Abstract
Dehalococcoides strains respire a wide variety of chloro-organic compounds and are important for the bioremediation of toxic, persistent, carcinogenic, and ubiquitous ground water pollutants. In order to better understand metabolism and optimize their application, we have developed a pan-genome-scale metabolic network and constraint-based metabolic model of Dehalococcoides. The pan-genome was constructed from publicly available complete genome sequences of Dehalococcoides sp. strain CBDB1, strain 195, strain BAV1, and strain VS. We found that Dehalococcoides pan-genome consisted of 1118 core genes (shared by all), 457 dispensable genes (shared by some), and 486 unique genes (found in only one genome). The model included 549 metabolic genes that encoded 356 proteins catalyzing 497 gene-associated model reactions. Of these 497 reactions, 477 were associated with core metabolic genes, 18 with dispensable genes, and 2 with unique genes. This study, in addition to analyzing the metabolism of an environmentally important phylogenetic group on a pan-genome scale, provides valuable insights into Dehalococcoides metabolic limitations, low growth yields, and energy conservation. The model also provides a framework to anchor and compare disparate experimental data, as well as to give insights on the physiological impact of “incomplete” pathways, such as the TCA-cycle, CO2 fixation, and cobalamin biosynthesis pathways. The model, referred to as iAI549, highlights the specialized and highly conserved nature of Dehalococcoides metabolism, and suggests that evolution of Dehalococcoides species is driven by the electron acceptor availability.
Author Summary
Dehalococcoides are strictly anaerobic bacteria capable of detoxifying widespread and harmful ground water pollutants — chlorinated ethenes, chlorinated benzenes, polychlorinated biphenyls and dioxins — largely originated from industry and agriculture sectors. However, how this unique niche has been acquired by these microbes is not well understood, specifically at the level of metabolism. In addition, these bacteria harness energy from the pollutant detoxification process — reductive dechlorination — for their growth; but they grow very slowly. Their slow growth rate, as well as their low growth yield, can hamper bioremediation processes. Thus, in order to obtain an improved understanding of Dehalococcoides' metabolism and the factors limiting their growth, we developed a constraint-based metabolic model of Dehalococcoides from publicly accessible genome sequences of 4 strains. This model, in addition to creating a valuable knowledgebase on Dehalococcoides, offers researchers in the bioremediation community a valuable tool for generating experimentally testable hypothesis for improving the efficacy of bioremediation by Dehalococcoides.
Introduction
Genome sequencing has enabled the characterization of biological systems in a more comprehensive manner. Recent research in bioinformatics and systems biology has resulted in the development of numerous systematic approaches for the analysis of cellular physiology that have been reviewed elsewhere [1]–[4]. However, constraint-based reconstruction and analysis (COBRA), a mathematical framework for integrating sequence data with a plethora of experimental ‘omics’ data has been shown to be successful in the genome-wide analysis of cellular physiology [5]–. In addition, this approach has also been utilized to explore the metabolic potential, as well as the gene essentiality analysis of several organisms across different kingdoms of life [8]–[13]; however, the COBRA approach has not yet been implemented for Dehalococcoides, or any other known dechlorinating bacterium.
Using acetate as a carbon source and hydrogen as an electron donor, small, disc-shaped anaerobic bacteria Dehalococcoides are capable of dehalogenating a variety of halogenated organic compounds as electron acceptors, of which many are problematic ground water pollutants [14]–[17]. Dehalococcoides ethenogenes strain 195 (strain 195) is the first member of this phylogenetic branch that was grown as an isolate [18]. Subsequently, a number of Dehalococcoides strains were isolated: strain CBDB1 [19], strain BAV1 [20], strain FL2 [21], [22], strain GT [23], and strain VS [24]. The strains respire through a membrane-bound electron transport chain (ETC) [25]–[27], which is incompletely defined. Reductive dehalogenases (RDases), encoded by reductive dehalogenase homologous (rdh) genes, are pivotal membrane-associated enzymes of the ETC [26], [27]. Genome sequencing has revealed the presence of multiple non-identical putative rdh genes in each strain [28]–[31]. Since these microbes respire chlorinated pollutants by RDase-catalyzed reductive dechlorination reaction, rdh genes determine a significant part of Dehalococcoides' phenotypes. Functional characterization of only 5 of the over 190 rdh genes reveals that cobalamin — a corrinoid compound — is an essential cofactor for the corresponding RDases [32]–[36]. Hydrogenase (H2ase) is another key enzyme of Dehalococcoides ETC [26], [27], [29], [30]. Interestingly, the genomes of Dehalococcoides strains encode 5 different types of H2ases: membrane-bound hup, ech, hyc, hym, and cytoplasmic vhu [29], [30], [37], [38]. The presence of multiple types of H2ases clearly emphasizes the importance of H2 in their energy metabolism [18]–[21]. This multiplicity of H2ases and RDases further highlights redundancy in the organisms' energy conservation process that may ensure a rapid and efficient response of their energy metabolism towards changing growth conditions [39], [40].
In addition to RDase and H2ase, the ETC likely requires an in vivo electron carrier to mediate electron transport between these enzymes. Previous studies have shown that the reductive dechlorination reaction requires an in vivo electron donor of redox potential (E0 ′) ≤−360 mV [25], [27], similar to other dechlorinating bacteria [17], [41], [42]. The cob(II)alamin of corrinoid cofactor in the RDase enzyme is reduced to cob(I)alamin during the reductive dechlorination reaction; hence, necessitating a low-potential donor because the redox potential (E0′) of Co(II)/Co(I) couple is between −500 and −600 mV [17], [41], [43]. While quinones, such as menaquinone or ubiquinone could act as electron carriers in anaerobes [44]–[46], experimental evidence suggests this is not the case in Dehalococcoides [27], [47]. Moreover, the redox potentials for quinones (Menaquinone ox/red E0 ′ = −70 mV, Ubiquinone ox/red E0 ′ = +113 mV; [48]) are not compatible with the RDases' requirement of a low potential donor. Furthermore, cytochrome b — a typical donor for the quinones to participate in the redox reactions of anaerobic ETCs [49], [50] — appears to be absent in the genomes of Dehalococcoides [29], [30]. However, the genomes have ferredoxin, an iron-sulphur protein, which can act as the low-potential donor for RDases because ferredoxin is the most electronegative electron carrier yet found in the bacterial ETCs [42], [48], [51]–[55].
Although, Dehalococcoides are capable of harnessing free energy from the RDase catalyzed exergonic reductive dechlorination reactions by coupling to ATP generation for growth [14], [17], their pure culture growth is much less robust than their growth in mixed cultures [24], [56], [57]; even in mixed cultures, their growth yield is not as high as that predicted from the free energy of reductive dechlorination [26], [58]. Thus, in order to better understand dechlorination-metabolism, and given that to-date sequenced Dehalococcoides genomes are more than 85% identical at the amino acid level [38], [59], we developed a pan-genome-scale constraint-based in silico metabolic model of Dehalococcoides. The model was constructed from the complete genome sequences of 4 geographically distinct strains: strain CBDB1 from the Saale river near Jena, Germany [60], [61], strain BAV1 from Oscoda, Michigan, USA [62], [63], strain 195 from a wastewater treatment plant in Ithaca, New York, USA [18], [64], [65], and strain VS from Victoria, Texas, USA [24], [66]. Although the model comprises multiple genomes, it analyzed the outcome of metabolic genes only. Also, it did not include information about cellular regulation due to the lack of adequate knowledge about Dehalococcoides regulatory networks. Nonetheless, the model was primarily used to investigate the intrinsic metabolic limitations, in addition to addressing open questions regarding Dehalococcoides physiology, such as the incomplete nature of various metabolic pathways, and attendant implications on metabolism and growth. We also identified the environmental conditions from the model simulations that resulted in faster in silico growth of Dehalococcoides. Furthermore, the constraint-based model, along with the comparative analysis of 4 genomes, clarifies both similarities and differences among the strains in terms of their core metabolism and other biosynthetic processes leading to an improved understanding of metabolism and evolution in Dehalococcoides.
Results/Discussion
Dehalococcoides Metabolic Network
Pan-metabolic-genes of Dehalococcoides
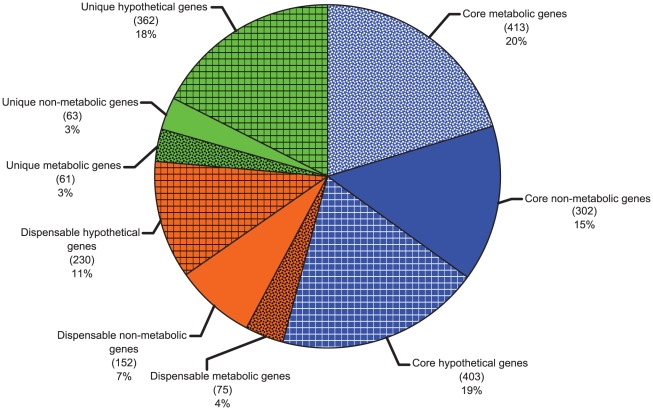
The concept of a pan-genome was first investigated by Tettelin and colleagues for the 8 isolates of common human pathogen Streptococcus agalactiae [67]. While pan-genome analyses for other organisms have been reported [68], no such analysis has been performed to-date for any dechlorinating bacterium, or any other microbe of bioremediation importance. In addition, most of the reported pan-genome analyses were conducted on pathogenic isolates for designing vaccines by assessing their virulence evolution and diversity. Here, we developed the Dehalococcoides pan-genome from the complete sequenced genomes of four Dehalococcoides strains. Method details are provided in the Materials and Methods and in Figures 1–4 in Text S2. The pan-genome comprises 2061 genes (Figure 1). Of these 2061 genes, 1118 genes are in the core, 457 are dispensable, and 486 are unique (Figure 1). The genes are further classified as metabolic, non-metabolic, and hypothetical based on information obtained from the literature and various biochemical databases, such as SWISSPROT [69], UniProt [70], IMG [71], and PDB [72]. We defined metabolic genes as those that are exclusively related to metabolic processes such as carbon and energy metabolism of Dehalococcoides. Genes that are involved in DNA repair and metabolism, as well as encoding putative transposable elements and insertion elements [29], [30] are classified as non-metabolic. Putative genes with a non-specific metabolic function or genes without any function or annotation are categorized as hypothetical (Figure 1).
Figure 1. Composition of the Dehalococcoides pan-genome.
Core, dispensable, and unique genomes are represented by blue, green, and orange, respectively. Genes in these genomes are also categorized as metabolic (spotted pattern), non-metabolic (plain), and hypothetical (grid pattern) on the basis of various bioinformatic analyses (see text for details).
