Abstract
We discovered novel catalytic activities of two atypical NADPH-dependent oxidoreductases (EhNO1/2) from the enteric protozoan parasite Entamoeba histolytica. EhNO1/2 were previously annotated as the small subunit of glutamate synthase (glutamine:2-oxoglutarate amidotransferase) based on similarity to authentic bacterial homologs. As E. histolytica lacks the large subunit of glutamate synthase, EhNO1/2 were presumed to play an unknown role other than glutamine/glutamate conversion. Transcriptomic and quantitative reverse PCR analyses revealed that supplementation or deprivation of extracellular l-cysteine caused dramatic up- or down-regulation, respectively, of EhNO2, but not EhNO1 expression. Biochemical analysis showed that these FAD- and 2[4Fe-4S]-containing enzymes do not act as glutamate synthases, a conclusion which was supported by phylogenetic analyses. Rather, they catalyze the NADPH-dependent reduction of oxygen to hydrogen peroxide and l-cystine to l-cysteine and also function as ferric and ferredoxin-NADP+ reductases. EhNO1/2 showed notable differences in substrate specificity and catalytic efficiency; EhNO1 had lower Km and higher kcat/Km values for ferric ion and ferredoxin than EhNO2, whereas EhNO2 preferred l-cystine as a substrate. In accordance with these properties, only EhNO1 was observed to physically interact with intrinsic ferredoxin. Interestingly, EhNO1/2 also reduced metronidazole, and E. histolytica transformants overexpressing either of these proteins were more sensitive to metronidazole, suggesting that EhNO1/2 are targets of this anti-amebic drug. To date, this is the first report to demonstrate that small subunit-like proteins of glutamate synthase could play an important role in redox maintenance, l-cysteine/l-cystine homeostasis, iron reduction, and the activation of metronidazole.
Keywords: Drug Action, Iron, Metabolism, Oxidation-Reduction, Protozoan, Glutamate Synthase, l-Cysteine
Introduction
Glutamate synthase (glutamine:2-oxoglutarate amidotransferase, GOGAT3) is an iron sulfur flavoprotein that catalyzes the transfer of the amide group of l-glutamine to 2-oxoglutrate to yield l-glutamate and is a key enzyme in the nitrogen assimilation pathway. In eubacteria, this enzyme is dependent on the pyridine nucleotide NAD(P)H for its reducing equivalents and is composed of large 150-kDa (α) and small 50-kDa (β) subunits that together form the active αβ protomer (1). The structural genes encoding the α and β subunit polypeptides are commonly designated gltB and gltD, respectively, and lie adjacent on the chromosome with the α subunit preceding the β subunit, except in γ-proteobacteria, where the gene order is reversed. The small subunit of eubacterial glutamate synthase shows sequence similarity to several other protein domains and enzyme subunits (2, 3) and is, therefore, proposed to represent a prototype domain used in many different cellular processes to transfer electrons from NAD(P)H to an acceptor protein or protein domain of unknown function (4). In concord with this view, numerous organisms have been recently identified to possess glutamate synthase β subunit-like genes based on DNA sequence homology (4, 5); however, the organisms often lack a gene encoding the corresponding α subunit, or the β subunit is not present adjacent to the α subunit and is, therefore, transcribed independently (5, 6). To our knowledge, among the organisms lacking a putative GOGAT α subunit, only the GOGAT β subunit from Thermococcus kodakaraensis (renamed from Pyrococcus sp. KOD1) has been functionally associated with independent GOGAT activity (7).
Entamoeba histolytica, the causative agent of human amebiasis, is an enteric protozoan parasite responsible for amebic colitis and extraintestinal abscesses in approximately 50 million inhabitants of endemic areas (8). As is the case with other microaerophilic parasitic infections, such as giardiasis and trichomoniasis, the 5-nitroimidazole drug metronidazole has been established as the most effective treatment of amebiasis. Because of the high prevalence of these infections (9) and because of its role as a second-line defense against Helicobacter pylori infections (10), metronidazole has been included in the list of “essential medicines” by the World Health Organization (11). Metronidazole is a prodrug that requires reduction of the nitro group to generate the cytotoxic nitroradical anion that undergoes further reduction resulting in the generation of nitrosoimidazole (12, 13). This active form can then react with sulfhydryl groups (14) and DNA (15) while being further reduced to an amine via a hydroxylamine intermediate. Here, we report for the first time multiple novel roles of two GOGAT β subunit-like proteins in E. histolytica. We demonstrated that they are not associated with glutamate synthase activity but instead exhibit robust reductase activities against l-cystine, ferredoxin, and ferric ion and are also involved in the response to oxidative stress. In addition, we showed that these enzymes can be capable of reducing and activating metronidazole and, thus, are responsible for its observed toxicity against E. histolytica. We designated the novel NADPH-dependent oxidoreductases as EhNO1 and -2.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
l-Cysteine, l-cystine, trans-epoxysuccinyl-l-leucylamido-(4-guanidino) butane, cytochrome c, iodonitrotetrazolium (INT), and metronidazole were purchased from Sigma. Nickel-nitrilotriacetic acid-agarose was purchased from Merck. All other chemicals of analytical grade were purchased from Wako Pure Chemical (Osaka, Japan) unless otherwise stated.
Microorganisms and Cultivation
Trophozoites of the E. histolytica clonal strain HM1:IMSS cl 6 were maintained axenically in Diamond's BI-S-33 medium at 35.5 °C as described previously (16, 17). Trophozoites were harvested in the late logarithmic growth phase for 2–3 days after inoculation of to of the total culture volume. After the cultures were chilled on ice for 5 min, trophozoites were collected by centrifugation at 500 × g for 10 min at 4 °C and washed twice with ice-cold PBS (pH 7.4). Escherichia coli BL21 (DE3) strain was purchased from Invitrogen.
Quantitative Real-time PCR
Trophozoites were cultured in BI-S-33 medium supplemented with or without 10 mm l-cysteine (18 or 8 mm final, respectively). After placing the culture on ice for 5 min, the trophozoites were harvested by centrifugation at 500 × g for 5 min at 4 °C. Polyadenylated RNA was extracted from ∼6 × 106 tropozoites with an mRNA isolation kit (Stratagene, La Jolla, CA) and then treated with deoxyribonuclease I (Invitrogen). cDNA was reverse-transcribed with 4 μg of isolated polyadenylated RNA, the SuperScript III First-Strand Synthesis System, and an oligo(dT)20 primer (Invitrogen). PCR was performed with the resulting cDNA as a template and specific oligonucleotide primers using the ABI PRISM 7300 Sequence Detection System (Applied Biosystems, Japan). The primers used were 5′-AGCTGCACCAGTTCCAATTC-3′ and 5′-CAATCCCCAGCTGCATATAA-3′ (EhNO1), 5′-CAGTTCCAATTCCAGGCAGT-3′ and 5′-TTGGTCCTGTAACACAATCTCCT-3′ (EhNO2), and 5′-GATCCAACATATCCTAAAACAACA-3′ and 5′-TCAATTATTTTCTGACCCGTCTTC-3′ (RNA polymerase II 15-kDa subunit, GenBankTM accession number XM_643999). The parameters for PCR were as follows: an initial step of denaturation at 95 °C for 9 min followed by 40 cycles of denaturation at 94 °C for 30 s, annealing at 50 °C for 30 s, and extension at 65 °C for 1 min and a final step at 95 °C for 9 s, 60 °C for 9 s, and 95 °C for 9 s was used to remove primer dimers.
Amino Acid Comparison and Phylogenetic Analysis
Amino acid sequences of the GOGAT β subunit and β subunit-like proteins from 40 other organisms were obtained from the DDBJ/EBI/GenBankTM data base using BLASTP searches with the novel amebic NADPH-dependent oxidoreductases (EhNO1 and EhNO2) described in this paper as queries. Sequence alignments of these proteins were generated using the ClustalW program (18). The alignments obtained by ClustalW were inspected and manually corrected using the Genedoc program (19). After the removal of all gaps, 326 unambiguously aligned residues were selected for phylogenetic analyses. The neighbor-joining and maximum parsimony methods were used to construct a final phylogenetic tree for 32 sequences using the MEGA4.1 program (20). The branch lengths and bootstrap values of 1000 replicates (in percentage) in these trees were obtained from the neighbor-joining analysis.
