Abstract
Metagenomic and bioinformatic approaches were used to characterize plant biomass conversion within the foregut microbiome of Australia's “model” marsupial, the Tammar wallaby (Macropus eugenii). Like the termite hindgut and bovine rumen, key enzymes and modular structures characteristic of the “free enzyme” and “cellulosome” paradigms of cellulose solubilization remain either poorly represented or elusive to capture by shotgun sequencing methods. Instead, multigene polysaccharide utilization loci-like systems coupled with genes encoding β-1,4-endoglucanases and β-1,4-endoxylanases—which have not been previously encountered in metagenomic datasets—were identified, as were a diverse set of glycoside hydrolases targeting noncellulosic polysaccharides. Furthermore, both rrs gene and other phylogenetic analyses confirmed that unique clades of the Lachnospiraceae, Bacteroidales, and Gammaproteobacteria are predominant in the Tammar foregut microbiome. Nucleotide composition-based sequence binning facilitated the assemblage of more than two megabase pairs of genomic sequence for one of the novel Lachnospiraceae clades (WG-2). These analyses show that WG-2 possesses numerous glycoside hydrolases targeting noncellulosic polysaccharides. These collective data demonstrate that Australian macropods not only harbor unique bacterial lineages underpinning plant biomass conversion, but their repertoire of glycoside hydrolases is distinct from those of the microbiomes of higher termites and the bovine rumen.
Keywords: cellulases, marsupials, metagenomics, plant biomass conversion, polysaccharide utilization loci
Australia possesses the largest share of the world's extant marsupial species, which diverged from other eutherian mammals ≈150 million years ago. Most likely, the most widely recognized members of this group are the macropods (kangaroos and wallabies). The macropods also evolved in geographical isolation of other eutherian herbivores, and although they are often compared with ruminants, the various macropod species show a wide range of unique adaptations to herbivory. These differences include their dentition and mastication of food, as well as the anatomical adaptations of the forestomach that supports a cooperative host–microbe association that efficiently derives nutrients from plant biomass rich in lignocellulose (1). Compared with ruminant species, the hydrolytic and fermentative processes these microbes provide must be relatively rapid because of the continuous transit of plant biomass through the herbivore gut (2, 3). There is also a widespread belief—developed from several studies during the late 1970s—that Australian macropods generate less methane during feed digestion than ruminant herbivores (4, 5), indicative of some novel host and microbe adaptations of the macropods to herbivory. Indeed, the limited studies published to date suggest the foregut microbiomes of macropods possess unique protozoal, bacterial, and archaeal microorganisms (6–8); however, very little is currently known about the genetic potential and structure–function relationships intrinsic to these microbiomes.
Metagenomics offers new opportunities to interrogate and understand this interesting host–microbe association. We present here a compositional and comparative analysis of metagenomic data pertaining to plant biomass hydrolysis by the foregut microbiome of Australia's model marsupial: the Tammar wallaby (Macropus eugenii). Several unique bacterial lineages were identified and nucleotide composition-based sequence binning using Phylopythia facilitated the production of a 2.3 Mbp assemblage of DNA representing one of the unique Lachnospiraceae clades present in this community. Further in silico analysis revealed this clade harbors numerous putative glycoside hydrolases (GHs) specifically targeting the side chains attached to noncellulosic polysaccharides.
Results and Discussion
Microbial Diversity Resident in the Tammar Wallaby Foregut.
An inventory of the various metagenomic resources created and analyzed as part of this study are summarized in Table S1. The rrs gene library is comprised of 663 bacterial sequences and included 236 phylotypes (using a 97% sequence identity threshold). Rarefaction analysis showed the bovine and macropod datasets afforded a similar degree of coverage of the biodiversity present in these microbiomes (Fig. S1). The overall community profile at a phylum-level is similar to that of other vertebrate herbivores, with representatives of the Firmicutes and Bacteroidetes being predominant (Fig. S2A). However, the majority of these phylotypes were only distantly related to any of the cultivated species from other gut microbiomes (Table S2). Furthermore, the comparison of these datasets via unweighted measures of β diversity, UniFrac analysis, and operational taxonomic unit (OTU) network maps clearly showed host-specificity, with only a small number of OTUs shared between the bovine and macropod microbiomes, and no OTUs shared with the termite sample (Fig. 1). We were also able to separate the macropod rrs gene library data with respect to time of collection, which revealed that the microbiome appeared to be more diverse in spring, most likely because of the availability of forb species during spring offering a greater amount of soluble carbohydrates, as compared with highly lignocellulosic biomass present in drier times of the year (Fig. 1 and Table S2). There were also five distinctive phylotypes identified from the rrs gene libraries: two of these were assigned as deeply branching, unique members of the γ-subdivision of Proteobacteria (hereafter referred to as Wallaby Group-1, WG-1); two more were positioned as a deeply branching and unique lineage within the Lachnospiraceae (hereafter referred to as Wallaby Group 2, WG-2); the last of these was a unique member of the Erysipelotrichaceae (Mollicutes), and is hereafter referred to as Wallaby Group-3, (WG-3) (Fig. S3 and Table S2). The archaeal populations have already been described in an earlier study (8) and are considerably smaller than those typically encountered in ruminants, and might be a key reason explaining the apparent differences between ruminants and macropods in terms of methane production during feed digestion (2, 4, 5).
