Abstract
The application of RNA interference (RNAi), either in the clinic or laboratory, requires safe and effective delivery methods. Here we develop a combinatorial approach to synthesize a library of delivery vectors based on two lipid-like substrates with known siRNA delivery capabilities. Members of this library have a mixture of lipid-like tails and feature appendages containing hydroxyl, carbamate, ether or amine functional groups as well as variations in alkyl chain length and branching. Using a luciferase reporter system in HeLa cells, we study the relationship between lipid chemical modification and delivery performance in vitro. The impact of the functional group was shown to vary depending on the overall amine content and tail number of the delivery vector. Additionally, in vivo performance was evaluated using a Factor VII knockdown assay. Two library members, each containing ether groups, were found to knock down the target protein at levels comparable to the parent delivery vector. These results demonstrate that small chemical changes to the delivery vector impact knockdown efficiency and cell viability both in vitro and in vivo. The work described here identifies new materials for siRNA delivery, as well as provides new insight into the parameters for optimized chemical makeup of lipid-like siRNA delivery materials.
Introduction
Over the last thirty years, RNA has been shown to perform a variety of activities within the cell that go beyond its traditional role as a translation template for protein synthesis. In particular, the recent discovery of RNA-mediated regulation of gene expression has spawned widespread interest (1). This technology, termed RNA interference (RNAi), is based on the ability of short duplexes of RNA, termed siRNA, to induce the specific cleavage of complementary mRNA, leading to a silencing of gene expression. RNAi has potential both for use as a tool capable of elucidating cellular mechanisms, as well as a therapeutic for treating disease at the genetic level (2, 3).
Despite the opportunities afforded by siRNA, a safe and effective delivery system that can transport the material across the cell membrane for incorporation into the necessary cellular machinery is required for use. Both the size and negative charge of siRNA render active transport across the cell membrane difficult. Furthermore, these RNAs are vulnerable to enzymatic degradation. To circumvent these issues, researchers have made extensive efforts to rationally design chemically modified siRNA (4, 5). Covalent attachment of peptides, cholesterol and aptamers have all been reported to increase cellular uptake (6–11). Additionally, variants such as substitution at the 2’ position (eg. 2’-fluoro, 2’-O-methyl, 2’-methoxyethyl) and modified phosphate backbone (eg. phosphorothioate and methylphosphonate) provide for improved efficacy by imparting a greater resistance to nuclease degradation (12). While these methods have demonstrated promise in vivo, the development of improved delivery methods are required to allow for the fullest application of RNAi in the clinic.
The application of materials previously used for non-viral DNA delivery to siRNA delivery has enabled the rapid development of siRNA for use in vivo. For example, both polymer and liposome delivery systems have demonstrated efficacy in animal models (13–15). However, structural differences between RNA and DNA, as well as differences in their method of action, are likely responsible for the fact that delivery systems for DNA do not always work for siRNA and visa versa (16). Therefore, the synthesis of new materials that complex, protect and deliver siRNA is currently an active area of research. As a result, polymers, polymersomes, dendrimers, lipid-based formulations and rationally designed systems specific for siRNA delivery have been developed, with carrying degrees of success in vitro and in vivo (17–26).
Recently a new approach to the synthesis of lipid-like materials for siRNA delivery vectors using combinatorial methods was reported (27). In this study, a library of over 1200 lipid-like materials was generated through the conjugate addition of an amine to an α,β-unstaurated carbonyl and evaluated for siRNA delivery performance. The resulting materials, termed lipidoids, were structurally distinct from other classes of lipid delivery vectors in that they contained multiple protonable amine groups connected to relatively short alkyl chains. In these studies, materials with good in vitro and in vivo performance were identified, including a lead material 98N12-5 that was shown efficacious in primates.
In this study, we build upon the first library of lipidoids to incorporate further diversity into two promising siRNA delivery candidates. These library members feature systematic variation of select side chains with different capacities for hydrogen bonding, hydrophobic interactions and protonation states, enabling the exploration of heterogeneously functionalized lipidoids for siRNA delivery.
Materials and Methods
Library synthesis
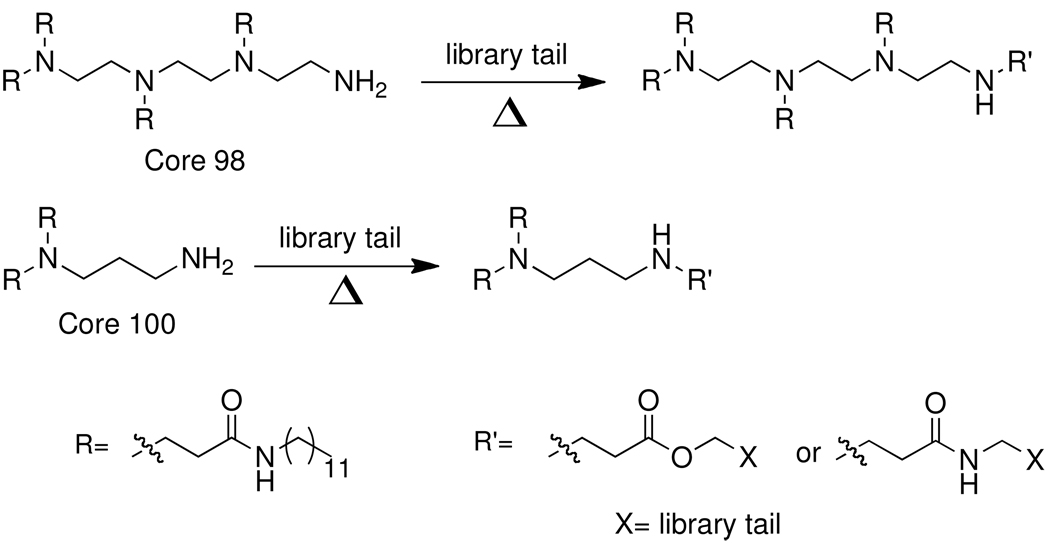
Lipidoid library members were synthesized by addition of acrylamides or acrylates to a partially substituted amine-modified core lipidoid. A detailed synthesis of the core lipidoids is found in the Supporting Information. Dodecylacrylamide was purchased from TCI America. Other acrylamides were synthesized as previously described (27). Acrylates were purchased from Sigma-Aldrich, Alfa Aesar and TCI America. Amines were purchased from Sigma-Aldrich. All library reactions were carried out in 2ml Teflon-lined glass screw-top vials containing a magnetic stir bar. 1.1eq of the library acrylate was added to 120mg of core amine and the mixture was stirred at 90°C for 24–48 hours. After cooling, the lipidoid mixtures were used without purification for initial siRNA transfection screening. Lead compounds were purified for further testing.
