Abstract
Mixed linear model (MLM) methods have proven useful in controlling for population structure and relatedness within genome-wide association studies. However, MLM-based methods can be computationally challenging for large datasets. We report a compression approach, called ‘compressed MLM’, that decreases the effective sample size of such datasets by clustering individuals into groups. We also present a complementary approach, ‘population parameters previously determined’ (P3D), that eliminates the need to re-compute variance components. We applied these two methods both independently and combined in selected genetic association datasets from human, dog and maize. The joint implementation of these two methods markedly reduced computing time and either maintained or improved statistical power. We used simulations to demonstrate the usefulness in controlling for substructure in genetic association datasets for a range of species and genetic architectures. We have made these methods available within an implementation of the software program TASSEL.
Although genome-wide association studies (GWAS) have the potential to pinpoint genetic polymorphisms underlying human diseases and agriculturally important traits, false discoveries are a major concern1 and can be partially attributed to spurious associations caused by population structure and unequal relatedness among individuals in a given cohort. Population stratification was initially addressed using general linear model (GLM)-based methods such as structured association2, genomic control3 and family-based tests of association4. The introduction of MLM approaches has more recently been demonstrated as an improved method to simultaneously account for population structure and unequal relatedness among individuals5.
In the MLM-based methods, population structure2,6 is fit as a fixed effect, whereas kinship among individuals is incorporated as the variance-covariance structure of the random effect for the individuals. Regardless of the applied statistical method, GWAS require large sample sizes to achieve sufficient statistical power7, especially in order to detect the small effect polymorphisms that underlie most complex traits8. For the MLM approach, datasets with these large sample sizes create a heavy computational burden because the computing time for solving a MLM increases with the cube of the number of individuals fit as a random effect. The earliest effort to reduce the size of the random effect in an MLM can be traced back to the sire model approach used in animal breeding9–12, which replaces an individual’s genetic effect with that of its sire. Consequently, the sire-model approach requires pedigrees, which are not always available, and which in particular are often not available in plant studies. Even with available pedigrees, the use of a marker-based kinship is preferred because of its higher accuracy13,14. The computing time is further increased because iteration is needed to estimate population parameters, such as variance components15, for each tested marker. Even though a number of studies have sought to improve the speed of the iteration process, including development of the recent efficient mixed-model association (EMMA) algorithm16, solving an MLM for a large number of individuals and markers remains computationally intensive. To address this issue, a residual approach was proposed based on a two-step strategy17. The first step optimized a reduced MLM with the genetic marker effect excluded. In the second step, the residual from the reduced MLM was fit as the dependent variable to test each marker in a GLM. Because the random genetic effect was not fit in the second step, iteration was not required when testing markers. This residual approach can be performed much faster than the one-step MLM with full optimization for all unknown parameters, but it has a statistical power equivalent to that of the full optimization approach only for traits with low heritability. We propose here methods to reduce the size of the random genetic effect in the absence of pedigree information and eliminate iterations to re-estimate the population parameters for each marker without compromising statistical power. We show that the joint use of these two methods greatly reduces computing time and maintains or even increases statistical power.
The total computing time for a GWAS with a standard MLM is mpn3, where m is the total number of markers, p is the number of iterations to solve the MLM and n is the total number of individuals assessed. Conducting a GWAS with a large sample size becomes computationally intensive because each iteration takes an amount of time that is proportional to the cube of the number of individuals in the random effect15,18. An approach for reducing this computational burden is to reduce the size of the random effect. We achieve this by substituting n individuals with a smaller number of groups, s (s ≤ n), clustered based on the kinship among individuals. Consequently, the kinship between pairs of groups replaces the kinship between pairs of individuals for the random effect of an MLM. If c = n/s is the average number of individuals per group, referred to hereafter as the compression level, this approach will reduce computing time by a factor of c3. We named this approach compression, referring to how the random effect in a MLM is compressed from individuals to groups. An MLM that uses compression is called a compressed MLM.
Because in this method individuals are clustered into groups based on kinship estimates, we consider the compressed MLM to be an extension of the pedigree-based sire model9–12 with notable advancements. The groups used in the compressed MLM can be clustered from kinship calculated from either markers or pedigrees. In addition, the number of groups in the compressed MLM can vary from n to 1, whereas the number of sires is fixed in the traditional method for a particular pedigree. This flexibility in the number of groups allows the accuracy of the group mean and number of groups to be optimized, which is a method similar to choosing the numbers of sires and progeny per sire to maximize genetic improvement in a breeding program19–21. The ability to optimize the number of groups could lead to increased statistical power in GWAS.