Construction of Plasmids
Standard techniques were used for cloning and plasmid construction, as previously described (21). Genes encoding EhNO1 and EhNO2 were cloned to produce a fusion protein containing a histidine tag (provided by the vector) at the amino terminus. The cDNA corresponding to the open reading frames of EhNO1 and EhNO2 was amplified by PCR using an E. histolytica cDNA library (22) as a template and oligonucleotide primers. The sense and antisense oligonucleotide primers used to amplify EhNO1 and EhNO2 were 5′-CTTATAAGGATCCATGAAGAGTTTCAACATTA-3′ and 5′-ATAGTCGACTTAATCTTGTTCCATTGGG-3′ (EhNO1) and 5′-CTTATAAGGATCCATGGCTGCTAATTATAATA-3′ and 5′-ATAGTCGACTTATTCTTCATTTTTTTTACCC-3′ (EhNO2) (bold letters indicate BamHI and SalI restriction sites). PCR was performed with Platinum Pfx DNA polymerase (Invitrogen) and the following parameters: an initial incubation at 94 °C for 2 min followed by 30 cycles of denaturation at 94 °C for 15 s, annealing at 45 °C for 30 s, and elongation at 68 °C for 2 min and a final extension at 68 °C for 10 min. The PCR fragments were digested with BamHI and SalI, subjected to gel electrophoresis, excised, purified with the Gene clean kit II (BIO 101, Vista, CA), and then ligated into BamHI- and SalI-digested pCOLD I (Takara Bio, Otsu, Japan) in the identical orientation as the T7 promoter to generate pCOLD1-EhNO1 and pCOLD1-EhNO2. The nucleotide sequences of the cloned EhNO1 and EhNO2 genes were verified by sequencing to be identical to the putative protein coding regions of XP_656997 and XP_653573, respectively, in E. histolytica.
Bacterial Expression and Purification of Recombinant EhNO (rEhNO)
The pCOLD1-EhNO1 and pCOLD1-EhNO2 expression constructs were introduced into competent E. coli BL21 (DE3) cells by heat shock at 42 °C for 30 s, and the resulting transformants were grown at 37 °C in 100 ml of Luria Bertani medium in the presence of 50 μg/ml ampicillin. The overnight culture was then used to inoculate 500 ml of fresh medium, which was further cultured at 37 °C with shaking at 180 rpm. When the A600 reached 0.6, 1 mm isopropyl β-d-thiogalactopyranoside was added to induce protein expression, and cultivation was continued for 24 h at 15 °C. The E. coli cells were then harvested by centrifugation at 4050 × g for 20 min at 4 °C, and the resulting cell pellet was washed with PBS (pH 7.4) and re-suspended in 20 ml of lysis buffer (50 mm Tris-HCl (pH 8.0), 300 mm NaCl, and 10 mm imidazole) containing 0.1% Triton X-100 (v/v), 100 μg/ml lysozyme, and 1 mm phenylmethylsulfonyl fluoride. After a 30-min incubation at room temperature, the cells were sonicated on ice and centrifuged at 25,000 × g for 15 min at 4 °C. The supernatant was mixed with 1.2 ml of a 50% nickel-nitrilotriacetic acid His-bind slurry (Qiagen, Tokyo, Japan) and incubated for 1 h at 4 °C with gentle shaking. The rEhNO-bound resin was washed three times with buffer A (50 mm Tris-HCl (pH 8.0), 300 mm NaCl, and 0.1% Triton X-100, v/v) containing 10–50 mm imidazole, and bound proteins were then eluted with buffer A containing 100–300 mm imidazole. After the integrity and purity of the rEhNO proteins were confirmed by 12% SDS-PAGE analysis and Coomassie Brilliant Blue staining, they were extensively dialyzed twice against a 300-fold volume of 50 mm Tris-HCl, 150 mm NaCl, pH 8.0, containing 10% glycerol (v/v) and the Complete Mini Protease Inhibitor Mixture (Roche Applied Science) for 18 h at 4 °C. The concentrations of the dialyzed proteins were spectrophotometrically determined by the Bradford method using bovine serum albumin as a standard as previously described (23). The rEhNO proteins were stored at −80 °C in 20% glycerol in small aliquots until needed.
Analysis of Prosthetic Groups
UV-visible absorption spectra of rEhNO1 (400 μg) and rEhNO2 (200 μg) were measured under both non-reducing and sodium dithionite-reducing conditions. The purified recombinant proteins were reduced with a 10-fold molar excess of sodium dithionite in 200 μl. Flavin was liberated from the recombinant enzymes by boiling samples for 10 min and then separated from proteins by centrifugation at 14,000 × g for 10 min. To determine whether FAD or FMN formed a prosthetic group, the fluorescence with excitation and emission wavelengths of 450 and 535 nm, respectively, was measured at pH 2.6 and 7.7 according to the method of Faeder and Siegel (24) using a fluorescence spectrophotometer (model F-2500; Hitachi).
Iron Assay
The iron content of EhNOs was determined by the O-phenanthroline method as previously described (25). Briefly, 60 μl samples of rEhNO1 and rEhNO2 were mixed with 4 μl of concentrated HCl and then diluted with distilled water to 0.2 ml. After the resulting mixtures were heated to 80 °C for 10 min and cooled to room temperature, they were then mixed with 0.6 ml of water, 40 μl of 10% hydroxylamine hydrochloride, and 0.2 ml of 0.1% O-phenanthroline and further incubated at room temperature for 30 min. The absorbances at 512 nm (A512) were then measured, and the iron concentrations were determined by comparison to a standard curve generated with 0–100 μm ferrous sulfate.
Enzyme Assays
Glutamate synthase activity was assayed spectrophotometrically by measuring the rate of NADPH or NADH oxidation at 340 nm with slight modifications of the procedure described by Jongsareejit et al. (7). The 200-μl assay mixture contained 20 mm potassium phosphate buffer, pH 7.5, 5 mm concentrations each of l-glutamine and 2-oxoglutarate, 0.4 mm cofactor (NADPH or NADH), and varying concentrations of rEhNO proteins. The reaction was initiated by the addition of cofactor and was performed at 37 °C. To test for ammonia-dependent activity, glutamine was replaced with 100 mm NH4Cl.
Oxidoreductase Activity
The NADPH-dependent reduction of menadione was monitored in a coupling assay under aerobic conditions. The rate of reduction of cytochrome c by menadione was monitored by the absorbance at 550 nm (ϵ550 = 21.1 mm−1 cm−1). Measurements were made in 50 mm Tris-HCl (pH 7.5), 200 μm NADPH, 1 μm menadione, and 30 μm cytochrome c. The reactions were initiated by the addition of 2 μg of rEhNO1/2.
For the other electron acceptors tested, a standard mixture containing 0.1 mm NADPH, 50 mm Tris-HCl (pH 7.5), and either 0.5 mm INT, 1 mm potassium ferricyanide, or 10 mm paraquat was used. The reactions were initiated by the addition of 2 μg of rEhNO1/2 enzyme, and the reduction of the acceptors was monitored spectrophotometrically at 490 nm for INT (ϵ = 18.5 mm−1 cm−1), 410 nm for potassium ferricyanide (ϵ = 1 mm−1 cm−1), and 340 nm for paraquat (NADPH oxidation, ϵ = 6.22 mm−1 cm−1). One unit of enzyme activity was defined as the formation of 1 μmol of product/min/mg of protein.
Metronidazole reduction activity was determined by measuring the oxidation of NADPH at 340 nm (ϵ340 = 6.22 mm−1 cm−1) or the reduction of metronidazole at 360 nm (ϵ360 = 9.2 mm−1 cm−1), as described by Chen and Blanchard (26). Assays were conducted at room temperature under strict anaerobic conditions. The reactions were initiated by the addition of 2 μg of rEhNO protein to a mixture comprising 50 mm Tris-HCl (pH 8.0), 0.5 mm metronidazole, and 0.2 mm NADPH.
The cystine reductase activity was calculated as μmol of NADPH oxidized per min at 340 nm. The assay mixture contained 0.1 m potassium phosphate (pH 7.5), 2 mm EDTA, 0.05–0.2 mm NADPH, and 0.1–5 mm l-cystine. Approximately 2 μg of rEhNO1/2 was added to initiate the reaction, and the change in absorbance at 340 nm was monitored. The effects of sulfydryl-dependent inhibitors were examined by preincubation of 2 μg of rEhNO1 and rEhNO2 with 0.1–5 mm N-ethylmaleimide for 10 min before the various assays. All sample reactions were performed in triplicate at a minimum.
NAD(P)H:flavin oxidoreductase activity was assayed by measuring the initial rate of NAD(P)H oxidation at 340 nm (ϵ = 6.22 mm−1cm−1) at 25 °C as described by Lo and Reeves (27). One unit of NAD(P)H:flavin oxidoreductase activity was defined as the amount of enzyme that catalyzed the oxidation of 1 μmol of NAD(P)H/min.
Ferric reductase activity was determined by measuring the difference of NAD(P)H consumption at 340 nm in the presence and absence of Fe(III) ammonium citrate. Reaction mixtures containing 0.2 mm NADPH, 100 mm Tris-HCl (pH 7.5), 0.005–1 mm ferric ammonium citrate, and 2 μg of rEhNO1 and -2 were used for the assays.
Ferredoxin-NADP+ reductase activity was determined by measuring ferredoxin-dependent reduction of cytochrome c (28). Activity was measured by monitoring cytochrome c reduction at 550 nm (ϵ = 21.1 mm−1 cm−1) in a reaction mixture containing 0.1 mm NADPH, 0.01–0.5 μm ferredoxin, 10 μm cytochrome c, and 50 mm Tris-HCl buffer (pH 7.5). The reactions were initiated by the addition of 1 μg of rEhNO1 and -2.