In vitro siRNA transfection assay
To facilitate high throughput screening, all transfections were performed using a multichannel pipet and serial dilutions in a 96-well plate format. HeLa cells stably expressing firefly luciferase and Renilla luciferase were seeded at 15,000 cells/well into each well of an opaque white 96-well plate (Corning-Costar) and allowed to attach overnight in growth medium. Growth medium was composed of 90% phenol red-free DMEM, 10% FBS, 100 units/ml penicillin and 100 mg/ml streptomycin (Invitrogen). Cells were transfected with 50 ng of firefly-specific siLuc complexed with lipidoid at lipidoid:siRNA ratios of 2.5:1, 5:1, 10:1 and 15:1 (wt/wt).
Transfections were performed in quadruplicate. Working dilutions of each lipid were prepared in 25-mM sodium acetate buffer (pH 5). 25 ul of the diluted lipid was added to 25 ul of 2.5ug/ml siRNA in a well of a 96-well plate. The mixtures were incubated for 20 min to allow for complex formation, and then 30 ul of each of the lipidoid/siRNA solutions was added to 200 ul of fresh growth medium in 96-well polystyrene plates. The growth medium was removed from the cells using a 12-channel aspirating wand (V&P Scientific) after which 150 ml of the DMEM/lipidoid/siRNA solution was immediately added. Cells were allowed to grow for 24 hours at 37 °C, 5% CO2 and were then analyzed for luciferase expression. Control experiments were performed with Lipofectamine 2000, as instructed by the vendor (Invitrogen). Firefly and Renilla luciferase expression was analyzed using the Dual-Glo® Assay System (Promega). Luminescence was measured using a Victor3 luminometer (Perkin Elmer). Firefly luminescence was normalized by the internal Renilla control luminescence and treated wells were compared against the untreated control for assessment of knockdown efficacy.
Viability assessment
Cell viability testing was performed as previously described (27). Briefly, HeLa cells were seeded at 15,000 cells/ well in a 96 well plate and allowed to adhere overnight. Cells were then transfected with 2.5, 5, 10 or 15 wt/wt ratio of lipidoid to 50ng siRNA. After incubation for 24 hours, the media was replaced and CellTiter 96® Aqueous One Solution (Promega) was added. After incubating for one hour, the absorbance at 490nm was read and viability calculated relative to an untransfected control.
In vivo lipidoid-siRNA formulation
Lipidoid-based siRNA formulations comprised lipidoid, cholesterol, polyethylene glycol-lipid (PEG-lipid) and siRNA. Formulations were prepared as described previously. Stock solutions of lipidoid, cholesterol MW 387 (Sigma-Aldrich), and mPEG2000-Ceramide C16 (Avanti Polar Lipids) MW 2634 were prepared in ethanol and mixed to yield a molar ratio of 42:48:10, respectively. Mixed lipids were added to 200 mM sodium acetate buffer pH 5 to yield a solution containing 35% ethanol by volume, resulting in spontaneous formation of empty lipidoid nanoparticles. The resulting nanoparticles were extruded through an 80nm membrane. siRNA in 35% ethanol and 50 mM sodium acetate pH 5 was added to the nanoparticles and incubated at 37 °C for 30 min. Ethanol removal and buffer exchange of siRNA-containing lipidoid nanoparticles was achieved by dialysis against PBS using a 3,500 MWCO membrane. Particle size was determined using a Malvern Zetasizer NanoZS (Malvern). siRNA content and entrapment efficiency was determined using the Quant-iT™ RiboGreen® RNA reagent (Invitrogen).
In vivo mouse Factor VII silencing experiments
All procedures used in animal studies conducted at MIT were approved by the Institutional Animal Care and Use Committee (IACUC) and were consistent with local, state and federal regulations as applicable. C57BL/6 mice (Charles River Labs) received either saline or siRNA in lipidoid formulations via tail vein injection at a volume of 0.01 ml/g. 72 hours after administration, animals were anesthetized by isofluorane inhalation and blood was collected into serum separator tubes by retroorbital bleed. Serum levels of Factor VII protein were determined in samples using a chromogenic assay (Biophen FVII, Aniara Corporation) according to manufacturers’ protocols. A standard curve was generated using serum collected from saline-treated animals.
Results
Synthesis of heterogeneous tail lipidoid library
The core lipidoid starting materials for this library were synthesized using a three or four step process (Scheme 1 and supporting information). Briefly, diethylenetriamine was heated neat at 90°C with dodecylacrylamide and purified to yield the desired four-tailed isomer. Coupling of 2-(Boc-amino)ethyl bromide followed by deprotection under acidic conditions yielded the amine functionalized library starting material, Core 98. Alternatively, 1,3-diaminopropane was mono-protected with Boc anhydride, heated neat at 90°C with dodecylacrylamide to add the alkyl tails, and deprotected under acidic conditions to yield the amine functionalized library starting material, Core 100.
Scheme 1.
Synthesis of lipidoid library
Library synthesis was accomplished by reaction of 120 mg of each building block with 1.1 equivalents of the acrylates or acrylamides shown in Table 1. Core 98 reactions yielded compound mixtures that contained mainly the fully substituted (n) and partially substituted (n-1) compounds. Compound mixtures from Core 100 contained a residual starting material in addition to the n and n-1 products. Select lipidoids were purified to isolate the n-1 isomer. The identity of these compounds was confirmed by mass spectrometry (see supporting info).
Table 1.