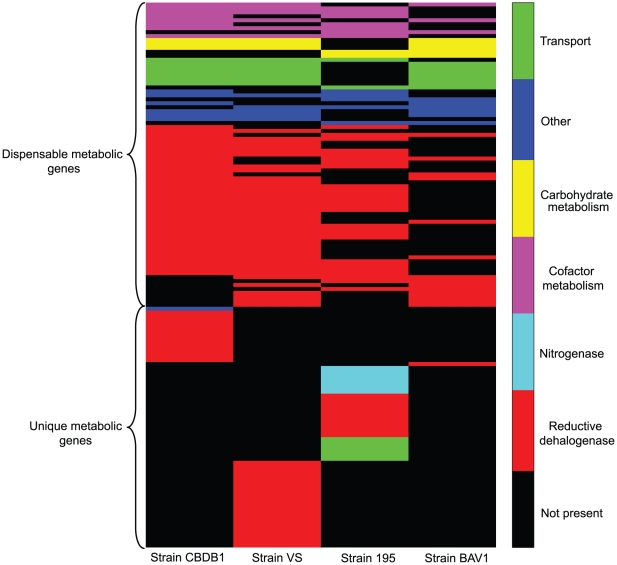
Most of the metabolic genes (413 out of 549) were found in the core genome while only a small number of those were identified in the dispensable (75) and unique (61) genomes. This abundance of core metabolic genes in the pan-genome indicate that the central metabolism of Dehalococcoides is very well conserved across strains since core genes are shared by all [2], [67], [73]–[75]. We further categorized the metabolic genes in the dispensable and unique genomes based on both function and strain (Figure 2). Clearly, the majority of differences among the strains (45 out of the 75 dispensable genes and 47 out of the 61 unique genes) are due to the rdh genes (Figure 2). In addition, only strain195 has nitrogen fixing genes and associated transporters related to the nitrogen fixation process. As a result of these genes, together with unique rdhs, strain 195 has the most unique genes of the 4 genomes compared. Due to the presence of a suite of multiple non-identical rdh genes, each strain metabolizes a unique set of specific chlorinated substrates [38], [76], [77]. Hence, the differences in rdh genes largely define the strain specific phenotypes of Dehalococcoides.
Figure 2. Distribution of dispensable and unique metabolic genes in different Dehalococcoides strains.
Colors are assigned to further categorize the genes according to their function identified from annotation and verified by different bioinformatic analyses. Each color except black signifies the presence of a corresponding metabolic gene while black indicates the absence of the corresponding gene. Genes belonging to amino acid metabolism, lipid metabolism and nucleotide metabolism are small in number; hence, included in ‘other’ category. This heat map essentially describes the differences among Dehalococcoides strains from the context of metabolic genes.
Though there were differences in rdh genes, most of these were found in the dispensable genome (Figure 2 and Table 14 in Text S1) while only 9 rdhs (5 rdhA and 4 rdhB genes with >35% amino acid sequence identity) were shared by all strains and found in the core genome (Table 13 in Text S1). The presence of majority of rdh genes in the dispensable genome further supports the hypothesis that they were acquired through lateral gene transfer events [29], [59], [78].
Features of the reconstructed metabolic network of Dehalococcoides
The reconstructed metabolic network of Dehalococcoides, denoted as iAI549 according to the established naming convention [79], accounted for 549 open reading frames (ORF) or protein coding genes (27% of the total 2061 genes). Metabolic genes were identified from the genome annotations which were verified with various bioinformatic analyses (see Materials and Methods). In addition, we annotated or revised the annotation for 70 ORFs based on information obtained from different biochemical databases (Table 2 in Text S1 provides a full list of reannotated genes). General features of Dehalococcoides metabolic network (iAI549) are provided in Table 1.
Table 1. General features of Dehalococcoides metabolic network (iAI549).
| Genes | |
| Total number of genes | 2,061 |
| Number of included genes | 549 |
| Number of excluded genes | 1,512 |
| Proteins | |
| Total number of proteins | 356 |
| Intra-system Reactions | |
| Total number of model reactions | 518 |
| Gene associated model reactions | 497 |
| Non-gene associated model reactions | 21 |
| Exchange Reactions | |
| Total number of exchange reactions | 36 |
| Input-output reactions | 35 |
| Demand reactions | 1 |
| Metabolites | |
| Total number of metabolites | 549 |
| Number of extracellular metabolites | 31 |
| Number of intracellular biomass metabolites | 110 |
iAI549 includes 518 model reactions and 549 metabolites where 497 reactions are gene associated and 21 (4%) are non-gene associated (Table 1). The non-gene associated reactions (Table 1 in Text S1) were added in order to fill gaps in the reconstructed network based on simulations. Although no gene associations were identified for these reactions, we provided a list of core hypothetical genes (Table 16 in Text S1) which potentially could contain genes associated with these reactions and are prime candidates for further biochemical testing. The network also comprises 36 exchange reactions, including one demand reaction called the biomass synthesis reaction (BIO_DHC_DM_61), to facilitate the transport of various metabolites into and out of the cell. The composition of the in silico minimal medium is shown in Table 2, while detailed composition of BIO_DHC_DM_61 is available in Text S2. We further categorized the genes and reactions of iAI549 into 7 different functional categories or subsystems based on the associated metabolic pathways (Figures 7 and 8 in Text S2). The differences among the strains are mainly observed in the energy metabolism category, which includes 51 dispensable and 54 unique metabolic genes, and most of these are rdhs (Figure 7 in Text S2). However, almost all the reactions of iAI549 (96% of the total 518) are core, which again indicates that the basic central metabolism of Dehalococcoides is strictly conserved (Figure 8 in Text S2). Although a number of dispensable metabolic genes are found in different subsystems, most of these genes are actually paralogs of the core metabolic genes. This relationship explains why, for example, there are 13 dispensable genes in the transport subsystem, 3 genes each in the lipid and nucleotide metabolism, but no corresponding dispensable reactions (Figure 8 in Text S2). Since rdhs were found in core, unique and dispensable genomes, we assigned the reductive dechlorination reaction as a core reaction. Therefore, the truly unique metabolic reactions of iAI549 are the nitrogen fixing reaction (EC-1.18.6.1) and the molybdate (required for synthesizing cofactor for the nitrogenase) transport reaction (TC-3.A.1.8) belonging to strain 195 only.
Table 2. Composition of the in silico minimal medium of Dehalococcoides.
| Abbreviation | Exchange reaction | Equation |
| Acetate exchange | EX_ac(e) | ac < = = > |
| Vitamin B12 or cobalamin exchange | EX_cbl1(e) | cbl1 < = = > |
| Chloride exchange | EX_cl(e) | cl < = = > |
| Carbon dioxide exchange | EX_co2(e) | co2 < = = > |
| Proton exchange | EX_h(e) | h < = = > |
| Hydrogen exchange | EX_h2(e) | h2 < = = > |
| Water exchange | EX_h2o(e) | h2o < = = > |
| Dichlorobenzene exchange | EX_dcb(e) | dcb < = = > |
| Ethene exchange | EX_etl(e) | etl < = = > |
| Tetrachloroethene exchange | EX_pce(e) | pce < = = > |
| Hexachlorobenzene exchange | EX_hcb(e) | hcb < = = > |
| Ammonium exchange | EX_nh4(e) | nh4 < = = > |
| Inorganic phosphate exchange | EX_pi(e) | pi < = = > |
| Sulphate exchange | EX_so4(e) | so4 < = = > |
Furthermore, we compared iAI549 to a number of in silico genome-scale models of other Bacteria and Archaea (Table 3): iAF1260 for Escherichia coli [80], iYO844 for Bacillus subtilis [81], iRM588 for Geobacter sulfurreducens [82], and iAF692 for Methanosarcina barkeri [83]. We found that iAI549 had the lowest number of total reactions because of the limited scope of Dehalococcoides' metabolism. In addition, these numbers also suggest that facultative anaerobes (E. coli and B. subtilis) are more versatile in their lifestyle and metabolism compared to obligate anaerobes (Dehalococcoides, Geobacter and Methanosarcina). These differences are further supported by the presence of a high number of transporters in iAF1260 and iYO844 compared to the presence of only 32 transporters in iAI549 (Table 3). A large number of reactions of iAI549 are found to be involved in the amino acid metabolism since the genes for de novo synthesis of all the amino acids except methionine are identified to be present in the genomes [29], [30]. Also, iAI549 comprises only 41 reactions for the central carbon metabolism — glycolysis, gluconeogenesis, TCA-cycle, pentose phosphate pathway, carbohydrate metabolism — compared to 262 reactions in iAF1260; an incomplete TCA-cycle and an inactive glycolysis pathway explain this low number for iAI549. Since Dehalococcoides lack a typical bacterial cell wall [18]–[20], iAI549 has only 81 reactions for the lipid metabolism category. Furthermore, the cofactor and prosthetic group biosynthesis comprises 101 reactions of iAI549 compared to 162 reactions of iAF1260 because the pathways for synthesizing vitamin B12 and quinones are predicted to be incomplete in Dehalococcoides [29], [30].
Table 3. Comparison of various in silico genome-scale models with iAI549.
| In silico models (Organisms) | iAI549 (Dehalococcoides) | iRM588 (G. sulfurreducens) | iAF692 (M. barkeri) | iAF1260 (E. coli) | iYO844 (B. subtilis) |
| Total reactions | 518 | 522 | 619 | 2077 | 1020 |
| Amino acid metabolism | 139 | 119 | 150 | 198 | 207 |
| Cofactor and prosthetic group biosynthesis | 102 | 100 | 153 | 162 | 83 |
| Nucleotide metabolism | 83 | 58 | 75 | 155 | 123 |
| Lipid metabolism | 81 | 93 | 46 | 522 | 126 |
| Central carbon metabolism | 41 | 64 | 72 | 252 | 196 |
| Energy metabolism | 40 | 37 | 41 | 90 | 41 |
| Transport | 32 | 51 | 82 | 698 | 244 |
Model-Based Simulations of Dehalococcoides Physiology
Exploring the central metabolism of Dehalococcoides
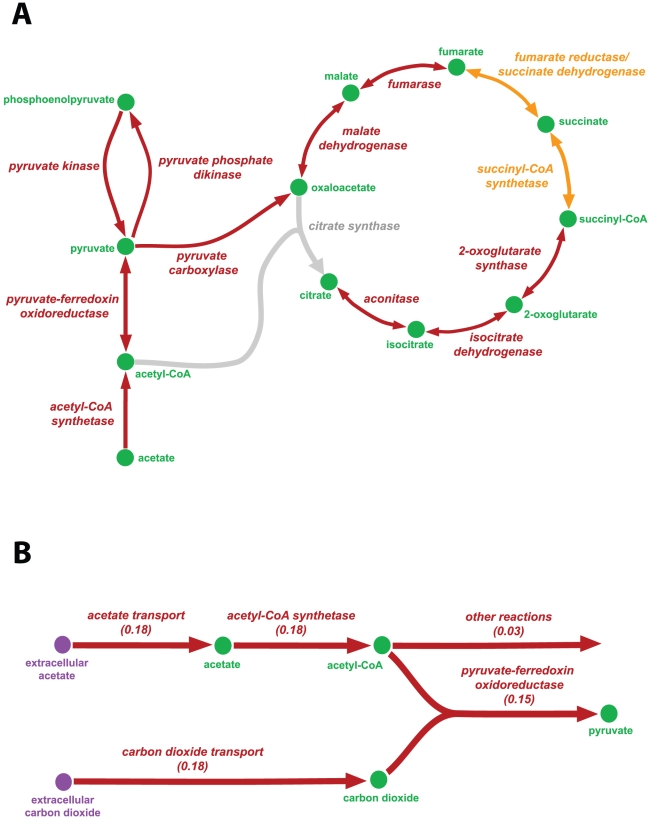
The reconstructed network for glycolysis, gluconeogenesis, the TCA-cycle and the pentose phosphate pathway of iAI549 highlighted some of the key limitations of Dehalococcoides central metabolism. Although putative genes for glycolysis and gluconeogenesis were identified, no gene for a glucose or fumarate transporter was found in any of the genomes, explaining the inability of Dehalococcoides to use glucose or fumarate as a carbon source. The TCA-cycle of Dehalococcoides (Figure 3A) is incomplete, as previously reported [29], [30]. We could identify putative genes for 2-oxoglutarate synthase and succinyl Co-A synthetase (with 26% amino acid sequence identity to the Methanococcus jannaschii gene), and fumarate reductase/succinate dehydrogenase (with 31–33% amino acid sequence identity to the E. coli gene) (Table 3 in Text S1), but we could not find a gene encoding the citrate synthase (CS) in Dehalococcoides. In a scenario without CS, carbon assimilation could occur using a reductive TCA-cycle. However, the biosynthetic formation of citrate by Dehalococcoides ethenogenes strain 195 was recently demonstrated using 13C-labeled isotopomer experiments, although the gene encoding the putative Re-type CS enzyme was not identified [84]. The two Dehalococcoides genes that are most similar to the only biochemically characterized Re-type CS gene from Clostridium kluyveri DSM555 [85] are annotated as isopropyl malate and homocitrate synthase; however, these genes share only 27% amino acid sequence identity with CS gene from C. kluyveri. Hence, further experiments are required to establish the role of these genes, as well as the aforementioned putative TCA-cycle genes in Dehalococcoides. Nonetheless, these isotope labeling studies suggest the formation of 2-oxoglutarate from citrate through the oxidative branch of the TCA-cycle.
