Abstract
Diapause is a widespread adaptation to seasonality across invertebrate taxa. It is critical for persistence in seasonal environments, synchronizing life histories with favorable, resource-rich conditions and mitigating exposure to harsh environments. Despite some promising recent progress, however, we still know very little about the molecular modifications underlying diapause. We used transcriptional profiling to identify key groups of genes and pathways differentially regulated during pupal diapause, dynamically regulated across diapause development, and differentially regulated after diapause was pharmacologically terminated in the flesh fly Sarcophaga crassipalpis. We describe major shifts in stress axes, endocrine signaling, and metabolism that accompany diapause, several of which appear to be common features of dormancy in other taxa. To assess whether invertebrates with different diapause strategies have converged toward similar transcriptional profiles, we use archived expression data to compare the pupal diapause of S. crassipalpis with the adult reproductive diapause of Drosophila melanogaster and the larval dauer of Caenorhabditis elegans. Although dormant invertebrates converge on a few similar physiological phenotypes including metabolic depression and stress resistance, we find little transcriptional similarity among dormancies across species, suggesting that there may be many transcriptional strategies for producing physiologically similar dormancy responses.
Keywords: dormancy, insulin signaling, metabolic depression, Sarcophaga crassipalpis, stress tolerance
Insects, like a wide range of organisms, use an environmentally programmed period of dormancy, or diapause, to synchronize their life histories to exploit favorable seasons and mitigate the stresses of unfavorable seasons. Diapause is distinct from quiescence, a state induced directly by environmental stress (e.g., cold or hypoxia). Insects enter diapause before inclement conditions using seasonally predictable cues (1). Diapause is not just an arrest of development. Rather, it is a dynamic alternative developmental pathway following a stereotypic trajectory including (i) a preparatory period, (ii) initiation and maintenance of developmental arrest, and (iii) termination and transition to active development (2). Although most insect species go through diapause in one particular life stage, a variety of diapause strategies exist among species including embryonic, larval, pupal, and adult diapause. A critical question is which cellular events and biochemical pathways are conserved as a core part of diapause across these diverse strategies and which are specific to one type of diapause or another.
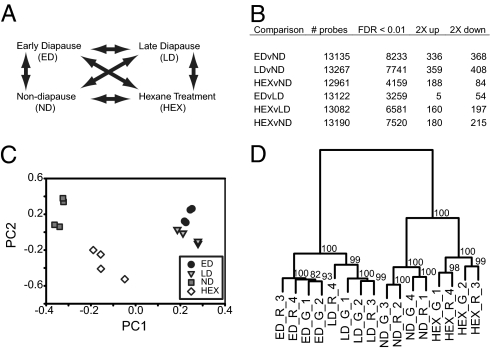
Here we use microarrays to compare transcriptome-wide gene expression between diapausing and nondiapausing flesh fly pupae and compare several stages of pupal diapause: early diapause, late diapause, and individuals treated pharmacologically with hexane to terminate diapause (Fig. 1A). In addition to identifying global differences in transcript abundance among common biochemical pathways, we directly tested a priori hypotheses about transcript abundance for several important components of the diapause program including endocrine regulation (insulin signaling and ecdysteroid signaling), metabolic depression, and stress tolerance (cold stress, heat stress, and oxidative stress). Further, we use published datasets to assess evolutionary conservation of dormancy regulation among pupal diapause in the flesh fly, adult reproductive diapause of the fly Drosophila melanogaster, and the larval dauer stage of the nematode Caenorhabditis elegans.
Fig. 1.
Comparisons of transcript abundance across diapause states in the pupae of the flesh fly, S. crassipalpis. (A) Hybridization design for all comparisons. (B) Total number of nonredundant probes and number of probes that differed in each comparison after FDR adjustment to the 0.01 level. (C) Principal components clustering. (D) Hierarchical clustering with bootstrap support.
Results and Discussion
All four developmental states—nondiapause pupae, early diapause pupae, late diapause pupae, and late diapause pupae treated with hexane to terminate diapause—were transcriptionally distinct. As expected from the dramatic physiological differences between diapausing and nondiapausing animals, 62% of ESTs differed between nondiapausing and early diapause pupae after false discovery rate (FDR) correction (Fig. 1B). Of differentially expressed ESTs, 368 were twofold or more down in early diapause versus nondiapause pupae and 336 were more than twofold up. Change in both directions, as well as our observation that approximately 25% of ESTs differed between early versus late diapause pupae, reinforces that the diapause state itself is not static, but is a physiologically dynamic alternative developmental pathway (2). For those interested in querying results for specific transcripts, a summary of the gene-by-gene analyses for each comparison is available on the corresponding author's Web site (http://entnemdept.ufl.edu/hahn/lab/index.htm).
The four developmental phenotypes each showed distinct transcriptional profiles in principal components analysis (Fig. 1C). Further, discriminate function analysis suggests that each phenotype is correctly classified with 100% accuracy using as few as two ESTs, one unannotated (EUA37Q302G01AX) and one predicted to have proteolytic activity (FLY.8601.C1; annotated to CG11956), as well as other, larger combinations (Dataset S1). Hierarchical clustering showed that early and late diapause are most transcriptionally alike, although there are clear differences between the two states (Fig. 1D). Pharmacological treatment with hexane and other organic solvents can terminate pupal diapause in several insect species within 24 h by stimulating ecdysteroid production through unknown mechanisms (3). Hexane-treated pupae are distinct from late diapause pupae and cluster with nondiapause pupae (Fig. 1D), consistent with hexane treatment causing rapid termination. Although hexane treatment yields a transcriptional phenotype consistent with diapause termination, additional work will be needed to separate expression differences resulting from diapause termination from any potential pharmacological effects of hexane treatment.
