Abstract
Toxoplasma gondii is an intracellular protozoan parasite that can cause devastating disease in fetuses and immune-compromised individuals. We previously reported that the α subunit of the host cell transcription factor, hypoxia-inducible factor-1 (HIF-1), is up-regulated by infection and necessary for Toxoplasma growth. Under basal conditions, HIF-1α is constitutively expressed but rapidly targeted for proteasomal degradation after two proline residues are hydroxylated by a family of prolyl hydroxylases (PHDs). The PHDs are α-ketoglutarate-dependent dioxygenases that have low Km values for oxygen, making them important cellular oxygen sensors. Thus, when oxygen levels decrease, HIF-1α is not hydroxylated, and HIF-1 is activated. How Toxoplasma activates HIF-1 under normoxic conditions remains unknown. Here, we report that Toxoplasma infection increases HIF-1α stability by preventing HIF-1α prolyl hydroxylation. Infection significantly decreases PHD2 abundance, which is the key prolyl hydroxylase for regulating HIF-1α. The effects of Toxoplasma on HIF-1α abundance and prolyl hydroxylase activity require activin-like receptor kinase signaling. Finally, parasite growth is severely diminished when signaling from this family of receptors is inhibited. Together, these data indicate that PHD2 is a key host cell factor for T. gondii growth and represent a novel mechanism by which a microbial pathogen subverts host cell signaling and transcription to establish its replicative niche.
Keywords: Gene Expression, Hypoxia, Parasitology, Signal Transduction, Transforming Growth Factor β (TGFβ)
Introduction
Toxoplasma gondii is an obligate intracellular protozoan parasite that can cause devastating disease in immune-compromised people and fetuses (1, 2). The ability for the parasite to cause disease is directly linked to its replication inside a parasitophorous vacuole within its host cell. From this vacuole, parasites scavenge nutrients from its host cell while causing reorganization of host organelles and cytoskeletal elements, preventing host cell apoptosis, and altering host gene expression (3–5). Therefore, Toxoplasma has developed mechanisms to manipulate host cell processes that permit it to grow. Identifying the parasite factors that cause these changes and the specific host pathways modulated by these factors is critical for developing new drugs to treat and prevent Toxoplasma infections and disease.
Previously, our laboratory demonstrated that Toxoplasma activates the host cell transcription factor hypoxia-inducible factor-1 (HIF-1)3 (6). HIF-1 is activated by a parasite-derived, secreted factor independent of direct contact between the parasite and host cell. This is in contrast to other Toxoplasma factors that control host cell signaling by virtue of their secretion into the host cell cytoplasm (7–10). Significantly, HIF-1 is important for parasite growth at both normoxic and physiological O2 levels but dispensable for host cell replication at either O2 levels (6).
HIF-1 is a heterodimer composed of α and β subunits and is the master regulator of the cellular response to decreased O2 availability (11). Although both subunits are constitutively expressed, the α subunit has a very short half-life (∼2 min) because it is targeted in an O2-dependent fashion to the proteasome by the von Hippel-Lindau E3 ubiquitin ligase (12–14). HIF-1α recognition by von Hippel-Lindau is dependent on the hydroxylation of HIF-1α on two proline residues (Pro402 and Pro564) in a domain termed the oxygen-dependent degradation domain (ODD) (15–19). Three HIF-1α prolyl hydroxylases (PHD1–3) have been identified. Gene knockout and siRNA-based studies indicate that PHD2 is the primary isoform responsible for regulating HIF-1α (20, 21). HIF-1-dependent transcription is regulated by an asparaginyl hydroxylase named factor inhibiting HIF (FIH) that hydroxylates Asn803 in the C terminus of HIF-1α (22–24). The PHDs and FIH are oxygen and α-ketoglutarate-dependent dioxygenases that use ascorbic acid and Fe2+ as cofactors. The affinity of these enzymes for O2 is low (Km = 90–250 μm), which renders them important cellular O2 sensors (25, 26). Thus, when O2 levels decrease, PHD and FIH activities are reduced, and two independent events take place: HIF-1α protein increases, and HIF-1α binds p300/CBP.
Increases in HIF-1α protein levels and activity are not restricted to hypoxic stress. Besides Toxoplasma, other bacterial, viral, and protozoan pathogens activate HIF-1 (27–33). HIF-1 is also activated by cytokines, growth factors, and other secreted factors (34–36). Although some of these agents increase HIF-1α protein by increasing its rate of translation, others act by stabilizing the protein. How HIF-1α is stabilized at normoxic O2 levels is largely unknown. Given its key role in regulating HIF-1α, dysregulation of PHD2 may be one way to stabilize HIF-1α under normoxic conditions. For example, HIF-1α can be activated by decreased availability of PHD2 cofactors and substrates (37). In addition, PHD2 activity can also be impacted by its abundance. Recent studies have highlighted two post-transcriptional mechanisms that control PHD2 protein levels. First, PHD2 binds the peptidyl prolyl cis/trans-isomerase FKBP38, and siRNA-mediated depletion of FKBP38 leads to increases in PHD2 protein abundance (38). TGFβ signaling through the Type I TGFβ receptor is a second mechanism to decrease PHD2 protein levels (39). The Type I TGFβ receptor is a member of the activin-like receptor kinase (ALK) family of receptors that bind several distinct ligand families including TGFβs, activins, and bone morphogenic proteins (40, 41). The cytoplasmic domains of the ALK receptors are serine/threonine kinases that are activated when the ligand induces the Type I receptor to dimerize with a Type II receptor. Seven Type I receptors (ALK1–7) have been identified and can be grouped based upon the homology of their cytoplasmic domains. The high degree of homology between ALK5 (Type I TGFβ receptor), ALK4, and ALK7 renders them similarly sensitive to a class of highly selective small molecule inhibitors that have very limited affects on other kinases including p38 MAPKs (42).
The goal of this work is to determine the mechanism by which Toxoplasma increases HIF-1α protein levels. We report that Toxoplasma infection increases HIF-1α stability by decreasing levels of HIF-1α hydroxylation. Alterations in HIF-1α stability are due to decreased PHD2 expression. Decreased PHD2 protein and HIF-1 activation was dependent on ALK signaling. Finally, we demonstrate that Toxoplasma growth was dependent on ALK signaling.
EXPERIMENTAL PROCEDURES
Cells and Parasites
The Toxoplasma strain RH was used for all experiments and maintained by serial passage in human foreskin fibroblast (HFF) cells in DMEM supplemented with 10% fetal bovine serum, glutamine, and penicillin/streptomycin as described (6). All other cells (HIF-1α WT, HIF-1α KO, and HeLa cells) were also grown in this medium. For all assays, the parasites were harvested from infected but nonlysed HFFs. The parasites were released from their host cells by passing them three times through a 27-gauge syringe needle. All of the cells and parasites were routinely tested with a mycoplasma detection kit (Lonza, Basel, Switzerland) and found to be negative for mycoplasma contamination.