Figure 3. The reconstructed TCA-cycle and CO2 fixation pathway of Dehalococcoides.
The arrows show the directionality of the reactions. (A) Grey: citrate synthase gene currently not identified in iAI549, but the pathway is suggested to be present by Tang et al. [84]; Orange: pathways for which homologous putative genes (∼30% amino acid sequence identity) were tentatively identified in Dehalococcoides, but are suggested to be absent by Tang et al. [84]; Red: pathways for which putative genes are confirmed to be present by both iAI549 and Tang et al. [84]. In all cases, the TCA-cycle of Dehalococcoides is not closed which explains their inability to use acetate as an energy source. (B) Dehalococcoides' requirement of CO2 in addition to acetate for their in silico growth. The numbers are flux values in mmol.gDCW−1.h−1. During pyruvate synthesis, Dehalococcoides require 67% carbon (molar basis) from acetate and 33% (molar basis) from CO2. Thus, Dehalococcoides fix carbon via the pyruvate-ferredoxin oxidoreductase or pyruvate synthase (POR) pathway.
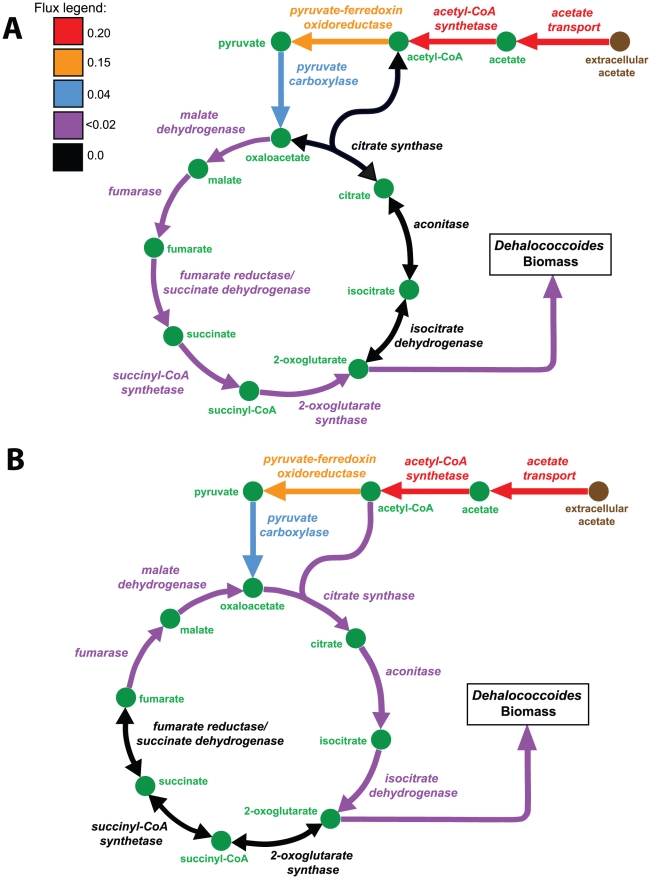
In order to analyze the effect of the presence of the CS reaction on Dehalococcoides growth, we conducted growth simulations with and without this reaction in iAI549. Only a subtle difference in the growth rate (0.0137 h−1 vs. 0.014 h−1) and yield (0.72 gDCW/eeq vs. 0.71 gDCW/eeq) was observed (Figures 4A, 4B and Table 32 in Text S2). Hence, regardless of whether the TCA- cycle is oxidative (Figure 4B) or reductive (4A), the fact that it is incomplete explains why Dehalococcoides are unable to use acetate as their energy source. Interestingly, iAI549 has one anaplerotic reaction — pyruvate carboxylase (PC) — which produces oxaloacetate from pyruvate (Figures 3A, 4A, and 4B). Generally, anaplerotic reactions generate intermediates of a TCA-cycle, but in the absence of a CS reaction, PC is essentially the sole pathway for producing oxaloacetate in the TCA-cycle of iAI549.
Figure 4. Analysis of the citrate synthase (CS) reaction on Dehalococcoides growth.
(A) In the absence of the CS reaction, the TCA-cycle operates reductively via succinyl-CoA synthetase and 2-oxoglutarate synthase for producing biomass precursors for Dehalococcoides to grow. (B) The oxidative TCA-cycle operates when the CS reaction is present, but succinyl-CoA synthetase and 2-oxoglutarate synthase are absent, as suggested by the carbon labeling experiments [84]. However, Dehalococcoides growth remains almost unchanged with and without the CS reaction (0.0137 h−1 vs. 0.014 h−1) as represented by the flux values obtained from the growth simulations of iAI549.
CO2-fixation by Dehalococcoides
Analysis of iAI549 also revealed the presence of a carbon fixation step via pyruvate-ferredoxin oxidoreductase or pyruvate synthase (POR) enzyme encoded by 4 putative Dehalococcoides genes (gene number 181, 182, 183, 184; Table 3 in Text S1). Anaerobes such as Geobacter sulfurreducens and Methanosarcina barkeri are also reported to utilize this step in their central metabolism [82], [86]. POR is essential for the in silico growth of Dehalococcoides using iAI549 since it is the only pathway for producing pyruvate from acetate (Figures 3A, 4A, and 4B). Growth simulations of iAI549 further predict that 33% of the total moles of carbon fixed into the biomass is from extracellular CO2 via POR, and the balance (67%) is from extracellular acetate through acetyl-CoA synthetase (Figure 3B); thus, clearly highlighting the important requirement for extracellular CO2 in addition to acetate as a carbon source for Dehalococcoides.
Moreover, the presence of both POR and carbon-monoxide dehydrogenase enzymes (CODHr) encoded by 4 putative genes of iAI549 (gene number 170, 171, 172, 174; Table 3 in Text S1) initially suggested that the Wood-Ljungdahl pathway [87] of CO2 fixation might be active in Dehalococcoides. However, the absence of several key enzyme encoding genes, such as the methylenetetrahydrofolate reductase and a methyltransferase in the folate-dependant branch of the Wood-Ljungdahl pathway [88]–[90] indicated that the pathway was incomplete in Dehalococcoides (Figure 5 in Text S2). All of these observations are consistent with the carbon labeling studies by Tang et al. [84].
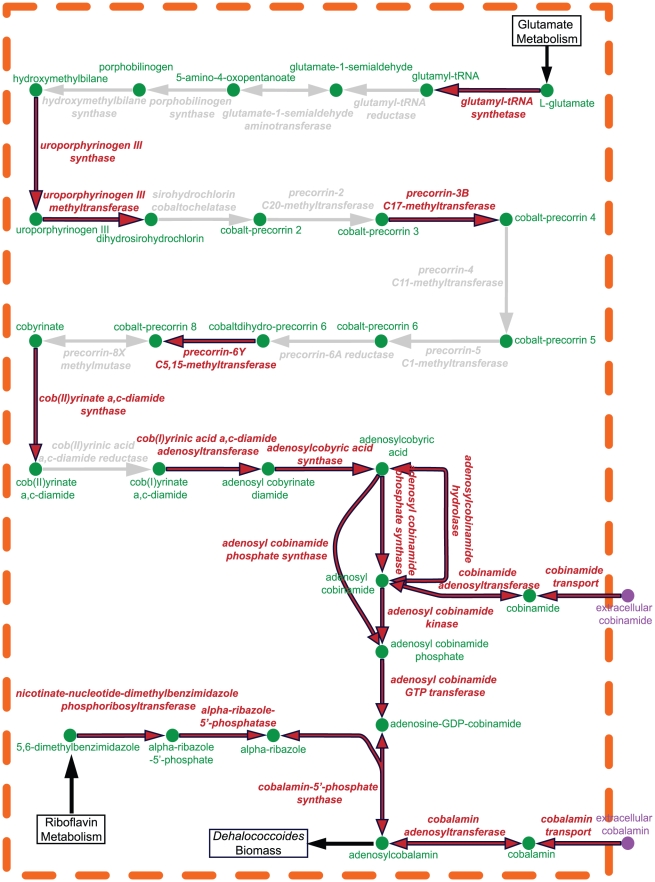
Implications of the incomplete cobalamin synthesis pathway in Dehalococcoides
Cobalamin or vitamin B12 is essential for RDase activity; however, the pathway for producing cobalamin is incomplete in Dehalococcoides [29], [30], [91] (Figure 5). The complete de novo biosynthesis (aerobic or anaerobic) of vitamin B12 requires around 30 genes [92], of which only 18 (Tables 3–7 in Text S1) are identified in Dehalococcoides. Seven (7) of these genes belong to the “anaerobic” pathway while 2 are found to be involved in the “aerobic” pathway of cobalamin biosynthesis. Several key enzyme encoding genes required for the precorrin ring formation, cobalt insertion, and methylation were not found in Dehalococcoides genomes (Figure 5). However, 7 genes of iAI549 (3 core, 1 dispensable, and 3 unique genes: 161, 162, 163, 433, 524, 525, 526; Tables 3, 5, and 7 in Text S1) that encode a putative cobalamin transporter were identified; thus, indicating that Dehalococcoides could uptake vitamin B12 from the medium in the form of either cobinamide or cobalamin [93]. In fact, vitamin B12 has been shown to be required for the growth of pure cultures, and its addition to the medium has been reported to enhance the growth rate of Dehalococcoides [91].