We used two different strategies for gene class enrichment analysis to organize transcriptional patterns in three focal comparisons: (i) early diapause versus nondiapause (ED vs. ND), (ii) early diapause versus late diapause (ED vs. LD), and (iii) hexane-treated, postdiapause versus late diapause (HEX vs. LD). First, we used the DAVID functional annotation database to perform an unguided analysis of enrichment, primarily among Gene Ontology (GO) categories and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways identified in D. melanogaster. Second, to complement our DAVID analyses, we performed Gene Set Analysis (GSA) on all S. crassipalpis ESTs that yielded BLAST hits with e < 10−5 to the D. melanogaster transcriptome. We applied GSA to test for (i) enrichment of KEGG pathways annotated for D. melanogaster and (ii) a priori gene lists assembled from previous transcriptome studies of candidate pathways involved in stress responses and endocrine signaling (Table S1).
Unguided DAVID analysis identified GO categories involved in stress responses and metabolic pathways as important players in diapause (Table 1). Unguided GSA analysis also showed enrichment in KEGG pathways like metabolism and DNA synthesis and repair (Table 2). We also observed enrichment in several a priori lists (Table 2), including enrichment for transcripts implicated in responses to cold stress (4), oxidative stress (5), hyperoxia (6), hypoxia (7), and ecdysteroid treatment (8).
Table 1.
Categories enriched in microarray comparisons through several stages of diapause using DAVID enrichment analysis
| Category | Term | Enrichment | FDR | Functional category |
| Enriched in early diapause vs. nondiapause | ||||
| KEGG pathway | dme00620: pyruvate metabolism | 4.66 | 0.028 | Metabolism |
| GO: cellular component | GO:0005739~mitochondrion | 1.54 | 0.076 | Metabolism |
| GO: biological process | GO:0006457~protein folding | 2.91 | <0.001 | DNA replication/development |
| GO: biological process | GO:0006952~defense response | 2.07 | 0.006 | Stress response |
| GO: biological process | GO:0006986~response to unfolded protein | 6.74 | <0.001 | Stress response |
| GO: molecular function | GO:0008483~transaminase activity | 3.97 | 0.101 | Stress response |
| GO: biological process | GO:0009266~response to temperature stimulus | 5.34 | <0.001 | Stress response |
| GO: biological process | GO:0009408~response to heat | 5.77 | <0.001 | Stress response |
| GO: biological process | GO:0009607~response to biotic stimulus | 2.36 | 0.014 | Stress response |
| GO: biological process | GO:0009628~response to abiotic stimulus | 2.49 | 0.008 | Stress response |
| GO: molecular function | GO:0016769~ nitrogenous transferase activity | 3.85 | 0.092 | Stress response |
| GO: biological process | GO:0042221~response to chemical stimulus | 2.18 | <0.001 | DNA replication/development |
| GO: biological process | GO:0044262~cellular carbohydrate metabolic process | 1.88 | 0.038 | Metabolism |
| GO: biological process | GO:0050896~response to stimulus | 1.42 | 0.066 | DNA replication/development |
| GO: molecular function | GO:0051082~unfolded protein binding | 3.53 | 0.002 | Stress response |
| GO: biological process | GO:0051789~response to protein stimulus | 6.74 | <0.001 | Stress response |
| Enriched in early diapause vs. late diapause | ||||
| GO: cellular component | GO:0005791~rough endoplasmic reticulum | 29.65 | 0.059 | DNA replication/development |
| GO: cellular component | GO:0005832~chaperonin-containing T-complex | 82.59 | <0.001 | Stress response |
| GO: biological process | GO:0006457~protein folding | 11.50 | <0.001 | Stress response |
| GO: cellular component | GO:0030867~rough endoplasmic reticulum membrane | 35.04 | 0.052 | DNA replication/development |
| GO: molecular function | GO:0051082~unfolded protein binding | 15.58 | 0.008 | Stress response |
| Enriched in diapause termination (hexane treated) vs. late diapause | ||||
| GO: cellular component | GO:0005656~prereplicative complex | 8.58 | 0.076 | DNA replication/development |
| GO: molecular function | GO:0043138~3′-5′ DNA helicase activity | 9.71 | <0.001 | DNA replication/development |
| GO: cellular component | GO:0044422~organelle part | 1.27 | 0.099 | DNA replication/development |
| GO: cellular component | GO:0044428~nuclear part | 1.64 | 0.044 | DNA replication/development |
Table 2.
Categories enriched in microarray comparisons through several stages of diapause using GSA enrichment analysis
| Category | Term | FDR | Functional category |
| Enriched in early diapause vs. nondiapause | |||
| KEGG pathway | DNA replication | <0.001 | DNA replication/development |
| KEGG pathway | Mismatch repair | <0.001 | DNA replication/development |
| KEGG pathway | p53 signaling | <0.001 | DNA replication/development |
| KEGG pathway | Glycerolipid metabolism | <0.001 | Metabolism |
| KEGG pathway | Ether lipid metabolism | <0.001 | Metabolism |
| KEGG pathway | Pyruvate metabolism | <0.001 | Stress response |
| A priori list from literature | Drosophila cold stress (4) | <0.001 | Stress response |
| A priori list from literature | Drosophila oxidative stress (5) | <0.001 | Stress response |
| A priori list from literature | Drosophila hyperoxia stress (6) | 0.007 | Stress response |
| A priori list from literature | Drosophila hypoxia stress (7) | 0.028 | Stress response |
| A priori list from literature | Drosophila ecdysone signaling (8) | <0.001 | Endocrine signaling |
| Enriched in early diapause vs. late diapause | |||
| KEGG pathway | p53 signaling | <0.001 | DNA replication/development |
| KEGG pathway | Napthalene and anthracene degradation | <0.001 | Metabolism |
| A priori list from literature | Drosophila reproductive diapause (15) | <0.001 | Diapause |
| Enriched in diapause termination (hexane treated) vs. late diapause | |||
| KEGG pathway | Citrate cycle | <0.001 | Metabolism |
| KEGG pathway | Pyruvate metabolism | <0.001 | Metabolism |
| KEGG pathway | Riboflavin metabolism | <0.001 | Metabolism |
Metabolism.
