Abstract
A unique chromosomal translocation involving the genes PAX3 and FKHR is characteristic of most human alveolar rhabdomyosarcomas. The resultant chimeric protein fuses the PAX3 DNA-binding domains to the transactivation domain of FKHR, suggesting that PAX3-FKHR exerts its role in alveolar rhabdomyosarcomas through dysregulation of PAX3-specific target genes. Here, we have produced transgenic mice in which PAX3-FKHR expression was driven by mouse Pax3 promoter/enhancer sequences. Five independent lines expressed PAX3-FKHR in the dorsal neural tube and lateral dermomyotome. Each line exhibited phenotypes that correlated with PAX3-FKHR expression levels and predominantly involved pigmentary disturbances of the abdomen, hindpaws, and tail, with additional neurological related alterations. Phenotypic severity could be increased by reducing Pax3 levels through matings with Pax3-defective Splotch mice, and interference between PAX3 and PAX3-FKHR was apparent in transcription reporter assays. These data suggest that the tumor-associated PAX3-FKHR fusion protein interferes with normal Pax3 developmental functions as a prelude to transformation.
The activation of cellular oncogenes through chromosomal translocation is common in human leukemias and lymphomas (1, 2). In contrast, specific translocations occur less frequently in solid tumors so that their role in carcinogenesis is less clear. One example is the pediatric tumor, alveolar rhabdomyosarcoma (ARMS).
ARMSs are associated with unique chromosomal translocations t(2;13) and t(1;13) that arise from fusion of PAX3 on chromosome 2 or PAX7 on chromosome 1, respectively, to the FKHR gene on chromosome 13 (3–7). The resulting fusion proteins contain the N-terminal region of the PAX proteins, including their intact paired-box and homeodomain DNA-binding elements, and the potent C-terminal transactivation domain of FKHR (5, 8). Interestingly, PAX3 and PAX7 share the greatest degree of similarity in protein structural domains among PAX family members, exhibit somewhat overlapping expression in the developing mouse embryo, and can recognize similar canonical target sequences in vitro (9, 10). Because the forkhead DNA-binding domain of FKHR is truncated in the fusion gene and is shown not to influence target-gene recognition (11), it has been proposed that PAX3-FKHR exerts its carcinogenic role through dysregulation of PAX3-specific target genes. Indeed, PAX3-FKHR can transactivate canonical PAX3-binding sites in vitro, as well as the PAX3-binding site within the MET promoter, and is a more potent transcriptional activator than the normal PAX3 protein (8, 11–14).
Pax3 function has been defined mainly through studying its expression during mouse development and understanding the defects found in the Pax3-deficient mouse mutant, Splotch. Splotch mice have been used extensively as a model for both neural tube and neural crest defects (15, 16), as heterozygous animals display pigmentary disturbances of the abdomen, feet, and tail. Splotch homozygous embryos, however, typically exhibit neural tube closure defects (spina bifida and exencephaly), neural crest-derived tissue defects (small or absent dorsal root ganglia, persistent truncus arteriosus), and problems in the normal formation of limb musculature. These phenotypes correspond to Pax3 expression within the developing brain, dorsal region of the neural tube, various neural crest-derived tissues including the dorsal root ganglia and Schwann cells, and the developing somites (17).
Here, we have produced and analyzed transgenic mice expressing PAX3-FKHR. We find that PAX3-FKHR acts in a dosage-dependent manner to interfere with Pax3 function in vivo and in vitro but does not lead to tumor formation. Thus, PAX3-FKHR may play its carcinogenic role through influencing cellular or tissue differentiation and development.
Materials and Methods
Transgene Constructs and Transgenic Mice Production.