Structure of library acrylates and acrylamides
| Core 98 | Core 100 | Library tail |
|---|---|---|
| 1a | 1b | 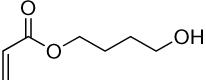 |
| 2a | 2b | 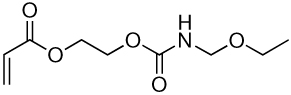 |
| 3a | 3b | 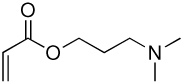 |
| 4a | 4b | 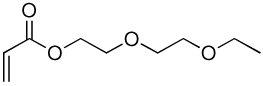 |
| 5a | 5b | 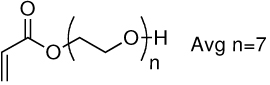 |
| 6a | 6b | 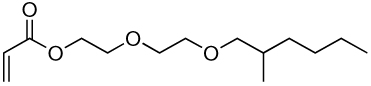 |
| 7a | 7b | 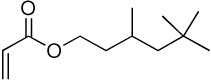 |
| 8a | 8b | 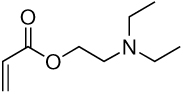 |
| 9a | 9b | 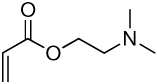 |
| 10a | 10b | 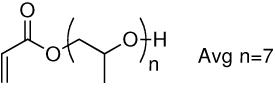 |
| 11a | 11b | 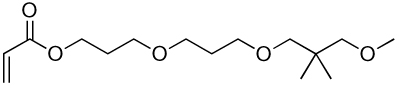 |
| 12a | 12b | 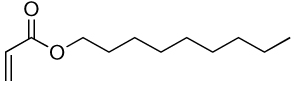 |
| 13a | 13b | 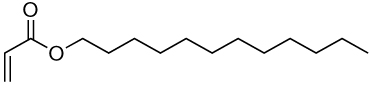 |
| 14a | 14b | 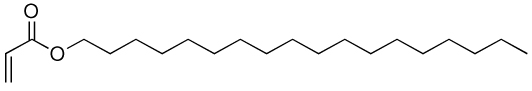 |
| 15a | 15b | 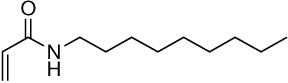 |
| 16a | 16b | 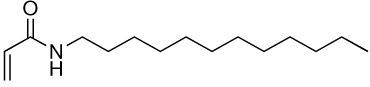 |
| 17a | 17b |  |
Library screening for siRNA delivery
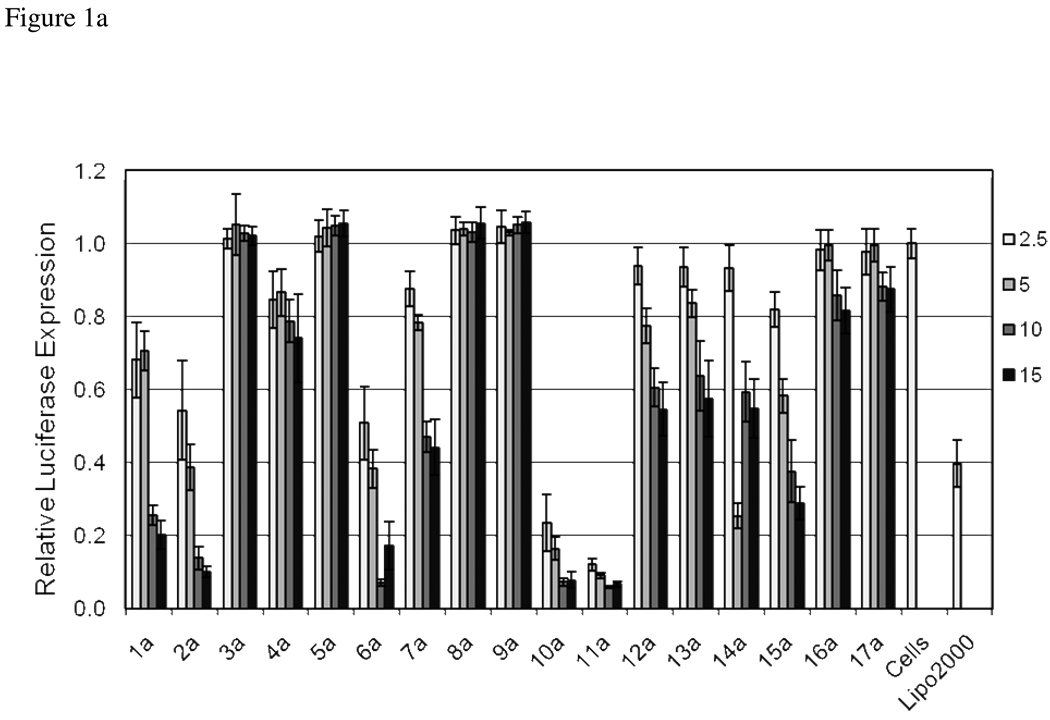
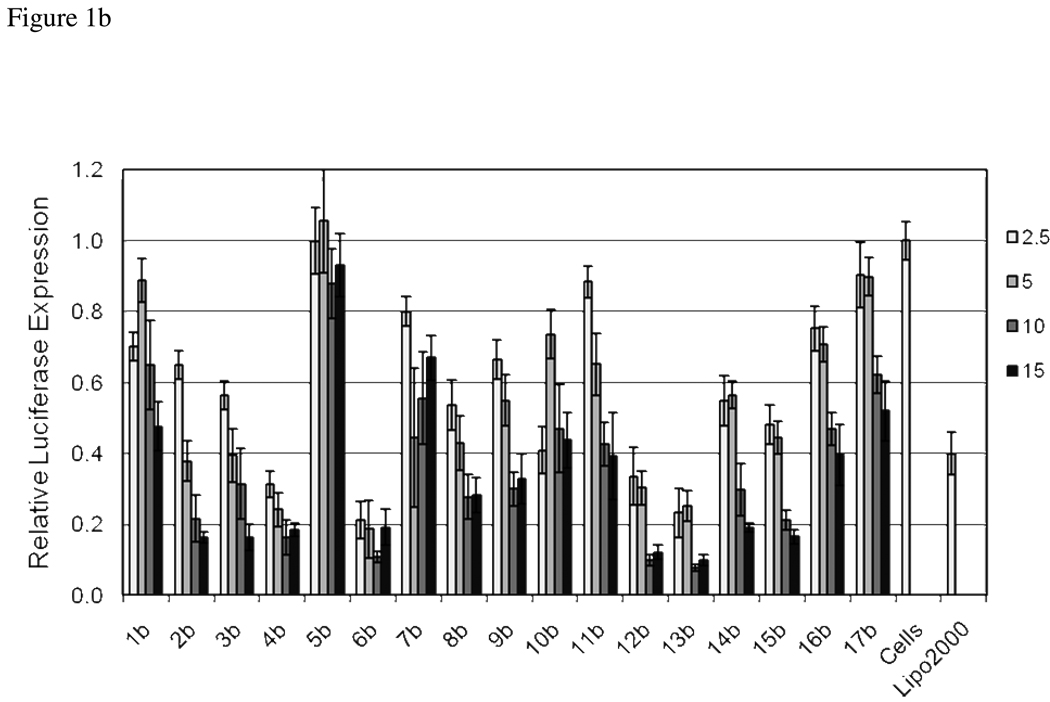
Evaluation of the library for delivering siRNA was performed using a HeLa cell line that is stably transfected with both Renilla and Firefly luciferase. In this assay, siRNA specific for firefly luciferase is used to measure knockdown, while the Renilla luciferase is monitored as a control. Lipofectamine2000, a commonly used commercially available reagent, was also included in the screen. Lipidoids were complexed with the siRNA at weight ratios of 2.5, 5, 10 and 15:1 lipidoid: siRNA. As shown in Figure 1, the knockdown of firefly luciferase was dependent on the ratio of lipidoid used and was affected by the modification with members of the library. In general, the knockdown at a 15:1 weight ratio of lipidoid to siRNA was highest and the efficiency decreased as the ratio of lipidoid to RNA was decreased. In cases where the knockdown was efficient or where none was observed, the efficiency with the ratios tested was less varied.