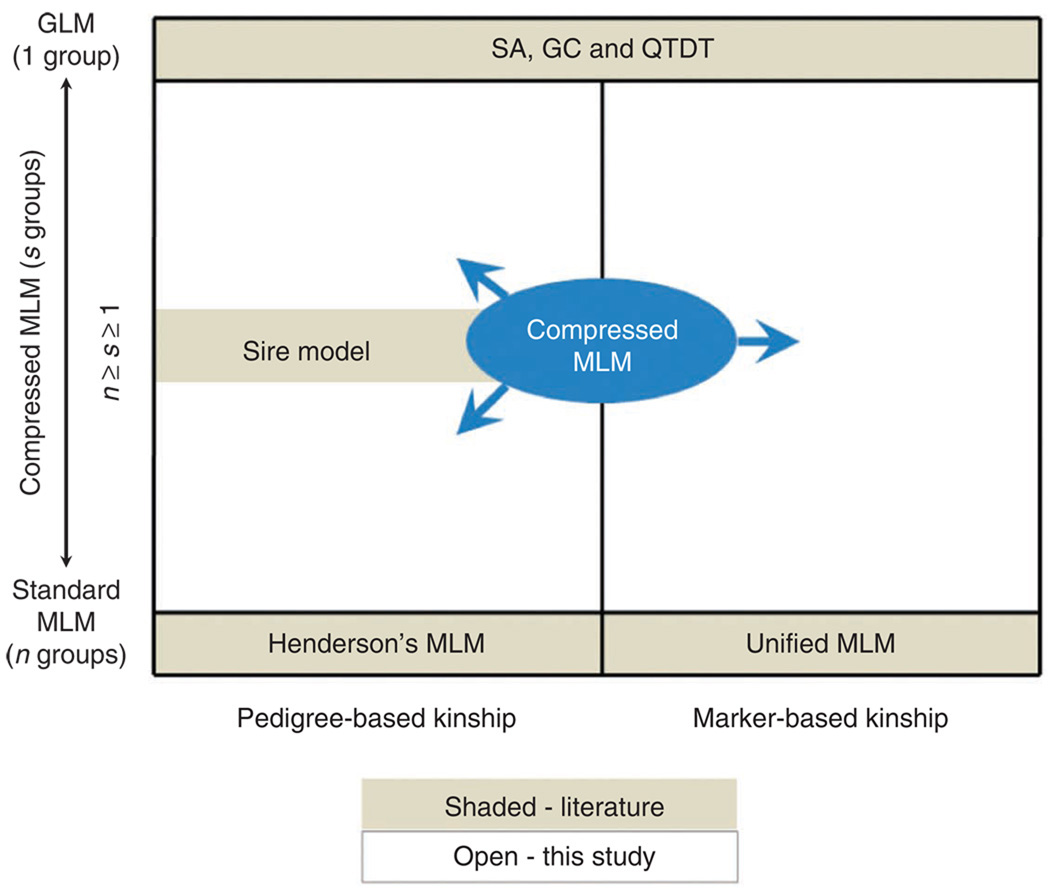
Compressed MLM crosses the boundary between GLM and MLM because GLM and MLM can both be considered extreme cases of compressed MLM (Fig. 1). MLM is equivalent to compressed MLM when each individual is treated as a single group (that is, s = n), whereas GLM is equivalent to compressed MLM when all individuals are in one group (s = 1). The latter causes the random effect to have a single level, thereby preventing the separate estimation of the random effect and residual variance components. In addition, the random effect and the overall mean are linearly dependent and thus cannot be estimated separately.
Figure 1.
The forms of MLM classified by the random effect size and types of kinship. The GLM and standard MLM are the two extremes of the compressed MLM with the number of groups determined as 1 and n (number of individuals), respectively. The sire model is a special case of the compressed MLM, with the groups determined as the sires derived from pedigrees. Kinship used in Henderson’s MLM15 was calculated from the pedigrees. It was extended to marker-based kinship in the unified MLM5. The GLM approach appears in many formats in various GWAS, including structure association (SA)2, genomic control (GC)3 and the quantitative transmission disequilibrium test (QTDT)4. The compressed MLM can be flexibly applied to the entire area by varying the number of groups (s), including the area investigated previously (shaded area) and the area proposed in this study (open area).
