Abstract
Wrapping DNA into chromatin provides a wealth of regulatory mechanisms that ensure normal growth and development in eukaryotes. Our understanding of chromatin structure, including nucleosomes and non-histone protein-DNA interactions, has benefited immensely from nuclease and chemical digestion techniques. DNA-bound proteins, such as histones or site-specific factors, protect DNA against nuclease cleavage and generate large nucleosomal or small regulatory factor footprints. Chromatin subject to distinct modes of regulation often coincides with sites of nuclease hypersensitivity or nucleosome positioning. An inherent limitation of cleavage-based analyses has been the inability to reliably analyze regions of interest when levels of digestion depart from single-hit kinetics. Moreover, cleavage-based techniques provide views that are averaged over all the molecules in a sample population. Therefore, in cases of occupancy of multiple regulatory elements by factors, one cannot define whether the factors are bound to the same or different molecules in the population. The recent development of DNA methyltransferase-based, single-molecule MAP-IT technology overcomes limitations of ensemble approaches and has opened numerous new avenues in chromatin research. Here, we review the strengths, limitations, applications and future prospects of MAP-IT ranging from structural issues to mechanistic questions in eukaryotic chromatin regulation.
Keywords: chromatin remodeling, DNA methyltransferases, positioned nucleosomes, protein-DNA interactions, transcription
Chromatin is central to transcriptional regulation and other DNA-directed processes in eukaryotes. As such, detailed biochemical knowledge of chromatin structure and its dynamics is needed to understand normal cellular function and disease states. Chromatin fibers consist of DNA-bound regulatory factors and nucleosomes, comprising 1.7 turns of DNA wrapped around the core histone octamer [Luger et al., 1997], repeated at a characteristic number of nucleotides in each eukaryote. Difficulties in analysis of chromatin are inherent to its highly heterogeneous nature. Heterogeneity in the state of chromatin among copies of the same region in different or the same cell(s) arises from: 1) combinatorial occupancy of multiple cis-acting elements; 2) random or quasi-random organization of nucleosomes [Bernstein et al., 2004; Lee et al., 2004; Yuan et al., 2005] 3) nucleosome occupancy at thermodynamically minor as well as favored locations in regions with strong positioning sequences [Pennings et al., 1991; Fragoso et al., 1995] 4) differential deposition or exchange of linker histones and histone variants (e.g. H2A.Z and H3.3) at the same region in different cells [Kusch and Workman, 2007] and 5) differential post-translational modification of histones, e.g. acetylation, which alters higher-order chromatin folding and recruitment of chromatin remodelers [Tse et al., 1998; Hassan et al., 2001; Hassan et al., 2002]. Thus, at a given locus, considerable diversity in chromatin states stems from a myriad of mechanisms.
Conventional probing methods cannot detect the inherent heterogeneity in chromatin because they average the contributions of all molecules within a sample population. In addition, nuclease-based techniques physically damage DNA, breaking apart the individual elements or modules within chromatin that form functional units. In this review, we describe a novel, powerful, single-molecule approach based on DNA methylation protection, termed MAP-IT, which holds great potential for addressing many formerly unapproachable issues confronting the field of chromosome biology.
ENSEMBLE VERSUS SINGLE-MOLECULE TECHNIQUES
Interactions of histone or non-histone proteins with DNA are most commonly resolved by using footprinting techniques. Classic strategies for chromatin probing rely on enzymatic (micrococcal nuclease (MNase), DNase I, DNase II, restriction endonucleases) or chemical (dimethyl sulfate, methidiumpropyl-EDTA, psoralens) agents that damage DNA and subsequent detection of resultant cleavage patterns by primer extension or indirect end-labeling [Simpson, 1998]. Comparison of chromatin and naked DNA samples digested by MNase, for example, is commonly used to infer changes in chromatin organization due to alterations in the presence, absence or specific positions of nucleosomes. In addition, based on the rate and extent of digestion by DNase I, the openness or accessibility of relatively large regions is revealed, thereby locating regulatory elements such as promoters, enhancers or insulators. The accessibility of these regions compared to repressed genes is often explained by significant depletion of histones due to nucleosome disassembly [Boeger et al., 2003; Reinke and Hörz, 2003].
Mapping the myriad of dynamic interactions of regulatory factors in chromatin has been difficult because of technical caveats imposed by these conventional, population-based approaches. First, probes that cause DNA strand scission disrupt the physical linkage between cis-acting elements that orchestrate gene regulatory programs. This precludes detection of multiple footprints due to cooperative or sequential binding of factors to the same DNA molecule. Second, nucleases, especially MNase, exhibit strong cleavage preferences that limit their probing resolution [Flick et al., 1986]. Third, while conventional footprinting techniques provide a qualitative view of where factors are bound to a chromosome, their ability to quantitatively assess promoter occupancy requires strict adherence to the principles of single-hit kinetics. To satisfy these conditions, according to Poisson distribution theory, random digestion and zero or one DNA cleavage(s) in ≥90% of the molecules must apply. This is because available primer extension and indirect end-labeling procedures can only detect the first cut site proximal to a hybridized probe molecule. In practice, however, the inherent complexity of biological systems presents non-random scenarios that preclude adherence to single-hit conditions. Regions displaying high levels of nuclease hypersensitivity are likely to contain multiple cuts that cannot be quantitatively mapped by conventional methods. Moreover, when distinct sub-populations of chromatin are present (e.g. bimodality), averaging due to population-ensemble methods causes misrepresentation of the true chromatin structures.
The advent of single-molecule technologies has the potential to revolutionize biochemical studies of chromatin and its related processes. Distributions in certain properties of single chromatin molecules can now be monitored in real-time, obtaining mechanistic insights that were previously unattainable with population-based assays. Currently, most single-molecule chromatin studies rely on force-based techniques that measure the physical properties of chromatin in real time [Bustamante et al., 2003]. These techniques have unveiled several key features of ATP-dependent chromatin remodeling, such as the translocation speed, force, processivity and step size in remodeler-catalyzed nucleosome movements [Cairns, 2007]. Readers interested in single-molecule biophysical aspects of chromatin are urged to consult recent reviews on the topic [Zlatanova and Leuba, 2003; Leuba et al., 2004; Cairns, 2007].