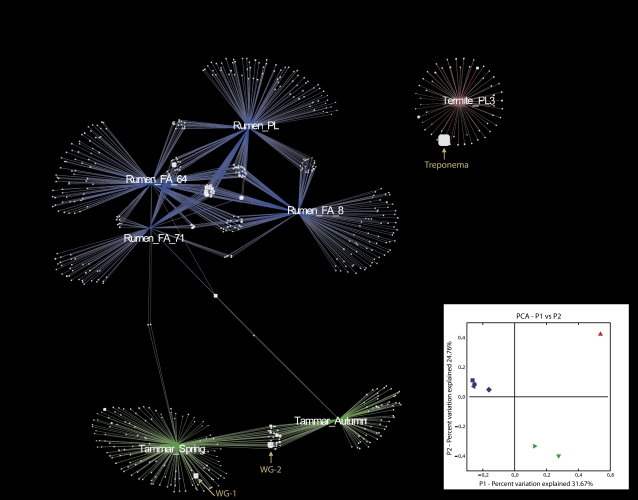
Fig. 1.
OTU network map showing OTU interactions between all rarefied samples from the Tammar wallaby (spring and autumn), rumen, and termite. Lines radiating from samples Rumen _FA_8, Rumen_FA_64, Rumen_FA_71, and Rumen_PL are colored blue (fiber-associated fraction and pooled liquid-associated, respectively, from ref. 13), Termite_PL3 colored red [termite lumen study (12)] and Tammar_Spring (T1) and Tammar_Autumn (T2) colored green (present study) are weighted with respect to contribution to the OTU. OTU size is weighted with respect to sequence counts within the OTU. (Inset) The first two principal coordinate axes (PCoA) for the unweighted UniFrac analysis colored by host animal: Rumen (FA_8, ■; FA_64, •; FA_71, ◆; PL,  ) blue; Termite (▲) red and Tammar (Spring,
) blue; Termite (▲) red and Tammar (Spring,  ; Autumn,▼) green. For complete inventory and comparisons between the two Tammar wallaby sample dates at an OTU definition (SONS analysis), see Table S2.
; Autumn,▼) green. For complete inventory and comparisons between the two Tammar wallaby sample dates at an OTU definition (SONS analysis), see Table S2.
Similar conclusions were drawn from phylogenetic analysis of the Sanger shotgun sequence data. First, MEGAN (9) was used to perform a phylogenetic assignment of the first round of metagenomic data generated, which represented ≈30% of the total data produced (Fig. S2B). The majority of these reads were assigned to the Firmicutes, Bacteroidetes, and the γ-subdivision of the Proteobacteria. We subsequently developed a collaborative partnership with the McHardy group and used the composition-based classifier Phylopythia (10) to examine the complete dataset, once it was produced. The fosmid libraries produced as part of this study were used to provide ~2.5 Mbp of training sequence for Phylopythia, which resulted in the classification of 76% of the contigs to at least the phylum level (Table 1). Again, the assignments favored the Firmicutes, Bacteroidetes, and γ-subdivision of the Proteobacteria, and confirmed the predominance of the WG-1, WG-2, and WG-3 populations in the metagenomic data (Table 1). Indeed, these three groups, which comprise ~34% of the sequences that comprise the rrs gene libraries (Table S2), also accounted for ~22% of the total Phylopythia assignments. A small number of reads were also assigned to the Euryarchaeota and, in particular, Methanobrevibacter sp., consistent with the small population size of archaea measured for these same animals in ref. 8. Similar to the results presented for the termite and bovine microbiomes (12, 13), a very small number of sequences were also assigned to the Cyanobacteria.
Table 1.
Phylogenetic profile of the Tammar wallaby metagenome sequence dataset, based on sequence composition-based binning using Phylopythia
| Taxonomic group | # Fragments | %* | #bp | %* |
| Bacteria | 9,648 | 76 | 19,776,692 | 86 |
| Actinobacteria | 110 | 1 | 169,998 | 1 |
| Bacteroidetes | 1,046 | 8 | 2,161,259 | 9 |
| Bacteroidales | 715 | 6 | 1,668,287 | 7 |
| Cyanobacteria | 14 | <1 | 25,871 | <1 |
| Firmicutes | 3,714 | 29 | 9,171,159 | 39 |
| Bacilli | 41 | <1 | 75,915 | <1 |
| Clostridia | 2,757 | 22 | 7,110,958 | 30 |
| Lachnospiraceae | 1,860 | 17 | 5,681,224 | 24 |
| Uncultured Lachnospiraceae bacterium (WG-2) | 482 | 4 | 2,266,276 | 10 |
| Erysipelotrichi | 700 | 5 | 1,651,819 | 7 |
| Erysipelotrichaceae | 694 | 5 | 1,638,325 | 7 |
| Uncultured Erysipelotrichaceae bacterium (WG-3) | 355 | 3 | 881,678 | 4 |
| Fusobacteria | 17 | <1 | 21,277 | <1 |
| Proteobacteria | 854 | 7 | 2,890,009 | 12 |
| Gammaproteobacteria | 450 | 4 | 2,180,396 | 9 |
| Aeromonadales | 425 | 3 | 2,120,188 | 8 |
| Uncultured bacterium (WG-1) | 366 | 3 | 1,995,748 | 8 |
| Spirochaetes | 34 | <1 | 60,615 | <1 |
| Tenericutes | 111 | 1 | 163,215 | 1 |
| Archaea | 752 | 6 | 970,797 | 4 |
| Euryarchaeota | 325 | 3 | 431,247 | 2 |
| Eukaryota | 77 | <1 | 133,072 | 1 |
| Other | 135 | 1 | 162,443 | 1 |
| Unclassified | 2,187 | 17 | 1,709,141 | 7 |
| TOTAL | 12,664 | 22,961,806 |
*Percentages are given at different taxonomic levels, therefore add up to more than 100%; data in shaded rows assigned to the sample-specific classes for the WG-1 (uncultured γ-Proteobacteria bacterium), WG-2 (uncultured Lachnospiraceae bacterium) and WG-3 (uncultured Erysipelotrichaceae bacterium) clades of the model; all other assignments are to classes trained from publicly available data.