Diapausing flesh fly pupae depress respiratory rates by approximately 90% compared with nondiapause pupae (3–9). But how does intermediary metabolism change with metabolic depression? Previous metabolomic studies comparing diapausing flesh fly pupae with nondiapause pupae showed more glucose, pyruvate, and alanine in diapausing pupae, suggesting heavy reliance on glycolytic and gluconeogenic pathways (10, 11).
DAVID analysis comparing early diapause with nondiapause pupae showed diapause enrichment in carbohydrate and pyruvate metabolism, and GSA analysis showed enrichment in glycerolipid metabolism, ether lipid metabolism, and pyruvate metabolism (Table 1). Examination of enriched gene lists shows up-regulation of key members of glycolysis and gluconeogenesis and changes in members of the citric acid [tricarboxylic acid (TCA)] cycle and PEPCK–succinate pathway (Figs. S1 and S2). We constructed pathway diagrams that indeed show concordance of our transcriptomic data with previous metabolomic results (10, 11) (Figs. S3 and S4).
Taken together, the transcriptomic and metabolomic results suggest that diapausing flesh fly pupae undergo substantially more glycolysis and gluconeogenesis than nondiapause pupae. Several important members of the TCA cycle show increased transcript abundance in diapausing pupae. However, the initial steps in the TCA cycle involving the conversion of pyruvate from glycolysis to acetyl-CoA appear to be suppressed, concordant with pyruvate recycling into gluconeogenesis (Fig. S3). The members of the TCA pathway that are up-regulated in early diapause are largely members of the pathway that are involved directly in oxidative metabolism in the mitochondrion or involved in alternative mitochondrial or cytosolic reactions that support glycolysis and gluconeogenesis, such as the PEPCK–succinate pathway that can regenerate reducing equivalents for use in glycolysis (e.g., NADH to NAD+; Fig. S4), the glyoxylate cycle which facilitates the flow of carbon from fatty acids into gluconeogenesis in invertebrates, or in routes of amino acid metabolism. Whole-body assessments do not distinguish where within the animal specific transcripts occurred. However, we expect that the majority of gluconeogenesis occurs in the fat body, after which glucose could be transported as trehalose to other tissues to fuel catabolism or possibly converted to glycerol, an important cryoprotectant in diapausing flesh fly pupae (12). Transcript abundance also does not necessarily predict protein abundance, location, or activation by secondary modification, thus our suggestions will require careful physiological and biochemical studies. However, a switch to anaerobic metabolism is an important component of both seasonal metabolic depression and hypoxia-induced metabolic depression in numerous animals from snails to turtles (13, 14). Enrichment in genes involved in anaerobic metabolism during dormancy has also been shown by other microarray studies in D. melanogaster (15), C. elegans (16, 17), and the pitcher-plant mosquito Wyeomyia smithii (18), reinforcing the centrality of a shift to anaerobic metabolism.
Previous studies suggest that diapausing animals can switch between favoring some metabolic substrates early in diapause and then favoring others later in diapause (19, 20). We did not detect metabolism-related enrichment between early and late diapausing pupae by using either DAVID or GSA analysis (Tables 1 and 2 and Figs. S1 and S2). However, several KEGG metabolic pathways were enriched between late diapausing pupae and pupae forced to terminate diapause with hexane (Tables 1 and 2). Upon diapause termination, flesh flies increase metabolic rates, preparing to resume development. Previous studies have shown that oxygen consumption increases dramatically within 24 h in both natural and hexane-induced diapause termination (3). Consistent with the observed increase in oxygen consumption, hexane-treated individuals showed enrichment in the TCA cycle and pyruvate metabolism KEGG pathways. Inspection of enrichment lists shows hexane treatment leads to increased transcript abundance for key enzymes in aerobic metabolism and aerobic processing of anaerobic end products such as pyruvate kinase, cytosolic malate dehydrogenase, lactate dehydrogenase, and ATP synthase (Figs. S1 and S2). Relative abundance of transcripts for key enzymes involved in anaerobic metabolism such as cytosolic malate dehydrogenase (malic enzyme) and phosphoenolpyruvate carboxykinase showed a relative decrease after hexane treatment, essentially mirroring the pattern observed between nondiapausing and early diapause pupae.
Increased anaerobic respiration in diapausing flesh fly pupae is consistent with overwintering biology, wherein pupae overwinter 1 to 2 cm down in the soil where they may be exposed to cold and oxygen limitation as a result of inundation by rain or snowmelt. Even when kept normoxic before stress exposure, as in this study, diapausing pupae are much more resistant to hypoxia and anoxia stress than nondiapause pupae (10). We found diapause enrichment of gene sets identified in previous transcriptional studies of hypoxia (7), oxidative stress (5), and hyperoxia (6) in D. melanogaster (Table 2). Examination of gene lists with the greatest affects on GSA enrichment in each category suggests the overlap between diapause and these oxidative stress responses occurs in three major areas: metabolism genes involved in glycolysis/gluconeogenesis, general stress response elements including HSPs and antioxidant/detoxification genes (e.g., GstD1), and mechanisms that suppress anabolic synthetic activity, like the translational repressor Thor (Figs. S1 and S2).
Byproducts of glycolysis and gluconeogenesis including carbohydrates, polyols, and amino acids are also known cryoprotectants (12). Increased cold tolerance is a preprogrammed aspect of flesh fly diapause (12), and increased concentrations of glucose, glycerol, pryuvate, and alanine are associated with both diapause and cold-hardening responses (11). Therefore, increased glycolysis and gluconeogenesis and decreased aerobic metabolism may simultaneously facilitate metabolic depression and stress resistance.
