Abstract
The release of the Entamoeba histolytica genome has facilitated the development of techniques to survey rapidly and to relate gene expression with biology. The association and potential contribution of differential gene expression to the life cycle and the virulence of this protozoan parasite of humans are reviewed here.
Infection and disease
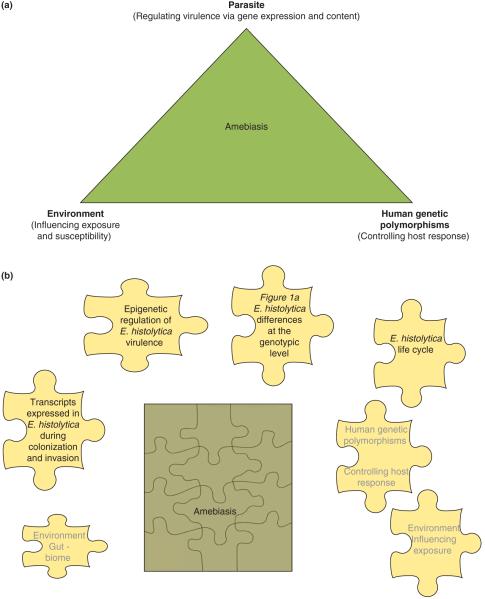
Entamoeba histolytica is the causative agent of amebic dysentery and liver abscess. Surprisingly, however, most infections with E. histolytica are asymptomatic, and only one of every five infections leads to disease [1-3]. The parasite and host factors that control the outcome of this infection (asymptomatic infection versus amebic dysentery and/or liver abscess) are not well understood, although there is emerging evidence of host, parasite and environmental factors influencing the outcome of infection [1-3] (Figure 1a). Alteration in transcription of certain crucial genes is also likely to contribute to the outcome of infection. The latent period between infection and disease in humans suggests adaptation of the parasite to the host via altered gene expression [4]. That this is the case is perhaps best illustrated by the ability to select for increased virulence of an axenic strain of E. histolytica by multiple rounds of passage through animals [5].
Figure 1.
Pathogenesis of amebiasis. The clinical manifestations of amebiasis are the result of interactions of the parasite, host and environment. These include, for example, parasite genotypes that differ in virulence, human leukocyte antigen system Dr/Dq alleles that are associated with resistance to infection and environmental exposure to the parasite. Topics that are covered in this review are shown in black (Figure 1b).
The ability to test for a contribution of altered gene expression to virulence has been provided by the release of the E. histolytica genome and the consequent development of genomic-wide mRNA abundance determinations. The amebic transcriptome has been measured with spotted long oligonucleotide and genomic microarrays, as well as Affymetrix one-color microarray platforms [6-8]. These techniques represent pivotal tools for understanding the biology and pathogenicity of E. histolytica via comparisons of mRNAs expressed in various conditions and trophozoite strains.
Protein family databases, such as Pfam (http://pfam.janelia.org/) and Prosite (http://ca.expasy.org/prosite/), are bioinformatic resources that enable the comparison of the Entamoeba coding sequences against those of well-studied metazoans. Comparisons not only highlight the transcripts unique to the ameba parasite but also promote rapid identification of potential roles from the conserved counterparts characterized in other systems. The tools now available permit the characterization of ‘hypothetical proteins of unknown function’. Although similarity at sequence should not lead to an automatic and untested assumption of functional concordance, the databases currently available improve the odds of correctly speculating about the role of a protein of interest.
To study the basis of the changes in parasite virulence, the transcripts of strains isolated from symptomatic and asymptomatic individuals and the transcripts expressed in the animal models of infection versus culture have been compared. In addition, gene-expression changes during the formation of the infectious cyst stage of the parasite have been studied (Figure 1b).
Comparison of mRNA profiles from Entamoeba strains
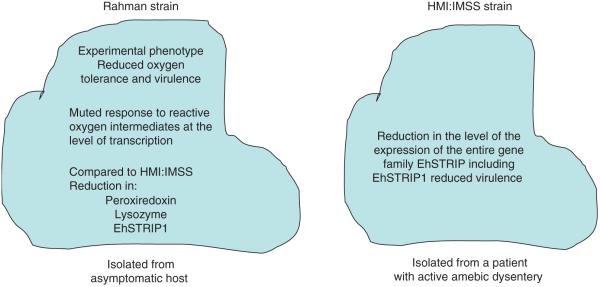
Clinical studies in Bangladesh have showed that certain parasite genotypes are more likely to cause disease [2,9]. The large number of genotypes identified demonstrates a high level of genetic diversity, and some genotypes are associated with increased virulence in humans, as well as a propensity to invade the liver [10]. Studies comparing the transcriptomes of different genotypes are limited at present. The Rahman strain (isolated from an asymptomatic carrier) exhibits a reduced virulence phenotype in multiple in vitro assays of cytotoxicity, as well as a decreased ability to form lesions in the human colonic xenograft model of amebiasis, compared with HM1:IMSS (isolated from a patient with active amebic dysentery) [11]. A potential drawback to this comparison is the early derivation of the HMI:IMSS and Rahman axenized strains (1979 and 1980, respectively) and the different geographical location of the infected hosts (Mexico or England, respectively, with unknown etiological origin); therefore, differences might not only be related to disease outcome but also reflect parasite speciation. Nevertheless, the transcriptome profiles of the HM1:IMSS and Rahman strain [6,7,11,12] revealed some interesting differences between the two isolates, most notably changes in genes involved in oxygen defense and protein degradation (summarized in Figure 2).