Determination of H2O2 Formation
The ferrithiocyanate method (29) was used to measure H2O2 formation at various time points (1–30 min) during the NADPH:favin oxidoreductase reaction. After the reactions were terminated by the addition of 0.125 volumes of 50% trichloroacetic acid, the samples were centrifuged at 12,000 × g, and 0.2 volumes of 10 mm ferrous ammonium sulfate and 0.1 volumes of 2.5 m potassium thiocyanate were then added. In the presence of H2O2, Fe2+ is oxidized, resulting in a colored thiocyanate-Fe3+ complex that can be measured by its absorption at 480 nm. The quantity of H2O2 formed was determined by comparison of the A480 values to standard curves generated using known amounts of H2O2.
Metabolite Extraction
Approximately 1.5 × 106 E. histolytica cells were harvested after 48 h of cultivation and washed twice with 5% mannitol. The cells were then resuspended in 1.6 ml of methanol containing 20 μm concentrations of the internal standard methionine sulfone acid and mixed with 1.6 ml of chloroform and 640 μl of deionized water. After vortexing, the mixture was centrifuged at 4600 × g at 4 °C for 5 min, and the aqueous layer (1.6 ml) was filtrated using an Amicon Ultrafree-MC ultrafilter (Corporation, Billerica, MA) by centrifugation at 9100 × g at 4 °C for ∼2 h. The filtrate was dried and preserved at −80 °C until mass spectrometric analysis.
Instrumentation and Capillary Electrophoresis (CE)-Time of Flight Mass Spectrometry (TOFMS) Conditions
CE-TOFMS was carried out using an Agilent CE Capillary Electrophoresis System equipped with an Agilent 6210 Time-of-Flight mass spectrometer, Agilent 1100 isocratic HPLC pump, Agilent G1603A CE-MS adapter kit, and Agilent G1607A CE-ESI-MS sprayer kit (Agilent Technologies, Waldbronn, Germany). The system was controlled by Agilent G2201AA ChemStation software for CE. Data acquisition was performed by Analyst QS Build: 7222 software for Agilent TOF (Applied Biosystems/MDS Sciex, Ontario, Canada). Instrumental conditions for the separation and detection of metabolites were as follows. The metabolites were separated on a fused silica capillary (50 μm × 100 cm) using 1 m formic acid as the electrolyte, and the applied voltage was set at +30 kV. A solution of 50% (v/v) methanol-water was delivered as the sheath liquid at 10 μl/min (30, 31). Electrospray ionization-TOFMS was conducted in the positive ion mode (4000 V). The pressure of dry nitrogen gas was maintained at 10 p.s.i. Exact mass data were acquired over a 50–1000 m/z range (32, 33). Before analysis, the sample was dissolved in 20 μl of deionized water containing 200 μm concentrations of the internal standard 3-aminopyrrolidine.
Generation of E. histolytica Transformants Overexpressing EhNO
The protein coding regions of EhNO1 and EhNO2 were amplified by PCR from cDNA using sense and antisense oligonucleotides containing appropriate restriction sites at the 5′ end. The sense and antisense oligonucleotide primers used for EhNO1 and EhNO2 were 5′-CTACCCGGGATGAAGAGTTTCAACATTACA-3′ and 5′-CAACTCGAGTTAATCTTGTTCCATTGGGGT-3′ (EhNO1) and 5′-CTACCCGGGATGGCTGCTAATTATAATAGA-3′ and 5′-CAACTCGAGTTATTCATTTTTTTTACC-3′ (EhNO2) (bold letters indicate restriction sites). The PCR-amplified DNA fragments were digested with SmaI and XhoI and ligated into SmaI and XhoI sites of the expression vector pKT-MR (34) to produce pKT-MR-NO1 and pKT-MR-NO2. Wild-type trophozoites were transformed with pKT-MR by liposome-mediated transfection as previously described (35). Transformants were initially selected in the presence of 3 μg/ml Geneticin (Invitrogen), which was then gradually increased to 6–20 μg/ml during the subsequent 2 weeks before subjecting the transformants to analyses.
Assay for Metronidazole Sensitivity of E. histolytica Trophozoites
To determine sensitivity to metronidazole, E. histolytica transfectants harboring pKT-MR-NO1, pKT-MR-NO2, or pKT-MR (control) were cultured at 37 °C in BI-S-33 medium containing 20 μg/ml Geneticin. For the assay, varying concentrations (0–16 μm) of metronidazole were added to samples containing an initial density of 104 cells/ml. After 48 h, the number of viable cells was counted on a hemocytometer using trypan blue to identify dead cells. The assays were performed five times in duplicate.
In Vitro Interaction of EhNO1/2 with Ferredoxin
Protein cross-linking was performed as described previously (36). Briefly, EhNO1/2 and ferredoxin (4 and 20 μm, respectively) were cross-linked by treatment with 5 mm N-ethyl-3-(3-dimethylaminopropyl)carbodiimide in 25 mm sodium phosphate, pH 7.5. The resulting complexes were analyzed by SDS-PAGE and Western blotting using anti-His antibody.
Immunoblot Analysis
Cell lysates and culture supernatants were separated on 12% (w/v) SDS-PAGE gels and subsequently electro-transferred onto nitrocellulose membranes (Hybond-C Extra; Amersham Biosciences) as previously described (37). Nonspecific binding was blocked by incubating the membranes for 1.5 h at room temperature in 5% nonfat dried milk in TBST (50 mm Tris-HCl (pH 8.0), 150 mm NaCl, and 0.05% Tween 20). The blots were then reacted with primary antibodies specific for EhNO1 and EhNO2 and mannose 6-phosphate receptor 1 (38) and cysteine synthase 1 (22) as controls for the membrane and cytosolic fractions, respectively, at dilutions of 1:500 to 1:100. Antisera against purified rEhNO1 and rEhNO2 were raised in rabbits commercially (Operon, Tokyo, Japan). The membranes were washed with TBST and further reacted with horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG antisera (1:20,000) (Invitrogen) at room temperature for 1.5 h. After further washing with TBST, specific proteins were visualized and measured with a chemiluminescence detection system (Millipore) using Scion Image software (Scion Corp., Frederick, MD) (39).
RESULTS
Identification of a GOGAT Small β Subunit Gene in E. histolytica upon l-Cysteine Supplementation
Upon analysis of the transcriptome of E. histolytica trophozoites cultured in medium supplemented with l-cysteine, a highly up-regulated gene (XM_648481) was previously identified.4 Although the entire transcriptome data is described elsewhere, we attempted to characterize this gene in detail in the present study. The identified gene and a gene (XM_651905) that appeared to be very closely related in the E. histolytica genome data base (40) were predicted to encode proteins showing high similarity to the small β subunit of GOGAT from bacteria. The genes were designated as E. histolytica NADPH-dependent oxidoreductase 1 and 2 [EhNO1 (XM_651905) and EhNO2 (XM_648481)] because although the encoded proteins lacked glutamate synthase activity, they showed robust NADPH-dependent oxidoreductase activity (described below). The EhNO1 and EhNO2 genes consisted of 1347- and 1338-bp open reading frames, respectively, which were predicted to encode proteins of 448 and 445 amino acids with predicted molecular masses of 49.3 and 49.0 kDa and isoelectric points of 6.31 and 7.02, respectively.
Features of the Deduced Protein Sequence of EhNOs
The predicted amino acid sequences of the two EhNOs shared 80% mutual identity and demonstrated 20–60% identities to the small β subunit of GOGAT from Archaea, bacteria, animals, and plants. EhNO1 had the highest amino acid identities to the GOGAT β subunit-like proteins of Chlorobium tepidum (green sulfur bacteria) and Methanosarcina mazei (Archaea) (62 and 59%, respectively), whereas EhNO2 showed 61–62% identities to the small subunit of Pyrococcus abyssi GOGAT and the β subunit chain of formate dehydrogenase from Moorella thermoacetica (Archaea). Although a multiple alignment of 32 GOGAT and GOGAT-like sequences was generated using ClustalW, the comparisons of representative sequences from E. coli, Clostridium saccharobutylicum, Azospirillum brasilense, and E. histolytica were sufficient to highlight the important similarities and differences among GOGAT proteins from these organisms and between the two EhNO isotypes (supplemental Fig. S1).
