Abstract
This article reviews the application of single-molecule fluorescence resonance energy transfer (smFRET) methods to the study of protein synthesis catalyzed by the ribosome. smFRET is a powerful new technique that can be used to investigate dynamic processes within enzymes spanning many orders of magnitude. The application of wide-field smFRET imaging methods to the study of dynamic processes in the ribosome offers a new perspective on the mechanism of protein synthesis. Using this technique, the structural and kinetic parameters of tRNA motions within wild-type and specifically mutated ribosome complexes have been obtained that provide valuable new insights into the mechanism and regulation of translation elongation. The results of these studies are discussed in the context of current knowledge of the ribosome mechanism from both structural and biophysical perspectives.
Keywords: single-molecule, FRET, ribosome, translation, tRNA
INTRODUCTION
The era of structural genomics has generated atomic resolution models of many enzyme structures that have had profound impact throughout the biological sciences. While this precise information is essential to understanding enzymatic function, extracting mechanistic information from static structures alone is speculative. Enzymes are intrinsically dynamic entities on timescales spanning many orders of magnitude.1,2 Therefore, the synthesis of structure-function relationships into experimentally-validated trajectories of enzyme motion is of widespread and significant importance. Here we discuss briefly our most recent efforts to understand ribosome function at the single-molecule scale using Fluorescence (or Förster) Resonance Energy Transfer (FRET) and synergistic computational efforts.
The ribosome is a dynamic molecular machine that converts genetic information in the form of messenger RNA (mRNA) into protein using aminoacylated transfer RNA (aa-tRNA) substrates. Single-molecule studies of protein synthesis benefit from prior discovery of the components necessary and sufficient to support accurate and expeditious expression of mRNA by the ribosome and decades of biochemical and structural interrogation.3–8 tRNA binding sites on the ribosome have been defined through chemical footprinting experiments,9,10 and later visualized by cryo-electron microscopy (cryo-EM).11–13 The precise interactions of tRNA with the T. thermophilus ribosome have been recently elucidated in atomic detail using X-ray crystallography.14–18
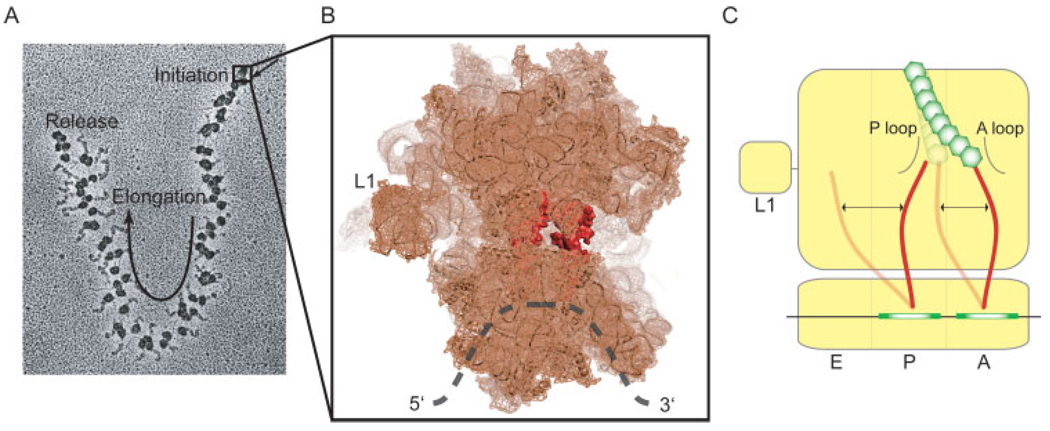
To synthesize a protein, the ribosome must first localize to the start site of protein synthesis (Initiation, ca. ∼1–10 s). This multibody reaction requires unfolding of the mRNA at the start site, contemporaneous with assembly of the initiation complex containing mRNA, the two unequally sized ribosomal subunits (30S and 50S in bacteria), and initiator tRNA (fMet-tRNAfMet in E. coli). This process, facilitated by three initiation factors (IF-1, -2, and -3), places fMet-tRNAfMet in the peptidyl (P) site. The initiation complex then directionally transits the mRNA open reading frame in discrete codon increments (elongation, ca. ∼100–500 ms per step) during which tRNAs move rapidly through the ribosome. Elongation occurs via a series of repetitive aa-tRNA selection and translocation processes catalyzed by Elongation Factors, EF-Tu and -G, respectively. aa-tRNA enter the aminoacyl (A) site in a ternary complex with EF-Tu and GTP in a GTP hydrolysis dependent manner19 (tRNA selection). Its accommodation into the A site is followed immediately by ribosome-catalyzed peptide bond formation,20 which transfers the amino acid linked to tRNAfMet to the alpha amino group of the incoming aa-tRNA thus creating peptidyl-tRNA in the A site and leaving a deacylated tRNAfMet in the P site.20 EF-G, also a GTPase, catalyzes the translocation of tRNA from the A site to the P site and then to the Exit (E) site in subsequent elongation steps while maintaining the correct reading frame.21 Deacylated tRNA dissociates from the ribosome via the E site. Upon reaching a stop codon, the ribosome is then separated from the protein product, mRNA, and tRNA (termination and recycling, ∼ca. 0.1–10 s). Synthesizing a single 50 KDa protein may therefore span several minutes- the vast majority during elongation. To speed the rate of protein production, a single mRNA is translated by many ribosomes simultaneously, forming a polysome (Figure 1A).
FIGURE 1.
Single-molecule imaging brings static structural models to life. (A) The ribosome is a processive enzyme that moves along mRNA in 3-nucleotide steps. In vivo, many ribosomes may translate the same mRNA concurrently, forming a polysome (Electron micrograph from nobelprize.org). (B) High resolution structural models of the ribosome provide detailed visualizations of the protein synthesis machinery.15 (C) Single-molecule methods aim at understanding how dynamic processes of tRNA within the A and P sites of the ribosome relate to the mechanism of translocation.
Although the rate limiting step in translation is often considered initiation, regulation of gene expression during elongation is drawing increased attention. “Dynamic recoding” events relevant to the regulation of elongation include frame-shifting, translational pausing, programmed miscoding, as well as premature termination and stop codon read through.22,23 Our group’s present focus on elongation processes stems from the understanding that even small perturbations of the ribosome mechanism during elongation steps can have significant cumulative impact at the systems level.
A Paradigm for Understanding the Ribosome Structure-Function Relationship
Structural and functional interrogations of the ribosome span nearly half a century. Today there are more than 50 published ribosome structures at varying resolution including recent atomic resolution structures of 70S complexes of T. thermophilus15,16 and E. coli18 ribosomes that include A-, P-, and E-site ligands (Figure 1B). These high- and low-resolution snapshots of ribosome conformation correspond to local minima,4–6 or an average over many local minima, on a complex free energy landscape.2
The energy landscape interpretation of enzyme conformation24,25 dictates that the ribosome can intrinsically access all conformations relevant, or irrelevant, to catalysis. The probability of the system spontaneously achieving a catalytically active state at equilibrium is determined by the height of the energy barrier(s) separating that state from all others with respect to the energy of the environment (kBT). When multiple conformational events separate the system from the catalytically active state, its average rate of formation would scale inversely as a function of each event’s probability. An important validation of this view is that the ribosome is able to catalyze translation in the absence of translation factors and energy consumption.26–28
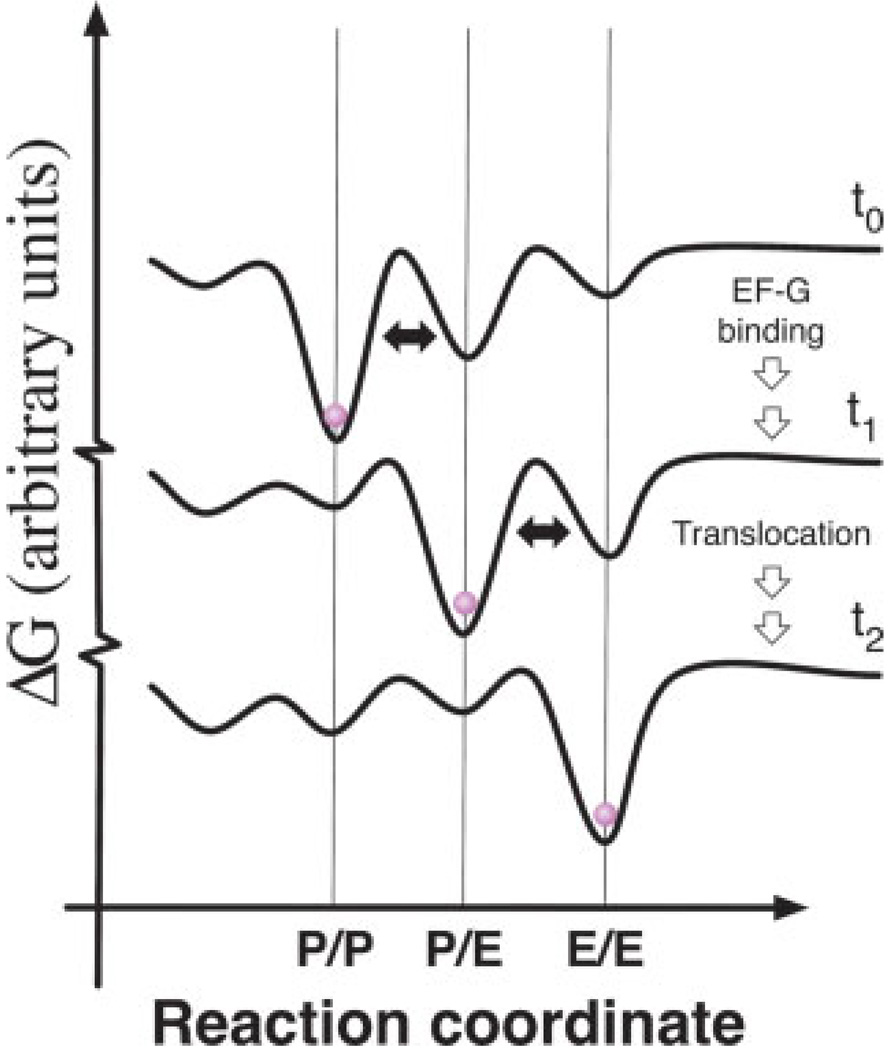
Correspondingly, cryo-EM data obtained on homogeneous ribosome preparations show distinct conformational heterogeneity, presumably reflecting a snapshot of the equilibrium distribution of states.29 Ligand binding and changes in environmental conditions, such as temperature or ionic strength, may affect ribosome function by biasing the energy landscape in favor of distinct conformational states.30,31 In this view, specific antibiotics that promote or inhibit certain translation processes32,33 may operate by remodeling the intrinsic ribosome energy landscape to favor catalytically active or inactive conformations. A schematic representation of how modulation of the ribosome energy landscape may be used to drive directional translocation is shown in Figure 2. Forward progression through the reaction coordinate is promoted by progressively shifting the global energy minimum of system towards the endpoint. This highly-simplified representation of the energy landscape is, in reality, multidimensional in nature due to coupled and uncoupled motions of the ribosome, factors, and substrates, and significantly more rugged than shown due to conformational substates of the system.