Plasmids
The HIF-1 luciferase reporter pHRE-luc and pTK-Rel were previously described (6). The GAL4 luciferase reporter (p5xGRE-luc) and the GAL4 DNA-binding domain fused to amino acids 737–826 of HIF-1α (pGBD-HIF-1α737–826 or pGBD as a empty vector control) were from Dr. Dan Peet (University of Adelaide) (22). FLAG-tagged PHD2 expression plasmid (p3XFLAG-PHD2) was from Dr. Richard Bruick (University of Texas Southwestern Medical Center). The ODD Renilla luciferase reporter (pTK-ODD-Rel) was generated by replacing the Renilla luciferase gene from pTK-Rel with the ODD-Rel fusion from pCMV-ODD-Rel kindly provided by Dr. Joan Conaway (Stowers Institute). pCDF1-SMAD7 was generated by amplifying full-length SMAD7 from MGC clone gi50960790 (Open Biosystems, Huntsville, AL) using primers containing SCA1/NOT1 restriction sites. The amplified fragment was cloned into pCDF1 (Systems Bioscience, Mountain View, CA) digested with SCA1 and NOT1.
Northern Blotting
Host cells were infected with parasites at a multiplicity of infection of 10:1 (parasites: host cells), and 18 h later total RNA was purified with the Stratagene Absolutely RNA Microprep kit (La Jolla, CA) and Northern blotted as previously described (43). Murine β-actin and HIF-1α probes were generated by reverse transcriptase PCR from total RNA using the following primers: β-actin forward, 5′-GAGGTCTTTACGGATGTCAA-3′; β-actin reverse, 5′-CCAGATCATGTTTGAGACCT-3′; HIF-1α forward, 5′-GAGTTCTGAACGTCGAAAAG-3′; and HIF-1α reverse, 5′-ACTTGATGTTCATCGTCCTC-3′. The probes were labeled with [α-32P]dGTP with the random primed DNA labeling kit (Roche Applied Science) and hybridized with Express-Hyb (Clontech, Palo Alto, CA). The blots were exposed to film and analyzed using the ImageQuaNT program (Molecular Dynamics, Sunnyvale, CA).
Western Blotting
Parasites were added to host cells at a multiplicity of infection of 10:1 (parasites:host cells). The cells were washed with ice-cold PBS and lysed in ice-cold radioimmune precipitation assay lysis buffer supplemented with EDTA-free protease inhibitor mixture (Roche Applied Science). The lysates were centrifuged to remove any remaining cellular debris, and protein concentration was determined with the DC protein assay (Bio-Rad). The samples were separated by SDS-PAGE, transferred to a PVDF membrane, blocked with 5% BSA, and Western blotted with anti-HIF-1α (BD Biosciences, San Jose, CA), anti-PHD1 (catalog no. NB100–310; Novus, Littleton, CO), anti-PHD2 (catalog no. NB100–113; Novus), anti-PHD3 (catalog no. NB100–303; Novus), anti-actin (Ambion, Austin, TX), anti-hydroxylated HIF-1α P402 and P564 (44), or anti-FKBP38 (Abcam, Cambridge, MA) antibodies. The proteins were detected using ECL Western blotting detection kit (GE Healthcare) and imaged using FluroChemQ (Cell Biosciences, Santa Clara, CA). Densitometry was performed using the AlphaInnotech software, and protein amounts were determined by normalizing to β-actin levels.
Luciferase Assays
HIF-1α WT cells were transfected using Lipofectamine (Invitrogen) as previously described (45). When infected cells were exposed to 3% O2, the parasites were allowed to invade for 1 h before transferring the infected cells to a hypoxia work station (Ruskinn, Cincinnati, OH). Unless otherwise indicated, the cells were harvested 24 h post-infection, which preceded parasite egress. The cells were then washed with PBS, lysed, and assayed using the Dual-Glo luciferase assay kit (Promega, Madison, WI). For all of the assays, the appropriate empty vector controls were included.
Toxoplasma Growth and Invasion Assays
Toxoplasma growth was measured using β-galactosidase-expressing parasites from Dr. Gustavo Arrizabalaga (University of Idaho) (46, 47). Briefly, parasites were added to host cells, and after 72 h the medium was removed. Z-buffer (100 μl) containing 20 μm chlorophenolred-β-d-galactopyranoside was then added to each well for 15 min, and the A570 was measured (48). The numbers of parasites in each well were determined by linear regression analysis from a standard curve generated in each plate. To specifically measure parasite replication, parasites were added to confluent HFFs on 12-mm2 glass coverslips at a multiplicity of infection of 1:4 (parasites:host cells). After 12 or 24 h, the cells were fixed and stained with a rabbit polyclonal antibody against the tachyzoite surface protein SAG1 (from Dr. John Boothroyd, Stanford University). Fifty vacuoles were counted for each time point. Synchronized parasite invasion assays were performed with GFP+ parasites using a high potassium buffer as described (49). Sixty minutes after adding invasion buffer, the cells were fixed by adding formaldehyde (final concentration, 3%) directly to the wells. The cells were stained with rabbit anti-SAG1 antisera and detected with anti-rabbit AlexaFluor 594 (Invitrogen) without permeabilizing the cells. Intracellular parasites were scored as GFP+/SAG1−, and extracellular parasites were scored as GFP+/SAG1+. A total of 200 parasites were counted in each experiment.
Flow Cytometry
HIF-1α WT cells were mock infected or infected with tachyzoites at a multiplicity of infection of 1:1 in the absence or presence of SB505124. After 6 h, 1,000 units/ml of IFNγ was added for an additional 18 h. The cells were harvested by scraping, blocked with Fc Block (BD Biosciences), and stained with rat anti-mouse PD-L1 or Rat IgG2A (eBioscience, San Diego, CA) for 2 h at room temperature. The cells were then fixed with 1% formaldehyde, washed, and analyzed on a FACSCaliber (BD Biosciences). Single color controls were used for compensation and gating.
West Nile Virus Growth Assay
HFFs grown to 80% confluence were inoculated with West Nile virus NY99 strain (AAF20092.2) at a multiplicity of infection of 1:1 (virus:host cell) in the absence or presence of SB505124. The virus was allowed to be absorbed for 2 h at 37 °C, and then the cells were washed with PBS, after which fresh medium containing either vehicle or drug was added to the monolayers. After 24 h, the medium was harvested, and the West Nile virus numbers were determined by quantitative RT-PCR detection of viral mRNA as previously described (50).
RESULTS
Toxoplasma Infection Stabilizes HIF-1α Protein
Toxoplasma up-regulates host HIF-1α protein levels and requires HIF-1α for growth at both normoxic and physiological oxygen tensions. Because increasing HIF-1α protein levels is a key step in activating the HIF-1 transcription factor complex, we sought to define the mechanism underlying how Toxoplasma up-regulates HIF-1α protein levels. These experiments were performed under normoxic conditions because examining parasite regulation of HIF-1 at decreased O2 tension is complicated because of O2-dependent HIF-1α stabilization. Thus, we examined whether infection leads to changes in HIF-1α transcription, translation, or stability. First, HIF-1α wild type or knockout cells were mock or parasite-infected for 18 h, after which total RNA was isolated and probed by Northern blotting to detect HIF-1α or β-actin (as a normalization control) mRNAs. The data indicated that infection did not significantly alter HIF-1α transcript levels (Fig. 1A). As expected, full-length HIF-1α mRNA was undetectable in HIF-1α knockout cells, demonstrating the specificity of the HIF-1α probe. These data are also consistent with previous microarray studies showing that HIF-1α transcript abundance was not altered in Toxoplasma-infected HFFs (8, 43).
