be converted to the GTP-bound state (Ruggieri et al., 1992). Expression of RSR1G12V also results in random bud-site selection. This is likely because the GTP-locked Rsr1 binds constitutively to the polarity proteins including Cdc24, so that these proteins cannot be targeted to a proper bud site (Ruggieri et al., 1992; Benton et al., 1993; Park et al., 1997).
Expression of the double mutant RSR1G12V, KS or RSR1K16N, KS from a multicopy plasmid caused random bud-site selection in wild-type strain, as did the single mutant RSR1G12V or RSR1K16N at both 30 and 37°C (Figure 6C). These results support the idea that the rsr1-7 mutation does not interfere with the interaction between Rsr1-GTP and Cdc24 (see below). In addition, these data suggest that the rsr1-7 mutation does not affect the interaction between Rsr1-GDP and Bud5. Therefore, the rsr1-7 mutant seems to encode a functional Rsr1 but with a specific defect in the homotypic and heterotypic GTPase interactions.
Because the rsr1-7 mutant was also defective in polarity establishment (see below), we wanted to obtain the separation-of-function alleles of RSR1. We thus examined two additional PBR mutants, rsr1-8 and rsr1-9, which carry substitutions of the first and the last two Lys residues to Ser, respectively (Figure 6A). Both rsr1-8 and rsr1-9 mutants exhibited the axial budding pattern at 25 and 30°C, but not at 37°C, although the defect at 37°C was less severe than that of rsr1-7 (Figure 6B). These Rsr1 mutant proteins, except Rsr1-9, were present at a level about equal to that of the wild-type protein at all temperatures tested (Supplemental Figure S2). These data thus suggest that the intact PBR of Rsr1 is necessary for proper bud-site selection.
The PBR of Rsr1 Is Involved in Polarity Establishment
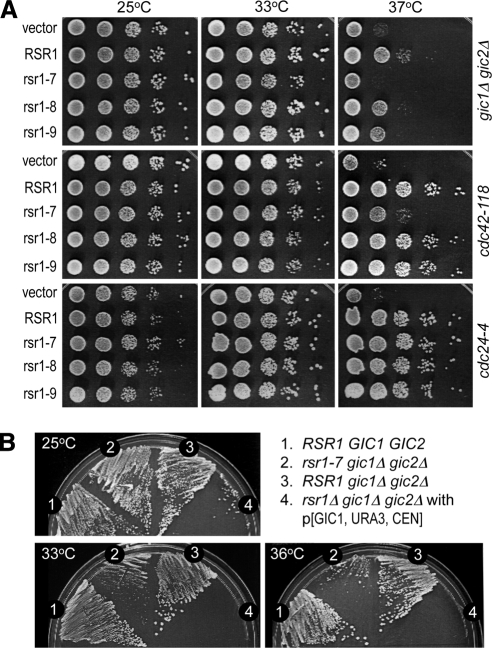
Rsr1 has also been implicated in polarity establishment, and this role of Rsr1 becomes evident in cells with compromised polarity establishment including the cdc42-118 or gic1Δ gic2Δ double mutants (Kawasaki et al., 2003; Kozminski et al., 2003). Because a multicopy plasmid carrying the wild-type RSR1 can suppress temperature-sensitive growth of cdc42-118 and gic1Δ gic2Δ, we utilized these mutants to determine how the rsr1 PBR mutations affect polarity establishment. When the gic1Δ gic2Δ mutant was transformed with a multicopy plasmid expressing RSR1 or rsr1 mutants, the rsr1-7 plasmid could not suppress the gic1Δ gic2Δ mutant at 37°C, whereas the rsr1-8 and rsr1-9 plasmids were only slightly less efficient than wild type in suppression of gic1Δ gic2Δ (Figure 7A, top panel). Similarly, the rsr1-7 plasmid poorly suppressed cdc42-118 (Kozminski et al., 2003), whereas the rsr1-8 and rsr1-9 plasmid were able to suppress cdc42-118 at 37°C (Figure 7A, middle panel). In contrast, all of the PBR mutant plasmids were able to suppress the temperature-sensitive growth of cdc24-4 equally as well as the wild-type RSR1 plasmid (Figure 7A, bottom panel; see also Figure 6C), suggesting that rsr1-7 is specifically defective at a step involving the function of CDC42 or GIC1/GIC2 during polarity establishment.