To further reduce computing time, we developed the P3D algorithm, a two-step approach that does not require iteration to estimate population parameters such as genetic variance and residual variance for each marker. The first step in the algorithm is to optimize the reduced MLM with the marker effect excluded. If compression is incorporated in the model, the population parameters also include the clustering algorithm and compression level. Taken from a similar approach that was applied to marker-assisted breeding22, the second step of the algorithm continues to fit the random genetic effect in the MLM with the previously determined population parameters fixed as empirical Bayesian priors23. Subsequently, the non-population parameters, including the marker effect and the random genetic effect, are estimated for each marker.
P3D is similar to the two-step residual approach17, but it also has notable differences. The residual approach fits the residuals from the reduced MLM as the dependent variable in the second step, whereas the original phenotype is fit as the dependent variable in the second step of P3D. In addition, the residual approach does not fit the random genetic effect and uses a GLM when testing markers, whereas P3D fits the random genetic effect with previously determined population parameters fixed in an MLM framework.
To evaluate compression and P3D relative to the standard MLM with full optimization of all unknown parameters for each marker, we conducted a series of association studies between observed or simulated phenotypes and observed markers in human, dog and maize. For the observed phenotypes, we evaluated the fit of compressed MLMs at different compression levels and with different clustering algorithms. Under the assumption that there is no association between the observed phenotypes and the observed genetic markers, we investigated the distribution of false positives by using the compressed MLM. The simulated phenotypes were used to evaluate statistical power by considering the potential true associations between the observed phenotypes and the observed markers. The simulated phenotypes were generated from the observed SNPs by adding the genetic effects. The SNPs with assigned genetic effects are called quantitative trait nucleotides (QTNs). Total number of QTNs, heritability and dominance and epistatic effects were varied to validate the robustness of P3D for phenotypes with different genetic architectures. We used the distribution of the F statistics from the association tests between the simulated phenotypes and the non-QTN markers to determine an empirical threshold5 at a significance level of 5% (P < 0.05). We then calculated the statistical power as the proportion of QTNs with F values greater than the thresholds.
RESULTS
Compression
We examined the fit of the compressed MLM on human height with eight hierarchical clustering algorithms24,25: unweighted pair group method with arithmetic average (UPGMA); unweighted pair-group centroid; complete linkage; Lance-Williams flexible-beta method; McQuitty’s similarity analysis (weighted pair-group method using arithmetic averages); weighted pair-group centroid median; single linkage (nearest neighbor); and Ward’s method. The fit of each model varied considerably with the use of different combinations of clustering algorithms and compression levels. For each clustering algorithm, at least one compression level had a better fit with the data than the standard MLM, with the exception of the unweighted and weighted pair-group centroid median algorithms in the human dataset (Supplementary Fig. 1). The variation in model fit among clustering algorithms suggests that additional research is needed to better understand the relationship between clustering algorithms and compression levels; however, this is beyond the scope of our study. Because UPGMA produced models that were generally equivalent to or better than other clustering algorithms, we chose to use that in the rest of the work presented here, including the examination of model fit for different phenotypes within the same population (Supplementary Fig. 2).
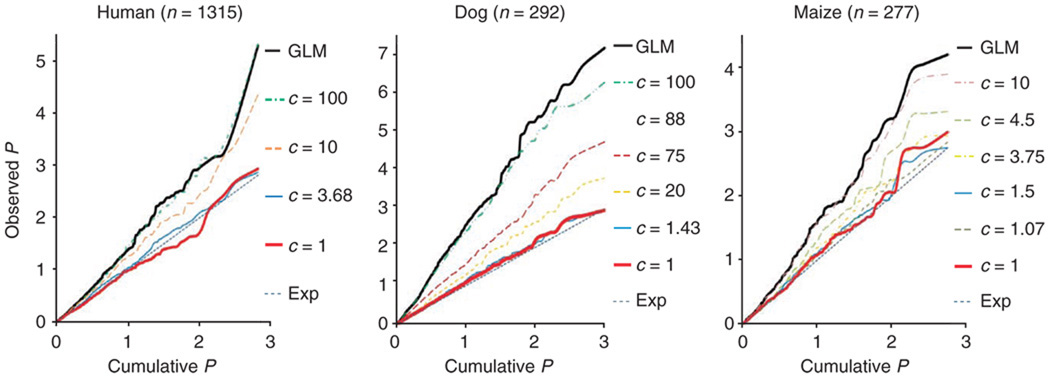
Under the assumption that there is no association between the observed phenotypes and the tested markers, the P values from the association tests should follow a uniform [0,1] distribution. This distribution is shown in the quantile-quantile plot in Figure 1. Notably, compressed MLM controlled the false positive rate better than the standard MLM when the compression levels were within the range of 1.5 to 10 (Fig. 2). At these same compression levels, the compressed MLM had a better model fit than the standard MLM when marker effects were excluded (the top panel in Fig. 3).