Integration of biophysical and biochemical data is necessary to unveil a complete picture of chromatin structure and its reorganization. However, few single-molecule tools are available to address the biochemical properties of these reactions in solution. A first generation, genome-wide mapping technique (GMAT), enriched various post-translational modifications by chromatin immunoprecipitation (ChIP) followed by sequencing of individual SAGE tags [Velculescu et al., 1995; Roh et al., 2004]. More recently, ChIP-Seq technology has employed massively parallel, deep sequencing of the resultant ChIP libraries [Johnson et al., 2007; Robertson et al., 2007]. Deep sequencing of the ends of nucleosome core particle-length (147 bp) fragments obtained from a nearly complete digestion of chromatin with MNase has yielded genome-wide maps of nucleosome positions [Schones et al., 2008]. Inclusion of a ChIP step after MNase digestion but prior to isolation of nucleosome core particle-length DNA and deep sequencing has been used to map genome-wide locations of post-translationally modified histones and histone H2A.Z in both yeast and humans [Albert et al., 2007; Barski et al., 2007; Schones et al., 2008]. In each of these instances, assuming comparable efficiencies for cross-linking and/or immunoprecipitation, the frequencies with which sequenced molecules map to the genome are thought to reflect relative (not absolute) occupancies for factors and modified histones. Questions involving co-occupancy of different factors along a region are precluded, again because cleavage or shearing destroys linkage between the independent modules along a chromosomal region.
DNA METHYLTRANSFERASES (DMTases) AS SINGLE-MOLECULE PROBES
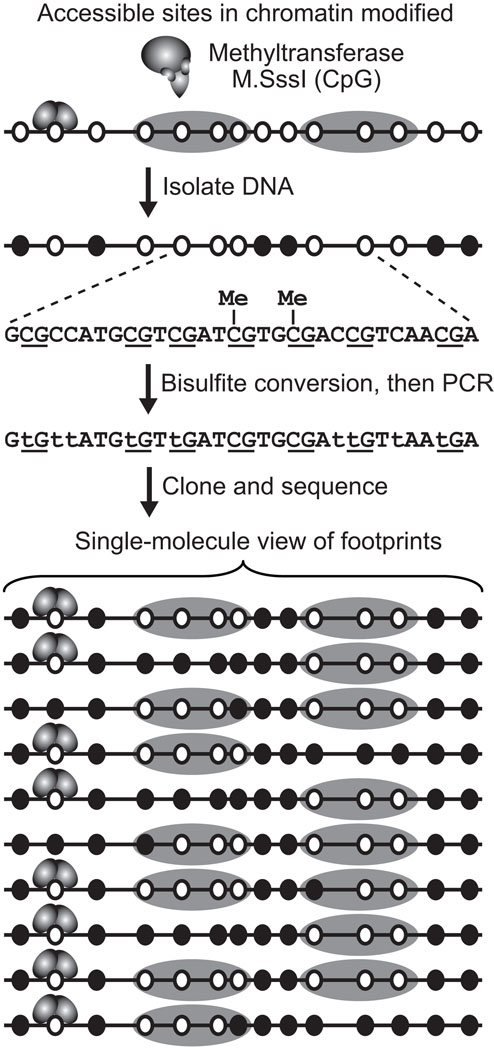
DMTases offer an attractive single-molecule strategy for exploring chromatin architecture and remodeler-catalyzed reorganization (Fig. 1). These enzymes transfer a methyl group from the co-factor S-adenosyl-L-methionine to a target cytosine or adenine on one or both strands within a specific 2–6 bp sequence. Enzymes that modify carbon 5 of cytosine (C-5) enable the mapping of chromatin structure in single molecules as this modification can be detected by positive chemical display using bisulfite genomic sequencing (BGS) [Frommer et al., 1992]. Bisulfite ion deaminates or converts unmethylated C to U, which undergoes transition to T following PCR amplification of a region of interest. By contrast, bisulfite-resistant m5C is propagated as C in the PCR product. The methylation status of every DMTase target site on an individual DNA strand is assessed by sequencing individual molecules cloned from the PCR amplicon. Probing chromatin structure with a C-5 DMTase followed by BGS is a technique called methyltransferase accessibility protocol for individual templates (MAP-IT). The success of MAP-IT stems from our earlier studies demonstrating that positioned nucleosomes and bound proteins hinder accessibility of DMTases to their cognate sites in DNA [Kladde and Simpson, 1994; Kladde et al., 1996]. From a structural perspective, methylation of DNA is relatively innocuous, and may explain the ability to detect protein-DNA interactions in systems where footprinting with nucleases had failed [Dong et al., 1999; Duan et al., 1999]. Avoidance of DNA damage also makes DMTases the least invasive of available chromatin probes and well suited for studies in the setting of living cells.
Fig. 1.
Single-molecule MAP-IT assay for footprinting protein-DNA interactions. A region of chromatin with two positioned nucleosomes (solid gray ellipses) and DNA-bound transcription factor (gray homodimer at left) is probed with a DMTase, e.g. M.SssI that recognizes CpG sites (open ellipses). After isolation of protein-free DNA, the lack of methylation at DMTase sites that were inaccessible (open ellipses) or accessible and hence methylated (filled ellipses) in chromatin is determined by BGS. The enlarged region of sequence shows CpG sites (underscored; number of nucleotides between each CpG not to scale) that were methylated (Me) or not. Following bisulfite treatment and amplification by PCR, C is converted to T (emphasized in lower case), whereas m5C (C marked by Me) amplifies as C. In the last step of BGS, single molecules are cloned from the bulk PCR amplicon and sequenced to obtain footprints on single molecules (proteins formerly bound in vivo are inserted for reference). Note the occasional methylation at the ends of some nucleosomes due to nucleosome site exposure [Polach and Widom, 1995; Anderson and Widom, 2000; Li et al., 2005].