Figure 7.
The rsr1-7KS mutant is defective in polarity establishment. (A) The gic1Δ gic2Δ mutant (HPY1523) (top panel) and the cdc42-118 mutant (DDY1326) (middle panel) carrying each RSR1 plasmid (on YEplac195) or vector control were grown at 25, 33, and 37°C for 3 d on SC-URA. Similarly, the cdc24-4 mutant (Y147) carrying the same set of the RSR1 plasmids was plated on SC-URA containing 1 M sorbitol (bottom panel). Each spot of cells represented a 10-fold serial dilution from left to right (starting from OD600 = 0.5). (B) The rsr1-7 allele expressed from the RSR1 locus can rescue the lethality of rsr1Δ gic1Δ gic2Δ at 25 but not at 36°C. All strains are isogenic to HPY210: (1) wild type (HPY210); (2) rsr1-7 gic1Δ gic2Δ (HPY1609); (3) RSR1 gic1Δ gic2Δ (YKT342); and (4) rsr1Δ gic1Δ gic2Δ (YKT401) carrying pKT1276 [GIC1, URA3, CEN] plasmid. Each strain was streaked on SC plates containing 5-FOA and incubated at 25, 33, and 36°C for 3 d.
Because the rsr1-7 mutant exhibited a temperature-sensitive defect in bud-site selection, we wondered whether Rsr1-7 is functional in polarity establishment at a lower temperature. We thus carried out a genetic test to characterize the rsr1-7 allele in another strain background. The phenotype of gic1Δ gic2Δ is less severe in this strain background, so that the double mutant grows up to 36°C, whereas cells deleted for RSR1, GIC1, and GIC2 cannot undergo bud emergence (Kawasaki et al., 2003). The rsr1-7 allele was introduced into the RSR1 locus in the gic1Δ gic2Δ rsr1Δ mutant, which was kept alive with the GIC1 plasmid carrying the URA3 marker. When these cells were streaked on plates containing 5-FOA (5-fluoroorotic acid) to select Ura− cells, the rsr1-7 gic1Δ gic2Δ mutant grew as well as wild type or gic1Δ gic2Δ at 25°C, whereas rsr1Δ gic1Δ gic2Δ could not grow as expected (Figure 7B). In contrast, the rsr1-7 gic1Δ gic2Δ mutant grew much more slowly than wild type or gic1Δ gic2Δ at 33°C and barely grew at 36°C except for some spontaneous suppressors (Figure 7B), indicating that rsr1-7 exhibits a temperature-dependent defect in polarity establishment in the absence of GIC1 and GIC2. Taken together, these results indicate that the PBR of Rsr1 is necessary for polarity establishment in cells with compromised Cdc42 or Gic1/Gic2 function.
DISCUSSION
Several GTPases including Ras, Rho, and Arf form dimers and oligomers (Zhang and Zheng, 1998; Inouye et al., 2000; Zhang et al., 2001; Beck et al., 2008), although the physiological significance of GTPase oligomerization is not fully understood. In this study, we found by in vitro and in vivo assays that Rsr1 forms a homodimer. This homotypic interaction of Rsr1 is unlikely to be a simple consequence of the enrichment of Rsr1, because it was not observed at other sites where Rsr1 became concentrated during polarized growth. Despite similar localization patterns and clustering of Rsr1 and Cdc42 (Park et al., 2002; Richman et al., 2002), we also did not observe homodimerization of Cdc42 by both BiFC and gel filtration methods (this study; data not shown).