All of the functional domains characteristic of GOGAT β subunits were conserved between these β subunit-like proteins (supplemental Fig. S1). Two amino-terminal cysteine clusters, CX2CX4CX3CP (residues 40–53 for EhNO1 and residues 41–54 for EhNO2) and CX3CX3CX3C (residues 87–99, EhNO1; 88–100, EhNO2), matched the conserved cysteine-rich patterns proposed to be involved in the formation of [4Fe-4S] clusters (2). Similarly, two regions (residues 137–165 and 264–293 of EhNO1, labeled “FAD-I” and “NAD(P)H”, respectively) matched the conserved sequences of an ADP binding fold for the binding of FAD and NAD(P)H. Both EhNO1 and -2 shared features in the NAD(P)H binding domain with the A. brasilense GOGAT β-protein (41), which has been proposed to confer specificity for NADPH, rather than NADH. The presence of alanine in place of glycine in the last residue of the motif GXGXX(G/A/P) (residues 269–274 of EhNO1 and 270–275 of EhNO2, shown in bold in supplemental Fig. S1) and a conserved arginine in the NAD(P)H binding domain (Arg-293 of EhNO1 and Arg-294 of EhNO2) (42) suggested that the two EhNOs prefer NADPH to NADH as a cofactor. Furthermore, a region in the carboxyl terminus (residues 401–411 of EhNO1 and 402–412 of EhNO2) matched the second FAD binding consensus sequence (TX8GD).
Phylogenetic Analysis
Phylogenetic reconstruction was performed using neighbor-joining and maximum parsimony programs using 32 GOGAT β subunit or β subunit-like protein sequences from various organisms. The phylogenetic tree constructed using the neighbor-joining method revealed (supplemental Fig. S2) that EhNOs are more closely related to other β subunit-like homologs (supplemental Fig. S2, Group I) than to known GOGAT β subunit proteins (supplemental Fig. S2, Group II). This conclusion was also supported by the phylogenetic reconstruction using the maximum parsimony method (data not shown). Although these data did not clearly indicate the origin of the amebic GOGAT-like proteins, they suggested that EhNOs were most likely obtained by lateral gene transfer from an ancestral organism possessing a Group I-type gene, as reported previously for several other glutamate synthase β subunit-like genes (43).
Regulation of Gene Expression of EhNO Isotypes by l-Cysteine Concentration
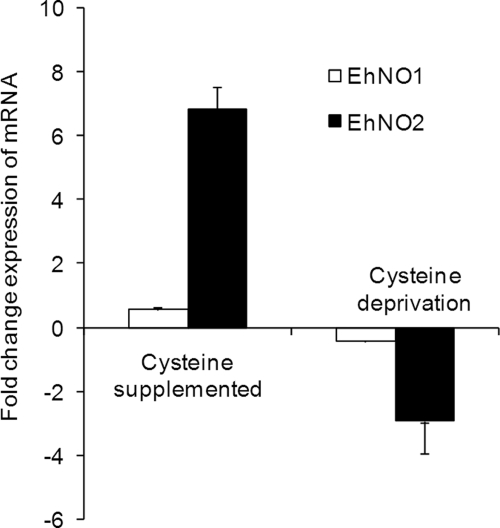
To verify the transcriptomic data and confirm that the expression of EhNO1 and -2 was regulated by l-cysteine, the relative steady-state mRNA levels of the EhNO isotypes in E. histolytica trophozoites cultivated under l-cysteine-enriched or deprived conditions were measured by quantitative real-time PCR (Fig. 1). Using the RNA polymerase II 15-kDa subunit as an internal control, EhNO2 mRNA increased by 7-fold when cultured in the presence of 18 mm l-cysteine for 48 h compared with the control condition (8 mm l-cysteine), whereas it was down-regulated by 4-fold when cultured in the absence of l-cysteine. In contrast, the level of EhNO1 remained unchanged in either the presence or absence of l-cysteine.
FIGURE 1.
Regulation of gene expression of EhNO isotypes in E. histolytica by extracellular l-cysteine concentration. E. histolytica trophozoites were cultured in normal (8 mm), l-cysteine-supplemented (18 mm), or deprived medium. The expression levels of the EhNO transcripts under l-cysteine-supplemented or -deprived conditions were normalized against those of RNA polymerase II and are shown as the -fold change expression of mRNA relative to that of trophozoites from the control (normal) culture. Error bars represent the S.E. of three independent experiments.
Expression of EhNO Proteins under l-Cysteine-supplemented and -deprived Conditions
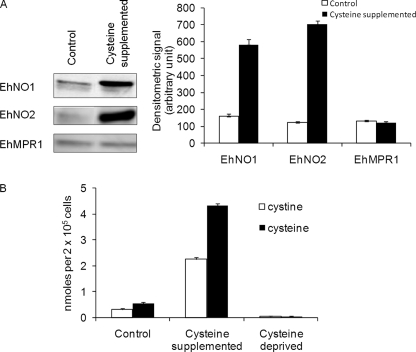
To confirm the changes of EhNO transcripts determined by the transcriptomic and real-time PCR analyses, we also examined EhNO expression at the protein level under l-cysteine-supplemented conditions. Immunoblot analysis using anti-rEhNO2 antibody showed that EhNO2 was induced by 6-fold when E. histolytica cultures were supplemented with l-cysteine (Fig. 2A). Although the RT-PCR results indicated that EhNO1 was not up-regulated under this condition, the protein recognized by anti-EhNO1 antibody was also found to be induced by 3.5-fold. Because anti-EhNO1 and anti-EhNO2 antibodies exhibited cross-reactivity (data not shown), the increased signal of the band recognized by anti-EhNO1 antibody was likely due to cross-reactivity with EhNO2. Alternatively, the level of EhNO1 may have increased by post-transcriptional mechanisms.
FIGURE 2.
Effects of extracellular l-cysteine concentrations on the amount of EhNO isotypes and intracellular l-cystine/l-cysteine concentrations. A, an immunoblot analysis of EhNO1 and -2 is shown. After trophozoites were cultured under normal or l-cysteine-supplemented conditions for 48 h, ∼15 μg of total cell lysate was electrophoresed on a 12% SDS-PAGE gel and subjected to an immunoblot assay with antibodies raised against EhNO1, EhNO2, or EhMPR1 as a control. The densitometric quantification of the reacted bands, shown in the right graph, was performed by Scion Image software, and the level of EhNO1, EhNO2, and EhMPR1 proteins was expressed in arbitrary units. Error bars represent the S.E. of three independent experiments. B, shown is intracellular l-cystine/cysteine concentrations under normal, l-cysteine-supplemented, and deprived conditions. l-Cystine/cysteine concentrations of the trophozoites cultivated for 48 h under the indicated conditions were analyzed by CE-MS. Error bars represent the S.E. of three independent experiments.
Changes in Intracellular l-Cysteine/Cystine Concentrations under l-Cysteine-supplemented and -deprived Conditions
To examine changes in intracellular l-cysteine and l-cystine concentrations caused by l-cysteine supplementation and deprivation, we quantitated their levels using a CE-MS-based approach in trophozoites maintained under normal (8 mm l-cysteine), enriched (18 mm l-cysteine), or deprived conditions. Under normal conditions, the l-cysteine to l-cystine ratio (1.70 ± 0.08) was deviated toward the reduced status. Upon l-cysteine supplementation, the intracellular levels of l-cysteine and l-cystine increased 8.1- and 7.3-fold, respectively, whereas under l-cysteine deprivation, both l-cysteine and l-cystine decreased to undetectable levels (Fig. 2B). Under l-cysteine-enriched conditions, the l-cysteine to l-cystine ratio (1.90 ± 0.14) slightly shifted toward the more reduced status (p = 0.031). Because several systems regulate the cellular redox reactions and electrochemical potential of the cell, the observed changes in the l-cysteine/cystine ratio were considered small. Taken together, these data clearly showed that although the extracellular l-cysteine concentration largely affects intracellular l-cysteine/l-cystine levels, its redox equilibrium is not severely affected. Furthermore, it appeared that a significant proportion of the l-cysteine incorporated into the cell was oxidized to l-cystine, which is supported by a previous finding (44). Alternatively, extracellular l-cysteine may be oxidized before uptake and reduced to l-cysteine intracellularly.
Expression and Purification of Recombinant EhNO Isotypes
To determine the biochemical properties of the two EhNO isoenzymes, recombinant proteins were first produced in E. coli. SDS-PAGE of the purified rEhNO1 and -2 proteins showed apparently single homogenous bands with molecular weights of 52.2 and 51.9 kDa, respectively, under reducing conditions (supplemental Fig. S3). The observed mobilities of rEhNO1 and -2 were consistent with the predicted sizes of the monomeric EhNO proteins with an extra 3.0-kDa histidine tag added at the amino terminus. The purity of the rEhNOs was estimated to be greater than 95% as judged by densitometric scanning of the stained gel. The rEhNO proteins were stable and retained their full activity for at least 3 months when stored in 10–15% glycerol at -30 or −80 °C.