Figure 1.
Library screening for siRNA transfection efficiency. HeLa cells stably expressing firefly and Renilla luciferase were transfected with 50 ng of firefly specific siRNA complexed at lipidoid/ siRNA ratios of 2.5:1, 5:1, 10:1 and 15:1 (wt/wt). Lipofectamine 2000 was used as instructed by the vendor. Luminescence was measured after 24 hour incubation, and the results presented relative to an untransfected control set of cells. a) Core 98 library. b) Core 100 library.
While the efficacy of both amine cores were affected by the tail modifications, Core 100 was generally more tolerant of the addition of the functional groups found in the library tails. At the highest ratio of lipidoid:siRNA, 14 of the 17 members of the library showed luciferase knockdown of at least 50%, the most active of which contained ether, carbamate or amine functionality in the added tail. At the lowest ratio, 9 of the 17 members showed luciferase knockdown of at least 40%. Only two members, 5b and 17B, containing PEG or a long alkyl chain respectively, were rendered essentially inactive, having less than 10% knockdown at two of the ratios tested.
Fewer derivatives of Core 98 had significant delivery efficiency when compared with the number of Core 100 derivatives. Only 4 of the 17 derivatives had luciferase knockdown levels of at least 50% at the 15:1 lipidoid: siRNA ratio and of at least 40% at the 2.5:1 ratio. These active library members contained either a carbamate group or ether functionalities. Core 98 was particularly sensitive to the appendage of other library members. Specifically, 3a, 5a, 8a, 9a, 16a and 17a showed little ability to deliver siRNA at any lipidoid: siRNA ratio. Interestingly, the ablation of activity due to the addition of a tertiary amine was specific to the Core 98 system, while the addition of PEG or long hydrophobic tails generally disrupted delivery in both Core 98 and Core 100 systems.
Dose response of siRNA in vitro
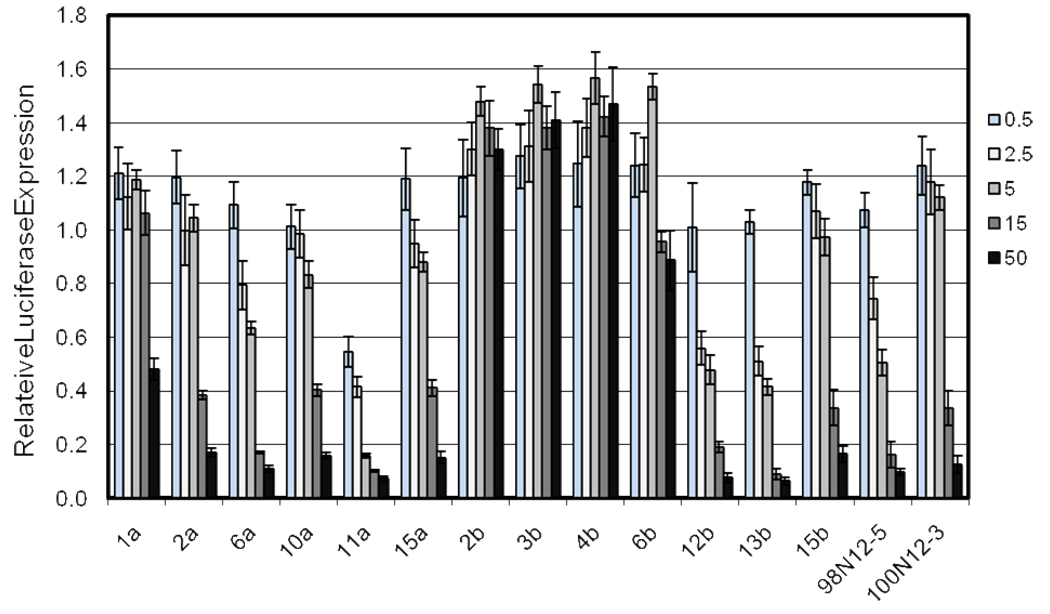
Based on the results of our initial screen, we chose 13 of the most promising compound mixtures to purify for further investigation of siRNA delivery efficiency (Figure 2). Two were derivatives obtained through reaction with an acrylamide while the remaining compounds were all generated via reaction with an acrylate moiety. Earlier screens had identified the most effective compounds as having the n-1 identity. After purification, the identity of the purified compounds was confirmed using mass spectrometry (see Supporting Information).
Figure 2.
Luciferase knockdown at low doses of siRNA. HeLa cells stably expressing firefly and Renilla luciferase were transfected using selected lipidoids at 15:1 lipidoid/siRNA ratio (wt/wt) at 0.5, 2.5, 5, 15 and 50 ng siRNA. Luminescence was measured after 24 hour incubation, and the results presented relative to an untransfected control set of cells. Lipidoids were purified isolate the n-1 isomer prior to transfection.
In this comparison, the purified library members were compared to 98N12-5 and 100N12-3, two members of the original library that were structurally similar to the library members derived from Core 98 and Core 100, but solely contained alkyl tails of uniform length. The majority of the purified set of library compounds, 8 of the 13, had a knockdown of at least 50% using 50ng of siRNA, and at least 60% using 15 ng of siRNA. Decreasing the dose of siRNA to 5 ng led to a significant separation amongst the library members. Only three members, 11a, 12b and 13b, retained knockdown efficiency greater than or equal to the parent compound at this level (50% knockdown). Interestingly, 11a was the most efficient compound for delivering low doses of siRNA in this assay, achieving 50% knockdown using only 0.5 ng siRNA.
