Abstract
Sirtuin 1 (SIRT1) is known to deacetylate histones and non-histone proteins including transcription factors thereby regulating metabolism, stress resistance, cellular survival, cellular senescence/aging, inflammation-immune function, and endothelial functions, and circadian rhythms. Naturally occurring dietary polyphenols, such as resveratrol, curcumin, quercetin, and catechins, have antioxidant and anti-inflammatory properties via modulating different pathways, such as NF-κB- and mitogen activated protein kinase-dependent signaling pathways. In addition, these polyphenols have also been shown to activate SIRT1 directly or indirectly in a variety of models. Therefore, activation of SIRT1 by polyphenols is beneficial for regulation of calorie restriction, oxidative stress, inflammation, cellular senescence, autophagy/apoptosis, autoimmunity, metabolism, adipogenesis, circadian rhythm, skeletal muscle function, mitochondria biogenesis and endothelial dysfunction. In this review, we describe the regulation of SIRT1 by dietary polyphenols in various cellular functions in response to environmental and pro-inflammatory stimuli.
Keywords: Polyphenols, SIRT1, oxidants, resveratrol, inflammation
INTRODUCTION
Sirtuins, the class III histone deacetylases (HDACs), are widely distributed and have been shown to regulate a variety of physiopathological processes, such as inflammation, cellular senescence/aging, cellular apoptosis/proliferation, metabolism, and cell cycle regulation. There are seven mammalian enzymes belonging to class III HDACs: SIRT1 to SIRT7. The details of localization, main functions, and substrate of SIRT1 to SIRT7 are given in Table 1. The best characterized and well-studied among the human sirtuins is sirtuin1 (SIRT1), a nuclear protein reported to regulate critical metabolic and physiological processes [1–5] (Figure 1). SIRT1, a mammalian ortholog of yeast silent information regulator 2 (Sir2), plays an important role in regulation of pathogenesis of chronic diseases including diabetes, chronic inflammatory pulmonary diseases, neurodegenerative diseases, cardiovascular diseases, and chronic renal diseases. Sir2 is the first to be reported to extend lifespan up to 70% in yeast, fly and the nematode via maintaining silent chromatin by deacetylating core histones [6–10]. The mechanism in regulation of SIRT1 or Sir2 on these processes is due to its ability to deacetylate histones and non-histone proteins, such as nuclear factor (NF)-κB, forkhead box class O (FOXO) 3, p53, peroxisome proliferator activated receptor (PPAR)-γ, PPAR-γ coactivator 1α (PGC-1α), and endothelial nitric oxide synthase (eNOS) [2–4].
Table 1.
Mammalian SIRTs family: cellular functions and polyphenol effects
| Sirtuin | Intracellular Location |
Substrate | Function | Polyphenol effect |
Ref. |
|---|---|---|---|---|---|
| SIRT1 | Nucleus and cytoplasm |
NF-κB, p53, FOXO |
Cell survival, differentiation, inflammation, and metabolism |
Increase by resveratrol and quercetin |
[12–14, 16, 39, 224] |
| SIRT2 | Cytoplasm | α-tublin, Histone H4 |
Cell cycle | Activation by resveratrol |
[225] |
| SIRT3 | Nucleus and mitochondria |
Acetyl-CoA synthetase |
Metabolism | Increase by kaempferol |
[226] |
| SIRT4 | Mitochondria | ADP ribosyl transferase |
Metabolism | Not known | [227] |
| SIRT5 | Mitochondria | Mitochondrial protein |
Mitochondrial function |
Not known | [228] |
| SIRT6 | Nucleus | ADP-ribosyl transferase and Histone H3 |
DNA repair | Not known | [229] |
| SIRT7 | Nucleus | RNA polymerase I |
rRNA transcription | Increase by resveratrol |
[230] |
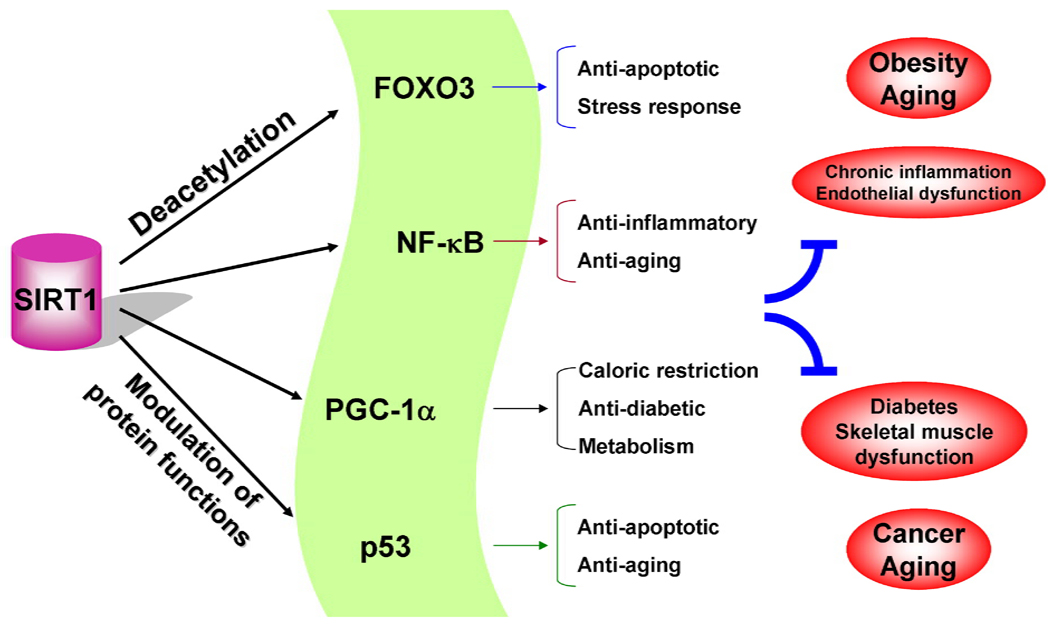
Figure 1. A schematic diagram for the role of SIRT1 in cell functions.
SIRT1 regulates a variety of cellular processes via deacetylating transcription factors and proteins thereby controlling the progression of chronic inflammatory and metabolic diseases as well as cancer.
Polyphenols, such as resveratrol, quercetin and catechins, have been shown to activate SIRT1 either directly or indirectly in vitro and in vivo [11–16]. Hence, the activation of SIRT1 by polyphenols would be beneficial in therapeutic intervention of a variety of chronic diseases. This review focuses on cellular and biological functions of SIRT1 and its regulation by polyphenols. Understanding the role and mechanisms of polyphenols in SIRT1 regulation and cellular functions will help in identification of pharmacological agents for their possible use as nutraceuticals in management of chronic diseases.
SIRT1 REGULATION
SIRT1 regulation by nicotinamide adenine dinucleotide (NAD+)
Unlike class I and II HDACs, SIRT1 activity requires NAD+ as cofactor and is not inhibited by trichostatin A [17]. SIRT1 removes acetyl groups from proteins by transferring the acetyl group to NAD+, generating two metabolites; 2′-O-acetyl-ADPribose and nicotinamide (NAM). Thus, the deacetylating activity of SIRT1 can be inhibited by the reaction product, NAM [8, 9, 18, 19]. There are two different routes of NAD+ biosynthesis in yeast and mammalian cells i.e. de novo production and salvage pathway [20]. In mammals, NAM can be converted into NAD+ through the salvage pathway; first converted to nicotinamide mononucleotide (NMN) by a rate-limiting enzyme nicotinamide phosphoribosyltransferase (NAMPT) and then to NAD+ by nicotinamide mononucleotide adenylyltransferase (NMNAT). Mammalian SIRT1 or Sir2 activation by NAD+ salvage pathway is shown to confer protection against high glucose environment in endothelial cells as well as myocardial infarction and ischemia/reperfusion injury in the heart [21–23]. It has also been shown that activation of NAD+ salvage pathway extends replicative lifespan in vascular smooth muscle cells by activating SIRT1 [24]. Therefore, activation of the enzymes (e.g. NAMPT or NMNAT) involved in NAD+ salvage pathway plays an important role in regulating SIRT1 activity [25, 26]. Resveratrol and quercetin are indirect activators of SIRT1 and have been to shown to activate the expression/activity of NAMPT and AMP-activated kinase (AMPK) [27–30]. AMPK is also shown to activate NAMPT thereby increasing intracellular level of NAD+ [24, 31]. Furthermore, activation of AMPK by 5-aminoimidazole-4-carboxamide-1-β-D-ribofuranoside (AICAR) increased the SIRT1 protein in skeletal muscle and attenuated LPS-induced lung inflammation [32, 33]. Interestingly, SIRT1 and AMPK are reported to mutually affect the functions of each other [24, 29, 30]. Therefore, further studies are required to investigate whether resveratrol’s real target is SIRT1, NAMPT, or AMPK since recent studies showing that SIRT1 is not a direct target of resveratrol [34, 35].
Interplay of SIRT1 with poly(ADP-ribose) polymerase 1 (PARP-1): NAD+ as a common substrate
In addition to SIRT1, PARP-1 also requires NAD+ as a substrate particularly during the DNA strand breaks to form ADP-ribose polymers, which non-covalently modifies targeting proteins. PARP-1 is a nuclear protein that is involved in DNA repair and chromatin remodeling. PARP-1 has been shown to possess a dual role in cell survival and necrosis via inducing autophagy and ATP depletion, respectively [36]. PARP-1 and SIRT1 were first shown to crosstalk in apoptosis-inducing factor (AIF)-mediated apoptosis [37]. Although the inhibition of SIRT1 by PARP-1 activation and subsequent NAD+ depletion has been determined in response to H2O2-induced cell senescence [38], the effect of oxidative/genotoxic stimuli on PARP-1 activation and SIRT1 activity still remains to be shown. Exposure of environmental and oxidative/genotoxic stresses leads to NAD+ depletion by concomitant decrease in SIRT1 activity and PARP-1 activation [38–41]. Furthermore, treatment with benzo[a]pyrene, a component of cigarette smoke (CS), causes intracellular NAD+ depletion by PARP-1 activation leading to cellular necrosis [42]. Over-activation of PARP-1 results in NAD+ depletion along with cellular senescence or death [43]. Recently, it has been shown that a methylxanthine derivative (a component of tea polyphenols) theophylline, an anti-inflammatory agent, protects against NAD+ depletion via PARP-1 inhibition and associated sparing of SIRT1 activity in macrophages and lung cells of patients with chronic obstructive pulmonary disease (COPD) [44–46]. Hence, a functional link may exist between PARP-1 and SIRT1 through NAD+ cofactor availability, and any changes in levels of intracellular NAD+ and/or PARP-1 activity, particularly in response to oxidants and environmental stimuli/inhaled pollutants, may influence SIRT1 activity. Surprisingly, inhibition of PARP-1 did not restore CS-induced decrease in SIRT1 activity in lung epithelial cells even in the present of resveratrol. This may be due to SIRT1 post-translational modifications, such as carbonylation/alkylation, thereby impairing the restoration of SIRT1 activity even the levels of NAD+ is elevated after PARP-1 inhibition [47]. Recent studies in double mutating PARP-1 and SIRT1 showed genome instability and chromatin modifications and increased lethality in mice [48], suggesting the key roles of PARP-1 and SIRT1 in cellular functions. It is interesting to note that PARP-1 can be subjected to acetylation and subsequent activation in the stress conditions (UV irradiation and genotoxic stresses), which is regulated by SIRT1 [49]. Therefore, it seems that a negative feedback loop exists between SIRT1 and PARP-1 in regulating cellular function.
SIRT1 regulation by oxidative stress and redox modification
SIRT1 has been shown to reduce cellular oxidative stress burden indirectly through deacetylation of FOXO3a (deacetylation of FOXO3a leads to up-regulation of catalase and MnSOD) [50]. SIRT1 also regulates aging and oxidative stress in the cardiomyocytes and endothelial cells, and oxidative stress leads to a redistribution of SIRT1 on chromatin [51–53]. Thus, SIRT1 either directly or indirectly can influence the redox property of the cell. In addition to reduce cellular oxidative stress burden, SIRT1 is also regulated by oxidative stress. We have recently shown that the levels of SIRT1 are decreased in vitro in lung epithelial cells, endothelial cells, and macrophages in response to cigarette smoke extract (CSE), as well as in lungs of patients with COPD [47, 54–56]. SIRT1 also undergoes oxidative/nitrosative post-translational modifications as shown by increased nitration of tyrosine residue and carbonylation (acrolein/4-hydroxy-2-nonenal-adducts formation) on cysteine residue in lungs of smokers and patients with COPD compared with nonsmokers as well as in human monocyte/macrophage cells and endothelial cells treated with aldehyde and CSE [39, 47, 54]. Furthermore, SIRT1 is shown to be a redox-sensitive molecule since intracellular thiols regulate its level and activity. Treatment with buthionine sulfoximine, an inhibitor for glutathione biosynthesis, further decreases SIRT1 levels in response to oxidative/carbonyl stress, whereas elevation of intracellular thiols pool by N-acetyl-L-cysteine rescues oxidant/carbonyl-mediated depletion of SIRT1 levels in epithelial cells. Furthermore, aldehydes cause carbonyl adducts formation on SIRT1 on cysteine residues, which is decreased by increasing intracellular thiols [39].