Prosthetic Groups of rEhNOs
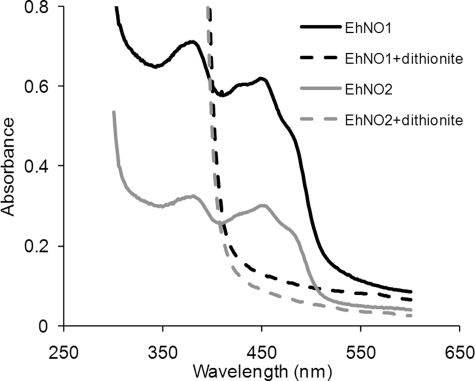
The UV-visible spectra of the purified rEhNOs showed absorbance maxima at 484, 450, 430, 378, and 280 nm (Fig. 3), which are characteristic of iron sulfur flavoproteins (45). The dithionite-reduced rEhNOs showed, in contrast, a relatively featureless spectrum, with the increased absorbance at shorter wavelengths attributable to dithionite. Denaturation of the rEhNOs by boiling resulted in the release of flavin, indicating that it formed a non-covalent association with the protein. The fluorescence intensity of the free flavin at pH 2.6 was ∼4-fold higher than that at pH 7.7 (data not shown). This indicates that FAD, rather than FMN, formed the prosthetic group in rEhNOs. It was calculated that 1.13 ± 0.32 mol of FAD (ϵ of FAD at 450 nm = 11.4 × 103 m−1 cm−1) is associated per molecule of rEhNO1, whereas 0.89 ± 0.21 mol of FAD bound per molecule of rEhNO2. The iron analysis using the O-phenanthroline method indicated that rEhNO1 contained 7.8 ± 0.62 irons per molecule of rEhNO1, whereas rEhNO2 contained 7.4 ± 0.71 irons per molecule of rEhNO2. These results indicate that two [4Fe-4S] clusters are present per subunit, which is consistent with the CX2CX4CX3CP and CX3CX3CX3C motifs present in EhNO1 and -2. These data together with the stability of the enzymatic activity of rEhNOs also support the premise that rEhNOs retain most, if not all, of the features of the native EhNOs.
FIGURE 3.
Absorption spectra of rEhNO1 and rEhNO2 proteins. UV-visible absorption spectra of rEhNO1 (400 μg of protein) and rEhNO2 (200 μg) under non-reducing (solid lines) and sodium dithionite-reducing conditions (broken lines) are shown. The samples were reduced with a 10-fold molar excess of sodium dithionite.
Kinetics Properties of rEhNO
The rEhNOs were devoid of glutamate synthase and glutamate dehydogenase activity in both directions at either pH 7.5 or 9.5. However, both proteins oxidized NADPH and transferred electrons to several alternative electron acceptors, including INT, ferricyanide, and menadione (Table 1). In the presence of NADPH, the reduction rate of INT by rEhNO1 (specific activity 19.42 ± 3.25 μmol min−1 mg−1) was >30-fold higher than that by rEhNO2 (0.62 ± 0.10 μmol min−1 mg−1), whereas rEhNO2 showed a 2.8-fold higher ferricyanide reducing activity (86.64 ± 8.33 μmol min−1 mg−1) than rEhNO1 (31.50 ± 6.21 μmol min−1 mg−1). The menadione-reducing activities of rEhNO1 and -2 were comparable (2.48 ± 0.39 and 2.25 ± 0.41 μmol min−1 mg−1, respectively). Both rEhNO1 and -2 were highly specific toward NADPH and did not reduce the above-tested electron acceptors with NADH as the electron donor. In the NADPH:flavin oxidoreductase reaction under aerobic conditions, rEhNO1 and -2 produced H2O2 at comparable levels (Table 1). Significantly, the two enzymes were also capable of reducing the anti-amebic drug metronidazole and the herbicide paraquat.
TABLE 1.
Specific activity of purified EhNO1 and EhNO2 with various electron acceptors
Values are expressed as the means ± S.D. of three independent experiments as described under “Experimental Procedures”
| Substrate | Specific activity |
|
|---|---|---|
| rEhNO1 | rEhNO2 | |
| μmol/min/mg | ||
| INT | 19.42 ± 3.25 | 0.62 ± 0.10 |
| Ferricyanide | 31.50 ± 6.21 | 86.64 ± 8.33 |
| Menadione | 2.48 ± 0.39 | 2.25 ± 0.41 |
| Metronidazole | 1.75 ± 0.42 | 1.41 ± 0.46 |
| Paraquat | 19.73 ± 4.23 | 10.60 ± 2.12 |
| Oxygen | 8.31 ± 2.12 | 3.42 ± 0.81 |
In addition to these properties, rEhNO1 and -2 could catalyze the reduction of disulfides, such as l-cystine, which was also dependent on NADPH. The Km and kcat/Km values of rEhNO1 and -2 for l-cystine and NADPH were significantly different (Table 2). At substrate-saturating concentrations, the Km values of rEhNO1 for l-cystine and l-NADPH were 3.3- and 2.3-fold higher, respectively, than those of rEhNO2. The kcat/Km value of rEhNO2 for l-cystine (measured at saturating concentrations of NADPH) was ∼4-fold higher than that of rEhNO1. The addition of N-ethylmaleimide, which is commonly used to inhibit sulfydryl-dependent reactions, inhibited the disulfide reducing activities of both rEhNO1 and rEhNO2 (0.5 mm N-ethylmaleimide caused 50% inhibition), whereas the presence of up to 5 mm N-ethylmaleimide had no effect on the reduction of INT. These results indicate that the two EhNOs contain thiol(s) groups that are involved in disulfide reduction but are not required for their observed oxidoreductase activity (46, 47).
TABLE 2.
Kinetic parameters for cystine, ferric, and ferredoxin NADP+ reductase reactions catalyzed by EhNO1 and EhNO2
Values are expressed as the means ± S.D. of three independent experiments.
| Substrate | rEhNO1 |
rEhNO2 |
||||||
|---|---|---|---|---|---|---|---|---|
| Km | Vmax | kcat | kcat/Km | Km | Vmax | kcat | kcat/Km | |
| μm | μmol/min/mg | min−1 | min−1·μm−1 | μm | μmol/min/mg | min−1 | min−1·μm−1 | |
| Cystine | 910 ± 20 | 0.88 ± 0.06 | 45.86 ± 7.52 | 0.05 ± 0.01 | 276 ± 23 | 1.15 ± 0.17 | 59.36 ± 3.81 | 0.22 ± 0.13 |
| NADPH (cystine) | 9.2 ± 2.1 | 1.70 ± 0.13 | 88.38 ± 5.83 | 9.61 ± 1.34 | 4.2 ± 0.8 | 1.33 ± 0.43 | 68.62 ± 5.40 | 16.33 ± 2.12 |
| Fe(III) citrate | 31.3 ± 3.4 | 9.10 ± 2.51 | 471.2 ± 17.5 | 15.1 ± 3.20 | 98.4 ± 4.5 | 0.26 ± 0.09 | 12.18 ± 2.81 | 0.12 ± 0.01 |
| NADPH (ferric) | 70.3 ± 4.9 | 10.12 ± 3.16 | 526.6 ± 21.2 | 7.49 ± 2.12 | 66.7 ± 8.3 | 0.72 ± 0.31 | 37.06 ± 6.28 | 0.56 ± 0.23 |
| Ferredoxin | 0.16 ± 0.05 | 2.28 ± 0.46 | 118.8 ± 7.4 | 742.5 ± 34.1 | 0.23 ± 0.07 | 0.43 ± 0.18 | 21.71 ± 4.51 | 94.39 ± 8.91 |
We also found that rEhNO1 and -2 catalyzed the reduction of ferric to ferrous ion. In the presence of NADPH, the reduction rate of Fe(III) by rEhNO1 (kcat/Km 15.1 ± 3.20 min−1 μm−1) was ∼116-fold higher than that by rEhNO2 (kcait/Km 0.12 ± 0.01 min−1 μm−1) (Table 2). In addition, both rEhNOs also acted as ferredoxin:NADP+ reductases capable of catalyzing the reduction of NADP+ to NADPH through the utilization of the electrons provided by reduced ferredoxin, although the observed activity of EhNO1 was again higher (7.8-fold) than that of EhNO2. These data suggest that EhNO1 is mainly involved in the reduction of ferric ion and ferrodoxin:NADP+, whereas EhNO2 primarily catalyzes the reduction of l-cystine. The uncatalyzed reaction rate (without enzyme) of each reaction was as follows: 22.5 ± 2.4 pmol/min, INT; 250 ± 32 pmol/min, ferricyanide; 29.2 ± 6.1 pmol/min, menadione; 307 ± 66 pmol/min, paraquat; 22.5 ± 7.8 pmol/min, cystine; 28.9 ± 6.9 pmol/min, ferric ammonium citrate; 6.16 ± 2.7 pmol/min, ferredoxin.