Figure 2.
Quantile-quantile plots of type I error (false positive) rates of association tests using the compressed MLM under different compression levels. The observed phenotypes are height in humans, hip dysplasia (Norberg angle) in dogs and flowering time (days to pollination) in maize. The distributions of P values are shown by plotting the observed P values against the cumulative P values in the negative log10 scale. Under the assumption that this set of genetic markers are unlinked to the polymorphism controlling the phenotypes, the P values of the association tests have a uniform distribution, indicated by the expected diagonal line (Exp)5. A statistical approach that has a distribution closer to the diagonal line indicates a better control for type I errors. The GLM that is equivalent to the compressed MLM at the maximum compression level had the most type I errors. For all the species, at least one compression level was found at which the compressed MLM performed better than the standard MLM, which is equivalent to the compressed MLM with compression level of 1.
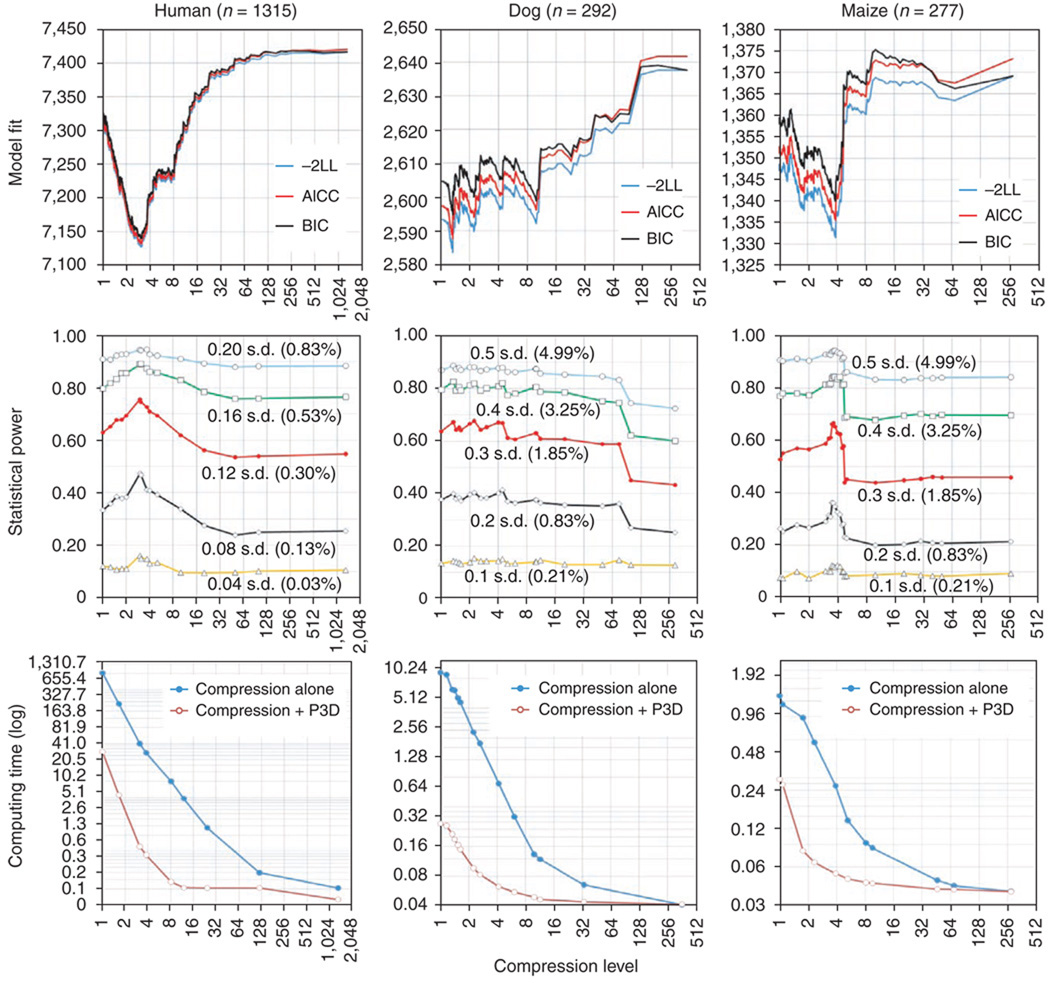
Figure 3.