Figure 2.
Comparison of mRNA-expression data from two Entamoeba histolytica isolates. The Rahman strain was isolated from an individual who was colonized and HM1:IMSS was cultured from the rectal biopsy of a patient with amebic colitis. Differences in transcripts encoding proteins involved in oxygen detoxification and virulence were observed.
Peroxiredoxin was one of the enzymes involved in oxygen metabolism that was expressed at higher levels in HM1:IMSS [6,13]. When this transcript was artificially increased in the Rahman strain, the transfected trophozoites had both greater resistance to killing by H2O2 and an increased pro-inflammatory phenotype in the human colonic xenograft model of amebiasis [11]. This protein is recruited to the host–parasite interface by the galactose- and N-acetyl-galactosamine-inhibitable (Gal/GalNAc) lectin. Gal/GalNAc lectin mediates amebic adherence to and contact-dependent killing of host cells. Therefore, the recruitment of peroxiredoxin could well protect the trophozoites against the reactive oxygen intermediates (ROS) generated by the host [14].
The later spatial regulation could be important in the normal role of peroxiredoxin in response to oxidative stress in particular strains or conditions. In contrast to the earlier work [15], Vicente et al. [16] report that exposure of either HM1:IMSS or Rahman to H2O2 did not cause a change in peroxiredoxin mRNA levels or upregulate transcripts encoding previously characterized enzymes involved in oxygen detoxification.
This work highlights the need to minimize confounding variables when comparing the results from different laboratories. Preparation of culture media in different laboratories might be important when studying oxidative stress because the cysteine included in the E. histolytica culture media acts as a reducing agent and has an essential role in its oxygen tolerance during in vitro culture.
Vicente et al. [16] hypothesize that in their in vitro growth conditions, the conventional pathways involved in oxygen defense might be expressed at the maximum rate and that the upregulation of a new pathway or pathways occurs during the E. histolytica response to stress. Although there is little overlap of mRNAs modulated by heat stress, as described by Weber et al. [17], this hypothesis is supported by the substantial overlap between the oxygen- and nitric-oxide-stress-responsive transcripts and mRNAs reported by Hackney et al. [18] to be modulated by heat stress.
The annotated transcripts identified by Vicente et al. [16] were the minority of the modulated transcripts but indicate an important role in cell signaling in response to stress (e.g. Rabl1 and Ram M1 were upregulated and RhoGEF and ArfGAP were downregulated) and the repair or metabolic pathways (e.g. deoxyuridine 5′-triphosphate nucleotidohydrolase was upregulated). The majority of the stress-modulated transcripts, however, encoded proteins of unknown function [18].
Comparison of transcripts regulated in response to ROS in Rahman and HM1:IMSS indicated that there was a decrease in both number and amplitude of change occurring in Rahman. Although this correlated with the increased sensitivity of Rahman to H2O2, this difference could also reflect the variation in growth and culture conditions, as discussed above.
Another transcript highly expressed in the HM1:IMSS microarray data, EhSTRIP1 (one of a family of E. histolytica serine-, threonine- and isoleucine-rich proteins), was apparently absent in Rahman [7]. Analyzing the functional role of the EhSTIRP gene family and its potential role in virulence was accomplished by decreasing the expression of the EhSTIRP family, including EhSTRIP1, in HM1:IMSS by using RNA interference. As expected, this led to a decrease in two in vitro assays of ameba virulence, adherence and cytolysis of Chinese hamster ovary cells, supporting the hypothesis that EhSTRIP1 has a role in amebic virulence [19].
Surprisingly, one confounding aspect is the variation in the results of two independent microarray comparisons of Rahman and HM1:IMSS, performed by two different laboratories. Although both groups used a two-color spotted microarray platform, the first group arrayed 2110 genomic amplicons and the second group used 70-base oligonucleotides to generate an array representing 6242 genes, based on the then-current genome assembly [20]. Different techniques of normalization (median adjustment to give a net change between arrays of zero or Loess the local estimation of weighted moving averages) [21] and differences in programs used to identify significantly modulated transcripts (Student’s t test p-value of <0.01 versus the Significance Analysis of Microarrays) [22] might explain some of the apparent differences. A cross-platform comparison using a standardized battery of bioinformatic and statistical analyses would be useful to assess the impact of array design on array sensitivity and specificity [23].