Despite these encouraging results, ≈60% of the Sanger reads subjected to MEGAN analysis and 61% of the Phylopythia assignments could not be extended deeper than an Order-level of classification, with ~20% having no assignments at any level. This “shallow” level of binning by both methods confirms the wallaby foregut microbiome is comprised of unique bacterial lineages, with only limited similarity to the (meta)genomic data derived from other microbial habitats and cultured isolates.
Tammar Foregut Microbiome Possesses a Different Repertoire of GH Genes and Related Modules, Compared with Other Herbivore Microbiomes.
The Tammar wallaby is a small macropod (4–10 kg), and primarily utilizes grasses and forbs as its principal source of energy nutrition (2). For many months of the year, such plant material is characteristically rich in lignocellulose and noncellulosic polysaccharides. The metagenomic data were subjected to automated annotation using Joint Genome Institute-Department of Energy's (JGI-DOE) integrated microbial genomes with microbiome samples (IMG/M) system; next, select functional categories were manually compared with the global hidden Markov models (HMMs) available via Pfam. These analyses recovered over 600 genes and modules from 53 different CAZy (Carbohydrate-Active EnZymes) families (11) (Table S3), but relatively few of these produced strong matches with endo- or exo-acting β-1,4-glucanases. Only 24 GH5 β-1,4-endoglucanases were identified from the metagenomic data, along with a smaller number of gene modules assigned to the GH6 and GH9 families (Table 2 and Table S3). In addition to these presumptive “cellulases,” the metagenomic data produced 25 sequences matching GH94 (cellobiose phosphorylase) catalytic modules. The number of “xylanase” genes identified in the metagenomic dataset was evenly distributed among the GH10, GH26, and GH43 families (Table 2). Furthermore, genes matching CE4 and AXE1 (Pfam) acetyl xylan esterases, “accessory enzymes” that are part of the xylanolytic system responsible for the complete hydrolysis of xylan, were also identified (Table S3). Interestingly, the GH11 xylanases, which are found in abundance among members of the Firmicutes, especially Clostridium and Ruminococcus spp., as well as specialist cellulolytic bacteria from other gut microbiomes, were absent from our datasets.
Table 2.
GH profiles targeting plant structural polysaccharides in three herbivore metagenomes
| Macropod | Termite | Bovine | Fosmids | |
| Cellulases | ||||
| GH5 | 10 | 56 | 8 | 14 |
| GH6 | 0 | 0 | 0 | 1 |
| GH7 | 0 | 0 | 0 | 0 |
| GH9 | 0 | 9 | 6 | 2 |
| GH44 | 0 | 6 | 0 | 0 |
| GH45 | 0 | 4 | 0 | 0 |
| GH48 | 0 | 0 | 0 | 0 |
| Total | 10 (2) | 75 (11) | 14 (2) | 17 |
| Endohemicellulases | ||||
| GH8 | 1 | 5 | 4 | 1 |
| GH10 | 11 | 46 | 7 | 3 |
| GH11 | 0 | 14 | 1 | 0 |
| GH12 | 0 | 0 | 0 | 0 |
| GH26 | 5 | 15 | 5 | 8 |
| GH28 | 2 | 6 | 5 | 1 |
| GH53 | 9 | 12 | 17 | 0 |
| Total | 28 (5) | 98 (14) | 39 (4) | 13 |
| Debranching enzymes | ||||
| GH51 | 12 | 18 | 64 | 1 |
| GH54 | 0 | 0 | 1 | 0 |
| GH62 | 0 | 0 | 0 | 0 |
| GH67 | 5 | 10 | 0 | 0 |
| GH78 | 25 | 0 | 34 | 0 |
| Total | 42 (8) | 18 (3) | 99 (10) | 1 |
| Oligosaccharide-degrading enzymes | ||||
| GH1 | 61 | 22 | 10 | 1 |
| GH2 | 24 | 23 | 186 | 4 |
| GH3 | 72 | 69 | 176 | 3 |
| GH29 | 2 | 0 | 74 | 1 |
| GH35 | 3 | 3 | 12 | 1 |
| GH38 | 3 | 11 | 17 | 0 |
| GH39 | 1 | 3 | 2 | 0 |
| GH42 | 8 | 24 | 11 | 1 |
| GH43 | 10 | 16 | 61 | 9 |
| GH52 | 0 | 3 | 0 | 0 |
| Total | 184 (33) | 174 (25) | 549 (57) | 20 |
| % ORFS | 0.71 | 0.78 | 0.78 | |
Data are presented using the format described in ref. 40, with the GHs grouped according to their major functional role in the degradation of plant fiber. The numbers in parentheses represent the percentages of these groups relative to the total number of GH’s identified in the metagenomic datasets [557 for macropod, 704 for termite (ref. 12), and 957 for bovine (ref. 13)]. A complete inventory of the GHs recovered from the Tammar wallaby foregut microbiome is presented in Table S3. The column annotated as fosmids represents the number of additional GH genes identified from sequencing fosmid clones, as described in the materials and methods and results.