The performance of the compressed MLM under different compression levels (horizontal axis). The two extremes of the compression level at 1 and n (the number of individuals) correspond to the standard MLM and the GLM, respectively. Performances were examined based on model fit, statistical power and computing time (s). The observed phenotypes are height in humans, hip dysplasia (Norberg angle) in dogs and flowering time (days to pollination) in maize. Individuals in each of the datasets were clustered into groups according to kinship by using the UPGMA algorithm implemented by proc cluster in SAS26. Model fit was evaluated using negative log likelihood (–2LL), adjusted Akaike information criterion (AICC) and Bayesian information content (BIC). Smaller values of –2LL, AICC and BIC indicate better fit. The statistical power was evaluated for QTNs with different size effect. The size of QTN effect is expressed in the unit of phenotypic standard deviation (s.d.). The average computing time was calculated from the observed CPU time for association tests on 647 markers in human datasets; 1,000 markers in dog datasets; and 553 markers in maize datasets. The computations were performed by proc mixed in SAS26 on a computer from Dell (Optiplex 755) with two physical CPUs (E6850 @ 3.00 GHz) and 3.25 GB RAM operated under Windows XP.
To deal with the risk that reducing the number of false positives might affect the ability to detect true positives (that is, statistical power), especially in the case of assuming that no association is violated, we examined the performance of the compressed MLM by simulation studies. After QTN effects were added to the observed phenotypes, tests of association between these simulated phenotypes and markers showed that the statistical power (that is, the ability to detect the simulated QTN) and model fit followed the same trend. The compression level that best fit a model without markers also provided the highest power to detect QTN (middle, Fig. 3). Compared to the standard MLM, equivalent power was achieved using compressed MLM with as much as 5- to 10-fold compression. The compression level with the best-fitting model increased statistical power by 34%, 42% and 20% for human, dog and maize for a QTN that explained 0.12, 0.30 and 0.30 units of the phenotypic standard deviation, respectively.
P3D
We compared P values obtained from using P3D to P values from using full optimization for testing the association between observed phenotypes and markers in human, dog and maize. The coefficient of determination (r2; Pearson’s correlation coefficient squared) between corresponding P values obtained from the two approaches were all greater than 0.96. Therefore, we concluded that the association tests obtained from the P3D and full optimization methods were approximately the same.
To evaluate the performance of P3D using phenotypes with different genetic architectures, we performed association tests on simulated phenotypes. Different numbers of QTNs with various levels of heritability, dominance and epistatic effects were simulated. Similarly, strong correlations (r2 > 0.97) between the corresponding P values from the P3D and full optimization approaches were observed for both QTN and non-QTN SNPs (top two panels in Fig. 4 and Supplementary Figs. 3 and 4).