FIGURE 2.
A schematic representation of how the ribosome energy landscape may be modulated to promote translocation. In this highly-simplified view, each curve schematically represents the energy landscape of the ribosome at discrete steps in translocation. The free energy of the system is shown as a function of the P-site tRNA position on the ribosome before EF-G binding (t0), after EF-G binding (t1) and after translocation (t2). This schematic is meant to portray only how the global minimum of the system may evolve to promote forward progression. The absolute height of the energy barriers depicted at t0, t1, and t2 are not drawn to scale and should not be compared to one another. Curves are off-set on the Y-axis for visualization purposes. Arrows linking each of the three states are shown to indicate multiple steps may separate t0, t1, and t2.
Subtle changes in the relative stabilities of different enzyme conformations may have profound impacts at the systems level, especially for processive enzymes such as the ribosome, and high-turnover kinases where numerous targets are rapidly phosphorylated. In the context of protein synthesis, it is well established that small changes in factor and substrate availability, mRNA structure, intracellular ionic strength, and growth temperature have significant impact on global translation levels.22 This model predicts that cell survival hinges on the ability to “fine tune” the ribosome’s energy landscape according to specific growth demands, altering the rate and/or efficiency of specific mRNA translation. Such mechanisms may be facilitate quick responses to changes in external stimuli, the cell cycle, or events in the differentiated state.34 In accordance with this model, post translational modifications and specific protein factors are present in ribosomes translating mRNAs with Internal Ribosome Entry Sites.35 Recent observations that p53 and p14ARF, tumor suppressor proteins, bind directly to the ribosome may be indicative that these proteins regulate translation by altering the ribosome energy landscape to suppress or promote specific translation events.36,37
Single-Molecule FRET Brings Static Structural Models to Life
Given this framework, questions of translation regulation and mechanism distill to understanding the order and timing of transitions between local minima on the energy landscape. Elucidation of the translation reaction trajectory from this perspective by such methods as single-molecule FRET is the first essential step in converting static snapshots of enzyme structure into movies depicting enzyme function. Information of this nature is not generally accessible using traditional bulk methods where asynchronous and stochastic processes are masked in the ensemble. 10,38,39 In bulk studies, important information may be obtained regarding how a system is driven from one equilibrium condition to another, however, information pertaining to the starting and endpoint equilibrium of states is sacrificed. Furthermore, intermediate states along the reaction coordinate may be too unstable to identify easily. In contrast, single-molecule measurements provide a reaction trajectory for each individual complex, allowing for the observation of specific pathways through the energy landscape40,41 and the full distribution of kinetic behavior.42 Single-molecule observations, presently limited by the time resolution and signal-to-noise ratio (SNR) of data acquisition, and total fluorescence lifetime, afford a direct means of determining the structural43–45 and compositional heterogeneities potentially obscured in bulk measurements. Investigations at the single-molecule scale also assist in the exploration of mechanistic questions in systems where only limited or incompletely labeled sample can be obtained.
Site-Specific Labeling of Biomolecules for Single-Molecule Imaging
The technologies that enable site-specific introduction of donor and acceptor fluorophores into complex biomolecules are essential to single-molecule FRET experiments. Here structural knowledge is paramount. Traditional labeling approaches that rely on amine- and sulfur-reactive fluorophores are not generally useful in large systems like the ribosome due to the abundance of reactive, solvent accessible side-chains.46 To circumnavigate this issue in the case of the ribosome, total reconstitution strategies have been employed.47 Alternatively, orthogonal chemistries including the introduction of non-natural amino acids48 and C-terminal intein ligation,49 offer powerful new solutions to these limitations. Nucleic acid containing enzymes, such as the ribosome, may also be labeled either by hybridization strategies46,50 or by exploiting the presence of naturally occurring modified nucleotides.51,52 In all cases, maintenance of biological function is essential.
The first single-molecule FRET investigations of DNA, RNA, and protein systems were reported over a decade ago.53–59 Its application to more complex systems was soon to follow.46,56,59,60 Many of the instruments employed were configured for wide-field total internal reflection fluorescence (TIRF) microscopy. Wide-field TIRF has the advantage that fluorescence and FRET trajectories can be obtained from up to ∼1000 surface-immobilized biomolecules simultaneously using a CCD array. With current CCD technology, timescales of ∼10 ms are readily achieved. By contrast, confocal imaging, which benefits similarly from S/N enhance- ments by confining the illuminated region to a tiny volume (ca. ∼0.5 µm3), is better suited for observations of just one molecule at a time. An advantage of confocal imaging, however, is that timescales commensurate with nuclear magnetic resonance (NMR) spectroscopy techniques (ca. ∼10 ns) can be achieved using avalanche photodiodes.61 The optimal experimental approach must be chosen based on the biological question at hand.
While many elaborations of the theme have been implemented by us and others, much of the credit belongs to the early pioneers of single-molecule imaging. The contributions of these early works included surface immobilization methods to extend observation times beyond the limits of diffusion, the intellectual framework for understanding the structurally dynamic properties of enzymes, and the first computational methods for analyzing dynamic FRET trajectories. More comprehensive reviews of the history of single-molecule methods applied to biological systems, the nature of stochastic processes, the repertoire of single-molecule imaging solutions, and considerations that must be given to the interpretation of FRET data, can be found elsewhere.40,61–65
The timescales of translation processes (spanning milliseconds to minutes) are well-suited to relatively slow wide-field imaging methods because SNR increases proportionally with integration time and photon count. Whereas other single-molecule methods, such as force spectroscopy using optical trapping, atomic force microscopy, or ion channel patch clamp methods, are generally limited by the number of molecules observed, single-molecule fluorescence resonance energy transfer (smFRET) studies are often limited by the number of FRET transitions that can be recorded during the lifetime of fluorescence. Although significant advancements have been made in the area of enhancing the lifetime and photophysical properties of organic dye molecules,66,67 in our experience, fluorophores generally last ∼5–10 s under the intense illumination that is requisite for high SNR measurements, even in idealized conditions (Dave et al. in preparation). Given the probabilistic nature of photobleaching rates, one solution to this problem is to image large numbers of single molecule FRET trajectories (ca. ∼104 ) in order to obtain meaningful numbers (ca. ∼103 ) of long-lived dye pairs. From this subset, a full distribution of kinetic behavior, reflecting the complete free energy landscape, can be obtained.
smFRET: A Nanoscopic Imaging Tool
Excitation of the donor fluorophore attached directly to the biological sample results in energy transfer to nearby acceptor fluorophores. The efficiency of this dipole–dipole interaction is determined principally by the distance between the two fluorophores according to the Förster relationship E = 1/(1 + (R/R0)6 ), where R is the distance and R0 is the distance around which FRET is a sensitive measure of motion. The R0 value is specific to the dye pair used (∼50–60 Å for the Cy3/ Cy5 dye pair) and is a function of the spectral overlap integral (J), dye mobility (κ), and the refractive index (n) according to the equation R0 = 0.211(κ2 n−4 QDJ(λ))1/6. Given fast, isotropic rotational sampling of the fluorophores during the observation period, FRET efficiency, determined by the ratio of acceptor fluorescence to total fluorescence, E = Iacceptor/(Iacceptor + Idonor), allows accurate measurement of the distance between donor and acceptor fluorophores on the nanometer scale. However, because other considerations do apply that are less readily determined, such as fluorescence lifetime and spectral drift, FRET measurements are not typically used for absolute distance determination.61 Molecular standards, such as oligonucleotides of known end-to-end distance, may be used to generate experimentally-derived Förster curves where deviations from theory can be assessed.40 However, it is a generally more cautious approach to report changes in FRET efficiency.
