Abstract
A bottleneck in drug discovery is the identification of the molecular targets of a compound (mode of action, MoA) and of its off-target effects. Previous approaches to elucidate drug MoA include analysis of chemical structures, transcriptional responses following treatment, and text mining. Methods based on transcriptional responses require the least amount of information and can be quickly applied to new compounds. Available methods are inefficient and are not able to support network pharmacology. We developed an automatic and robust approach that exploits similarity in gene expression profiles following drug treatment, across multiple cell lines and dosages, to predict similarities in drug effect and MoA. We constructed a “drug network” of 1,302 nodes (drugs) and 41,047 edges (indicating similarities between pair of drugs). We applied network theory, partitioning drugs into groups of densely interconnected nodes (i.e., communities). These communities are significantly enriched for compounds with similar MoA, or acting on the same pathway, and can be used to identify the compound-targeted biological pathways. New compounds can be integrated into the network to predict their therapeutic and off-target effects. Using this network, we correctly predicted the MoA for nine anticancer compounds, and we were able to discover an unreported effect for a well-known drug. We verified an unexpected similarity between cyclin-dependent kinase 2 inhibitors and Topoisomerase inhibitors. We discovered that Fasudil (a Rho-kinase inhibitor) might be “repositioned” as an enhancer of cellular autophagy, potentially applicable to several neurodegenerative disorders. Our approach was implemented in a tool (Mode of Action by NeTwoRk Analysis, MANTRA, http://mantra.tigem.it).
Keywords: computational drug discovery, drug repurposing, systems biology, chemotherapy
Identifying molecular pathways targeted by a compound (drug effects), and the specific compound-substrate interactions (drug mode of action—MoA), is of paramount importance for the development of new drugs, and also for new clinical applications of already existing drugs (1–3). Systems biology approaches are naturally suited to capture the complexity of drug activity in cells (4–6). Prediction of drug MoA has been attempted by using gene expression profiles following drug treatment (7–13), by comparing side-effect similarities (14), by text-mining literature (15), or by applying chemoinformatic tools to search for small molecules similarities (16, 17). Most of these approaches are applicable only to well-characterized molecules (e.g., when the structure is available, or side effects are documented). On the other hand, expression profile-based methods are the most general ones, because they do not require any prior information on the compound being analyzed. Among the most promising approaches are the ones based on “gene signatures” (11, 12), i.e., subset of genes whose differential expression can be used as a marker of the activity of a given pathway, disease or compound. Gene signatures can be used to discover “connections” among drugs, pathways, and diseases (8, 11, 12, 18) using a large collection of transcriptional responses following compound treatments, such as the “Connectivity Map” (11, 12). These compound-specific expression profiles can be queried with a gene signature to recover a subset of compounds connected to the signature of interest. A compound is selected if genes in the signature are significantly modulated in the compound-specific transcriptional response. If a gene signature for a new compound is available, it is then possible to search the collection of transcriptional responses with that signature to identify well-characterized drugs, which behave similarly, and thus infer the MoA of the new compound. The problems affecting gene signature-based methods are in the choice of the subset of genes composing the signature, and in the proper handling of multiple expression profiles obtained by treating different cell lines, with the same compound. A wrong selection of genes in the signature will lead to capture similarities in the experimental settings (i.e., same cell line) rather than in the drug MoAs (SI Methods). Because of these limitations, the analysis of the transcriptional response to a new compound is usually performed by mapping the most differentially expressed genes, following compound treatment, onto known biological pathways, in order to detect the most perturbed pathways. Such attempts are met with limited success due to the complexity of “backtracking” expression changes to primary causes (i.e., molecular targets).
Inspired by these considerations, we developed a general approach, with a matched online tool, to identify and classify the pathway targeted by a compound and its MoA. We computed for each drug a “consensus” synthetic transcriptional response summarizing the transcriptional effect of the drug across multiple treatments on different cell lines and/or at different dosages. We then constructed a “drug network” (DN) in which two drugs are connected to each other if their consensus responses are similar according to a similarity measure that we developed (drug distance). We divided the DN into interconnected modules termed “communities” and “rich clubs” (19). By analyzing these modules, we were able to capture similarities and differences in pharmacological effects and MoAs; we were able to predict MoA of anticancer compounds still being studied and to discover previously unreported MoAs for well-known drugs.
We developed a Web-based tool to explore the DN and query it for classification of previously undescribed compounds (Mode of Action by Network Analysis—MANTRA, http://mantra.tigem.it).
Results
Drug Network and Communities.