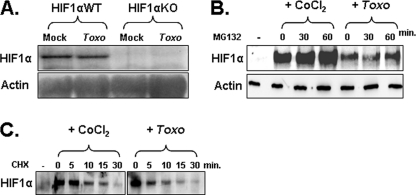
FIGURE 1.
Toxoplasma infection increases HIF-1α stability. A, total RNA from mock or Toxoplasma-infected (Toxo) cells was Northern blotted to detect HIF-1α and β-actin as a loading control. B, HFFs were mock infected, CoCl2-treated, or Toxoplasma-infected for 18 h and then treated with MG132 for the indicated times. Lysates were prepared and Western blotted to detect HIF-1α and actin as a loading control. Densitometry indicated no significant increase in HIF-1α levels in the Toxoplasma-infected cells. C, HFFs were mock infected, CoCl2-treated, or Toxoplasma-infected for 18 h and then treated with cycloheximide (CHX) for the indicated times. Lysates were prepared and Western blotted to detect anti-HIF-1α. Shown is a representative blot from three independent experiments.
We next addressed how infection affected HIF-1α translation by comparing its rate of synthesis in Toxoplasma-infected cells and cells treated with the hypoxia mimetic CoCl2, which stabilizes HIF-1α protein by inhibiting its hydroxylation (51). Eighteen hours after they were either infected or treated with CoCl2, the cells were exposed for increasing times to the proteasome inhibitor MG132. Lysates were prepared, separated by SDS-PAGE, and then immunoblotted with anti-HIF-1α antibodies. Densitometric analysis indicated that relative to CoCl2-treated cells, HIF-1α protein levels did not significantly increase after Toxoplasma-infected host cells were MG132-treated (Fig. 1B), indicating that Toxoplasma did not increase the rates of HIF-1α protein synthesis.
Finally, we assessed whether infection affected HIF-1α stability by treating parasite-infected or CoCl2-treated cells with the protein synthesis inhibitor cycloheximide. The data indicated that the half-lives of HIF-1α in Toxoplasma-infected and CoCl2-treated cells were 11.8 and 12.2 min, respectively, which is significantly higher than the half-life of HIF-1α under basal conditions (∼2 min) (52). Taken together, these data indicate that HIF-1α protein levels increase in Toxoplasma-infected host cells by increasing HIF-1α protein stability.
Toxoplasma Infection Reduces HIF-1α Prolyl Hydroxylation
HIF-1α stability is regulated by HIF-1α prolyl hydroxylation that allows the protein to be recognized and ubiquitylated by the von Hippel-Lindau ubiquitin ligase protein. The finding that Toxoplasma infection increased HIF-1α stability led us to test the hypothesis that Toxoplasma impairs HIF-1α prolyl hydroxylation. To monitor this modification, the cells were transfected with pTK-ODD-Rel, which contains the luciferase gene fused downstream to the ODD of HIF-1α. The transfected host cells were then either mock or parasite-infected or exposed to 3% O2 as a positive control. Luciferase activity was measured 18 h later, and the data indicated that luciferase activity was increased in parasite-infected cells to levels similar to those when uninfected cells were exposed to 3% O2 (Fig. 2A). Interestingly, we observed a reproducible additive effect on luciferase activity in parasite-infected cells at 3% O2.
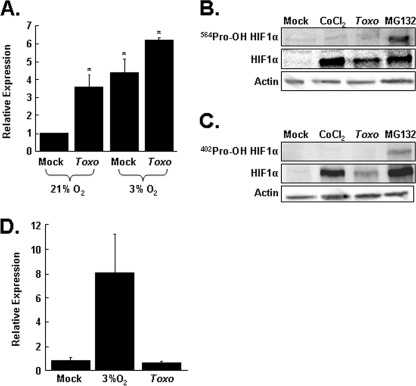
FIGURE 2.
Toxoplasma decreases prolyl hydroxylase activity. A, Renilla-ODD transfected cells were mock or parasite-infected and grown at 21 or 3% O2. Lysates were collected 24 h later, and luciferase activity was measured. Shown are the averages and standard deviations of six independent experiments. *, p < 0.05 Student's t test. B, HFFs were mock infected, CoCl2-treated, Toxoplasma-infected (Toxo) for 18 h, or MG132-treated for 2 h. C, the lysates were prepared and Western blotted with antibodies against either total HIF-1α, Pro402-OH HIF-1α, or Pro564-OH HIF-1α or actin as a loading control. Shown are representative blots from three independent experiments. D, cells cotransfected with pGBD-HIF-1α737–826 and the p5xGRE-luc reporter were either mock infected, Toxoplasma-infected, or shifted to 3% oxygen for 24 h. The lysates were collected, and luciferase activity was measured. Shown are the averages and standard deviations of three independent experiments. *, p < 0.05 Student's t test.
Next, we directly examined the prolyl hydroxylation state of HIF-1α using antibodies that specifically recognize prolyl hydroxylated HIF-1α. Lysates from mock infected, parasite-infected, CoCl2-treated, or MG132-treated cells were separated by SDS-PAGE and Western blotted with antibodies to detect either total HIF-1α or HIF-1α hydroxylated at Pro402 or Pro564. As expected, proteasome inhibition by MG132 led to increases in both total and prolyl-hydroxylated HIF-1α. This was in contrast to CoCl2-treated cells, in which only total HIF-1α could be detected because CoCl2 blocks HIF-1α prolyl hydroxylation. Similar to CoCl2, total HIF-1α protein was increased in parasite-infected host cells, but hydroxylated HIF-1α was undetectable (Fig. 2, B and C). Together, these data indicate that Toxoplasma reduces HIF-1α-directed PHD activity.
It is possible that the effect of Toxoplasma on HIF-1α prolyl hydroxylation was due to a global defect in the activity of O2/α-ketoglurate-dependent hydroxylases. To address this issue, we examined how infection affected the HIF-1α asparaginyl hydroxylase FIH that, like the PHDs, requires iron, ascorbate, O2, and α-ketoglutarate as substrate and cofactors. Thus, cells were transfected with the p5XGRE-luciferase reporter and a plasmid encoding either the GalDBD-HIF-1α737–826, which contains the asparagine residue targeted by FIH, or pGBD as a control. As expected, exposure of the transfected cells to hypoxia potently increased luciferase expression because of a reduction in FIH activity. In contrast, luciferase expression was unchanged in Toxoplasma-infected cells, indicating that infection had no apparent effect on FIH activity (Fig. 2D). Together, these data indicate that Toxoplasma specifically decreases the HIF-1α PHD.
Toxoplasma Infection Specifically Decreases PHD2 Protein Levels
We next hypothesized that Toxoplasma affected HIF-1α PHD activity by decreasing the expression of the prolyl hydroxylase enzymes. Thus, lysates from mock or parasite-infected cells were prepared 6 or 18 h post-infection and Western blotted with antibodies to detect the three HIF-1α PHDs: PHD1, PHD2, and PHD3. Densitometric analysis indicated that there was no detectable decrease in PHD1 protein levels in Toxoplasma-infected cells, and we failed to reproducibly detect PHD3 protein in either mock or parasite-infected cells (Fig. 3A and data not shown). In contrast, PHD2 protein levels were decreased as early as 6 h post-infection, and by 18 h the protein was reduced by 64%. As a second approach, the cells were transfected with a plasmid encoding FLAG-tagged PHD2 and then either mock or parasite-infected. Eighteen hours later, lysates were prepared and analyzed by Western blotting. The data showed that the levels of both the endogenous and epitope-tagged PHD2 were significantly reduced in parasite-infected cells by 67 and 55%, respectively (Fig. 3B). These data indicate that Toxoplasma specifically reduces expression of PHD2, which is the key prolyl hydroxylase in regulating HIF-1α levels.