The structural and kinetic features of distinct conformational states in a system, observed as time-dependent changes in FRET, may be quantified by measuring the FRET value and duration of each FRET state. Finding a correspondence between FRET states observed and specific configurations of the biomolecule relies heavily on physical frameworks provided by established structural models of the system. This effort is further aided by molecular modeling of the exact dye locations. Cryo-EM or NMR data on enzyme solution-structure, and molecular dynamics (MD) simulations, can be of great use in assigning structural configurations to the observed FRET states. Alternatively, as will be discussed below, distinct FRET states may be stabilized through mutagenic strategies, or the addition of pharmaceutical agents and exogenous factors of known function.
tRNA Motions are an Intrinsic Feature of the Translation Reaction Coordinate
Here we elaborate on recent progress in single-molecule investigations of the ribosome energy landscape and the identification of novel intermediate states along the elongation reaction coordinate using smFRET Particular attention will be drawn to the mechanistic significance of intermediate states and their potential relation to previous studies of translocation.41
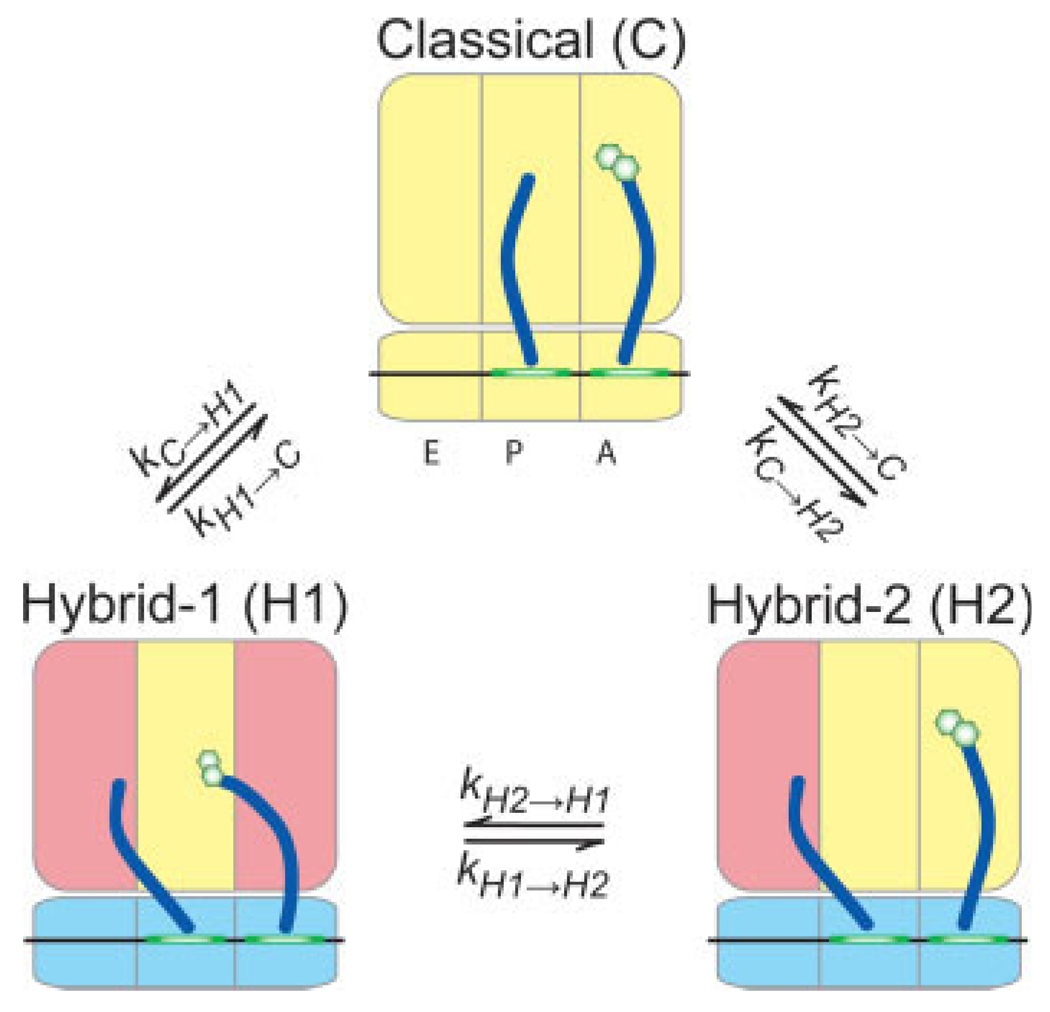
The three structurally-defined tRNA binding sites (the A, P, and E sites) represent stable configurations and local minima on the energy landscape of ribosome-tRNA interactions. Initially it was proposed that tRNAs translocate through the small and large subunits of the ribosome simultaneously.68 Chemical probing experiments clarified this hypothesis, indicating that tRNAs spontaneously adopt hybrid configurations on the ribosome whereby their 3′-acceptor ends transition to adjacent sights on the large subunit, while the codon-anticodon helices remain fixed in the small subunit69 (Figure 1C). These hybrid tRNA configurations were therefore termed the A/P and P/E hybrid states, in reference to peptidyl and deacylated tRNA, respectively. Kinetic studies suggested that the energy required for spontaneous formation of hybrid states stems from the favorable ΔG of forming a peptide bond.70 Consistent with the hybrid state model, a low, residual reactivity of pretranslocation complexes towards puromycin, an analogue of the 3′-aminoacyl terminus of A-site tRNA, supported the notion that peptidyl-tRNA transiently and spontaneously vacates the large sub-unit A site in the absence of EF-G.71
Further structural description of the hybrid states was provided by low resolution cryo-EM microscopy reconstructions of ribosomes with deacylated P-site tRNA stabilized in a P/E configuration by nonphysiological buffer conditions.72 Later, cryo-EM models of the ribosome carrying deacylated P-site tRNA and EF-G in its GTP form in the A-site, revealed in more detail, that formation of the P/E hybrid state73–77 is likely coupled to the ratchet-like rotation of the large and small ribosomal subunits.73–75,77,78 This notion is now supported by recent bulk biochemical and FRET studies suggesting that EF-G binding induces large-scale reorganizations between P-site tRNA and the ribosome as well as intersubunit ratcheting.47,,79–82
smFRET Observations of the Translocation Reaction Coordinate
The establishment of single-molecule FRET methods for translation studies allowed for the first kinetic and structural interpretations of hybrid tRNA configurations.83,84 These smFRET studies took advantage of earlier work showing that tRNAfMet and tRNAPhe could be labeled with fluorescent dyes at naturally occurring modified nucleotides without impairing their function in translation (including aminoacylation, formylation, ternary complex formation with EF-Tu, or their interaction with the ribosome51,52,83).
These initial data,83 and our more recent studies taken at 2.5-fold higher time resolution,85 were performed using sucrose gradient purified ribosome complexes carrying biotinylated mRNA, and donor fluorophore (Cy3) labeled fMet-tRNAfMet(Cy3-s4U8) in the P site. Thus only fully assembled ribosome complexes, localized through a biotin-streptavidin bridge to polyethylene glycol (PEG)-passivated (doped with biotin-PEG) quartz surface, are visualized. The A site of surface-immobilized ribosome complexes can then be enzymatically filled with high efficiency (ca. >90%) by incubation with nanomolar concentrations of acceptor fluorophore (Cy5) labeled Phe-tRNAPhe (Cy5-acp3U47) ternary complex.
On the ribosome, tRNAs in the A and P site predominantly occupy a stable high-FRET configuration that is consistent with a dye separation of ∼42 Å. This distance is in excellent agreement with current structural models of classically-bound A and P site tRNA14 (Figure 3B). Short-lived fluctuations to lower FRET states were also observed in both the earlier and more recent studies that correspond to an increase in distance between the dye pairs of ∼5–15 Å. Based on chemical footprinting studies,69 and experimental evidence of rapid isotropic dye mobility of tRNA-linked fluorophores on the ribosome, these transient fluctuations were initially assigned to the established P/E and A/P hybrid states. Consistent with previous chemical footprinting,69 and ki- netic studies of translocation,70,86 similar complexes bearing deacylated tRNAfMet (Cy3-s4U8) in the P site, and Phe-tRNA Phe-(Cy5-acp3U47) in the A site underwent less frequent transitions to lower FRET states. It was also shown that the rates of tRNA motion depended on Mg2+ concentration, consistent with rearrangements of RNA-RNA interactions.87,88 The role of Mg2+ ions in modulating the ribosome energy landscape has recently received more attention, and it will continue to be an important area of research for understanding ribosome regulation.89 While these studies showed unequivocally that tRNAs are in dynamic exchange between unique configurations on the ribosome, because of limitations in time resolution, multiple hybrid states were not resolved.