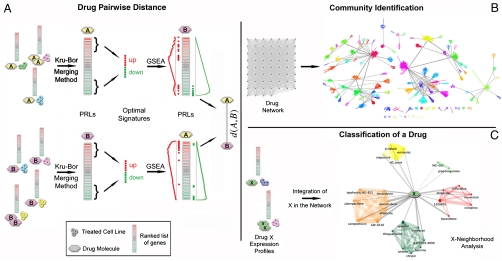
We quantified the degree of similarity in the transcriptional responses among drugs. To this end, we exploited a repository of transcriptional responses to compounds: the Connectivity Map (cMap) (11, 12) containing 6,100 genome-wide expression profiles obtained by treatment of five different human cell lines at different dosages with a set of 1,309 different molecules. We represented the similarity between two drugs as a “distance” and computed it as summarized in Fig. 1A: For each compound, we considered all the transcriptional responses following treatments, across different cell lines and/or at different concentrations. Each transcriptional response was represented as a list of genes ranked according to their differential expression. We then computed a single “synthetic” ranked list of genes, the Prototype Ranked List (PRL), by merging all the ranked lists referring to the same compound. In order to equally weight the contribution of each of the cell lines to the drug PRL, rank merging was achieved with a procedure (detailed in SI Methods) based on a hierarchical majority-voting scheme, where genes consistently overexpressed/down-regulated across the ranked lists are moved at the top/bottom of the PRL (18). The rank-merging procedure first compares, pairwise, the ranked lists obtained with the same drug using the Spearman’s Footrule similarity measure (20). Then, it merges the two lists that are the most similar to each other, following the Borda Merging Method (21), thus obtaining a single ranked list. This new ranked list replaces the two lists, and then the procedure is repeated until only one ranked list remains (the PRL of the drug). The PRL thus captures the consensus transcriptional response of a compound across different experimental settings, consistently reducing nonrelevant effects due to toxicity, dosage, and cell line (SI Methods).
Fig. 1.
Methodology overview. (A) A distance value for each couple of drugs is computed. (B) Each drug is considered as a node in a network with weighted edges (proportional to distances) connecting pairs of drugs. Network communities are identified. (C) Ranked list of differentially expressed genes, following treatment with a previously undescribed drug X are merged together, and the distance d(X,Y) is computed for each drug Y in the reference dataset. X is connected to drugs whose distance is below a significant threshold.
The distance between a pair of compounds is computed by comparing the two PRLs. To this end, we extracted an “optimal” gene signature for each of the two compounds by selecting the first 250 genes at the top of the PRL (most overexpressed) and the last 250 genes at the bottom of the PRL (most down-regulated). The size of these optimal signatures was heuristically determined as described in SI Methods.
We then checked if the genes in the optimal gene signature of the first compound ranked consistently at the top/bottom of the PRL of the second compound, and vice versa, using the Gene Set Enrichment Analysis (GSEA) (22). We computed the GSEA enrichment score of the optimal gene signature of compound A in the PRL of compound B, and vice versa. We then combined the two scores to obtain a single value quantifying the distance between compound A and B (SI Methods). The smaller the distance, the more similar the two compounds are. We computed the distance for each pair of the 1,309 compounds in the cMap dataset for a total of 856,086 pairwise comparisons. We then considered each compound as a node in a network and connected two nodes with a weighted edge (where the weight is proportional to their distance), if their distance was below a significant threshold value (Fig. 1B). Drugs that were not connected to any other compound by at least one edge were excluded from the DN (SI Methods).
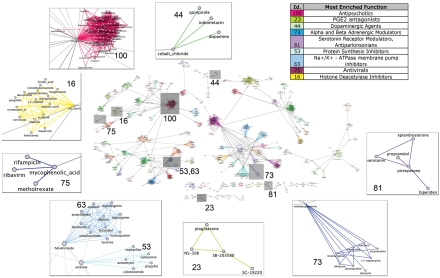
The resulting DN has a giant connected component with 1,302 nodes (i.e., drugs) out of 1,309 and 41,047 edges, corresponding to 5% of a fully connected network with the same number of nodes (856,086 edges). In order to analyze and visualize the DN, we identified its communities via a recent clustering algorithm (23) (SI Methods and Fig. 1B). A community is defined as a group of nodes densely interconnected with each other and with fewer connections to nodes outside the group (24). As shown in Fig. 2, we identified 106 communities [online Supporting Information (SI) at http://mantra.tigem.it]. Each community was coded with a numerical identifier, a color, and one of its nodes was identified as the “exemplar” of the community, i.e., the drug whose effect best represents the effects of the other drugs in the community. We assessed that the tendency of our method to group drugs in the same community was not due to trivial chemical commonalities (SI Methods).
Fig. 2.
The Drug network. Communities and rich clubs are highlighted. (Insets) Some communities are magnified, and the enriched Mode of Actions are provided.