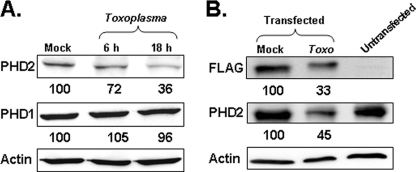
FIGURE 3.
Toxoplasma decreases PHD2 protein levels. A, HFFs were mock or Toxoplasma-infected (Toxo) for 6 or 18 h. Lysates were collected and Western blotted with antibodies against PHD1, PHD2, or actin as a loading control. Shown are representative blots from three independent experiments. B, mock or FLAG-tagged PHD2-transfected HeLa cells were mock or Toxoplasma-infected for 18 h. Lysates were collected and Western blotted with antibodies to detect the FLAG epitope, PHD2, and actin as a loading control. Shown are representative blots from three independent experiments. The number listed under each band in A and B represents the amount of protein remaining relative to the mock infected samples.
Toxoplasma Reduces Prolyl Hydroxylase Activity by Signaling through an Activin-like Receptor Kinase
The peptidyl prolyl cis/trans-isomerase FKBP38 binds PHD2, and its up-regulation results in decreased PHD2 protein levels (38). We therefore tested whether decreased PHD2 protein levels were due to the infection up-regulating FKBP38 protein levels. Thus, lysates from mock or parasite-infected cells were Western blotted with anti-FKBP38 antibodies. The data indicated that infection led to a reproducible decrease in FKBP38 expression (supplemental Fig. S1). This result strongly suggests that FKBP38-mediated degradation of PHD2, which is mediated by increases in FKBP38 protein, is likely not the mechanism controlling PHD2 expression in Toxoplasma-infected host cells.
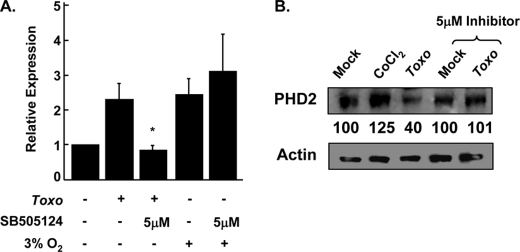
TGFβ activates HIF-1 by decreasing PHD2 protein levels (39). This decrease in PHD2 protein levels is dependent on signaling from the ALK5 Type I TGFβ receptor because highly specific pharmacological inhibitors prevent TGFβ-stimulated decreases in PHD2. To test whether Type I ALK receptor signaling regulates HIF-1 activation in Toxoplasma-infected host cells, pHRE-luc-transfected cells were mock or parasite-infected in the presence of increasing concentrations of SB505124, which is a highly specific ALK4,5,7 inhibitor (42). Lysates were prepared 18 h later, and luciferase activity was measured. The data indicated that the inhibitor potently blocked Toxoplasma induced pHRE-luc activity (Fig. 4A). The effect of SB505124 on parasite stimulation of HIF-1 was not a general consequence of altering HIF-1 signaling because the drug did not significantly affect pHRE-luc activation in cells exposed to decreased O2 tension (Fig. 4B).
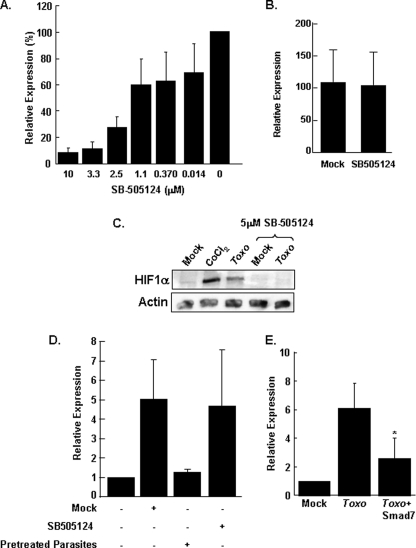
FIGURE 4.
Type I TGFβ receptor signaling is required for Toxoplasma stabilization of HIF-1α. A, pHRE-luc-transfected cells were infected in the presence of increasing concentrations of SB505124. After 24 h, the cells were lysed, and luciferase activity was measured. Shown are the means and standard deviations of three independent assays. B, pHRE-luc-transfected cells were exposed to 21 or 3% O2 in the absence or presence of SB505124. After 24 h, the cells were lysed, and luciferase activity was measured. Shown are the means and standard deviations of three independent assays. C, vehicle-treated or 5 μm SB505124-treated cells were infected with parasites or treated with CoCl2. After 18 h, the cells were lysed, and the lysates were Western blotted to detect HIF-1α or β-actin protein levels. D, mock or SB505124-treated (5 μm) pHRE-luc transfected host cells were infected with parasites, and luciferase activity was measured 18 h later. Pretreated parasites were prepared by incubating tachyzoites with 5 μm SB505124 or for 1 h and then washing the parasites with drug-free medium before adding them to pHRE-luc transfected host cells. E, pHRE-luc-transfected cells were cotransfected with either SMAD7 or empty vector as a control. The cells were mock or parasite-infected, and the luciferase activity was measured 18 h later. Shown are the averages and standard deviations from five independent experiments. *, p < 0.05 Student's t test. Toxo, Toxoplasma-infected.
Next, we tested whether the affects of the drug on parasite activation of pHRE-luc activity corresponded to a concomitant decrease in HIF-1α protein levels in Toxoplasma-infected cells. Thus, vehicle- or SB505124-treated cells were mock or parasite-infected. The lysates were prepared 18 h later and Western blotted to examine HIF-1α protein levels. As we previously demonstrated, infection dramatically increased HIF-1α abundance (6). HIF-1α protein levels were, however, significantly reduced in Toxoplasma-infected cells treated with SB505124 (Fig. 4C).
It is possible that the drug impacts Toxoplasma induction of HIF-1 by having a general affect on the ability of parasites to signal to their host cell. We therefore tested whether SB505124 blocked Toxoplasma inhibition of IFNγ signaling (53–55). Hence, vehicle- or drug-treated HIF-1α wild type cells were first infected with GFP+ parasites and 6 h later were treated with IFNγ for an additional 18 h. Expression of the costimulatory molecule, PD-L1, was then examined by flow cytometry because in fibroblasts as well as macrophages its expression is critically dependent on IFNγ (56–59) and because it is up-regulated more rapidly and robustly than MHC Class II.4 Similar to the effect of Toxoplasma on other IFNγ-regulated genes, PD-L1 expression was significantly reduced on the surface of Toxoplasma-infected cells (>95% versus 35%). This inhibition of PD-L1 expression was not affected by the presence of SB505124, indicating that the drug does overtly interfere with Toxoplasma signaling to its host cell (supplemental Fig. S2).
Caveats associated with using pharmacological inhibitors are that they may target the parasite and not the host and may affect cellular targets besides the Type I ALK4,5,7 receptor family. We therefore compared Toxoplasma up-regulation of HRE-luc activity when either the host cells or parasites were pretreated with the SB505124. The data indicated that HRE-luc activity was reduced by pretreating the host cells, but not the parasites, indicating that the drug did not have an unexpected, irreversible effect on the parasite (Fig. 4D).