Binary Complexes of EhNO1/2 with Ferredoxins
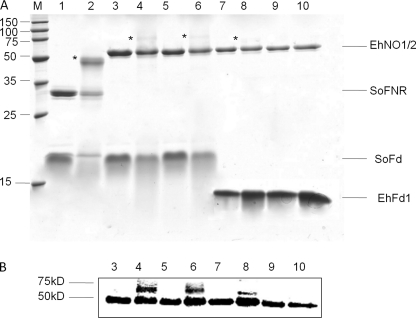
To examine whether electron transfer between reduced ferredoxin and NADP+ by EhNO1 was dependent on the physical interaction between these two proteins (48), as reported for the spinach leaf redox couple (36), we investigated whether EhNO1 and -2 physically interacted with homologous and heterologous (Spinacia oleracea) ferredoxin (SoFd) using a carbodiimide-promoted cross-linking (50). We first cloned and purified a representative [3Fe4S] ferredoxin found in the E. histolytica genome data base (EhFd1; TIGR ID 128.m00136). For the assay, rEhNO1 and -2 were mixed with either purified EhFd1 or SoFd, cross-linked, and then analyzed by SDS-PAGE (Fig. 4A). It was observed that both rEhNO1 and -2 formed a complex with spinach ferredoxin, as shown by the appearance of a large 60-kDa band in the gel and more easily observed in the Western blot analysis using an anti-histidine antibody (Fig. 4B, lanes 4 and 6). However, only EhNO1 formed a complex with E. histolytica ferredoxin (EhFd1) (Fig. 4, A and B, lanes 8 and 10).
FIGURE 4.
In vitro interactions of EhNO with ferredoxin. A, SDS-PAGE analysis of the complex of EhNO and ferredoxin is shown. Protein mixtures were incubated for 30 min with (even lane numbers) and without (odd lane numbers) 5 mm carbodiimide, electrophoresed on a 15% SDS-PAGE gel under reducing conditions, and then stained with Coomassie Brilliant Blue R250. The examined protein mixtures were as follows: lanes 1 and 2, spinach (S. oleracea) ferredoxin:NADP+ reductase (SoFNR) + spinach ferredoxin (SoFd); lanes 3 and 4, EhNO1+SoFd; lanes 5 and 6, EhNO2+SoFd; lanes 7 and 8, EhNO1+EhFd1; lanes 9 and 10, EhNO2+EhFd1. Protein bands corresponding to the cross-linked proteins are indicated by an asterisk. The positions of the purified proteins are indicated on the right side of the gel. B, shown is an immunoblot analysis of the cross-linked samples using an anti-His antibody.
Cellular Distribution of EhNO
We also examined the cellular distribution of EhNO1 and -2 in trophozoites. The immunofluorescence imaging using antiserum raised against the corresponding recombinant protein revealed that the two isotypes were distributed throughout the cytosol (data not shown). We also verified the localization of EhNOs by immunoblotting using lysates produced by a Dounce glass homogenizer followed by sonication and centrifugation at 100,000 × g at 4 °C for 1 h. Both EhNO1 and -2 fractionated into the soluble fraction (data not shown).
Increased Metronidazole Sensitivity by EhNO Overexpression
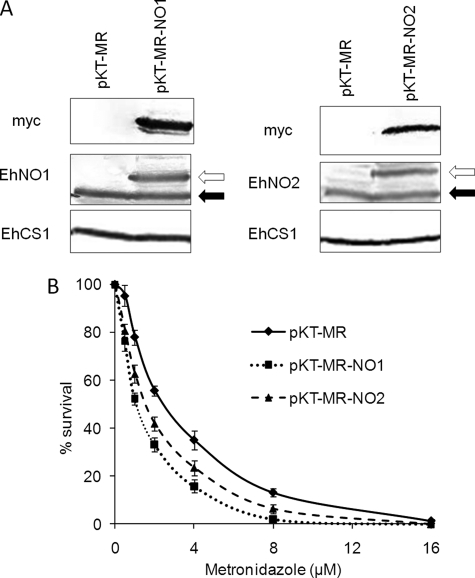
To confirm that EhNO is the target of metronidazole in E. histolytica, stable transformants that overexpressed Myc-tagged EhNO1 or 2 were generated. Both the transformants expressing Myc-tagged EhNO1 or EhNO2 expressed ∼2-fold higher levels of the corresponding enzymes than the control (Fig. 5A) and were more sensitive to metronidazole. The 50% growth inhibitory concentrations (IC50) of metronidazole for the Myc-EhNO1- and Myc-EhNO2-overexpressing transformants were 1.03 ± 0.05 and 1.42 ± 0.12 μm, respectively (Fig. 5B), whereas the control showed an IC50 of 2.24 ± 0.33 μm.
FIGURE 5.
Changes in the sensitivity of E. histolytica to metronidazole by overexpression of EhNOs. A, shown is an immunoblot analysis of EhNOs in the transformants expressing Myc-tagged EhNO1 and -2. Approximately 40 μg of total lysate from the pKT-MR (control), EhNO1 (pKT-MR-NO1), and EhNO2 (pKT-MR-NO2)-overexpressing transformants was electrophoresed on a SDS-PAGE gel under reducing conditions and subjected to immunoblot analysis using anti-EhNO1, anti-EhNO2, anti-Myc, and anti-EhCS1 (control) antibodies. Black and white arrows indicate endogenous and Myc-tagged EhNO1 and -2, respectively. B, susceptibility of transformed trophozoites to metronidazole is shown. Trophozoites (104 cells/ml) were cultivated in the presence of 0–16 μm metronidazole for 48 h, and the number of viable cells was then counted. The percentages of living cells are shown relative to those of unexposed control cells. Error bars represent the S.E. of five independent experiments.
DISCUSSION
In the present study we demonstrated novel enzymatic reactions catalyzed by a new class of FAD- and 2[4Fe-4S]-containing NADPH-dependent oxidoreductases from E. histolytica, which had been initially discovered by virtue of tightly regulated gene expression in correlation with l-cysteine concentrations. Although the two EhNOs characterized in this study had been annotated before this study based on their high degree of homology with GOGAT β subunit and β subunit-like genes from a variety of organisms, their biochemical function was unknown. The fact that the two EhNOs shared significant similarity with homologs from archaeal organisms raised the question of whether they represented a prototype GOGAT protein, similar to the β subunit protein from Pyrococcus, which was reported to possess NADPH-dependent GOGAT activity and be capable of both glutamine and ammonia-dependent synthesis in the absence of the α subunit (7). However, we were unable to observe glutamate synthase activity of the EhNOs under similar conditions used for the Pyrococcus glutamate synthase. Furthermore, the expression of the GOGAT β subunit failed to restore glutamate auxotrophy in an E. coli GOGAT α subunit-deficient strain (5, 51). In addition, it was somewhat puzzling how the Pyrococcus GOGAT β subunit functioned in substrate binding and catalysis without the α subunit, which has been shown to be responsible for substrate recognition (52). Thus, it was thought that the EhNO β subunit-like proteins may be involved in reactions other than glutamate synthesis.
The physiological roles of EhNO1 and EhNO2 have not been unequivocally demonstrated because our attempt to repress EhNO expression by gene silencing (53) failed (data not shown), and gene knock-out has not been accomplished in E. histolytica. Nevertheless, our present enzymological characterization revealed the physiological significance of the presence of the two isotypes of EhNOs. EhNO2 appears to play an important role in the reduction of cystine to l-cysteine. Because l-cysteine is partially present in the oxidized form inside cells (CE-MS analysis, Fig. 2B), l-cystine reduction is necessary before the utilization of l-cysteine, which has been implicated in the attachment to matrix, elongation, motility, growth, and anti-oxidative defense (35, 54, 55). Transcriptomic analysis demonstrated that the transcription of EhNO2, but not EhNO1, is tightly regulated by extracellular l-cysteine concentrations. Furthermore, the measured kinetic parameters indicate that EhNO2 possesses 4-fold higher l-cystine reduction efficiency than EhNO1.
The acquisition of iron and subsequent assimilation into cellular proteins are ubiquitously essential for life. However, at physiological pH under aerobic conditions, iron is present as Fe3+ hydroxides and oxyhydroxides or in a complex with ferric-specific chelators, e.g. siderophores (56). Subsequent reduction of complexed Fe3+ is accomplished by ferric reductases using NAD(P)H as the electron donor (57), with the resulting Fe2+ being subsequently released and incorporated into iron-containing proteins (58). We showed that rEhNO1 catalyzes the reduction of ferric ion >100-fold more efficiently than rEhNO2 (Table 2), suggesting that EhNO1 is mainly involved in ferric reduction. We also confirmed that EhNO1 functions as a ferredoxin:NADP+ reductase, similar to the recently reported ferric reductase from Pseudomonas putida (59), by catalyzing reversible electron transfer between one molecule of NADP+/NADPH and two molecules of ferredoxin. In vitro cross-linking of the two EhNOs with ferredoxin indicate that only EhNO1 forms a stable complex with E. histolytica ferredoxin (EhFd1), whereas both EhNO1 and EhNO2 physically interact with spinach ferredoxin (Fig. 4B), indicating that the specificity toward ferredoxin differs between these two proteins. The E. histolytica genome encodes four types of ferredoxins which are highly divergent at the primary sequence level and also in the Fe-S clusters. We, therefore, hypothesize that EhNO2 interacts with a ferredoxin(s) other than EhFd1 in E. histolytica, a speculation that is supported by the observed differential binding of photosynthetic and non-photosynthetic maize ferredoxins to root Zea mays ferredoxin:NADP+ reductase (60).