We next determined whether drugs within a community shared a common MoA. We collected for each drug the Anatomical Therapeutic Chemical (ATC) code, the known direct target genes, and other literature-based evidences. ATC codes (25, 26) are alphanumerical strings assigned by the World Health Organization to group drugs according to their therapeutic and chemical profiles. ATC codes were available for 59% of the drugs (768 out of 1,309). We retrieved the known target genes for 535 out of 1,309 (41%) drugs from two public repositories, DrugBank (27) and ChemBank (28). We thus assigned a known MoA to 804 drugs out of 1,309 (61%) (Dataset S1). For each community, we counted the number of drugs with the same MoA. We then divided this number by the number one would expect had the drugs been randomly grouped, to compute “odds ratios” and p values (SI Methods). We found that 52 out of 95 assessable communities (i.e., those containing at least two compounds with known MoA) were significantly enriched (p value < 0.05) for compounds with similar MoA. Specifically, 3 communities were enriched for a direct target gene, 28 for one ATC code, whereas 21 were enriched for both a direct target gene and an ATC code. Additionally, by searching the literature for supporting evidences, we found 43 communities including several compounds with similar MoA, 9 of which were composed by compounds with no ATC codes and no known target genes. So the total number of enriched communities was 61 (52 + 9) (Fig. S1). This number goes up to 77, considering as significant communities, those with a corresponding significant odds ratio greater than 1 (SI Methods).
We further checked if compounds in the same community impinge on common biological pathways. We developed a Fuzzy-Logic-based approach (SI Methods) to identify a common set of genes that was consistently up-, or, down-regulated in the PRLs of the compounds in the same community. We thus associated significant gene ontology (GO) terms to 57 communities by performing a GO enrichment analysis on the common set of genes (see online SI: http://mantra.tigem.it).
For example, in community n.3, mainly composed of cell cycle blockers (Resveratrol, Ciclopirox, Etoposide, Deferoxamine, Kaempferol, Colforsin, and Quercetin), the most enriched GO terms associated to down-regulated genes in this community were cell cycle process (p value 2.31 × 10-13), mitotic cell cycle (p value 1.12 × 10-12), and M phase (p value 1.49 × 10-10). These terms are strictly related to the MoA shared by the drugs in this community. Other examples are reported in SI Methods.
We then assessed the opposite tendency, i.e., whether compounds characterized by the same MoA end up in the same community. We considered in the set of 804 compounds with known MoA (i.e., with an ATC code or a known target gene) a subset of 698 drugs. This subset contained only the drugs sharing their MoA with at least another drug and was divided in 429 groups (not mutually disjointed) of drugs with the same MoA (SI Methods). We verified that the MoA of 512 drugs (out of 698) was enriched for a specific community (p value < 0.05). This number goes up to 586 drugs, considering those with a significant odds ratio greater than 1 (Dataset S1 and online SI Fig. 9).
Prediction of Drug Mode of Action.
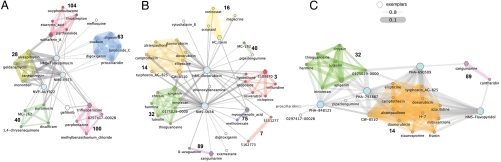
We assessed the ability of the DN to predict the MoA of anticancer compounds whose gene expression profiles were not included in the original cMap dataset. As summarized in Fig. 1C, we measured expression profiles derived from different cell lines treated with anticancer compounds still being studied developed at Nerviano Medical Sciences (NMS) and reference drugs already present in the cMap dataset. Nine compounds were considered for a total amount of 39 microarray hybridizations. We computed a PRL for each of the tested compounds, and their distances from the 1,309 drugs in the cMap dataset. We then integrated the compounds in the DN by connecting them to the other drugs, if their distance was below the significant threshold (Fig. 3). Additionally, we computed a “drug-to-community” distance (SI Methods), which quantifies how close the tested compound is to each of the communities. This distance was defined as the weighted geometric average of the distances between the tested compound and the drugs belonging to the same community. The most similar compounds and the closest communities in the DN are provided in Table S1 and Dataset S1 for each of the tested compounds.
Fig. 3.
Classification of drugs. Subnetworks connected to the tested compounds (cyan nodes) once they have been integrated in the drug network. For clarity we included only compounds whose distances from the tested compounds were less than 0.8 (A and C) or 0.72 (B). Edge thickness is inversely proportional to the distance between the drugs; edge and node colors indicate communities. Hexagonal-shaped nodes represent community exemplars. (A) HSP90 inhibitors; (B) Topo inhibitors; (C) CDK inhibitors.