Figure 5. Reconstructed cobalamin biosynthesis pathway of Dehalococcoides.
Dashed orange lines indicate cell membrane, grey lines indicate missing pathways, and red lines indicate existing pathways, putative genes of which are identified in Dehalococcoides during the reconstruction of iAI549. The arrows are denoting the directionality of the reactions. Since the genomes encode a putative cobalamin transporter, Dehalococcoides may salvage vitamin B12 either in the form of cobinamide or cobalamin from the environment as indicated by ‘cobinamide transport’ and ‘cobalamin transport’ reactions in the figure. The adenosylcobalamin, which is the end product of the entire pathway, is a biomass constituent and is assumed to take part in Dehalococcoides cell formation.
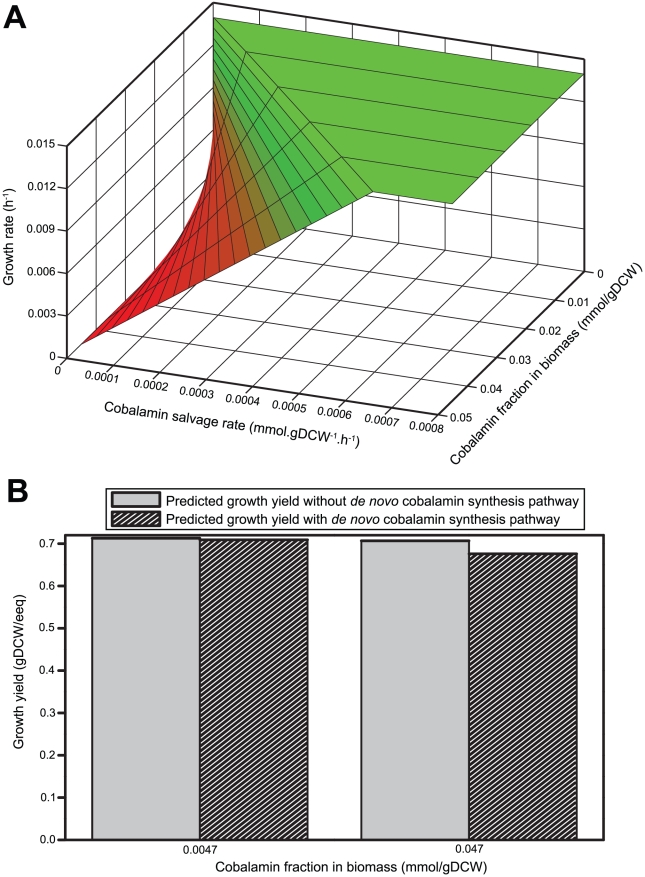
Therefore, in order to examine the influence of cobalamin on the growth of Dehalococcoides, we conducted growth simulations for two scenarios using iAI549: 1) Dehalococcoides growth rate as a function of weight fraction of cobalamin in the biomass and cobalamin salvage rate from the medium (Figure 6A), and 2) Dehalococcoides growth yield assuming it could synthesize its own cobalamin (i.e., adding all the reactions to iAI549 required for de novo cobalamin synthesis) compared to the yield when B12 is salvaged from the medium (Figure 6B). Predictably, the growth rate decreases to zero at low cobalamin salvage rates (Figure 6A). Also, the cobalamin salvage rate at which metabolism becomes limited by B12 is a strong function of the cobalamin fraction in the biomass, which has never been experimentally measured for Dehalococcoides (Figure 6A). From the second simulation, it is clear that the energetic cost for synthesizing cobalamin de novo is not very significant since the predicted yield with and without a cobalamin synthesis pathway is almost identical (Figure 6B). Only if one assumes a biomass cobalamin fraction 10 times higher than the maximum reported, a small (4%) reduction in the growth yield (from 0.72 gDCW/eeq to 0.69 gDCW/eeq) is predicted as a penalty for synthesizing cobalamin de novo (Figure 6B and Table 31 in Text S2). This low synthesis cost, along with the fact that cobalamin is essential, yet its synthesis pathway is incomplete in Dehalococcoides suggests perhaps that Dehalococcoides might have evolved syntrophically with cobalamin secreters and never faced significant evolutionary pressure to acquire a complete cobalamin synthesis pathway in their genomes.
Figure 6. Influence of cobalamin on the growth rate and yield of Dehalococcoides.
(A) Growth rate of Dehalococcoides is simulated as a function of both cobalamin salvage rate and cobalamin fraction in the biomass equation. It shows the role of cobalamin in limiting the growth rate of Dehalococcoides. Clearly, the cobalamin uptake or salvage rate at which Dehalococcoides growth is limiting increases with the increase of cobalamin fraction in the biomass. (B) The cost of de novo cobalamin synthesis in terms of Dehalococcoides growth yield is compared (see text for details). The predicted yield of Dehalococcoides with and without the de novo cobalamin synthesis pathway remains almost identical for the reported maximum cobalamin fraction in the biomass. However, the predicted yield decreased only by 4% (from 0.72 gDCW/eeq to 0.69 gDCW/eeq) with 10 fold increase of cobalamin fraction in the biomass indicating the low cost of de novo cobalamin synthesis.
Does carbon or energy limit the in silico growth of Dehalococcoides?
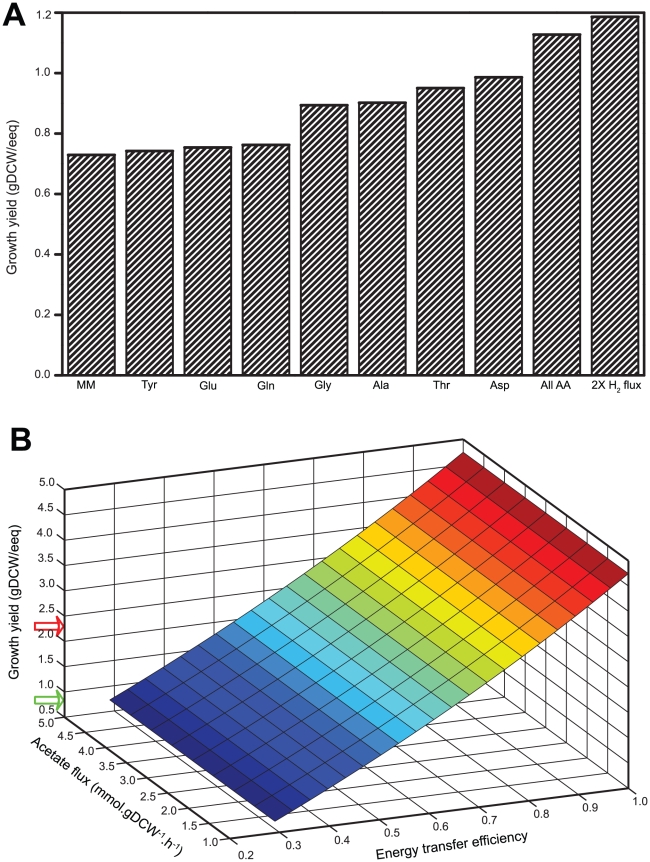
Growth of Dehalococcoides is more rapid in mixed microbial communities than in pure cultures [24], [57], [60], [94] although the reasons for this discrepancy are not entirely clear. The difference in reported growth yields between pure and mixed cultures is more significant (p = 0.0005 at 95% confidence level) than the difference in reported growth rates (p = 0.05 at 95% confidence level) (Tables 25, 26 in Text S2). Thus, in order to examine the growth-limiting conditions, we simulated Dehalococcoides growth yields under two different conditions: 1) allowing unlimited flux of amino acids in the medium at a hydrogen flux of 10 mmol.gDCW−1.h1 (equivalent to the dechlorination rate obtained from average pure-culture growth yields and rates; Tables 24, 25 in Text S2), and 2) doubling the hydrogen flux (20 mmol. gDCW−1.h−1) without allowing any amino acid flux in the medium. The first condition mimics a carbon-rich environment while the second one represents an energy-rich situation. The model predicts that adding unlimited amount of any or all of the amino acids in the growth medium (obviating the need for the cell to synthesize these amino acids) increased the growth yield by a maximum of 55% (1.13 gDCW/eeq) compared to the case with no amino acids in the medium (0.72 gDCW/eeq) (Figure 7A). However, doubling only the hydrogen flux enhanced the growth yield by 65% (from 0.72 gDCW/eeq to 1.19 gDCW/eeq) (Figure 7A).
Figure 7. Effect of carbon and energy sources on the growth yield of Dehalococcoides.
(A) The experimental growth yield of Dehalococcoides in the minimal medium (0.69 gDCW/eeq) is compared with increased growth yields achieved by allowing unlimited fluxes of all amino acids at a H2 flux of 10 mmol.gDCW−1.h−1 (corresponding to the experimental dechlorination rate), as well as doubling the H2 flux (20 mmol.gDCW−1.h−1). It shows that unlimited flux of amino acids (carbon source) increased the in silico growth yield of Dehalococcoides by 55%, whereas doubling the H2 flux (electron donor or energy source) alone enhanced the yield by 65%. (B) Analysis of the energy limited growth of Dehalococcoides. Since the growth yield of Dehalococcoides varies linearly with the energy transfer efficiency, their yield can be improved by increasing the flux of their energy source or electron donor to generate more ATP per electron. However, the variation in acetate fluxes has no effect on growth yields. Red and green arrows show growth yields and corresponding efficiencies for Dehalococcoides growth in mixed and pure cultures, respectively. ‘MM’ = minimal medium; ‘Tyr’ = tyrosine; ‘Glu’ = glutamate; ‘Gln’ = glutamine; ‘Gly’ = glycine; ‘Ala’ = alanine; ‘Thr’ = threonine; ‘Asp’ = aspartate; ‘All AA’ = all amino acids; ‘2X H2 flux’ = 20 mmol H2.gDCW−1.h−1.
To further analyze this aspect of energy limitation, we simulated in silico growth yields of Dehalococcoides as a function of both acetate flux (carbon availability) and energy flux, represented by the energy transfer efficiency. This analysis (Figure 7B) shows that the growth of Dehalococcoides is energy-limited but not carbon-limited since growth yield increases proportionally to increase in energy transfer efficiency regardless of acetate flux. Moreover, simulations also reveal that growth yield of Dehalococcoides in a pure culture is only 30% efficient (corresponding to the green arrow) compared to 65% efficient (corresponding to the red arrow) in a mixed culture (Figure 7B). These simulations point towards electron flux from hydrogen to the RDase as the rate-limiting step, which is somehow more efficient in mixed cultures. It is possible that interspecies hydrogen transfer, such as in a mixed culture is more direct than hydrogen provided in the medium (as for pure cultures). Electrons supplied from an electrode that was polarized to a very low potential were shown to stimulate Dehalococcoides metabolism [95], possibly illustrating such an effect; if true, these results suggest a mechanism for the enhanced growth of Dehalococcoides and to a faster dechlorination of pollutants.