SIRT1 is also subjected to phosphorylation, which affects its activity and protein levels through proteasome-dependent or independent degradation [39, 54, 57, 58]. Various serine phosphorylation sites on SIRT1 (S27, S47, S659, and S661) were identified which is regulated by protein kinase CK2 and JNK under basal physiological conditions [57–61]. We have recently showed that oxidants/aldehydes derived from tobacco smoke caused SIRT1 phosphorylation in macrophages, epithelial cells as well as in mouse lungs [39]. Proteasome inhibitors inhibited phosphorylation of SIRT1 suggesting that phosphorylation in addition to covalent oxidative/nitrosative modifications of SIRT1 cause irreversible modifications of SIRT1 and subsequent proteasomal degradation [39, 47]. Taken together, it may be surmised here that SIRT1 is a novel redox-sensitive protein, which can be regulated by post-translational modifications, such as carbonylation and phosphorylation [39, 47, 54]. Oxidants/electrophiles covalently modify SIRT1 post-translationally, decreasing enzymatic activity and marking the protein for proteasomal degradation [39]. We and others have shown that SIRT1 is also subject to S-glutathionylation and its enzymatic activity is modulated by intracellular redox GSH status [39, 62]. Oxidant/carbonyl stress-induced reduction of SIRT1 may have implications in chronic inflammatory conditions. Recent study indicated that resveratrol and other dietary polyphenols attenuate mitochondrial oxidative stress in endothelial cells via activation of SIRT1 [63]. We and others have shown that resveratrol and SRT1720 can activate SIRT1 in a variety of human cell lines [47, 64], but it remains to be seen whether the similar activation of SIRT1 can occur in vivo by polyphenols in response to oxidative and pro-inflammatory stimuli.
POLYPHENOLS AND SIRT1 ACTIVATORS
Polyphenols are secondary metabolites of plants and represent a vast group of compounds having aromatic ring(s), characterized by presence of one or more hydroxyl groups with varying structural complexities. The most widely distributed group of plant phenolics are flavonoids. The flavonoids subclasses comprise of flavonols, flavones, isoflavones, antocyanidins, and others. The commonly studied dietary polyphenols, such as resveratrol, quercetin, and catechins, have been reported to possess antioxidant and anti-inflammatory properties. Resveratrol (3,5,4’-trihydroxystilbene) is a phytoalexin found in red wine and grapes, which has two phenolic rings connected by a double bond and has two isoforms; trans-resveratrol and cis-resveratrol, the former being more stable in its biological activity. Quercetin (3,3’,4’,5,7-pentahydroxylflavone) is a plant-derived flavonol found in apples, tea, capers, and onion, used as a nutritional supplement. Catechins are monomeric flavanols comprising of chemically similar compounds, such as epicatechin, epigallocatechin, epicatechin gallate (EGC), and epigallocatechin gallate (EGCG). EGCG predominates among the various tea polyphenols and is considered to be the major bioactive and well-studied catechin. Several reports highlighted that dietary supplementation of polyphenols may protect against neurodegenerative, cardiovascular, inflammatory, metabolic diseases, and cancer by enhancing SIRT1 deacetylase activity. However, the therapeutic and pharmacological potential of these natural compounds remains to be translated in humans in clinical conditions. This is in part due to the lack of knowledge of their mode of action as well as their multiple signaling targets, non-specificity, complex pharmacokinetic properties (e.g. absorption, biotransformation, and bioavailability). Furthermore, these polyphenols may act as pre-emptying or prophylactic agents in terms of dietary intake/interventions in susceptible conditions rather than as therapeutic agents.
Resveratrol is the first polyphenolic compound which has been shown to activate SIRT1 [13–16]. However, the mechanism of SIRT1 activation by resveratrol has been debated, and recent studies show that resveratrol is not specific activator of SIRT1 [34, 35]. In addition, there are some scanty reports available that quercetin and catechins also activate mammalian SIRT1 or yeast Sir2 albeit to a lesser extent as compared to resveratrol [6, 11, 12]. However, the separate studies show that polyphenols, such as EGCG and quercetin did not exhibit any ability to activate SIRT1 in cellular system [12, 65, 66]. On the contrary, these polyphenols inhibit SIRT1 activity [12]. This is due to their instability to form oxidized form and produce reactive oxygen species in the medium (i.e. EGCG) or the formation of SIRT1-inhibitory metabolites (i.e. quercetin and its metabolites) [12].
A number of other compounds (mainly analogs of resveratrol) have been reported to activate SIRT1 activity in mammals [67–69]. A newly identified and more potent SIRT1 activator, SRT1720, has been reported to be 800–1000-fold more effective than resveratrol [69]. Milne and their colleagues identified SRT1460 and SRT2183 which can increase SIRT1 enzyme activity by about 3–5 fold [69]. SRT2172 has been shown to be more effective in inhibiting matrix metalloproteinase-9 (MMP-9) production in monocytes as compared to resveratrol [64]. Interestingly, recent studies also showed that SRT1720, SRT2183, and SRT1460 are nonspecific for SIRT1 activation [34]. Therefore, development of specifically pharmacological SIRT1 activator is crucial in understanding the role of SIRT1 in cellular function and potentially clinical application of SIRT1 activators.
CELLULAR AND BIOLOGICAL FUNCTIONS OF SIRT1: ROLE OF POLYPHENOLS
In calorie restriction (CR)
Sir2 has been identified as one of the key proteins in not only establishing the transcriptional silencing via deacetylating histone H4 at lys16 (K16), but also extending the lifespan in yeast, C. elegans, and drosophila [9, 70]. CR is the major mechanism known to extend the lifespan of organisms. Activation and regulation of SIRT1 has been extensively studied in understanding the underlying mechanism of CR-mediated lifespan extension. Activation of Sir2 or mammalian SIRT1 has been shown to contribute to the beneficial effects of CR in yeast and more complex organisms [16, 50, 71–75]. A recent study has shown enhanced renal cell adaption to hypoxia in condition of CR, which is mediated by activating SIRT1-FOXO3 pathway [75]. CR not only increases SIRT1 levels and activity, but also skews the NAD+/NADH ratio towards higher NAD+ levels as well as decreased NAM [31]. Under ad-libitum condition, glucose is converted to pyruvate by glycolysis whereas glycolysis is decreased and there is a shift towards respiration under CR condition [72, 76]. This shift increases the NAD+/NADH ratio leading to SIRT1 activation. SIRT1 is also shown to increase gluconeogenesis in the liver leading to hepatic glucose output, which is a characteristic under CR [77]. Indeed, SIRT1 transgenic mice have the phenotypes resembling CR [78]. It is interesting to note that Sir2-independent pathway also exists in CR-mediated the lifespan extension [79–81]. The effect of resveratrol on lifespan extension has been tested in C. elegans, yeast, fruitfly, mice, and human cells [9, 13, 82–84]. These synthetic compounds or analogs of resveratrol may provide a potential therapeutic benefit in mimicking CR by activating SIRT1 [85–87].
Polyphenols have been shown to activate SIRT1 which in turn mimic the CR, and hence halt the aging process. The role of SIRT1 is also highlighted in chronic lung diseases, such as asthma and COPD, where lung levels of SIRT1 are altered [55, 64, 88]. This is in particular important as COPD is now considered a disease of accelerated aging where SIRT1 levels are dramatically reduced as the disease progresses. Hence, using SIRT1 activators either by novel synthetic resveratrol derivatives or other polyphenolic compounds may halt the progression of accelerated aging and/or reverse abnormal cellular immune-inflammatory functions including steroid resistance to inhibit inflammatory response in chronic lung disorders. It is interesting to note that a dietary supplementation of 0.1% quercetin significantly reduced the lifespan of mice [89]. This may be due to the SIRT1 inhibition by quercetin metabolite quercetin-3-O-glucuronide in vivo [12]. Similar possibilities may exist with other polyphenols and their interactions with quinones and aldehydes or other biochemical molecules which needs to be determined particularly in vivo.
In metabolism
SIRT1 activity requires NAD+ as a cofactor whereas it can be inhibited by NADH which reflect the energy status of a cell [72, 90]. Both NAD+ and NADH play an important role in cellular metabolism and survival [91]. It is therefore proposed that SIRT1 acts as a molecular link between cellular metabolism via the redox equivalents, NAD+/NADH levels, mitochondria biogenesis, and cellular functions. Decreased SIRT1 activity by altered NAD+ level has effects on its substrate, PGC-1α. As an important mitochondrial activity regulator, PGC-1α modulates energy production by regulating mitochondrial biogenesis and function [87]. Recent in vivo studies have revealed that resveratrol supplementation leads to increased mitochondrial activity and PGC-1α activity with a concomitant decrease in their acetylation status [87, 92]. SIRT1-mediated deacetylation of PGC-1α by resveratrol acts as regulator of mitochondrial energy balance and biogenesis. Increasing lines of evidence suggests that the activation of SIRT1 might be effective in intervention and prevention of type 2 diabetes [93, 94]. SIRT1 modulates glucose-ATP signaling and insulin secretion from pancreatic β-cells mainly via regulating uncoupling protein 2 (UCP2), FOXO2 and NAD+-dependent pathway [95]. Overexpression of SIRT1 in pancreatic β-cells improves glucose tolerance and enhances insulin secretion in response to glucose, while SIRT1 knockdown exhibits impaired glucose-stimulated insulin secretion [95, 96]. Furthermore, SIRT1 overexpression attenuates the high fat-induced hepatic steatosis by induction of manganese superoxide dismutase (MnSOD) and NF-E2-related factor-1 (Nrf1) as well as by lowering the activity of TNF-α and IL-6 via downregulation of NF-κB activity [97]. Deficiency of SIRT1 enhances liver steatosis with increased liver lipid contents and inflammation as compared to wild-type mice in response to moderate and high fat diets [98]. These observations suggest that polyphenol-dependent activation of SIRT1 may be a potentially therapeutic approach for management of metabolic diseases.
In energetics
The dependence on NAD+ as a cofactor links SIRT1 activity to the energetic state of the cell. AMPK has been shown to regulate energy expenditure by modulating NAD+ metabolism and SIRT1 activity suggesting a direct involvement of AMPK in regulating SIRT1 and energy metabolism [24]. Indeed, mice deficient with SIRT1 are hypermetabolic but lethargic, and they can not utilize ingested food inefficiently. Further investigation shows that SIRT1 knockout mice are defective in energy generation system, which is reflected by interrupted generation of ATP in liver mitochondria during food deprivation [90]. These results indicate both AMPK and SIRT1 might act as energy sensors that regulate energy metabolism [99]. It has been shown that oxidative stress and CS reduce SIRT1 level/activity, and implicate in altered cellular energetics. This is confirmed by the study that CSE lowers ATP levels by blocking the mitochondrial respiratory chain leading to cellular necrosis [100]. However, it is unknown whether this decreased ATP level is associated with reduced SIRT1 in response to CS/oxidative stress.
In addition to antioxidant and anti-inflammatory effect, polyphenols have been shown to regulate energy metabolism. Polyphenol EGCG is shown to reduce energy absorption and increase fat oxidation in diet-induced obesity of mice [101]. This is associated with SIRT1 and PGC-1α activation thereby improving mitochondria function and protecting subsequent metabolic diseases [87].
In inflammation
SIRT1 protein is associated directly with RelA/p65 subunit of NF-κB, and deacetylates lys310 residue of RelA/p65, a site that is critical for NF-κB transcriptional activity [102]. Recent studies have shown that SIRT1 also deacetylates and suppresses the transcription activity of activator protein-1 (AP-1) leading to down-regulation of cyclooxygenase-2 (COX-2) gene expression [103–105]. SIRT1 knockout or knockdown leads to increased NF-κB activation and proinflammatory cytokine release whereas activation by SIRT1 activators (e.g. SRT1720 and resveratrol) inhibits NF-κB-mediated inflammatory mediators release in vitro and in vivo suggesting the role of SIRT1 in regulation of inflammation, and activation of SIRT1 by activators or polyphenols would be an approach for the intervention of various chronic inflammatory diseases [55, 69, 84, 106–109]. Furthermore, SIRT1 level is reduced in rat lung and human macrophage cells exposed to CS as well as in lungs of smokers and patients with COPD [55, 56], implicating a pivotal role of SIRT1 in the pathogenesis of COPD. Recently, it has been shown that SIRT1 activators also inhibit NF-κB-mediated inflammatory mediators release and possibly overcome steroid-resistance in response to oxidative stress [55, 56, 64, 110]. However, the role of endogenous SIRT1 in the development of emphysema/COPD and resistance to steroid therapy and whether or polyphenols (resveratrol, quercetin, cucumin, and catechins) can reverse steroid-resistance is not known. Similarly, SIRT1 activation by resveratrol leads to down-regulation of NF-κB which was associated with abrogation of colitis [108]. Overall, it can be surmised that activation of SIRT1 might act as a novel immunomodulatory approach in intervention of chronic inflammatory diseases, such as COPD, diabetes, and colitis via modulating NF-κB-dependent pathway.