The BiFC assays allowed the visualization of the homotypic interaction of Rsr1 and the heterotypic interaction of Rsr1 with Cdc42 in vivo. The Rsr1–Rsr1 bimolecular fluorescent complex was observed mainly at the cell division site. These static images suggest that the homotypic interaction of Rsr1 is transient and reversible, unlike that of Rsr1K16N, which exhibits more persistent BiFC signal even after bud emergence. An alternative possibility is that the Rsr1 bimolecular fluorescent complex is unstable. However, because the steady-state levels of Rsr1 and Rsr1K16N are about the same (Park et al., 1997; data not shown), protein stability cannot explain the difference between the bimolecular fluorescent complexes of Rsr1 and Rsr1K16N. Instead, Rsr1K16N, which cannot be converted to the GTP-bound state, may remain continuously self-associated once the dimer is formed at the division site. Thus, despite the potential caveats of the BiFC approach (see below), our results suggest the transient nature of the Rsr1 bimolecular fluorescent complex.
A potential caveat of the BiFC approach is that formation of the bimolecular fluorescent complex might be irreversible as seen in vitro and under some in vivo conditions (Hu et al., 2002; Kerppola, 2008). However, spatial and temporal formation of a bimolecular fluorescent complex has been observed in several other cases (Schmidt et al., 2003; Blondel et al., 2005; Guo et al., 2005; Cole et al., 2007; Sung and Huh, 2007), suggesting that the reversibility of a bimolecular fluorescent complex is dependent on the cellular context or the stability of the complex. The BiFC signal of the Rsr1 bimolecular fluorescent complex was mainly observed during late M and early G1 phases of the cell cycle in the cells, which coexpress YFPN-Rsr1 and YFPC-Rsr1 at the endogenous level. This timing coincides with enrichment of Rsr1 and the localization of its GEF Bud5 and the cell type–specific landmarks including Bud3 and Bud4 to the mother-bud neck (Park and Bi, 2007). However, the exact timing of the homotypic interaction in the cell remains unknown because it is not known how fast fluorophore maturation occurs once YFPN-Rsr1 and YFPC-Rsr1 associate with each other. We thus cannot completely rule out the possibility that the homotypic interaction of Rsr1 occurs overall plasma membrane, but the bimolecular fluorescent complex may not be easily detectable until it becomes enriched at the division site.
Localization of both YFP-Rsr1 and the Rsr1 bimolecular fluorescent complex to the division site are similarly dependent on Bud5, suggesting that Bud5 plays a critical role in recruiting Rsr1 to the division site. This conclusion is supported by the analyses of the dominant RSR1 mutants that are expected to encode the Rsr1 mutant protein locked in either the GTP- or GDP-bound state. A much higher percentage of cells showed the BiFC signal at the division sites when Rsr1K16N was expressed, whereas cells expressing Rsr1G12V failed to show such homotypic interaction. Consistent with these results, we previously found that YFP-Rsr1 and YFP-Rsr1K16N localized to the division site in newly born G1 cells, whereas YFP-Rsr1G12V did not show such discrete enrichment at the division site (Park et al., 2002). Interestingly, the BiFC signal was largely lacking when Rsr1 (and Rsr1K16N) was examined in bud5Δ cells (in which Rsr1 is expected to be mostly in the GDP-bound state because of the lack of its GEF Bud5). These apparently counterintuitive observations can be explained by the physical requirement of Bud5, rather than its GEF activity, for the recruitment of Rsr1 to the division site. Rsr1K16N is expected to strongly interact with Bud5, whereas Rsr1G12V does not. Similarly, it is thought that the dominant-inhibitory Ras mutants bind more tightly to Ras GEFs than does the wild-type Ras and thus prevent activation of endogenous Ras (Feig, 1999).
Consistent with its role in recruitment of Rsr1 to the mother-bud neck and the division site, Bud5 localizes to these sites (Kang et al., 2001; Marston et al., 2001) by interacting with cell type–specific landmarks such as Axl2 and Bud9 (Kang et al., 2001; Krappmann et al., 2007). It remains unknown, however, whether Bud5 functions only in the initial recruitment of Rsr1 or whether Bud5 is also directly involved in Rsr1 dimerization. Although the Bud5 GEF activity is not necessary for the homotypic interaction for Rsr1, it is essential for a subsequent step in bud-site selection. Despite efficient dimerization of Rsr1K16N, expression of Rsr1K16N (and Rsr1G12V) cannot rescue the bud-site selection defect of an rsr1Δ mutant (Ruggieri et al., 1992) nor suppress the polarity defect of cdc42-118, unlike the wild-type RSR1 (Kozminski et al., 2003). It is thus likely that transient, reversible homotypic interaction of Rsr1 is critical for its function. The GEF-dependent recruitment and homotypic interaction of Rsr1 suggest that Rsr1 needs to maintain the ability to pass through its GDP-bound state to carry out its role, consistent with previous findings (Ruggieri et al., 1992; Park et al., 1993; Park et al., 1997).