Comparative analysis of the repertoire of GH families recovered from the Tammar, termite hindgut, and bovine rumen metagenomes revealed some interesting similarities and differences. The GH5 cellulases were numerically most abundant in the wallaby and termite metagenomes, with less representation of the GH9 family (Table 2). In contrast, the bovine metagenomic dataset was more evenly balanced with respect to these two GH families (Table 2) (12, 13). Similar to the rumen, the wallaby foregut microbiome possessed a large number of reads matching GH families specific for xylooligosaccharides and the side chains attached to noncellulosic polysaccharides (Table 2). The most abundant were GH1, GH2, and GH3 β-glycosidases, as well as matches with GH51 and GH67 enzymes, which typically target glucuronic acid and arabinose-containing side chains, respectively. The Tammar metagenome also contained a range of carbohydrate-active enzymes targeting pectic polysaccharides, plant pigments, gums, glycolipids, and other glycosides, including GH78 rhamnosidases, CE8 pectin methylesterases, several GH28 rhamnogalacturonases, and a pectate lyase (PL1) (Table S3). These findings were not entirely unexpected, given the dietary profiles of the macropod (predominantly grass and forbs, with a small amount of a commercial pellet mix) compared with termites (wood); the findings also partially explain the higher abundance of GH genes that catalyze the hydrolysis of the side chains of noncellulosic plant polysaccharides in the grass/legume feeding herbivores, compared with wood-eating termites (13).
However, and despite the differences in nutritional ecology, gut anatomy, and microbiome structure, probably the most notable observation drawn from all these datasets is the virtual absence of genes encoding GH6, GH7, and GH48 β-1,4-exoglucanases (Table 2), which are essential in virtually all cultured bacteria and fungi for cellulose solubilization, as well as the dearth of cellulosome-associated modules, such as cohesins and dockerins (Table S3). Although the wallaby metagenome dataset does contain 42 Type I dockerin modules, all these modules were linked to hypothetical sequences of unknown function, with no examples linked to recognized GH catalytic modules, other carbohydrate-active enzymes, or serpins. Such findings suggest there is still much to learn about cellulose hydrolysis and dockerin-cohesin-mediated complex assemblies in gut microbiomes.
Identification of Unique Polysaccharide Utilization Loci-Like Gene Clusters Associated with Cellulase Genes in the Sequenced Fosmid Clones.
There were 33 fosmids selected for 454 pyrosequencing on the basis that their inserts encode gene products resulting in carboxymethylcellulase or xylanase activity visualized in plate-screen assays. Phylopythia assigned the majority of the scaffolds produced from these clones to the Bacteroidales or Lachnospiraceae (Table S4). Twelve of these scaffolds possess genes encoding a GH5 catalytic module, two more encode a gene with a GH9 catalytic module, and one encodes a gene with a GH6 catalytic module. Interestingly, half of the scaffolds assigned to the Bacteroidales also possessed genes homologous to the polysaccharide utilization loci (PULs) present in the genomes of Bacteroides and related genera (14–18). The presumptive PUL-like gene arrangement borne by one of these fosmids (annotated in Table S4 as part of scaffold 78) is shown as an example in Fig. 2, along with a hypothetical functional model of the cluster. In brief detail, the PUL-like gene cluster consists of an AraC-like regulatory protein, a putative acetylxylan esterase and two genes with homology to the Bacteroides thetaiotaomicron susC (tonB) and susD genes. These latter two genes were initially defined as part of the starch utilization system (sus) of B. thetaiotaomicron (19, 20). The SusC protein is a tonB-dependent receptor family member, a group of outer membrane-spanning proteins that can import solutes and macromolecules into the periplasm (21, 22); the SusD protein coordinates polysaccharide binding at the cell surface (20). Two genes located directly downstream from the susC and susD homologs were predicted to be outer membrane lipoproteins and therefore might play a role similar to the B. thetaiotaomicron SusE and SusF proteins, whose functional role is currently unknown. The remaining six genes in this cluster encode putative glycoside hydrolases and a putative inner-membrane-bound “sugar transporter.” Although PULs were not readily assembled from our Sanger sequence data, there were 36 susC and 42 susD genes identified in the dataset (Table S3). For these reasons, we propose that the sus-like PULs represent a key adaptation to growth on cellulose and other polysaccharides by the large number of Bacteroidetes resident in the wallaby foregut. Interestingly, sus gene homologs were not identified in the termite hindgut and bovine rumen data, presumably because of the lower representation of Bacteroidetes in the termite and the short read lengths in the bovine dataset.