Polyphenols (e.g. catechins and curcumin) have been shown to induce hypoacetylation of RelA/p65 by directly inhibiting the activity of histone acetyltransferase (HAT) enzymes leading to the down-regulation of NF-κB function and associated inflammatory response [65]. It is possible that dietary polyphenols could induce protein deacetylase activity, such as HDAC2/SIRT1, which in turn may inhibit the transcription of pro-inflammatory genes (Figure 2). This is supported by the report that resveratrol inhibits inflammatory cytokine expression in response to lipopolysaccharide challenge in rat lungs [109]. Furthermore, the expression of various genes encoding other pro-inflammatory factors, such as COX-2, MMPs, adhesion molecules, and inducible nitric oxide synthase (iNOS) can be significantly inhibited by resveratrol, quercetin, curcumin and catechins via down-regulating NF-κB and AP-1 transcription factors [111–114]. Green tea catechin EGCG has been shown to inhibit CSE-induced pro-inflammatory cytokine release in lung epithelial cells, which is associated with inhibition of NF-κB and AP-1 [115–117]. Similar to curcumin, quercetin and catechins also modulate a myriad of inflammatory signaling pathways [11, 118, 119]. However, the role of quercetin and catechins in modulation of SIRT1 and hence controlling the inflammatory response is currently interesting areas of research. In light of the above observations, it appears that polyphenols (e.g. resveratrol, quercetin, and catechins) can modulate a variety of proinflammatory pathways via activating SIRT1 thereby inhibits NF-κB and/or AP-1 pathways [63, 120, 121].
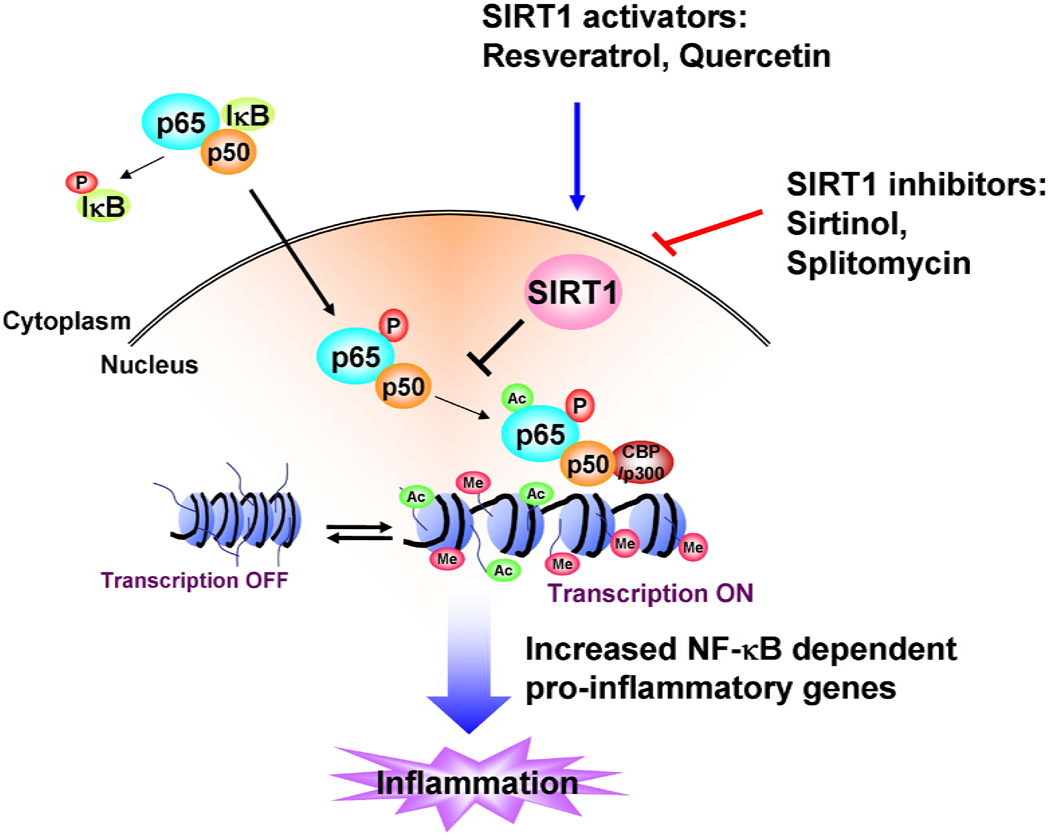
Figure 2. Role of SIRT1 in inflammation.
SIRT1 suppresses NF-κB transcription factor by deacetylation of RelA/p65 subunit. Acetylation of NF-κB increases the transcription of proinflammatory mediators. Polyphenols, such as resveratrol and quercetin, can increase the activity of SIRT1 leading to repression of inflammation.
In immune function
SIRT1 has been shown to be associated with the regulation of immune function since SIRT1 is expressed at high levels in the thymus including CD4+ and CD8+ thymocytes, and knockout of SIRT1 enhances thymocytes apoptosis after ionizing radiation in mice [122]. This is supported by a recent finding that SIRT1 inhibits T cell activation and the loss of SIRT1 function results in abnormally increased T cell activation and a breakdown of CD4+ T cell tolerance [105]. In addition, SIRT1-deficient mice are unable to maintain T cell tolerance and are susceptible to autoimmune diseases [105, 122]. Furthermore, SIRT1 deacetylates forkhead box class P3 (FOXP3) thereby increasing the number and function of regulatory T cell [123]. Sera from adult SIRT1-null mice contained antibodies that reacted with nuclear antigens leading to formation of immune complexes, which were deposited in the livers and kidneys of these animals. Some of the SIRT1-null animals developed a disease resembling diabetes insipidus when they approached 2 years of age [124]. These observations are consistent with a role for SIRT1 in sustaining normal immune function and thus delaying the onset of autoimmune diseases, and highlights the role of novel SIRT1 activators in amelioration or prevention of these diseases [105]. Resveratrol is shown to have anti-asthmatic effects in mouse model [125]. On the contrary, administration of sirtinol, a SIRT1 inhibitor, significantly attenuated antigen-induced airway inflammation and hyperresponsiveness that are characteristics of asthma [88]. Therefore, further studies are required to study whether immune function is altered in these sirtinol- or resveratrol-treated mice as compared to control although immune dysfunction plays an important role in pathogenesis of asthma. Accumulating evidence has shown that polyphenols, such as resveratrol, catechins, and quercetins, have a regulatory role in immune function in vitro and in vivo [126–131]. Therefore, polyphenols may have beneficial effects to in prevention of the progression of chronic diseases where immune dysfunction occurs. Nevertheless, the role of polyphenols in SIRT1-mediated regulation in immune function remains to be studied.
In cellular apoptosis and tumorigenesis
SIRT1 regulates a variety of processes that alter cell response to genotoxicity, including the detoxification of reactive oxygen species (ROS) by up-regulation of MnSOD, DNA repair mechanisms (cyclin D, GADD45, p27/Kip1) and sensitivity of cells to apoptosis [53, 84, 132–135] (Figure 3). This is due to the deacetylation and activation of a transcription factor FOXO3a [133, 136]. Pharmacological inhibition of SIRT1 decreases cellular resistance to stress and hence promote cellular apoptosis due to reduced constraint on FOXO3/4 otherwise inhibited by SIRT1 [133]. FOXO3a activity is regulated by phosphorylation and acetylation [137]. These modifications lead to the loss of its transactivation properties; whereas transcriptional activity of FOXO3a is restored by deacetylation carried out by SIRT1 [136, 138]. Thus, SIRT1 regulates the ability of FOXO3a to induce cell cycle arrest; and high SIRT1 activity promotes cell survival suggesting SIRT1 tips FOXO-dependent response away from cell death and towards stress resistance [133]. FOXO1 is also involved in protection of SIRT1 against apoptosis in cardiomyocytes [139]. Furthermore, SIRT1-mediated COX-2 expression reduces oxidative stress-induced renal medullary interstitial cell apoptosis [140]. SIRT1 is also shown to regulate the expression of genes involved in apoptosis, such as Fas ligand, pro-apoptotic BH3-only proteins, Bcl-2 interacting mediators of cell death (BIM) and TNF-related apoptosis-inducing ligand [133, 136]. A separate study showed that specific overexpression of NAMPT in heart reduced the size of myocardial infarction and apoptosis via increasing the contents of NAD+ in response to prolonged ischemia and reperfusion [22]. It is not known if SIRT1-mediated regulation of FOXO3a or other factors plays a role in oxidant/carbonyl stress-induced apoptosis and whether polyphenols, such as resveratrol, regulate apoptosis via activating SIRT1 or NAMPT.
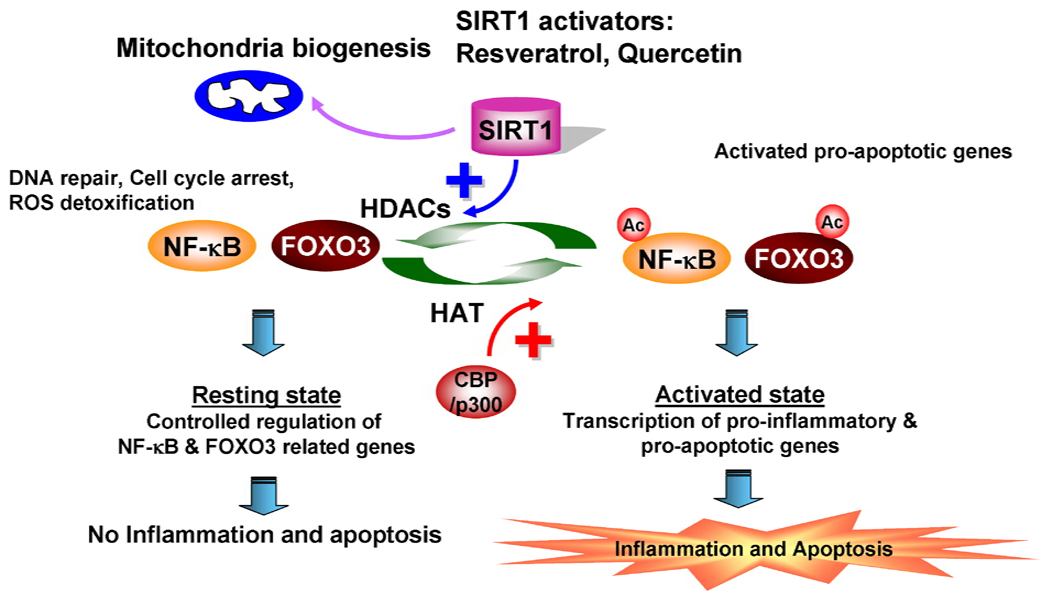
Figure 3. Role of FOXO and SIRT1 in cell functions.
FOXO transcription factor is shown to regulate cellular signaling involved in DNA repair, cell cycle, oxidative stress, and apoptosis. Acetylation of FOXO is associated with inflammatory cell activation and cellular apoptosis. Under resting conditions, inflammation-related genes are controlled by SIRT1 deacetylase. However, when NF-κB and FOXO are acetylated by CBP/p300, transcription of proinflammatory and apoptotic genes are initiated.
SIRT1 also interacts with p53 and deacetylates its C-terminal regulatory domain thereby decreasing its ability to induce cellular apoptosis [141–143]. SIRT1 inhibitors have ability to induce cancer cell damage by sensitizing the cells to p53-dependent apoptosis [6]. Therefore, oxidative stress/CS-mediated SIRT1 reduction may acetylate p53 leading to sustained lung cell apoptosis. Although it has been shown that nuclear SIRT1 levels are decreased in vivo and in vitro in response to CS/oxidants exposure [56], it is however, not known if SIRT1-mediated regulation of p53 (acetylation) plays a role in CS/oxidants-mediated apoptosis and senescence and whether polyphenols can reverse this via SIRT1. Several other SIRT1 protein substrates involved in cell stress response signaling have also been identified, including Ku70, a pro-apoptotic factor that is down-regulated and deacetylated by SIRT1 [77, 144]. However, the role of polyphenols in SIRT1-mediated these proteins deacetylation and cellular apoptosis is unclear and needs further investigation.
A role of SIRT1 in tumorigenesis is still controversial because SIRT1 has been shown to act as both tumor promoter and/or tumor suppressor. The levels of SIRT1 are increased in cancer and SIRT1 transgenic mice develop tumors suggesting a tumor-promoting role of SIRT1 [145–147]. This is attributed to the deacetylation of tumor-suppressor protein p53 on lysine 382 by SIRT1 leading to its inhibition and subsequent tumorigenesis [19, 142]. Thus, the inhibition of SIRT1 by its inhibitors would induce cell death of cancer cells by activating and acetylating p53. Interestingly, SIRT1 also acts as a tumor suppressor in vitro and in vivo. SIRT1 is known to sensitize tumor cells to TNF-α-induced cell death via inhibiting transactivation of NF-κB [84]. It is proposed that the ability of SIRT1 to induce either apoptosis or cell survival depends on the apoptotic stimuli and on whether this deacetylase is inhibiting NF-κB or p53. In mouse colon cancer model, SIRT1 suppresses intestinal tumor formation in vivo through deacetylation of β-catenin [148]. Breast cancer associated gene 1 (BRCA1) is the most frequently mutated tumor suppressor gene found in familial breast cancers. Wang et al. revealed that SIRT1 expression is much lower in the BRCA1-associated breast cancer than BRCA1-wildtype breast cancer in human [149]. They further showed that activation of SIRT1 by resveratrol induced apoptosis in BRCA-1 deficient cancer cells and strongly inhibited tumor formation. A separate study demonstrated that knockout or overexpression of SRIT1 did not exhibit any effect on incidence and tumor load of skin papillomas in mice [145]. Therefore, the effect of SIRT1 on tumorigenesis is complicated, and its effect may be cancer-specific and is related with the status of cancer cells (e.g. deactivation and/or activation of tumor suppressor genes).