Although Rsr1G12V exhibited little homotypic interaction in vivo as discussed above, in vitro studies revealed the nucleotide-independent self-association of Rsr1 (see Figures 4, A and B, and 5B). This apparently contradictory observation is likely due to the in vitro conditions in which Rsr1 was the only protein present, and it was also at a relatively higher concentration than in vivo. It appears that the Rsr1 homotypic interaction is different from those between a GTPase and its typical downstream effectors such as Cdc24, which specifically interacts with Rsr1-GTP (Zheng et al., 1995; Park et al., 1997). These observations suggest that the Rsr1 homotypic interaction is unlikely to involve the region of Rsr1 that undergoes drastic conformational change upon its conversion to the GTP-bound state. Consistent with this idea, we found that the PBR of Rsr1 is important for its homotypic and heterotypic interactions (this study), whereas the Ras-like effector domain of Rsr1 is involved in interaction with Cdc24 (Park et al., 1997).
The PBR of GTPases has been shown to be involved in interaction with acidic phospholipids of the plasma membrane, as in the case of the Rho1 GTPase in budding yeast (Yoshida et al., 2009). Rho1 is targeted to the division site by two different mechanisms—a GEF-dependent mechanism operating during anaphase and another PBR-dependent mechanism during cell separation and abscission (Yoshida et al., 2009). In the case of Rsr1, its recruitment to the division site occurs after anaphase and requires both its GEF Bud5 and its PBR (this study). Because recombinant Rsr1 purified from bacteria could form dimers in vitro, its geranylgeranylation at the C terminus and its membrane association is not essential for the homotypic interaction, although these factors may contribute to these interactions in vivo to minor extent.
The phenotypes of the rsr1 PBR mutants suggest that the intact PBR is necessary for bud-site selection and polarity establishment. The rsr1-7KS mutant, which was defective in the formation of bimolecular fluorescent complexes with itself and with Cdc42, exhibited defects in both bud-site selection and polarity establishment (Park et al., 2002; Kozminski et al., 2003; this study). We thus suggest that the Rsr1 PBR is involved in both GTPase interactions, although the interface residues in the Rsr1 homodimer or Rsr1-Cdc42 heterodimer remain unknown. However, GFP-Rsr1KS also showed reduced membrane association (Park et al., 2002). Further studies are thus necessary to clearly resolve the issue of whether the defect in bud-site selection or in polarity establishment is due to the reduced association of Rsr1KS with itself, Cdc42 or membrane, although these may not be separable functions of Rsr1. Interestingly, genetic data suggest that Rsr1KS maintains its interaction with Cdc24 or Bud5. The role of Rsr1 in polarity establishment is thus likely to be specific to the step at which Cdc42 or the Cdc42 targets Gic1/Gic2 function.
This study uncovers the GEF-dependent recruitment of Rsr1 to the division site and the homotypic and heterotypic interactions of Rsr1 during yeast budding. The homotypic interaction of Rsr1 and subsequent heterotypic interaction of Rsr1 with Cdc42 may contribute to efficient polarization of Rsr1 and Cdc42. In response to a spatial landmark, Bud5 may recruit Rsr1 monomers to a single spot on the plasma membrane and may promote Rsr1 dimerization. We postulate that Rsr1 dimerization may facilitate the polarization of Rsr1 at the division site perhaps by counteracting lateral diffusion or internalization from the site. Although the details of the mechanism remain unknown, dimerization of GTPases may provide an efficient mechanism to set up cellular asymmetry.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Konz, K. Singh, D. Berdysz, and M. Lee for their assistance in constructing plasmids and strains; Drs. K. Tanaka (Institute for Genetic Medicine, Japan), W.-K. Huh (Seoul National University, Korea), R. Tabtiang (University of California-San Francisco, CA), M.-N. Simon (Genome Instability and Carcinogenesis, CNRS, France), and K. Kozminski (University of Virginia, Charlottesville, VA) for providing strains and plasmids; and Dr. H. Chamberlin, E. Oakley, and the anonymous reviewers for their valuable comments on the manuscript. This work was supported by a research grant (R01-GM76375) from the National Institutes of Health, National Institute of General Medical Sciences to H.-O. P.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-03-0232) on July 14, 2010.