Stress.
In addition to hypoxia/anoxia, overwintering animals are exposed to myriad environmental insults including thermal stress and desiccation. Diapausing flesh fly pupae are more resistant to cold and desiccation stress (1), and our results suggest diapause enrichment of several stress-based GO categories and D. melanogaster cold responsive genes (Figs. S1 and S2). Heat shock proteins are involved in many stress responses, and up-regulation of HSPs during dormancy is a common pattern across species (1). A recent study showed that RNAi knockdown of chaperones in the HSP-70 and HSP-23 families reduced cold tolerance in diapausing flesh fly pupae, reinforcing the importance of HSPs in resisting overwintering cold stress (21). Inspection of ESTs in stress response pathways (Figs. S1 and S2) identified members of the HSP-70 and small heat shock (HSP-23, HSP-28) families up-regulated in diapause. The HSP-90 family showed no differences, concordant with numerous studies in flesh flies (1, 21). Our results extend previous HSP studies by screening several previously unidentified chaperone response members (22). Putative flesh fly orthologues of Hsf, DnaJ, and Hop, three key proteins in HSP-70 function, are increased in early diapause. Interestingly, DAVID analysis of early versus late diapause showed enrichment in protein folding and chaperone activity GO categories (Table 1 and Figs. S1 and S2), suggesting that some aspects of HSP-based stress resistance may be diminished late in diapause.
Diapausing flesh fly pupae are highly resistant to hypoxia/anoxia stress, but we do not observe increased transcript quantities for the antioxidant enzymes most typically associated with oxidative stress such as catalase, glutathione peroxidase, or superoxide dismutases (Figs. S1 and S2). Other authors have also noted no change or a decrease in some of these canonical antioxidant enzymes in diapausing insects (23, 24). However, diapausing pupae are enriched in antioxidant and detoxification transcripts such as ferroredoxin, GstD1, Cyp12a, and Cyp6g1 (Figs. S1 and S2), suggesting that metalloproteins may provide alternative oxidative stress protection. Transcripts for metalloproteins with antioxidant capacity were increased in diapausing flesh fly pupae in a previous SSH study (25). However, metalloproteins and detoxification enzymes can be involved in many stress responses including immunity. Diapausing flesh fly pupae spend several months in the soil, where they may encounter pathogens, so an increased immune response may be important. Inspection of DAVID and GSA lists shows several immunity ESTs increased during diapause (Figs. S1 and S2), including the signaling proteins cactus and dorsal and defense, an antimicrobial peptide (26). Interestingly, the cold-tolerance candidate gene Frost, increased in early diapause (4, 27), has also been implicated in the immune response (28), suggesting overlap between cold tolerance, antioxidant, and immune-defense responses. Many genes, like Frost, can differ in function and interactions based on the immediate cellular context (e.g., cold-stressed cells vs. immune-challenged cells) and new minor functions have recently been discovered even for well characterized genes in model organisms like D. melanogaster. Therefore, additional studies are clearly needed to determine functions for candidate ESTs, with particular focus on proteins that may act in multiple stress resistance pathways.
Endocrine.
A halt in ecdysteroid production is a key component of developmental arrest in diapausing flesh fly pupae, and diapause is terminated upon application of ecdysteroids (3). Consistent with this view, transcripts implicated in developmental responses to ecdysteroids in D. melanogaster were differentially regulated in nondiapause compared with early diapause pupae (Tables 1 and 2 and Fig. S2). Previous work in flesh flies suggests that late diapause pupae may be more responsive to ecdysteroids and therefore more likely to terminate diapause than early diapause pupae (1). We did not detect enrichment in ecdysteroid responsive transcripts (8) between early diapause pupae and late diapause pupae, or between late diapause pupae and pupae treated with hexane to terminate diapause. Because hexane treatment promotes ecdysteroid production approximately 24 h after treatment (3), we expected ecdysteroid responsive transcripts to be differentially regulated between late diapause and individuals terminating diapause. However, 24 h may not have been sufficient time after hexane treatment to manifest a full transcriptional response to ecdysteroids. Similarly, we found no enrichment in lists including members of the insulin signaling pathway (29) or lists of transcripts expected to be downstream of the insulin signaling pathway in D. melanogaster (30, 31).
Reduced insulin signaling is thought to be an important component of diapause across several species, including the larval dauer of C. elegans (32), the pupal diapause of the moth Pieris brassicae (33), and the adult reproductive diapause of Culex pipens mosquitoes and D. melanogaster flies (34, 35). We had expected to detect nondiapause versus early-diapause enrichment in our insulin-related lists. Gene set enrichment approaches normalize for gene set size, but this normalization may not be accurate for very small lists (e.g., insulin signaling, n = 12) and very large lists (e.g., FOXO response, n = 989; TOR response, n = 890; GSEA User Guide, http://www.broadinstitute.org/gsea/doc/GSEAUserGuideFrame.html). Further, because of the way the test statistic is constructed (based on ranks of differential expression), lists containing genes with only modest expression differences are more difficult to detect. Finally, enrichment approaches do not take into account a priori expectations of directionality. Considering that small differences in expression may be important in signaling pathways but hard to detect by enrichment methods, we applied an alternative approach, directly mapping the expression patterns of several key members of the insulin signaling pathway (29) (Fig. S5). Nine of 12 insulin-pathway transcripts differed in expression (FDR < 0.001) between early diapause and nondiapause pupae. Counter to our predictions of decreased insulin signaling, however, eight of the nine insulin-signaling pathway members were relatively increased in early diapause compared with nondiapause pupae, whereas the other, PTEN, a negative regulator, was decreased (Fig. S5). Only one of 12 insulin-pathway transcripts differed between early diapause and late diapause pupae, and we conclude there was no change in insulin-pathway transcripts during diapause. However, a comparison of insulin-pathway transcripts in hexane-treated pupae terminating diapause with late diapause pupae shows concordance with the nondiapause and early diapause comparisons, wherein nine of 12 transcripts differ in the opposite direction compared with the nondiapause to early diapause comparison (Fig. S5).