Mouse Pax3 promoter and enhancer sequences were derived from P1 15392 (Genome Systems, St. Louis), which was identified by using a pair of primers (mP-1, 5′-AGCAGAAGAAAGCGAGAAAA; mP-2, 3′-CATGGCGGTGGGAGGGAATC) that lie within the Pax3 promoter region (18). A 337-bp HindIII–BstEII fragment containing the mouse Pax3 promoter, less the initiating methionine, was subcloned into pBluescript (Stratagene). A PAX3-FKHR cDNA fragment containing the entire ORF, as well as the bovine growth hormone polyadenylation signal derived from pcDNA3 (Invitrogen), was inserted downstream of the HindIII–BstEII promoter sequences (pMP3-D13). pMP3-D13(HA) included a PAX3-FKHR cDNA with three tandem C-terminal hemagglutinin (HA) tags. An approximately 14-kb HindIII fragment containing mouse Pax3 enhancer sequences, which have been shown previously to mimic the spatial and temporal expression pattern of Pax3 (18), was cloned from P1 15392 and inserted into the HindIII site just 5′ to the promoter region. The HindIII enhancer-containing fragment is normally contiguous with the HindIII–BstEII sequences in the mouse genome. NotI and/or SfiI cloning sites had been engineered to flank the entire transgene region. All DNAs were verified by direct sequencing. Digestion removed vector sequences from pMP3-D13 and pMP3-D13(HA), respectively, and transgene DNA was purified through a sucrose gradient. The linearized constructs were injected into the pronucleus of fertilized C57BL/6 × BALB/c (CB6F1) oocytes, which were then implanted into pseudopregnant foster mothers at the University of California, San Diego, Cancer Center Transgenic Mouse Core Facility. Founders were identified by Southern blot analysis, and germ-line transmission was achieved by breeding to inbred C57BL/6 mice (Charles River Breeding Laboratories). All animals were maintained on a 12-h light/dark cycle with food and water provided ad libitum. The colony was cared for according to the guidelines of the Institute of Laboratory Animal Resources.
Genotyping and Transgene Copy Number Determination.
Routine genotyping was done by Southern blot analysis of genomic DNA isolated from tail biopsies, or from yolk sac and amniotic membranes, by standard methods by using a 612-bp EcoRI FKHR-specific probe (probe B) on genomic DNA digested with either BamHI or PstI (19). Transgene copy number in each of the independent mouse lines was determined by Southern blot analysis by comparing the relative intensity of transgene and endogenous Pax3-specific bands that hybridize to a mouse Pax3-derived probe (probe A) after BamHI digestion. Autoradiographic quantitation was carried out on a model 445 SI PhosphorImager (Molecular Dynamics) by using IMAGEQUANT 1.1 software. Splotch mice, which are maintained on a C57BL/6 background, were purchased from The Jackson Laboratory. The offspring of transgene × Splotch mice were genotyped by PCR as described (20).
Whole-Mount in Situ Hybridization.
Timed matings were routinely achieved by caging transgenic males with inbred C57BL/6 females. Females were checked each morning for the presence of a vaginal plug, and embryonic day 0.5 (E0.5) was designated as noon on the day that a plug was detected. Pregnant females were killed by cervical dislocation, and the embryos were dissected free from the uterus in a Petri dish containing PBS. The yolk sac and amnion were removed, DNA was isolated for genotyping, and embryos were placed in 4% paraformaldehyde in PBS and fixed overnight at 4°C. Embryos were then washed free of fixative and dehydrated stepwise with increasing concentrations of methanol, and then stored in 100% methanol at −20°C before processing. Probe preparation and whole-mount in situ hybridization was carried out essentially as described (21), with a few modifications (22). Nonradioactive antisense and sense riboprobes were synthesized as in vitro run-off transcripts by using digoxigenin-UTP. The mouse Pax3 probe was derived from a 516-bp HindIII–PstI cDNA fragment cloned into pBluescript. Two separate transgene probes [pMP3-D13 and pMP3-D13(HA) specific] were derived after HindIII digestion and included the last 50 bp of FKHR through to the bovine growth hormone polyadenylation signal derived from pcDNA3.
Muscle Sectioning and Staining.
Fresh vastus lateralis muscle biopsies were removed from adult wild-type and transgenic mice, flash-frozen in isopentane precooled in liquid nitrogen, and stored at −80°C until processed. Unfixed cryostat sections (8 μm) were air-dried and stained with hematoxylin and eosin (H&E), modified Gomori trichrome, and reacted with acid-stable ATPase (23).
Transient Transfections and Chloramphenicol Acetyltransferase Assays.
NIH 3T3 cells were grown in DMEM supplemented with 10% FCS, and transfections were performed by using a modification of the calcium phosphate precipitation method (24). Briefly, cells were transfected with 2.5 μg of the PRS-9 reporter plasmid, 2.5 μg of pcDNA3-lacZ, 100 ng of pcPAX3-FKHR(HA), varying amounts of pcPAX3(HA) that ranged from 50 ng to 1 μg, and pcDNA3 to bring the amount of DNA to 10 μg. After exposure to DNA for 24 h, the medium was changed, and cells were incubated for an additional 48 h before being harvested. β-galactosidase activity was determined by using standard procedures, and chloramphenicol acetyltransferase activity was tested on heat-treated extracts by using the phase exchange assay (25) with commercially available reagents (Promega). Chloramphenicol acetyltransferase activity was normalized to β-galactosidase activity, and experiments were performed within the linear range of the assays and done three times in triplicate.