A key feature of MAP-IT is that it enables visualization of the occupancy of multiple sites on single molecules that make up the overall population (Fig. 1). At a first approximation, short consecutive spans or patches of unmethylated DNA in each sequenced molecule, ranging from a few nucleotides to a few helical turns, can be attributed to bound non-histone proteins. Longer regions of protection of ~140 bp punctuated by runs of m5C are inferred to be nucleosomes with intervening accessible linkers. An advantage of MAP-IT over all nuclease-based footprinting methods is that the constraints of single-hit kinetics do not apply, as the methylation status of every C residue in each molecule is scored by BGS. This allows the DMTase concentration or time of probing to be increased to minimize false negatives, namely, sites located in accessible regions that are not methylated due to insufficient DMTase activity.
In contrast, elevated enzyme concentration and incubation time will increase the probability of accessing the ends or termini of nucleosomes. This is because nucleosomes undergo rapid, short-lived conformational fluctuations whereby DNA on the histone octamer is unwrapped and rewrapped, a process termed spontaneous site exposure [Polach and Widom, 1995; Anderson and Widom, 2000; Li et al., 2005]. Sequences at nucleosome termini are preferentially exposed as fewer histone-DNA contacts must be broken. Experimentally, a gradient of decreasing accessibility to restriction endonucleases and DMTases at internal superhelical locations has been observed in the population-based analyses performed to date [Kladde and Simpson, 1994; Polach and Widom, 1995; Kladde et al., 1996; Xu et al., 1998b; Anderson and Widom, 2000]. Hence, in MAP-IT, individual nucleosomes would be expected to display variable extents of protection against methylation due to DMTases gaining differential access to one or more helical turns at their termini. In the probing reactions, the rate of 'breathing' at nucleosome ends is extremely rapid compared to the rate of DNA methylation. Therefore, under conditions where methylation exhibits first-order kinetics, the frequencies of observing single nucleosomes with unwrapped sequences and the extent of unwrapping will increase as a function of DMTase concentration and incubation time. Summing the frequency of m5C at each site over a cohort of sequenced single molecules should recapitulate the accessibility gradient observed in population studies. To obtain the positions of individual nucleosomes in a region of interest most accurately, the DMTase concentration should be titrated until mostly 147-bp nucleosomal-length patches of protection are obtained. Under these conditions, the single-molecule view provided by MAP-IT will reveal if a region contains well-positioned or more disorganized nucleosomes.
We consider recent advances provided by the versatility of C-5 DMTases as single-molecule probes of chromatin in the next section.
NON-RANDOM DISTRIBUTION OF REMODELED NUCLEOSOMES AT THE INDUCED PHO5 PROMOTER
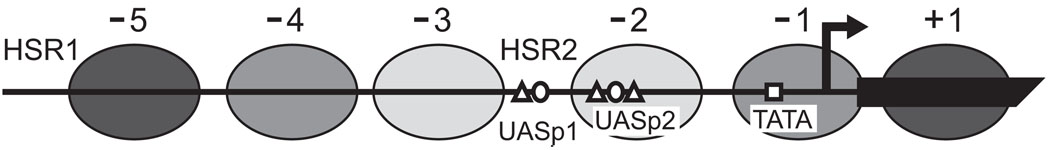
The absence of detectable de novo DNA methylation in budding yeast makes it an excellent host system for probing protein-DNA interactions with exogenously supplied DMTases. The S. cerevisiae PHO5 promoter is a proven model system for elucidating mechanisms of transcriptional activation within chromatin. Under repressive conditions of high environmental phosphate, the PHO5 promoter is incorporated into an array of well-positioned nucleosomes, numbered −5 to +3 relative to the translational start codon (Fig. 2). Upstream activating sequence (UAS) p1 is localized to a DNase I hypersensitive region (HSR2) between nucleosomes −3 and −2, whereas UASp2 and the TATA box are packaged into nucleosomes −2 and −1, respectively [Almer and Hörz, 1986]. When phosphate availability diminishes, the transactivators Pho4 and Pho2 bind cooperatively to UASp1 and UASp2 and recruit distinct multi-protein complexes with histone acetyltransferase (SAGA and NuA4) and ATP-dependent chromatin remodeling (SWI/SNF and INO80) activity [Barbaric et al., 2003; Steger et al., 2003; Nourani et al., 2004; Dhasarathy and Kladde, 2005].
Fig. 2.
Chromatin structure of the PHO5 promoter. The transcriptionally inactive promoter consists of an array of positioned nucleosomes (filled ellipses), numbered −5 to +3 (+2 and +3 not shown). HSR1 and HSR2, DNase I hypersensitive sites 1 and 2; UASp1 and UASp2, low and high affinity Pho4 binding sites (open ellipses), respectively; Pho2 sites (open triangles); TATA box (unfilled square); major transcription start site (bent arrow) and PHO5 coding region (filled polygon). The shading intensity of each nucleosome is inversely proportional to its accessibility to M.HhaI as determined by MAP-IT analysis of the transcriptionally active promoter [Jessen et al., 2006]. These data agree well with anti-histone ChIP results showing less histone density at the activated core promoter as compared to nucleosome −5 [Adkins et al., 2004].
In yeast, nucleosome remodeling is frequently confined to one, sometimes two, nucleosome(s) in the vicinity of TATA boxes [Kuo et al., 1998; Bernstein et al., 2004; Lee et al., 2004; Yuan et al., 2005]. However, at PHO5, sensitivity to nucleases mapped to at least four positioned nucleosomes (−4 to −1) after promoter activation [Almer and Hörz, 1986; Almer et al., 1986]. These early studies led to the prevailing view that nucleosomes −4 to −1 were disrupted at similar frequencies in every cell across the population. In contrast, using a different approach, it was concluded that only one-half of the expected number of nucleosomes were lost from the PHO5 promoter on activation [Boeger et al., 2003; Boeger et al., 2004]. These studies used topological analysis to measure the average number of nucleosomes lost over a population of promoter molecules that had been excised from the genome and circularized.