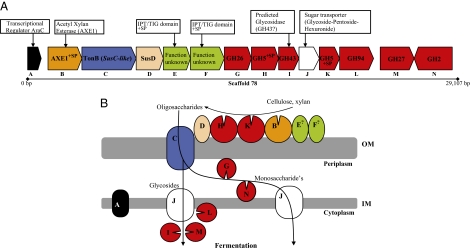
Fig. 2.
Gene arrangement in the Bacteroidales-affiliated fosmid and a hypothetical model of polysaccharide-adhesion and hydrolysis coordinated by this gene cluster. (A) Phylopythia affiliated the fosmid clone from which scaffold 78 is derived to the order Bacteroidales, as described in the text. The putative PUL gene cluster consists of an AraC family transcriptional regulator (geneA), an acetylxylan esterase (geneB), susC and susD gene homologs (genes C and D, respectively), and two genes encoding outer membrane-targeted lipoproteins (genes E and F). Genes G, H, and I encode proteins containing GH26, GH5, and GH43 catalytic modules, respectively. Gene J encodes a putative inner-membrane bound “sugar transporter” followed by genes K and L, which encode proteins containing GH5 and GH94 catalytic modules, respectively. (B) The hypothetical model predicts that polysaccharides are bound by the outer membrane-associated components, principally via the SusD homolog in a complex with the SusC, and the two lipoproteins. The GH5-containing proteins generate oligosaccharides, which are transported across the outer membrane, principally via the protein complex described above. These oligosaccharides may be further hydrolyzed by periplasmic GHs or transported to the cytoplasm via a glycoside sugar transporter (encoded by gene J), before hydrolysis by glycoside hydrolases (gene M and I) or terminal phosphorolytic cleavage by the GH94 glycoside phosphorylase (encoded by gene L).
Phylopythia-Supported Metabolic Reconstruction of the WG-2 Population.
Phylopythia supported a 2.3 Mbp assemblage of metagenome fragments assigned to the WG-2 population (Fig. S4 and Table S5). The current assemblage includes 20 different families of carbohydrate-active enzymes principally involved with the hydrolysis of noncellulosic polysaccharides and pectin. However, none of the sequences encoding dockerin modules were assigned to WG-2, suggesting no cellulosome complex assembly by this population. The assemblage includes genes encoding homologs of GH1, GH2, GH3, GH27, and GH42 catalytic modules, as well as several GH5 endoglucanases and GH94 cellobiose phosphorylases. Five GH43 arabinoxylosidases and several acetyl esterases genes (CE12) contiguous with GH78 rhamnosidases were also assigned to WG-2 (Table S5). Interestingly, arabinose-rich rhamnogalacturonan side chains have been speculated to play an essential role for some plant species to tolerate severe desiccation (23, 24). Given that many of Australia's native plant species are drought-tolerant or drought-resistant, WG-2 might have evolved to specialize in the hydrolysis and use of these types of poly- and oligosaccharides for growth. Indeed, Phylopythia also assigned genes encoding xylose isomerase and xylulokinase enzymes to the WG-2 assemblage, as well as acetate and butyrate kinases. From these data, we propose that the WG-2 population plays a quantitatively important role in both the degradation and fermentation of the pentoses derived from noncellulosic polysaccharides, and produces acetate and butyrate as fermentation end-products.
Australia's native herbivores are recognized throughout the world for their unique attributes in diversity, form, and function, but our understanding of their evolutionary adaptations for niche occupation has been compromised because we had virtually no understanding of their gut microbiomes, which contribute greatly to the nutrition and well-being of these animals. Our metagenomic analyses of the Tammar wallaby foregut microbiome clearly shows these animals are the host for unique bacterial lineages that are numerically predominant within the microbiome. For example, the WG-2 lineage appears to play a key role in the deconstruction of noncellulosic poly- and oligosaccharides by producing a large number of enzymes targeting both heteroxylans and pectins. Furthermore, the functional screening of the fosmid libraries for cellulases and xylanases recovered clones assigned to the Bacteroidetes encoding PUL-like gene clusters, including susC and susD gene homologs linked with GH5 and/or GH10 genes. Such findings distinguish the Tammar wallaby foregut microbiome from that of the bovine rumen (predominantly Clostridiales and Prevotellas) and the termite hindgut (Fibrobacteres and Spirochetes). The collective findings from this and other metagenomic studies also still need to be reconciled with the extensive literature developed from the biochemical, molecular, and genomic analyses of specialist gut bacteria and fungi, which have created the cellulosome and free enzyme paradigms of cellulose solubilization. These paradigms are underpinned by a restricted number of known GH families, which remain poorly represented in metagenomic data. Much still remains to be learned about the structure-function relationships of these interesting microbiomes.
Materials and Methods
Wallaby Sampling.