E. histolytica is anaerobic/microaerophilic and possesses highly degenerated mitochondria that are incapable of oxidative phosphorylation and ATP generation. A crucial step in energy production via glycolysis and fermentation in E. histolytica involves the decarboxylation of pyruvate to acetyl CoA that is catalyzed by pyruvate:ferredoxin oxidoreductase (61). Concomitant with the decarboxylation of pyruvate, an electron is transferred to oxidized ferredoxin. Generally, reduced ferredoxin subsequently donates an electron to NAD(P) by the action of ferredoxin:NADP reductase, which serves to regenerate the intracellular pools of NAD(P)H and oxidized ferredoxin. However, as the Entamoeba genome does not contain an ferredoxin:NADP reductase homolog, it was unclear how NAD(P)H was regenerated. Our enzymological study indicates that EhNOs, and EhNO1 in particular, function as ferredoxin:NADP reductases and are involved in the regeneration of NADPH and oxidized ferredoxin required for continuous energy production.
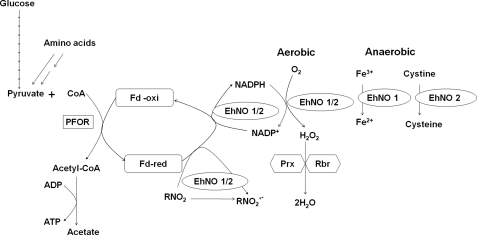
As stated above, E. histolytica possesses highly divergent mitochondria (62) and lacks a functional tricarboxylic acid cycle, cytochromes, and a conventional respiratory electron transport chain terminating in the reduction of oxygen to water. However, amebae can still tolerate up to 5% of oxygen in the gas phase (63, 64) and consume oxygen (65). As shown here, EhNOs are flavoproteins containing 1 mol of FAD as a prosthetic group per mol of enzyme. During the NADPH:flavin oxidoreductase reaction, NADPH binds to EhNOs, and two electrons are transferred to FAD to yield FADH2, which is immediately dissociated from the enzyme (66). Under aerobic conditions, FADH2 is rapidly oxidized by molecular oxygen to yield H2O2 and FAD (67). As E. histolytica amebae do not produce detectable amounts of H2O2 (27), it is possible that H2O2 is further converted to water by peroxiredoxin (68) and rubrerythrin (69) to overcome oxidative stress. Under anaerobic conditions, EhNO1 catalyzes ferric ion reduction, whereas EhNO2 catalyzes cystine reduction. Based on the demonstrated reactions catalyzed by the two EhNOs, we have proposed functional roles for these two proteins in E. histolytica that are summarized in Fig. 6.
FIGURE 6.
Proposed in vivo reactions catalyzed by EhNO1 and -2. PFOR, pyruvate:ferredoxin oxidoreductase; Fd-red and Fd-oxi, reduced and oxidized form of ferredoxin; Prx, peroxiredoxin; Rbr, rubrerythrin; RNO2/RNO2°, metronidazole/reduced metronidazole.
Metronidazole is a prodrug currently used to treat a number of microbial infections, and its activation requires intracellular reduction to produce cytotoxic short-lived radicals and other reactive species (70). Entamoeba electron transport proteins, which have been reported to provide the source of electrons for the reductive activation of metronidazole, include ferredoxin (71), thioredoxin reductase (72), and nitroreductase (49). We demonstrated that both EhNO1 and -2 catalyze metronidazole reduction in vitro (Table 1), and their overexpression confers increased sensitivity to this drug (Fig. 5B). This finding suggests that in addition to ferredoxin (71), pyruvate:ferredoxin oxidoreductase (71), thioredoxin (72), and nitroreductase (49), EhNO1 and -2 are also involved in metronidazole activation in E. histolytica.
In conclusion, we have demonstrated for the first time that two novel NADPH-dependent GOGAT small subunit-like proteins of E. histolytica function, at least in vitro, as cystine/ferric/ferredoxin:NADP+ reductase. We propose that they play a role in maintaining intracellular redox potential and may be responsible for metronidazole activation in this parasite. The physiological substrates and biological roles of the majority of oxidoreductases discovered by genome mining remain largely unknown. Vigorous attempts to discover the substrates and functions of individual oxidoreductases should unveil novel cellular metabolic processes in pathogens and cancer cells that may lead to the development of new chemotherapeutics.
Supplementary Material
Acknowledgments
We thank Kumiko Nakada-Tsukui, Fumika Mi-ichi, Takashi Makiuchi, and all other members of our laboratory for technical assistance and valuable discussions.
This work was supported by Grants-in-aid for Scientific Research 18GS0314, 18050006, and 18073001 (to T. N.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, a grant for research on emerging and re-emerging infectious diseases from the Ministry of Health, Labour, and Welfare of Japan (H20-Shinkosaiko-016), and a grant for research to promote the development of anti-AIDS pharmaceuticals from the Japan Health Sciences Foundation (to T. N.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
The nucleotide sequence(s) reported in this paper has been submitted to the Gen-BankTM/EBI Data Bank with accession number(s) AB521132 and AB521133.
A. Husain, D. Sato, G. Jeelani, M. Suematsu, T. Soga, and T. Nozaki, unpublished information.
- GOGAT
- glutamine:2-oxoglutarate amidotransferase
- EhNO
- E. histolytica NADPH-dependent oxidoreductase
- rEhNO
- recombinant EhNO
- INT
- iodonitrotetrazolium
- CE
- capillary electrophoresis
- SoFd
- S. oleracea ferredoxin
- EhFd1
- E. histolytica ferredoxin.
REFERENCES
- 1.Ratti S., Curti B., Zanetti G., Galli E. (1985) J. Bacteriol. 163, 724–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosenbaum K., Jahnke K., Curti B., Hagen W. R., Schnackerz K. D., Vanoni M. A. (1998) Biochemistry 37, 17598–17609 [DOI] [PubMed] [Google Scholar]
- 3.Tóth A., Takács M., Groma G., Rákhely G., Kovács K. L. (2008) FEMS Microbiol. Lett. 282, 8–14 [DOI] [PubMed] [Google Scholar]
- 4.Vanoni M. A., Curti B. (1999) Cell Mol. Life Sci. 55, 617–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stutz H. E., Reid S. J. (2004) Biochim. Biophys. Acta 1676, 71–82 [DOI] [PubMed] [Google Scholar]
- 6.Saum S. H., Sydow J. F., Palm P., Pfeiffer F., Oesterhelt D., Müller V. (2006) J. Bacteriol. 188, 6808–6815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jongsareejit B., Rahman R. N., Fujiwara S., Imanaka T. (1997) Mol. Gen. Genet. 254, 635–642 [DOI] [PubMed] [Google Scholar]
- 8.World Health Organization (1997) WHO/PAHO/UNESCO Report: A consultation with experts on amebiasis. Mexico City, Mexico: 28–29January, 1997. Epidemiol. Bull 18, 13–14 [PubMed] [Google Scholar]
- 9.Upcroft P., Upcroft J. A. (2001) Clin. Microbiol. Rev. 14, 150–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffman J. S., Cave D. R. (2001) Curr. Opin. Gastroenterol. 17, 30–34 [DOI] [PubMed] [Google Scholar]
- 11.World Health Organization (2007March) WHO Model List of Essential Medicines, 15th Ed [Google Scholar]
- 12.Müller M. (1983) Surgery 93, 165–171 [PubMed] [Google Scholar]
- 13.Moreno S. N., Docampo R. (1985) Environ. Health Perspect. 64, 199–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.West S. B., Wislocki P. G., Fiorentini K. M., Alvaro R., Wolf F. J., Lu A. Y. (1982) Chem. Biol. Interact. 41, 265–279 [DOI] [PubMed] [Google Scholar]
- 15.Ludlum D. B., Colinas R. J., Kirk M. C., Mehta J. R. (1988) Carcinogenesis 9, 593–596 [DOI] [PubMed] [Google Scholar]
- 16.Diamond L. S., Harlow D. R., Cunnick C. C. (1978) Trans. R. Soc. Trop. Med. Hyg. 72, 431–432 [DOI] [PubMed] [Google Scholar]