We employed MAP-IT as a means independent of population averaging to resolve the controversy in the number of evicted nucleosomes, determining the distribution of remodeled nucleosomes on single copies of the PHO5 promoter from different cells [Jessen et al., 2006]. We also obtained mechanistic insight into how localized recruitment of co-activator complexes to UASp1 and UASp2 by Pho4-Pho2 leads to reorganization of multiple nucleosomes situated further upstream.
For analysis by MAP-IT of chromatin remodeling associated with transactivation of PHO5, the C-5 MTase M.HhaI was selected as the chromatin probe, because the core promoter encompassed by nucleosome −1 contained one natural HhaI site. This side-stepped the need to make mutations that might affect transcriptional regulation of the promoter. Base changes were made to mark each of the remaining positioned nucleosomes (and HSR2) with a single HhaI site as close to the pseudodyad as possible to minimize potential differences in site accessibility due to nucleosome site exposure. Control experiments verified that the induction kinetics and chromatin structure of the altered PHO5 promoter were indistinguishable from wild-type, validating its use as a surrogate system. Lastly, the M.HhaI probe was placed under conditional regulatory control so that changes in PHO5 chromatin during the initial times of induction following phosphate starvation could be monitored. The use of M.HhaI (GCGC) simplified data analysis for the debut MAP-IT study as the methylation status of only 6 sites per molecule had to be scored. Dinucleotide-recognition probes, M.CviPI (GC) [Xu et al., 1998a] and M.SssI (CG or, historically, CpG) [Renbaum et al., 1990], could clearly be used to increase resolution and avoid the need to introduce mutations.
MAP-IT analysis of cloned molecules, each representing the chromatin state of a full PHO5 promoter from a single cell, revealed that the number of nucleosomes remodeled varied substantially from one cell to another. Despite this pronounced heterogeneity of chromatin states, it was clear that nucleosomes −3 and −2 flanking UASp1, the initial site of transactivator binding [Venter et al., 1994; Carvin et al., 2003; Adkins et al., 2007], were remodeled preferentially compared to more distal nucleosomes. These data suggested that chromatin remodeling initiates at the UASs, propagates or spreads outwardly and diminishes with distance. This model predicts a higher frequency of disruption of adjacent nucleosomes, especially near the UASs. Indeed, the single-molecule view of MAP-IT offered a unique way to test this model further; compared to methylation of a naked DNA control, there was a statistically significant preference for disruption of neighboring or contiguous nucleosomes on single, activated PHO5 promoters.
In addition to this non-random behavior among single PHO5 promoter molecules in vivo, even at a very early stage of activation, the majority of molecules had accumulated multiple m5C residues or ‘hits’. As more hits are expected with the use of probes with higher spatial resolution (M.CviPI, M.SssI or nucleases), our data strongly suggest that the concept of achieving single-hit-kinetic levels of nuclease cutting in complex biological systems needs to be critically re-evaluated. This is important because only the first, probe-proximal cut site can be detected in each molecule; information regarding accessibility at all distal sites is lost. By comparison, as no signal is discarded in MAP-IT, we were able to detect significant probe access as far upstream as nucleosome −5 for the first time. Therefore, by avoiding the limitations of single-hit kinetics, MAP-IT provides both a more sensitive and quantitative view of chromatin accessibility than nucleases. The combined attributes of MAP-IT have established that active PHO5 chromatin is highly heterogeneous with respect to nucleosome occupancy and encompasses a larger domain of positioned nucleosomes than previously appreciated. Mechanistically, the results suggest that chromatin remodeling at a euchromatic locus initiates at the UASs, spreads outwardly and falls off as a function of distance.
SINGLE-MOLECULE FOOTPRINTING OF MAMMALIAN NUCLEI
In vertebrates, DNA methylation occurs predominantly, if not exclusively, at CpG dinucleotides to yield m5CpG [Bird, 2002]. CpG methylation is closely associated with heritable gene silencing and formation of heterochromatin. In disease-free cells, clusters of CpG sites, densely localized to regions called CpG islands at gene promoters, are present in an unmethylated state. Hypermethylation of CpG islands is now firmly established as a correlative or causative mechanism for inappropriate silencing of tumor suppressor genes in carcinogenesis [Robertson, 2005; Jones and Baylin, 2007]. Overall, aberrant genomic methylation patterns have serious implications for proper growth and development.
The mechanism of epigenetic silencing has been shrouded in debate. Accumulating evidence indicates that DNA methylation affects nucleosome structure and positioning in a context-dependent manner. Nonetheless, the relationship between DNA methylation, chromatin dynamics and gene activity at the endogenous promoters remains enigmatic. Recently, MAP-IT using the CpG methyltransferase M.SssI, dubbed methylation-sensitive promoter analysis (MSPA), was used to address nucleosome occupancy at promoters bearing unmethylated CpG islands in cultured mammalian cells. Nuclei extracted from two different cell lines with markedly different levels of p16 gene expression were probed with M.SssI followed by BGS analysis of cloned promoter molecules. Analysis of single promoter molecules revealed runs or patches of methylation protection [Fatemi et al., 2005]. A definition for a patch was adopted so that the majority of patches corresponded to the 147-bp length of a nucleosome. However, a significant number of patches on single molecules were sub-nucleosomal in length, possibly due to site exposure at nucleosome ends [Polach and Widom, 1995; Anderson and Widom, 2000; Li et al., 2005]. Importantly, as a different number of helical turns of DNA can be accessed on each side of a given single nucleosome, the center of a protected region may frequently not correspond to the pseudodyad center at superhelix location 0. Despite this, patches corresponding to nucleosomes were evident on single p16 promoters from cells with low-level expression of the gene, and this pattern appeared more disorganized in highly-expressing cells. An overall ~2-fold increase in methylation of DNA in linkers versus nucleosome centers was observed at repressed promoters. In such cases, titration of DMTase concentration might improve the analysis and more accurately assess the positions of individual nucleosomes.