As described earlier, experimental studies clearly illustrate the favorable growth of Dehalococcoides in syntrophic microbial consortia compared to their isolated pure cultures. This obviously points towards the existence of some undefined beneficial metabolic interactions among the consortia members. Although iAI549 simulations suggested more efficient electron transfer and energy utilization in a mixed culture, this result requires further experimental validation. Because these microbes harness energy for their growth from reductive dechlorination reactions, their increased growth will certainly accelerate the bioremediation process. Hence, the current challenge is to understand the reason behind their favorable growth in a mixed microbial community prevailing in their natural habitat. Therefore, a genome-scale metabolic model of a syntrophic community of dechlorinating bacteria, where Dehalococcoides are the dominant members, can be useful to understand the factors influencing their growth. This information may also help to develop a defined bacterial community with enhanced bioremediation capability, in addition to developing effective strategies for exploiting these microbes for effective bioremediation of contaminated sites around the world.
Conclusions
Although genome-scale constraint-based models are available for several microbes from all three forms of life, iAI549 is the first such endeavor for dechlorinating bacteria. This constraint-based flux balance model is consistent with the specialized nature of Dehalococcoides metabolism. The model supports the idea that evolution of the chlorinated compound specific rdh genes conferred the strain-specific metabolic phenotype to Dehalococcoides. In addition to cataloguing significant metabolic similarities among Dehalococcoides strains, the model also provides valuable insights regarding physiological and metabolic bottlenecks of these microbes. Reconstructed central metabolic pathways, for example, identified underlying reasons for Dehalococcoides' requirement of a separate energy source in addition to a carbon source for growth, as well as a carbon fixation step. Also, growth simulations revealed the energy-limited rather than carbon or cobalamin-limited growth of these organisms. In the process of developing the model, detailed tables of metabolic gene correspondences among 4 genomes, reannotations based on pathway analysis, and intrinsic kinetic and stoichiometric parameters were developed for the user community. We also created lists of core hypothetical genes and non-gene associated model reactions; these lists will be useful for designing enzyme assays for functional annotation of the hypothetical genes. Finally and most importantly, this pan-genome-scale metabolic model now provides a common and scalable framework as well as a knowledgebase, which can be used for visualization and interpretation of various omics-scale data from transcriptomics, proteomics and metabolomics for any Dehalococcoides strain; such analysis will further our understanding of these environmentally important organisms so that the outcome of bioremediation can be improved.
Materials and Methods
Dehalococcoides Pan-Genome
In order to develop the pan-genome of Dehalococcoides, we obtained strain CBDB1 genome sequence from JCVI (http://cmr.jcvi.org/tigr-scripts/CMR/CmrHomePage.cgi) while strain 195 and strain BAV1 genome sequences were downloaded from the IMG database (http://img.jgi.doe.gov/cgi-bin/pub/main.cgi). Strain VS genome sequence was obtained from Alfred Spormann at Stanford University, CA. The genome sequences were compared using OrthoMCL [96], a widely accepted method for finding orthologs across different genomes [97]. OrthoMCL is based on reciprocal best BLAST hit (RBH), but recognizes co-ortholog groups using a Markov graph clustering (MCL) algorithm [98]. The Dehalococcoides pan-genome was developed following a previously described approach [67], [68] outlined in Figures 1–4 in Text S2 and in the following section.
First, we identified putative orthologs between a reference genome and a subject genome which were selected arbitrarily from the 4 genomes compared. The analysis was conducted by OrthoMCL keeping the parameters of the algorithm in default settings. Subsequently, those genes that were present only in subject genome 1 were identified and combined with the reference genome to create the augmented genome 1 (Figure 1 in Text S2). Then, the augmented genome 1 was compared and analyzed with subject genome 2 as described above to construct the augmented genome 2. The pan-genome was obtained by comparing the augmented genome 2 and subject genome 3. The number of genes in a pan-genome was reported to depend on both the order of genomes analyzed and the reference genome [67]; hence, we constructed 6 pan-genomes for 6 different genome-order combinations. Of these 6 pan-genomes, we selected the one with the highest number of genes (2061) as Dehalococcoides pan-genome in order to capture the entire gene repertoire of Dehalococcoides species [74]. We also identified the core, dispensable and unique genomes for Dehalococcoides pan-genome using methods described previously [2], [67], (Figures 2–4 in Text S2).
Reconstructing the Metabolic Network of Dehalococcoides
The pan-genome was used to reconstruct the pan-genome-scale metabolic network, and the constraint-based model of Dehalococcoides metabolism was developed from this reconstruction. Since the strains of Dehalococcoides share a high degree of sequence identity, we arbitrarily chose strain CBDB1 genome as a reference and constructed the metabolic network from its annotated genome sequence [29], publications regarding its physiology, and various genomic and biochemical databases [7]. Then, we included other metabolic genes from the pan-genome into the reconstructed network that were missing from strain CBDB1 genome. Five gene correspondence tables for the four genomes were prepared (Tables 3–7 in Text S1) for facilitating gene identification and cross-reference regardless of the genome of interest. We developed and manually curated the reconstructed network using the procedures described previously [3], [7], [99], [100] with the SimPheny platform (Genomatica Inc., San Diego, CA). Since genome annotations are error prone [101], annotated genes of strain CBDB1, as well as the pan-genome genes with defined metabolic functions were verified by identifying their homologs in other well characterized organisms, including Escherichia coli, Bacillus subtilis, Geobacter sulfurreducens and Saccharomyces cerevisiae with BLAST [102]. Subsequently, confidence levels were assigned based on the degree of sequence identities or reciprocal best BLAST hits. Dehalococcoides genes, for instance, having >40% amino acid sequence identity with homologs in the protein databases (SWISSPROT [103], IMG [71], PDB [72], GO [104]) were given a confidence level of 3, and genes with >30% and <30% identity were assigned a confidence level of 2 and 1, respectively. In addition, these genes were also evaluated on the basis of both gene order or conserved synteny [105], along with phylogenetic analysis with the updated versions of biological databases, such as UniProt [70], IMG [71], GO [104], and PDB [72]. Afterwards, both elementally and charge balanced biochemical reactions were assigned to the genes to create the gene-protein-reaction (GPR) associations [3]. These reactions were further verified by biochemical literature as well as enzyme databases, such as KEGG [106], BRENDA [107], MetaCyc [108], and ENZYME [109]. In some instances, genes required for some biosynthetic reactions essential for producing all the precursor metabolites for cell biomass were not identified. Such reactions (21 in number detailed in Table 1 in Text S1) were added to the reconstructed network as non-gene associated reactions.
Estimation of Biomass Composition and Maintenance Energy Requirements
The biomass composition (dry basis) of 1 gram of Dehalococcoides cells was calculated from various published and experimental data, and expressed in mmol (millimoles)/g DCW (dry cell weight) (Tables 19–24 in Text S2). Due to the lack of detailed experimental data on the cellular composition of Dehalococcoides, the weight fractions of protein, lipid, carbohydrate, soluble pools and ions of the cell were estimated from the published genome-scale model of Methanosarcina barkeri [83]. We choose to use data from M. barkeri model — an archaeon — because Dehalococcoides cells are enclosed by the archaeal S-layer like protein instead of a typical bacterial cell wall [18]–[20]. The weight percent of DNA was estimated from the cell morphology, length of the genome sequence [110] and molar mass of the DNA, and the weight percent of RNA was calculated from the experimental data on a Dehalococcoides containing mixed microbial culture (see Text S2 for details). In addition, the detailed composition of each macromolecule as well as the composition of cofactors, and other soluble pools and ions are presented in Tables 19–24 in Text S2. The distribution of amino acids, nucleotides and cofactors in the biomass was calculated from the data reported previously [111], [112] while the weight fractions of different fatty acids were estimated from White et al. [113]. These compositions were then integrated into the model as a biomass synthesis reaction, BIO_DHC_DM_61 (see Text S2 for additional details).
Maintenance energy accounts for the ATP requirements of cellular processes, such as turnover of the amino acid pools, polymerization of cellular macromolecules, and ion transport, which are not included in the biomass synthesis reaction [114]–[116]. These ATP requirements can be either growth associated (GAM), i.e., related to assembly and polymerization of macromolecules (eg. proteins, DNA, etc.), or non-growth associated (NGAM) that corresponds to maintaining membrane potential for keeping cellular integrity [114], [115], [117]. Due to the lack of experimental chemostat data required for calculating both maintenance parameters [118], the NGAM for a Dehalococcoides cell (1.8 mmol ATP.gDCW−1.h−1) was calculated from the experimental decay rate (0.09 day−1) [24] and the average of pure-culture experimental growth yields (0.69 g DCW/eeq; Table 25 in Text S2) following the procedures described previously [114], [116]. The GAM was estimated by regression, using an initial estimate of 26 mmol ATP/g DCW for a typical bacterial cell (Table 28 in Text S2) [111]. The initial estimate of GAM and the calculated NGAM were then used to simulate (using flux balance analysis, described below) the average of reported pure-culture experimental growth rates (0.014 h−1; Table 26 in Text S2). A GAM of 61 mmol ATP/g DCW gave the best prediction of the experimental growth rate.
In Silico Analysis of Dehalococcoides Metabolism
Flux Balance Analysis (FBA) relies on the imposition of a series of constraints including stoichiometric mass balance constraints derived from the metabolic network, thermodynamic reversibility constraints and any available enzyme capacity constraints [3], [119], [120]. The imposition of these constraints leads to a linear optimization (Linear Programming, LP) problem formulated to maximize a cellular objective function such as the growth rate. Hence, the biomass synthesis reaction is assumed to be the objective function to be maximized to solve the LP problem in SimPheny. In addition, a number of reversible reactions were added in the network for exchanging external metabolites, such as acetate (ac), chloride (Cl−), carbondioxide (CO2), sulphate (SO4 −2) etc., to represent the in silico minimal medium (Table 2) for Dehalococcoides. As discussed earlier, cobalamin is essential for Dehalococcoides growth, but they are unable to synthesize it de novo; hence, they salvage cobalamin from the medium. In order to analyze whether cobalamin flux can limit Dehalococcoides growth, we performed a robustness analysis on the cobalamin exchange reaction for different weight fractions of cobalamin in the biomass. We also simulated growth rates by incorporating all the pathways required for de novo cobalamin synthesis in iAI549 for analyzing cobalamin synthesis cost, and its effect on Dehalococcoides growth. Finally, to identify whether the growth of Dehalococcoides was carbon or energy limited, the growth simulations were conducted by varying acetate fluxes and energy transfer efficiencies since acetate and H2 are the carbon and energy sources of these microbes, respectively [18]–[21], . Energy transfer efficiencies were calculated by normalizing the ATP fluxes to the maximum ATP that could be generated from H2 based on Gibb's free energy of H2 oxidation and the energetic cost of ATP synthesis (mol ATP/mol H2) (see Table 30 and Text S2 for additional details). The constraints set used to simulate Dehalococcoides growth is listed in Table 18 in Text S1, and the SBML file for the reconstructed network (iAI549) is presented as Dataset S1.
