Abstract
Cholesterol homeostasis is critical for cell viability and proliferation. The sterol regulatory element-binding protein (SREBP) pathway is crucial for the maintenance of cholesterol homeostasis. This pathway is controlled by cholesterol and cholesterol-derived oxysterols. J774 cells cannot convert desmosterol into cholesterol, a defect resulting from the absence of mRNA for sterol-Δ24-reductase. Using J774 cells, we addressed the capacity of desmosterol to replace cholesterol in sustaining cell proliferation and regulating the SREBP pathway. J774 cells were able to grow indefinitely after the virtually total replacement of cholesterol by desmosterol (J774-D cells). Inhibition of sterol biosynthesis with lovastatin suppressed J774-D cell proliferation. Desmosterol prevented this effect but its analogue, cholest-5,22-trans-dien-3β-ol, did not. Addition of desmosterol inhibited processing of SREBP-1 and -2 as well as reduced the expression of SREBP-targeted genes. As occurs in cholesterol-containing cells, 25-hydroxycholesterol was more potent than desmosterol or cholesterol in suppressing these processes. Moreover, desmosterol addition enhanced the expression of Abca1 and Srebf1c, two liver X receptor (LXR)-targeted genes. To test the ability of endogenously produced desmosterol to regulate gene expression, J774-D cells were pretreated with lovastatin to inhibit sterol biosynthesis. After removal of the inhibitor the expression of SREBP-targeted genes decreased and that of an LXR-targeted gene increased, reaching control levels. Our results demonstrate that the virtually complete replacement of cholesterol by desmosterol is compatible with cell growth and the functioning of the SREBP pathway. In these cells, desmosterol suppresses SREBP processing and targeted gene expression, and it is especially effective activating LXR-targeted genes.
Keywords: cell proliferation; sterol biosynthesis; cholest-5,22-trans-dien-3β-ol; 25-hydroxycholesterol; SREBP-targeted genes; LXR-targeted genes
INTRODUCTION
As a key molecule for maintaining cell membrane architecture and biophysical properties, cholesterol is essential for mammalian cell viability and function. Cell proliferation requires membrane synthesis and this, in turn, depends critically on the availability of cholesterol. We have demonstrated that cholesterol is required for the G2-to-M phase transition and completion of mitosis [1-4]. On the other hand, excess unesterified cholesterol is cytotoxic [5]. Therefore, cholesterol homeostasis is subjected to strict control, and cells have developed a complex regulatory system to maintain the cellular content of cholesterol within a narrow range [6]. Moreover, in order to preserve the biophysical properties of cell membranes, the metabolism of cholesterol is regulated in concert with that of fatty acids and acylglycerols. The sterol regulatory element-binging protein (SREBP) family of transcription factors is crucial for the control of lipid homeostasis [7, 8] and a functional SREBP pathway is required for cell survival and proliferation [8, 9]. Cholesterol itself and certain cholesterol-derived oxysterols, such as 25-hydroxycholesterol (25HC), control this regulatory system. In the present study we focussed on these two essential and tightly linked functions of cholesterol, namely, support of cell proliferation and regulation of the SREBP pathway.
The cholesterol required for cell proliferation is either obtained from plasma lipoproteins or synthesized intracellularly from acetyl-coenzyme A. 3-hydroxy-3-methylglutaryl coenzyme A (HMGCoA) reductase—the target of the hypocholesterolemic statin drugs —catalyzes the conversion of HMG-CoA into mevalonate, the rate-limiting step in cholesterol biosynthesis. There is a stringent requirement for cholesterol to support optimal mammalian cell growth [3, 10-12]. Some sterols, such as certain cholesterol precursors and phytosterols, can support cell growth provided that a suboptimal amount of cholesterol is also available [3, 12-14], a feature that is reminiscent of the synergism between sterols first observed in lower eukaryotes [15, 16]. This is interpreted as a dual role of sterols in cell growth: a bulk membrane requirement, which can be satisfied by different sterols with broad specificity, and a regulatory role that is highly specific and fulfilled by small amounts of the selected sterol, cholesterol in mammalian cells.