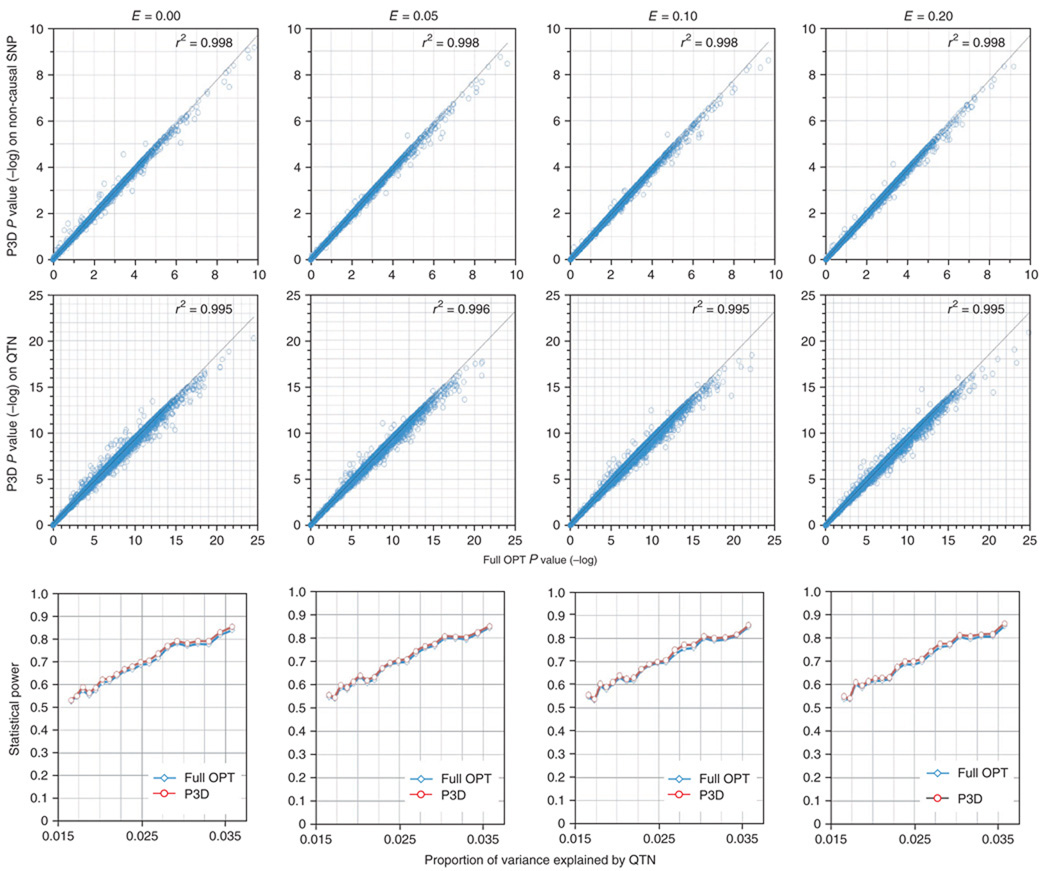
Figure 4.
The P values and statistical power of association tests obtained by using the one-step MLM with the full optimization (full OPT) for all unknown parameters compared to P3D on a maize phenotype simulated with different epistatic effects (E). The phenotype was controlled by 20 QTNs, which were randomly assigned to the SNPs from the maize dataset5. Heritability was defined as the proportion of additive genetic variance over the total variance (the sum of additive genetic variance, epistatic variance and residual variance) and was set at 0.5. Because all maize used here belonged to inbred lines, no dominance effect was included. The experiment was repeated 1,000 times. For each replicate, the number of non-causal SNPs that were randomly sampled was the same as the number of causal QTNs. The top two panels display the P values using the full OPT (x axis) and P3D (y axis). Each dot represents a test on a non-causal SNP (top) and a causal QTN (middle). The P values from P3D are highly correlated with the ones from the full OPT for the non-causal SNPs and causal QTNs (r2 > 99%). The empirical statistical power for detecting the causal QTNs is displayed (bottom) as a function of the proportion of the total variation explained (x axis). The P3D approach and the full OPT had approximately the same statistical power for detecting the causal QTNs.
For a simulated phenotype with a heritability of 50% and that is controlled by 20 QTNs randomly sampled from the SNPs in the human dataset, we used four compression levels. At each compression level, association tests were performed using both the P3D and full optimization approach. Strong correlations between the corresponding P values from P3D and the full optimization were also observed (r2 > 0.99) for both QTN and non-QTN SNPs across the different compression levels (top two panels in Supplementary Fig. 5).
We used the distribution of the F statistics for the non-QTN SNPs to derive the empirical threshold for evaluating F values at each compression level. We calculated the empirical statistical power as the proportion of QTNs with F values greater than the threshold corresponding to a significance level of 5% (P < 0.05). The empirical statistical power of the P3D and full optimization approaches were approximately the same in all tested scenarios (bottom panels in Fig. 4 and Supplementary Figs. 3–5).