We tested three HSP90 inhibitors: Tanespimycin (already present in the cMap, used as control), the second-generation HSP90 inhibitors NVP-AUY922 (29) and NMS-E973 (30). Tanespimycin is close to all four HSP90 inhibitors present in the database, as well as, to the protein synthesis inhibitor Puromycin, and to the proteasome inhibitors Withaferin A and Parthenolide; a similar list was also obtained for NVP-AUY922 and NMS-E973. Fig. 3A shows the position of the three compounds in the DN. The closest community to the three tested compounds is n. 28, composed by the HSP90 inhibitors present in cMap, as well as the antiestrogen drug Fulvestrant, known to bind the estrogen receptor, dissociate HSP90, and trigger its intracellular degradation. The second closest community common to all the three compounds (n. 40) is enriched for proteasome inhibitors, ubiquitin proteasome system modulators (Celastrol, MG-132, MG-262, Thapsigargin, Disulfiram, Mometasone), and protein synthesis inhibitors (Puromycin and Primaquine). Another interesting surrounding community is n.104, which contains the proteasome/NF-kB inhibitors Withaferin A, Parthenolide, Thiostrepton, and Etacrynic acid. Weaker edges connect two of the three tested compounds to community n. 63, consisting of Na+/K+-ATPase membrane pump inhibitors. This proximity might be explained by the fact that inhibition of Na+/K+-ATPases by cardiac glycosides has been shown to affect NF-kB signaling (31). Fuzzy GO term enrichment analysis showed that genes involved in the response to unfolded proteins are up-regulated in community n. 28 and community n. 104, whereas community n. 40 is enriched for GO terms relative to endoplasmatic reticulum overload and stress. Therefore, the DN approach correctly predicted, with multiple evidences, the MoA of the tested compounds by identifying them as HSP90 inhibitors.
We also tested four cyclin-dependent kinases (CDKs) inhibitors. CDKs are key regulators of cell cycle progression: CDK2 and CDK4 are responsible for phosphorylation of the Retinoblastoma (RB) protein, causing activation of the E2F transcription factor and transcription of genes involved in G1/S transition and initiation of DNA replication (32). Several CDK inhibitors are being developed as anticancer agents, including Flavopiridol, currently in Phase III clinical trials (33). The cMap includes a limited number of molecules associated with this MoA. Therefore, we sought to probe the DN with the transcriptional profile of Flavopiridol, as well as those of PHA-690509, PHA-793887, and PHA-848125, three ATP-competitive CDK inhibitors developed at NMS, with different selectivity profiles within the CDK family, which have completed Phase I clinical trials (34) (Table S2 reports a selectivity profile of the four CDK inhibitors). The closest neighboring drugs and communities in the DN to each of the tested compound are listed in Table S1. All four CDK inhibitors were positioned in the DN in close vicinity to community n. 14, which includes a mixture of CDK and Topoisomerase inhibitors, altogether accounting for about 80% of this community (Fig. 3C). The other closest community was n. 32, also containing several CDK and/or Topoisomerase inhibitors, such as the CDK2 inhibitors Chrysin, Harmine, Harman, and Harmol, the CDK2/Topo II inhibitor Apigenin, the CDK2/Topo I inhibitor Luteolin, and the Topo I inhibitors Irinotecan and Skimmianine.
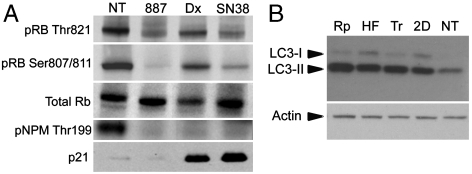
The intermixing of CDK and Topoisomerase inhibitors in communities n. 14 and n. 32, as well as the identification of several Topoisomerase inhibitors as the closest neighbors of the CDK inhibitors, implies a similarity of their effects at the transcriptional level, despite their different intracellular protein targets. To confirm this transcriptional similarity, we probed the DN with in-house generated transcriptional profiles following treatment with SN-38, the active metabolite of Irinotecan (a prototypic Topo I inhibitor) and with Doxorubicin (a prototypic Topo II inhibitor). SN-38 and Doxorubicin were positioned, as expected, close to communities n. 14 and n. 32, containing their counterparts in the database (Fig. 3B). The 10 closest neighbors for both compounds are found in Table S1 and include a mixture of CDK and Topo I or II inhibitors. Whereas most CDK inhibitors act by competitively binding to the ATP pocket of kinases, and given that Topo II uses ATP hydrolysis for its function, we verified that there was no direct biochemical inhibition of CDKs by SN-38 and Doxorubicin, and that Flavopiridol was not able to interfere with the ATPase activity of Topo II (Fig. S2). Another possible way to induce functional inhibition of CDKs is through the induction of their universal inhibitor p21. Indeed, DNA damage induced by Topoisomerase inhibitors causes p21 up-regulation activating both p53-dependent and independent apoptosis (35, 36). We hypothesized that p21 inhibition of the endogenous CDKs, and in particular CDK2, elicited an effect on RB-mediated transcription and might thus explain the similarity at the gene expression level. To confirm this, we treated MCF7 cells for 6 h with PHA-793887 (used as reference CDK inhibitor), Doxorubicin, or SN-38, at the same doses previously used, and analyzed the protein cell lysates by Western blot (WB). Following treatment with both Topoisomerase inhibitors, we observed induction of p21 resulting in inhibition of CDK2, as measured by decreased phosphorylation of the CDK2 substrates, RB. and nucleophosmin (Fig. 4A). Although we cannot exclude that induction of other genes, such as p27, in addition to p21, may also contribute to this effect. It was recently proposed that Camptothecin treatment would directly inhibit CDK9 activity by disrupting its complex with the activating Cyclin T partner, inducing a functional effect similar to that observed after ATP-competitive inhibition of CDK9 by Flavopiridol (37). To test this hypothesis, we analyzed the protein cell lysates used in the previous experiment for inhibition of RNA polymerase II, as measured by decreased phosphorylation of its carboxy-terminal domain and diminished MCL1 (myeloid cell leukemia sequence 1) levels. After treatment with PHA-793887 (CDK7 inhibition IC50 = 10 nM; CDK9 inhibition IC50 = 140 nM), a decrease of phosphoserine 5, and to a minor extent also of phosphoserine 2, was detected and resulted in diminished levels of MCL1. However, no effect on RNA Polymerase II phosphorylation or MCL1 levels was observed after treatment with the Topo inhibitors, suggesting that this pathway was not affected (Fig. S3). Taken together, these data prove that the transcriptional effects observed with the Topo I and Topo II inhibitors are due to an (indirect) inhibition of CDK2 (and possibly other CDKs such as CDK4) mediated by p21 induction, highlighting a previously unreported similarity that provides a strong rationale for the DN classification results.