REFERENCES
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Struhl K. New York: John Wiley & Sons; 1999. Current Protocols in Molecular Biology. [Google Scholar]
- Bailly E., Cabantous S., Sondaz D., Bernadac A., Simon M.-N. Differential cellular localization among mitotic cyclins from Saccharomyces cerevisiae: a new role for the axial budding protein Bud3 in targeting Clb2 to the mother-bud neck. J. Cell Sci. 2003;116:4119–4130. doi: 10.1242/jcs.00706. [DOI] [PubMed] [Google Scholar]
- Beck R., Sun Z., Adolf F., Rutz C., Bassler J., Wild K., Sinning I., Hurt E., Brügger B., Béthune J., Wieland F. Membrane curvature induced by Arf1-GTP is essential for vesicle formation. Proc. Natl. Acad. Sci. USA. 2008;105:11731–11736. doi: 10.1073/pnas.0805182105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender A. Genetic evidence for the roles of the bud-site-selection genes BUD5 and BUD2 in control of the Rsr1p (Bud1p) GTPase in yeast. Proc. Natl. Acad. Sci. USA. 1993;90:9926–9929. doi: 10.1073/pnas.90.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender A., Pringle J. R. Multicopy suppression of the cdc24 budding defect in yeast by CDC42 and three newly identified genes including the ras-related gene RSR1. Proc. Natl. Acad. Sci. USA. 1989;86:9976–9980. doi: 10.1073/pnas.86.24.9976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton B., Tinkelenberg A. H., Jean D., Plump S. D., Cross F. R. Genetic analysis of Cln/Cdc28 regulation of cell morphogenesis in budding yeast. EMBO J. 1993;12:5267–5275. doi: 10.1002/j.1460-2075.1993.tb06222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel M., Bach S., Bamps S., Dobbelaere J., Wiget P., Longaretti C., Barral Y., Meijer L., Peter M. Degradation of Hof1 by SCFGrr1 is important for actomyosin contraction during cytokinesis in yeast. EMBO J. 2005;24:1440–1452. doi: 10.1038/sj.emboj.7600627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. L., Jaquenoud M., Gulli M. P., Chant J., Peter M. Novel Cdc42-binding proteins Gic1 and Gic2 control cell polarity in yeast. Genes Dev. 1997;11:2972–2982. doi: 10.1101/gad.11.22.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chant J., Corrado K., Pringle J. R., Herskowitz I. Yeast BUD5, encoding a putative GDP-GTP exchange factor, is necessary for bud site selection and interacts with bud formation gene BEM1. Cell. 1991;65:1213–1224. doi: 10.1016/0092-8674(91)90016-r. [DOI] [PubMed] [Google Scholar]
- Chant J., Herskowitz I. Genetic control of bud site selection in yeast by a set of gene products that constitute a morphogenetic pathway. Cell. 1991;65:1203–1212. doi: 10.1016/0092-8674(91)90015-q. [DOI] [PubMed] [Google Scholar]
- Chen G. C., Kim Y. J., Chan C. S. The Cdc42 GTPase-associated proteins Gic1 and Gic2 are required for polarized cell growth in Saccharomyces cerevisiae. Genes Dev. 1997;11:2958–2971. doi: 10.1101/gad.11.22.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole K. C., McLaughlin H. W., Johnson D. I. Use of bimolecular fluorescence complementation to study in vivo interactions between Cdc42p and Rdi1p of Saccharomyces cerevisiae. Eukaryot. Cell. 2007;6:378–387. doi: 10.1128/EC.00368-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville S. Cdc42–the centre of polarity. J. Cell Sci. 2004;117:1291–1300. doi: 10.1242/jcs.01115. [DOI] [PubMed] [Google Scholar]
- Feig L. A. Tools of the trade: use of dominant-inhibitory mutants of Ras-family GTPases. Nat. Cell Biol. 1999;1:E25–E27. doi: 10.1038/10018. [DOI] [PubMed] [Google Scholar]