Verification of some of the observed changes was performed by northern blot [7] and reverse transcription quantitative PCR [6]. Therefore, at least some of the differences observed between the two studies could be due to biological differences in the laboratory cultures of Rahman and HM1:IMSS strain. More work is required to resolve these differences, as well as to compare different genotypes of varying virulence from the same geographic location.
Transcripts expressed in E. histolytica during colonization and invasion
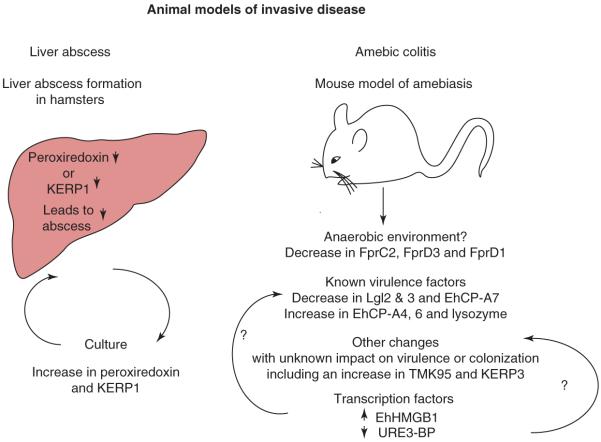
Trophozoites cultured axenically are less virulent, both in the hamster model of amebic liver abscess and in the mouse model of amebic colitis [5,24]. RNA from amebae grown axenically was compared with the transcripts of amebae passed through hamster liver abscesses to retain virulence [25]. Upregulated transcripts included those involved in both oxidative and stress defense and included the peroxiredoxin transcripts discussed previously [11], as well as calcium-binding proteins 1 and 2 [26,27].
An important validation of the microarray approach, and its ability to identify transcripts of interest for further study, was the demonstration that decreased expression of peroxiredoxin (EHI_122310, XM_642754 or X70996, the gEh29 gene for alkyl-hydroperoxidase reductase) by the use of an antisense RNA in HM1:IMSS decreased trophozoite survival during oxidative stress and led to a decrease in liver abscess formation in hamsters [28].
In addition to the increase in peroxiredoxins, the hamster-passed HM1:IMSS amebae had an increase in transcripts encoding a novel family of seven lysine-rich hypothetical proteins [25]. This family included the previously identified surface proteins lysine- and glutamic-acid-rich protein (KERP)1 and KERP2 [29]. Under axenic culture conditions, a decrease in KERP1 transcript expression was observed, whereas a significant increase occurred during liver abscess formation; this indicates that regulation of this transcript occurs in response to the liver abscess environment. A decrease in liver abscess formation in hamsters was observed when KERP1 mRNA translation was inhibited by microRNAs that functioned in a condition-dependent manner and were effective only in culture conditions that include 70% nonheat-inactivated serum and in vivo, which perhaps reflect a stress-dependent phenotype [25].
Comparison of in-vitro-cultured mouse-passaged HM1:IMSS with the trophozoites isolated from the mouse model of amebiasis
To identify changes in parasite gene expression that occur when amebae colonize and invade the host, a transcriptional analysis was conducted in the murine model of amebic colitis [8]. Adaptation to the intestinal environment was accompanied by increases in a subset of cell-signaling genes including transmembrane kinases, Ras and Rho family GTPases, and calcium-binding proteins. Significant decreases in mRNA abundance for genes involved in glycolysis and concomitant increases in lipases were consistent with a change in energy metabolism. Decreases in oxygen-detoxification pathways were observed, as expected, in the anaerobic colonic lumen. Three iron–sulfur flavoproteins (FprC2, FprD3 and FprD1) that are downregulated in the anaerobic luminal trophozoites were, as expected, upregulated in response to H2O2 [16]. However, the response to reactive oxygen included the downregulation of the second transcript encoding the FprC2 protein, coordinately downregulated in vivo. Other transcripts changed in both sets of arrays did not show inverse concordance [16]. This could either reflect input from other stimuli in the complex in vivo environment or simply reflect the high basal expression of the transcripts encoding enzymes involved in oxygen defense in vitro, as discussed earlier.
Of the known virulence factors, the most remarkable changes were a 20–35-fold increase in the cysteine proteinase A subfamily (EhCP-A) member EhCP-A4, a 6–10-fold increase in a second EhCP-A transcript 6 and a 2–3-fold decrease in two members of the Gal/GalNAc lectin light subunit family (Figure 3).
Figure 3.
Comparison of in vivo and in vitro mRNA-expression data for Entamoeba histolytica. Experimental models of amebic liver abscess and colitis have facilitated the identification of transcripts expressed in E. histolytica during colonization and invasion.