Polyphenols are well known chemopreventive agents which are known to induce apoptosis and cell cycle arrest in cancer cells. Resveratrol has been shown to have anti-carcinogenic effect in vitro and in vivo [145, 149–152]. Similarly, quercetin inhibits cell proliferation in different cancer cells [153]. The underlying mechanism of these polyphenols’ anti-carcinogenic effect is associated with the regulation of SIRT1 [145, 149]. Other mechanisms such as antioxidant and anti-inflammatory properties involved in anti-carcinogenic effect of polyphenols can not be excluded because of the complicated effect of SIRT1 on tumorigenesis [154].
In cellular proliferation/senescence
SIRT1 has been shown to control cell cycle, proliferation and senescence due to its regulation on FOXO and p53 proteins. Senescent mouse embryonic fibroblasts and human endothelial cells have decreased levels of SIRT1 [155, 156]. SIRT1 can interact, deacetylate and activate FOXO3, a transcription factor promoting a variety of cellular responses, such as cell cycle arrest, cellular senescence, proliferation, and resistance to oxidative stress and apoptosis (Figure 3) [133]. The mechanism of SIRT1/FOXO3 or SIRT1/p53 pathway in cellular proliferation and senescence is associated with altered transcription of downstream cell cycle inhibitors (e.g. p16, p21, and p27) [38, 157]. These cyclin-dependent kinase inhibitors are the biological markers for cell cycle arrest and cellular senescence. We have shown that FOXO3 and p53 are acetylated when SIRT1 is reduced in response to oxidants/CS exposure in mouse lung [158]. Oxidative stress/CS exposure has been shown to induce senescence in lung epithelial cells and fibroblasts, and cellular senescence occurs in mouse lung exposed to CS and lungs of patients with COPD [159]. Therefore, studies on SIRT1-FOXO3 pathway will provide more insight into imbalance of cellular proliferation/senescence in response to oxidative stress, and whether polyphenols have effect on this pathway although polyphenols (e.g. resveratrol) regulate cellular senescence [160, 161]. Accumulating evidence shows that SIRT1 interacts eNOS-nitric oxide system thereby regulating vascular senescence and dysfunction as well as atherosclerosis [162]. Moreover, it is possible that SIRT1 regulates the function of p21 by posttranslational modification since p21 itself can be subject to acetylation/deacetylation [163]. However, it remains to be seen whether SIRT1 is also involved in p21-mediated regulation of cell proliferation/senescence and whether polyphenols can inhibit acetylation of p21 via activating SIRT1.
In autophagy
Autophagy is a dynamic process which is responsible for the turnover of cellular organelles and proteins thereby maintaining cell homeostasis and conferring adaption to adverse environmental stimuli. Excessive autophagy will lead to cell death. Inhibition of gene required for autophagy prevents CR-induced lifespan extension [164]. This is associated with the negative regulation of SIRT1 on the mammalian target of rapamycin (mTOR) [165], an evolutionarily-conserved protein kinase for modulating autophagy. Resveratrol reduces the activity of mTOR in a SIRT1 dependent manner [165]. SIRT1 can interact and deacetylate pro-autophagic Atg5, Atg7, and Atg8 [166]. Furthermore, SIRT1-deficient mice have accumulation of damaged organelles and disruption of homeostasis which resembles to Atg5 knockout mice [166]. NAMPT is also shown to regulate cell survival through autophagy in cardiomyocytes, which is associated with the alteration of NAD+ [167]. These results suggest that SIRT1 plays an important role in regulating autophagy. Interestingly, inhibition of AMPK by adenine 9-beta-d-arabinofuranoside or dominant negative AMPK significantly reduced autophagy in cardiac myocytes [168, 169]. Both SIRT1 and AMPK regulate each other [170], therefore, the differential roles of these molecules in autophagy is an interesting area of research particularly in response to oxidative stress. SIRT1 is shown to deacetylate the major AMPK kinase LKB1, thereby increasing its activity and ability to activate AMPK [171]. Thus, the SIRT1-LKB1-AMPK pathway may be involved in regulating autophagy. In addition to activate SIRT1, resveratrol is also shown to inhibit p70 S6 kinase thereby suppressing autophagy [172]. Other polyphenols, such as quercetin can also inhibit LPS-induced type-II microtubule-associated protein 1A/1B-light chain 3 production and aggregation [173]. It seems that multiple target molecules are involved in the regulation of polyphenols effects on autophagy. Investigations of these molecular pathways would identify the molecular target(s) in regulation of autophagy in chronic inflammatory diseases, such as COPD and diabetes, where autophagy is dysregulated [174–177].
In skeletal muscle function
Skeletal muscle dysfunction including function and structural alteration is an important feature in a variety of chronic inflammatory diseases, such as diabetes and COPD [178–180]. PGC-1α has been shown to regulate the expression of genes involved in fatty acid oxidation to increase lipid utilization thereby allowing skeletal muscle cells to survive under conditions of nutrient restriction [181]. It is interesting to note that PGC-1α mRNA is reduced while PGC-1α is acetylated in skeletal muscles from mice lacking SIRT1 [181, 182], suggesting that SIRT1 regulates PGC-1α by transcriptional and post-translational mechanisms. Treatment with nicotinamide, an inhibitor of SIRT1, decreased the expression of PGC-1α-target genes for mitochondrial and fatty acid utilization in primary skeletal muscle cells [181]. Therefore, regulation of SIRT1/PGC-1α pathway using SIRT1 activators would modulate or induce mitochondrial antioxidant genes thereby restoring skeletal muscle function. It is likely that SIRT1 activators including resveratrol increase AMPK thereby induce PGC1-α and restore mitochondrial function or increase mitochondrial biogenesis in skeletal muscle [183, 184].
Dietary polyphenols, such as resveratrol, quercetin, and catechin, in green tea are beneficial in improving exercise endurance by regulating lipid metabolism [185]. A variety of polyphenols including resveratrol activate SIRT1 and PGC-1α leading to reprogramming of muscle gene expression and improvement of mitochondrial function thereby protecting against metabolic stresses [87]. Further study suggests that activation and deacetylation of PGC-1α by resveratrol is mediated via enhanced SIRT1 activity [87]. Recently, a role of quercetin in exercise performance and skeletal muscle function via SIRT1 has also been reported [186]. Skeletal muscle atrophy is one of the key characteristics in cancer cachexia, which has a negative impact on prognosis, leading to asthenia, immobility and early death. Treatment with polyphenols (i.e. resveratrol, quercetin and curcumin) significantly attenuated total protein degradation in mouse myotubes in vitro, and administration of resveratrol attenuated weight loss and protein degradation in skeletal muscle of mice bearing the MAC16 tumor [187]. Hence, the development of specific SIRT1 activators with low toxicity and high bioavailability will pave an avenue to intervene the progression of chronic inflammatory and metabolic diseases by ameliorating skeletal muscle dysfunction.
In adipogenesis
Adipocytes are highly specialized cells that play a major role in adipogenesis. Excess adipocyte number is a hallmark of obesity and major risk factor for development of type 2 diabetes, cardiovascular diseases, and hypertension. SIRT1 is identified as a regulator of biological process including adipogenesis and adipolysis [188, 189]. SIRT1 represses PPAR-γ, an adipose specific nuclear hormone receptor by interacting with its cofactors, nuclear receptor co-repressor and silencing mediator of retinoid and thyroid hormone receptors (SMRT), thereby reducing adipogenesis and promoting lipolysis [190]. It is interesting to note that SIRT1 activates FOXO1 and CCAAT/enhancer-binding protein α interaction leading to increased adiponectin gene transcription, which is an essential factor for regulation of adipogenesis [189]. Moreover, treatment with resveratrol significantly up-regulates the mRNA and protein levels of adiponectin, which is associated with increased levels of FOXO1 in adipose tissue [191]. Other polyphenols, such as quercetin and catechins, also have anti-adipogenesis activity, which is related with increased phosphorylation of AMPK [192–194]. Resveratrol treatment also increased the SIRT1 expression, thus in turn, resulted in decreased lipid accumulation by repressing PPAR-γ [190]. Furthermore, SIRT1 positively regulates liver X receptor via deacetylation at lysine K432 and thus may play an important role in lipid homeostasis [195]. Thus, due to its biological and cellular regulatory mechanisms, SIRT1 merits particular attention for pharmacological intervention for obesity and age-related disease. Recently, activation of SIRT1 by resveratrol was shown to regulate sterol regulatory element binding protein 1 (SREBP1), which is involved in modulation of lipogenic genes, such as fatty acid synthase, suggesting that SIRT1 plays an important role in attenuation of fat deposition by inhibiting SREBP1 expression [196].
In endothelial function and cardioprotection
Impairment of endothelial function critically contributes to the pathogenesis of several diseases including diabetes, arteriosclerosis, cardiovascular and pulmonary diseases. SIRT1 is highly expressed in vasculature, which plays a critical role in regulating endothelial cell-mediated vascular homeostasis and remodeling (Figure 4) [197]. However, the level/activity of SIRT1 is significantly decreased associated with increased acetylation of eNOS in endothelial cells in response to oxidant/aldehyde treatments [54]. Endothelial specific over-expression of SIRT1 significantly blunted high fat-induced atherogenesis in apoE−/− mice via improving endothelial cell survival and function [198]. This may be due to deacetylation of the calmodulin-binding domain of eNOS (on lysine residues at K496 and K506) leading to enhanced nitric oxide production [199]. On the other hand, inhibition of SIRT1 down-regulates eNOS level leading to premature senescence-like phenotype in endothelial cells [157, 200]. Moreover, SIRT1 activation attenuates oxidative stress-induced apoptosis and NF-κB gene expression and thereby improves endothelial dysfunction [201, 202].
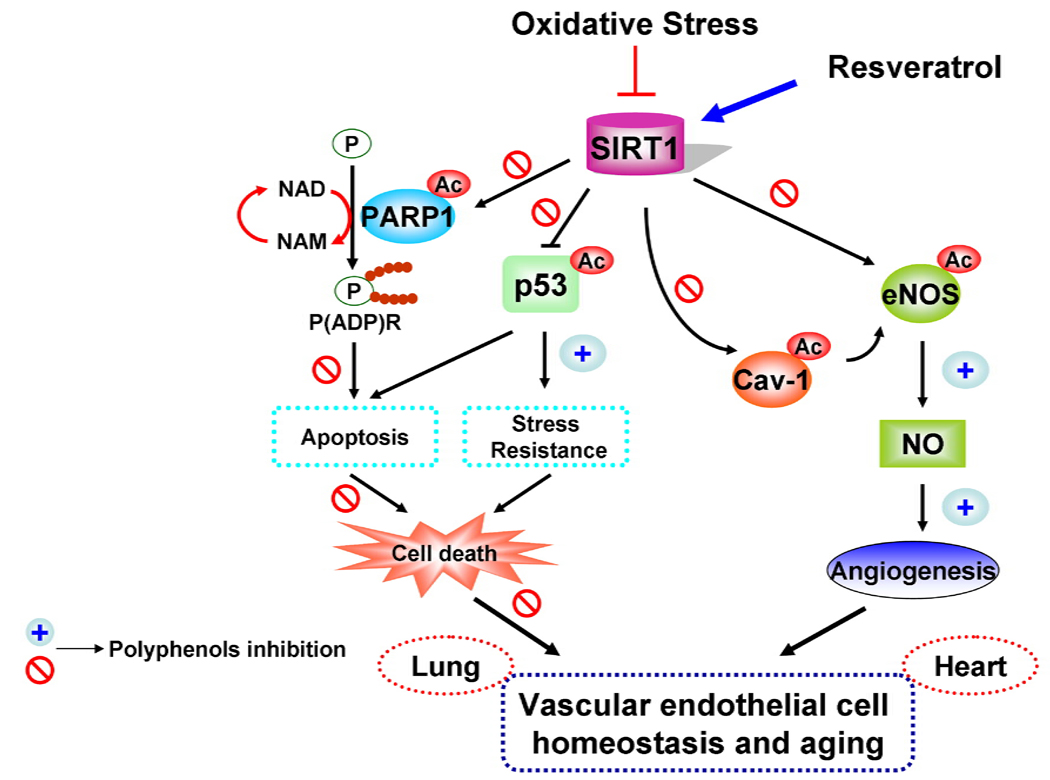
Figure 4. Oxidative stress-induced alteration in cellular survival and angiogenic signaling pathways.
Oxidative stress down-regulates SIRT1 thereby modulating the endothelial cell survival and angiogenic signaling by acetylating eNOS, p53, and PARP-1. SIRT1 and PARP utilize NAD+ as a common substrate. Oxidants activate PARP-1 and deplete intracellular NAD+ pool associated with activation of various cellular pathways leading to apoptosis and stress response. Polyphenols, such as resveratrol and quercetin, attenuate oxidative stress-mediated modification of SIRT1 either by increasing the activity of SIRT1 or reversing the posttranslational modifications of SIRT1. Activation of SIRT1 leads to angiogenesis and the inhibition of inflammatory/apoptotic signaling.