Supporting Information
SBML File for Dehalococcoides Metabolic Model, iAI549.
(0.48 MB XML)
This file contains the detailed list of genes, proteins, reactions and metabolites included in the Dehalococcoides pan-metabolic-model, iAI549. Tables 3–7 are gene correspondences where a unique gene number is provided for each gene in the model (Model gene number) so that a gene or corresponding reaction can be located conveniently irrespective of the 4 genomes of interest. Strain VS gene locus names are obtained from Alfred Spormann at Stanford University, CA. A reaction can be associated with more than one gene where some genes are core, some are dispensable and others can be unique. For such instances, 2 additional gene correspondence tables (Table 5 and Table 7) are created for dispensable and unique genes respectively.
(5.41 MB PDF)
This file contains the tables of detailed macromolecular composition of a gram of Dehalococcoides cell, experimental values of various pan-model parameters and the detailed procedure to calculate those parameters, as well as supplemental text regarding energy conservation process of Dehalococcoides. In addition, all the supplemental figures are included at the end of this document.
(0.31 MB PDF)
Acknowledgments
The authors would like thank Dr. R. K. Thauer of MPI fuer Terrestrische Mikrobiologie, Marburg, Germany, and Dr. Alison Waller, Laura Hug and Laurence Yang of the University of Toronto for their insightful discussions, as well as Dr. Alfred Spormann of Stanford University, CA, for giving access to the genome sequence of strain VS.
Footnotes
The authors have declared that no competing interests exist.
This research was funded by the University of Toronto, the Natural Sciences and Engineering Research Council of Canada (NSERC), the Government of Canada through Genome Canada and the Ontario Genomics Institute (2009-OGI-ABC-1405) and the United States Department of Defense Strategic Environmental Research and Development Program (SERDP). MAI was funded by the Ontario Graduate Scholarship (OGS), the SERDP and Genome Canada funds to EAE and the departmental faculty start-up funds to RM. We would also like to acknowledge Dr. Derek Lovley and funding from the US Department of Energy (Cooperative Agreement No. DE-FC02-02ER63446) for enabling access to the SimPheny program. After acceptance of the paper, the authors acknowledge Ontario Genomics Institute Genomics Publication Fund for defraying the open-access publication costs. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Covert MW, Xiao N, Chen TJ, Karr JR. Integrating metabolic, transcriptional regulatory and signal transduction models in Escherichia coli. Bioinformatics. 2008;24:2044–2050. doi: 10.1093/bioinformatics/btn352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Medini D, Serruto D, Parkhill J, Relman DA, Donati C, et al. Microbiology in the post-genomic era. Nat Rev Microbiol. 2008;6:419–430. doi: 10.1038/nrmicro1901. [DOI] [PubMed] [Google Scholar]
- 3.Reed JL, Famili I, Thiele I, Palsson BO. Towards multidimensional genome annotation. Nature Reviews: Genetics. 2006;7:130–141. doi: 10.1038/nrg1769. [DOI] [PubMed] [Google Scholar]
- 4.Young JD, Henne KL, Morgan JA, Konopka AE, Ramkrishna D. Integrating cybernetic modeling with pathway analysis provides a dynamic, systems-level description of metabolic control. Biotechnol Bioeng. 2008;100:542–559. doi: 10.1002/bit.21780. [DOI] [PubMed] [Google Scholar]
- 5.Becker SA, Feist AM, Mo ML, Hannum G, Palsson BO, et al. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox. Nat Protoc. 2007;2:727–738. doi: 10.1038/nprot.2007.99. [DOI] [PubMed] [Google Scholar]
- 6.Becker SA, Palsson BO. Context-specific metabolic networks are consistent with experiments. PLoS Computa Biol. 2008;4(5):e1000082. doi: 10.1371/journal.pcbi.1000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feist AM, Herrgard MJ, Thiele I, Reed JL, Palsson BO. Reconstruction of biochemical networks in microorganisms. Nat Rev Microbiol. 2009;7:129–143. doi: 10.1038/nrmicro1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heinemann M, Kümmel A, Ruinatscha R, Panke S. In silico genome-scale reconstruction and validation of the Staphylococcus aureus metabolic network. Biotechnol Bioeng. 2005;92:850–864. doi: 10.1002/bit.20663. [DOI] [PubMed] [Google Scholar]
- 9.Joyce AR, Palsson BO. Predicting gene essentiality using genome-scale in silico models. In: Osterman AL, Gerdes SY, editors. Microbial Gene Essentiality: Protocols and Bioinformatics. Humana Press; 2008. pp. 433–457. [DOI] [PubMed] [Google Scholar]
- 10.Nookaew I, Jewett MC, Meechai A, Thammarongtham C, Laoteng K, et al. The genome-scale metabolic model iIN800 of Saccharomyces cerevisiae and its validation: a scaffold to query lipid metabolism. BMC Syst Biol. 2008 doi: 10.1186/1752-0509-2-71. doi: 10.1186/1752-0509-2-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schilling CH, Covert MW, Famili I, Church GM, Edwards JS, et al. Genome-scale metabolic model of Helicobacter pylori 26695. J Bacteriol. 2002;184:4582–4593. doi: 10.1128/JB.184.16.4582-4593.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teusink B, Wiersma A, Molenaar D, Francke C, Vos WMMd, et al. Analysis of growth of Lactobacillus plantarum WCFS1 on a complex medium using a genome-scale metabolic model. J Biol Chem. 2006;281:40041–40048. doi: 10.1074/jbc.M606263200. [DOI] [PubMed] [Google Scholar]
- 13.Kim H-S, Lee J-Y. Molecular bioremediation: metabolic engineering for biodegradation of recalcitrant pollutants. In: Lee SY, Papoutsakis ET, editors. Metabolic Engineering. New York: Marcel Dekker, Inc; 1999. [Google Scholar]
- 14.de Vos WM, Smidt H. Anaerobic microbial dehalogenation. Annu Rev Microbiol. 2004;58:43–73. doi: 10.1146/annurev.micro.58.030603.123600. [DOI] [PubMed] [Google Scholar]
- 15.El Fantroussi S, Naveau H, Agathos SN. Anaerobic dechlorinating bacteria. Biotechnol Prog. 1998;14:167–188. doi: 10.1021/bp980011k. [DOI] [PubMed] [Google Scholar]
- 16.Haggbloom MM, Bossert ID. Halogenated organic compounds-a global perspective. In: Haggbloom MM, Bossert ID, Wulder MA, editors. Microbial Processes and Environmental Applications. Boston: Kluwer Academic Publishers; 2003. [Google Scholar]
- 17.Holliger C, Wohlfarth G, Diekert G. Reductive dechlorination in the energy metabolism of anaerobic bacteria. FEMS Microbiol Rev. 1999;22:383–398. [Google Scholar]
- 18.Maymo-Gatell X, Chien Y-t, Gossett JM, Zinder SH. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science. 1997;276:1568–1571. doi: 10.1126/science.276.5318.1568. [DOI] [PubMed] [Google Scholar]
- 19.Adrian L, Szewzyk U, Wecke J, Gorisch H. Bacterial dehalorespiration with chlorinated benzenes. Nature. 2000;408:580–583. doi: 10.1038/35046063. [DOI] [PubMed] [Google Scholar]
- 20.He J, Ritalahti KM, Yang K-L, Koenigsberg SS, Loffler FE. Detoxification of vinyl chloride to ethene coupled to growth of an anaerobic bacterium. Nature. 2003;424:62–65. doi: 10.1038/nature01717. [DOI] [PubMed] [Google Scholar]
- 21.He J, Sung Y, Brown RK, Ritalahti KM, Loffler FE. Isolation and characterization of Dehalococcoides sp. strain FL2, a trichloroethene (TCE)- and 1,2- dechloroethene-respiring anaerobe. Environ Microbiol. 2005;7:1442–1450. doi: 10.1111/j.1462-2920.2005.00830.x. [DOI] [PubMed] [Google Scholar]
- 22.Loffler FE, Sun Q, Li JR, Tiedje JM. 16S rRNA gene-based detection of tetrachloroethene dechlorinating Desulfuromonas and Dehalococcoides species. Appl Environ Microbiol. 2000;66:1369–1374. doi: 10.1128/aem.66.4.1369-1374.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sung Y, Ritalahti KM, Apkarian RP, Loffler FE. Quantitive PCR confirms purity of strain GT, a novel trichloroethene-to-ethene-respiring Dehalococcoides isolate. Appl Environ Microbiol. 2006;72:1980–1987. doi: 10.1128/AEM.72.3.1980-1987.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cupples AM, Spormann AM, McCarty PL. Growth of a Dehalococcoides - like microorganism on vinyl chloride and cis - dichloroethene as electron acceptors as determined by competitive PCR. Appl Environ Microbiol. 2003;69:953–959. doi: 10.1128/AEM.69.2.953-959.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holscher T, Gorisch H, Adrian L. Reductive dehalogenation of chlorobenzene congeners in cell extracts of Dehalococcoides sp. strain CBDB1. Appl Environ Microbiol. 2003;69:2999–3001. doi: 10.1128/AEM.69.5.2999-3001.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jayachandran G, Gorisch H, Adrian L. Studies on hydrogenase activity and chlorobenzene respiration in Dehalococcoides sp. strain CBDB1. Arch Microbiol. 2004;182:498–504. doi: 10.1007/s00203-004-0734-9. [DOI] [PubMed] [Google Scholar]
- 27.Nijenhuis I, Zinder SH. Characterization of hydrogenase and reductive dehalogenase activities of Dehalococcoides ethenogenes strain 195. Appl Environ Microbiol. 2005;71:1664–1667. doi: 10.1128/AEM.71.3.1664-1667.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holscher T, Brown RK, Ritalahti KM, Wintzingerode Fv, Gorisch H, et al. Multiple nonidentical reductive-dehalogenase-homologous genes are common in Dehalococcoides. Appl Environ Microbiol. 2004;70:5290–5297. doi: 10.1128/AEM.70.9.5290-5297.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kube M, Beck A, Zinder SH, Kuhl H, Reinhardt R, et al. Genome sequence of the chlorinated compound respiring bacterium Dehalococcoides species strain CBDB1. Nat Biotechnol. 2005;23:1269–1273. doi: 10.1038/nbt1131. [DOI] [PubMed] [Google Scholar]
- 30.Seshadri R, Adrian L, Fouts DE, Eisen JA, Phillippy AM, et al. Genome sequence of the PCE-dechlorinating bacterium Dehalococcoides ethenogenes. Science. 2005;307:105–108. doi: 10.1126/science.1102226. [DOI] [PubMed] [Google Scholar]