Control of the observed changes in mRNA abundance in the intestine might potentially be due to a subset of encoded proteins containing DNA-binding domains that were regulated in the intestinal environment [8]. Characterization of the proteins encoded by one of these transcripts, which was upregulated more than twofold at day 1 of infection, confirmed that it encoded a functional high-mobility group B (HMGB) protein. Recombinant EhHMGB1 was able to bend DNA in vitro, a characteristic of HMGB proteins. Core conserved residues required for DNA-bending activity in other HMGB proteins were demonstrated (by mutational analysis) to be essential for EhHMGB1 activity. EhHMGB1 was also able to enhance the binding of human p53 to its cognate DNA sequence in vitro, as expected for an HMGB1 protein. Overexpression of EhHMGB1 in HM1:IMSS trophozoites led to modulation of 33 transcripts involved in a variety of cellular functions. Of these, 20 were also modulated in the mouse model of intestinal amebiasis at either day 1 or day 29 (when EhHMGB1 transcripts were increased sixfold). However, this change, which was possibly due to a dominant-negative effect from the overexpression of EhHMGB1 (at least 100-fold over the basal level), was in the converse direction of the change in vivo [30].
The well-characterized transcription factor upstream regulatory element 3-binding protein (URE3-BP) was also modulated in the mouse model [31,32]. URE3-BP is a calcium-responsive regulator of two E. histolytica virulence genes, hgl5 and fdx1. Transient overexpression of a dominant-active mutant of URE3-BP resulted in the identification of additional genes regulated by this factor, including several novel amebic membrane proteins [33]. These changes in expression were accompanied by an increase in parasite motility (measured by migration towards serum through a trans-well apparatus), indicating a possible role for URE3-BP in linking the regulation of cellular motility with the response to calcium signaling. Motility and attachment are both considered components in E. histolytica virulence. Motility might affect the capacity of trophozoites to invade human tissues but, conversely, adherence to the host extracellular matrix in the motile gut might be required for colonization. URE3-BP, which potentially represses attachment and increases motility, might, therefore, have an important role during infection.
Both motility and attachment might respond to a complex interplay of signals, as evidenced by the lack of correlation between the identified URE3-BP targets and the transcripts upregulated by the phosphoinositide-3-kinases-dependent motility occurring in response to the pro-inflammatory cytokine tumor necrosis factor-α (TNF-α) [34]. Blazquez et al. [34] used a dedicated array to assay cytoskeletal- and signal-related transcripts. Modest increases between 1.2–1.9-fold were observed in transcripts encoding proteins involved in actin cytoskeleton dynamics when amebae that chemotaxed towards TNF-α were compared to non-responders.
Using microarrays to study the E. histolytica life cycle
E. histolytica has a simple two-stage life cycle and exists either as cysts (the infectious form) or as vegetative, ameboid trophozoites (the form responsible for host invasion) [35].
Recent isolates of E. histolytica, grown under xenic conditions in Robinson’s complex diphasic media for less than eight weeks, contain calcofluor-staining amebae, indicative of cyst chitin. Comparing the transcriptome of the laboratory-cultured strain of E. histolytica-HM1:IMSS to recent clinical isolates has identified a distinct gene-expression profile (Figure 4).
Figure 4.
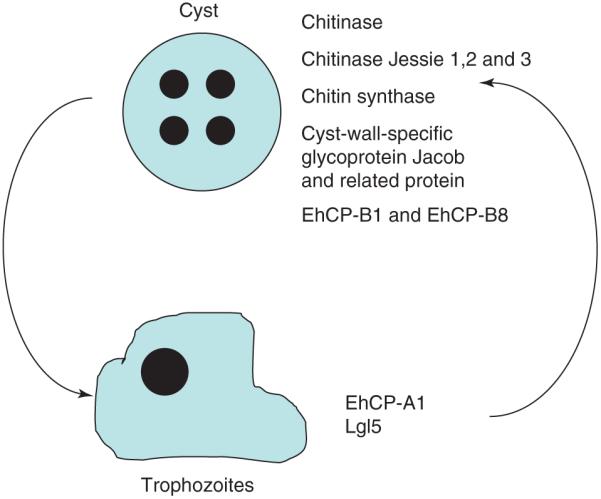
Comparison of Entamoeba histolytica mRNA expression in trophozoites and cysts. Measurements of changes in gene expression as the parasite transforms from a trophozoite to a cyst have identified several cyst-specific genes.
In the E. histolytica analysis, experimental material was only available in small quantities and was, therefore, amplified. Arrays from several different studies were combined in the statistical analysis, and these included samples amplified using differing methods, which result in arrays with different biases in the resulting data [12,36-38]. To find out whether the differences introduced by amplification were greater than experimental differences, the variations in microarrays both within and between experimental conditions were compared using a Pearson’s correlation (scale −1 to +1) that reflects the degree of linear relationship [36,38,39]. The variation between the laboratory strains in this analysis was 0.95–0.88 and the difference between laboratory strains and recent clinical isolates was 0.84–0.77, indicating that the array results did group within experimental conditions [23]. Because the Pearson’s correlation was still close to 1.00, standard microarray analytical tools (which assume that most transcripts remain unchanged) could be used in the analysis of this data [37].