Moderate overexpression (∼2.5 folds) of SIRT1 protects the heart from oxidative stress through FOXO-dependent mechanism [1]. PARP activation, which can lead to cardiomyocyte cell death through a p53-dependent mechanism, is also modulated by SIRT1 activity [203, 204]. Resveratrol showed considerable protection against cardiac dysfunction [205–207], which confirms the beneficial role of polyphenols in cardioprotection. It is likely that polyphenols may mimic the effect of preconditioning of heart by activation of SIRT1 to protect against ischemia-reperfusion in heart.
Activation of SIRT1 by resveratrol regulates endothelial vasoprotective phenotype via up-regulation of Krüppel-like factor 2 in human vascular endothelial cells [208]. Recent studies also showed that resveratrol therapy ameliorates endothelial dysfunction by regulating vascular endothelial growth factor, eNOS, caveolin-1, and heme oxygenase (HO)-1 leading to neovascularization of the hypercholesterolemic myocardium and confers protection against myocardial injury caused by ischemia-reperfusion [206, 209]. It is also known that resveratrol protects ischemia-reperfusion injury in mice via activation of SIRT1 in myocytes and endothelium. In addition to resveratrol, quercetin and catechins also have shown to protect against endothelial dysfunction in vitro and in vivo [210–213]. Therefore, SIRT1 activation through nutraceutical polyphenols may be beneficial in intervention treatment of diabetes, arteriosclerosis, cardiovascular and pulmonary diseases where endothelial dysfunction occurs.
In circadian rhythm
The circadian clock controls intrinsic daily rhythms of physiology and behavior through negative feedback of transcriptional-translational loops [214]. The genes encoding these proteins are regulated by the heterodimers of transcription activator core CLOCK and BMAL1 via the binding to E-box elements in their promoters. This regulatory pathway is modulated by posttranslational modifications, such as acetylation and phosphorylation, that affect the activity and stability of circadian core proteins [215, 216]. CLOCK has intrinsic HAT activity [217], and it can acetylate histone H3, H4, BMAL1, and Per2, leading to transcriptional inhibition and specific gene expression [214]. SIRT1 has been reported to counteract the HAT activity of CLOCK [218, 219]. In light of tight coupling between circadian rhythms and metabolic regulation [220], the NAD+ substrate dependency of SIRT1 suggests that it might constitute a functional link between metabolic activity and circadian proteins [216]. Recent reports have shown that NAD+-dependent SIRT1 deacetylates histone H3, H4, BMAL1, Per, and Cry and thereby regulates the transcription of circadian proteins and hence circadian cycle [215, 219, 221, 222]. It is also known that inflammation and circadian rhythm disorder are intimately associated with fatigue and diminished locomotor activity [223]. The role of polyphenols in regulating these circadian proteins is still not clear. However, the possibility of regulation of circadian rhythm by polyphenols via increasing SIRT1 activity is fascinatingly therapeutic modes against various metabolic and chronic inflammatory diseases.
CONCLUSIONS AND FUTURE DIRECTIONS
The yeast Sir2 or mammalian ortholog SIRT1 has been identified as key regulator of lifespan in several model organisms. Activation of SIRT1 by polyphenols have beneficial effects on regulation of CR, oxidative stress, inflammation, adipogenesis, cellular senescence, autophagy, apoptosis, circadian rhythm, autoimmunity, skeletal muscle function, metabolism, mitochondria biogenesis and endothelial dysfunction. However, the molecular mechanism of these SIRT1-mediated processes is not well understood. Remarkably, the expression level and activity of SIRT1 are reduced in several chronic diseases, including diabetes, chronic inflammatory lung diseases, neurodegenerative diseases and cardiovascular diseases. Activation of SIRT1 results in the multiple beneficial health outcomes, such as improvement in insulin sensitivity, decreased adiposity, increased mitochondrial functions, decreased glucose levels, and enhanced physiological functions. Many of these improvements are a direct consequence of enhanced SIRT1 deacetylase activity suggesting that SIRT1 would be a pharmacologically therapeutic target in various metabolic, proliferative, and inflammatory diseases. Dietary polyphenols (e.g. resveratrol, quercetin, and catechins) have been shown to increase SIRT1 level/activity in several systems along with their other well-known properties, such as activation of Nrf2 (antioxidant response) and inhibition of NF-κB (anti-inflammatory response). However, most polyphenols are poorly absorbed, rapidly metabolized and oxidized and undergo sulfation and glucuronidation and also lead to formation of their own oxidation products. The biochemical mode of action of dietary polyphenols on activation of SIRT1 is an important area for further research. Future studies are required to understand on the mechanisms of the in vivo effects of polyphenols are required to understand the SIRT1-mediated improvement of various abnormal cellular and biological functions. Additional aspects about bioactivity, absorption, and stability of dietary polyphenols with respect to modulation of SIRT1 activity are also important areas of further research. Overall, regulation of SIRT1 activity by dietary polyphenols is a promising therapeutic strategy against many chronic inflammatory diseases including the diseases which are associated with inflammaging.
ACKNOWLEDGEMENTS
This study was supported by the NIH 1R01HL085613, 1R01HL097751, 1R01HL092842 and NIEHS Environmental Health Science Center grant P30-ES01247.
ABBREVIATIONS
- AIF
apoptosis-inducing factor
- AMPK
AMP-activated kinase
- AP
activator protein
- BIM
Bcl-2 interacting mediators of cell death
- BRCA1
breast cancer associated gene 1
- COPD
chronic obstructive pulmonary disease
- COX-2
cyclooxygenase-2
- CR
calorie restriction
- CS
cigarette smoke
- CSE
cigarette smoke extract
- EGC
epicatechin gallate
- EGCG
epigallocatechin gallate
- eNOS
endothelial nitric oxide synthase
- FOXO
forkhead box class O
- FOXP
forkhead box class P
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- iNOS
inducible nitric oxide synthase
- mTOR
mammalian target of rapamycin
- MMPs
metalloproteinases
- MnSOD
manganese superoxide dismutase
- NAD
nicotinamide adenine dinucleotide
- NAM
nicotinamide
- NAMPT
nicotinamide phosphoribosyltransferase
- NF-κB
nuclear factor kappa B
- NMN
nicotinamide mononucleotide
- NMNAT
nicotinamide mononucleotide adenylyltransferase
- Nrf1
NF-E2-related factor-1
- PARP
poly(ADP-ribose) polymerase
- PGC
peroxisome proliferator-activated receptor γ coactivator
- PPAR
peroxisome proliferator activated receptor
- ROS
reactive oxygen species
- SIRT1
Sirtuin1
- SMRT
silencing mediator of retinoid and thyroid hormone receptors
- SREBP1
sterol regulatory element binding protein 1
- UCP2
uncoupling protein 2
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Alcendor RR, Gao S, Zhai P, Zablocki D, Holle E, Yu X, Tian B, Wagner T, Vatner SF, Sadoshima J. Circ Res. 2007;100:1512–1521. doi: 10.1161/01.RES.0000267723.65696.4a. [DOI] [PubMed] [Google Scholar]
- 2.Michan S, Sinclair D. Biochem J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lavu S, Boss O, Elliott PJ, Lambert PD. Nat Rev Drug Discov. 2008;7:841–853. doi: 10.1038/nrd2665. [DOI] [PubMed] [Google Scholar]
- 4.Finkel T, Deng CX, Mostoslavsky R. Nature. 2009;460:587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang T, Sauve AA. AAPS J. 2006;8:E632–E643. doi: 10.1208/aapsj080472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, L Zhang L, Scherer B, Sinclair DA. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 7.Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA. Nature. 2003;423:181–185. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. J Biol Chem. 2002;277:45099–45107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- 9.Tissenbaum HA, Guarente L. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 10.Lin SJ, Defossez PA, Guarente L. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 11.Davis JM, Murphy EA, Carmichael MD, Davis B. Am J Physiol Regul Integr Comp Physiol. 2009;296:R1071–R1077. doi: 10.1152/ajpregu.90925.2008. [DOI] [PubMed] [Google Scholar]
- 12.de Boer VC, de Goffau MC, Arts IC, Hollman PC, Keijer J. Mech Ageing Dev. 2006;127:618–627. doi: 10.1016/j.mad.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 13.Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, Pistell PJ, Poosala S, G Becker K, Boss O, Gwinn D, Wang M, Ramaswamy S, Fishbein KW, G Spencer R, Lakatta EG, Le Couteur D, Shaw RJ, Navas P, Puigserver P, Ingram DK, de Cabo R, Sinclair DA. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaeberlein M, McDonagh T, Heltweg B, Hixon J, Westman EA, Caldwell SD, Napper A, Curtis R, DiStefano PS, Fields S, Bedalov A, Kennedy BK. J Biol Chem. 2005;280:17038–17045. doi: 10.1074/jbc.M500655200. [DOI] [PubMed] [Google Scholar]
- 15.Borra MT, Smith BC, Denu JM. J Biol Chem. 2005;280:17187–17195. doi: 10.1074/jbc.M501250200. [DOI] [PubMed] [Google Scholar]
- 16.Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, Sinclair DA. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 17.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 18.Borra MT, O'Neill FJ, Jackson MD, Marshall B, Verdin E, Foltz KR, Denu JM. J Biol Chem. 2002;277:12632–12641. doi: 10.1074/jbc.M111830200. [DOI] [PubMed] [Google Scholar]
- 19.Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 20.Sauve AA, Wolberger C, Schramm VL, Boeke JD. Annu Rev Biochem. 2006;75:435–465. doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- 21.Borradaile NM, Pickering JG. Aging Cell. 2009;8:100–112. doi: 10.1111/j.1474-9726.2009.00453.x. [DOI] [PubMed] [Google Scholar]
- 22.Hsu CP, Oka S, Shao D, Hariharan N, Sadoshima J. Circ Res. 2009;105:481–491. doi: 10.1161/CIRCRESAHA.109.203703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Revollo JR, Grimm AA, Imai S. J Biol Chem. 2004;279:50754–50763. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- 24.Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang T, Berrocal JG, Frizzell KM, Gamble MJ, DuMond ME, Krishnakumar R, Yang T, Sauve AA, Kraus WL. J Biol Chem. 2009;284:20408–20417. doi: 10.1074/jbc.M109.016469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho C, van der Veer E, Akawi O, Pickering JG. FEBS Lett. 2009;583:3081–3085. doi: 10.1016/j.febslet.2009.08.031. [DOI] [PubMed] [Google Scholar]