As an alternative approach to inhibiting ALK4,5,7 signaling, we examined the effect of SMAD7 overexpression on pHRE-luc activity in Toxoplasma-infected cells. SMAD7 is an endogenous inhibitor of ALK4,5,7 by preventing activation of downstream effectors and by recruiting E3 ubiquitin ligases to the activated receptors (60–62). Host cells cotransfected with SMAD7 and the HRE-luc reporter were mock or parasite-infected, and 18 h later luciferase activity was measured. The data indicated that similar to SB505124, SMAD7 significantly reduced HRE-luc expression in Toxoplasma-infected cells by ∼60% (Fig. 4E).
Having established a requirement for Type I ALK4,5,7 receptor signaling in Toxoplasma activation of HIF-1, we next examined whether this receptor signaling pathway mediated decreases in PHD2 expression in Toxoplasma-infected cells. Hence, pTK-ODD-Rel-transfected cells were pretreated with vehicle or SB505124 and then mock or parasite-infected for 18 h, at which time luciferase activity was measured. We found that consistent with the HIF-1 data, SB505124 blocked Toxoplasma-induced ODD-luc expression (Fig. 5A).
FIGURE 5.
Inhibition of ALK4,5,7 signaling blocks Toxoplasma-induced decrease in PHD2. A, vehicle- or drug-treated pTK-ODD-Rel transfected host cells were treated as indicated. Lysates were prepared 24 h later, and luciferase activity was measured. Shown are the averages and standard deviations from three independent experiments. B, vehicle-treated or 5 μm SB505124-treated cells were infected with parasites or treated with CoCl2. After 18 h, the cells were lysed, and lysates were Western blotted to detect PHD2 and β-actin protein levels. The number listed under each band represents the amount of protein remaining relative to the mock infected samples. Toxo, Toxoplasma-infected.
Next, we tested whether the drug also impacted decreased PHD2 protein levels in Toxoplasma-infected cells. Thus, vehicle- or SB505124-treated cells were mock or parasite-infected. The lysates were prepared 18 h later and Western blotted to examine PHD2 protein levels. Densitometric analysis indicated that PHD2 protein levels decreased 60% in Toxoplasma-infected cells, but infection-induced decreases in PHD2 levels were abrogated by treating the cells with SB505124 (Fig. 5B).
ALK4,5,7 Receptor Inhibition Reduces Toxoplasma Replication
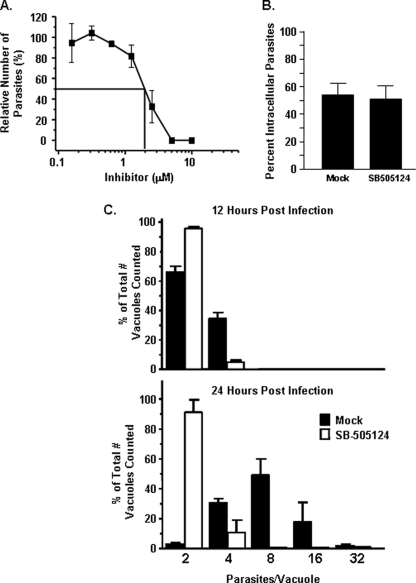
We next examined the effect of SB505124 on parasite growth. Thus, cells treated with increasing concentrations of the drug were infected with β-galactosidase-expressing parasites, and 3 days later the parasite numbers were determined. The data indicated that the drug reduced parasite growth with an apparent IC50 of 2 μm (Fig. 6A). Decreases in Toxoplasma growth in the drug-treated cells were likely not a consequence of decreased cell viability because previous work demonstrated that cell viability is not affected by SB505124 at the concentrations we used (42). In addition, 5 μm SB505124 had no detectable impact on the growth of an unrelated intracellular pathogen, West Nile virus (not shown).
FIGURE 6.
ALK4,5,7 signaling is required for Toxoplasma replication. A, parasite growth was measured 72 h after infected cells with β-galactosidase-expressing parasites in the presence of increasing levels of Type I Inhibitor. B, parasite invasion was measured by pretreating host cells with 5 μm SB505124 for 60 min. GFP-expressing parasites were then added, and the cells were fixed but not permeabilized 60 min later. Invasion was determined by differential SAG1 staining. C, parasite replication was measured by infecting mock or 5 μm SB505124-treated cells and then fixing the cells 12 or 24 h later. The parasites were detected by staining with anti-SAG1 antibody, and the numbers of parasites in individual vacuoles were counted. Shown are the averages and standard deviations from three independent experiments.
To determine how the drug reduced parasite growth, Toxoplasma invasion was assessed with GFP-expressing parasites using a potassium-based synchronized invasion assay (63). No apparent difference in parasite invasion between mock and drug-treated cells was observed, indicating that ALK4,5,7 signaling is not required for parasite invasion (Fig. 6B). We next tested whether the drug impacted parasite replication by counting numbers of parasites/vacuole in vehicle- or drug-treated cells 12 or 24 h post-infection. We found that relative to mock treated cells, parasite replication was severely reduced in SB505124-treated cells (Fig. 6C). Together, these data indicate that SB505124 reduces tachyzoite growth by reducing its rate of replication.
DISCUSSION
As all intracellular parasites must do, Toxoplasma modifies its host cell to create an environment permissive for its own growth. These changes include rearrangement of host cytoskeleton (64–66); relocalization of host mitochondria, endoplasmic reticulum, and lysosomes to the parasitophorous vacuole (67, 68); inhibition of host cell apoptosis (69–71); and modulation of host cell transcription (8, 43, 72, 73) by a variety of transcription factors including HIF-1, STAT3, EGR2, AP-1, and NF-κB (6–8, 45, 74). In this report, we defined the mechanism by which Toxoplasma activates HIF-1: increasing the stability, but not synthesis, of the HIF-1α subunit. This was surprising because our experiments were performed under normoxic conditions and because Toxoplasma activates host phosphatidylinositol 3-kinases (75, 76), which regulates HIF-1α protein synthesis (34, 77). Moreover, we found that Toxoplasma infection significantly decreased prolyl hydroxylase activity and expression of PHD2, which is the key regulator of HIF-1α protein degradation. Finally, we showed that Toxoplasma-dependent increases in HIF-1α protein and activity and decreases in PHD2 protein and prolyl hydroxylase activity were dependent on Type I activin-like kinase receptor signaling (supplemental Fig. S3).
In many cases, neither the biological significance of the parasite-induced changes to its host cell nor the molecular mechanisms mediating these changes are known. HIF-1 is one of the few host factors modulated by infection that is established to be necessary for parasite growth (6). Thus, the data in this report are significant because they define PHD2 as a key host cell factor targeted by infection. Moreover, parasite regulation of PHD2 via Type I ALK4,5,7 receptor family signaling indicates that this signaling pathway is important for infection and represents a novel drug target to treat Toxoplasma infections.