DISCUSSION
Compression decreases computing time in proportion to the inverse of the cube of the compression level. For instance, a compression level of 2 will reduce the computing time by about 87%. The standard MLM with each individual as a single group has a compression level of 1 and requires the most computing time. The GLM, equivalent to the highest compression level with all individuals assigned to a single group, requires the least computing time. In our analyses, both model fit and statistical power improved as the compression level increased from one. After reaching the optimum compression level, further compression reduced model fit and statistical power, which eventually became the same as the power with the GLM at the maximum compression.
The fit of the reduced model (that is, the model without markers) under different compression levels followed the same trend as the statistical power of the full model for testing markers. Because the reduced model did not include marker effects, the computing time required to find the compression level with the best-fitting model was independent of the number of markers. For these reasons, the P3D model used an efficient strategy that determined the optimal clustering algorithm and compression level only once.
Similar to the residual approach, P3D eliminates the need to estimate population parameters separately for every marker. The advantage of P3D is that it does not lower statistical power regardless of the genetic architecture of the phenotypes. The P3D method works well for different numbers of QTNs and with various levels of heritability, dominance or epistatic effects.
Compressed MLM and P3D can be applied either separately or jointly and can also be used in combination with other approaches, such as the EMMA algorithm, to speed up the iteration process in the first step of P3D. The compressed MLM improves both computing speed and statistical power, whereas P3D improves computing speed without sacrificing statistical power. In addition, compressed MLMs can be applied at various compression levels. For an analysis in which statistical power is the top priority, the compression level with the best model-fit should be chosen; otherwise, a higher compression level may be chosen to reduce computing time. It should be noted that no trend has been identified to determine the compression level with the best model-fit across different datasets. The compression level that generated the best model-fit varied among phenotypes in the same population when the same kinship was used (Supplementary Fig. 2). Thus, for each new study, the compression level needed to be optimized using the reduced MLM.
The theoretical computing time reduction is faster by a factor of pc3 for the joint use of compressed MLM and P3D, where p is the number of iterations and c is the compression level. When using proc mixed and proc cluster in SAS26 on the three datasets, we showed that the computing time for the human dataset (largest sample size) decreased 19-fold with compressed MLM alone and 877-fold with compression with P3D at the compression level with the greatest statistical power (Fig. 3, bottom). Choosing a compression level that had power equivalent to that of the standard MLM reduced the computing time even more: computing time was 103-fold faster with compression alone and 7,582-fold faster with compression with P3D, respectively. For the human dataset with 1,315 individuals, the standard MLM (no compression, no P3D) took 821 s to screen one marker. (Fig. 3) At this speed, it would take 9,502 d (26 years) to analyze a GWAS with 1 million markers. The current methods (compression with P3D) took 0.34 s to screen a marker at the compression level of 3.8, which showed the highest statistical power, and at this speed, it would take only 2.7 d to screen one million markers. The increased speed is even more important for larger datasets (for example, one containing 5,000 individuals). This suggests that current GWAS datasets on several thousand of individuals at 500,000–1,000,000 markers could be analyzed by our methods within several days. We have made these methods available within an implementation of the software program TASSEL27.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturegenetics/.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the US National Science Foundation (NSF)–Plant Genome Program (DBI-0321467, 0703908 and 0820619), NSF–Plant Genome Comparative Sequencing Program (DBI-06638566), US National Institutes of Health (1R21AR055228-01A1), National Heart, Lung, and Blood Institute (U 01 HL72524, HL54776 and 5U01HL072524-06), US Department of Agriculture Research Service (53-K06–5-10 and 58–1950-9–001), USDA–Cooperative State Research, Education and Extension Service National Research Initiative (2006-35300-17155), Morris Animal Foundation (D04CA-135), WALTHAM Centre for Pet Nutrition, Cornell Advanced Technology in Biotechnology and the Collaborative Research Program in the Cornell Veterinary College. The authors would like to thank K. Zhao for providing the source code to compute kinship and L. Rigamer Lirette, A.L. Ingham and S. Myles for editing of the manuscript.
Footnotes
Note: Supplementary information is available on the Nature Genetics website.
AUTHOR CONTRIBUTIONS: Z.Z. conceptualized the study, performed the data analyses and wrote the manuscript. E.E., M.A.G. and J.Y. participated in the data analyses and wrote the manuscript. P.J.B. implemented the two new methods in the TASSEL software package. C.L., H.K.T., D.K.A. and J.M.O. provided the human data and supervised its analyses. R.J.T. provided the dog data and supervised its analyses. E.S.B designed and supervised the project. All authors edited the manuscript.