- 31.Waller AS, Brown RK, Loffler FE, Edwards EA. Multiple reductive-dehalogenase-homologous genes are simultaneously transcribed during dechlorination by Dehalococcoides-containing structures. Appl Environ Microbiol. 2005;71:8257–8264. doi: 10.1128/AEM.71.12.8257-8264.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adrian L, Rahnenfuhrer J, Gobom J, Holscher T. Identification of a chlorobenzene reductive dehalogenase in Dehalococcoides sp. strain CBDB1. Appl Environ Microbiol. 2007;73:1–7. doi: 10.1128/AEM.01649-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Müller J, Rosner B, Von Abendroth G, Meshulam-Simon G, McCarty P, et al. Molecular identification of the catabolic vinyl chloride reductase from Dehalococcoides sp. strain VS and its environmental distribution. Appl Environ Microbiol. 2004;70:4880–4888. doi: 10.1128/AEM.70.8.4880-4888.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Magnuson J, Romine M, Burris D, Kingsley M. Trichloroethene reductive dehalogenase from Dehalococcoides ethenogenes: sequence of tceA and substrate range characterization. Appl Environ Microbiol. 2000;66:5141–5147. doi: 10.1128/aem.66.12.5141-5147.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Magnuson J, Stern R, Gossett J, Zinder S, Burris D. Reductive dechlorination of tetrachloroethene to ethene by a two-component enzyme pathway. Appl Environ Microbiol. 1998;64:1270–1275. doi: 10.1128/aem.64.4.1270-1275.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krajmalnik-Brown R, Holscher T, Thomson I, Saunders F, Ritalahti K, et al. Genetic identification of a putative vinyl chloride reductase in Dehalococcoides sp. strain BAV1. Appl Environ Microbiol. 2004;70:6347–6351. doi: 10.1128/AEM.70.10.6347-6351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morris RM, Sowell S, Barofsky D, Zinder S, Richardson R. Transcription and mass-spectroscopic proteomic studies of electron transport oxidoreductases in Dehalococcoides ethenogenes. Environ Microbiol. 2006;8:1499–1509. doi: 10.1111/j.1462-2920.2006.01090.x. [DOI] [PubMed] [Google Scholar]
- 38.Morris RM, Fung JM, Rahm BG, Zhang S, Freedman DL, et al. Comparative proteomics of Dehalococcoides spp. reveals strain-specific peptides associated with activity. Appl Environ Microbiol. 2007;73:320–326. doi: 10.1128/AEM.02129-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meyer J. [FeFe] hydrogenases and their evolution: a genomic perspective. Cell Mol Life Sci. 2007;64:1063–1084. doi: 10.1007/s00018-007-6477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vignais PM, Billoud B, Meyer J. Classification and phylogeny of hydrogenases. FEMS Microbiol Rev. 2001;25:455–501. doi: 10.1111/j.1574-6976.2001.tb00587.x. [DOI] [PubMed] [Google Scholar]
- 41.Krasotkina J, Walters T, Maruya KA, Ragsdale SW. Characterization of the B12 and iron-sulfur-containing reductive dehalogenase from Desulfitobacterium chlororespirans. J Biol Chem. 2001;276:40991–40997. doi: 10.1074/jbc.M106217200. [DOI] [PubMed] [Google Scholar]
- 42.Miller E, Wohlfarth G, Diekert G. Studies on tetrachloroethene respiration in Dehalospirillum multivorans. Arch Microbiol. 1997;166:379–387. doi: 10.1007/BF01682983. [DOI] [PubMed] [Google Scholar]
- 43.Banerjee R, Ragsdale SW. The many faces of vitamin B12: catalysis by cobalamin-dependent enzymes. Annu Rev Biochem. 2003;72:209–247. doi: 10.1146/annurev.biochem.72.121801.161828. [DOI] [PubMed] [Google Scholar]
- 44.Kroger A, Biel S, Simon J, Gross R, Unden G, et al. Fumarate respiration of Wolinella succinogens: enzymology, energetics and coupling mechanism. Biochim Biophys Acta. 2002;1553:23–38. doi: 10.1016/s0005-2728(01)00234-1. [DOI] [PubMed] [Google Scholar]
- 45.Louie TM, Mohn WW. Evidence for a chemiosmotic model of dehalorespiration in Desulfomonile tiedjei DCB-1. J Bacteriol. 1999;181:40–46. doi: 10.1128/jb.181.1.40-46.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schumacher W, Holliger C. The proton/electron ration of the menaquinone-dependent electron transport from dihydrogen to tetrachloroethene in “Dehalobacter restrictus”. J Bacteriol. 1996;178:2328–2333. doi: 10.1128/jb.178.8.2328-2333.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jayachandran G, Gorisch H, Adrian L. Dehalorespiration with hexachlorobenzene and pentachlorobenzene by Dehalococcoides sp. strain CBDB1. Arch Microbiol. 2003;180:411–416. doi: 10.1007/s00203-003-0607-7. [DOI] [PubMed] [Google Scholar]
- 48.Thauer RK, Jungermann K, Decker K. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol Rev. 1977;41:100–180. doi: 10.1128/br.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dross F, Geisler V, Lenger R, Theis F, Krafft T, et al. The quinone-reactive Ni/Fe-hydrogenase of Wolinella succinogenes. Eur J Biochem. 1992;206:93–102. doi: 10.1111/j.1432-1033.1992.tb16905.x. [DOI] [PubMed] [Google Scholar]
- 50.Menon AL, Mortenson LE, Robson RL. Nucleotide sequences and genetic analysis of hydrogen oxidation (hox) genes in Azotobacter vinelandii. J Bacteriol. 1992;174:4549–4557. doi: 10.1128/jb.174.14.4549-4557.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bruschi M, Guerlesquin F. Structure, function and evolution of bacterial ferredoxins. FEMS Microbiol Rev. 1988;54:155–176. doi: 10.1111/j.1574-6968.1988.tb02741.x. [DOI] [PubMed] [Google Scholar]
- 52.Eisenstein KK, Wang JH. Conversion of light to chemical free energy. J Biol Chem. 1969;244:1720–1728. [PubMed] [Google Scholar]
- 53.Sterner R. Ferredoxin from Thermotoga maritima. Methods Enzymol. 2001;334:23–30. doi: 10.1016/s0076-6879(01)34454-3. [DOI] [PubMed] [Google Scholar]
- 54.Valentine RC. Bacterial ferredoxin. Bacteriol Rev. 1964;28:497–517. doi: 10.1128/br.28.4.497-517.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Valentine RC, Wolfe RS. Role of ferredoxin in the metabolism of molecular hydrogen. J Bacteriol. 1963;85:1114–1120. doi: 10.1128/jb.85.5.1114-1120.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adrian L, Szewzyk U, Gorisch H. Bacterial growth based on reductive dechlorination of trichlorobenzenes. Biodegradation. 2000;11:73–81. doi: 10.1023/a:1026504605443. [DOI] [PubMed] [Google Scholar]
- 57.Duhamel M, Edwards EA. Growth and yields of dechlorinators, acetogens, and methanogens during reductive dechlorination of chlorinated ethenes and dihaloelimination of 1,2-Dichloroethane. Environ Sci Technol. 2007;41:2303–2310. doi: 10.1021/es062010r. [DOI] [PubMed] [Google Scholar]
- 58.Jayachandran G. Physiological and enzymatic studies of respiration in Dehalococcoides species strain CBDB1. 2004. PhD Thesis. [PhD]: Technischen Universität Berlin, Germany. [DOI] [PubMed]
- 59.Krajmalnik-Brown R, Sung Y, Ritalahti KM, Saunders FM, Loffler FE. Environmental distribution of the trichloroethene reductive dehalogenase gene (tceA) suggests lateral gene transfer among Dehalococcoides. FEMS Microbiol Ecol. 2006;59:206–214. doi: 10.1111/j.1574-6941.2006.00243.x. [DOI] [PubMed] [Google Scholar]
- 60.Adrian L, Manz W, Szewzyk U, Gorisch H. Physiological characterization of a bacterial consortium reductively dechlorinating 1,2,3 - and 1,2,4- trichlorobenzene. Appl Environ Microbiol. 1998;64:496–503. doi: 10.1128/aem.64.2.496-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nowak J, Kirsch NH, Hegemann W, Stan HJ. Total reductive dechlorination of chlorobenzenes to benzene by a methanogenic mixed culture isolated from Saale river sediment. Appl Microbiol Biotechnol. 1996;45:700–709. [Google Scholar]
- 62.He J, Sung Y, Dollhopf ME, Fathepure BZ, Tiedje JM, et al. Acetate versus hydrogen as direct electron donors to stimulate the microbial reductive dechlorination process at chloroethene-contaminated sites. Environ Sci Technol. 2002;36:2945–3952. doi: 10.1021/es025528d. [DOI] [PubMed] [Google Scholar]
- 63.Lendvay JM, Loffler FE, Dollhopf M, Aiello MR, Daniels G, et al. Bioreactive barriers: bioaugmentation and biostimulation for chlorinated solvent remediation. Environ Sci Technol. 2003;37:1422–1431. [Google Scholar]
- 64.Distefano TD, Gossett J, Zinder S. Reductive dechlorination of high concentrations of tetrachloroethene to ethene by an anaerobic enrichment culture in the absence of methanogenesis. Appl Environ Microbiol. 1991;57:2287–2292. doi: 10.1128/aem.57.8.2287-2292.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Freedman DL, Gossett JM. Biological reductive dechlorination of tetrachloroethylene and trichloroethylene to ethylene under methanogenic conditions. Appl Environ Microbiol. 1989;55:2144–2151. doi: 10.1128/aem.55.9.2144-2151.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cupples AM, Spormann AM, McCarty PL. Vinyl chloride and cis-Dichloroethene dechlorination kinetics and microorganism growth under substrate limiting conditions. Environ Sci Technol. 2004;38:1102–1107. doi: 10.1021/es0348647. [DOI] [PubMed] [Google Scholar]
- 67.Tettelin H, Masignani V, Cieslewicz M, Donati C, Medini D, et al. Genome analysis of multiple pathegenic isolates of Streptococcus agalactiae: Implications for the microbial “pan-genome”. Proc Natl Acad Sci U S A. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tettelin H, Riley D, Cattuto C, Medini D. Comparative genomics: the bacterial pan-genome. Curr Opin Microbiol. 2008;12:472–477. doi: 10.1016/j.mib.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 69.Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, et al. The Swiss-Prot protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu CH, Apweiler R, Bairoch A, Natale DA, Barker WC, et al. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Markowitz VM, Szeto E, Palaniappan K, Grechkin Y, Chu K, et al. The integrated microbial genomes (IMG) system in 2007: data content and analysis tool extensions. Nucleic Acids Res. 2007:1–6. doi: 10.1093/nar/gkm846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tettelin H. The bacterial pan-genome and reverse vaccinology. Genome Dyn. 2009;6:35–47. doi: 10.1159/000235761. [DOI] [PubMed] [Google Scholar]
- 74.Muzzi A, Masignani V, Rappuoli R. The pan-genome: towards a knowledge-based discovery of novel targets for vaccines and antibacterials. Drug Discov Today. 2007;12:429–439. doi: 10.1016/j.drudis.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 75.Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R. The microbial pan-genome. Curr Opin Genet Dev. 2005;15:589–594. doi: 10.1016/j.gde.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 76.Adrian L, Hansen SK, Fung JM, Gorisch H, Zinder SH. Growth of Dehalococcoides strains with chlorophenols as electron acceptors. Environ Sci Technol. 2007;41:2318–2323. doi: 10.1021/es062076m. [DOI] [PubMed] [Google Scholar]