Previous work suggested that TGFβ decreases PHD2 protein levels by reducing PHD2 mRNA abundance (39). However, microarray (8, 43) and RT-PCR data5 have failed to detect decreased PHD2 mRNA levels in Toxoplasma-infected cells. Thus, Toxoplasma post-transcriptionally regulates PHD2 levels by affecting PHD2 protein synthesis and/or stability. We favor the latter mechanism for two reasons. First, PHD2 is a relatively long-lived enzyme with a half-life of >20 h (38), but a significant decrease in PHD2 protein levels was observed within 6 h post-infection. Hence, the effect that reduced PHD2 would have on total PHD2 protein levels would only be apparent at later time points. Second, the data in Fig. 3B were generated using an epitope-tagged PHD2 construct that contains the full-length PHD2 open reading frame without either endogenous 5′- or 3′-untranslated regions. This is significant because changes in untranslated region secondary structure are commonly associated with translational regulation (78). It is, however, possible that a microRNA targeting the PHD2 coding sequence may regulate PHD2 translation. Our future work will focus on defining the mechanism by which Toxoplasma affects PHD2 levels.
Signaling by the three Type I receptors (ALK4, ALK5, and ALK7) targeted by SB505124 is complex for a variety of reasons. First, the three receptors are expressed throughout the body and often are coexpressed by the same cell (79, 80). In addition, each of the receptors can bind and transduce signaling from numerous ligands e.g. TGFβ and GDF11 bind ALK5, activin A, and mysostatin bind ALK4, and activin AB and activin B interact with ALK7 (81, 82). Moreover, some ligands interact with more than one receptor. Nodal, which functions in embryonic development, binds both ALK4 and ALK7 (83). Finally, some ALK4,5,7 ligands are multimeric complexes whose individual subunits can be part of different multimeric complexes that bind to similar as well as different receptors. As an example, activin A is a homodimer of Inhibin βa (INHBA) that binds ALK4. INHBA is also a subunit of activin AB (the other subunit is Inhibin βb) that binds ALK5. To add to this complexity, INHBA can also interact with Inhibin α to form a complex named Inhibin, which is an antagonist of activin signaling. The function of INHBA expression may be particularly relevant to Toxoplasma infections because INHBA mRNA is up-regulated in Toxoplasma-infected cells (8, 43). Interestingly, TGFβ is also up-regulated in Toxoplasma infections (84–87) and in some cases can increase host cell susceptibility to infection (88). Thus, it is quite likely that signaling by more than one ALK4,5,7 ligand can signal through one or more receptors to activate HIF-1 in Toxoplasma-infected cells. It is also possible that the parasite expresses and secretes its own ligand that binds to ALK4,5 or 7. Our future work will focus on determining which of the Type I receptors are important for Toxoplasma infections and whether host- and/or parasite-derived ligands activate this receptor(s).
HIF-1 was originally identified as a critical player in cell response to hypoxic stress (89). It is, however, becoming increasingly apparent that HIF-1 is activated by diverse stimuli under normoxic conditions. A key question resulting from these studies is whether HIF-1α functions similarly when it is activated at either O2-rich or -poor conditions. HIF-1α has two transactivation domains that regulate distinct and common sets of genes (90). The C-terminal transactivation domain is regulated by FIH, which in vitro has a higher Km for O2 than the PHDs. Thus, the N-terminal transactivation domain is most likely to be active under O2-replete conditions such as Toxoplasma-infected cells. This conclusion is consistent with our data that although Toxoplasma could stimulate a HIF-1-regulated luciferase reporter, it could not overcome FIH-mediated inhibition of the C-TAD. Thus, future work aimed at identifying HIF-1 target genes that promote parasite growth most likely can exclude those regulated by the C-TAD.
At 21% O2, parasite growth was decreased in HIF-1α KO cells by ∼70% (6), which was in contrast to the dramatic effect that SB505124 had on parasite replication. This could be due to the fact that other host proteins regulated by ALK4,5,7 can compensate for the loss of HIF-1α at 21% O2. Alternatively, it is possible that Type I receptor signaling regulates several pathways that act independently to promote parasite growth. Thus, whereas HIF-1α/PHD2 may be one target of Type I receptor signaling, other pathways may also contribute to parasite growth (supplemental Fig. S3). Possible targets may include MAPK and phosphatidylinositol 3-kinase-dependent signaling, which are activated in Toxoplasma-infected cells and are also regulated by these receptors (40, 91, 92), but whether the loss of other individual pathways will impact parasite growth to the same extent as the loss of HIF-1α is unknown and will need to be tested as additional host factors that are regulated by Toxoplasma are identified.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant AI069986. This work was also supported by American Cancer Society Grant MBC-114461 (to I. J. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
K. M. Brown and I. J. Blader, unpublished data.
M. Wiley and I. J. Blader, unpublished data.
- HIF-1
- hypoxia-inducible factor 1
- PHD
- prolyl hydroxylase
- ALK
- activin-like receptor kinase
- GBD
- GAL4 DNA-binding domain
- ODD
- oxygen degradation domain
- E3
- ubiquitin-protein isopeptide ligase
- FIH
- factor inhibiting HIF
- HFF
- human foreskin fibroblast.
REFERENCES
- 1.Kim K., Weiss L. M. (2004) Int. J. Parasitol. 34, 423–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Montoya J. G., Liesenfeld O. (2004) Lancet 363, 1965–1976 [DOI] [PubMed] [Google Scholar]
- 3.Blader I. J., Saeij J. P. (2009) Acta Pathol. Microbiol. Immunol. Scand. 117, 458–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coppens I., Joiner K. A. (2001) Expert. Rev. Mol. Med. 2001, 1–20 [DOI] [PubMed] [Google Scholar]
- 5.Zeiner G. M., Norman K. L., Thomson J. M., Hammond S. M., Boothroyd J. C. (2010) PLoS ONE 5, e8742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spear W., Chan D., Coppens I., Johnson R. S., Giaccia A., Blader I. J. (2006) Cell. Microbiol. 8, 339–352 [DOI] [PubMed] [Google Scholar]