The eight adult females (aged between 1.5 and 4 y) sampled for this study were all from the same colony maintained near Canberra, Australia. Three animals were sampled in November 2006 (late spring: T1) and another five in May 2007 (late autumn: T2). During this period the animals were provided free range access to pastures composed predominantly of Timothy Canary grass (Phalaris angusta) and were also provided with a commercial pellet mix containing wheat, bran, pollard, canola, soy, salt, sodium bicarbonate, bentonite, lime, and a vitamin premix (Young Stockfeeds). Animals were killed with an overdose of pentobarbitone sodium (Commonwealth Scientific and Industrial Research Organization Sustainable Ecosystems Animal Ethics Approval Number 06–20) and foregut contents were either transferred to sterile containers and immediately frozen at −20 °C, or mixed 1:1 with phenol:ethanol (5%:95%).
Cell Dissociation and DNA Extraction.
Before cell dissociation and DNA extraction, a subsample of each digesta sample was pooled and hereafter is referred to as T1 (November 2006) and T2 (May 2007). To desorb and recover those microbes adherent to plant biomass, 5 to 10 g of the pooled samples was centrifuged at 12,000 × g for 2 min, and the pellet was resuspended in dissociation buffer and subjected to a dissociation procedure described by ref. 25 (details provided in SI Materials and Methods). HMW DNA was extracted using a gentle enzymatic lysis procedure (details provided in SI Materials and Methods).
16S rRNA Gene PCR Clone Libraries.
Two rrs clone libraries were prepared from the metagenomic DNA samples extracted from T1 and T2 by using two different primer pairs broadly targeting the bacterial domain: 27F (5′-AGA GTT TGA TCC TGG CTC AG-3′) and 1492R (5′-GGT TAC CTT GTT ACG ACT T-3′); and GM3 (5′-AGA GTT TGA TCM TGG C-3′) and GM4 (5′-TAC CTT GTT ACG ACT T-3′) (12) (details provided in SI Materials and Methods). Similar attempts produced archaeal rrs gene libraries with results described in ref. 8. A total of 663 near-complete bacterial rrs gene sequences passed the quality and chimera filters and were used in the subsequent analyses (details provided in SI Materials and Methods).
Phylogenetic Analysis of 16S rRNA Gene Sequences.
The 663 sequences were aligned using the NAST aligner (26) and imported into an ARB database with the same alignment (http://greengenes.lbl.gov/) (27). Fifty-one partial and near complete 16S sequences were extracted from the Tammar metagenomic data set aligned using NAST aligner and also imported into ARB (28). Sequences were initially assigned to phylogenetic groups using the ARB Parsimony insertion tool. Phylogenetic trees (Fig. S2A and Fig. S3) were constructed from masked ARB alignments (to remove ambiguously alignable positions) using RAxML (29) and bootstrap analysis using parsimony and neighbor-joining was performed using 100 replicates. The phylum-level trees (Fig. S3) were reconstructed using TREE-PUZZLE (30) in ARB. The rrs gene sequences from the two libraries were assigned to phylotypes (OTUs) at 97 and 99% sequence identity thresholds using the DOTUR package (31) (Table S1) and comparisons at an OTU definition was calculated as a percentage using SONS (32) (Table S2). Additional phylogenetic comparisons and diversity estimates were performed using the QIIME package (Quantitative Insights Into Microbial Ecology) (33), with OTUs at the 97% sequence identity threshold used (Fig. S1). Sample heterogeneity was removed by rarefaction before comparison of Tammar rrs gene sequences with rumen and termite samples. The OTU network maps were generated using QIIME and visualized with Cytoscape (34). In addition, α diversity [PD_Whole_Tree (35), observed species count and Chao1 richness estimators] and β diversity (unifrac weighted and unweighted) metrics along with rarefaction plots were also calculated using QIIME.
Metagenome Processing: Shotgun Library Preparation, Sequencing, and Assembly.
Shotgun libraries from the Tammar genomic DNA were prepared from each of the pooled samples T1 and T2: a 2- to 4-kb insert library cloned into pUC18 and a roughly 36 kb insert fosmid library cloned in pCC1Fos (Epicentre Corp.). Libraries were sequenced with BigDye Terminators v3.1 and resolved with ABI PRISM 3730 (ABI) sequencers. Subsequent sequences were assembled with the Paracel Genome Assembler (PGA version 2.62, www.paracel.com) (details provided in SI Materials and Methods).
Full Fosmid Sequencing and Assembly.
Based on a number of functional and hybridization-based screens, 98 fosmids were chosen for sequencing. The individual fosmids were induced to increase their copy number following Epicentre protocols, and the fosmid DNA purified using Qiagen MiniPrep columns. Equimol amounts of the fosmids were pooled together (~20 μg total DNA) and both a 3-kb paired-end library and a 454 standard shotgun library were constructed. Both libraries were directly sequenced with the 454 Life Sciences Genome Sequencer GS FLX and assembled using Newbler (details provided in SI Materials and Methods).
Gene Prediction.
Putative genes in the Tammar wallaby gut microbiome metagenome were called with GeneMark (36) and putative genes in the fosmid assemblies were called with a combination of MetaGene (37) and BLASTX. All called genes were annotated via the IMG/M-ER annotation pipeline and loaded as independent data sets into IMG/M-ER (38) (http://img.jgi.doe.gov/cgi-bin/m/main.cgi), a data-management and analysis platform for genomic and metagenomic data based on IMG (39).
Binning.