As anticipated, upregulated transcripts included those with known cyst-associated functions (e.g. chitinase and the cyst-wall-specific glycoprotein Jacob). Interestingly, members of the large EhCP-B family of E. histolytica cysteine proteases EhCP-B1 and EhCP-B8 were also upregulated. It has been speculated previously that some members of this large gene family, which are not expressed during trophozoite tissue culture, might play an important part during encystation and excystation of E. histolytica [40]. The reptilian parasite Entamoeba invadens is the model organism in which Entamoeba cyst development has been studied in depth. In work that complements the E. histolytica data, Ebert et al. [41] have observed the upregulation of EiCP-B9 at both RNA and protein levels during E. invadens encystations. The EiCP-A3 and 11 transcripts were downregulated during encystations. In the chitin-producing E. histolytica strains, the EhCP-A1 transcript was downregulated [12,42]. Intriguingly, also downregulated was one of the transcripts encoding the light subunit of the E. histolytica Gal/GalNAc lectin. During E. invadens studies, high levels of Gal-terminated ligands (10 mM) inhibited the ameba aggregation that precedes encystation and prevented formation of mature cysts [43].
Impact of epigenetic changes on virulence
In metazoan cells, transcription is regulated, in part, at the chromatin level by the histone code. This consists of modifications of the histone tail (such as acetylation, methylation, ubiquitylation and phosphorylation) that affect DNA accessibility for transcription. Short-chain fatty acids, which are known modulators of histone acetylation, are anticipated to be present in the intestine and in the media of recent clinical isolates [44], and Ramakrishnan et al. have demonstrated the existence of a Tricostatin-A-inhibitable histone deacetylase [45].
Using the 200:NIH strain, Ehrenkaufer et al. [44] tested whether histone acetylation played a part in the unique gene-expression profile of clinical isolates by treating this strain with both short-chain fatty acids and 150 mM of Tricostatin A. Although Ehrenkaufer et al. observed minimal changes in response to short-chain fatty acids, 122 transcripts were upregulated and 41 downregulated in response to Tricostatin A. More than half of these transcripts were similarly modulated in the recent clinical isolates, indicating that acetylation could play a part in the transcription profile [44]. The upregulated transcripts included heat-shock and cell-signaling genes. Downregulated genes include two cysteine proteases and the light subunit of the galactose-inhibitable lectin. In contrast to the results of Ehrenkaufer et al., Isakov et al. [46] demonstrated that treating HM1:IMSS with Tricostatin A causes an increase in the transcripts of peroxiredoxin and the light subunit of the galactose-inhibitable lectin. Amebae treated with 50 mM Tricostatin A showed an increase in in vitro virulence in cytotoxicity assays and increased survival when undergoing oxidative stress.
These different results might indicate that amebae are extremely sensitive to acetylation levels and reflect the different amounts of drug added (50 mM versus 150 mM). However, the two papers also differ greatly in the reported sensitivity of E. histolytica strains to Tricostatin A – Ehrenkaufer et al. found only a modest effect on growth when trophozoites were treated with 150nM Tricostatin A, whereas Isakov et al. report less than 50% survival after treatment with 100 nM Tricostatin A. This strongly indicates important differences in the E. histolytica strains, which might provide a valuable insight into the function of histone acetylation in E. histolytica. Ehrenkaufer et al. have already described strain-specific differences in the expression of transcripts encoding genes involved in histone acetylation in HM1:IMSS and 200:NIH trophozoites [44]. In summary, although these results are obviously in conflict, this indicates important strain-to-strain differences in histone acetylation and strongly indicates the need for additional studies (Figure 5).
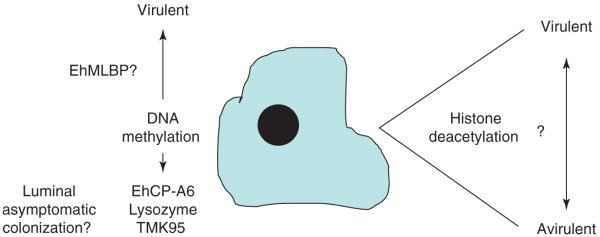
Figure 5.
Impact of epigenetic regulation of gene expression on virulence. Still unknown is the role of the histone code, DNA methylation and other epigenetic mechanisms in control of virulence.