β-Galactosidase Staining.
Staining for Wnt-1/LacZ was carried out essentially as described (26, 27). Embryos were routinely fixed for 20 min before being stained for 45 min to 1 h at 37°C. Embryos were always postfixed in 4% paraformaldehyde before being photographed.
Results
Generation of PAX3-FKHR Transgenic Mice.
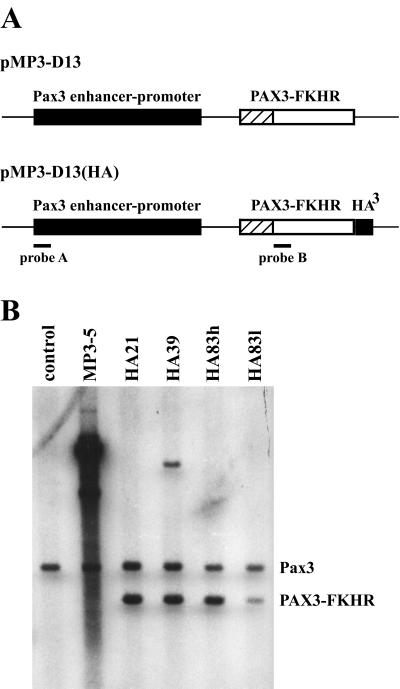
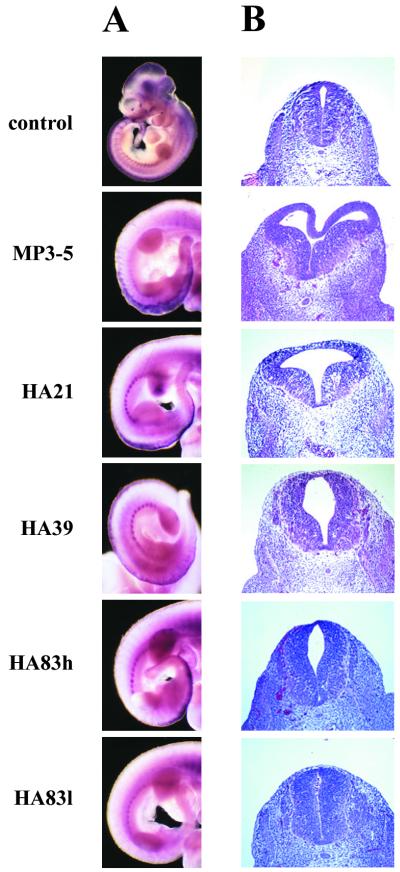
The critical PAX3 intron where all chromosome t(2;13) translocations occur in human ARMS is missing in the mouse so that an endogenous Pax3-Fkhr translocation would be unlikely (17). We therefore sought to test the effects of ectopic PAX3-FKHR expression under the control of Pax3 regulatory sequences (18) on organismal development and cancer predisposition (Fig. 1A). Transgene expression was controlled by 14 kb of mouse Pax3 promoter/enhancer sequences, which have been shown to direct correct temporal and spatial Pax3 expression in the dorsal neural tube and developing somites. Among 138 potential pMP3-D13(HA) founders, four of eight transmitted to the germ line (Fig. 1) and showed correct expression (Fig. 2A). Embryos from the four MP3-D13(HA) lines exhibited varying degrees of expansion of the dorsal midline of the neural tube centering about the hindlimb buds (Fig. 2B). Histological sections stained with H&E through this region showed that expansion of the neural tube resulted in a convolution of the dorsal midline, the widening and enlargement of the central canal, and asymmetry. These complex phenotypes were common among the different lines, although their severity varied. Whole-mount in situ hybridization of PAX3-FKHR showed expression in the dorsal neural tube and lateral dermomyotome (Fig. 2A). As might be expected, expression in the neural tube overlapped with the dorsal midline expansion, with expression extending as anterior as the spinal dysmorphology (Fig. 2B). The distribution and extent of signal in these embryos suggested a hierarchy in the level of neural tube expression among the lines such that HA21>HA39≈HA83h>HA83l.
Figure 1.