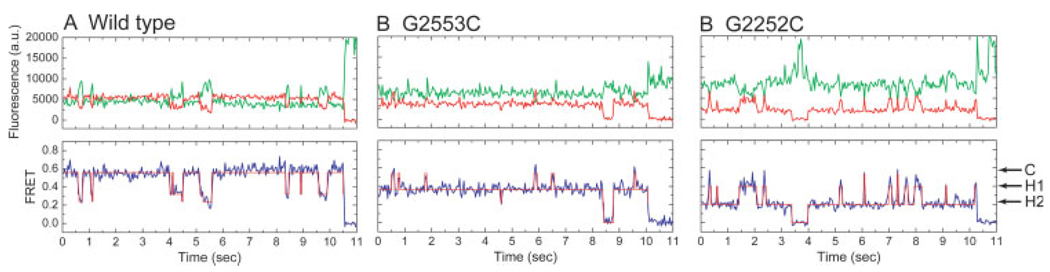
FIGURE 3.
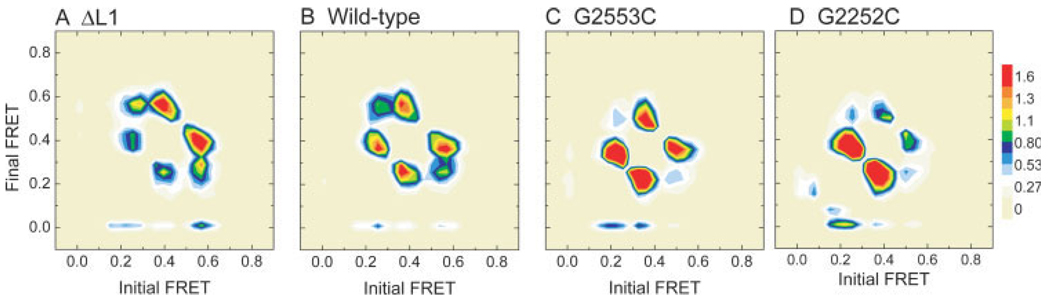
Wide-field, single-molecule FRET measurements provide direct observations of conformational dynamics of tRNAs on individual ribosomes. Anticorrelated changes in donor (green) and acceptor (red) fluorescence of dyes linked to tRNA report on the dynamics of tRNA within the ribosome. The efficiency of FRET (blue) reports on the distance separating the two dyes. Hidden Markov modeling, resulting in the idealization of the FRET trajectories (overlaid in red), is used to identify the FRET value and lifetimes of each FRET state observed as well as the order of transitions in the system. Here, FRET trajectories have been idealized to a model containing 3 states corresponding to the classical state (C), hybrid state 1 (H1), and hybrid state 2 (H2). (A) tRNAs on the wild-type ribosome are found to occupy predominantly a high-FRET classical configuration with transient excursions to the lower FRET hybrid states. (B) Mutation of residue G2553 of the A loop, and (C) G2252 of the P loop dramatically alter the stability of tRNA configurations on the ribosome, changing the energy landscape to favor intermediate- (B) and low- (C) FRET hybrid states.
The Identification and Stabilization of Two Distinct Hybrid State Intermediates on the Ribosome
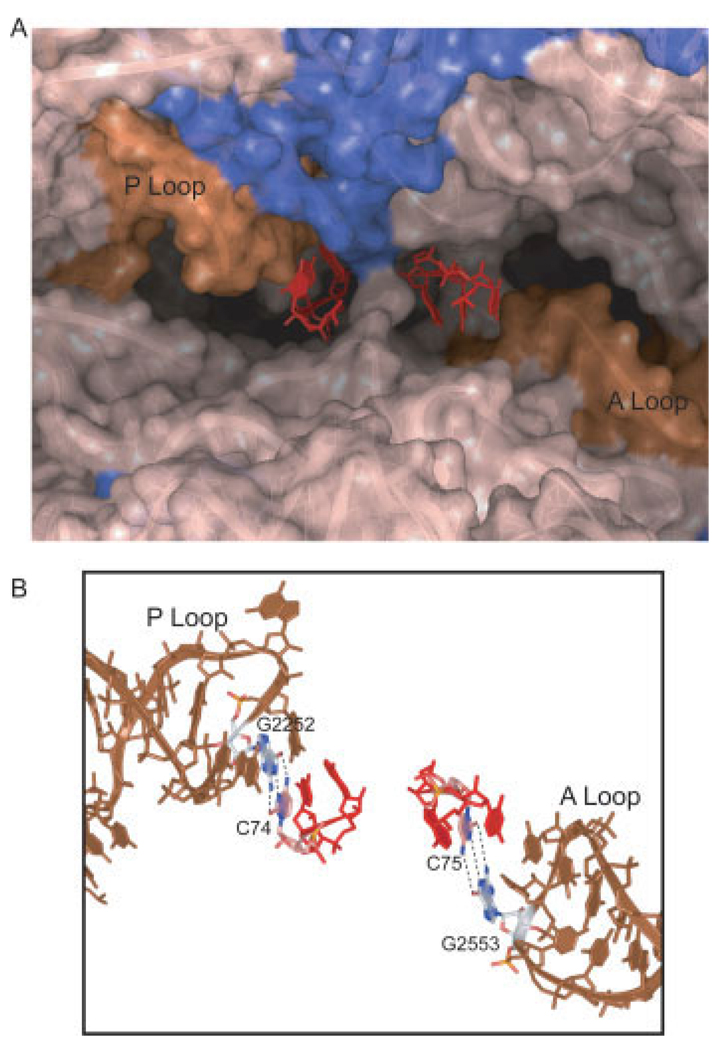
At increased time resolution, visual inspection of FRET trajectories revealed fluctuations to multiple low-FRET states with values ∼0.24 and ∼0.38 (Figure 3A). Previous ribosome interrogations have demonstrated base pairing between the 3′-CCA ends of tRNA and the large subunit as well as atomic resolution models of tRNA-ribosome interactions. 15,16,90–92 The C75 residue of A-site tRNA base pairs with G2553 within the conserved A loop domain; C74 and C75 of P-site tRNA base pair with G2552 and G2551 of the P loop (Figure 4). The prediction was that if FRET fluctuations corresponded to hybrid state transitions, then mutation of the tRNA binding sites should alter the energy landscape underpinning tRNA motions and promote hybrid state formation. Using recent technologies affording the isolation of ribosomes containing lethal point mutations,93 mutations were made in the A and P loops of the 23S rRNA. To disrupt tRNA interactions with the E site, ribosomal protein L1 was removed from the E. coli ribosome by genetic means.
FIGURE 4.
Mutations in key ribosome-tRNA interaction sites aided in identifying and stabilizing hybrid state intermediates on the ribosome. The 3′-CCA termini of A and P site tRNA make critical contacts with the A and P loops of 23S rRNA in the peptidyl transferase center of the 50S subunit.92–94 (A) Here the CCA termini shown in red, of the A- (right) and P- (left) site tRNA are shown in a surface-rendered crystal structure of the peptidyl transferase center.15 rRNA is shown in tan, with the A and P loops highlighted in brown. Protein components (mainly L27) are shown in blue. (B) Mutations G2553C in the A loop and G2252C in the P loop interrupted base-pairing interactions between C75 and C74 of the A- and P-site tRNA, respectively. Mutation in the A loop promoted formation of the A/P hybrid state, giving rise to an A/P-P/E hybrid state configuration. The P loop mutation promoted formation of the previously unidentified A/A-P/E hybrid state, which leave the 50S P site vacant.85
Whereas tRNA on a wild-type ribosome adopted a configuration with stable high FRET, as previously observed,83 a G2553C mutation in the A loop promoted formation of an intermediate-FRET state (mean value ∼0.38) (Figure 3B). Because the classical tRNA configuration should be destabilized by mutation of the 50S A site, we tentatively assigned the 0.38 FRET state to the previously described hybrid state configuration where deacylated P-site tRNA occupies a P/E configuration and A-site peptidyl-tRNA occupies an A/P configuration [hybrid state-1 (H1)] (Figure 5). A G2252C mutation promoted formation of a low-FRET state (mean value ∼0.24) (Figure 3B). This state was assigned to P-site tRNA adopting a P/E configuration while A-site tRNA remains classically configured [hybrid state-2 (H2)] (Figure 5). Depletion of ribosomal protein L1, a known E-site component that contributes to rapid translocation, destabilized both H1 and H2 tRNA configurations consistent with disruption of deacylated tRNA binding in the large subunit E site and an earlier report that contacts between the CCA tail of deacylated tRNA and 23S rRNA in the ribosomal E site stimulates translocation.94 These FRET state assignments were corroborated by measurements puromycin reactivity, an assay that tests for A site vacancy. Puromycin assays on wild-type and mutant ribosome complexes indicated that the G2553C mutant reacted faster than wild-type with puromycin, consistent with the promotion of the H1 state shifting the peptidyl moiety to the large subunit P site. The G2252C mutant reacted with puromycin slower than the wild-type ribosome consistent with the promotion of an H2 state in which binding at the P site is destabilized. While these experiments support the conclusion that the H1 state contains a puromycin-reactive, peptidly-tRNA in an A/P hybrid state, more detailed mechanistic studies are required to determine more precisely its reactivity relative to peptidyl-tRNA in the classical P/P state.