Previous studies have associated suppression of insulin-like signaling with diapause in invertebrates including worms, flies, mosquitoes, and moths, but we found that most insulin pathway members have greater relative transcript abundance in diapausing pupae relative to nondiapause pupae. How can these two views be reconciled? We propose that members of the insulin signaling pathway are themselves present and maintained during pupal diapause in flesh flies, but that they remain inactive. Maintaining the members of the insulin signaling pathway may be important for rapidly enhancing downstream insulin-like signaling when cues signal diapause termination, by activating ecdysteroid production and initiating adult morphogenesis. Indeed, insulin signaling has been shown to promote ecdysteroid production in mosquito and blow fly ovaries (36), and treatment with exogenous bovine insulin terminates pupal diapause and is associated with ecdysteroid production during diapause termination in the moth P. brassicae (33). Our observation of a change in relative transcript abundance of insulin pathway genes but not ecdysteroid-responsive genes 24 h after hexane treatment is concordant with insulin signaling being the initial trigger that terminates diapause by enhancing ecdysteroid production. Furthermore, our results are concordant with expression studies in D. melanogaster (15) and C. elegans (16, 17), wherein dormant individuals have mostly higher transcript abundance for multiple insulin-signaling pathway members, such as insulin receptor and AKT, compared with nondormant individuals at the same life stage. However, transcript abundance alone does not indicate whether the insulin signaling pathway is potentially functional or whether the activity of the insulin signaling pathway itself is increased or suppressed during pupal diapause in flesh flies. Like many other biochemical pathways involved in dormancy and stress resistance (14), signaling activity of key members of the insulin signaling pathway (e.g., AKT and FOXO) is regulated after translation by phosphorylation reactions, and thus transcript abundance does not necessarily reflect pathway activity. Testing whether insulin signaling capacity is maintained while insulin signaling activity is suppressed during diapause will require protein quantification and activity assays coupled with in vivo manipulation across diapause development.
Detailed examination of other gene networks will undoubtedly reveal new and interesting processes in flesh fly diapause that our enrichment analyses have missed. However, detailed pathways do not exist for many biological processes, and many genes, even in D. melanogaster, have not been stringently assigned to particular pathways/networks; in addition, genes can participate in multiple pathways/networks, making it difficult to assess them in this context. For example, a recent QTL study in D. melanogaster implicated the gene couch potato (cpo) as important in adult reproductive diapause (37). An EST on our array matching cpo has substantially lower transcript abundance in diapausing compared with nondiapausing pupae or pupae terminating diapause (Fly.2469.c1, ED vs. ND FDR < 0.001 and log2FC = −1.30, HEX vs. LD FDR <0.001 and log2FC = 1.03). However, the function of this gene is currently unknown and no annotated pathway information is available, limiting our ability to place it into a broader gene class or pathway context. Similar limitations apply to many of the thousands of ESTs we found differing among our comparisons.
Comparing Diapause Responses Across Taxa.
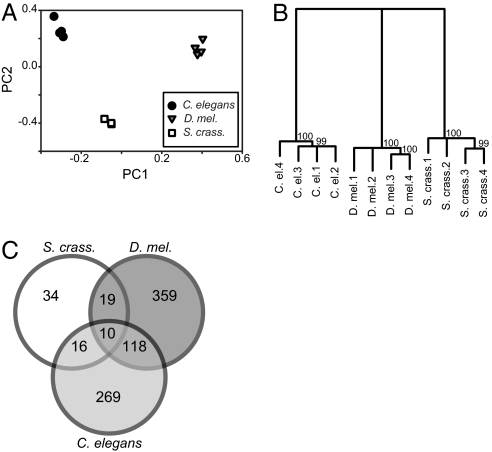
The transcriptional basis of dormancy appears to differ substantially among S. crassipalpis, D. melanogaster, and C. elegans. Arrays comparing dormancy to nondormancy phenotypes cluster most closely within species (Fig. 2A), but hierarchical clustering suggests that, although they are both dipterans, S. crassipalpis and D. melanogaster diapause gene expression patterns are no more similar to each other than to C. elegans dauer (i.e., there is a polytomy at the base of the cluster tree; Fig. 2B). Further, there is little overlap in lists of highly differentially regulated genes (twofold or greater ratios of dormant vs. nondormant phenotype), with only 10 twofold differentially regulated genes common to all three species (Fig. 2C and Table S2). An additional test using discrimate function analysis shows that the most differentially regulated dormancy versus nondormancy genes in all three species are also the genes whose expression patterns differ most among the species (Fig. S6).
Fig. 2.
Comparisons of transcriptional profiles in dormancy among S. crassipalpis, D. melanogaster, and C. elegans, including principal components analysis of all identified orthologues (A), hierarchical clustering with bootstrap support (B), and overlap of genes twofold or greater differentially regulated between dormancy/nondormancy phenotypes.