Rothblat et al reported that L-cell mouse fibroblasts, which are defective in cholesterol biosynthesis and accumulate desmosterol, are able to survive and proliferate indefinitely in lipoprotein-deficient serum [17]. This indicated that in L-cells cholesterol can be replaced by desmosterol. Desmosterol, the immediate unsaturated precursor, is converted to cholesterol through a reaction catalyzed by sterol-Δ24-reductase (DHCR24, also known as Seladin-1). Desmosterol only differs from cholesterol in the presence of a double bond in the side chain at C24. Nevertheless, it has been suggested that, aside from the structural stringency of sterols, the ability of cells to compensate through changes in the composition of membrane lipids is also important in order to adapt to growth without cholesterol [13]. Whether this adaptability is a peculiarity of certain cells, such as fibroblasts, or it is a common phenomenon is unknown.
The SREBP family consists of three members, designated SREBP-1a, SREBP-1c and SREBP-2, which are encoded by two genes, Srebf1 – that gives rise to SREBP-1a and SREBP-1c - and Srebf2. The SREBPs are synthesized as precursor proteins that are embedded in the membrane of the endoplasmic reticulum (ER). In this location, SREBPs bind to the SREBP cleavage-activating protein (Scap), which serves as both a cholesterol sensor and an escort protein. When cells are depleted of cholesterol, Scap escorts SREBP from the ER to the Golgi apparatus via COPII-coated vesicles. In the Golgi apparatus, SREBPs are cleaved sequentially by the Site-1 and Site-2 proteases, releasing the NH2-terminal domain of the protein, which is the active transcription factor. This mature form of SREBPs enters the nucleus and binds to specific nucleotide sequences known as sterol regulatory elements (SRE) located in the promoters of target genes, thus activating their transcription. When the cholesterol content of the ER membrane rises, cholesterol binds to Scap and induces a conformational change that allows Scap to bind to Insig-1 or Insig-2, which are resident membrane proteins of the ER. This interaction results in the retention of Scap/SREBP complexes in the ER by preventing their incorporation into transport vesicles and subsequent SREBP cleavage in the Golgi apparatus [8, 18]. Different sterols can induce a conformational change in Scap in cell membrane suspensions [19] and/or can inhibit SREBP processing in intact cells [20]. Certain cholesterol-derived oxysterols, such as 25HC, are potent inhibitors of SREBP processing. In this case, the inhibition is initiated by its binding to Insig but not to Scap [19, 21], causing Insig to bind to Scap and thereby blocking the export of Scap/SREBP complexes from the ER [20, 22].
As master regulators of lipid homeostasis, SREBPs induce the transcription of many genes involved in both cholesterol and fatty acid metabolism. SREBP-1a and, especially, SREBP-1c preferentially activate genes for fatty acid biosynthesis and their incorporation into triglycerides and phospholipids, whereas SREBP-2 principally activates genes involved in cholesterol biosynthesis and uptake [23, 24]. Moreover, SREBPs stimulate the expression of genes of the regulatory machinery, such as those encoding SREBP-1, SREBP-2 and Insig-1 [18, 23, 24].
Recently, DHCR24 and its substrate, desmosterol, have generated great interest due to the down-regulation of DHCR24 in regions of the brain affected by Alzheimer's disease [25]. By contrast with other cholesterol analogues, desmosterol is an abundant component of the membrane of certain mammalian cells such as spermatozoa [26] and astrocytes [27], and high desmosterol levels have been detected in developing brain [28]. Mice with a targeted disruption of DHCR24 (Dhcr24−/−) were viable but infertile, although they had no gross morphological defects [29]. This is in marked contrast to the multiple severe inborn defects appearing in human subjects with desmosterolosis, who carry a mutation in DHCR24 [30, 31]. It should be noted, however, that analysis of sterol content in embryos of Dhcr24−/− mice revealed that the animals represent a low-cholesterol, high-desmosterol model rather than being cholesterol-free [29, 32]. Furthermore, there is no evidence that either of the two cases of desmosterolosis reported to date were cholesterol-free during development [30, 31]. Thus, a possible contribution of cholesterol to cell proliferation and other cellular processes in these situations can not be ruled out. J774 mouse monocyte/macrophages have been reported to be unable to convert desmosterol into cholesterol [33]. Using this cell line, in the present study we addressed the capacity of desmosterol to replace cholesterol in sustaining cell proliferation and regulating the SREBP pathway.