- Guo Y., Rebecchi M., Scarlata S. Phospholipase Cβ2 binds to and inhibits phospholipase Cδ1. J. Biol. Chem. 2005;280:1438–1447. doi: 10.1074/jbc.M407593200. [DOI] [PubMed] [Google Scholar]
- Guthrie C., Fink G. R. San Diego: Academic Press; 1991. Guide to Yeast Genetics and Molecular Biology. [Google Scholar]
- Hu C. D., Chinenov Y., Kerppola T. K. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell. 2002;9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- Inouye K., Mizutani S., Koide H., Kaziro Y. Formation of the Ras dimer is essential for Raf-1 activation. J. Biol. Chem. 2000;275:3737–3740. doi: 10.1074/jbc.275.6.3737. [DOI] [PubMed] [Google Scholar]
- Kang P. J., Sanson A., Lee B., Park H.-O. A GDP/GTP exchange factor involved in linking a spatial landmark to cell polarity. Science. 2001;292:1376–1378. doi: 10.1126/science.1060360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki R., Fujimura-Kamada K., Toi H., Kato H., Tanaka K. The upstream regulator, Rsr1p, and downstream effector, Gic1p and Gic2p, of the Cdc42p small GTPase coordinately regulate initiation of budding in Saccharomyces cerevisiae. Genes Cells. 2003;8:235–250. doi: 10.1046/j.1365-2443.2003.00629.x. [DOI] [PubMed] [Google Scholar]
- Kerppola T. K. Bimolecular fluorescence complementation: visualization of molecular interactions in living cells. Methods Cell Biol. 2008;85:431–470. doi: 10.1016/S0091-679X(08)85019-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski K. G., Beven L., Angerman E., Tong A.H.Y., Boone C., Park H.-O. Interaction between a Ras and a Rho GTPase couples selection of a growth site to the development of cell polarity in yeast. Mol. Biol. Cell. 2003;14:4958–4970. doi: 10.1091/mbc.E03-06-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski K. G., Chen A. J., Rodal A. A., Drubin D. G. Functions and functional domains of the GTPase Cdc42p. Mol. Biol. Cell. 2000;11:339–354. doi: 10.1091/mbc.11.1.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krappmann A.-B., Taheri N., Heinrich M., Mosch H.-U. Distinct domains of yeast cortical tag proteins Bud8p and Bud9p confer polar localization and functionality. Mol. Biol. Cell. 2007;18:3323–3339. doi: 10.1091/mbc.E06-10-0899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston A. L., Chen T., Yang M. C., Belhumeur P., Chant J. A localized GTPase exchange factor, Bud5, determines the orientation of division axes in yeast. Curr. Biol. 2001;11:803–807. doi: 10.1016/s0960-9822(01)00230-5. [DOI] [PubMed] [Google Scholar]
- Park H.-O., Bi E. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol. Mol. Biol. Rev. 2007;71:48–96. doi: 10.1128/MMBR.00028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.-O., Bi E., Pringle J. R., Herskowitz I. Two active states of the Ras-related Bud1/Rsr1 protein bind to different effectors to determine yeast cell polarity. Proc. Natl. Acad. Sci. USA. 1997;94:4463–4468. doi: 10.1073/pnas.94.9.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.-O., Chant J., Herskowitz I. BUD2 encodes a GTPase-activating protein for Bud1/Rsr1 necessary for proper bud-site selection in yeast. Nature. 1993;365:269–274. doi: 10.1038/365269a0. [DOI] [PubMed] [Google Scholar]
- Park H.-O., Kang P. J., Rachfal A. W. Localization of the Rsr1/Bud1 GTPase involved in selection of a proper growth site in yeast. J. Biol. Chem. 2002;277:26721–26724. doi: 10.1074/jbc.C200245200. [DOI] [PubMed] [Google Scholar]