Despite these interspecific differences in transcript profiles, there are some clear commonalities suggesting evolutionary convergence on a few important physiological phenotypes. Dormancy in nearly all species with any appreciable physiological and/or gene expression data involves similar up-regulation of stress resistance axes (1). Dormancy almost always involves metabolic depression as well (9, 13, 14). For the three species in our comparative analysis, the general metabolic depression phenotype is accompanied by an increase in glycolysis and gluconeogenesis. Enzymes associated with irreversible reactions in both pathway directions are significantly up-regulated. During dormancy in all three species the gluconeogeneis enzymes phosphoenolpyruvate carboxykinase and pyruvate carboxylase are at least ninefold and twofold up-regulated. In fact, these are the only two genes that are twofold or greater up-regulated in dormant compared with nondormant individuals in all three species (Fig. S6). Dormancy-associated increases in the glycolytic enzymes hexokinase and phosphofructokinase were also consistent, but a bit more modest (minimum of 1.3-fold increase). S. crassipalpis, D. melanogaster, and C. elegans all seem to make this similar metabolic switch during dormancy; however, this convergence on increased glycolysis/gluconeogenesis is likely reached through different transcriptional strategies.
Dormancy is an evolutionarily labile trait, and phylogenetic patterns are consistent with a diversity of regulatory strategies. The relative lack of conservation of gene expression during dormancy in C. elegans, D. melanogaster, and S. crassipalpis may therefore reflect multiple evolutionary origins and life history strategies. Adult reproductive arrest in D. melanogaster is relatively shallow (38), i.e., not very recalcitrant to terminating cues, and diapausing adults remain mobile and capable of feeding. C. elegans dauer occurs in the larval stage; larvae are mobile but nonfeeding and quickly transition to active development under favorable conditions (16, 17). In contrast, diapausing S. crassipalpis pupae are completely nonmotile, nonfeeding, and highly recalcitrant to transient environmental cues (1). We have identified transcriptional correlates of two common physiological phenotypes, metabolism and stress resistance, across these three species with very different diapause strategies. Studies of additional species with diverse diapause strategies will be needed to more broadly define which components of the diapause program are conserved and which are labile, but our prediction from this comparative study is that similar diapause phenotypes will be derived from diverse transcriptional mechanisms.
Methods
Flies were maintained under diapause-averting conditions (15 h light and 9 h dark at 25 °C for adults and 20 °C for larvae) or diapause-inducing conditions (10 h light and 14 h dark at 25 °C for adults and 20 °C for larvae). Nondiapause, early diapause, and late diapause pupae were sampled 5, 20, and 50 d after pupariation (diapause typically terminates after approximately 60 d at 20 °C). Late-diapause pupae were treated topically with 5 μL of hexane to pharmacologically stimulate diapause termination and sampled 24 h later (3).
RNA was extracted from pools of four individuals using the Ambion RiboPure Kit, and assessed using a NanoDrop spectrophotometer and the Agilent 2100 Bioanlyzer. cDNA was synthesized from 500 ng of total RNA using the Agilent Low RNA Input Linear Amplification Kit. Samples were hybridized to custom Agilent 4 × 44k arrays, washed, and scanned on an Agilent G2505B scanner, and data were extracted using Feature Extraction 9.5 software (Agilent Technologies).
Oligonucleotide (60mer) probes were designed using Agilent eArray 4.0 from a S. crassipalpis EST project (22). We chose two different probes in the sense strand for 11,656 target ESTs that had good BLAST hits to the National Center for Biotechnology Information (NCBI)–NR database (e < 1 × 10−4) and an additional 26 previously sequenced transcripts available in the NCBI database for S. crassipalpis. We included one sense strand probe for another 10,636 ESTs that had predicted ORFs but poorer BLAST hits to NCBI-NR (1 < e < 1 × 10−4). We also chose one probe from the well annotated group in the antisense to generate a negative control distribution for incidental background hybridization. Original EST sequences and contigs used to design probes can be accessed at NCBI-SRA SRR005065 and SRR006884 and NCBI-TSA EZ596711–EZ617705 (22). Raw array results and design files can be accessed at NCBI-GEO GSE20526.
Images were corrected to internal Agilent control probes and data were preprocessed and normalized using the Linear Models for Microarray Data package in R (39). Raw mean signal intensities were background-corrected and normalized using a within-array lowess approach and log-transformed. Histograms, box plots, and pair-wise scatter plots were used to examine data quality. A linear modeling approach and the empirical Bayes statistics as implemented in Linear Models for Microarray Data were used for differential expression analysis. P values were adjusted using the Benjamini and Hochberg FDR method (40). Probe-set detection calls were estimated as “present” or “absent” according to a mixture Gaussian model based on normalized signal values on each channel compared with our antisense negative control pool of probes (41). Probe sets with absent calls were excluded from data analysis.
For multivariate analyses, we removed redundancy by calculating mean signal intensities for multiple probes of the same gene. Principal components analysis and discriminant analysis were performed using the JMP Genomics 4 package. We used the R package “pvclust” to perform hierarchical clustering on the phenotypic classes, with the “average” agglomerative method, correlation as the distance metric, and 1,000 bootstrap replicates to estimate Approximately Unbiased bootstrap support for each node (42). For enrichment analysis, we created lists of nonredundant genes annotated to the D. melanogaster transcriptome (http://www.flybase.org). Lists were submitted to the DAVID functional annotation database (http://david.abcc.ncifcrf.gov/), providing a broad, unguided test against primarily GO groups and KEGG pathways. Next, we performed Gene Set Analysis using the R package “GSA” (43) to test more specifically for differential regulation of (i) D. melanogaster KEGG pathways (http://www.genome.jp/kegg/) and (ii) a priori lists of genes differentially regulated in D. melanogaster in response to stress, hormone signaling, or diapause (Table S1). We performed GSA on full, nonredundant data sets using 1,000 phenotypic permutations to estimate FDR.
Gene expression data comparing diapause to nondiapause D. melanogaster females (15) and dauer to nondauer C. elegans (16) were extracted from the NCBI GEO database to perform comparative analysis of gene expression during dormancy. Both data sets contained four array equivalents comparing dormancy to nondormancy, balancing our set of four competitive hybridizations comparing S. crassipalpis early diapause to nondiapause. We performed analyses on log(2) fold change values only for genes found to be orthologous across all species (see SI Methods for details). Principal-components, discriminant, and hierarchical clustering analysis were performed as described earlier.