Creation of PAX3-FKHR transgenic mice. (A) The two transgene constructs contain mouse Pax3 promoter/enhancer sequences upstream of the PAX3-FKHR cDNA. (B) Transgene copy number assessment by Southern blot analysis. Probe A detects an endogenous Pax3-specific band, as well as band(s) specific to the transgene. The profiles of pMP3-D13(HA) derived lines differ from MP3–5 because of a BamHI restriction site in the HA tag. Comparison between the relative band intensities allowed for copy number determination.
Figure 2.
PAX3-FKHR transgene expression correlates with neural tube dysmorphology. (A) Whole-mount in situ hybridization shows transgene expression in the dorsal neural tube and lateral dermomyotome. Expression of mouse Pax3 in an entire control embryo is shown for comparison. With the exception of MP3–5, expression in the developing head region was absent in the other lines. (B) H&E-stained sections of E11.5 neural tubes taken at the level of the hindlimb buds.
Among 24 potential pMP3-D13 founders, five were shown to harbor transgene DNA, and although all exhibited germ-line transmission, only MP3–5 (Fig. 1) showed transgene expression by whole-mount in situ hybridization (Fig. 2A). The MP3–5 line harbored an exceptionally large number of integrated transgene copies (Fig. 1B) and showed the greatest expression level both by visual inspection (Fig. 2A) and semiquantitative reverse transcription–PCR (not shown). Backcrossing MP3–5 parental mice yielded some animals with spinal dysraphism and kyphosis, severe paralysis and reduced sensory perception of the hindlimbs and tail, reduced or absent macroscopic muscle of the hindlimbs, and premature death. Midgestational embryos from 24 MP3–5 × C57BL/6 backcrosses were analyzed for embryonic lethality. Whereas nearly all E10.5-E12.5 transgenic embryos were alive, 23% and nearly 50% of E13.5 and E15.5 embryos, respectively, were dead. Almost half of all transgenic embryos displayed exencephaly. Although the E15.5 dead embryos were being resorbed, their fore/hindplates were still paddle-shaped, characteristic of developmental age E13.5–E14 (28). The graduated death observed here resembles that of Pax3-deficient Splotch mice (15, 16). Compared with the four other HA lines, these embryos also displayed the greatest neural tube dysmorphology (Fig. 2B), extending anterior to the forelimb buds. In these series of backcrosses, some embryos showed lesser affects, and there was some suggestion of transgene silencing in these cases.
Hindlimb Skeletal Muscle Defects.
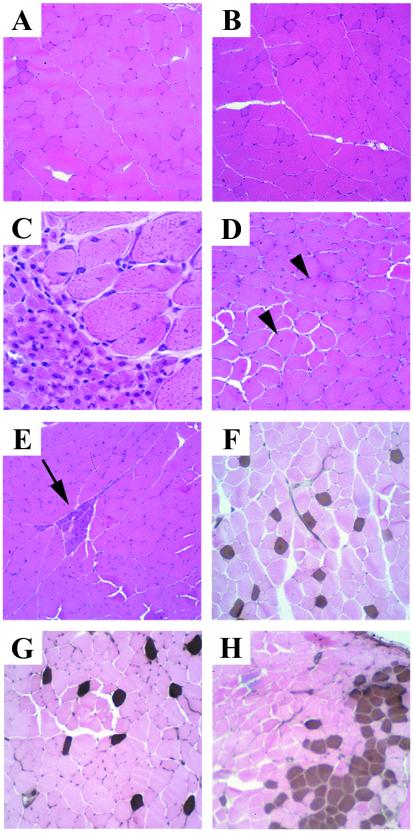
To determine whether the irregular gait observed in these lines of mice was due in part to abnormal muscle development in the hindlimbs, vastus lateralis muscle was examined. Those MP3–5 mice with the most severe symptoms had readily observable neuropathic abnormalities including variable muscle fiber size, increased numbers of centrally nucleated fibers, small angulated fibers, small groups of atrophic fibers, and fiber-type grouping (Fig. 3 C–E and H). The small angulated fibers and fiber-type grouping, or neurogenic rearrangement, are indicative of denervation of muscle fibers followed by reinnervation. Such morphological abnormalities were not easily observed in the lines of mice with less severe phenotypes (Fig. 3 B and G), which had characteristics in this regard more resembling control samples (Fig. 3 A and F). Thus, the abnormalities of the hindlimbs and tail in these strains are likely primarily neuropathic in origin and were dosage sensitive.
Figure 3.