EhMLBP, an Entamoeba protein identified on the basis of its capacity to bind to methylated repetitive DNA, is apparently important for ameba growth and cytotoxicity [47]. Ali et al. [48] have shown by treatment with an inhibitor of demethylases (5-azacytidine) that 68 genes were upregulated and 131 genes downregulated in response to 5-azacytidine. The downregulated transcripts encoded two potential virulence factors, the cysteine proteinase EhCP-A6 and a lysozyme enzyme, both of which were upregulated in the mouse model of amebiasis. Other downregulated transcripts encoded proteins with potential roles in signaling (e.g. transmembrane kinase 95, or TMK95, and a Rho family GTPase). TMK95 is one of a large family of transmembrane kinases and, like EhCP6, was upregulated in the mouse model of amebiasis. The transmembrane kinases (TMKs) have been grouped into nine distinct families based on motifs present on both extracellular and kinase domains [49]. The extracellular domains had considerable similarity to the intermediate subunit (Igl) of the parasite Gal/GalNAc lectin. TMK95 has also been named subgroup B1.II protein 4 [50]. The B1 group of TMKs have been analyzed in more depth by Mehra et al. [50]. The expression of different receptor kinases in the plasma membrane might alter the ability of the parasite to respond to the host environment.
Concluding remarks
This is only the beginning of the journey to understand the biology of E. histolytica. Community annotation of the genome is required in a process of collaborative knowledge discovery and input, as approximately half of the genome with potential parasite-specific information is not annotated. The development of a comprehensive ‘bottom-up’ Gene Wiki approach, such as the community curation tool at the Pathema_Entamoeba website (http://pathema.jcvi.org/cgi-bin/entamoeba/pathemahomepage.cgi), will enable us to move from a small number of contributors to a more dynamic curation model. In addition, a common Entamoeba nomenclature would enable cross-comparison of array results. The new Gene Expression Omnibus database at the National Center for Biotechnology is the beginning of a standardized databank that can enable access to the microarray data by all the community, to take advantage of the data generated by high-throughput techniques [51].
Although much remains to be learned from experiments in cultured amebae, the HMI:IMSS strain has been in culture for a long time and it is becoming increasingly obvious that intra-laboratory comparisons should use a common reference stock. Growth of E. histolytica trophozoites in undefined media might generate considerable differences in gene-expression profiles between laboratories. A cultured ameba might provide a transcriptome profile that has an unclear relationship with the trophozoite growth conditions in vivo.
In future, genotypic isolates from the same geographical location that have different clinical presentations should be compared to identify potential virulence determinants. The transcriptome profile of invading ameba trophozoites should be compared to that of luminal amebae to determine the transcripts regulated in vivo. The role of regulators of transcription needs to be explored in relation to the modulation of the virulence phenotype and the therapeutic potential of proteins encoded by transcripts modulated during encystation and excystation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Duggal P, et al. Influence of human leukocyte antigen class II alleles on susceptibility to Entamoeba histolytica infection in Bangladeshi children. J. Infect. Dis. 2004;189:520–526. doi: 10.1086/381272. [DOI] [PubMed] [Google Scholar]
- 2.Ali IK, et al. Evidence for a link between parasite genotype and outcome of infection with Entamoeba histolytica. J. Clin. Microbiol. 2007;45:285–289. doi: 10.1128/JCM.01335-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mondal D, et al. Entamoeba histolytica-associated diarrheal illness is negatively associated with the growth of preschool children: evidence from a prospective study. Trans. R. Soc. Trop. Med. Hyg. 2006;100:1032–1038. doi: 10.1016/j.trstmh.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 4.Haque R, et al. Amebiasis. N. Engl. J. Med. 2003;348:1565–1573. doi: 10.1056/NEJMra022710. [DOI] [PubMed] [Google Scholar]
- 5.Houpt ER, et al. The mouse model of amebic colitis reveals mouse strain susceptibility to infection and exacerbation of disease by CD4+ T cells. J. Immunol. 2002;169:4496–4503. doi: 10.4049/jimmunol.169.8.4496. [DOI] [PubMed] [Google Scholar]