Also of interest is the deleterious effect that purification of the n-1 isomer had on certain compound mixtures. The delivery ability of 1a, 2b, 3b, 4b and 6b were all significantly attenuated after they were removed from the crude mixture. Of these, only 1a is derived from Core 98, while the other four compounds are derived from Core 100. Only one condition, 50ng of siRNA with 1a, showed any luciferase knockdown at all. Activity was completely abolished in all other cases.
Viability after siRNA delivery with selected library members
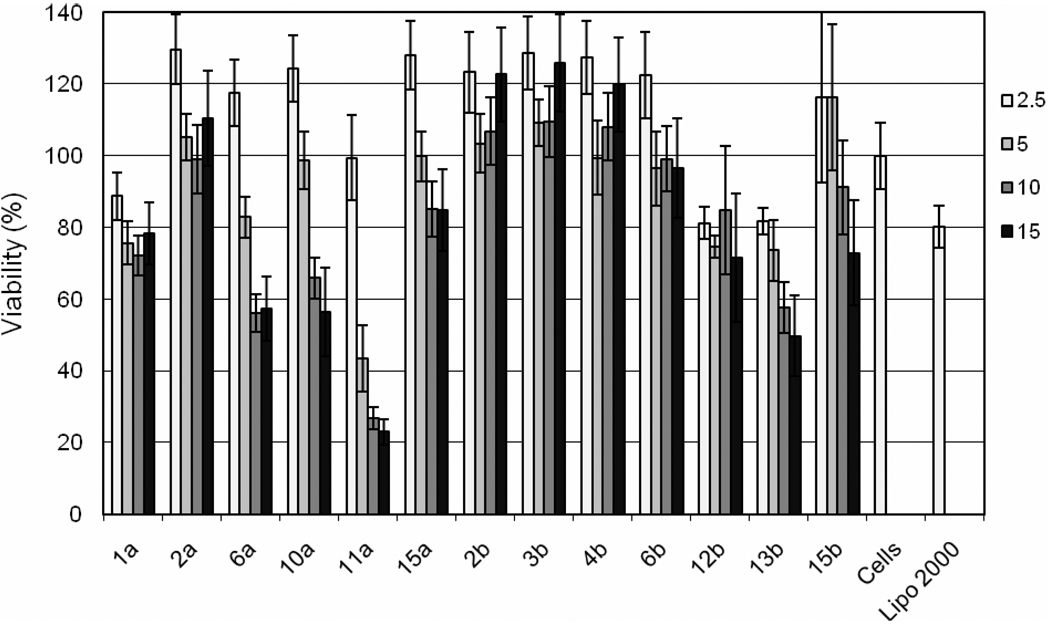
Purified library members were evaluated for their effect on cellular viability using an MTS assay (Figure 3). All compounds had at least one weight ratio that gave viability values above 80%, the value also observed using Lipofectamine2000. At a weight ratio of 15:1, 6a, 10a, 11a and 13b all had cell viability rates of less than 60%, while the remainder of the purified compounds were at or above 80%.
Figure 3.
Viability of HeLa cells post-transfection with selected lipidoids. HeLa cells were transfected with 50 ng siRNA complexed at lipidoid/ siRNA ratios of 2.5:1, 5:1, 10:1 and 15:1 (wt/wt). Lipofectamine 2000 was used as instructed by the vendor. After incubation for 24 hours, the media was replaced and CellTiter viability solution (Promega) was added. The absorbance at 490nm was read after incubating for 1 hour. Values are reported relative to an untransfected set of control cells.
Factor VII knockdown in vivo
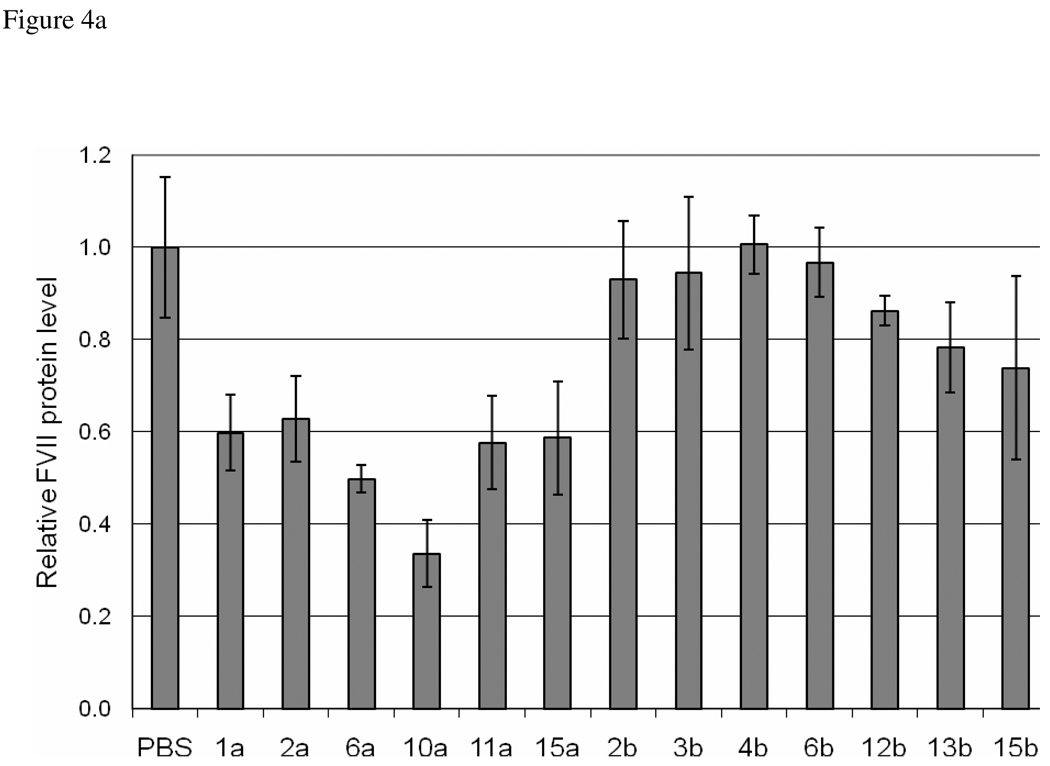
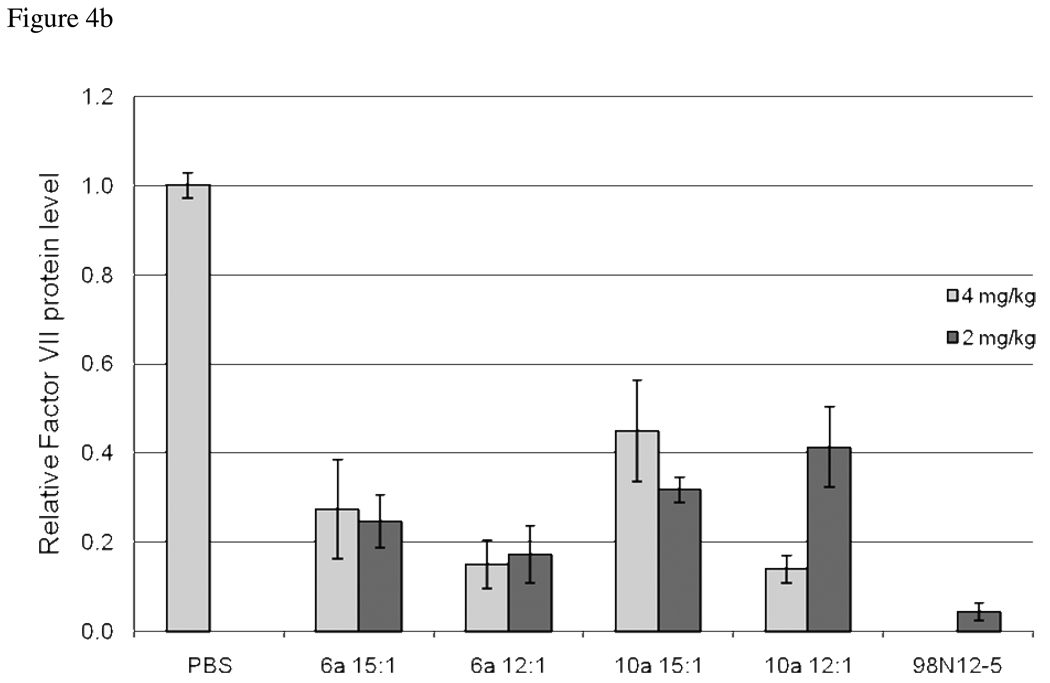
We further assessed the ability of these selected library members to deliver siRNA in vivo. For increased serum stability and enhanced shielding of the payload, lipidoids were formulated with cholesterol and PEG-ceramide prior to complexation with the duplexes. The entrapment efficiency and particle sizes resulting from these formulations varied from 68% to 98% and 45nm to 138nm, respectively (See Supporting Information). siRNA directed against the hepatocyte-specific blood-clotting protein Factor VII was administered via tail vein injection. At 72 hours post injection, mice were bled and assayed for protein levels. Notably, six compounds, 1a, 2a, 6a, 10a, 11a and 15a, all derivatives of Core 98, were shown to decrease the serum levels of Factor VII by at least 40% when compared to a saline control (Figure 4a). In contrast, none of the Core 100 based compounds had a knockdown efficiency greater than 20%. The two most effective compounds, 6a and 10a were screened in this same assay at two different doses of siRNA (Figure 4b). 6a, when complexed at 12:1 wt/wt at doses of 2 or 4 mg/kg of siRNA resulted in over 80% knockdown of factor VII at 72 hours post injection. Using a higher ratio of 15:1 wt/wt led to a slightly decreased knockdown efficiency. 10a achieved over 80% knockdown when complexed at 12:1 wt ratio with 4 mg/kg siRNA. Complexes of 10a at 15:1 wt ratio and a dose of 4mg/kg, or either ratio at 2 mg/kg led to approximately 60% knockdown at 72 hours. These compounds compared favorably to the previously identified lead compound 98N12-5, which, while slightly more efficacious, contained no heterogeneity in the tails.