FIGURE 5.
Single-molecule FRET studies resulted in a new model of tRNA dynamics on the ribosome. The large activation energies associated with transitions between tRNA hybrid states indicate that these transitions are coupled to large-scale conformational rearrangements of the ribosome. A key observation was that tRNA can move independently on the ribosome. The motions of each tRNA appear to be governed by distinct conformational changes of the ribosome as indicated by the coloration of the ribosomal domains.
A confirmation of these preliminary assignments was made through detailed kinetic analysis of more than 3000 smFRET trajectories representative of wild-type and mutated systems. This exercise was carried out using Hidden Markov modeling (HMM) procedures akin to those introduced for ion channel patch clamp recordings.95–98 This method results in idealization of each smFRET trajectory (as seen in Figure 3) that can be further analyzed to determine the distribution of transitions present in each system, the transition density plot (Figure 6).99 Concurrently, HMM analytical procedures were also introduced for different systems.99,100 An important outcome of our HMM analysis was the ability to fit multiple kinetic models to the experimental FRET data using a maximum likelihood optimization approach.98 In so doing, a log-likelihood per event score can be obtained for each model. After adjusting the optimized likelihood for different numbers of model parameters using the Akaike information criterium,101 it was shown that a model with two hybrid states provided the best fit to the data. This result was supported by simulations of single-molecule FRET data which indicated that a model with two hybrid states was needed to recapitulate the experimental data.
FIGURE 6.
Transition density plots depict the remodeling of the free energy landscape due to mutation in tRNA binding sites. Plotting the initial and final FRET values of each transition indicates the existence of three distinct FRET states corresponding to the classical state and two hybrid states. The distribution of transitions shifts as a result of alterations in the energy landscape of the ribosome-tRNA complex due to removal of ribosomal protein L1 (A), or mutations made in key rRNA residues (B–D).
The nature of the hybrid states was more deeply probed by employing transition state theory where the free energies of activation for specific transitions are estimated according to the equation: ΔGij‡ = −RTln(hkij/kBT), where kij is the rate of transition from state i to state j, h is Planck’s constant, kB is Boltzman’s constant, R is the gas constant, and T is the temperature.102 This analysis led to the conclusion that two large activation barriers independently control the rate of A- and P-site tRNA movements on the ribosome. Moreover, the analysis showed that ribosome and tRNA motions could occur independently, or through a strongly coupled process (Figure 5). The observation that tRNA can move independently allowed us to probe specifically A- and P-site tRNA motions. The A-loop mutation had only a small impact on the activation energy for the isolated A/P state transition (kH1−H2), ΔΔG‡ ≈ −0.93 kJ/mol. Similarly, the P loop mutant led to a small change in activation energy for the isolated P/E transition (kC−H2), ΔΔG‡ ≈ −22.4 kJ/mol. Although both energy changes were consistent with the disruption of a single base-pairing interaction (ca. ≤1 kBT/mol), dramatic alterations in transition probabilities could be observed (Figure 6).
Concurrently, it was shown that titration of the peptide length carried by A-site tRNA showed even more subtle, but quantifiable, perturbations in the ribosome energy landscape. Lengthening of the peptide by a single amino acid (from Phe to Met-Phe) resulted in a ΔΔG‡ ≈ −0.91 kJ/mol for the H2 to H1 transition (∼0.4 kBT/mol). Similarly, addition of the formyl group onto the Met-Phe dipeptide resulted in a change in activation energy for the same transition of ΔΔG‡ ≈ 0.58 kJ/mol. The surprising sensitivity of the energy landscape of the ∼2.5 MDa ribosome to ∼100 Da changes at the 3′-CCA end of A-site peptidyl-tRNA requires further consideration and experimentation. What purpose does sensing the peptide serve? What is the dynamic range of this signal? We have speculated that growing peptide length may play a particularly important role during the early stages of translation, perhaps in regulation of the transition from initiation to elongation. At this stage the nascent peptide chain does not have the ability to form potentially stabilizing contacts with the peptide exit tunnel. A stable classical state may therefore be preferred given that hybrid state configurations are prone to dissociation70 (our unpublished data). Given previous observations that the nascent peptide can promote translational pausing103 and that binding of deacylated tRNA to A site during amino acid starvation leads to a transcriptional signaling,104 the biological implications of the classical-hybrid equilibrium needs deeper investigation.
Hybrid States and the Translocation Reaction Coordinate
Previous experiments demonstrate that peptide bond formation lowers the activation energy for translocation.70 Interpreted in the context of our data, and those of others, 105 there is now strong evidence that hybrid state configurations lie on the translocation reaction coordinate. 105 The free energy of activation for EF-G-catalyzed translocation ranges from ΔG‡ ≈ 67–72 kJ/mol, depending on buffer conditions.86,105,106 Spontaneous translocation has an estimated free energy of activation of ΔG‡ ≈ ∼95 k/mol.106,107 Thus, while EF-G significantly lowers the activation energy for translocation, ΔΔG‡ ≈ −29 kJ/mol (∼67–95 kJ/mol), compelling evidence exists for at least one rate limiting conformational process in translocation that has an activation energy of ∼68 kJ/mol. The movement of A- and P-site tRNA, and mRNA to the P- and E-sites doubtless accounts for the vast majority of this activation barrier. However, the estimated barrier for hybrid states formation, determined from the sum of rates exiting the classical state (kC−H1 + kC−H2), ∼70 kJ/mol, suggests that although perhaps different in the presence of EF-G, conformational processes within the ribosome leading to hybrid states formation may contribute substantially to the activation barrier of translocation.
Speculating on cryo-EM73–75 and chemical footprinting data,108 we have posited that the activation barrier observed for P/E hybrid state formation may report on the energy for intersubunit ratcheting, the downstream consequence of “unlocking” in the nomenclature of Valle et al.73 A direct link between P/E hybrid state formation and subunit ratcheting is supported by recent bulk FRET measurements.47,79 Contemporary models of translocation based on bulk fluorescence data contain a single rate limiting conformational transition.109,110 The most recent study includes the notion of two independent hybrid states where EF-G-mediated GTP hydrolysis first induces a global conformational change in the ribo-some placing deacylated tRNA in the P/E hybrid state, followed by A site peptidyl-tRNA rapidly adopting an A/P hybrid state.110 Translocation is completed with the release of inorganic phosphate by EF-G and tRNA-mRNA movement in the 30S subunit. We speculate that P/E hybrid state formation, coupled with subunit ratcheting, must precede productive EF-G reactions. In this view, hybrid state configurations, which occur spontaneously, are the most likely substrates for EF-G catalyzed GTP hydrolysis. The observed classical-hybrid equilibrium then represents the ribosome oscillating between conformations that are competent and incompetent for productive EF-G binding, perhaps explaining why EF-G has such an apparently low affinity for the ribosome particle. 21, 111
Molecular Dynamics Simulations and an Atomic Description of the P/E Hybrid State
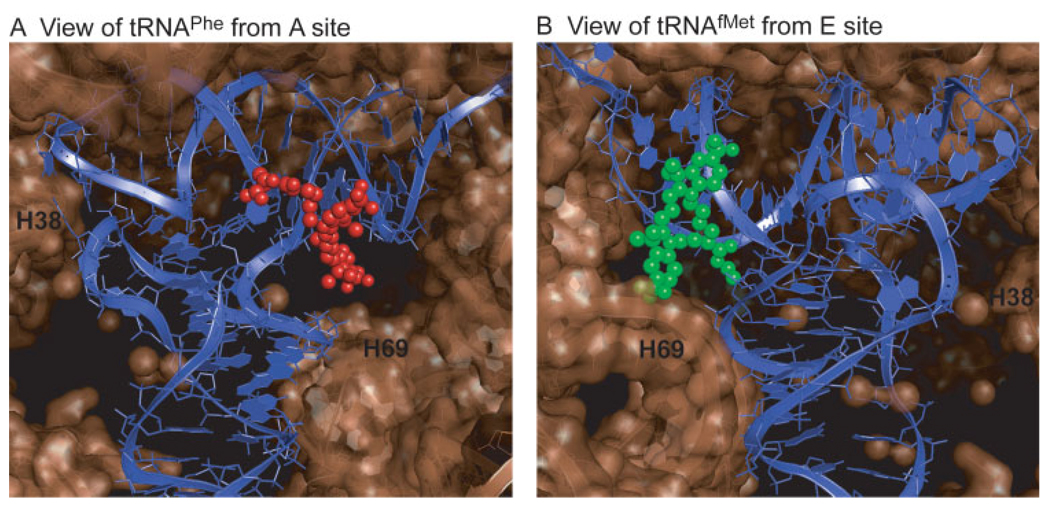
These first glimpses of the structural and kinetic determinants of hybrid state tRNA configurations provided by our smFRET data must now be reconciled with atomistic representations of ribosome dynamics during translation. This effort begins with understanding the physical locations of the dye molecules linked to tRNA on the ribosome. Using MD simulations the locations of the dyes linked to A- and P-site tRNA-bound dyes have been modeled on the ribosome (K. Sanbonmatsu, unpublished data) (Figure 7). Here it is shown that the donor fluorophore linked to tRNAfMet resides in close proximity to the E site near the subunit interface. The acceptor fluorophore linked to tRNAPhe is located on the solvent side of the A site. Consistent with anisotropy measurements,83 both dyes are solvent exposed with sufficient room to tumble freely. The dye separation in this preliminary model is in good agreement with our smFRET data (∼42 Å).