Supplementary Material
Acknowledgments
We thank the University of Florida Genetics Core Facility for technical help. We appreciate the thoughtful reviews provided by Marla Sokolowski and Karen Williams (University of Toronto) and Paul Schmidt (University of Pennsylvania). This work was supported by US Department of Agriculture (USDA) National Research Initiative (NRI) Grant 2004-35302-745994 (to D.A.H.), National Science Foundation (NSF) Grant IOS-641505 (to D.A.H.), NSF Integrative Organismal Systems Grant 0840772 (to D.L.D.), USDA NRI Grant 2006-35607-16582 (to D.L.D.), and National Institutes of Health Grant R01-AI1058279 (to D.L.D.); and by the Florida State Agricultural Experiment Station (D.A.H.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA), Transcriptome Shotgun Assembly (TSA; www.ncbi.nlm.nih.gov/genbank/TSA.html), and Gene Expression Omnibus (GEO; www.ncbi.nlm.nih.gov/geo) databases (accession nos. NCBI-SRA SRR005065 and SRR006884, NCBI-TSA EZ596711–EZ617705, NCBI-GEO GSE20526).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1007075107/-/DCSupplemental.
References
- 1.Denlinger DL, Yocum GD, Rinehart JL. In: Hormonal Control of Diapause: Comprehensive Molecular Insect Science. Gilbert LI, Iatrou K, Gill SS, editors. Vol. 3. Amsterdam: Elsevier Press; 2005. pp. 615–650. [Google Scholar]
- 2.Kostál V. Eco-physiological phases of insect diapause. J Insect Physiol. 2006;52:113–127. doi: 10.1016/j.jinsphys.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 3.Denlinger DL, Campbell JJ, Bradfield JY. Stimulatory effect of organic solvents on initiating development in diapausing pupae of the flesh fly Sarcophaga crassipalpis and the tobacco hornworm Manduca sexta. Physiol Entomol. 1980;5:7–15. [Google Scholar]
- 4.Qin W, Neal SJ, Robertson RM, Westwood JT, Walker VK. Cold hardening and transcriptional change in Drosophila melanogaster. Insect Mol Biol. 2005;14:607–613. doi: 10.1111/j.1365-2583.2005.00589.x. [DOI] [PubMed] [Google Scholar]
- 5.Girardot F, Monnier FV, Tricoire H. Genome wide analysis of common and specific stress responses in adult Drosophila melanogaster. BMC Genomics. 2004;5:16. doi: 10.1186/1471-2164-5-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Landis GN, et al. Similar gene expression patterns characterize aging and oxidative stress in Drosophila melanogaster. Proc Natl Acad Sci USA. 2004;101:7663–7668. doi: 10.1073/pnas.0307605101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu GW, Roy J, Johnson EA. Identification and function of hypoxia-response genes in Drosophila melanogaster. Physiol Genomics. 2006;25:134–141. doi: 10.1152/physiolgenomics.00262.2005. [DOI] [PubMed] [Google Scholar]
- 8.Beckstead RB, Lam G, Thummel CS. The genomic response to 20-hydroxyecdysone at the onset of Drosophila metamorphosis. Genome Biol. 2005;6:R99. doi: 10.1186/gb-2005-6-12-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hahn DA, Denlinger DL. Meeting the energetic demands of insect diapause: Nutrient storage and utilization. J Insect Physiol. 2007;53:760–773. doi: 10.1016/j.jinsphys.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 10.Kukal O, Denlinger DL, Lee RE., Jr. Developmental and metabolic changes induced by anoxia in diapausing and non-diapausing flesh fly pupae. J Comp Physiol B. 1991;160:683–689. [Google Scholar]
- 11.Michaud MR, Denlinger DL. Shifts in the carbohydrate, polyol, and amino acid pools during rapid cold-hardening and diapause-associated cold-hardening in flesh flies (Sarcophaga crassipalpis): A metabolomic comparison. J Comp Physiol B. 2007;177:753–763. doi: 10.1007/s00360-007-0172-5. [DOI] [PubMed] [Google Scholar]
- 12.Denlinger DL, Lee RE. Physiology of Cold Sensitivity. In: Hallman GJ, Denlinger DL, editors. Temperature Sensitivity in Insects and Application in Integrated Pest Management. Oxford: Westview Press; 1998. pp. 55–95. [Google Scholar]
- 13.Guppy M, Withers P. Metabolic depression in animals: Physiological perspectives and biochemical generalizations. Biol Rev Camb Philos Soc. 1999;74:1–40. doi: 10.1017/s0006323198005258. [DOI] [PubMed] [Google Scholar]
- 14.Storey KB, Storey J. Life in the slow lane: Molecular mechanisms of estivation. Comp Biochem Physiol A Mol Integr Physiol. 2002;133:733–754. doi: 10.1016/s1095-6433(02)00206-4. [DOI] [PubMed] [Google Scholar]
- 15.Baker DA, Russell S. Gene expression during Drosophila melanogaster egg development before and after reproductive diapause. BMC Genomics. 2009;10:242. doi: 10.1186/1471-2164-10-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, Kim SK. Global analysis of dauer gene expression in Caenorhabditis elegans. Development. 2003;130:1621–1634. doi: 10.1242/dev.00363. [DOI] [PubMed] [Google Scholar]