- 27.Suchankova G, Nelson LE, Gerhart-Hines Z, Kelly M, Gauthier MS, Saha AK, Ido Y, Puigserver P, Ruderman NB. Biochem Biophys Res Commun. 2009;378:836–841. doi: 10.1016/j.bbrc.2008.11.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mukherjee S, Lekli I, Gurusamy N, Bertelli AA, Das DK. Free Radic Biol Med. 2009;46:573–578. doi: 10.1016/j.freeradbiomed.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 29.Hou X, Xu S, Maitland-Toolan KA, Sato K, Jiang B, Ido Y, Lan F, Walsh K, Wierzbicki M, Verbeuren TJ, Cohen RA, Zang M. J Biol Chem. 2008;283:20015–20026. doi: 10.1074/jbc.M802187200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Um JH, Park SJ, Kang H, Yang S, Foretz M, McBurney MW, Kim MK, Viollet B, Chung JH. Diabetes. 2010;59:554–563. doi: 10.2337/db09-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fulco M, Cen Y, Zhao P, Hoffman EP, McBurney MW, Sauve AA, Sartorelli V. Dev Cell. 2008;14:661–673. doi: 10.1016/j.devcel.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao X, Zmijewski JW, Lorne E, Liu G, Park YJ, Tsuruta Y, Abraham E. Am J Physiol Lung Cell Mol Physiol. 2008;295:L497–L504. doi: 10.1152/ajplung.90210.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suwa M, Nakano H, Radak Z, Kumagai S. Metabolism. 2010 doi: 10.1016/j.metabol.2010.03.003. doi:10.1016/j.metabol.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 34.Pacholec M, Bleasdale JE, Chrunyk B, Cunningham D, Flynn D, Garofalo RS, Griffith D, Griffor M, Loulakis P, Pabst B, Qiu X, Stockman B, Thanabal V, Varghese A, Ward J, Withka J, Ahn K. J Biol Chem. 2010;285:8340–8351. doi: 10.1074/jbc.M109.088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beher D, Wu J, Cumine S, Kim KW, Lu SC, Atangan L, Wang M. Chem Biol Drug Des. 2009;74:619–624. doi: 10.1111/j.1747-0285.2009.00901.x. [DOI] [PubMed] [Google Scholar]
- 36.Huang Q, Shen HM. Autophagy. 2009;5:273–276. doi: 10.4161/auto.5.2.7640. [DOI] [PubMed] [Google Scholar]
- 37.Kolthur-Seetharam U, Dantzer F, McBurney MW, de Murcia G, Sassone-Corsi P. Cell Cycle. 2006;5:873–877. doi: 10.4161/cc.5.8.2690. [DOI] [PubMed] [Google Scholar]
- 38.Furukawa A, Tada-Oikawa S, Kawanishi S, Oikawa S. Cell Physiol Biochem. 2007;20:45–54. doi: 10.1159/000104152. [DOI] [PubMed] [Google Scholar]
- 39.Caito S, Rajendrasozhan S, Cook S, Chung S, Yao H, Friedman AE, Brookes PS, Rahman I. FASEB J. 2010 doi: 10.1096/fj.09-151308. doi:10.1096/fj.09-151308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hong EH, Lee SJ, Kim JS, Lee KH, Um HD, Kim JH, Kim SJ, Kim JI, G Hwang S. J Biol Chem. 2010;285:1283–1295. doi: 10.1074/jbc.M109.058628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu X, Conner H, Kobayashi T, Kim H, Wen F, Abe S, Fang Q, Wang X, Hashimoto M, Bitterman P, Rennard SI. Am J Respir Cell Mol Biol. 2005;33:121–129. doi: 10.1165/rcmb.2003-0341OC. [DOI] [PubMed] [Google Scholar]
- 42.Lin T, Yang MS. Toxicology. 2008;245:147–153. doi: 10.1016/j.tox.2007.12.020. [DOI] [PubMed] [Google Scholar]
- 43.Hassa PO, Hottiger MO. Front Biosci. 2008;13:3046–3082. doi: 10.2741/2909. [DOI] [PubMed] [Google Scholar]
- 44.Moonen HJ, Geraets L, Vaarhorst A, Bast A, Wouters EF, Hageman GJ. Biochem Biophys Res Commun. 2005;338:1805–1810. doi: 10.1016/j.bbrc.2005.10.159. [DOI] [PubMed] [Google Scholar]
- 45.Hageman GJ, Larik I, Pennings HJ, Haenen GR, Wouters EF, Bast A. Free Radic Biol Med. 2003;35:140–148. doi: 10.1016/s0891-5849(03)00237-5. [DOI] [PubMed] [Google Scholar]
- 46.Weseler AR, Geraets L, Moonen HJ, Manders RJ, van Loon LJ, Pennings HJ, Wouters EF, Bast A, Hageman GJ. J Nutr. 2009;139:952–957. doi: 10.3945/jn.108.102756. [DOI] [PubMed] [Google Scholar]
- 47.Caito S, Hwang JW, Chung S, Yao H, Sundar IK, Rahman I. Biochem Biophys Res Commun. 2010;392:264–270. doi: 10.1016/j.bbrc.2009.12.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.El Ramy R, Magroun N, Messadecq N, Gauthier LR, Boussin FD, Kolthur-Seetharam U, Schreiber V, McBurney MW, Sassone-Corsi P, Dantzer F. Cell Mol Life Sci. 2009;66:3219–3234. doi: 10.1007/s00018-009-0105-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rajamohan SB, Pillai VB, Gupta M, Sundaresan NR, G Birukov K, Samant S, Hottiger MO, Gupta MP. Mol Cell Biol. 2009;29:4116–4129. doi: 10.1128/MCB.00121-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elliott PJ, Jirousek M. Curr Opin Investig Drugs. 2008;9:371–378. [PubMed] [Google Scholar]
- 51.Grimsrud PA, Xie H, Griffin TJ, Bernlohr DA. J Biol Chem. 2008;283:21837–21841. doi: 10.1074/jbc.R700019200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nystrom T. EMBO J. 2005;24:1311–1317. doi: 10.1038/sj.emboj.7600599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tanno M, Kuno A, Yano T, Miura T, Hisahara S, Ishikawa S, Shimamoto K, Horio Y. J Biol Chem. 2010;285:8375–8382. doi: 10.1074/jbc.M109.090266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arunachalam G, Yao H, Sundar IK, Caito S, Rahman I. Biochem Biophys Res Commun. 2010;393:66–72. doi: 10.1016/j.bbrc.2010.01.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rajendrasozhan S, Yang SR, Kinnula VL, Rahman I. Am J Respir Crit Care Med. 2008;177:861–870. doi: 10.1164/rccm.200708-1269OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang SR, Wright J, Bauter M, Seweryniak K, Kode A, Rahman I. Am J Physiol Lung Cell Mol Physiol. 2007;292:L567–L576. doi: 10.1152/ajplung.00308.2006. [DOI] [PubMed] [Google Scholar]
- 57.Kang H, Jung JW, Kim MK, Chung JH. PLoS One. 2009;4:e6611–e6618. doi: 10.1371/journal.pone.0006611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sasaki T, Maier B, Koclega KD, Chruszcz M, Gluba W, Stukenberg PT, Minor W, Scrable H. PLoS One. 2008;3:e4020–e4032. doi: 10.1371/journal.pone.0004020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 60.Zschoernig B, Mahlknecht U. Biochem Biophys Res Commun. 2009;381:372–377. doi: 10.1016/j.bbrc.2009.02.085. [DOI] [PubMed] [Google Scholar]
- 61.Nasrin N, Kaushik VK, Fortier E, Wall D, Pearson KJ, de Cabo R, Bordone L. PLoS One. 2009;4:e8414–e8422. doi: 10.1371/journal.pone.0008414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zee RS, Yoo CB, Pimentel DR, Perlman DH, Burgoyne JR, Hou X, McComb ME, Costello CE, Cohen RA, Bachschmid M. Antioxid Redox Signal. 2010 doi: 10.1089/ars.2010.3251. doi: 10.1089/ars. 2010.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ungvari ZI, Labinskyy N, Mukhopadhyay P, Pinto JT, Bagi Z, Ballabh P, Zhang C, Pacher P, Csiszar A. Am J Physiol Heart Circ Physiol. 2009;297:H1876–H1881. doi: 10.1152/ajpheart.00375.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nakamaru Y, Vuppusetty C, Wada H, Milne JC, Ito M, Rossios C, Elliot M, Hogg J, Kharitonov S, Goto H, Bemis JE, Elliott P, Barnes PJ, Ito K. FASEB J. 2009;23:2810–2819. doi: 10.1096/fj.08-125468. [DOI] [PubMed] [Google Scholar]
- 65.Choi KC, Jung MG, Lee YH, Yoon JC, Kwon SH, Kang HB, Kim MJ, Cha JH, Kim YJ, Jun WJ, Lee JM, Yoon HG. Cancer Res. 2009;69:583–592. doi: 10.1158/0008-5472.CAN-08-2442. [DOI] [PubMed] [Google Scholar]
- 66.Feng Y, Wu J, Chen L, Luo C, Shen X, Chen K, Jiang H, Liu D. Anal Biochem. 2009;395:205–210. doi: 10.1016/j.ab.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 67.Yamazaki Y, Usui I, Kanatani Y, Matsuya Y, Tsuneyama K, Fujisaka S, Bukhari A, Suzuki H, Senda S, Imanishi S, Hirata K, Ishiki M, Hayashi R, Urakaze M, Nemoto H, Kobayashi M, Tobe K. Am J Physiol Endocrinol Metab. 2009 doi: 10.1152/ajpendo.90997.2008. doi: 10.1152/ajpendo.90997.2008. [DOI] [PubMed] [Google Scholar]
- 68.Smith JJ, Kenney RD, Gagne DJ, Frushour BP, Ladd W, Galonek HL, Israelian K, Song J, Razvadauskaite G, Lynch AV, Carney DP, Johnson RJ, Lavu S, Iffland A, Elliott PJ, Lambert PD, Elliston KO, Jirousek MR, Milne JC, Boss O. BMC Syst Biol. 2009;3:31–44. doi: 10.1186/1752-0509-3-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, Gagne DJ, Jin L, Boss O, Perni RB, Vu CB, Bemis JE, Xie R, Disch JS, Ng PY, Nunes JJ, Lynch AV, Yang H, Galonek H, Israelian K, Choy W, Iffland A, Lavu S, Medvedik O, Sinclair DA, Olefsky JM, Jirousek MR, Elliott PJ, Westphal CH. Nature. 2007;450:712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rogina B, Helfand SL. Proc Natl Acad Sci U S A. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen D, Steele AD, Lindquist S, Guarente L. Science. 2005;310:1641. doi: 10.1126/science.1118357. [DOI] [PubMed] [Google Scholar]
- 72.Bordone L, Guarente L. Nat Rev Mol Cell Biol. 2005;6:298–305. doi: 10.1038/nrm1616. [DOI] [PubMed] [Google Scholar]
- 73.Sinclair DA, Guarente L. Sci Am. 2006;294:48–51. doi: 10.1038/scientificamerican0306-48. 54–47. [DOI] [PubMed] [Google Scholar]
- 74.Lu SP, Lin SJ. Biochim Biophys Acta. 2009 doi: 10.1016/j.bbapap.2009.09.030. [Google Scholar]
- 75.Kume S, Uzu T, Horiike K, Chin-Kanasaki M, Isshiki K, Araki SI, Sugimoto T, Haneda M, Kashiwagi A, Koya D. J Clin Invest. 2010;120:1043–1055. doi: 10.1172/JCI41376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guarente L. Cell. 2008;132:171–176. doi: 10.1016/j.cell.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 78.Bordone L, Cohen D, Robinson A, Motta MC, van Veen E, Czopik A, Steele AD, Crowe H, Marmor S, Luo J, Gu W, Guarente L. Aging Cell. 2007;6:759–767. doi: 10.1111/j.1474-9726.2007.00335.x. [DOI] [PubMed] [Google Scholar]
- 79.Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. PLoS Biol. 2004;2:E296–E302. doi: 10.1371/journal.pbio.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kaeberlein M, Steffen KK, Hu D, Dang N, Kerr EO, Tsuchiya M, Fields S, Kennedy BK. Science. 2006;312:1312. doi: 10.1126/science.1124608. author reply 1312. [DOI] [PubMed] [Google Scholar]
- 81.Kaeberlein M, Powers RW., 3rd Ageing Res Rev. 2007;6:128–140. doi: 10.1016/j.arr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 82.G Wood J, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, Sinclair D. Nature. 2004;430:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- 83.Valenzano DR, Terzibasi E, Genade T, Cattaneo A, Domenici L, Cellerino A. Curr Biol. 2006;16:296–300. doi: 10.1016/j.cub.2005.12.038. [DOI] [PubMed] [Google Scholar]
- 84.Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW. EMBO J. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tang BL, Chua CE. Biochem Biophys Res Commun. 2010;391:6–10. doi: 10.1016/j.bbrc.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 86.Baur JA. Biochim Biophys Acta. 2009 doi: 10.1016/j.bbapap.2009.10.025. [Google Scholar]
- 87.Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, Geny B, Laakso M, Puigserver P, Auwerx J. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 88.Kim SR, Lee KS, Park SJ, Min KH, Choe YH, Moon H, Yoo WH, Chae HJ, Han MK, Lee YC. J Allergy Clin Immunol. 2010;125:449–460. doi: 10.1016/j.jaci.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 89.Jones E, Hughes RE. Exp Gerontol. 1982;17:213–217. doi: 10.1016/0531-5565(82)90027-4. [DOI] [PubMed] [Google Scholar]
- 90.Boily G, Seifert EL, Bevilacqua L, He XH, Sabourin G, Estey C, Moffat C, Crawford S, Saliba S, Jardine K, Xuan J, Evans M, Harper ME, McBurney MW. PLoS One. 2008;3:e1759–e1770. doi: 10.1371/journal.pone.0001759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xia W, Wang Z, Wang Q, Han J, Zhao C, Hong Y, Zeng L, Tang L, Ying W. Curr Pharm Des. 2009;15:12–19. doi: 10.2174/138161209787185832. [DOI] [PubMed] [Google Scholar]
- 92.Baur JA, Sinclair DA. Nat Rev Drug Discov. 2006;5:493–506. doi: 10.1038/nrd2060. [DOI] [PubMed] [Google Scholar]