Figure 4.
In Vivo Silencing of Factor VII in Mice. Lipidoids shown to be efficacious in in vitro studies were formulated for intravenous delivery of anti-Factor VII siRNA to hepatocytes via tail vein injection. a) Mice were administered a single dose of 2.5 mg/kg siRNA and blood samples were drawn at 72 hours for detection of Factor VII levels in serum. b) Selected lipidoids are formulated with varied weight ratios of lipiods to siRNA and injected at doses of 4 and/ or 2mg/kg. Factor VII levels in all treated groups are expressed relative to PBS injected mice.
Discussion
The application of siRNA in both laboratory and clinical settings requires safe and effective delivery methods. While a number of materials and methods for siRNA delivery have been investigated, non-viral chemical approaches continue to receive widespread attention due to their relative ease of production and favorable safety profile. To increase the diversity of delivery molecules, the development of alternative lipid-like structures for siRNA delivery has been pursued (16, 18, 26, 28–32). Here we systematically investigate the effect of the chemical modification of lipid-like materials.
Most siRNA delivery reagents utilize the overall negative charge of the nucleic acid to facilitate binding and particle formation. Many of these materials feature various degrees of protonable amine functionalities. For example, cationic lipids generally feature one cationic amine, while polymeric vectors such as polylysine and polyethyleneimine (PEI) have multiple amine groups. Previous studies identified the best lipidoid delivery molecules to contain three or four secondary or tertiary amines (26, 27). The results here offer further evidence for this optimal number of amines. Compounds 3b, 8b and 9b all feature three amine functional groups and were among the top performers in the initial library screen. Alternatively, compounds 3a, 8a and 9a, featuring the same tertiary amine tails conjugated to a tetraamine core, were less efficient. In other polyamine delivery systems, such as PEI, high amine content can lead to cytotoxicity (21). In this system, it appears that the increased amine content serves to disrupt the particle formation and delivery, as little toxicity was observed in those particular molecules.
Other modifications featuring tails of varying degrees of hydrophobicity or hydrogen bonding capacities were generally well tolerated. For example, appendages featuring hydroxyl, ether and carbamate functional groups predominantly led to increased siRNA delivery efficiency, indicating that these moieties enhanced particle formation for intracellular delivery. Some biophysical characterization of siRNA complexes with nonviral delivery vectors has been reported (33–35). While electrostatic forces are generally accepted as the driving force behind cationic lipids binding to nucleic acids, entropy, hydrophilic and hydrophobic forces have been shown to contribute to the complex formation (36–38). The delivery systems described in this work could provide further insight into the contributions of these parameters to siRNA complexation.