MEGAN was used to determine the phylogenetic distribution of the first batch of 30,000 Sanger reads generated by the CSP program. BLASTX was used to compare all reads against the NCBI-NR (“non-redundant”) protein database. Results of the BLASTX search were subsequently uploaded into MEGAN (9) for hierarchical tree constructions which uses the BLAST bit-score to assign taxonomy, as opposed to using percentage identity. Assembled metagenomic contigs were binned (classified) using PhyloPythia (10). Generic models for the ranks of domain, phylum, and class were combined with sample-specific models for the clades “uncultured γ-Proteobacteria bacterium” (WG-1), “uncultured Lachnospiraceae bacterium” (WG-2), and “uncultured Erysipelotrichaceae bacterium” (WG-3) (details provided in SI Materials and Methods).
GH and Carbohydrate-Binding Modules: Annotation and Phylogenetic Analysis.
Searches for GHs and carbohydrate-binding modules (CBMs) were performed as described in ref. 12. Briefly, database searches were performed using HMMER hmmsearch with Pfam_Is HMMs (full-length models) to identify complete matches to the family, which were named in accordance with the CAZy nomenclature scheme (11). All hits with E-values less than 10−4 were counted and their sequences further analyzed. For those GH and CBM families for which there is currently no Pfam HMM, the representative sequences selected from the CAZy Web site and described by ref. 12 were used in BLAST searches of the metagenomic data to identify these GH and CBM families. An E-value cutoff of 10−6 was used in these searches. For phylogenetic analysis of selected GH families (GH5: Fig. S5), sequence alignments were first produced using HMMER hmmalign and to the corresponding Pfam HMM; next, a protein maximum likelihood program used with the Jones-Taylor-Thornton probability model of change between amino acids was applied to these data.
Identification of Fosmid Clones Bearing GH Genes.
Fosmid clones bearing β-1,4-endoglucanase and/or β-1,4-xylanase activity were detected by plating the Escherichia coli library on LB-chloramphenicol agar plate medium containing either 0.2% (wt/vol) carboxymethylcellulose or birchwood xylan (Sigma). Approximately 20,000 recombinant strains were plated in a 384-well format and incubated overnight at 37 °C. The plates were then stained with Congo red dye and destained with 1 M NaCl to reveal zones of hydrolysis. Positive colonies were isolated and reexamined to confirm activity. Twenty-seven fosmid clones positive for carboxymethylcellulose hydrolysis and six positive for xylan hydrolysis were selected for 454 pyrosequencing and assembly.
Supplementary Material
Acknowledgments
We are especially grateful to the support from Lyn Hinds (Commonwealth Scientific and Industrial Research Organization Australia) who assisted in sample collection. The Tammar wallaby project is partially supported by the Commonwealth Scientific and Industrial Research Organization's Office of the Chief Executive Science Leader program (M.M.), a Commonwealth Scientific and Industrial Research Organization Office of the Chief Executive Postdoctoral Fellowship (to P.B.P.), and the US Department of Energy-Joint Genome Institute Community Sequencing Program. This work was performed in part under the auspices of the US Department of Energy's Office of Science, Biological, and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under Contract DE-AC02-05CH11231, Lawrence Livermore National Laboratory under Contract DE-AC52-07NA27344, and Los Alamos National Laboratory under contract DE-AC02-06NA25396.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: 16S rRNA gene sequences are deposited under GenBank accession numbers GQ358225–GQ358517. Metagenome is deposited under GenBank accession number ADGC00000000.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1005297107/-/DCSupplemental.
References
- 1.Hume ID. Microbial fermentation in herbivorous marsupials. Bioscience. 1984;34:435–440. [Google Scholar]
- 2.Smith JA. Macropod nutrition. Vet Clin North Am Exot Anim Pract. 2009;12:197–208, xiii. doi: 10.1016/j.cvex.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 3.Flint HJ. The rumen microbial ecosystem—some recent developments. Trends Microbiol. 1997;5:483–488. doi: 10.1016/S0966-842X(97)01159-1. [DOI] [PubMed] [Google Scholar]
- 4.Kempton TJ, Murray RM, Leng RA. Methane production and digestibility measurements in the grey kangaroo and sheep. Aust J Biol Sci. 1976;29:209–214. doi: 10.1071/bi9760209. [DOI] [PubMed] [Google Scholar]
- 5.Engelhardt W, Wolter S, Lawrenz H. Production of methane in two non-ruminant herbivores. Comp Biochem Physiol. 1978;60:309–311. [Google Scholar]
- 6.Dehority BA. A new family of entodiniomorph protozoa from the marsupial forestomach, with descriptions of a new genus and five new species. J Eukaryot Microbiol. 1996;43:285–295. doi: 10.1111/j.1550-7408.1996.tb03991.x. [DOI] [PubMed] [Google Scholar]
- 7.Ouwerkerk D, Klieve AV, Forster RJ, Templeton JM, Maguire AJ. Characterization of culturable anaerobic bacteria from the forestomach of an eastern grey kangaroo, Macropus giganteus. Lett Appl Microbiol. 2005;41:327–333. doi: 10.1111/j.1472-765X.2005.01774.x. [DOI] [PubMed] [Google Scholar]
- 8.Evans PN, et al. Community composition and density of methanogens in the foregut of the Tammar wallaby (Macropus eugenii) Appl Environ Microbiol. 2009;75:2598–2602. doi: 10.1128/AEM.02436-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McHardy AC, Martín HG, Tsirigos A, Hugenholtz P, Rigoutsos I. Accurate phylogenetic classification of variable-length DNA fragments. Nat Methods. 2007;4:63–72. doi: 10.1038/nmeth976. [DOI] [PubMed] [Google Scholar]