- 93.Westphal CH, Dipp MA, Guarente L. Trends Biochem Sci. 2007;32:555–560. doi: 10.1016/j.tibs.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 94.Imai S, Kiess W. Front Biosci. 2009;14:2983–2995. doi: 10.2741/3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bordone L, Motta MC, Picard F, Robinson A, Jhala US, Apfeld J, McDonagh T, Lemieux M, McBurney M, Szilvasi A, Easlon EJ, Lin SJ, Guarente L. PLoS Biol. 2006;4:e31. doi: 10.1371/journal.pbio.0040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Moynihan KA, Grimm AA, Plueger MM, Bernal-Mizrachi E, Ford E, Cras-Meneur C, Permutt MA, Imai S. Cell Metab. 2005;2:105–117. doi: 10.1016/j.cmet.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 97.Pfluger PT, Herranz D, Velasco-Miguel S, Serrano M, Tschop MH. Proc Natl Acad Sci U S A. 2008;105:9793–9798. doi: 10.1073/pnas.0802917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xu F, Gao Z, Zhang J, Rivera CA, Yin J, Weng J, Ye J. Endocrinology. 2010 doi: 10.1210/en.2009-1013. doi:10.1210/en.2009-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Canto C, Auwerx J. Curr Opin Lipidol. 2009;20:98–105. doi: 10.1097/MOL.0b013e328328d0a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.van der Toorn M, Slebos DJ, de Bruin HG, Leuvenink HG, Bakker SJ, Gans RO, Koeter GH, van Oosterhout AJ, Kauffman HF. Am J Physiol Lung Cell Mol Physiol. 2007;292:L1211–L1218. doi: 10.1152/ajplung.00291.2006. [DOI] [PubMed] [Google Scholar]
- 101.Klaus S, Pultz S, Thone-Reineke C, Wolfram S. Int J Obes (Lond) 2005;29:615–623. doi: 10.1038/sj.ijo.0802926. [DOI] [PubMed] [Google Scholar]
- 102.Chen LF, Mu Y, Greene WC. EMBO J. 2002;21:6539–6548. doi: 10.1093/emboj/cdf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang R, Chen HZ, Liu JJ, Jia YY, Zhang ZQ, Yang RF, Zhang Y, Xu J, Wei YS, Liu DP, Liang CC. J Biol Chem. 2010;285:7097–7110. doi: 10.1074/jbc.M109.038604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gao Z, Ye J. Biochem Biophys Res Commun. 2008;376:793–796. doi: 10.1016/j.bbrc.2008.09.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang J, Lee SM, Shannon S, Gao B, Chen W, Chen A, Divekar R, McBurney MW, Braley-Mullen H, Zaghouani H, Fang D. J Clin Invest. 2009;119:3048–3058. doi: 10.1172/JCI38902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Milne JC, Denu JM. Curr Opin Chem Biol. 2008;12:11–17. doi: 10.1016/j.cbpa.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 107.Yoshizaki T, Schenk S, Imamura T, Babendure JL, Sonoda N, Bae EJ, Oh D, Lu M, Milne JC, Westphal C, Bandyopadhyay G, Olefsky JM. Am J Physiol Endocrinol Metab. 2010;298:E419–E428. doi: 10.1152/ajpendo.00417.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Singh UP, Singh N, Singh B, Hofseth LJ, Price BL, Nagarkatti M, Nagarkatti P. J Pharmacol Exp Ther. 2010;332:829–839. doi: 10.1124/jpet.109.160838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Birrell MA, McCluskie K, Wong S, Donnelly LE, Barnes PJ, Belvisi MG. FASEB J. 2005;19:840–841. doi: 10.1096/fj.04-2691fje. [DOI] [PubMed] [Google Scholar]
- 110.Amat R, Solanes G, Giralt M, Villarroya F. J Biol Chem. 2007;282:34066–34076. doi: 10.1074/jbc.M707114200. [DOI] [PubMed] [Google Scholar]
- 111.Donnelly LE, Newton R, Kennedy GE, Fenwick PS, Leung RH, Ito K, Russell RE, Barnes PJ. Am J Physiol Lung Cell Mol Physiol. 2004;287:L774–L783. doi: 10.1152/ajplung.00110.2004. [DOI] [PubMed] [Google Scholar]
- 112.Manna SK, Mukhopadhyay A, Aggarwal BB. J Immunol. 2000;164:6509–6519. doi: 10.4049/jimmunol.164.12.6509. [DOI] [PubMed] [Google Scholar]
- 113.Leiro J, Arranz JA, Fraiz N, Sanmartin ML, Quezada E, Orallo F. Int Immunopharmacol. 2005;5:393–406. doi: 10.1016/j.intimp.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 114.Biesalski HK. Curr Opin Clin Nutr Metab Care. 2007;10:724–728. doi: 10.1097/MCO.0b013e3282f0cef2. [DOI] [PubMed] [Google Scholar]
- 115.Dong Z, Ma W, Huang C, Yang CS. Cancer Res. 1997;57:4414–4419. [PubMed] [Google Scholar]
- 116.Nomura M, Ma W, Chen N, Bode AM, Dong Z. Carcinogenesis. 2000;21:1885–1890. doi: 10.1093/carcin/21.10.1885. [DOI] [PubMed] [Google Scholar]
- 117.Syed DN, Afaq F, Kweon MH, Hadi N, Bhatia N, Spiegelman VS, Mukhtar H. Oncogene. 2007;26:673–682. doi: 10.1038/sj.onc.1209829. [DOI] [PubMed] [Google Scholar]
- 118.Stewart LK, Soileau JL, Ribnicky D, Wang ZQ, Raskin I, Poulev A, Majewski M, Cefalu WT, Gettys TW. Metabolism. 2008;57:S39–S46. doi: 10.1016/j.metabol.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Busse WW, Kopp DE, Middleton E., Jr J Allergy Clin Immunol. 1984;73:801–809. doi: 10.1016/0091-6749(84)90450-0. [DOI] [PubMed] [Google Scholar]
- 120.Ramadori G, Gautron L, Fujikawa T, Vianna CR, Elmquist JK, Coppari R. Endocrinology. 2009;150:5326–5333. doi: 10.1210/en.2009-0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yang J, Kong X, Martins-Santos ME, Aleman G, Chaco E, Liu GE, Wu SY, Samols D, Hakimi P, Chiang CM, Hanson RW. J Biol Chem. 2009;284:27042–27053. doi: 10.1074/jbc.M109.047340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, Bronson R, Appella E, Alt FW, Chua KF. Proc Natl Acad Sci U S A. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.van Loosdregt J, Vercoulen Y, Guichelaar T, Gent YY, Beekman JM, van Beekum O, Brenkman AB, Hijnen DJ, Mutis T, Kalkhoven E, Prakken BJ, Coffer PJ. Blood. 2010;115:965–974. doi: 10.1182/blood-2009-02-207118. [DOI] [PubMed] [Google Scholar]
- 124.Sequeira J, Boily G, Bazinet S, Saliba S, He X, Jardine K, Kennedy C, Staines W, Rousseaux C, Mueller R, McBurney MW. Exp Cell Res. 2008;314:3069–3074. doi: 10.1016/j.yexcr.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 125.Lee M, Kim S, Kwon OK, Oh SR, Lee HK, Ahn K. Int Immunopharmacol. 2009;9:418–424. doi: 10.1016/j.intimp.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 126.Singh NP, Hegde VL, Hofseth LJ, Nagarkatti M, Nagarkatti P. Mol Pharmacol. 2007;72:1508–1521. doi: 10.1124/mol.107.038984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sharma S, Chopra K, Kulkarni SK, Agrewala JN. Clin Exp Immunol. 2007;147:155–163. doi: 10.1111/j.1365-2249.2006.03257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gao X, Deeb D, Media J, Divine G, Jiang H, Chapman RA, Gautam SC. Biochem Pharmacol. 2003;66:2427–2435. doi: 10.1016/j.bcp.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 129.Shim JH, Choi HS, Pugliese A, Lee SY, Chae JI, Choi BY, Bode AM, Dong Z. J Biol Chem. 2008;283:28370–28379. doi: 10.1074/jbc.M802200200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Song EK, Hur H, Han MK. Arch Pharm Res. 2003;26:559–563. doi: 10.1007/BF02976881. [DOI] [PubMed] [Google Scholar]
- 131.Park HJ, Lee CM, Jung ID, Lee JS, Jeong YI, Chang JH, Chun SH, Kim MJ, Choi IW, Ahn SC, Shin YK, Yeom SR, Park YM. Int Immunopharmacol. 2009;9:261–267. doi: 10.1016/j.intimp.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 132.Giannakou ME, Partridge L. Trends Cell Biol. 2004;14:408–412. doi: 10.1016/j.tcb.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 133.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 134.Dryden SC, Nahhas FA, Nowak JE, Goustin AS, Tainsky MA. Mol Cell Biol. 2003;23:3173–3185. doi: 10.1128/MCB.23.9.3173-3185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kops GJ, Dansen TB, Polderman PE, Saarloos I, Wirtz KW, Coffer PJ, Huang TT, Bos JL, Medema RH, Burgering BM. Nature. 2002;419:316–321. doi: 10.1038/nature01036. [DOI] [PubMed] [Google Scholar]
- 136.Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, Bultsma Y, McBurney M, Guarente L. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- 137.You H, Mak TW. Cell Cycle. 2005;4:37–38. doi: 10.4161/cc.4.1.1401. [DOI] [PubMed] [Google Scholar]
- 138.van der Horst A, Tertoolen LG, de Vries-Smits LM, Frye RA, Medema RH, Burgering BM. J Biol Chem. 2004;279:28873–28879. doi: 10.1074/jbc.M401138200. [DOI] [PubMed] [Google Scholar]
- 139.Chen CJ, Yu W, Fu YC, Wang X, Li JL, Wang W. Biochem Biophys Res Commun. 2009;378:389–393. doi: 10.1016/j.bbrc.2008.11.110. [DOI] [PubMed] [Google Scholar]
- 140.He W, Wang Y, Zhang MZ, You L, Davis LS, Fan H, Yang HC, Fogo AB, Zent R, Harris RC, Breyer MD, Hao CM. J Clin Invest. 2010;120:1056–1068. doi: 10.1172/JCI41563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chen WY, Wang DH, Yen RC, Luo J, Gu W, Baylin SB. Cell. 2005;123:437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 142.Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 143.Luo J, Li M, Tang Y, Laszkowska M, G Roeder R, Gu W. Proc Natl Acad Sci U S A. 2004;101:2259–2264. doi: 10.1073/pnas.0308762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Puigserver P, Spiegelman BM. Endocr Rev. 2003;24:78–90. doi: 10.1210/er.2002-0012. [DOI] [PubMed] [Google Scholar]
- 145.Boily G, He XH, Pearce B, Jardine K, McBurney MW. Oncogene. 2009;28:2882–2893. doi: 10.1038/onc.2009.147. [DOI] [PubMed] [Google Scholar]
- 146.Huffman DM, Grizzle WE, Bamman MM, Kim JS, Eltoum IA, Elgavish A, Nagy TR. Cancer Res. 2007;67:6612–6618. doi: 10.1158/0008-5472.CAN-07-0085. [DOI] [PubMed] [Google Scholar]
- 147.Liu T, Liu PY, Marshall GM. Cancer Res. 2009;69:1702–1705. doi: 10.1158/0008-5472.CAN-08-3365. [DOI] [PubMed] [Google Scholar]
- 148.Firestein R, Blander G, Michan S, Oberdoerffer P, Ogino S, Campbell J, Bhimavarapu A, Luikenhuis S, de Cabo R, Fuchs C, Hahn WC, Guarente LP, Sinclair DA. PLoS One. 2008;3 doi: 10.1371/journal.pone.0002020. e2020-e2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang RH, Zheng Y, Kim HS, Xu X, Cao L, Luhasen T, Lee MH, Xiao C, Vassilopoulos A, Chen W, Gardner K, G Man Y, Hung MC, Finkel T, Deng CX. Mol Cell. 2008;32:11–20. doi: 10.1016/j.molcel.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lee MH, Choi BY, Kundu JK, Shin YK, Na HK, Surh YJ. Cancer Res. 2009;69:7449–7458. doi: 10.1158/0008-5472.CAN-09-1266. [DOI] [PubMed] [Google Scholar]
- 151.Whyte L, Huang YY, Torres K, G Mehta R. Cancer Res. 2007;67:12007–12017. doi: 10.1158/0008-5472.CAN-07-2464. [DOI] [PubMed] [Google Scholar]
- 152.van Ginkel PR, Sareen D, Subramanian L, Walker Q, Darjatmoko SR, Lindstrom MJ, Kulkarni A, Albert DM, Polans AS. Clin Cancer Res. 2007;13:5162–5169. doi: 10.1158/1078-0432.CCR-07-0347. [DOI] [PubMed] [Google Scholar]
- 153.Paliwal S, Sundaram J, Mitragotri S. Br J Cancer. 2005;92:499–502. doi: 10.1038/sj.bjc.6602364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Murakami A, Ashida H, Terao J. Cancer Lett. 2008;269:315–325. doi: 10.1016/j.canlet.2008.03.046. [DOI] [PubMed] [Google Scholar]
- 155.Orimo M, Minamino T, Miyauchi H, Tateno K, Okada S, Moriya J, Komuro I. Arterioscler Thromb Vasc Biol. 2009;29:889–894. doi: 10.1161/ATVBAHA.109.185694. [DOI] [PubMed] [Google Scholar]
- 156.Sasaki T, Maier B, Bartke A, Scrable H. Aging Cell. 2006;5:413–422. doi: 10.1111/j.1474-9726.2006.00235.x. [DOI] [PubMed] [Google Scholar]
- 157.Ota H, Akishita M, Eto M, Iijima K, Kaneki M, Ouchi Y. J Mol Cell Cardiol. 2007;43:571–579. doi: 10.1016/j.yjmcc.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 158.Rajendrasozhan S, Yang SR, Caito S, Rahman I. Am J Respir Crit Care Med. 2008;177:A266. doi: 10.1164/rccm.200708-1269OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.MacNee W, Tuder RM. Proc Am Thorac Soc. 2009;6:527–531. doi: 10.1513/pats.200905-027DS. [DOI] [PubMed] [Google Scholar]