Fig. 4.
Western blots (A) Western blot of total MCF7 cell lysates following 6-h treatment of MCF7 cells with Doxorubicin (Dx), SN38, and the CDK inhibitor PHA-793887 (887). Induction of p21 coupled to decreased phosphorylation of the CDK2 substrates Retinoblastoma (Rb) and Nucleophosmin (NPM) by the Topo inhibitors Dx and SN-38. (B) Evaluation of LC3 levels in human fibroblasts after treatment with drugs: (Rp, Rapamycin; HF, Fasudil; Tr, Trifluoperazine; 2D, 2-deoxy-D-glucose; NT, untreated). The experiments were performed in triplicate, and representative results are shown.
Prediction of Unique Clinical Applications for Known Drugs.
The DN approach can be used to find candidates for drug “repositioning,” i.e., to identify unique clinical applications of well-known drugs. We focused on identifying drugs that could enhance autophagy, a key biological process involved in cancer and in infectious and neurodegenerative diseases (38).
To this end, we searched the DN for drugs similar to 2-deoxy-D-glucose (2DOG), a molecule that is known for its ability to induce autophagy (39). 2-deoxy-D-glucose was found in community n. 1, which contained, in increasing order of distance to 2DOG, Fasudil, Sodium-phenylbutirate, Tamoxifen, Arachidonyltrifluoromethane, and Novobiocin (see Table S3). Note that, in this community, two drugs (2DOG and Tamoxifen) are known autophagy inducers (39, 40) and that Fasudil is the closest drug to 2DOG. In addition, by analyzing the distances of 2DOG from the other compounds in the network, independently of the community they belong to, we found that the closest compounds to 2DOG were, in order of similarity, Fasudil, Thapsigargin, Trifluoperazine, and Gossypol (the whole neighborhood is provided in Table S3). Of these, Thapsigargin, Trifluoperazine, and Gossypol are known inducers of autophagy (41–43).
Despite being a drug with a well-characterized MoA, Fasudil has never been previously linked to autophagy. To verify the effect of Fasudil on the induction of autophagic pathway, we evaluated the LC3-II levels in wild-type human fibroblasts treated with Fasudil, by WB with anti-LC3 antibody, a well-established assay for the activation of autophagy (44). We measured a marked increase in LC3-II levels in fibroblasts treated with Fasudil and Trifluoperazine identified by the DN, as well as, in cells treated with 2DOG and Rapamycin, two well-known inducers of autophagy (Fig. 4B). Immunostaining with LC3 antibody further confirmed the WB analysis, demonstrating a strong activation of autophagic degradation upon treatment with Fasudil (Fig. S4). The effect of Fasudil on autophagy enhancement was further confirmed in HeLa cells (Fig. S5).
Discussion
We developed a general procedure to predict the molecular effects and MoA of new compounds, and to find previously unrecognized applications of well-known drugs. We were able to exploit information hidden in the gene expression profiles following drug treatment to capture similarity in drug MoA. Previous attempts to use gene expression profiles following compound treatment in mammalian cells did not consider the variability in the transcriptional response to the compound due to cell-line effects, to different dosages, and to different experimental settings. Moreover, information embedded in the global structure of the network of similarities among drugs has not been fully exploited in the past. We removed unspecific effects by capturing the “consensus” transcriptional response to a compound across multiple cell lines and dosages. We then automatically extracted a gene signature for each compound and computed pairwise similarities between compounds using a gene signature-based approach.
We analyzed the resulting network to identify communities of drugs with similar MoA and to determine the biological pathways perturbed by these compounds. We remark that, differently from other methods, whose aim is to identify the specific drug substrates (2, 6), our approach also groups together compounds interacting with distinct members of the same pathway.
The DN can be used to infer the MoA and targeted pathways of anticancer compounds still being studied and to find candidates for “drug repositioning” (i.e., to suggest unique clinical application for well-known and approved drugs).