MATERIALS AND METHODS
Materials
J774A.1 mouse monocyte/macrophages (ATCC TIB-67) and HL-60 human promyeloblasts (ATCC CCL-240) were obtained from the American Type Culture Collection. Fetal bovine serum (FBS), RPMI, antibiotics and L-glutamine were purchased from Gibco BRL. Lipoprotein deficient serum (LPDS) was obtained by ultracentrifugation of FBS at a density of 1.21 kg/L. DCCM-1 was purchased from Biological Industries. Lovastatin was from Merck, Sharp and Dohme. Cholesterol, mevalonate and 5-fluorodeoxyuridine were purchased from Sigma and desmosterol, cholest-5,22-trans-dien-3β-ol and 25-hydroxycholesterol (25HC) were from Steraloids. Hydroxypropyl-β-cyclodextrin (HPCD) and methyl-β-cyclodextrin (MCD) were obtained from Sigma. [3H-methyl]thymidine (5 Ci/mmol) and [2-14C]acetate (53 mCi/mmol) were from Amersham Biosciences. Multiscreen-HV culture plates were purchased from Millipore. All other chemicals were of analytical grade.
Cell cultures
J774 and HL-60 cells were maintained in RPMI containing 10% FBS, 100 U/ml penicillin, 100 U/ml streptomycin and 10 μg/ml gentamicin at 37 °C in a humidified atmosphere containing 5% CO2. J774 cells were adapted to grow in the cholesterol-free medium DCCM-1 supplemented with 2 mM L-glutamine and the above mentioned antibiotics. For this, the regular maintenance medium was gradually replaced by increasing the proportion of DCCM-1 every two days, until complete withdrawal of the maintenance medium was achieved on the eighth day. Thereafter, the cells were grown in DCCM-1 and passaged every three days.
Proliferation assays
DCCM-1 was replaced with fresh medium and the cells were plated in sterile 96-well Multiscreen-HV plates at 2 × 104 cells in a final volume of 225 μL. The medium was supplemented with lovastatin (final concentration, 0.5 μM) dissolved in dimethylsulfoxide (final concentration, 0.044%), pure sterols dissolved in ethanol (final concentration, 0.44%), and/or the corresponding vehicles, as indicated. At 71 h of incubation, the medium was supplemented with 10 mM 5-fluorodeoxyuridine and 1 h later with 0.5 μCi [3H-methyl]thymidine. Following a further 18 h of incubation the cells were processed for the measurement of radioactivity incorporated into DNA, as described previously [1]. Cells (2.5 × 105/ml) were also incubated in 12-well plates in the indicated conditions for 72 h to determine cell number and viability by trypan blue dye exclusion using a hemocytometer.
Analysis of sterol biosynthesis and content by HPLC
To study sterol biosynthesis, 107 cells were incubated in 10 ml of the indicated medium supplemented with 40 μCi [2-14C]acetate for 10 h (in RPMI containing 10% LPDS) or 72 h (in DCCM-1). Subsequently, cells were washed twice with PBS and lysed with 0.5 ml of 10% KOH. Protein concentration was determined in an aliquot as described above. The nonsaponifiable lipid fraction was isolated and analyzed by HPLC and radioactivity counting as described previously [11]. Briefly, ergosterol was added to the samples as an internal standard and the lipid fraction was extracted with chloroform/methanol (2:1, v/v). Lipids were subjected to saponification and the nonsaponifiable fraction was extracted and resuspended in hexane. Sterol separation was accomplished by reverse-phase HPLC with a Luna 5 μm C18 column (Phenomenex). The eluate was monitored simultaneously by UV absorption using a Beckman 168 variable-wavelength detector (Beckman Instruments) and online radioactivity detection using an LB-506 C-1 detector (Berthold). Peaks were identified by comparing their retention time and UV spectrum with those of pure sterol standards. Sterols were quantified by measurement of the area under the peak at 206 nm. The lowest limit of detection (LOD) for cholesterol was less than 60 ng/ml.