- 6.Davis PH, et al. Transcriptomic comparison of two Entamoeba histolytica strains with defined virulence phenotypes identifies new virulence factor candidates and key differences in the expression patterns of cysteine proteases, lectin light chains, and calmodulin. Mol. Biochem. Parasitol. 2007;151:118–128. doi: 10.1016/j.molbiopara.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 7.MacFarlane RC, Singh U. Identification of differentially expressed genes in virulent and nonvirulent Entamoeba species: potential implications for amebic pathogenesis. Infect. Immun. 2006;74:340–351. doi: 10.1128/IAI.74.1.340-351.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilchrist CA, et al. Impact of intestinal colonization and invasion on the Entamoeba histolytica transcriptome. Mol. Biochem. Parasitol. 2006;147:163–176. doi: 10.1016/j.molbiopara.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 9.Ayeh-Kumi PF, et al. Entamoeba histolytica: genetic diversity of clinical isolates from Bangladesh as demonstrated by polymorphisms in the serine-rich gene. Exp. Parasitol. 2001;99:80–88. doi: 10.1006/expr.2001.4652. [DOI] [PubMed] [Google Scholar]
- 10.Ali IK, et al. Tissue invasion by Entamoeba histolytica: evidence of genetic selection and/or DNA reorganization events in organ tropism. PLoS Negl. Trop. Dis. 2008;2:e219. doi: 10.1371/journal.pntd.0000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis PH, et al. Comparative proteomic analysis of two Entamoeba histolytica strains with different virulence phenotypes identifies peroxiredoxin as an important component of amoebic virulence. Mol. Microbiol. 2006;61:1523–1532. doi: 10.1111/j.1365-2958.2006.05344.x. [DOI] [PubMed] [Google Scholar]
- 12.Ehrenkaufer GM, et al. Identification of developmentally regulated genes in Entamoeba histolytica: insights into mechanisms of stage conversion in a protozoan parasite. Cell. Microbiol. 2007;9:1426–1444. doi: 10.1111/j.1462-5822.2006.00882.x. [DOI] [PubMed] [Google Scholar]
- 13.Bruchhaus I, et al. Removal of hydrogen peroxide by the 29 kDa protein of Entamoeba histolytica. Biochem. J. 1997;326:785–789. doi: 10.1042/bj3260785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hughes MA, et al. Identification of Entamoeba histolytica thiol-specific antioxidant as a GalNAc lectin-associated protein. Mol. Biochem. Parasitol. 2003;127:113–120. doi: 10.1016/s0166-6851(02)00326-2. [DOI] [PubMed] [Google Scholar]
- 15.Akbar MA, et al. Genes induced by a high-oxygen environment in Entamoeba histolytica. Mol. Biochem. Parasitol. 2004;133:187–196. doi: 10.1016/j.molbiopara.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 16.Vicente JB, et al. Entamoeba histolytica modulates a complex repertoire of novel genes in response to oxidative and nitrosative stress: implications for amebic pathogenesis. Cell Microbiol. 2009;11:51–69. doi: 10.1111/j.1462-5822.2008.01236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weber C, et al. Stress by heat shock induces massive down regulation of genes and allows differential allelic expression of the Gal/GalNAc lectin in Entamoeba histolytica. Eukaryot. Cell. 2006;5:871–875. doi: 10.1128/EC.5.5.871-875.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hackney JA, et al. Identification of putative transcriptional regulatory networks in Entamoeba histolytica using Bayesian inference. Nucleic Acids Res. 2007;35:2141–2152. doi: 10.1093/nar/gkm028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macfarlane RC, Singh U. Identification of an Entamoeba histolytica serine, threonine, isoleucine, rich protein with roles in adhesion and cytotoxicity. Eukaryot. Cell. 2007;6:2139–2146. doi: 10.1128/EC.00174-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loftus B, et al. The genome of the protist parasite Entamoeba histolytica. Nature. 2005;433:865–868. doi: 10.1038/nature03291. [DOI] [PubMed] [Google Scholar]
- 21.Yang YH, et al. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tusher VG, et al. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U. S. A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Severgnini M, et al. Strategies for comparing gene expression profiles from different microarray platforms: application to a case-control experiment. Anal. Biochem. 2006;353:43–56. doi: 10.1016/j.ab.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 24.Bruchhaus I, et al. Differential gene expression in Entamoeba histolytica isolated from amoebic liver abscess. Mol. Microbiol. 2002;44:1063–1072. doi: 10.1046/j.1365-2958.2002.02941.x. [DOI] [PubMed] [Google Scholar]
- 25.Santi-Rocca J, et al. The lysine- and glutamic acid-rich protein KERP1 plays a role in Entamoeba histolytica liver abscess pathogenesis. Cell. Microbiol. 2008;10:202–217. doi: 10.1111/j.1462-5822.2007.01030.x. [DOI] [PubMed] [Google Scholar]
- 26.Sahoo N, et al. Calcium binding protein 1 of the protozoan parasite Entamoeba histolytica interacts with actin and is involved in cytoskeleton dynamics. J. Cell Sci. 2004;117:3625–3634. doi: 10.1242/jcs.01198. [DOI] [PubMed] [Google Scholar]
- 27.Chakrabarty P, et al. Identification and characterization of EhCaBP2. A second member of the calcium-binding protein family of the protozoan parasite Entamoeba histolytica. J. Biol. Chem. 2004;279:12898–12908. doi: 10.1074/jbc.M304716200. [DOI] [PubMed] [Google Scholar]
- 28.Sen A, et al. The 29-kilodalton thiol-dependent peroxidase of Entamoeba histolytica is a factor involved in pathogenesis and survival of the parasite during oxidative stress. Eukaryot. Cell. 2007;6:664–673. doi: 10.1128/EC.00308-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seigneur M, et al. A lysine- and glutamic acid-rich protein, KERP1, from Entamoeba histolytica binds to human enterocytes. Cell. Microbiol. 2005;7:569–579. doi: 10.1111/j.1462-5822.2005.00487.x. [DOI] [PubMed] [Google Scholar]