- Park H.-O., Sanson A., Herskowitz I. Localization of Bud2p, a GTPase-activating protein necessary for programming cell polarity in yeast to the presumptive bud site. Genes Dev. 1999;13:1912–1917. doi: 10.1101/gad.13.15.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle J. R. Staining of bud scars and other cell wall chitin with Calcofluor. In: Guthrie C., Fink G. R., editors. Methods in Enzymology. vol. 194. San Diego: Academic Press; 1991. pp. 732–735. [DOI] [PubMed] [Google Scholar]
- Raftopoulou M., Hall A. Cell migration: Rho GTPases lead the way. Dev. Biol. 2004;265:23–32. doi: 10.1016/j.ydbio.2003.06.003. [DOI] [PubMed] [Google Scholar]
- Richman T. J., Sawyer M. M., Johnson D. I. Saccharomyces cerevisiae Cdc42p localizes to cellular membranes and clusters at sites of polarized growth. Eukaryot. Cell. 2002;1:458–468. doi: 10.1128/EC.1.3.458-468.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas R. J., Kimple R. J., Rossman K. L., Siderovski D. P., Sondek J. Established and emerging fluorescence-based assays for G-protein function: Ras-superfamily GTPases. Comb. Chem. High Throughput Screen. 2003;6:409–418. doi: 10.2174/138620703106298509. [DOI] [PubMed] [Google Scholar]
- Ruggieri R., Bender A., Matsui Y., Powers S., Takai Y., Pringle J. R., Matsumoto K. RSR1, a ras-like gene homologous to Krev-1 (smg21A/rap1A): role in the development of cell polarity and interactions with the Ras pathway in Saccharomyces cerevisiae. Mol. Cell. Biol. 1992;12:758–766. doi: 10.1128/mcb.12.2.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt C., Peng B., Li Z., Sclabas G. M., Fujioka S., Niu J., Schmidt-Supprian M., Evans D. B., Abbruzzese J. L., Chiao P. J. Mechanisms of proinflammatory cytokine-induced biphasic NF-κB activation. Mol. Cell. 2003;12:1287–1300. doi: 10.1016/s1097-2765(03)00390-3. [DOI] [PubMed] [Google Scholar]
- Singh K., Kang P. J., Park H.-O. The Rho5 GTPase is necessary for oxidant-induced cell death in budding yeast. Proc. Natl. Acad. Sci. USA. 2008;105:1522–1527. doi: 10.1073/pnas.0707359105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung M.-K., Huh W.-K. Bimolecular fluorescence complementation analysis system for in vivo detection of protein-protein interaction in Saccharomyces cerevisiae. Yeast. 2007;24:767–775. doi: 10.1002/yea.1504. [DOI] [PubMed] [Google Scholar]
- Williams C. L. The polybasic region of Ras and Rho family small GTPases: a regulator of protein interactions and membrane association and a site of nuclear localization signal sequences. Cell Signal. 2003;15:1071–1080. doi: 10.1016/s0898-6568(03)00098-6. [DOI] [PubMed] [Google Scholar]
- Yoshida S., Bartolini S., Pellman D. Mechanisms for concentrating Rho1 during cytokinesis. Genes Dev. 2009;23:810–823. doi: 10.1101/gad.1785209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Gao Y., Moon S. Y., Zhang Y., Zheng Y. Oligomerization of Rac1 GTPase mediated by the carboxyl-terminal polybasic domain. J. Biol. Chem. 2001;276:8958–8967. doi: 10.1074/jbc.M008720200. [DOI] [PubMed] [Google Scholar]
- Zhang B., Zheng Y. Negative regulation of Rho family GTPases Cdc42 and Rac2 by homodimer formation. J. Biol. Chem. 1998;273:25728–25733. doi: 10.1074/jbc.273.40.25728. [DOI] [PubMed] [Google Scholar]
- Zheng Y., Bender A., Cerione R. A. Interactions among proteins involved in bud-site selection and bud-site assembly in Saccharomyces cerevisiae. J. Biol. Chem. 1995;270:626–630. doi: 10.1074/jbc.270.2.626. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.