Extensive runs of mostly consecutive m5C on single molecules from the cell line with high p16 expression suggested that regions of histone-free DNA were generated [Fatemi et al., 2005]. These results are reminiscent of early studies in yeast showing 80% methylation, on average, over nine sites within a 100 bp region of open chromatin [Kladde and Simpson, 1994]. At such high levels of methylation it can be deduced that many single molecules bear consecutive runs of modification. In more recent studies, extensive, consecutive runs of M.SssI-catalyzed m5C were also found in single molecules of the bidirectional EMP2AIP1-MLH1 and stress-induced GRP78 promoters [Gal-Yam et al., 2006; Lin et al., 2007]. ChIP analysis was used to demonstrate significant depletion of histones over the M.SssI-methylated regions in the promoters [Gal-Yam et al., 2006; Lin et al., 2007], consistent with abundant evidence that active promoters evict nucleosomes [Boeger et al., 2003; Reinke and Hörz, 2003; Bernstein et al., 2004; Lee et al., 2004; Yuan et al., 2005]. It seems probable that consecutive runs of m5C in MAP-IT will become synonymous with disassembled nucleosomes as probing with DMTases gains in usage.
In colorectal cancer cell lines that have epigenetically silenced EMP2AIP1-MLH1, the CpG island is heavily methylated by endogenous DMTases (Dnmts). Bulk analysis of the promoter with nucleases suggested higher nucleosome occupancy in the silenced cell lines [Lin et al., 2007]. Further analysis of the silenced chromatin by MAP-IT with M.SssI was precluded as both the chromatin probe and endogenous Dnmt enzymes methylate CpG sites. Therefore, DNA methylation in the silenced cell line was reversed with the Dnmt inhibitor 5-aza-2'-deoxycytidine to reactivate EMP2AIP1-MLH1 expression. BGS showed that single EMP2AIP1-MLH1 promoters from drug-treated cells populated three classes, ones with full, partial or no demethylation, in roughly equal proportions. Single-molecule MAP-IT analysis of chromatin in the first two classes was obtained by selective amplification of the bidirectional promoter with primers that annealed to unmethylated sequences. About half of the selectively amplified molecules contained nucleosomes and the other half had apparently evicted the nucleosome over core promoter sequences. The analysis is complicated because it is not possible to determine what fraction of molecules from the initial population was selectively amplified. Additionally, there is no way to ascertain if m5C on the nucleosome-containing promoters is due to M.SssI or endogenous Dnmts. Regardless, these single-molecule chromatin analyses have clearly demonstrated that heritable DNA methylation contributes to silencing by increasing nucleosome occupancy over promoter elements.
Single-molecule DMTase footprinting has also been combined with computational methods to reveal the dynamics of transcriptional activation at the CpG island of the stress-inducible GRP78 promoter [Gal-Yam et al., 2006]. Strikingly, 12 contiguous CpG sites in this island were found to be constitutively depleted of histones and hence accessible in nearly all of ~300 clonally analyzed promoters. Apart from analyzing the nucleosome positions, the single-molecule studies also detected constitutive binding of TATA-binding protein at the TATA element. The preservation of linkage between multiple cis elements on clonal promoters also provided a unique means to display factor footprints through autocorrelation analysis of co-protected CpG sites. An elegant further statistical analysis of combinatorial modes of factor occupancy supported a model for sequential loading of site-specific transactivators onto GRP78 stress-responsive elements during promoter activation.
CONCLUSIONS AND PERSPECTIVES
All DNA-templated processes in eukaryotic cells rely on the orchestration of activities of multiple proteins in the context of chromatin. Owing to the stochasticity of these processes, invariably, significant heterogeneity is introduced at the level of individual regions of chromosomes in different cells. Differential occupancy of multiple sites by nucleoprotein assemblies with varied composition and dynamic changes in nucleosome structure and position promote further complexity that has formerly posed a considerable hurdle to footprinting studies.
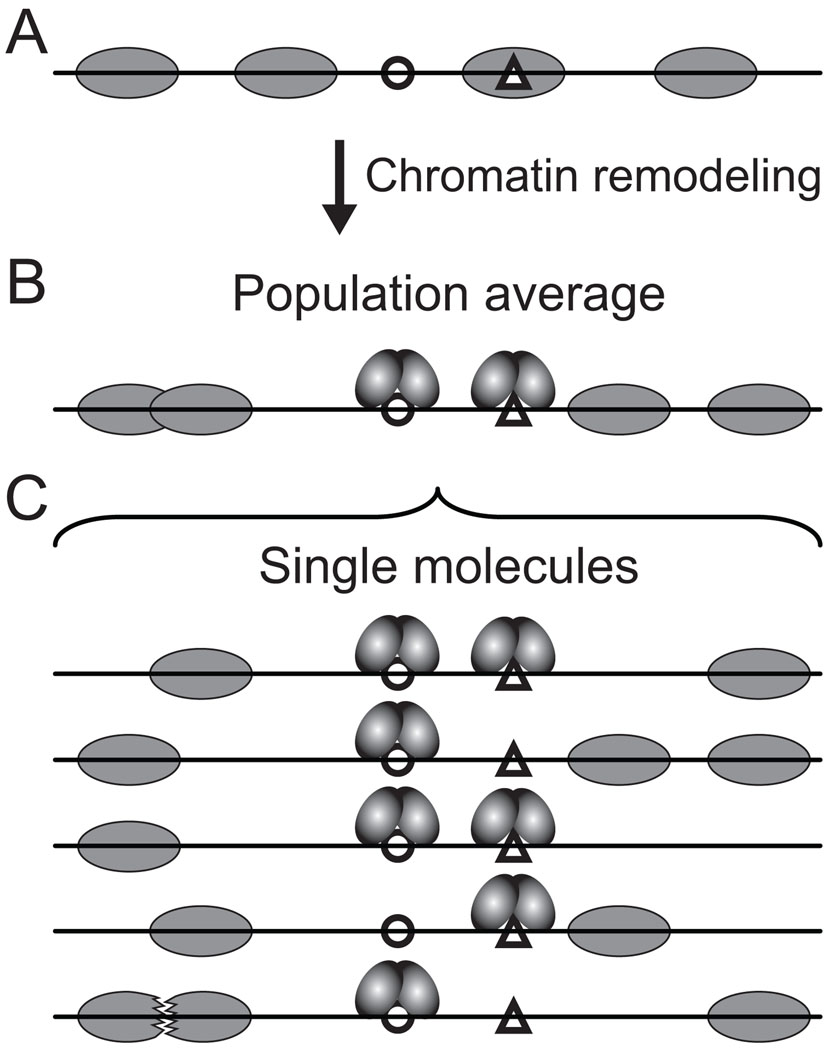
The single-molecule technology of MAP-IT offers a unique opportunity to tease apart this complexity as it avoids population-ensemble averaging over all molecules present in a sample. We envision that the commercial availability of DMTases with dinucleotide specificity will open up several exciting avenues of inquiry. For instance, MAP-IT can be used in mechanistic studies of chromatin remodeling in cell-free systems. A long-standing view has been that chromatin remodelers mobilize nucleosomes by propagating waves or loops around the histone octamer surface in response to ATP hydrolysis. It may be possible to capture and mark such loops or other intermediates on reconstituted mononucleosomes by using MAP-IT in combination with stop-flow technology for rapid mixing of reaction components. MAP-IT can also be extended to study remodeling of dinucleosomes and other more complex substrates. The ability to localize multiple nucleosomes on single molecules should distinguish if nucleosomes are mobilized to the same or distinct positions from one molecule to another (Fig. 3).