Modification of the lipidoid with ligands containing ethylene or propylene glycol units was shown to result in efficient delivery systems in vivo. Both polyethylene and polypropylene glycol are commonly used excipients in drug delivery formulations that stabilize particles in serum. Indeed, a recent report indicates successful siRNA delivery to primates using such methods (10). In this study, the best performing lipidoids from the in vitro screen were also investigated for their ability to deliver siRNA to mice in vivo using a Factor VII liver function assay. The range of activity observed in vivo indicates that alteration of the lipidoid molecular structure through simple tail modification can affect overall particle characteristics, and may have some effect on pharmacokinetic properties and biodistribution. Indeed, two members of this library containing a combination of alkyl tails and ether groups were particularly well tolerated in vivo, indicating that heterogeneous functionalized delivery vectors warrant further investigation. While more in depth studies will have to be conducted to support these ideas, the ability to tune such pharmacodynamic properties may provide a tool to optimize the biodistribution of delivery. Furthermore, the appendage of these stabilizing ligands suggests that other more specific targeting ligands could be utilized with this system to enable localization of siRNA delivery to tumors or specific areas in vivo.
Conclusion
Here, we have demonstrated the utility of combinatorial methods to generate a chemically diverse set of heterogeneously functionalized compounds for siRNA delivery. These compounds are useful as in vitro delivery vectors, and, importantly, have shown the ability to translate in vitro efficacy to in vivo utility. Furthermore, the range of activity observed through subtle changes in chemical functionality to the lipidoid provides insight into the relationship between chemical structure and cellular delivery performance.
Supplementary Material
Acknowledgements
This work was supported by NIH grants EB000244 and CA132091 and a grant from Alnylam.
Footnotes
Supporting Information Available
Detailed synthetic procedures and formulation parameters. This material is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Shrivastava N, Srivastava A. RNA interference: an emerging generation of biologicals. Biotechnol J. 2008;3:339–353. doi: 10.1002/biot.200700215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pushparaj PN, Melendez AJ. Short interfering RNA (siRNA) as a novel therapeutic. Clin Exp Pharmacol Physiol. 2006;33:504–510. doi: 10.1111/j.1440-1681.2006.04399.x. [DOI] [PubMed] [Google Scholar]
- 4.Chiu YL, Rana TM. siRNA function in RNAi: a chemical modification analysis. Rna. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Paula D, Bentley MV, Mahato RI. Hydrophobization and bioconjugation for enhanced siRNA delivery and targeting. Rna. 2007;13:431–456. doi: 10.1261/rna.459807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeong JH, Mok H, Oh YK, Park TG. siRNA conjugate delivery systems. Bioconjug Chem. 2009;20:5–14. doi: 10.1021/bc800278e. [DOI] [PubMed] [Google Scholar]
- 7.Deshayes S, Simeoni F, Morris MC, Divita G, Heitz F. Peptide-mediated delivery of nucleic acids into mammalian cells. Methods Mol Biol. 2007;386:299–308. doi: 10.1007/978-1-59745-430-8_11. [DOI] [PubMed] [Google Scholar]
- 8.McNamara JO, 2nd, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E, Sullenger BA, Giangrande PH. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol. 2006;24:1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- 9.Muratovska A, Eccles MR. Conjugate for efficient delivery of short interfering RNA (siRNA) into mammalian cells. FEBS Lett. 2004;558:63–68. doi: 10.1016/S0014-5793(03)01505-9. [DOI] [PubMed] [Google Scholar]
- 10.Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, Harborth J, Heyes JA, Jeffs LB, John M, Judge AD, Lam K, McClintock K, Nechev LV, Palmer LR, Racie T, Rohl I, Seiffert S, Shanmugam S, Sood V, Soutschek J, Toudjarska I, Wheat AJ, Yaworski E, Zedalis W, Koteliansky V, Manoharan M, Vornlocher HP, MacLachlan I. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 11.Wolfrum C, Shi S, Jayaprakash KN, Jayaraman M, Wang G, Pandey RK, Rajeev KG, Nakayama T, Charrise K, Ndungo EM, Zimmermann T, Koteliansky V, Manoharan M, Stoffel M. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat Biotechnol. 2007;25:1149–1157. doi: 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]
- 12.Zhang HY, Du Q, Wahlestedt C, Liang Z. RNA Interference with chemically modified siRNA. Curr Top Med Chem. 2006;6:893–900. doi: 10.2174/156802606777303676. [DOI] [PubMed] [Google Scholar]
- 13.Grzelinski M, Urban-Klein B, Martens T, Lamszus K, Bakowsky U, Hobel S, Czubayko F, Aigner A. RNA interference-mediated gene silencing of pleiotrophin through polyethylenimine-complexed small interfering RNAs in vivo exerts antitumoral effects in glioblastoma xenografts. Hum Gene Ther. 2006;17:751–766. doi: 10.1089/hum.2006.17.751. [DOI] [PubMed] [Google Scholar]
- 14.Lungwitz U, Breunig M, Blunk T, Gopferich A. Polyethylenimine-based non-viral gene delivery systems. Eur J Pharm Biopharm. 2005;60:247–266. doi: 10.1016/j.ejpb.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 15.Sato Y, Murase K, Kato J, Kobune M, Sato T, Kawano Y, Takimoto R, Takada K, Miyanishi K, Matsunaga T, Takayama T, Niitsu Y. Resolution of liver cirrhosis using vitamin A-coupled liposomes to deliver siRNA against a collagen-specific chaperone. Nat Biotechnol. 2008;26:431–442. doi: 10.1038/nbt1396. [DOI] [PubMed] [Google Scholar]