We correctly classified both known and previously undescribed HSP90 inhibitors. Interestingly, in addition to the HSP90 inhibitors present in the database (Alvespimycin, Geldanamycin, and Monorden), several drugs included in the top 10 closest neighbors for Tanespimycin and NMS-E973 were connected to inhibitors of the proteasome/NF-κB pathway, including Disulfiram (45), Withaferin A (46), and Parthenolide (47).
We also investigated the ability of the DN to classify well-known (Flavopiridol) and previously undescribed CDK inhibitors (PHA-690509, PHA-793887, and PHA-848125). These drugs were correctly classified as CDK inhibitors, distinct from the other kinase inhibitors in the database, and were also predicted to be very similar to Topoisomerase inhibitors. Although the induction of p21 by DNA damage-inducing agents was previously reported, here we showed that this is clearly detected at the transcriptional level, supporting the concept that gene modulations can be used as a biomarker to monitor the effect of DNA damage-inducing agents.
In addition, we experimentally verified a surprising prediction: Fasudil promotes cellular autophagy. Given the excellent safety profile, this newly recognized effect of Fasudil could be exploited for disorders due to protein misfolding, including neurodegenerative diseases.
The drug network can be useful for formulating hypotheses on the MoA of previously undescribed compounds by simply measuring multiple transcriptional responses in different cell lines. In addition, drug repositioning is the easiest way to find previously undescribed drug therapies for different conditions. We have shown that it is possible to find previously unrecognized MoAs of well-characterized drugs by simply looking for the drugs neighboring a drug of interest. In addition, by analyzing the PRLs associated to each drug in the network, we may identify the drug communities that consistently up-, or down-regulate a given set of genes, thus hinting to drug classes able to modulate a specific pathway of interest. The major limitation of our approach is in the limited number of compounds in the network. Because our approach is based on comparing how similar two drugs are, if a compound is not similar to any of the drugs in the network, no inference on its MoA or its biological effects can be done.
Moreover, for a compound having inconsistent effects on different cell lines (for example, due to a cell line with a mutated substrate–protein targeted by the compound) merging gene expression profiles from distinct cell lines may dilute the biological effects of the compound. Nevertheless, when no information on the drug MoA is available a priori, the best strategy is still to merge profiles from multiple cell lines. We have evidences, reported in the online SI Table 5 and SI Methods, that merging profiles coming from a sufficiently large, even if heterogeneous, pool of treated cell lines, provides a summary of the transcriptional response to the drug that can still be well classified by the DN.
We have made our approach publicly available as an online tool (http://mantra.tigem.it). The DN can be easily searched for a compound of interest, or queried with the transcriptional responses of a unique compound, thus providing a valuable tool to the research community.
Methods
Treatment with Test Drugs for Microarray Hybridizations.
A2780 cell lines (human ovary adenocarcinoma) were treated with Flavopiridol, PHA-848125, PHA-690509, and PHA-793887, whereas MCF7 cell lines (human mammary adenocarcinoma) were treated with PHA-848125, PHA-793887, Tanespimycin, NVP-AUY922, NMS-E973, SN-38, and Doxorubicin. The procedures following treatments and the data availability are exhaustively detailed in SI Methods.
Treatments for Western Blot of Total MCF7 Cell Lysates and Evaluation of Autophagy.
For the Western blot, MCF7 cells were treated with PHA-793887, Doxorubicin, or SN-38 at a dose equal to 5 times the IC50 for 6 h. For autophagy evaluation, synchronized wild-type human fibroblasts were treated with the following drugs: Fasudil dihydrochloride (Sigma) 10 μM, Trifluoperazine (Sigma) 1 μM, and 2DOG (Sigma) 100 μM for 48 h. The procedures following treatments are exhaustively detailed in SI Methods.
Supplementary Material
Acknowledgments.
The authors thank W. Pastori, A. Deponti, I. Fraietta, L. Raddrizzani, G. Locatelli, S. Healy, G. G. Galli, N. Avanzi, D. Volpi, F. Roletto, R. Baldi, B. Valsasina, M. Ciomei, G. Fogliatto, G. Raiconi, M. Lauria, G. Diez-Roux, J. D. Wichard, G. Gambardella, and J. Lamb. This work was supported by Fondazione Telethon Grants TDDP51TELC and TCBP37TELC (to D.d.B. and N.B.P) and by the Italian Bioinformatics Network grant of the Italian Ministry of Research (to D.d.B).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE18552).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1000138107/-/DCSupplemental and at http://mantra.tigem.it.