Analysis of Dhcr24 expression
Total RNA was extracted with TriReagent (Sigma). For Northern blot analysis, 80 μg of total RNA were subjected to electrophoresis in a 1.2% agarose gel containing 2.2 M formaldehyde, transferred to Hybond-N+ membranes (Amersham) and cross linked with UV light. The blots were then hybridized with Dhcr24 and Gapdh cDNA probes labelled with digoxygenin using the DIG High Prime DNA Labelling and Detection Starter Kit (Roche). The detection of the probes was carried out with an alkaline phosphatase-labelled antibody against digoxygenin and developed by chemiluminescence following the instructions given by the manufacturer. The cDNA probes were prepared by RT-PCR (see below) using primers no. 2 and 3 for Dhcr24 detection and primers no. 9 and 10 for Gapdh detection (Supplementary Table S1).
For RT-PCR analysis of Dhcr24, 2 μg of total RNA were reverse-transcribed with 200 U of M-MLV-RT enzyme (Promega) using random hexamers (Promega). Two μl of cDNA were then amplified by PCR with 2.5 U of TaqGold DNA Polymerase (Applied Biosystems) by using primers no. 1 and 2 (Supplementary Table S1). Gapdh was used as an internal control (primers no. 9 and 10, Supplementary Table S1). To amplify Dhcr24 cDNA fragments of increasing length, total RNA was reverse-transcribed as above, and cDNA was subjected to a set of PCRs in which a sense primer whose sequence was comprised in exon 1 of Dhcr24 mRNA (primer no. 3, Supplementary Table S1) was combined with different antisense primers whose sequences were located in exons 2 to 7 (primers no. 4-8, respectively; Supplementary Table S1). The PCR products were resolved in a 1% agarose gel and were photographed using a gel documentation system (Kodak 1D Software).
Analysis of SREBP processing
On day 0, cells were plated at 106 cells/well in 6-well plates. On day 1, they were washed with PBS and switched to medium containing 2.5% LPDS (J774 and L cells) or to fresh DCCM-1 (J774-D cells), both supplemented with 1% (HPCD, Sigma), 2.5 μM lovastatin and 0.1 mM mevalonate, and incubated for 1 h. Next, cells were washed twice with PBS and switched to the same medium but without HPCD in the absence or presence of 0.5 μg/mL 25HC dissolved in ethanol, 25 μg/mL cholesterol or desmosterol dissolved in ethanol or 11.6 μg/mL cholesterol or desmosterol complexed with MCD. The sterol-MCD complexes were prepared as described elsewhere [19]. After incubation for 5 h, cells were washed with PBS, harvested and processed for western blot analysis and RNA isolation.