Fig. 3.
Population versus single-molecule analysis of chromatin remodeling. A) Chromatin substrate with four positioned nucleosomes (gray ellipses) and two factor binding sites, one accessible (open circle) and one occluded by a nucleosome (open triangle). B) Population-ensemble averaging of nucleosome positions after factor-mediated chromatin remodeling. The overlapping nucleosomes correspond to a footprint between mono- and dinucleosome length [Barbaric et al., 1992]. On average, two pairs of homodimeric transactivators occupy the remodeled promoter. C) Hypothetical distribution of nucleosomes and transactivators on single chromatin substrates constituting the population in B. Note that the large footprint corresponding to the overlapping nucleosomes could result from two distinct, but overlapping, sub-populations of positioned nucleosomes (molecules 1–4), collided nucleosomes (molecule 5; jagged edges depicting partial nucleosome disassembly) or both.
MAP-IT is also likely to prove valuable in resolving numerous questions where one encounters cell-to-cell variation or distinct sub-populations of cells. An obvious example includes epigenetic studies employing continuous cell lines that have undergone genetic drift. Systems where significant stochasticity in basal or activated levels of transcription [Raser and O'Shea, 2004] or promoters are subject to binary or all-or-none modes of activation [Becskei et al., 2001; Biggar and Crabtree, 2001] are also good candidates for single-molecule probing. MAP-IT can potentially be applied either directly to the population to identify different subclasses of gene copies with inactive or active chromatin or, alternatively, after sorting cells into sub-populations using fluorescence-based methods.
In vertebrate cells, the importance of epigenetic regulation of chromatin and CpG methylation in normal cell function and development and its aberrations in disease have become increasingly evident [Robertson, 2005; Jones and Baylin, 2007]. Using the GC DMTase M.CviPI [Xu et al., 1998a] as the probe in MAP-IT enables direct comparison of endogenous CpG methylation and chromatin accessibility on the same molecule, excluding only overlapping GCG sites [Kilgore et al., 2007]. Due to their high GC content, CpG islands typically harbor an excellent lattice of M.CviPI sites.
Numerous studies can be contemplated in which simultaneous, single-molecule visualization of both of chromatin architecture and CpG methylation by MAP-IT using M.CviPI would be highly advantageous. Specific examples include the study of random inactivation of a single X chromosome in female mammals and epigenetic imprinting dependent on the maternal or paternal origin of chromosomes [Krueger and Morison, 2008]. A sizeable number of human autosomal genes also display allelic exclusion or random monoallelic expression of either the maternally- or paternally-derived gene copy [Gimelbrant et al., 2007]. MAP-IT could provide insights into the extent to which this diversity is due to endogenous DNA methylation, chromatin organization and regulatory single nucleotide polymorphisms (rSNPs) that impair transcription factor binding to DNA [Pampin and Rodriguez-Rey, 2007]. The latter would require either beforehand knowledge of rSNPs or the discovery by MAP-IT of A or G rSNPs (i.e. C is converted to T by BGS) that do not overlap with sites recognized by the DMTase probe. Lastly, MAP-IT could be used to study the coordinated reprogramming of chromatin and DNA demethylation that precede genetic rearrangement of only one allele of immunoglobulin and T-cell receptor genes in each lymphoid precursor cell [Bergman and Cedar, 2004]. Uniquely, MAP-IT avoids bulk averaging of the distinct sub-populations of chromosomal copies that are intrinsic to each of these systems.
In summary, we contend that single-molecule probing with C-5 DMTases in MAP-IT is a powerful technology that can be used in lieu of conventional nucleases for probing chromatin structure. We predict that single-molecule landscapes of chromosomal regions will gain a strong foothold in situations where diverse or distinct molecular populations are prevalent. Further increases in the probing resolution of MAP-IT in vertebrate systems will be realized if additional DMTases with short recognition sites are discovered that have limited or no overlap in methylation of CpG sites. Moreover, as the relatively innocuous methylation mark has facilitated footprinting in living yeast cells, similar studies may also be possible in cultured cells or perhaps animals, provided suitable means of enzyme regulation/delivery can be devised. The recent development of shotgun deep sequencing of bisulfite-converted DNA [Cokus et al., 2008; Lister et al., 2008] invites the use MAP-IT as a single-molecule platform for simultaneous genome-wide mapping of chromatin structure and endogenous CpG methylation.
ACKNOWLEDGEMENTS
We thank Russell Darst, Carolina Pardo and Jon Widom for helpful discussions and Russell Darst for critical reading of the manuscript.
Grant sponsor: NIH; Grant number: CA095525; Grant sponsor: Department of Defense; Grant number: BC062914.