- 7.Yamamoto M., Standley D. M., Takashima S., Saiga H., Okuyama M., Kayama H., Kubo E., Ito H., Takaura M., Matsuda T., Soldati-Favre D., Takeda K. (2009) J. Exp. Med. 206, 2747–2760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saeij J. P., Coller S., Boyle J. P., Jerome M. E., White M. W., Boothroyd J. C. (2007) Nature 445, 324–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saeij J. P., Boyle J. P., Coller S., Taylor S., Sibley L. D., Brooke-Powell E. T., Ajioka J. W., Boothroyd J. C. (2006) Science 314, 1780–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor S., Barragan A., Su C., Fux B., Fentress S. J., Tang K., Beatty W. L., Hajj H. E., Jerome M., Behnke M. S., White M., Wootton J. C., Sibley L. D. (2006) Science 314, 1776–1780 [DOI] [PubMed] [Google Scholar]
- 11.Semenza G. L. (2007) Sci. STKE 2007, cm8. [DOI] [PubMed] [Google Scholar]
- 12.Huang L. E., Gu J., Schau M., Bunn H. F. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 7987–7992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salceda S., Caro J. (1997) J. Biol. Chem. 272, 22642–22647 [DOI] [PubMed] [Google Scholar]
- 14.Maxwell P. H., Wiesener M. S., Chang G. W., Clifford S. C., Vaux E. C., Cockman M. E., Wykoff C. C., Pugh C. W., Maher E. R., Ratcliffe P. J. (1999) Nature 399, 271–275 [DOI] [PubMed] [Google Scholar]
- 15.Jaakkola P., Mole D. R., Tian Y. M., Wilson M. I., Gielbert J., Gaskell S. J., Kriegsheim A., Hebestreit H. F., Mukherji M., Schofield C. J., Maxwell P. H., Pugh C. W., Ratcliffe P. J. (2001) Science 292, 468–472 [DOI] [PubMed] [Google Scholar]
- 16.Ivan M., Kondo K., Yang H., Kim W., Valiando J., Ohh M., Salic A., Asara J. M., Lane W. S., Kaelin W. G., Jr. (2001) Science 292, 464–468 [DOI] [PubMed] [Google Scholar]
- 17.Masson N., Ratcliffe P. J. (2003) J. Cell Sci. 116, 3041–3049 [DOI] [PubMed] [Google Scholar]
- 18.Yu F., White S. B., Zhao Q., Lee F. S. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 9630–9635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bruick R. K., McKnight S. L. (2001) Science 294, 1337–1340 [DOI] [PubMed] [Google Scholar]
- 20.Appelhoff R. J., Tian Y. M., Raval R. R., Turley H., Harris A. L., Pugh C. W., Ratcliffe P. J., Gleadle J. M. (2004) J. Biol. Chem. 279, 38458–38465 [DOI] [PubMed] [Google Scholar]
- 21.Berra E., Benizri E., Ginouvès A., Volmat V., Roux D., Pouysségur J. (2003) EMBO J. 22, 4082–4090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lando D., Peet D. J., Whelan D. A., Gorman J. J., Whitelaw M. L. (2002) Science 295, 858–861 [DOI] [PubMed] [Google Scholar]
- 23.Lando D., Peet D. J., Gorman J. J., Whelan D. A., Whitelaw M. L., Bruick R. K. (2002) Genes Dev. 16, 1466–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hewitson K. S., McNeill L. A., Riordan M. V., Tian Y. M., Bullock A. N., Welford R. W., Elkins J. M., Oldham N. J., Bhattacharya S., Gleadle J. M., Ratcliffe P. J., Pugh C. W., Schofield C. J. (2002) J. Biol. Chem. 277, 26351–26355 [DOI] [PubMed] [Google Scholar]
- 25.Hirsilä M., Koivunen P., Günzler V., Kivirikko K. I., Myllyharju J. (2003) J. Biol. Chem. 278, 30772–30780 [DOI] [PubMed] [Google Scholar]
- 26.Koivunen P., Hirsilä M., Günzler V., Kivirikko K. I., Myllyharju J. (2004) J. Biol. Chem. 279, 9899–9904 [DOI] [PubMed] [Google Scholar]
- 27.Kempf V. A., Lebiedziejewski M., Alitalo K., Wälzlein J. H., Ehehalt U., Fiebig J., Huber S., Schütt B., Sander C. A., Müller S., Grassl G., Yazdi A. S., Brehm B., Autenrieth I. B. (2005) Circulation 111, 1054–1062 [DOI] [PubMed] [Google Scholar]
- 28.Hartmann H., Eltzschig H. K., Wurz H., Hantke K., Rakin A., Yazdi A. S., Matteoli G., Bohn E., Autenrieth I. B., Karhausen J., Neumann D., Colgan S. P., Kempf V. A. (2008) Gastroenterology 134, 756–767 [DOI] [PubMed] [Google Scholar]
- 29.Peyssonnaux C., Datta V., Cramer T., Doedens A., Theodorakis E. A., Gallo R. L., Hurtado-Ziola N., Nizet V., Johnson R. S. (2005) J. Clin. Invest. 115, 1806–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sodhi A., Montaner S., Patel V., Zohar M., Bais C., Mesri E. A., Gutkind J. S. (2000) Cancer Res. 60, 4873–4880 [PubMed] [Google Scholar]
- 31.Wakisaka N., Kondo S., Yoshizaki T., Murono S., Furukawa M., Pagano J. S. (2004) Mol. Cell. Biol. 24, 5223–5234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakamura M., Bodily J. M., Beglin M., Kyo S., Inoue M., Laimins L. A. (2009) Virology 387, 442–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arrais-Silva W. W., Paffaro V. A., Jr., Yamada A. T., Giorgio S. (2005) Exp. Mol. Pathol. 78, 49–54 [DOI] [PubMed] [Google Scholar]
- 34.Treins C., Giorgetti-Peraldi S., Murdaca J., Semenza G. L., Van Obberghen E. (2002) J. Biol. Chem. 277, 27975–27981 [DOI] [PubMed] [Google Scholar]
- 35.Thornton R. D., Lane P., Borghaei R. C., Pease E. A., Caro J., Mochan E. (2000) Biochem. J. 350, 307–312 [PMC free article] [PubMed] [Google Scholar]
- 36.Liu X. H., Kirschenbaum A., Lu M., Yao S., Dosoretz A., Holland J. F., Levine A. C. (2002) J. Biol. Chem. 277, 50081–50086 [DOI] [PubMed] [Google Scholar]
- 37.Pan Y., Mansfield K. D., Bertozzi C. C., Rudenko V., Chan D. A., Giaccia A. J., Simon M. C. (2007) Mol. Cell. Biol. 27, 912–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barth S., Nesper J., Hasgall P. A., Wirthner R., Nytko K. J., Edlich F., Katschinski D. M., Stiehl D. P., Wenger R. H., Camenisch G. (2007) Mol. Cell. Biol. 27, 3758–3768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McMahon S., Charbonneau M., Grandmont S., Richard D. E., Dubois C. M. (2006) J. Biol. Chem. 281, 24171–24181 [DOI] [PubMed] [Google Scholar]
- 40.Derynck R., Zhang Y. E. (2003) Nature 425, 577–584 [DOI] [PubMed] [Google Scholar]
- 41.Shi Y., Massagué J. (2003) Cell 113, 685–700 [DOI] [PubMed] [Google Scholar]
- 42.DaCosta Byfield S., Major C., Laping N. J., Roberts A. B. (2004) Mol. Pharmacol. 65, 744–752 [DOI] [PubMed] [Google Scholar]
- 43.Blader I. J., Manger I. D., Boothroyd J. C. (2001) J. Biol. Chem. 276, 24223–24231 [DOI] [PubMed] [Google Scholar]
- 44.Chan D. A., Sutphin P. D., Yen S. E., Giaccia A. J. (2005) Mol. Cell. Biol. 25, 6415–6426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Phelps E. D., Sweeney K. R., Blader I. J. (2008) Infect. Immun. 76, 4703–4712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fruth I. A., Arrizabalaga G. (2007) Int. J. Parasitol. 37, 1559–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seeber F., Boothroyd J. C. (1996) Gene 169, 39–45 [DOI] [PubMed] [Google Scholar]