- 77.Bunge M, Adrian L, Kraus A, Opel M, Lorenz WG, et al. Reductive dehalogenation of chlorinated dioxins by an anaerobic bacterium. Nature. 2003;421:357–360. doi: 10.1038/nature01237. [DOI] [PubMed] [Google Scholar]
- 78.McMurdie P, Behrens S, Muller J, Goke J, Ritalahti K, et al. Localized plasticity in the streamlined genomes of vinyl chloride respiring Dehalococcoides. PLoS Genet. 2009;5:e1000714. doi: 10.1371/journal.pgen.1000714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Reed JL, Vo TD, Schilling CH, Palsson BO. An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/GPR). Genome Biol. 2003;4:R54.51–R54.12. doi: 10.1186/gb-2003-4-9-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Feist AM, Henry CS, Reed JL, Krummenacker M, Joyce AR, et al. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol Syst Biol. 2007;3:121. doi: 10.1038/msb4100155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Oh Y-K, Palsson BO, Park SM, Schilling CH, Mahadevan R. Genome-scale reconstruction of metabolic network in Bacillus subtilis based on high-throughput phenotyping and gene essentiality data. J Biol Chem. 2007;10:M703759200. doi: 10.1074/jbc.M703759200. [DOI] [PubMed] [Google Scholar]
- 82.Mahadevan R, Bond DR, Butler JE, Esteve-Nuñez A, Coppi MV, et al. Characterization of metabolism in the Fe (III)-reducing organism Geobacter sulfurreducens by constraint-based modeling. Appl Environ Microbiol. 2006;72:1558–1568. doi: 10.1128/AEM.72.2.1558-1568.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Feist AM, Scholten JCM, Palsson BO, Brockman FJ, Ideker T. Modelling methanogenesis with a genome-scale metabolic reconstruction of Methanosarcina barkeri. Mol Syst Biol. 2006;2:1–14. doi: 10.1038/msb4100046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tang YJ, Yi S, Zhuang W-Q, Zinder SH, Keasling JD, et al. Investigation of carbon metabolism in “Dehalococcoides ethenogenes” strain 195 using isotopomer and transcriptomic analyses. J Bacteriol. 2009;191:5224–5231. doi: 10.1128/JB.00085-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li F, Hagemeier CH, Seedorf H, Gottschalk G, Thauer RK. Re-citrate synthase from Clostridium kluyveri is phylogenetically related to homocitrate synthase and isopropylmalate synthase rather than to Si-citrate synthase. J Bacteriol. 2007;189:4299–4304. doi: 10.1128/JB.00198-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bock AK, Kunow J, Glasemacher J, Schönheit P. Catalytic properties, molecular composition and sequence alignments of pyruvate: ferredoxin oxidoreductase from the methanogenic archaeon Methanosarcina barkeri (strain Fusaro). Eur J Biochem. 1996;237:35–44. doi: 10.1111/j.1432-1033.1996.0035n.x. [DOI] [PubMed] [Google Scholar]
- 87.Wood HG, Ljungdahl LG. Autotrophic character of acetogenic bacteria. In: Shively JM, Barton LL, editors. Variations in Autotrophic Life. San Diego, CA: Acdemic Press; 1991. [Google Scholar]
- 88.Drake HL, Gossner AS, Daniel SL. Old acetogens, new light. Ann N Y Acad Sci. 2008;1125:100–128. doi: 10.1196/annals.1419.016. [DOI] [PubMed] [Google Scholar]
- 89.Müller V. Energy conservation in acetogenic bacteria. Appl Environ Microbiol. 2003;69:6345–6353. doi: 10.1128/AEM.69.11.6345-6353.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ragsdale SW. Enzymology of the Wood-Ljungdahl pathway of acetogenesis. Ann N Y Acad Sci. 2008;1125:129–136. doi: 10.1196/annals.1419.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.He J, Holmes VF, Lee PK, Alvarez-Cohen L. Influence of vitamin B12 and cocultures on the growth of Dehalococcoides isolates in defined medium. Appl Environ Microbiol. 2007;73:2847–2853. doi: 10.1128/AEM.02574-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Warren MJ, Raux E, Schubert HL, Escalante-Semerena JC. The biosynthesis of adenosylcobalamin (vitamin B12). Nat Prod Rep. 2002;19:390–412. doi: 10.1039/b108967f. [DOI] [PubMed] [Google Scholar]
- 93.Escalante-Semerena JC. Conversion of cobinamide into adenosylcobamide in Bacteria and Archaea. J Bacteriol. 2007;189:4555–4560. doi: 10.1128/JB.00503-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Duhamel M, Mo K, Edwards EA. Characterization of a highly enriched Dehalococcoides-containing culture that grows on vinyl chloride and trichloroethene. Appl Environ Microbiol. 2004;70:5538–5545. doi: 10.1128/AEM.70.9.5538-5545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Aulenta F, Canosa A, Reale P, Rossetti S, Panero S, et al. Microbial reductive dechlorination of trichloroethene to ethene with electrodes serving as electron donors without the external addition of redox mediators. Biotechnol Bioeng. 2008;103:85–91. doi: 10.1002/bit.22234. [DOI] [PubMed] [Google Scholar]
- 96.Li L, Jr CJS, Roos DS. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen F, Mackey AJ, Vermunt JK, Roos DS. Assessing performance of orthology detection strategies applied to eukaryotic genomes. PLoS One. 2007;2:e383. doi: 10.1371/journal.pone.0000383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Van Dongen S. Graph Clustering by Flow Simulation. 2000. PhD Thesis: University of Utrecht, The Netherlands.
- 99.Covert M, Schilling CH, Famili I, Edwards JS, Goryanin II, et al. Metabolic modeling of microbial strains in silico. Trends Biochem Sci. 2001;26:179–186. doi: 10.1016/s0968-0004(00)01754-0. [DOI] [PubMed] [Google Scholar]
- 100.Francke C, Siezen RJ, Teusink B. Reconstructing the metabolic network of a bacterium from its genome. Trends Microbiol. 2005;13:550–558. doi: 10.1016/j.tim.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 101.Devos D, Valencia A. Intrinsic errors in genome annotation. Trends Genet. 2001;17:429–431. doi: 10.1016/s0168-9525(01)02348-4. [DOI] [PubMed] [Google Scholar]
- 102.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gasteiger, et al. ExPASY: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Poyatos JF, Hurst LD. The determinants of gene order conservation in yeasts. Genome Biol. 2007;8:R233. doi: 10.1186/gb-2007-8-11-r233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chang A, Scheer M, Grote A, Schomburg I, Schomburg D. BRENDA, AMENDA and FRENDA the enzyme information system: new content and tools in 2009. Nucleic Acids Res. 2009;37:588–592. doi: 10.1093/nar/gkn820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Caspi R, Foerster H, Fulcher CA, Kaipa P, Krummenacker M, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2008;36:623–631. doi: 10.1093/nar/gkm900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bairoch A. The ENZYME database in 2000. Nucleic Acids Res. 2000;28:304–305. doi: 10.1093/nar/28.1.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Borodina I, Krabben P, Nielsen J. Genome-scale analysis of Streptomyces coelicolor A3(2) metabolism. Genome Res. 2005;15:820–829. doi: 10.1101/gr.3364705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Neidhardt FC, Ingraham JL, Schaechter M. Physiology of the bacterial cell: a molecular approach. Sunderland, Massachusetts: Sinauer Associates, Inc; 1990. [Google Scholar]
- 112.Pramanik J, Keasling JD. Stoichiometric model of Escherichia coli metabolism: incorporation of growth-rate dependent biomass composition and mechanistic energy requirements. Biotechnol Bioeng. 1997;56:398–421. doi: 10.1002/(SICI)1097-0290(19971120)56:4<398::AID-BIT6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 113.White DC, Geyer R, Peacock AD, Hedrick DB, Koenigsberg SS, et al. Phospholipid furan fatty acids and Ubiquinone-8: lipid biomakers that may protect Dehalococcoides strains from free radicals. Appl Environ Microbiol. 2005;71:8426–8433. doi: 10.1128/AEM.71.12.8426-8433.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pirt SJ. The maintenance of bacteria in growing cultures. Proc R Soc Lond B Biol Sci. 1965;163:224–231. doi: 10.1098/rspb.1965.0069. [DOI] [PubMed] [Google Scholar]
- 115.Pirt SJ. Maintenance energy: a general model for energy-limited and energy-sufficient growth. Arch Microbiol. 1982;133:300–302. doi: 10.1007/BF00521294. [DOI] [PubMed] [Google Scholar]
- 116.Russell JB, Cook GM. Energetics of bacterial growth: balance of anabolic and catabolic reactions. Microbiol Rev. 1995;59:48–62. doi: 10.1128/mr.59.1.48-62.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Neijssel OM, Mattos MJTd, Tempest DW. Growth yield and energy distribution. In: Neidhardt FC, editor. Escherichia coli and Salmonella: cellular and molecular biology. Washington D C: ASM Press; 1996. [Google Scholar]
- 118.Varma A, Palsson BO. Metabolic flux balancing: basic concepts, scientific and practical use. Nat Biotechnol. 1994;12:994–998. [Google Scholar]
- 119.Price ND, Reed JL, Palsson BO. Genome-scale models of microbial cells: evaluating the consequences of contraints. Nat Rev Microbiol. 2004;2:886–897. doi: 10.1038/nrmicro1023. [DOI] [PubMed] [Google Scholar]
- 120.Reed JL, Palsson BO. Thirteen years of building constraint-based in silico models of Escherichia coli. J Bacteriol. 2003;185:2692–2699. doi: 10.1128/JB.185.9.2692-2699.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SBML File for Dehalococcoides Metabolic Model, iAI549.
(0.48 MB XML)
This file contains the detailed list of genes, proteins, reactions and metabolites included in the Dehalococcoides pan-metabolic-model, iAI549. Tables 3–7 are gene correspondences where a unique gene number is provided for each gene in the model (Model gene number) so that a gene or corresponding reaction can be located conveniently irrespective of the 4 genomes of interest. Strain VS gene locus names are obtained from Alfred Spormann at Stanford University, CA. A reaction can be associated with more than one gene where some genes are core, some are dispensable and others can be unique. For such instances, 2 additional gene correspondence tables (Table 5 and Table 7) are created for dispensable and unique genes respectively.
(5.41 MB PDF)
This file contains the tables of detailed macromolecular composition of a gram of Dehalococcoides cell, experimental values of various pan-model parameters and the detailed procedure to calculate those parameters, as well as supplemental text regarding energy conservation process of Dehalococcoides. In addition, all the supplemental figures are included at the end of this document.
(0.31 MB PDF)