References
- 1.Terstappen GC, Schlupen C, Raggiaschi R, Gaviraghi G. Target deconvolution strategies in drug discovery. Nat Rev Drug Discov. 2007;6:891–903. doi: 10.1038/nrd2410. [DOI] [PubMed] [Google Scholar]
- 2.di Bernardo D, et al. Chemogenomic profiling on a genome-wide scale using reverse-engineered gene networks. Nat Biotechnol. 2005;23:377–383. doi: 10.1038/nbt1075. [DOI] [PubMed] [Google Scholar]
- 3.Ambesi-Impiombato A, di Bernardo D. Computational biology and drug discovery: From single-tTarget to network drugs. Curr Bioinform. 2006;1:3–13. [Google Scholar]
- 4.Berger SI, Iyengar R. Network analyses in systems pharmacology. Bioinformatics. 2009;25:2466–2472. doi: 10.1093/bioinformatics/btp465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hopkins AL. Network pharmacology: The next paradigm in drug discovery. Nat Chem Biol. 2008;4:682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 6.Mani KM, et al. A systems biology approach to prediction of oncogenes and molecular perturbation targets in B-cell lymphomas. Mol Syst Biol. 2008;4:169. doi: 10.1038/msb.2008.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gardner TS, di Bernardo D, Lorenz D, Collins JJ. Inferring genetic networks and identifying compound mode of action via expression profiling. Science. 2003;301:102–105. doi: 10.1126/science.1081900. [DOI] [PubMed] [Google Scholar]
- 8.Hu G, Agarwal P. Human disease-drug network based on genomic expression profiles. PloS One. 2009;4(8):e6536. doi: 10.1371/journal.pone.0006536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hughes TR, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102(1):109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 10.Kohanski MA, Dwyer DJ, Wierzbowski J, Cottarel G, Collins JJ. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell. 2008;135(4):679–690. doi: 10.1016/j.cell.2008.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamb J. The Connectivity Map: A new tool for biomedical research. Nat Rev Cancer. 2007;7(1):54–60. doi: 10.1038/nrc2044. [DOI] [PubMed] [Google Scholar]
- 12.Lamb J, et al. The Connectivity Map: Using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 13.Yang K, Bai H, Ouyang Q, Lai L, Tang C. Finding multiple target optimal intervention in disease-related molecular network. Mol Syst Biol. 2008;4:228. doi: 10.1038/msb.2008.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campillos M, Kuhn M, Gavin AC, Jensen LJ, Bork P. Drug target identification using side-effect similarity. Science. 2008;321:263–266. doi: 10.1126/science.1158140. [DOI] [PubMed] [Google Scholar]
- 15.Li J, Zhu X, Chen JY. Building disease-specific drug-protein connectivity maps from molecular interaction networks and PubMed abstracts. PLoS Comput Biol. 2009;5(7):e1000450. doi: 10.1371/journal.pcbi.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keiser MJ, et al. Predicting new molecular targets for known drugs. Nature. 2009;462:175–181. doi: 10.1038/nature08506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller MA. Chemical database techniques in drug discovery. Nat Rev Drug Discov. 2002;1:220–227. doi: 10.1038/nrd745. [DOI] [PubMed] [Google Scholar]
- 18.Iorio F, Tagliaferri R, di Bernardo D. Identifying network of drug mode of action by gene expression profiling. J Comput Biol. 2009;16:241–251. doi: 10.1089/cmb.2008.10TT. [DOI] [PubMed] [Google Scholar]
- 19.McAuley JJ, Costa LF, Caetano TS. Rich-club phenomenon across complex network hierarchy. Appl Phys Lett. 2007;91:084103. [Google Scholar]
- 20.Diaconis P, Graham R. Spearman’s footrule as a measure of disarray. J R Stat Soc. 1977;39:262–268. [Google Scholar]
- 21.Lin S, et al. Space oriented rank-based data integration. Stat Appl Genet Mol Biol. 2010;9:Article20. doi: 10.2202/1544-6115.1534. [DOI] [PubMed] [Google Scholar]
- 22.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frey BJ, Dueck D. Clustering by passing messages between data points. Science. 2007;315:972–976. doi: 10.1126/science.1136800. [DOI] [PubMed] [Google Scholar]
- 24.Newman ME. Modularity and community structure in networks. Proc Natl Acad Sci USA. 2006;103:8577–8582. doi: 10.1073/pnas.0601602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pahor M, et al. Drug data coding and analysis in epidemiologic studies. Eur J Epidemiol. 1994;10(4):405–411. doi: 10.1007/BF01719664. [DOI] [PubMed] [Google Scholar]
- 26.Schwabe U. ATC-Code. Bonn, Germany: Wissenschaftliches Institut der AOK; 1995. [Google Scholar]
- 27.Wishart DS. DrugBank and its relevance to pharmacogenomics. Pharmacogenomics. 2008;9:1155–1162. doi: 10.2217/14622416.9.8.1155. [DOI] [PubMed] [Google Scholar]