FIGURE 7.
MD simulations predict the locations of the fluorophores bound to the A and P site tRNAs. (A) Cy5 bound to the A site tRNAPhe at acp3U47 is shown from the leading edge of the ribosomal A site. (B) The P-site tRNAfMet is shown with Cy3 bound at s4U8 as viewed from the E site. H69 and H38 (A-site finger) are identified as reference points (K. Sanbonmatsu, unpublished data). The structures predict ample space for the fluorophores to reside, consistent with measurements of fluorescence anisotropy.83
Recently, atomistic models of the P/E hybrid state have been proposed using MD simulations and available cryo-EM maps.112 These data allow for prediction of the key interactions between the P/E-site tRNA and the ribosome, and also suggest that subunit ratcheting, which opens the gate formed by the A790 loop and G1338-A1339 of the small subunit,15, 16 is required for P/E state formation.112 Further smFRET studies will be required to experimentally verify the relationship between hybrid states formation and subunit ratcheting.
It is now clear that hybrid state ribosome configurations containing both A- and P-site tRNAs have eluded structure determination efforts as a consequence of their transient nature. Yet a deeper structural knowledge of hybrid states is critical to understanding the translocation reaction coordinate. Mutationally- and/or drug-stabilized complexes may prove an essential route to such efforts as it is clear that the classical-hybrid equilibrium may be dramatically altered by even modest perturbations to the system.
Along this road, computational efforts will provide an essential tool for gaining a deeper understanding of the molecular determinants of the ribosome energy landscape. In particular, targeted MD simulations provide potential avenues for reconciling dynamic FRET information with atomistic views of specific translational events.113,114 Targeted MD simulations move the system from one state to another, producing all-atom trajectories with correct stereochemistry, electrostatic interactions, and solvation structure. Such studies on the intact ribosome have already been performed114 producing realistic distance trajectories between points in space in the ribosome complex that may be later experimentally validated. Including the FRET dye pairs in the experiment may produce a close agreement between smFRET data and the targeted MD simulation.
CONCLUSIONS
The Future of Single-Molecule FRET Studies of Translation
The timescales (ca. milliseconds to minutes), and the amplitudes of motions (ca. 5–40 Å) occurring during translation make wide-field smFRET TIRF imaging methods a powerful tool for monitoring ribosome function from the intuitive perspective of motion. The implementation of established HMM-based analytical methods for the treatment of smFRET data, complemented with genetic and biochemical strategies, have brought new insights into the kinetic and structural parameters of tRNA motions on the ribosome. These findings include the identification of two distinct hybrid state tRNA configurations that serve as intermediates in the process of translocation. Future investigations using mutated tRNAs, unique tRNAs containing varied sequence elements,115 “slippery” mRNA sequences that promote fra-meshifting, and altered peptide composition are sure to provide further insights into the parameters that regulate the energy landscape underpinning the regulation of ribosome function in translation. Moreover, investigations of tRNA dynamics on the ribosome from different structural perspectives will be important to draw conclusively direct relationships between tRNA movements and discrete conformational changes of the ribosome particle. To create realistic movies of directional ribosomal translocation along mRNA with accurate mechanistic detail, all of these data will need to be interpreted in an atomistic framework through the synthesis of new structure determination efforts and computational methods.
The ability of smFRET to identify intermediate states in enzyme function rests on the proper choice of experimental observation times and the placement of fluorophores in locations yielding unique FRET states in response to conformational change. These choices will always be profoundly aided by structural and functional knowledge of the experimental system under investigation. Numerous biological systems have already benefited from FRET studies at the single-molecule scale including simple RNA folding reactions,116 RNA polymerase,117 DNA binding proteins,100,118 Ras signaling proteins,119,120 and the ribosome.50,83–85,89,121,122 Given continued improvements in the photophysical properties of fluorophore molecules, CCD technologies, computational methods, and an ever expanding repertoire of site-specific labeling strategies and novel experimental approaches, the application of smFRET is sure to expand to an even more diverse range of biomolecular questions in the foreseeable future. When used in combination with structural techniques such as NMR, cryo-EM, and X-Ray crystallography, smFRET is an invaluable asset in the expanding structural toolbox for exploring the molecular basis of life.
Footnotes
This article was originally published online as an accepted preprint. The “Published Online” date corresponds to the preprint version. You can request a copy of the preprint by emailing the Biopolymers editorial office at biopolymers@wiley.com
REFERENCES
- 1.Frauenfelder H, Sligar S, Wolynes PG. Science. 1991;254:1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 2.Nienhaus GU, Young RD. Encyclopedia Appl Phys. 1996;15:163–184. [Google Scholar]
- 3.Green R, Noller HF. Annu Rev Biochem. 1997;66:679–716. doi: 10.1146/annurev.biochem.66.1.679. [DOI] [PubMed] [Google Scholar]
- 4.Wilson DN, Blaha G, Connell SR, Ivanov PV, Jenke H, Stelzl U, Teraoka Y, Nierhaus KH. Curr Protein Peptide Sci. 2002;3:1–53. doi: 10.2174/1389203023380846. [DOI] [PubMed] [Google Scholar]
- 5.Korostelev A, Noller HF. Trends Biochem Sci. 2007;32:434–441. doi: 10.1016/j.tibs.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 6.Mitra K, Frank J. Annu Rev Biophys Biomol Struct. 2006;35:299–317. doi: 10.1146/annurev.biophys.35.040405.101950. [DOI] [PubMed] [Google Scholar]
- 7.Ramakrishnan V. Cell. 2002;108:557–572. doi: 10.1016/s0092-8674(02)00619-0. [DOI] [PubMed] [Google Scholar]
- 8.Puglisi JD, Blanchard SC, Green R. Nat Struct Biol. 2000;7:855–861. doi: 10.1038/79603. [DOI] [PubMed] [Google Scholar]
- 9.Moazed D, Noller HF. Cell. 1989;57:585–597. doi: 10.1016/0092-8674(89)90128-1. [DOI] [PubMed] [Google Scholar]
- 10.Moazed D, Noller HF. Cell. 1986;47:985–994. doi: 10.1016/0092-8674(86)90813-5. [DOI] [PubMed] [Google Scholar]
- 11.Agrawal RK, Penczek P, Grassucci RA, Li Y, Leith A, Nierhaus KH, Frank J. Science. 1996;271:1000–1002. doi: 10.1126/science.271.5251.1000. [DOI] [PubMed] [Google Scholar]
- 12.Gabashvili IS, Agrawal RK, Spahn CM, Grassucci RA, Svergun DI, Frank J, Penczek P. Cell. 2000;100:537–549. doi: 10.1016/s0092-8674(00)80690-x. [DOI] [PubMed] [Google Scholar]
- 13.Stark H, Orlova EV, Rinke-Appel J, Junke N, Mueller F, Rodnina M, Wintermeyer W, Brimacombe R, van Heel M. Cell. 1997;88:19–28. doi: 10.1016/s0092-8674(00)81854-1. [DOI] [PubMed] [Google Scholar]
- 14.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JHD, Noller HF. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 15.Selmer M, Dunham CM, Murphy FVt, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 16.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 17.Cate JH, Yusupov MM, Yusupova GZ, Earnest TN, Noller HF. Science. 1999;285:2095–2104. doi: 10.1126/science.285.5436.2095. [DOI] [PubMed] [Google Scholar]