- 160.Zamin LL, Filippi-Chiela EC, Dillenburg-Pilla P, Horn F, Salbego C, Lenz G. Cancer Sci. 2009;100:1655–1662. doi: 10.1111/j.1349-7006.2009.01215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Stefani M, Markus MA, Lin RC, Pinese M, Dawes IW, Morris BJ. Ann N Y Acad Sci. 2007;1114:407–418. doi: 10.1196/annals.1396.001. [DOI] [PubMed] [Google Scholar]
- 162.Ota H, Eto M, Ogawa S, Iijima K, Akishita M, Ouchi Y. J Atheroscler Thromb. 2010 doi: 10.5551/jat.3525. [DOI] [PubMed] [Google Scholar]
- 163.Chen X, Chi Y, Bloecher A, Aebersold R, Clurman BE, Roberts JM. Mol Cell. 2004;16:839–847. doi: 10.1016/j.molcel.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 164.Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. PLoS Genet. 2008;4:e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ghosh HS, McBurney M, Robbins PD. PLoS One. 2010;5:e9199. doi: 10.1371/journal.pone.0009199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, Finkel T. Proc Natl Acad Sci U S A. 2008;105:3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hsu CP, Hariharan N, Alcendor RR, Oka S, Sadoshima J. Autophagy. 2009;5:1229–1231. doi: 10.4161/auto.5.8.10275. [DOI] [PubMed] [Google Scholar]
- 168.Matsui Y, Takagi H, Qu X, Abdellatif M, Sakoda H, Asano T, Levine B, Sadoshima J. Circ Res. 2007;100:914–922. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- 169.Takagi H, Matsui Y, Hirotani S, Sakoda H, Asano T, Sadoshima J. Autophagy. 2007;3:405–407. doi: 10.4161/auto.4281. [DOI] [PubMed] [Google Scholar]
- 170.Ruderman NB, Xu XJ, Nelson L, Cacicedo JM, Saha AK, Lan F, Ido Y. Am J Physiol Endocrinol Metab. 2010;298:E751–E760. doi: 10.1152/ajpendo.00745.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lan F, Cacicedo JM, Ruderman N, Ido Y. J Biol Chem. 2008;283:27628–27635. doi: 10.1074/jbc.M805711200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Armour SM, Baur JA, Hsieh SN, Land-Bracha A, Thomas SM, Sinclair DA. Aging (Albany NY) 2009;1:515–528. doi: 10.18632/aging.100056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Tang D, Kang R, Xiao W, Zhang H, Lotze MT, Wang H, Xiao X. Am J Respir Cell Mol Biol. 2009;41:651–660. doi: 10.1165/rcmb.2008-0119OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ryter SW, Chen ZH, Kim HP, Choi AM. Autophagy. 2009;5:235–237. doi: 10.4161/auto.5.2.7495. [DOI] [PubMed] [Google Scholar]
- 175.H Chen Z, Kim HP, Sciurba FC, Lee SJ, Feghali-Bostwick C, Stolz DB, Dhir R, Landreneau RJ, Schuchert MJ, Yousem SA, Nakahira K, Pilewski JM, Lee JS, Zhang Y, Ryter SW, Choi AM. PLoS One. 2008;3:e3316. doi: 10.1371/journal.pone.0003316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Marchetti P, Masini M. Autophagy. 2009;5:1055–1056. doi: 10.4161/auto.5.7.9511. [DOI] [PubMed] [Google Scholar]
- 177.Ost A, Svensson K, Ruishalme I, Brannmark C, Franck N, Krook H, Sandstrom P, Kjolhede P, Stralfors P. Mol Med. 2010 doi: 10.2119/molmed.2010.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kim HC, Mofarrahi M, Hussain SN. Int J Chron Obstruct Pulmon Dis. 2008;3:637–658. doi: 10.2147/copd.s4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Am J Respir Crit Care Med. 1999;159:S1–S40. doi: 10.1164/ajrccm.159.supplement_1.99titlepage. [DOI] [PubMed] [Google Scholar]
- 180.Kelley DE, He J, Menshikova EV, Ritov VB. Diabetes. 2002;51:2944–2950. doi: 10.2337/diabetes.51.10.2944. [DOI] [PubMed] [Google Scholar]
- 181.Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, Puigserver P. EMBO J. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Amat R, Planavila A, Chen SL, Iglesias R, Giralt M, Villarroya F. J Biol Chem. 2009;284:21872–21880. doi: 10.1074/jbc.M109.022749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Dasgupta B, Milbrandt J. Proc Natl Acad Sci U S A. 2007;104:7217–7222. doi: 10.1073/pnas.0610068104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Shin SM, Cho IJ, Kim SG. Mol Pharmacol. 2009;76:884–895. doi: 10.1124/mol.109.058479. [DOI] [PubMed] [Google Scholar]
- 185.Murase T, Haramizu S, Shimotoyodome A, Tokimitsu I, Hase T. Am J Physiol Regul Integr Comp Physiol. 2006;290:R1550–R1556. doi: 10.1152/ajpregu.00752.2005. [DOI] [PubMed] [Google Scholar]
- 186.Nieman DC, Williams AS, Shanely RA, Jin F, McAnulty SR, Triplett NT, Austin MD, Henson DA. Med Sci Sports Exerc. 2010;42:338–345. doi: 10.1249/MSS.0b013e3181b18fa3. [DOI] [PubMed] [Google Scholar]
- 187.Wyke SM, Russell ST, Tisdale MJ. Br J Cancer. 2004;91:1742–1750. doi: 10.1038/sj.bjc.6602165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Qiang L, Wang H, Farmer SR. Mol Cell Biol. 2007;27:4698–4707. doi: 10.1128/MCB.02279-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Qiao L, Shao J. J Biol Chem. 2006;281:39915–39924. doi: 10.1074/jbc.M607215200. [DOI] [PubMed] [Google Scholar]
- 190.Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Ajmo JM, Liang X, Rogers CQ, Pennock B, You M. Am J Physiol Gastrointest Liver Physiol. 2008;295:G833–G842. doi: 10.1152/ajpgi.90358.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Lin J, Della-Fera MA, Baile CA. Obes Res. 2005;13:982–990. doi: 10.1038/oby.2005.115. [DOI] [PubMed] [Google Scholar]
- 193.Park HJ, Yang JY, Ambati S, Della-Fera MA, Hausman DB, Rayalam S, Baile CA. J Med Food. 2008;11:773–783. doi: 10.1089/jmf.2008.0077. [DOI] [PubMed] [Google Scholar]
- 194.Ahn J, Lee H, Kim S, Park J, Ha T. Biochem Biophys Res Commun. 2008;373:545–549. doi: 10.1016/j.bbrc.2008.06.077. [DOI] [PubMed] [Google Scholar]
- 195.Li X, Zhang S, Blander G, Tse JG, Krieger M, Guarente L. Mol Cell. 2007;28:91–106. doi: 10.1016/j.molcel.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 196.Wang GL, Fu YC, Xu WC, Feng YQ, Fang SR, Zhou XH. Biochem Biophys Res Commun. 2009;380:644–649. doi: 10.1016/j.bbrc.2009.01.163. [DOI] [PubMed] [Google Scholar]
- 197.Potente M, Dimmeler S. Cell Cycle. 2008;7:2117–2122. doi: 10.4161/cc.7.14.6267. [DOI] [PubMed] [Google Scholar]
- 198.Zhang QJ, Wang Z, Chen HZ, Zhou S, Zheng W, Liu G, Wei YS, Cai H, Liu DP, Liang CC. Cardiovasc Res. 2008;80:191–199. doi: 10.1093/cvr/cvn224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Mattagajasingh I, Kim CS, Naqvi A, Yamamori T, Hoffman TA, Jung SB, DeRicco J, Kasuno K, Irani K. Proc Natl Acad Sci U S A. 2007;104:14855–14860. doi: 10.1073/pnas.0704329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Jin BY, Sartoretto JL, Gladyshev VN, Michel T. Proc Natl Acad Sci U S A. 2009;106:17343–17348. doi: 10.1073/pnas.0907409106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Csiszar A, Labinskyy N, Jimenez R, Pinto JT, Ballabh P, Losonczy G, Pearson KJ, de Cabo R, Ungvari Z. Mech Ageing Dev. 2009;130:518–527. doi: 10.1016/j.mad.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Csiszar A, Labinskyy N, Podlutsky A, Kaminski PM, Wolin MS, Zhang C, Mukhopadhyay P, Pacher P, Hu F, de Cabo R, Ballabh P, Ungvari Z. Am J Physiol Heart Circ Physiol. 2008;294:H2721–H2735. doi: 10.1152/ajpheart.00235.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Pillai JB, Gupta M, Rajamohan SB, Lang R, Raman J, Gupta MP. Am J Physiol Heart Circ Physiol. 2006;291:H1545–H1553. doi: 10.1152/ajpheart.01124.2005. [DOI] [PubMed] [Google Scholar]
- 204.Pillai JB, Isbatan A, Imai S, Gupta MP. J Biol Chem. 2005;280:43121–43130. doi: 10.1074/jbc.M506162200. [DOI] [PubMed] [Google Scholar]
- 205.Penumathsa SV, Thirunavukkarasu M, Koneru S, Juhasz B, Zhan L, Pant R, Menon VP, Otani H, Maulik N. J Mol Cell Cardiol. 2007;42:508–516. doi: 10.1016/j.yjmcc.2006.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Thirunavukkarasu M, Penumathsa SV, Koneru S, Juhasz B, Zhan L, Otani H, Bagchi D, Das DK, Maulik N. Free Radic Biol Med. 2007;43:720–729. doi: 10.1016/j.freeradbiomed.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Sulaiman M, Matta MJ, Sunderesan NR, Gupta MP, Periasamy M, Gupta M. Am J Physiol Heart Circ Physiol. 2010;298:H833–H843. doi: 10.1152/ajpheart.00418.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Gracia-Sancho J, Villarreal G, Jr, Zhang Y, Garcia-Cardena G. Cardiovasc Res. 2010;85:514–519. doi: 10.1093/cvr/cvp337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Penumathsa SV, Koneru S, Samuel SM, Maulik G, Bagchi D, Yet SF, Menon VP, Maulik N. Free Radic Biol Med. 2008;45:1027–1034. doi: 10.1016/j.freeradbiomed.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Loke WM, Proudfoot JM, Hodgson JM, McKinley AJ, Hime N, Magat M, Stocker R, Croft KD. Arterioscler Thromb Vasc Biol. 2010;30:749–757. doi: 10.1161/ATVBAHA.109.199687. [DOI] [PubMed] [Google Scholar]
- 211.Romero M, Jimenez R, Sanchez M, Lopez-Sepulveda R, Zarzuelo MJ, O'Valle F, Zarzuelo A, Perez-Vizcaino F, Duarte J. Atherosclerosis. 2009;202:58–67. doi: 10.1016/j.atherosclerosis.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 212.Sanchez M, Galisteo M, Vera R, Villar IC, Zarzuelo A, Tamargo J, Perez-Vizcaino F, Duarte J. J Hypertens. 2006;24:75–84. doi: 10.1097/01.hjh.0000198029.22472.d9. [DOI] [PubMed] [Google Scholar]
- 213.Ihm SH, Lee JO, Kim SJ, Seung KB, Schini-Kerth VB, Chang K, Oak MH. Atherosclerosis. 2009;206:47–53. doi: 10.1016/j.atherosclerosis.2009.01.036. [DOI] [PubMed] [Google Scholar]
- 214.Hirayama J, Sahar S, Grimaldi B, Tamaru T, Takamatsu K, Nakahata Y, Sassone-Corsi P. Nature. 2007;450:1086–1090. doi: 10.1038/nature06394. [DOI] [PubMed] [Google Scholar]
- 215.Grimaldi B, Nakahata Y, Kaluzova M, Masubuchi S, Sassone-Corsi P. Int J Biochem Cell Biol. 2009;41:81–86. doi: 10.1016/j.biocel.2008.08.035. [DOI] [PubMed] [Google Scholar]
- 216.Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Doi M, Hirayama J, Sassone-Corsi P. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 218.Nakahata Y, Grimaldi B, Sahar S, Hirayama J, Sassone-Corsi P. Curr Opin Cell Biol. 2007;19:230–237. doi: 10.1016/j.ceb.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 219.Belden WJ, Dunlap JC. Cell. 2008;134:212–214. doi: 10.1016/j.cell.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Rutter J, Reick M, McKnight SL. Annu Rev Biochem. 2002;71:307–331. doi: 10.1146/annurev.biochem.71.090501.142857. [DOI] [PubMed] [Google Scholar]
- 221.Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 222.Imai SI. Biochim Biophys Acta. 2009 doi:10.1016/j.bbapap.2009.10.024. [Google Scholar]
- 223.Hayashi M, Shimba S, Tezuka M. Biol Pharm Bull. 2007;30:621–626. doi: 10.1248/bpb.30.621. [DOI] [PubMed] [Google Scholar]
- 224.Tanno M, Sakamoto J, Miura T, Shimamoto K, Horio Y. J Biol Chem. 2007;282:6823–6832. doi: 10.1074/jbc.M609554200. [DOI] [PubMed] [Google Scholar]
- 225.Suzuki K, Koike T. Biochem Biophys Res Commun. 2007;359:665–671. doi: 10.1016/j.bbrc.2007.05.164. [DOI] [PubMed] [Google Scholar]
- 226.Marfe G, Tafani M, Indelicato M, Sinibaldi-Salimei P, Reali V, Pucci B, Fini M, Russo MA. J Cell Biochem. 2009;106:643–650. doi: 10.1002/jcb.22044. [DOI] [PubMed] [Google Scholar]
- 227.Argmann C, Auwerx J. Cell. 2006;126:837–839. doi: 10.1016/j.cell.2006.08.031. [DOI] [PubMed] [Google Scholar]
- 228.Nakagawa T, Lomb DJ, Haigis MC, Guarente L. Cell. 2009;137:560–570. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, Chang HY, Bohr VA, Ried T, Gozani O, Chua KF. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Vakhrusheva O, Smolka C, Gajawada P, Kostin S, Boettger T, Kubin T, Braun T, Bober E. Circ Res. 2008;102:703–710. doi: 10.1161/CIRCRESAHA.107.164558. [DOI] [PubMed] [Google Scholar]