Skeletal muscle morphology and histochemical staining in transgenic mice compared with control. (A–E) H&E-stained vastus lateralis muscle. (C, D, and E) Marked variation in fiber size, increased numbers of fibers with central nuclei (arrowheads), and basophilic staining regions (arrow) were observed in a severely affected MP3–5 mouse. Staining in control (A) and each of the other lines (B, represented here with an HA21 mouse) was normal. (F–H) Fiber-type grouping is shown by the acid-stable myosin ATPase reaction. In control muscle (F), type 1 (brown) fibers are uniformly distributed among type 2 (pink) fibers. This uniform staining was seen in HA21, HA39, and HA83 lines (G, represented here with an HA83h mouse). (H) Fiber-type grouping (clustering of brown-staining type 1 fibers) was found in a severely affected MP3–5 mouse. C was taken at a higher magnification than the others. (A, B, E–H, ×190; C, ×760; D, ×390.)
Neural Crest Migration Is Defective in PAX3-FKHR Animals.
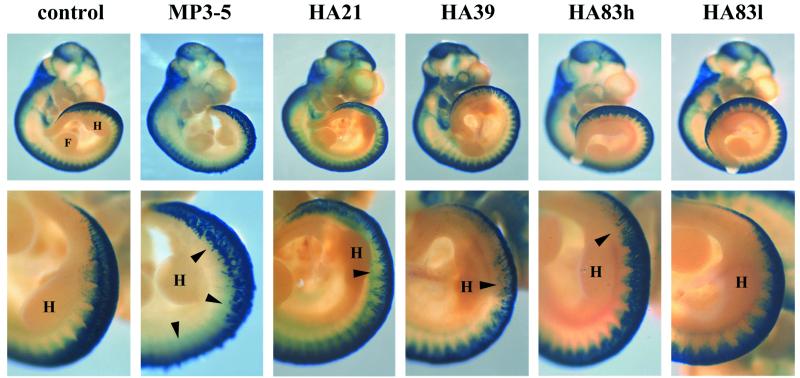
We next determined whether the pigmentary disturbances and embryonic lethality caused by PAX3-FKHR expression resulted from perturbation of the normal migration of neural crest cells and establishment of their derivative cell types. To test this, we crossed each of the PAX3-FKHR lines with Wnt-1/LacZ mice. The stable Wnt-1/LacZ transgenic line expresses the LacZ reporter gene under the control of Wnt-1 enhancer sequences (26). While mimicking Wnt-1 specific expression in cells of the dorsal neural tube, β-galactosidase activity can be observed in emigrating, migrating, and differentiating neural crest cells. E10.5 embryos were stained since dorsal neural tube expansion was first readily observed at this developmental stage. Whereas labeled cells were present in the dorsal neural tube of all embryos, fewer neural crest cells were emigrating from the dorsal neural tube, and with abnormal patterns in PAX3-FKHR mice compared with normal controls (Fig. 4). These effects corresponded to the level of PAX3-FKHR expression and the severity of neural tube dysmorphology among the lines of mice. Similar results were reported on crossing Splotch and Wnt-1/LacZ mice (27).
Figure 4.
Analysis of neural crest cell emigration from the dorsal neural tube. Whole-mount E10.5 Wnt-1/LacZ (control) and Tg/Wnt-1/LacZ embryos stained for β-galactosidase activity. Staining of cells in the cranial region, including the branchial arches and nasotemporal region around the eyes, was similar among each of the embryos. In the posterior region, centering on the hindlimb buds, fewer labeled cells in the transgenic lines can be seen emigrating from the neural tube (arrowheads). (Lower) A higher magnification of the hindlimb bud region. For orientation, the forelimb (F) and hindlimb (H) buds are labeled. (Upper, ×7; Lower, ×15.)
In Vivo Evidence That PAX3-FKHR and Pax3 Interfere with Each Other.