- 16.Gary DJ, Puri N, Won YY. Polymer-based siRNA delivery: perspectives on the fundamental and phenomenological distinctions from polymer-based DNA delivery. J Control Release. 2007;121:64–73. doi: 10.1016/j.jconrel.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 17.Kim WJ, Kim SW. Efficient siRNA delivery with non-viral polymeric vehicles. Pharm Res. 2009;26:657–666. doi: 10.1007/s11095-008-9774-1. [DOI] [PubMed] [Google Scholar]
- 18.Kim Y, Tewari M, Pajerowski JD, Cai S, Sen S, Williams J, Sirsi S, Lutz G, Discher DE. Polymersome delivery of siRNA and antisense oligonucleotides. J Control Release. 2009;134:132–140. doi: 10.1016/j.jconrel.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou J, Wu J, Hafdi N, Behr JP, Erbacher P, Peng L. PAMAM dendrimers for efficient siRNA delivery and potent gene silencing. Chem Commun (Camb) 2006:2362–2364. doi: 10.1039/b601381c. [DOI] [PubMed] [Google Scholar]
- 20.Patil ML, Zhang M, Betigeri S, Taratula O, He H, Minko T. Surface-modified and internally cationic polyamidoamine dendrimers for efficient siRNA delivery. Bioconjug Chem. 2008;19:1396–1403. doi: 10.1021/bc8000722. [DOI] [PubMed] [Google Scholar]
- 21.Waite CL, Sparks SM, Uhrich KE, Roth CM. Acetylation of PAMAM dendrimers for cellular delivery of siRNA. BMC Biotechnol. 2009;9:38. doi: 10.1186/1472-6750-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rozema DB, Lewis DL, Wakefield DH, Wong SC, Klein JJ, Roesch PL, Bertin SL, Reppen TW, Chu Q, Blokhin AV, Hagstrom JE, Wolff JA. Dynamic PolyConjugates for targeted in vivo delivery of siRNA to hepatocytes. Proc Natl Acad Sci U S A. 2007;104:12982–12987. doi: 10.1073/pnas.0703778104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolff JA, Rozema DB. Breaking the bonds: non-viral vectors become chemically dynamic. Mol Ther. 2008;16:8–15. doi: 10.1038/sj.mt.6300326. [DOI] [PubMed] [Google Scholar]
- 24.Li W, Szoka FC., Jr Lipid-based nanoparticles for nucleic acid delivery. Pharm Res. 2007;24:438–449. doi: 10.1007/s11095-006-9180-5. [DOI] [PubMed] [Google Scholar]
- 25.Tseng YC, Mozumdar S, Huang L. Lipid-based systemic delivery of siRNA. Adv Drug Deliv Rev. 2009 doi: 10.1016/j.addr.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Love KT, Mahon KP, Levins CG, Whitehead KA, Querbes W, Dorkin JR, Qin J, Cantley W, Qin LL, Racie T, Frank-Kamenetsky M, Yip KN, Alvarez R, Sah DW, de Fougerolles A, Fitzgerald K, Koteliansky V, Akinc A, Langer R, Anderson DG. Lipid-like materials for low-dose, in vivo gene silencing. Proc Natl Acad Sci U S A. 2010;107:1864–1869. doi: 10.1073/pnas.0910603106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Busini V, Hossain N, Bacallado SA, Nguyen DN, Fuller J, Alvarez R, Borodovsky A, Borland T, Constien R, de Fougerolles A, Dorkin JR, Narayanannair Jayaprakash K, Jayaraman M, John M, Koteliansky V, Manoharan M, Nechev L, Qin J, Racie T, Raitcheva D, Rajeev KG, Sah DW, Soutschek J, Toudjarska I, Vornlocher HP, Zimmermann TS, Langer R, Anderson DG. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat Biotechnol. 2008;26:561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baigude H, McCarroll J, Yang CS, Swain PM, Rana TM. Design and creation of new nanomaterials for therapeutic RNAi. ACS Chem Biol. 2007;2:237–241. doi: 10.1021/cb7000582. [DOI] [PubMed] [Google Scholar]
- 29.Ceballos C, Prata CA, Giorgio S, Garzino F, Payet D, Barthelemy P, Grinstaff MW, Camplo M. Cationic nucleoside lipids based on a 3-nitropyrrole universal base for siRNA delivery. Bioconjug Chem. 2009;20:193–196. doi: 10.1021/bc800432n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghosn B, Kasturi SP, Roy K. Enhancing polysaccharide-mediated delivery of nucleic acids through functionalization with secondary and tertiary amines. Curr Top Med Chem. 2008;8:331–340. doi: 10.2174/156802608783790947. [DOI] [PubMed] [Google Scholar]
- 31.Islam RU, Hean J, van Otterlo WA, de Koning CB, Arbuthnot P. Efficient nucleic acid transduction with lipoplexes containing novel piperazine- and polyamine-conjugated cholesterol derivatives. Bioorg Med Chem Lett. 2009;19:100–103. doi: 10.1016/j.bmcl.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 32.Kim Y, Tewari M, Pajerowski JD, Cai S, Sen S, Williams J, Sirsi S, Lutz G, Discher DE. Polymersome delivery of siRNA and antisense oligonucleotides. J Control Release. 2009;134:132–140. doi: 10.1016/j.jconrel.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bouxsein NF, McAllister CS, Ewert KK, Samuel CE, Safinya CR. Structure and gene silencing activities of monovalent and pentavalent cationic lipid vectors complexed with siRNA. Biochemistry. 2007;46:4785–4792. doi: 10.1021/bi062138l. [DOI] [PubMed] [Google Scholar]
- 34.Grayson AC, Doody AM, Putnam D. Biophysical and structural characterization of polyethylenimine-mediated siRNA delivery in vitro. Pharm Res. 2006;23:1868–1876. doi: 10.1007/s11095-006-9009-2. [DOI] [PubMed] [Google Scholar]
- 35.Mao S, Neu M, Germershaus O, Merkel O, Sitterberg J, Bakowsky U, Kissel T. Influence of polyethylene glycol chain length on the physicochemical and biological properties of poly(ethylene imine)-graft-poly(ethylene glycol) block copolymer/SiRNA polyplexes. Bioconjug Chem. 2006;17:1209–1218. doi: 10.1021/bc060129j. [DOI] [PubMed] [Google Scholar]
- 36.Marty R, N'Soukpoe-Kossi CN, Charbonneau D, Weinert CM, Kreplak L, Tajmir-Riahi HA. Structural analysis of DNA complexation with cationic lipids. Nucleic Acids Res. 2009;37:849–857. doi: 10.1093/nar/gkn1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pozharski E, MacDonald RC. Thermodynamics of cationic lipid-DNA complex formation as studied by isothermal titration calorimetry. Biophys J. 2002;83:556–565. doi: 10.1016/S0006-3495(02)75191-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prevette LE, Kodger TE, Reineke TM, Lynch ML. Deciphering the role of hydrogen bonding in enhancing pDNA-polycation interactions. Langmuir. 2007;23:9773–9784. doi: 10.1021/la7009995. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.