- 18.Berk V, Zhang W, Pai RD, Cate JH. Proc Natl Acad Sci USA. 2006;103:15830–15834. doi: 10.1073/pnas.0607541103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodnina MV, Wintermeyer W. Annu Rev Biochem. 2001;70:415–435. doi: 10.1146/annurev.biochem.70.1.415. [DOI] [PubMed] [Google Scholar]
- 20.Beringer M, Rodnina MV. Mol Cell. 2007;26:311–321. doi: 10.1016/j.molcel.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 21.Rodnina MV, Savelsbergh A, Katunin VI, Wintermeyer W. Nature. 1997;385:37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- 22.Parker J. Microbiol Rev. 1989;53:273–298. doi: 10.1128/mr.53.3.273-298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farabaugh PJ. Microbiol Rev. 1996;60:103–134. doi: 10.1128/mr.60.1.103-134.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frauenfelder H, Parak F, Young RD. Annual Review of Biophysics and Biophysical Chemistry. 1988;17:451–479. doi: 10.1146/annurev.bb.17.060188.002315. [DOI] [PubMed] [Google Scholar]
- 25.Kern D, Zuiderweg ERP. Curr Opin Struct Biol. 2003;13:748–757. doi: 10.1016/j.sbi.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 26.Gavrilova LP, Kostiashkina OE, Koteliansky VE, Rutke-vitch NM, Spirin AS. J Mol Biol. 1976;101:537–552. doi: 10.1016/0022-2836(76)90243-6. [DOI] [PubMed] [Google Scholar]
- 27.Bergemann K, Nierhaus KH. J Biol Chem. 1983;258:15105–15113. [PubMed] [Google Scholar]
- 28.Cabanas MJ, Modolell J. Biochemistry. 1980;19:5411–5416. doi: 10.1021/bi00564a040. [DOI] [PubMed] [Google Scholar]
- 29.Scheres SH, Gao H, Valle M, Herman GT, Eggermont PP, Frank J, Carazo JM. Nat Methods. 2007;4:27–29. doi: 10.1038/nmeth992. [DOI] [PubMed] [Google Scholar]
- 30.Messina TC, Talaga DS. Biophys J. 2007;93:579–585. doi: 10.1529/biophysj.107.103911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agrawal RK, Grassucci R, Penczek P, Heagle AB, Burkhardt N, Nierhaus KH, Frank J. Mol Biol Cell. 1998;9(SS):405. [Google Scholar]
- 32.Fredrick K, Noller H. Science. 2003;300:1159–1162. doi: 10.1126/science.1084571. [DOI] [PubMed] [Google Scholar]
- 33.Gromadski KB, Rodnina MV. Nat Struct Mol Biol. 2004;11:316–322. doi: 10.1038/nsmb742. [DOI] [PubMed] [Google Scholar]
- 34.Ruggero D, Pandolfi PP. Nat Rev Cancer. 2003;3:179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- 35.Yu Y, Ji H, Doudna JA, Leary JA. Protein Sci. 2005;14:1438–1446. doi: 10.1110/ps.041293005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rizos H, McKenzie HA, Ayub AL, Woodruff S, Becker TM, Scurr LL, Stahl J, Kefford RF. J Biol Chem. 2006;281:38080–38088. doi: 10.1074/jbc.M609405200. [DOI] [PubMed] [Google Scholar]
- 37.Yin X, Fontoura BM, Morimoto T, Carroll RB. J Cell Physiol. 2003;196:474–482. doi: 10.1002/jcp.10329. [DOI] [PubMed] [Google Scholar]
- 38.Min W, English BP, Luo G, Cherayil BJ, Kou SC, Xie XS. Acc Chem Res. 2005;38:923–931. doi: 10.1021/ar040133f. [DOI] [PubMed] [Google Scholar]
- 39.English BP, Min W, van Oijen AM, Lee KT, Luo G, Sun H, Cherayil BJ, Kou SC, Xie XS. Nat Chem Biol. 2006;2:87–94. doi: 10.1038/nchembio759. [DOI] [PubMed] [Google Scholar]
- 40.Michalet X, Weiss S, Jager M. Chem Rev. 2006;106:1785–1813. doi: 10.1021/cr0404343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ritort F. J Phys: Condens Matter. 2006;18:R531. doi: 10.1088/0953-8984/18/32/R01. [DOI] [PubMed] [Google Scholar]
- 42.Xie XS. J Chem Phys. 2002;117:11024–11032. [Google Scholar]
- 43.Kuzmenkina EV, Heyes CD, Nienhaus GU. J Mol Biol. 2006;357:313–324. doi: 10.1016/j.jmb.2005.12.061. [DOI] [PubMed] [Google Scholar]
- 44.Xie Z, Srividya N, Sosnick TR, Pan T, Scherer NF. Proc Natl Acad Sci USA. 2004;101:534–539. doi: 10.1073/pnas.2636333100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coban O, Lamb DC, Zaychikov E, Heumann H, Nien-haus GU. Biophys J. 2006;90:4605–4617. doi: 10.1529/biophysj.105.078840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ha T. Methods. 2001;25:78–86. doi: 10.1006/meth.2001.1217. [DOI] [PubMed] [Google Scholar]
- 47.Ermolenko DN, Majumdar ZK, Hickerson RP, Spiegel PC, Clegg RM, Noller HF. J Mol Biol. 2007;370:530–540. doi: 10.1016/j.jmb.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 48.Wang L, Schultz PG. Angew Chem Int Ed Engl. 2004;44:34–66. doi: 10.1002/anie.200460627. [DOI] [PubMed] [Google Scholar]
- 49.Muir TW. Annu Rev Biochem. 2003;72:249–289. doi: 10.1146/annurev.biochem.72.121801.161900. [DOI] [PubMed] [Google Scholar]
- 50.Dorywalska M, Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. Nucleic Acids Res. 2005;33:182–189. doi: 10.1093/nar/gki151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Plumbridge JA, Baumert HG, Ehrenberg M, Rigler R. Nucleic Acids Res. 1980;8:827–843. [PMC free article] [PubMed] [Google Scholar]
- 52.Watson BS, Hazlett TL, Eccleston JF, Davis C, Jameson DM, Johnson AE. Biochemistry. 1995;34:7904–7912. doi: 10.1021/bi00024a015. [DOI] [PubMed] [Google Scholar]
- 53.Rice S, Lin A, Safer D, Hart C, Naber N, Carragher B, Cain S, Pechatnikova E, Wilson-Kubalek E, Whittaker M, Pate E, Cooke R, Taylor E, Milligan R, Vale R. Nature. 1999;402:778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- 54.Ambrose W, Moerner W. Nature. 1991;349:225–227. [Google Scholar]
- 55.Ha T, Zhuang X, Kim HD, Orr JW, Williamson JR, Chu S. Proc Natl Acad Sci USA. 1999;96:9077–9082. doi: 10.1073/pnas.96.16.9077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ha T, Ting AY, Liang J, Caldwell WB, Deniz AA, Chemla DS, Schultz PG, Weiss S. Proc Natl Acad Sci USA. 1999;96:893–898. doi: 10.1073/pnas.96.3.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhuang X, Bartley LE, Babcock HP, Russell R, Ha T, Herschlag D, Chu S. Science. 2000;288:2048–2051. doi: 10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]
- 58.Zhuang X, Kim H, Pereira MJ, Babcock HP, Walter NG, Chu S. Science. 2002;296:1473–1476. doi: 10.1126/science.1069013. [DOI] [PubMed] [Google Scholar]
- 59.Ha T, Enderle T, Ogletree DF, Chemla DS, Selvin PR, Weiss S. Proc Natl Acad Sci USA. 1996;93:6264–6268. doi: 10.1073/pnas.93.13.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ha T, Rasnik I, Cheng W, Babcock HP, Gauss GH, Lohman TM, Chu S. Nature. 2002;419:638–641. doi: 10.1038/nature01083. [DOI] [PubMed] [Google Scholar]
- 61.Xie XS, Trautman JK. Ann Rev Phys Chem. 1998;49:441–480. doi: 10.1146/annurev.physchem.49.1.441. [DOI] [PubMed] [Google Scholar]
- 62.Myong S, Stevens BC, Ha T. Structure. 2006;14:633–643. doi: 10.1016/j.str.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 63.Weiss S. Science. 1999;283:1676–1683. doi: 10.1126/science.283.5408.1676. [DOI] [PubMed] [Google Scholar]
- 64.Peterman EJ, Sosa H, Moerner WE. Annu Rev Phys Chem. 2004;55:79–96. doi: 10.1146/annurev.physchem.55.091602.094340. [DOI] [PubMed] [Google Scholar]
- 65.Greenleaf WJ, Woodside MT, Block SM. Annu Rev Biophys Biomol Struct. 2007;36:171–190. doi: 10.1146/annurev.biophys.36.101106.101451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aitken CE, Marshall RA, Puglisi J. Biophys J. 2007 DOI: biophysj. 107.117689. [Google Scholar]
- 67.Rasnik I, McKinney SA, Ha T. Nat Meth. 2006;3:891–893. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]
- 68.Noller HF, Yusupov MM, Yusupova GZ, Baucom A, Cate JH. FEBS Lett. 2002;514:11–16. doi: 10.1016/s0014-5793(02)02327-x. [DOI] [PubMed] [Google Scholar]