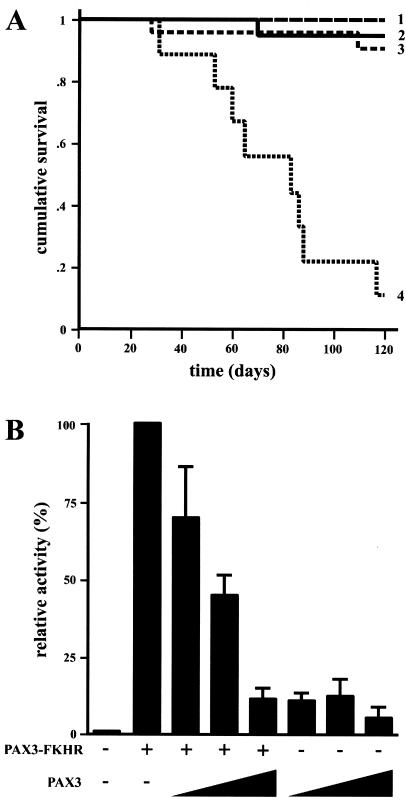
All of these results together suggested a correlation between PAX3-FKHR transgene number or expression levels and phenotypic alteration. To manipulate the levels of endogenous mouse Pax3, HA21 transgenic and Splotch heterozygous mice were mated. We hypothesized that if PAX3-FKHR interferes with endogenous Pax3, the compound heterozygotes would have exacerbated phenotypes. In fact, HA21/Splotch heterozygote animals had substantially worse phenotypes (Fig. 5 B and D), including a tendency toward premature death between 50 and 90 days after birth (Fig. 6A). Grossly distended bladders with particulate matter in most cases accompanied such deaths and in the severely affected urine accumulated in one or both kidneys. Premature death and bladder complications in compound HA83h/Splotch heterozygotes were also apparent but to a lesser extent. This obstructive uropathy likely arises from incomplete and/or improper innervation of the bladder.
Figure 5.
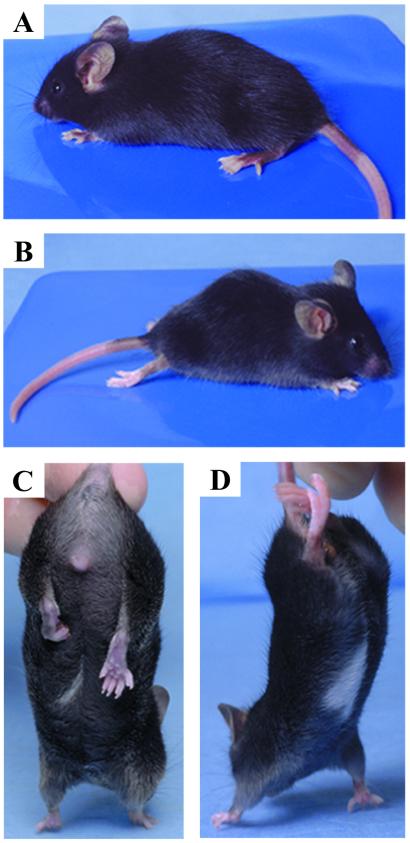
Representative phenotypes. (A and C) HA21 mouse showing overall appearance. Pigmentary disturbances of the tail and hindpaws were the most common features seen in all lines, except HA83l (A). A white belly patch was observed at a relatively low frequency only in HA21 and HA39 mice (C). HA21 and affected MP3–5 mice behaved abnormally when held by their tail, by pulling their hindlimb(s) to their body (C). (B and D) Compound HA21/Splotch heterozygotes displayed severe spinal dysraphism and kyphosis, paralysis and reduced sensory perception of their hindlimbs and tail, and exaggerated gait involving dragging their hindlimbs.
Figure 6.
Negative influence of PAX3-FKHR on Pax3 activity. Kaplan–Meier survival plots of offspring resulting from HA21 × Splotch matings show that HA21 Tg+Sp± mice had a significantly reduced lifespan (A). The survival of Tg−Sp+/+ (row 2, n = 20), Tg−Sp± (row 1, n = 20), Tg+Sp+/+ (row 3, n = 23), and Tg+Sp± (row 4, n = 9) mice was monitored over the course of 120 days. PAX3 and PAX3-FKHR compete for transcriptional activity (B). When increasing amounts of PAX3 were added (50 ng, 100 ng, and 1 μg), PAX3-FKHR transactivation of PRS-9 was blocked to the levels of PAX3 alone. The activity from PAX3-FKHR alone was arbitrarily set at 100%.
PAX3 and PAX3-FKHR Compete for Transcriptional Activity in Vitro.
Because PAX3-FKHR expression causes a neurogenic disorder that becomes more severe when Pax3 expression is reduced, PAX3-FKHR and Pax3 might be interfering with the transcriptional function of each other. To test this, transient transcription reporter assays were performed. It has been shown that the PRS-9 sequence, a variant of the e5 sequence from the Drosophila even-skipped promoter containing intact paired domain and homeodomain motifs, can be activated by PAX3-FKHR and PAX3 (11, 13, 17, 29). In fact, PAX3-FKHR activated nearly 10-fold better than did PAX3 (Fig. 6B). In the presence of increasing levels of PAX3, reporter transactivation by PAX3-FKHR was blocked in a dosage-dependent manner, reaching levels that were seen by PAX3 transactivation alone. Thus, PAX3-FKHR competes with PAX3 for transcriptional targets in vitro, mirroring its Pax3 dosage-dependent effects in vivo.