- 17.Jeong P-Y, Kwon M-S, Joo H-J, Paik Y-K. Molecular time-course and the metabolic basis of entry into dauer in Caenorhabditis elegans. PLoS ONE. 2009;4:e4162. doi: 10.1371/journal.pone.0004162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emerson KJ, Bradshaw WE, Holzapfel CM. Microarrays reveal early transcriptional events during the termination of larval diapause in natural populations of the mosquito, Wyeomyia smithii. PLoS ONE. 2010;5:e9574. doi: 10.1371/journal.pone.0009574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yocum GD, Kemp WP, Bosch J, Knoblett JN. Temporal variation in overwintering gene expression and respiration in the solitary bee Megachile rotundata. J Insect Physiol. 2005;51:621–629. doi: 10.1016/j.jinsphys.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 20.Zhou G, Miesfeld RL. Energy metabolism during diapause in Culex pipiens mosquitoes. J Insect Physiol. 2009;55:40–46. doi: 10.1016/j.jinsphys.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rinehart JP, et al. Up-regulation of heat shock proteins is essential for cold survival during insect diapause. Proc Natl Acad Sci USA. 2007;104:11130–11137. doi: 10.1073/pnas.0703538104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hahn DA, Ragland GJ, Shoemaker DD, Denlinger DL. Gene discovery using massively parallel pyrosequencing to develop ESTs for the flesh fly Sarcophaga crassipalpis. BMC Genomics. 2009;10:234. doi: 10.1186/1471-2164-10-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jovanovic-Galanovic A, et al. Antioxidant defense in mitochondria during diapause and post-diapause development of European corn borer. Arch Insect Biochem Physiol. 2007;64:111–119. doi: 10.1002/arch.20160. [DOI] [PubMed] [Google Scholar]
- 24.Krishnan N, Kodrík D, Turanli F, Sehnal F. Stage-specific distribution of oxidative radicals and antioxidant enzymes in the midgut of Leptinotarsa decemlineata. J Insect Physiol. 2007;53:67–74. doi: 10.1016/j.jinsphys.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 25.Rinehart JP, Robich RM, Denlinger DL. Isolation of diapause-regulated genes from the flesh fly, Sarcophaga crassipalpis by suppressive subtractive hybridization. J Insect Physiol. 2010;56:603–609. doi: 10.1016/j.jinsphys.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 26.Goving S. Innate immunity in Drosophila: Pathogens and pathways. Insect Sci. 2008;15:29–43. doi: 10.1111/j.1744-7917.2008.00185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goto SG. A novel gene that is up-regulated during recovery from cold shock in Drosophila melanogaster. Gene. 2001;270:259–264. doi: 10.1016/s0378-1119(01)00465-6. [DOI] [PubMed] [Google Scholar]
- 28.Apidianakis Y, et al. Profiling early infection responses: Pseudomonas aeruginosa eludes host defenses by suppressing antimicrobial peptide gene expression. Proc Natl Acad Sci USA. 2005;102:2573–2578. doi: 10.1073/pnas.0409588102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu Q, Brown MR. Signaling and function of insulin-like peptides in insects. Annu Rev Entomol. 2006;51:1–24. doi: 10.1146/annurev.ento.51.110104.151011. [DOI] [PubMed] [Google Scholar]
- 30.Guertin DA, Guntur KVP, Bell GW, Thoreen CC, Sabatini DM. Functional genomics identifies TOR-regulated genes that control growth and division. Curr Biol. 2006;16:958–970. doi: 10.1016/j.cub.2006.03.084. [DOI] [PubMed] [Google Scholar]
- 31.Gershman B, et al. High-resolution dynamics of the transcriptional response to nutrition in Drosophila: A key role for dFOXO. Physiol Genomics. 2007;29:24–34. doi: 10.1152/physiolgenomics.00061.2006. [DOI] [PubMed] [Google Scholar]
- 32.Fielenbach N, Antebi A. C. elegans dauer formation and the molecular basis of plasticity. Genes Dev. 2008;22:2149–2165. doi: 10.1101/gad.1701508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arpagaus M. Vertebrate insulin induces diapause termination in Pieris brassicae pupae. Roux's Arch Dev Biol. 1987;196:527–530. doi: 10.1007/BF00399877. [DOI] [PubMed] [Google Scholar]
- 34.Tatar M, Yin CM. Slow aging during insect reproductive diapause: Why butterflies, grasshoppers and flies are like worms. Exp Gerontol. 2001;36:723–738. doi: 10.1016/s0531-5565(00)00238-2. [DOI] [PubMed] [Google Scholar]
- 35.Sim C, Denlinger DL. Insulin signaling and FOXO regulate the overwintering diapause of the mosquito Culex pipiens. Proc Natl Acad Sci USA. 2008;105:6777–6781. doi: 10.1073/pnas.0802067105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brown MR, Sieglaff DH, Rees HH. Gonadal ecdysteroidogenesis in arthropoda: Occurrence and regulation. Annu Rev Entomol. 2009;54:105–125. doi: 10.1146/annurev.ento.53.103106.093334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt PS, et al. An amino acid polymorphism in the couch potato gene forms the basis for climatic adaptation in Drosophila melanogaster. Proc Natl Acad Sci USA. 2008;105:16207–16211. doi: 10.1073/pnas.0805485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saunders DS, Henrich VC, Gilbert LI. Induction of diapause in Drosophila melanogaster: Photoperiodic regulation and the impact of arrhythmic clock mutations on time measurement. Proc Natl Acad Sci USA. 1989;86:3748–3752. doi: 10.1073/pnas.86.10.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21:2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- 40.Benjamini Y, Hochberg Y. Controlling the false discovery rate – a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 41.Lee M-L, Kuo FC, Whitmore GA, Sklar J. Importance of replication in microarray gene expression studies: Statistical methods and evidence from repetitive cDNA hybridizations. Proc Natl Acad Sci USA. 2000;97:9834–9839. doi: 10.1073/pnas.97.18.9834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 2002;51:492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- 43.Efron B, Tibshirani R. On testing the significance of sets of genes. Ann Appl Stat. 2007;1:107–129. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.