- 17.Clark C. G., Diamond L. S. (2002) Clin. Microbiol. Rev. 15, 329–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leão-Helder A. N., Krikken A. M., Gellissen G., van der Klei I. J., Veenhuis M., Kiel J. A. (2004) FEBS Lett. 577, 491–495 [DOI] [PubMed] [Google Scholar]
- 20.Kumar S., Tamura K., Jakobsen I. B., Nei M. (2001) Bioinformatics 17, 1244–1245 [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J., Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 22.Nozaki T., Asai T., Kobayashi S., Ikegami F., Noji M., Saito K., Takeuchi T. (1998) Mol. Biochem. Parasitol. 97, 33–44 [DOI] [PubMed] [Google Scholar]
- 23.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 24.Faeder E. J., Siegel L. M. (1973) Anal. Biochem. 53, 332–336 [DOI] [PubMed] [Google Scholar]
- 25.Olson J. W., Agar J. N., Johnson M. K., Maier R. J. (2000) Biochemistry 39, 16213–16219 [DOI] [PubMed] [Google Scholar]
- 26.Chen J. S., Blanchard D. K. (1979) Anal. Biochem. 93, 216–222 [PubMed] [Google Scholar]
- 27.Lo H., Reeves R. E. (1980) Mol. Biochem. Parasitol. 2, 23–30 [DOI] [PubMed] [Google Scholar]
- 28.Ichikawa Y., Hiwatashi A., Yamano T., Kim H. J., Maruya N. (1980) in Flavins and Flavoproteins (Yagi K., Yamano T. eds) pp. 677–691, University Park Press, Baltimore, MD [Google Scholar]
- 29.Thurman R. G., Ley H. G., Scholz R. (1972) Eur. J. Biochem. 25, 420–430 [DOI] [PubMed] [Google Scholar]
- 30.Soga T., Heiger D. N. (2000) Anal. Chem. 72, 1236–1241 [DOI] [PubMed] [Google Scholar]
- 31.Soga T., Ohashi Y., Ueno Y., Naraoka H., Tomita M., Nishioka T. (2003) J. Proteome Res. 2, 488–494 [DOI] [PubMed] [Google Scholar]
- 32.Soga T., Baran R., Suematsu M., Ueno Y., Ikeda S., Sakurakawa T., Kakazu Y., Ishikawa T., Robert M., Nishioka T., Tomita M. (2006) J. Biol. Chem. 281, 16768–16776 [DOI] [PubMed] [Google Scholar]
- 33.Ohashi Y., Hirayama A., Ishikawa T., Nakamura S., Shimizu K., Ueno Y., Tomita M., Soga T. (2008) Mol. Biosyst. 4, 135–147 [DOI] [PubMed] [Google Scholar]
- 34.Nakada-Tsukui K., Okada H., Mitra B. N., Nozaki T. (2009) Cell. Microbiol. 11, 1471–1491 [DOI] [PubMed] [Google Scholar]
- 35.Nozaki T., Asai T., Sanchez L. B., Kobayashi S., Nakazawa M., Takeuchi T. (1999) J. Biol. Chem. 274, 32445–32452 [DOI] [PubMed] [Google Scholar]
- 36.Zanetti G., Morelli D., Ronchi S., Negri A., Aliverti A., Curti B. (1988) Biochemistry 27, 3753–3759 [Google Scholar]
- 37.Tokoro M., Asai T., Kobayashi S., Takeuchi T., Nozaki T. (2003) J. Biol. Chem. 278, 42717–42727 [DOI] [PubMed] [Google Scholar]
- 38.Nakada-Tsukui K., Saito-Nakano Y., Ali V., Nozaki T. (2005) Mol. Biol. Cell 16, 5294–5303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Srivastava M., Ahmad N., Gupta S., Mukhtar H. (2001) J. Biol. Chem. 276, 15481–15488 [DOI] [PubMed] [Google Scholar]
- 40.Loftus B., Anderson I., Davies R., Alsmark U. C., Samuelson J., Amedeo P., Roncaglia P., Berriman M., Hirt R. P., Mann B. J., Nozaki T., Suh B., Pop M., Duchene M., Ackers J., Tannich E., Leippe M., Hofer M., Bruchhaus I., Willhoeft U., Bhattacharya A., Chillingworth T., Churcher C., Hance Z., Harris B., Harris D., Jagels K., Moule S., Mungall K., Ormond D., Squares R., Whitehead S., Quail M. A., Rabbinowitsch E., Norbertczak H., Price C., Wang Z., Guillén N., Gilchrist C., Stroup S. E., Bhattacharya S., Lohia A., Foster P. G., Sicheritz-Ponten T., Weber C., Singh U., Mukherjee C., El-Sayed N. M., Petri W. A., Jr, Clark C. G., Embley T. M., Barrell B., Fraser C. M., Hall N. (2005) Nature 433, 865–868 [DOI] [PubMed] [Google Scholar]
- 41.Morandi P., Valzasina B., Colombo C., Curti B., Vanoni M. A. (2000) Biochemistry 39, 727–735 [DOI] [PubMed] [Google Scholar]
- 42.Pelanda R., Vanoni M. A., Perego M., Piubelli L., Galizzi A., Curti B., Zanetti G. (1993) J. Biol. Chem. 268, 3099–3106 [PubMed] [Google Scholar]
- 43.Andersson J. O., Roger A. J. (2002) Eukaryot. Cell 1, 304–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gillin F. D., Diamond L. S. (1981) Exp. Parasitol. 51, 382–391 [DOI] [PubMed] [Google Scholar]
- 45.Latimer M. T., Painter M. H., Ferry J. G. (1996) J. Biol. Chem. 271, 24023–24028 [DOI] [PubMed] [Google Scholar]
- 46.Fontecave M., Eliasson R., Reichard P. (1987) J. Biol. Chem. 262, 12325–12331 [PubMed] [Google Scholar]
- 47.Jablonski E., DeLuca M. (1978) Biochemistry 17, 672–678 [DOI] [PubMed] [Google Scholar]
- 48.Foust G. P., Mayhew S. G., Massey V. (1969) J. Biol. Chem. 244, 964–970 [PubMed] [Google Scholar]
- 49.Pal D., Banerjee S., Cui J., Schwartz A., Ghosh S. K., Samuelson J. (2009) Antimicrob. Agents Chemother. 53, 458–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zanetti G., Aliverti A., Curti B. (1984) J. Biol. Chem. 259, 6153–6157 [PubMed] [Google Scholar]
- 51.Deane S. M., Rawlings D. E. (1996) Gene 177, 261–263 [DOI] [PubMed] [Google Scholar]
- 52.Vanoni M. A., Fischer F., Ravasio S., Verzotti E., Edmondson D. E., Hagen W. R., Zanetti G., Curti B. (1998) Biochemistry 37, 1828–1838 [DOI] [PubMed] [Google Scholar]
- 53.Bracha R., Nuchamowitz Y., Anbar M., Mirelman D. (2006) PLoS Pathog. 2, e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gillin F. D., Diamond L. S. (1981) Exp. Parasitol. 52, 9–17 [DOI] [PubMed] [Google Scholar]
- 55.Gillin F. D., Diamond L. S. (1980) J. Protozool. 27, 474–478 [DOI] [PubMed] [Google Scholar]
- 56.Barchini E., Cowart R. E. (1996) Arch. Microbiol. 166, 51–57 [DOI] [PubMed] [Google Scholar]
- 57.Lesuisse E., Crichton R. R., Labbe P. (1990) Biochim. Biophys. Acta 1038, 253–259 [DOI] [PubMed] [Google Scholar]
- 58.Guerinot M. L. (1994) Annu. Rev. Microbiol. 48, 743–772 [DOI] [PubMed] [Google Scholar]
- 59.Yeom J., Jeon C. O., Madsen E. L., Park W. (2009) J. Bacteriol. 191, 1472–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Onda Y., Matsumura T., Kimata-Ariga Y., Sakakibara H., Sugiyama T., Hase T. (2000) Plant Physiol. 123, 1037–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kerscher L., Oesterhelt D. (1982) Trends Biol. Sci. 7, 371–374 [Google Scholar]
- 62.Tovar J., Fischer A., Clark C. G. (1999) Mol. Microbiol. 32, 1013–1021 [DOI] [PubMed] [Google Scholar]
- 63.Band R. N., Cirrito H. (1979) J. Protozool. 26, 282–286 [DOI] [PubMed] [Google Scholar]
- 64.Reeves R. E. (1984) Adv. Parasitol. 23, 105–142 [DOI] [PubMed] [Google Scholar]
- 65.Weinbach E. C., Diamond L. S. (1974) Exp. Parasitol. 35, 232–243 [DOI] [PubMed] [Google Scholar]
- 66.Inouye S. (1994) FEBS Lett. 347, 163–168 [DOI] [PubMed] [Google Scholar]
- 67.Gibson Q. H., Hastings J. W. (1962) Biochem. J. 83, 368–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bruchhaus I., Richter S., Tannich E. (1997) Biochem. J. 326, 785–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maralikova B., Ali V., Nakada-Tsukui K., Nozaki T., van der Giezen M., Henze K., Tovar J. (2010) Cell Microbiol. 12, 331–342 [DOI] [PubMed] [Google Scholar]
- 70.Goldman P., Koch R. L., Yeung T. C., Chrystal E. J., Beaulieu B. B., Jr., McLafferty M. A., Sudlow G. (1986) Biochem. Pharmacol. 35, 43–51 [DOI] [PubMed] [Google Scholar]
- 71.Müller M. (1986) Biochem. Pharmacol. 35, 37–41 [DOI] [PubMed] [Google Scholar]
- 72.Leitsch D., Kolarich D., Wilson I. B. H., Altmann F., Duchene M. (2007) PLoS Biol. 5, e211. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.