Discussion
Several reports have shown that PAX3-FKHR can transactivate PAX3-responsive sequences in vitro (8, 11–13). Because both the paired box and paired-type homeodomain of PAX3 are intact in PAX3-FKHR, it has been proposed to elicit its oncogenic transformation properties in part through up-regulation of PAX3-specific target genes. This would predict that developmental expression of PAX3-FKHR would perturb the normal role Pax3 plays in neural crest migration, as well as neural tube and somite development. The present results provide evidence for this, as well as demonstrating interference between PAX3-FKHR and Pax3. Moreover, it is interesting to note that despite the neuropathy and other developmental aberrancies, we observed no tumors in more than 300 mice. This suggests that, in contrast to similar types of translocations in leukemias and lymphomas (1, 2), PAX3-FKHR may not be sufficient to elicit tumorigenesis. Although this study was aimed at studying the results of PAX3-FKHR expression in the developing embryo, it is possible that tumors may arise only after constitutive expression within adult tissues. Recently, transgenic mice expressing the FUS-CHOP fusion protein have been shown to specifically induce liposarcomas despite ubiquitous expression in other adult tissues (30). It remains to be determined whether PAX3-FKHR must cooperate with other mutations such as p53 inactivation (31) to cause transformation, or whether it serves to cause developmental arrest and therefore an expanded pool of target cells from which to accumulate further genetic lesions.
Acknowledgments
We thank Martyn Goulding for the PRS-9 reporter constructs; Carrie Viars, Antonia Boyer, and Norma Prades for excellent technical assistance; and Phil Richter, Albee Messing, Charles De Smet, Frank Furnari, and Bill Biggs for helpful discussions. This work was supported in part by the Robert Steel Foundation for Pediatric Cancer Research.
Abbreviations
- ARMS
alveolar rhabdomyosarcoma
- HA
hemagglutinin
- H&E
hematoxylin and eosin
- En
embryonic day n
References
- 1.Kagan J, Croce C M. Ann Oncol. 1991;2:9–21. doi: 10.1007/978-1-4899-7305-4_2. [DOI] [PubMed] [Google Scholar]
- 2.Rabbitts T H. Nature (London) 1994;372:143–149. doi: 10.1038/372143a0. [DOI] [PubMed] [Google Scholar]
- 3.Turc-Carel C, Lizard-Nacol S, Justrabo E, Favrot M, Philip T, Tabone E. Cancer Genet Cytogenet. 1986;19:361–362. doi: 10.1016/0165-4608(86)90069-5. [DOI] [PubMed] [Google Scholar]
- 4.Douglass E C, Valentine M, Etcubanas E, Parham D, Webber B L, Houghton P J, Green A A. Cytogenet Cell Genet. 1987;45:148–155. doi: 10.1159/000132446. [DOI] [PubMed] [Google Scholar]
- 5.Galili N, Davis R J, Fredericks W J, Mukhopadhyay S, Rauscher F J, III, Emanuel B S, Rovera G, Barr F G. Nat Genet. 1993;5:230–235. doi: 10.1038/ng1193-230. [DOI] [PubMed] [Google Scholar]
- 6.Davis R J, D'Cruz C M, Lovell M A, Biegel J A, Barr F G. Cancer Res. 1994;54:2869–2872. [PubMed] [Google Scholar]
- 7.Arden K C, Anderson M J, Finckenstein F G, Czekay S, Cavenee W K. Genes Chromosomes Cancer. 1996;16:254–260. doi: 10.1002/(SICI)1098-2264(199608)16:4<254::AID-GCC5>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 8.Fredericks W J, Galili N, Mukhopadhyay S, Rovera G, Bennicelli J, Barr F G, Rauscher F J., III Mol Cell Biol. 1995;15:1522–1535. doi: 10.1128/mcb.15.3.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mansouri A, Hallonet M, Gruss P. Curr Opin Cell Biol. 1996;8:851–857. doi: 10.1016/s0955-0674(96)80087-1. [DOI] [PubMed] [Google Scholar]