- 28.Seiler KP, et al. ChemBank: A small-molecule screening and cheminformatics resource database. Nucleic Acids Res. 2008;36(Database issue):D351–359. doi: 10.1093/nar/gkm843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eccles SA, et al. NVP-AUY922: A novel heat shock protein 90 inhibitor active against xenograft tumor growth, angiogenesis, and metastasis. Cancer Res. 2008;68:2850–2860. doi: 10.1158/0008-5472.CAN-07-5256. [DOI] [PubMed] [Google Scholar]
- 30.Fogliatto G, et al. Identification of a potent and specific inhibitor of Hsp90 showing in vivo efficacy. American Association for Cancer Research—Annual Meeting. 2009. Poster 37 (Abstract #4685)
- 31.Yang Q, et al. Cardiac glycosides inhibit TNF-alpha/NF-kappaB signaling by blocking recruitment of TNF receptor-associated death domain to the TNF receptor. Proc Natl Acad Sci USA. 2005;102:9631–9636. doi: 10.1073/pnas.0504097102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malumbres M, Barbacid M. Mammalian cyclin-dependent kinases. Trends Biochem Sci. 2005;30:630–641. doi: 10.1016/j.tibs.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 33.Senderowicz AM. Flavopiridol: The first cyclin-dependent kinase inhibitor in human clinical trials. Invest New Drugs. 1999;17:313–320. doi: 10.1023/a:1006353008903. [DOI] [PubMed] [Google Scholar]
- 34.Brasca MG, et al. Identification of N,1,4,4-tetramethyl-8-{[4-(4-methylpiperazin-1-yl)phenyl]amino}-4,5-dihydr o-1H-pyrazolo[4,3-h]quinazoline-3-carboxamide (PHA-848125), a potent, orally available cyclin dependent kinase inhibitor. J Med Chem. 2009;52:5152–5163. doi: 10.1021/jm9006559. [DOI] [PubMed] [Google Scholar]
- 35.Abal M, et al. Enhanced sensitivity to irinotecan by Cdk1 inhibition in the p53-deficient HT29 human colon cancer cell line. Oncogene. 2004;23:1737–1744. doi: 10.1038/sj.onc.1207299. [DOI] [PubMed] [Google Scholar]
- 36.Liu W, Zhang R. Upregulation of p21WAF1/CIP1 in human breast cancer cell lines MCF-7 and MDA-MB-468 undergoing apoptosis induced by natural product anticancer drugs 10-hydroxycamptothecin and camptothecin through p53-dependent and independent pathways. Int J Oncol. 1998;12:793–804. [PubMed] [Google Scholar]
- 37.Amente S, Gargano B, Napolitano G, Lania L, Majello B. Camptothecin releases P-TEFb from the inactive 7SK snRNP complex. Cell Cycle. 2009;8:1249–1255. doi: 10.4161/cc.8.8.8286. [DOI] [PubMed] [Google Scholar]
- 38.Rubinsztein DC, Gestwicki JE, Murphy LO, Klionsky DJ. Potential therapeutic applications of autophagy. Nat Rev Drug Discov. 2007;6:304–312. doi: 10.1038/nrd2272. [DOI] [PubMed] [Google Scholar]
- 39.Ravikumar B, et al. Raised intracellular glucose concentrations reduce aggregation and cell death caused by mutant huntingtin exon 1 by decreasing mTOR phosphorylation and inducing autophagy. Hum Mol Genet. 2003;12:985–994. doi: 10.1093/hmg/ddg109. [DOI] [PubMed] [Google Scholar]
- 40.de Medina P, Silvente-Poirot S, Poirot M. Tamoxifen and AEBS ligands induced apoptosis and autophagy in breast cancer cells through the stimulation of sterol accumulation. Autophagy. 2009;5:1066–1067. doi: 10.4161/auto.5.7.9820. [DOI] [PubMed] [Google Scholar]
- 41.Criollo A, et al. Regulation of autophagy by the inositol trisphosphate receptor. Cell Death Differ. 2007;14:1029–1039. doi: 10.1038/sj.cdd.4402099. [DOI] [PubMed] [Google Scholar]
- 42.Lei X, et al. Gossypol induces Bax/Bak-independent activation of apoptosis and cytochrome c release via a conformational change in Bcl-2. FASEB J. 2006;20:2147–2149. doi: 10.1096/fj.05-5665fje. [DOI] [PubMed] [Google Scholar]
- 43.Ogata M, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mizushima N. Methods for monitoring autophagy. Int J Biochem Cell Biol. 2004;36:2491–2502. doi: 10.1016/j.biocel.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 45.Cvek B, Dvorak Z. The value of proteasome inhibition in cancer. Can the old drug, disulfiram, have a bright new future as a novel proteasome inhibitor? Drug Discov Today. 2008;13:716–722. doi: 10.1016/j.drudis.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 46.Yang H, Shi G, Dou QP. The tumor proteasome is a primary target for the natural anticancercompound Withaferin A isolated from “Indian winter cherry”. Mol Pharmacol. 2007;71:426–437. doi: 10.1124/mol.106.030015. [DOI] [PubMed] [Google Scholar]
- 47.Hehner SP, Hofmann TG, Droge W, Schmitz ML. The antiinflammatory sesquiterpene lactone parthenolide inhibits NF-kappa B by targeting the I kappa B kinase complex. J Immunol. 1999;163:5617–5623. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.