- 11.Cantarel BL, et al. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 2009;37(Database issue):D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warnecke F, et al. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature. 2007;450:560–565. doi: 10.1038/nature06269. [DOI] [PubMed] [Google Scholar]
- 13.Brulc JM, et al. Gene-centric metagenomics of the fiber-adherent bovine rumen microbiome reveals forage specific glycoside hydrolases. Proc Natl Acad Sci USA. 2009;106:1948–1953. doi: 10.1073/pnas.0806191105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bjursell MK, Martens EC, Gordon JI. Functional genomic and metabolic studies of the adaptations of a prominent adult human gut symbiont, Bacteroides thetaiotaomicron, to the suckling period. J Biol Chem. 2006;281:36269–36279. doi: 10.1074/jbc.M606509200. [DOI] [PubMed] [Google Scholar]
- 15.Xu J, et al. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003;299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- 16.Xu J, et al. Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 2007;5:e156. doi: 10.1371/journal.pbio.0050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie G, et al. Genome sequence of the cellulolytic gliding bacterium Cytophaga hutchinsonii. Appl Environ Microbiol. 2007;73:3536–3546. doi: 10.1128/AEM.00225-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martens EC, Koropatkin NM, Smith TJ, Gordon JI. Complex glycan catabolism by the human gut microbiota: The Bacteroidetes Sus-like paradigm. J Biol Chem. 2009;284:24673–24677. doi: 10.1074/jbc.R109.022848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho KH, Salyers AA. Biochemical analysis of interactions between outer membrane proteins that contribute to starch utilization by Bacteroides thetaiotaomicron. J Bacteriol. 2001;183:7224–7230. doi: 10.1128/JB.183.24.7224-7230.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koropatkin NM, Martens EC, Gordon JI, Smith TJ. Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure. 2008;16:1105–1115. doi: 10.1016/j.str.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferguson AD, Deisenhofer J. TonB-dependent receptors-structural perspectives. Biochim Biophys Acta. 2002;1565:318–332. doi: 10.1016/s0005-2736(02)00578-3. [DOI] [PubMed] [Google Scholar]
- 22.Schauer K, Rodionov DA, de Reuse H. New substrates for TonB-dependent transport: Do we only see the ‘tip of the iceberg’? Trends Biochem Sci. 2008;33:330–338. doi: 10.1016/j.tibs.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 23.Moore JP, et al. Response of the leaf cell wall to desiccation in the resurrection plant Myrothamnus flabellifolius. Plant Physiol. 2006;141:651–662. doi: 10.1104/pp.106.077701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore JP, Farrant JM, Driouich A. A role for pectin-associated arabinans in maintaining the flexibility of the plant cell wall during water deficit stress. Plant Signal Behav. 2008;3:102–104. doi: 10.4161/psb.3.2.4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang S, Denman SE, Morrison M, Yu Z, McSweeney CS. An efficient RNA extraction method for estimating gut microbial diversity by polymerase chain reaction. Curr Microbiol. 2009;58:464–471. doi: 10.1007/s00284-008-9345-z. [DOI] [PubMed] [Google Scholar]
- 26.DeSantis TZ, Jr, et al. NAST: A multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 2006;34(Web Server issue):W394–W399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeSantis TZ, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ludwig W, et al. ARB: A software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stamatakis A, Ludwig T, Meier H. RAxML-III: A fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics. 2005;21:456–463. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- 30.Schmidt HA, Strimmer K, Vingron M, von Haeseler A. TREE-PUZZLE: Maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- 31.Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol. 2005;71:1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schloss PD, Handelsman J. Introducing SONS, a tool for operational taxonomic unit-based comparisons of microbial community memberships and structures. Appl Environ Microbiol. 2006;72:6773–6779. doi: 10.1128/AEM.00474-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shannon P, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ley RE, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borodovsky M, Mills R, Besemer J, Lomsadze A. Prokaryotic gene prediction using GeneMark and GeneMark.hmm. Curr Protoc Bioinformatics. 2003 doi: 10.1002/0471250953.bi0405s01. Chapter 4: Unit 4.5. [DOI] [PubMed] [Google Scholar]
- 37.Noguchi H, Park J, Takagi T. MetaGene: Prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res. 2006;34:5623–5630. doi: 10.1093/nar/gkl723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Markowitz VM, et al. IMG/M: A data management and analysis system for metagenomes. Nucleic Acids Res. 2008;36(Database issue):D534–D538. doi: 10.1093/nar/gkm869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Markowitz VM, et al. The integrated microbial genomes (IMG) system. Nucleic Acids Res. 2006;34(Database Issue):D344–D348. doi: 10.1093/nar/gkj024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Allgaier M, et al. Targeted discovery of glycoside hydrolases from a switchgrass-adapted compost community. PLoS ONE. 2010;5:e8812. doi: 10.1371/journal.pone.0008812. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.