- 10.Schafer B W, Czerny T, Bernasconi M, Genini M, Busslinger M. Nucleic Acids Res. 1994;22:4574–4582. doi: 10.1093/nar/22.22.4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sublett J E, Jeon I-S, Shapiro D N. Oncogene. 1995;11:545–552. [PubMed] [Google Scholar]
- 12.Bennicelli J L, Fredericks W J, Wilson R B, Rauscher F J, III, Barr F G. Oncogene. 1995;11:119–130. [PubMed] [Google Scholar]
- 13.Bennicelli J L, Edwards R H, Barr F G. Proc Natl Acad Sci USA. 1996;93:5455–5459. doi: 10.1073/pnas.93.11.5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Epstein J A, Shapiro D N, Cheng J, Lam P Y P, Maas R L. Proc Natl Acad Sci USA. 1996;93:4213–4218. doi: 10.1073/pnas.93.9.4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Auerbach R. J Exp Zool. 1954;127:305–329. [Google Scholar]
- 16.Conway S J, Henderson D J, Kirby M L, Anderson R H, Copp A J. Cardiovasc Res. 1997;36:163–173. doi: 10.1016/s0008-6363(97)00172-7. [DOI] [PubMed] [Google Scholar]
- 17.Goulding M D, Chalepakis G, Deutsch U, Erselius J R, Gruss P. EMBO J. 1991;10:1135–1147. doi: 10.1002/j.1460-2075.1991.tb08054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Natoli T A, Ellsworth M K, Wu C, Gross K W, Pruitt S C. Development (Cambridge, UK) 1997;124:617–626. doi: 10.1242/dev.124.3.617. [DOI] [PubMed] [Google Scholar]
- 19.Anderson M J, Viars C S, Czekay S, Cavenee W K, Arden K C. Genomics. 1998;47:187–199. doi: 10.1006/geno.1997.5122. [DOI] [PubMed] [Google Scholar]
- 20.Daston G, Lamar E, Olivier M, Goulding M. Development (Cambridge, UK) 1996;122:1017–1027. doi: 10.1242/dev.122.3.1017. [DOI] [PubMed] [Google Scholar]
- 21.Wilkinson D G. In: In Situ Hybridization: A Practical Approach. Wilkinson D G, editor. Oxford: IRL; 1992. pp. 75–83. [Google Scholar]
- 22.Conlon R. In: Manipulating the Mouse Embryo: A Laboratory Manual. Hogan B, Beddington R, Costantini F, Lacy E, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. pp. 360–362. [Google Scholar]
- 23.Dubowitz V. In: Muscle Biopsy: A Practical Approach. Dubowitz V, editor. London: Bailliere Tindale; 1985. pp. 19–40. [Google Scholar]
- 24.Chen C A. In: Current Protocols in Molecular Biology. Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. New York: Wiley; 2000. pp. 9.1.7–9.1.11. [Google Scholar]
- 25.Seed B, Sheen J-Y. Gene. 1988;67:271–277. doi: 10.1016/0378-1119(88)90403-9. [DOI] [PubMed] [Google Scholar]
- 26.Echelard Y, Vassileva G, McMahon A P. Development (Cambridge, UK) 1994;120:2213–2224. doi: 10.1242/dev.120.8.2213. [DOI] [PubMed] [Google Scholar]
- 27.Serbedzija G N, McMahon A P. Dev Biol. 1997;185:139–147. doi: 10.1006/dbio.1997.8551. [DOI] [PubMed] [Google Scholar]
- 28.Kaufman M H. The Atlas of Mouse Development. San Diego: Academic; 1995. [Google Scholar]
- 29.Chalepakis G, Fritsch R, Fickenscher H, Deutsch U, Goulding M, Gruss P. Cell. 1991;66:873–884. doi: 10.1016/0092-8674(91)90434-z. [DOI] [PubMed] [Google Scholar]
- 30.Perez-Losada J, Pintado B, Gutierrez-Adan A, Flores T, Banares-Gonzalez B, Calabia del Campo J, Martin-Martin J F, Battaner E, Sanchez-Garcia I. Oncogene. 2000;19:2413–2422. doi: 10.1038/sj.onc.1203572. [DOI] [PubMed] [Google Scholar]
- 31.Mulligan L M, Matlashewski G J, Scrable H J, Cavenee W K. Proc Natl Acad Sci USA. 1990;87:5863–5867. doi: 10.1073/pnas.87.15.5863. [DOI] [PMC free article] [PubMed] [Google Scholar]