- 69.Moazed D, Noller HF. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 70.Semenkov YP, Rodnina MV, Wintermeyer W. Nat Struct Biol. 2000;7:1027–1031. doi: 10.1038/80938. [DOI] [PubMed] [Google Scholar]
- 71.Sharma D, Southworth DR, Green R. RNA. 2004;10:102–113. doi: 10.1261/rna.5148704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Agrawal RK, Penczek P, Grassucci RA, Burkhardt N, Nierhaus KH, Frank J. J Biol Chem. 1999;274:8723–8729. doi: 10.1074/jbc.274.13.8723. [DOI] [PubMed] [Google Scholar]
- 73.Valle M, Zavialov AV, Sengupta J, Rawat U, Ehrenberg M, Frank J. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 74.Frank J, Agrawal RK. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 75.Agrawal RK, Heagle AB, Penczek P, Grassucci RA, Frank J. Nat Struct Biol. 1999;6:643–647. doi: 10.1038/10695. [DOI] [PubMed] [Google Scholar]
- 76.Spahn CM, Gomez-Lorenzo MG, Grassucci R, Jorgen-sen R, Andersen GR, Beckmann R, Penczek P, Ballesta JP, Frank J. EMBO J. 2004;23:1008–1019. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stark H, Rodnina MV, Wieden HJ, van Heel M, Wintermeyer W. Cell. 2000;100:301–309. doi: 10.1016/s0092-8674(00)80666-2. [DOI] [PubMed] [Google Scholar]
- 78.Taylor DJ, Nilsson J, Merrill AR, Andersen GR, Nissen P, Frank J. EMBO J. 2007;26:2421–2431. doi: 10.1038/sj.emboj.7601677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ermolenko DN, Spiegel PC, Majumdar ZK, Hickerson RP, Clegg RM, Noller HF. Nat Struct Mol Biol. 2007;14:493–497. doi: 10.1038/nsmb1243. [DOI] [PubMed] [Google Scholar]
- 80.Savelsbergh A, Katunin VI, Mohr D, Peske F, Rodnina MV, Wintermeyer W. Mol Cell. 2003;11:1517–1523. doi: 10.1016/s1097-2765(03)00230-2. [DOI] [PubMed] [Google Scholar]
- 81.Peske F, Savelsbergh A, Katunin VI, Rodnina MV, Wintermeyer W. J Mol Biol. 2004;343:1183–1194. doi: 10.1016/j.jmb.2004.08.097. [DOI] [PubMed] [Google Scholar]
- 82.Zavialov AV, Ehrenberg M. Cell. 2003;114:113–122. doi: 10.1016/s0092-8674(03)00478-1. [DOI] [PubMed] [Google Scholar]
- 83.Blanchard SC, Kim HD, Gonzalez RL, Jr, Puglisi JD, Chu S. Proc Natl Acad Sci USA. 2004;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. Nat Struct Mol Biol. 2004;11:1008–1014. doi: 10.1038/nsmb831. [DOI] [PubMed] [Google Scholar]
- 85.Munro JB, Altman RB, O’Connor N, Blanchard SC. Mol Cell. 2007;25:505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Studer SM, Feinberg JS, Joseph S. J Mol Biol. 2003;327:369–381. doi: 10.1016/s0022-2836(03)00146-3. [DOI] [PubMed] [Google Scholar]
- 87.Kim HD, Nienhaus GU, Ha T, Orr JW, Williamson JR, Chu S. Proc Natl Acad Sci USA. 2002;99:4284–4289. doi: 10.1073/pnas.032077799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bokinsky G, Rueda D, Misra VK, Rhodes MM, Gordus A, Babcock HP, Walter NG, Zhuang X. Proc Natl Acad Sci USA. 2003;100:9302–9307. doi: 10.1073/pnas.1133280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kim HD, Puglisi JD, Chu S. Biophys J. 2007;93:3575–3582. doi: 10.1529/biophysj.107.109884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Moazed D, Noller HF. Proc Natl Acad Sci USA. 1991;88:3725–3728. doi: 10.1073/pnas.88.9.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Samaha RR, Green R, Noller HF. Nature. 1995;377:309–314. doi: 10.1038/377309a0. [DOI] [PubMed] [Google Scholar]
- 92.Kim DF, Green R. Mol Cell. 1999;4:859–864. doi: 10.1016/s1097-2765(00)80395-0. [DOI] [PubMed] [Google Scholar]
- 93.Youngman EM, Green R. Methods. 2005;36:305–312. doi: 10.1016/j.ymeth.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 94.Lill R, Robertson JM, Wintermeyer W. EMBO J. 1989;8:3933–3938. doi: 10.1002/j.1460-2075.1989.tb08574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Qin F, Auerbach A, Sachs F. Biophys J. 1996;70:264–280. doi: 10.1016/S0006-3495(96)79568-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sakmann B, Neher E. Single Channel Recording. New York: Springer; 2007. [Google Scholar]
- 97.Qin F. Biophys J. 2004;86:1488–1501. doi: 10.1016/S0006-3495(04)74217-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Qin F, Auerbach A, Sachs F. Proc Biol Sci. 1997;264:375–383. doi: 10.1098/rspb.1997.0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.McKinney SA, Joo C, Ha T. Biophys J. 2006;91:1941–1951. doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Joo C, McKinney SA, Nakamura M, Rasnik I, Myong S, Ha T. Cell. 2006;126:515–527. doi: 10.1016/j.cell.2006.06.042. [DOI] [PubMed] [Google Scholar]
- 101.Akaike H. IEEE Trans Auto Cont. 1974;19:716–723. [Google Scholar]
- 102.Fersht A. Structure and Mechanism in Protein Science. New York: W.H. Freeman and Company; 1999. [Google Scholar]
- 103.Gong M, Cruz-Vera LR, Yanofsky C. J Bacteriol. 2007;189:3147–3155. doi: 10.1128/JB.01868-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cashel M. J Biol Chem. 1969;244:3133–3141. [PubMed] [Google Scholar]
- 105.Dorner S, Brunelle JL, Sharma D, Green R. Nat Struct Mol Biol. 2006;13:234–241. doi: 10.1038/nsmb1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Katunin VI, Savelsbergh A, Rodnina MV, Wintermeyer W. Biochemistry. 2002;41:12806–12812. doi: 10.1021/bi0264871. [DOI] [PubMed] [Google Scholar]
- 107.Wintermeyer W, Savelsbergh A, Semenkov YP, Katunin VI, Rodnina MV. Cold Spring Harb Symp Quant Biol. 2001;66:449–458. doi: 10.1101/sqb.2001.66.449. [DOI] [PubMed] [Google Scholar]
- 108.Spiegel PC, Ermolenko DN, Noller HF. RNA. 2007;13:1473–1482. doi: 10.1261/rna.601507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pan D, Kirillov S, Zhang C-M, Hou Y-M, Cooperman BS. Nat Struct Mol Biol. 2006;13:354–359. doi: 10.1038/nsmb1074. [DOI] [PubMed] [Google Scholar]
- 110.Pan D, Kirillov S, Cooperman BS. Mol Cell. 2007;25:519–529. doi: 10.1016/j.molcel.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Matassova AB, Rodnina MV, Wintermeyer W. RNA. 2001;7:1879–1885. [PMC free article] [PubMed] [Google Scholar]
- 112.Li W, Frank J. Proc Natl Acad Sci USA. 2007;104:16540–16545. doi: 10.1073/pnas.0708094104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sanbonmatsu KY, Tung CS. J Struct Biol. 2006;157:470–480. doi: 10.1016/j.jsb.2006.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sanbonmatsu KY, Joseph S, Tung CS. Proc Natl Acad Sci USA. 2005;102:15854–15859. doi: 10.1073/pnas.0503456102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schultz DW, Yarus M. J Mol Biol. 1994;235:1395–1405. doi: 10.1006/jmbi.1994.1096. [DOI] [PubMed] [Google Scholar]
- 116.Zhuang X. Annu Rev Biophys Biomol Struct. 2005;34:399–414. doi: 10.1146/annurev.biophys.34.040204.144641. [DOI] [PubMed] [Google Scholar]
- 117.Margeat E, Kapanidis AN, Tinnefeld P, Wang Y, Mukho-padhyay J, Ebright RH, Weiss S. Biophys J. 2006;90:1419–1431. doi: 10.1529/biophysj.105.069252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Myong S, Rasnik I, Joo C, Lohman TM, Ha T. Nature. 2005;437:1321–1325. doi: 10.1038/nature04049. [DOI] [PubMed] [Google Scholar]
- 119.Arai Y, Iwane AH, Wazawa T, Yokota H, Ishii Y, Kataoka T, Yanagida T. Biochem Biophys Res Comm. 2006;343:809–815. doi: 10.1016/j.bbrc.2006.03.031. [DOI] [PubMed] [Google Scholar]
- 120.Sugawa M, Arai Y, Iwane AH, Ishii Y, Yanagida T. Biosystems. 2007;88:243–250. doi: 10.1016/j.biosystems.2006.09.041. [DOI] [PubMed] [Google Scholar]
- 121.Lee T-H, Blanchard SC, Kim HD, Puglisi JD, Chu S. Proc Natl Acad Sci USA. 2007;104:13661–13665. doi: 10.1073/pnas.0705988104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wang Y, Qin H, Kudaravalli RD, Kirillov SV, Dempsey GT, Pan D, Cooperman BS, Goldman YE. Biochemistry. 2007;46:10767–10775. doi: 10.1021/bi700657d. [DOI] [PubMed] [Google Scholar]