COMPETING FINANCIAL INTERESTS: The authors declare no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Abiola O, et al. The nature and identification of quantitative trait loci: a community’s view. Nat. Rev. Genet. 2003;4:911–916. doi: 10.1038/nrg1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 4.Abecasis GR, Cardon LR, Cookson WO. A general test of association for quantitative traits in nuclear families. Am. J. Hum. Genet. 2000;66:279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu J, et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006;38:203–208. doi: 10.1038/ng1702. [DOI] [PubMed] [Google Scholar]
- 6.Zhao K, et al. An Arabidopsis example of association mapping in structured samples. PLoS Genet. 2007;3:e4. doi: 10.1371/journal.pgen.0030004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balding DJ. A tutorial on statistical methods for population association studies. Nat. Rev. Genet. 2006;7:781–791. doi: 10.1038/nrg1916. [DOI] [PubMed] [Google Scholar]
- 8.Buckler ES, et al. The genetic architecture of maize flowering time. Science. 2009;325:714–718. doi: 10.1126/science.1174276. [DOI] [PubMed] [Google Scholar]
- 9.Henderson CR. Comparison of alternative sire evaluation methods. J. Anim. Sci. 1975;41:760–770. [Google Scholar]
- 10.Pollak EJ, Quaas RL. Definition of group effects in sire evaluation models. J. Dairy Sci. 1983;66:1503–1509. [Google Scholar]
- 11.Thompson R. Sire evaluation. Biometrics. 1979;35:339–353. [Google Scholar]
- 12.Quass RL, Pollak EJ. Mixed model methodology for farm and ranch beef cattle testing programs. J. Anim. Sci. 1980;51:1277–1287. [Google Scholar]
- 13.Myles S, et al. Association mapping: critical considerations shift from genotyping to experimental design. Plant Cell. 2009;21:2194–2202. doi: 10.1105/tpc.109.068437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu L, et al. The long (and winding) road to gene discovery for canine hip dysplasia. Vet. J. 2009;181:97–110. doi: 10.1016/j.tvjl.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henderson CR. Applications of Linear Models in Animal Breeding. Guelph, Ontario, Canada: University of Guelph; 1984. [Google Scholar]
- 16.Kang HM, et al. Efficient control of population structure in model organism association mapping. Genetics. 2008;178:1709–1723. doi: 10.1534/genetics.107.080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aulchenko YS, de Koning D-J, Haley C. Genomewide rapid association using mixed model and regression: a fast and simple method for genomewide pedigreebased quantitative trait loci association analysis. Genetics. 2007;177:577–585. doi: 10.1534/genetics.107.075614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Searle SR, Casella G, McCulloch CE. Variance Components. New York: Wiley & Sons; 1992. [Google Scholar]
- 19.Robertson A. Optimum group size in progeny testing and family selection. Biometrics. 1957;13:442–450. [Google Scholar]
- 20.Hannrup B, Jansson G, Danell Ö. Comparing gain and optimum test size from progeny testing and phenotypic selection in Pinus sylvestris. Can. J. For. Res. 2007;37:1227–1235. [Google Scholar]
- 21.de Oliveira HN, Lobo RB. Use of progeny testing in beef cattle: prediction of genetic gain in Nelore cattle breeding program. Rev. Bras. Genet. 1995;18:207–214. z. [Google Scholar]
- 22.Yu J, Arbelbide M, Bernardo R. Power of in silico QTL mapping from phenotypic, pedigree and marker data in a hybrid breeding program. Theor. Appl. Genet. 2005;110:1061–1067. doi: 10.1007/s00122-005-1926-7. [DOI] [PubMed] [Google Scholar]
- 23.Rutherford JR, Krutchkoff RG. The empirical Bayes approach: estimating the prior distribution. Biometrika. 1967;54:326–328. [PubMed] [Google Scholar]
- 24.Romesberg HC. Cluster Analysis for Researchers. Raleigh, North Carolina, USA: LULU Press; 2004. [Google Scholar]
- 25.Jain AK, Murty MN, Flynn PJ. Data clustering: a review. ACM Comput. Surv. 1999;31:264–323. [Google Scholar]
- 26.Statistical Analysis Software for Windows. North Carolina: Cary; 2002. SAS Institute Inc. [Google Scholar]
- 27.Bradbury PJ, et al. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.