- 48.Eustice D. C., Feldman P. A., Colberg-Poley A. M., Buckery R. M., Neubauer R. H. (1991) BioTechniques 11, 739–740, 742–743 [PubMed] [Google Scholar]
- 49.Sweeney K. R., Morrissette N. S., Lachapelle S., Blader I. J. (2010) Eukaryotic Cell, EC. 00079–00010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McMurtrey C. P., Lelic A., Piazza P., Chakrabarti A. K., Yablonsky E. J., Wahl A., Bardet W., Eckerd A., Cook R. L., Hess R., Buchli R., Loeb M., Rinaldo C. R., Bramson J., Hildebrand W. H. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 2981–2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salnikow K., Donald S. P., Bruick R. K., Zhitkovich A., Phang J. M., Kasprzak K. S. (2004) J. Biol. Chem. 279, 40337–40344 [DOI] [PubMed] [Google Scholar]
- 52.Wang G. L., Jiang B. H., Rue E. A., Semenza G. L. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 5510–5514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zimmermann S., Murray P. J., Heeg K., Dalpke A. H. (2006) J. Immunol. 176, 1840–1847 [DOI] [PubMed] [Google Scholar]
- 54.Kim S. K., Fouts A. E., Boothroyd J. C. (2007) J. Immunol. 178, 5154–5165 [DOI] [PubMed] [Google Scholar]
- 55.Lüder C. G., Lang T., Beuerle B., Gross U. (1998) Clin. Exp. Immunol. 112, 308–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dong H., Strome S. E., Salomao D. R., Tamura H., Hirano F., Flies D. B., Roche P. C., Lu J., Zhu G., Tamada K., Lennon V. A., Celis E., Chen L. (2002) Nat. Med. 8, 793–800 [DOI] [PubMed] [Google Scholar]
- 57.Lee S. K., Seo S. H., Kim B. S., Kim C. D., Lee J. H., Kang J. S., Maeng P. J., Lim J. S. (2005) J. Dermatol. Sci. 40, 95–103 [DOI] [PubMed] [Google Scholar]
- 58.Mazanet M. M., Hughes C. C. (2002) J. Immunol. 169, 3581–3588 [DOI] [PubMed] [Google Scholar]
- 59.Lázár-Molnár E., Gácser A., Freeman G. J., Almo S. C., Nathenson S. G., Nosanchuk J. D. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 2658–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hayashi H., Abdollah S., Qiu Y., Cai J., Xu Y. Y., Grinnell B. W., Richardson M. A., Topper J. N., Gimbrone M. A., Jr., Wrana J. L., Falb D. (1997) Cell 89, 1165–1173 [DOI] [PubMed] [Google Scholar]
- 61.Kavsak P., Rasmussen R. K., Causing C. G., Bonni S., Zhu H., Thomsen G. H., Wrana J. L. (2000) Mol. Cell 6, 1365–1375 [DOI] [PubMed] [Google Scholar]
- 62.Nakao A., Afrakhte M., Morén A., Nakayama T., Christian J. L., Heuchel R., Itoh S., Kawabata M., Heldin N. E., Heldin C. H., ten Dijke P. (1997) Nature 389, 631–635 [DOI] [PubMed] [Google Scholar]
- 63.Kafsack B. F., Beckers C., Carruthers V. B. (2004) Mol. Biochem. Parasitol. 136, 309–311 [DOI] [PubMed] [Google Scholar]
- 64.Halonen S. K., Weidner E. (1994) J. Eukaryot. Microbiol. 41, 65–71 [DOI] [PubMed] [Google Scholar]
- 65.Coppens I., Sinai A. P., Joiner K. A. (2000) J. Cell Biol. 149, 167–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Walker M. E., Hjort E. E., Smith S. S., Tripathi A., Hornick J. E., Hinchcliffe E. H., Archer W., Hager K. M. (2008) Microbes Infect. 10, 1440–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sinai A. P., Webster P., Joiner K. A. (1997) J. Cell Sci. 110, 2117–2128 [DOI] [PubMed] [Google Scholar]
- 68.Coppens I., Dunn J. D., Romano J. D., Pypaert M., Zhang H., Boothroyd J. C., Joiner K. A. (2006) Cell 125, 261–274 [DOI] [PubMed] [Google Scholar]
- 69.Nash P. B., Purner M. B., Leon R. P., Clarke P., Duke R. C., Curiel T. J. (1998) J. Immunol. 160, 1824–1830 [PubMed] [Google Scholar]
- 70.Goebel S., Lüder C. G., Gross U. (1999) Med. Microbiol. Immunol. 187, 221–226 [DOI] [PubMed] [Google Scholar]
- 71.Payne T. M., Molestina R. E., Sinai A. P. (2003) J. Cell Sci. 116, 4345–4358 [DOI] [PubMed] [Google Scholar]
- 72.Gail M., Gross U., Bohne W. (2001) Mol. Genet. Genomics. 265, 905–912 [DOI] [PubMed] [Google Scholar]
- 73.Chaussabel D., Semnani R. T., McDowell M. A., Sacks D., Sher A., Nutman T. B. (2003) Blood 102, 672–681 [DOI] [PubMed] [Google Scholar]
- 74.Molestina R. E., Payne T. M., Coppens I., Sinai A. P. (2003) J. Cell Sci. 116, 4359–4371 [DOI] [PubMed] [Google Scholar]
- 75.Kim L., Butcher B. A., Lee C. W., Uematsu S., Akira S., Denkers E. Y. (2006) J. Immunol. 177, 2584–2591 [DOI] [PubMed] [Google Scholar]
- 76.Wang Y., Weiss L. M., Orlofsky A. (2009) Cell Microbiol. 6, 983–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Harada H., Itasaka S., Kizaka-Kondoh S., Shibuya K., Morinibu A., Shinomiya K., Hiraoka M. (2009) J. Biol. Chem. 284, 5332–5342 [DOI] [PubMed] [Google Scholar]
- 78.Wilkie G. S., Dickson K. S., Gray N. K. (2003) Trends Biochem. Sci. 28, 182–188 [DOI] [PubMed] [Google Scholar]
- 79.Chang H., Brown C. W., Matzuk M. M. (2002) Endocr. Rev. 23, 787–823 [DOI] [PubMed] [Google Scholar]
- 80.Taylor A. W. (2009) J. Leukocyte Biol. 85, 29–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Andersson O., Reissmann E., Ibáñez C. F. (2006) EMBO Rep. 7, 831–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Harrison C. A., Gray P. C., Vale W. W., Robertson D. M. (2005) Trends Endocrinol. Metab. 16, 73–78 [DOI] [PubMed] [Google Scholar]
- 83.Reissmann E., Jörnvall H., Blokzijl A., Andersson O., Chang C., Minchiotti G., Persico M. G., Ibáñez C. F., Brivanlou A. H. (2001) Genes. Dev. 15, 2010–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bermudez L. E., Covaro G., Remington J. (1993) Infect. Immun. 61, 4126–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nagineni C. N., Detrick B., Hooks J. J. (2002) Clin. Exp. Immunol. 128, 372–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schlüter D., Deckert M., Hof H., Frei K. (2001) Infect. Immun. 69, 7889–7893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hunter C. A., Abrams J. S., Beaman M. H., Remington J. S. (1993) Infect. Immun. 61, 4038–4044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Barbosa B. F., Silva D. A., Costa I. N., Mineo J. R., Ferro E. A. (2008) Clin. Exp. Immunol. 151, 536–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang G. L., Semenza G. L. (1993) J. Biol. Chem. 268, 21513–21518 [PubMed] [Google Scholar]
- 90.Dayan F., Roux D., Brahimi-Horn M. C., Pouyssegur J., Mazure N. M. (2006) Cancer Res. 66, 3688–3698 [DOI] [PubMed] [Google Scholar]
- 91.Kim L., Denkers E. Y. (2006) J. Cell Sci. 119, 2119–2126 [DOI] [PubMed] [Google Scholar]
- 92.Mason N. J., Fiore J., Kobayashi T., Masek K. S., Choi Y., Hunter C. A. (2004) Infect. Immun. 72, 5662–5667 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.