- 30.Abhyankar MM, et al. Characterization of an Entamoeba histolytica high-mobility-group box protein induced during intestinal infection. Eukaryot. Cell. 2008;7:1565–1572. doi: 10.1128/EC.00123-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gilchrist CA, et al. Identification and characterization of an Entamoeba histolytica upstream regulatory element 3 sequence-specific DNA-binding protein containing EF-hand motifs. J. Biol. Chem. 2001;276:11838–11843. doi: 10.1074/jbc.M007375200. [DOI] [PubMed] [Google Scholar]
- 32.Gilchrist CA, et al. Calcium modulates promoter occupancy by the Entamoeba histolytica Ca2+-binding transcription factor URE3-BP. J. Biol. Chem. 2003;278:4646–4653. doi: 10.1074/jbc.M211271200. [DOI] [PubMed] [Google Scholar]
- 33.Gilchrist CA, et al. Targets of the Entamoeba histolytica transcription factor URE3-BP. PLoS Negl. Trop. Dis. 2008;2:e282. doi: 10.1371/journal.pntd.0000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blazquez S, et al. Chemotaxis of Entamoeba histolytica towards the pro-inflammatory cytokine TNF is based on PI3K signalling, cytoskeleton reorganization and the Galactose/N-acetylgalactosamine lectin activity. Cell Microbiol. 2008;8:1676–1686. doi: 10.1111/j.1462-5822.2008.01158.x. [DOI] [PubMed] [Google Scholar]
- 35.Stanley SL., Jr Amoebiasis. Lancet. 2003;361:1025–1034. doi: 10.1016/S0140-6736(03)12830-9. [DOI] [PubMed] [Google Scholar]
- 36.Goff LA, et al. Evaluation of sense-strand mRNA amplification by comparative quantitative PCR. BMC Genomics. 2004;5:76. doi: 10.1186/1471-2164-5-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hunter SM, et al. Optimization of minuscule samples for use with cDNA microarrays. J. Biochem. Biophys. Methods. 2008;70:1048–1058. doi: 10.1016/j.jprot.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 38.Puskas LG, et al. RNA amplification results in reproducible microarray data with slight ratio bias. Biotechniques. 2002;32:1330–1334. 1336, 1338, 1340. doi: 10.2144/02326mt04. [DOI] [PubMed] [Google Scholar]
- 39.Eisen MB, et al. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U. S. A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bruchhaus I, et al. The intestinal protozoan parasite Entamoeba histolytica contains 20 cysteine protease genes, of which only a small subset is expressed during in vitro cultivation. Eukaryot. Cell. 2003;2:501–509. doi: 10.1128/EC.2.3.501-509.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ebert F, et al. An Entamoeba cysteine peptidase specifically expressed during encystation. Parasitol. Int. 2008;57:521–524. doi: 10.1016/j.parint.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 42.Singh U, Ehrenkaufer GM. Recent insights into Entamoeba development: identification of transcriptional networks associated with stage conversion. Int. J. Parasitol. 2008;39:41–47. doi: 10.1016/j.ijpara.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eichinger D. A role for a galactose lectin and its ligands during encystment of Entamoeba. J. Eukaryot. Microbiol. 2001;48:17–21. doi: 10.1111/j.1550-7408.2001.tb00411.x. [DOI] [PubMed] [Google Scholar]
- 44.Ehrenkaufer GM, et al. Trichostatin A effects on gene expression in the protozoan parasite Entamoeba histolytica. BMC Genomics. 2007;8:216. doi: 10.1186/1471-2164-8-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramakrishnan G, et al. Histone acetyltransferases and deacetylase in Entamoeba histolytica. Mol. Biochem. Parasitol. 2004;138:205–216. doi: 10.1016/j.molbiopara.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 46.Isakov E, et al. Trichostatin A regulates peroxiredoxin expression and virulence of the parasite Entamoeba histolytica. Mol. Biochem. Parasitol. 2008;158:82–94. doi: 10.1016/j.molbiopara.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 47.Lavi T, et al. EhMLBP is an essential constituent of the Entamoeba histolytica epigenetic machinery and a potential drug target. Mol. Microbiol. 2008;69:55–66. doi: 10.1111/j.1365-2958.2008.06258.x. [DOI] [PubMed] [Google Scholar]
- 48.Ali IK, et al. Growth of the protozoan parasite Entamoeba histolytica in 5-azacytidine has limited effects on parasite gene expression. BMC Genomics. 2007;8:7. doi: 10.1186/1471-2164-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beck DL, et al. Identification and gene expression analysis of a large family of transmembrane kinases related to the Gal/GalNAc lectin in Entamoeba histolytica. Eukaryot. Cell. 2005;4:722–732. doi: 10.1128/EC.4.4.722-732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mehra A, et al. Expression and function of a family of transmembrane kinases from the protozoan parasite Entamoeba histolytica. Infect. Immun. 2006;74:5341–5351. doi: 10.1128/IAI.00025-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Edgar R, et al. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]