REFERENCES
- Adkins MW, Howar SR, Tyler JK. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol Cell. 2004;14:657–666. doi: 10.1016/j.molcel.2004.05.016. [DOI] [PubMed] [Google Scholar]
- Adkins MW, Williams SK, Linger J, Tyler JK. Chromatin disassembly from the PHO5 promoter is essential for the recruitment of the general transcription machinery and coactivators. Mol Cell Biol. 2007;27:6372–6382. doi: 10.1128/MCB.00981-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- Almer A, Hörz W. Nuclease hypersensitive regions with adjacent positioned nucleosomes mark the gene boundaries of the PHO5/PHO3 locus in yeast. EMBO J. 1986;5:2681–2687. doi: 10.1002/j.1460-2075.1986.tb04551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almer A, Rudolph H, Hinnen A, Hörz W. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 1986;5:2689–2696. doi: 10.1002/j.1460-2075.1986.tb04552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JD, Widom J. Sequence and position-dependence of the equilibrium accessibility of nucleosomal DNA target sites. J Mol Biol. 2000;296:979–987. doi: 10.1006/jmbi.2000.3531. [DOI] [PubMed] [Google Scholar]
- Barbaric S, Fascher KD, Hörz W. Activation of the weakly regulated PHO8 promoter in S. cerevisiae: chromatin transition and binding sites for the positive regulatory protein PHO4. Nucleic Acids Res. 1992;20:1031–1038. doi: 10.1093/nar/20.5.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbaric S, Reinke H, Hörz W. Multiple mechanistically distinct functions of SAGA at the PHO5 promoter. Mol Cell Biol. 2003;23:3468–3476. doi: 10.1128/MCB.23.10.3468-3476.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Becskei A, Seraphin B, Serrano L. Positive feedback in eukaryotic gene networks: cell differentiation by graded to binary response conversion. EMBO J. 2001;20:2528–2535. doi: 10.1093/emboj/20.10.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman Y, Cedar H. A stepwise epigenetic process controls immunoglobulin allelic exclusion. Nat Rev Immunol. 2004;4:753–761. doi: 10.1038/nri1458. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Liu CL, Humphrey EL, Perlstein EO, Schreiber SL. Global nucleosome occupancy in yeast. Genome Biol. 2004;5:R62. doi: 10.1186/gb-2004-5-9-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggar SR, Crabtree GR. Cell signaling can direct either binary or graded transcriptional responses. EMBO J. 2001;20:3167–3176. doi: 10.1093/emboj/20.12.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. Nucleosomes unfold completely at a transcriptionally active promoter. Mol Cell. 2003;11:1587–1598. doi: 10.1016/s1097-2765(03)00231-4. [DOI] [PubMed] [Google Scholar]
- Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol Cell. 2004;14:667–673. doi: 10.1016/j.molcel.2004.05.013. [DOI] [PubMed] [Google Scholar]
- Bustamante C, Bryant Z, Smith SB. Ten years of tension: single-molecule DNA mechanics. Nature. 2003;421:423–427. doi: 10.1038/nature01405. [DOI] [PubMed] [Google Scholar]
- Cairns BR. Chromatin remodeling: insights and intrigue from single-molecule studies. Nat Struct Mol Biol. 2007;14:989–996. doi: 10.1038/nsmb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvin CD, Dhasarathy A, Friesenhahn LB, Jessen WJ, Kladde MP. Targeted cytosine methylation for in vivo detection of protein-DNA interactions. Proc Natl Acad Sci USA. 2003;100:7743–7748. doi: 10.1073/pnas.1332672100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhasarathy A, Kladde MP. Promoter occupancy is a major determinant of chromatin remodeling enzyme requirements. Mol Cell Biol. 2005;25:2698–2707. doi: 10.1128/MCB.25.7.2698-2707.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L, Wang W, Wang F, Stoner M, Reed JC, Harigai M, Samudio I, Kladde MP, Vyhlidal C, Safe S. Mechanisms of transcriptional activation of bcl-2 gene expression by 17β-estradiol in breast cancer cells. J Biol Chem. 1999;274:32099–32107. doi: 10.1074/jbc.274.45.32099. [DOI] [PubMed] [Google Scholar]
- Duan R, Porter W, Samudio I, Vyhlidal C, Kladde M, Safe S. Transcriptional activation of c-fos protooncogene by 17β-estradiol: mechanism of aryl hydrocarbon receptor-mediated inhibition. Mol Endocrinol. 1999;13:1511–1521. doi: 10.1210/mend.13.9.0338. [DOI] [PubMed] [Google Scholar]
- Fatemi M, Pao MM, Jeong S, Gal-Yam EN, Egger G, Weisenberger DJ, Jones PA. Footprinting of mammalian promoters: use of a CpG DNA methyltransferase revealing nucleosome positions at a single molecule level. Nucleic Acids Res. 2005;33:e176. doi: 10.1093/nar/gni180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flick JT, Eissenberg JC, Elgin SC. Micrococcal nuclease as a DNA structural probe: its recognition sequences, their genomic distribution and correlation with DNA structure determinants. J Mol Biol. 1986;190:619–633. doi: 10.1016/0022-2836(86)90247-0. [DOI] [PubMed] [Google Scholar]
- Fragoso G, John S, Roberts MS, Hager GL. Nucleosome positioning on the MMTV LTR results from the frequency-biased occupancy of multiple frames. Genes Dev. 1995;9:1933–1947. doi: 10.1101/gad.9.15.1933. [DOI] [PubMed] [Google Scholar]
- Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal-Yam EN, Jeong S, Tanay A, Egger G, Lee AS, Jones PA. Constitutive nucleosome depletion and ordered factor assembly at the GRP78 promoter revealed by single molecule footprinting. PLoS Genet. 2006;2:e160. doi: 10.1371/journal.pgen.0020160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimelbrant A, Hutchinson JN, Thompson BR, Chess A. Widespread monoallelic expression on human autosomes. Science. 2007;318:1136–1140. doi: 10.1126/science.1148910. [DOI] [PubMed] [Google Scholar]
- Hassan AH, Neely KE, Workman JL. Histone acetyltransferase complexes stabilize SWI/SNF binding to promoter nucleosomes. Cell. 2001;104:817–827. doi: 10.1016/s0092-8674(01)00279-3. [DOI] [PubMed] [Google Scholar]
- Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, Carrozza MJ, Workman JL. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell. 2002;111:369–379. doi: 10.1016/s0092-8674(02)01005-x. [DOI] [PubMed] [Google Scholar]
- Jessen WJ, Hoose SA, Kilgore JA, Kladde MP. Active PHO5 chromatin encompasses variable numbers of nucleosomes at individual promoters. Nat Struct Mol Biol. 2006;13:256–263. doi: 10.1038/nsmb1062. [DOI] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilgore JA, Hoose SA, Gustafson TL, Porter W, Kladde MP. Single-molecule and population probing of chromatin structure using DNA methyltransferases. Methods. 2007;41:320–332. doi: 10.1016/j.ymeth.2006.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kladde MP, Simpson RT. Positioned nucleosomes inhibit Dam methylation in vivo. Proc Natl Acad Sci USA. 1994;91:1361–1365. doi: 10.1073/pnas.91.4.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kladde MP, Xu M, Simpson RT. Direct study of DNA-protein interactions in repressed and active chromatin in living cells. EMBO J. 1996;15:6290–6300. [PMC free article] [PubMed] [Google Scholar]
- Krueger C, Morison IM. Random monoallelic expression: making a choice. Trends Genet. 2008 doi: 10.1016/j.tig.2008.03.005. [DOI] [PubMed] [Google Scholar]
- Kuo MH, Zhou J, Jambeck P, Churchill ME, Allis CD. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 1998;12:627–639. doi: 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch T, Workman JL. Histone variants and complexes involved in their exchange. Subcell Biochem. 2007;41:91–109. [PubMed] [Google Scholar]
- Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet. 2004;36:900–905. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]
- Leuba SH, Bennink ML, Zlatanova J. Single-molecule analysis of chromatin. Methods Enzymol. 2004;376:73–105. doi: 10.1016/S0076-6879(03)76006-6. [DOI] [PubMed] [Google Scholar]
- Li G, Levitus M, Bustamante C, Widom J. Rapid spontaneous accessibility of nucleosomal DNA. Nat Struct Mol Biol. 2005;12:46–53. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- Lin JC, Jeong S, Liang G, Takai D, Fatemi M, Tsai YC, Egger G, Gal-Yam EN, Jones PA. Role of nucleosomal occupancy in the epigenetic silencing of the MLH1 CpG island. Cancer Cell. 2007;12:432–444. doi: 10.1016/j.ccr.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Nourani A, Utley RT, Allard S, Côté J. Recruitment of the NuA4 complex poises the PHO5 promoter for chromatin remodeling and activation. EMBO J. 2004;23:2597–2607. doi: 10.1038/sj.emboj.7600230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pampin S, Rodriguez-Rey JC. Functional analysis of regulatory single-nucleotide polymorphisms. Curr Opin Lipidol. 2007;18:194–198. doi: 10.1097/MOL.0b013e3280145093. [DOI] [PubMed] [Google Scholar]
- Pennings S, Meersseman G, Bradbury EM. Mobility of positioned nucleosomes on 5 S rDNA. J Mol Biol. 1991;220:101–110. doi: 10.1016/0022-2836(91)90384-i. [DOI] [PubMed] [Google Scholar]
- Polach KJ, Widom J. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J Mol Biol. 1995;254:130–149. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- Raser JM, O'Shea EK. Control of stochasticity in eukaryotic gene expression. Science. 2004;304:1811–1814. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke H, Hörz W. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol Cell. 2003;11:1599–1607. doi: 10.1016/s1097-2765(03)00186-2. [DOI] [PubMed] [Google Scholar]
- Renbaum P, Abrahamove D, Fainsod A, Wilson G, Rottem S, Razin A. Cloning, characterization, and expression in Escherichia coli of the gene coding for the CpG DNA methylase from Spiroplasma sp. strain MQ1 (M.SssI) Nucleic Acids Res. 1990;18:1145–1152. doi: 10.1093/nar/18.5.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, Thiessen N, Griffith OL, He A, Marra M, Snyder M, Jones S. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- Roh TY, Ngau WC, Cui K, Landsman D, Zhao K. High-resolution genome-wide mapping of histone modifications. Nat Biotechnol. 2004;22:1013–1016. doi: 10.1038/nbt990. [DOI] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson RT. Chromatin structure and analysis of mechanisms of activators and repressors. Methods. 1998;15:283–294. doi: 10.1006/meth.1998.0632. [DOI] [PubMed] [Google Scholar]
- Steger DJ, Haswell ES, Miller AL, Wente SR, O'Shea EK. Regulation of chromatin remodeling by inositol polyphosphates. Science. 2003;299:114–116. doi: 10.1126/science.1078062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse C, Sera T, Wolffe AP, Hansen JC. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol Cell Biol. 1998;18:4629–4638. doi: 10.1128/mcb.18.8.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- Venter U, Svaren J, Schmitz J, Schmid A, Hörz W. A nucleosome precludes binding of the transcription factor Pho4 in vivo to a critical target site in the PHO5 promoter. EMBO J. 1994;13:4848–4855. doi: 10.1002/j.1460-2075.1994.tb06811.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Kladde MP, Van Etten JL, Simpson RT. Cloning, characterization and expression of the gene coding for a cytosine-5-DNA methyltransferase recognizing GpC. Nucleic acids research. 1998a;26:3961–3966. doi: 10.1093/nar/26.17.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Simpson RT, Kladde MP. Gal4p-mediated chromatin remodeling depends on binding site position in nucleosomes but does not require DNA replication. Mol Cell Biol. 1998b;18:1201–1212. doi: 10.1128/mcb.18.3.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- Zlatanova J, Leuba SH. Chromatin fibers, one-at-a-time. J Mol Biol. 2003;331:1–19. doi: 10.1016/s0022-2836(03)00691-0. [DOI] [PubMed] [Google Scholar]