Western blot analysis
For LDL receptor detection cells were lysed in 20 mM Tris-HCl buffer, pH 8, containing 120 mM KCl, 1 mM DTT, 1 mM Na2-EDTA, 2 mM EGTA, 0,1 % Triton X-100, 0,5 % Nonidet P40, 1 mM benzamidine, 10 μg/mL antipain, 1 μg/mL leupeptin, 40 μg/mL aprotinin, 100 mM NaF, 20 mM sodium molibdate, 20 mM β-glycerophosphate, 2 mM sodium ortovanadate and 1 mM PMSF and subsequently sonicated. For SREBP processing analysis, the above mentioned lysis buffer was supplemented with 1 μg/mL of the caspase-3 inhibitor Ac-DMQD-CHO (Alexis), and cells were passed through a 22-gauge needle 10 times. For HMG-CoA reductase detection the lysis buffer was 1M PBS, pH 7.4, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 0.01% sodium azide, 5 mM Na2-EDTA and 0.1 M NaCl. After passing the cells through a 22-gauge needle 10 times, proteins were precipitated with trichloroacetic acid and resuspended in Laemmli sample buffer [34]. The protein concentration of each cell lysate was measured by using the BCA kit (Pierce), and equal amounts of protein were subjected to 10% SDS-PAGE. The proteins were transferred to a nitrocellulose filter and probed with rabbit polyclonal antibodies against LDL receptor [35], HMG-CoA reductase [36], SREBP-1 [37], SREBP-2 [38], or actin as the loading control (Amersham). Bound antibodies were visualized by chemiluminescence (ECL from Amersham or Inmun-Star HRP from Bio-Rad) and exposure to X-ray film (Hyperfilm, Amersham). Optical density of the bands was quantified by using the ImageQuant TL software (Amersham).
Quantitative real-time PCR
Total RNA was extracted with TriReagent (Sigma). Two μg of total RNA were reverse-transcribed with 200 U of M-MVL reverse transcriptase enzyme (Promega) using oligo(dT) primers (Promega). The cDNA was then subjected to quantitative real-time PCR using the FastStart DNA Master SYBR Green I kit and LightCycler 2.0 equipment, and data were analyzed with the manufacturer's software version 4.05 (Roche Diagnostics GmbH). Analyses were performed in duplicate using the cyclophilin B gene (Cypb) as the invariant control. Primer sequences for the different genes analyzed are shown in Supplementary Table S1.
Statistical analyses
Statistical comparisons were performed by paired t-test using SigmaStat, version 2.3 (Jandel Corporation, San Rafael, CA). Statistical significance was set at P < 0.05.
RESULTS
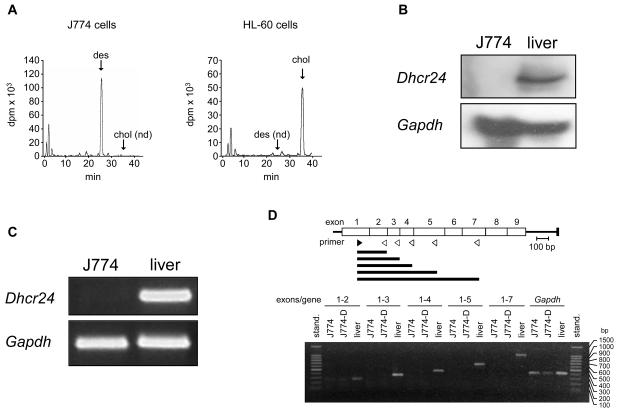
It has been reported that J774 cells are unable to synthesize cholesterol and instead accumulate desmosterol, suggesting a deficiency of DHCR24 activity [33]. This was confirmed in the J774 cells used in the present study by analyzing the incorporation of [14C]acetate into sterols. As shown in Figure 1A, HL-60 cells, which were used as a reference, accumulated most of the radioactivity in cholesterol. By contrast, no incorporation of 14C into cholesterol was detected in J774 cells; instead 14C accumulation was observed in desmosterol (Figure 1A).
Figure 1.
Sterol biosynthesis and Dhcr24 expression in J774 cells. (A) [14C]Acetate incorporation into sterols in J774 and HL-60 cells. Cells were incubated in RPMI containing 10% LPDS supplemented with 40 μCi of [14C]acetate for 10 h and the cell extracts containing the nonsaponifiable lipid fraction were analyzed by reverse-phase HPLC and in-line radioactivity counting. The arrows indicate the elution times of desmosterol (des), and cholesterol (chol). nd, not detected. (B) Northern blot analysis of RNA of J774 cells and mouse liver. Total RNA was subjected to electrophoresis and blot hybridization with digoxygenin-labelled cDNA probes for mouse Dhcr24 and mouse Gapdh. (C) RT-PCR analysis of RNA of J774 cells and mouse liver for detection of Dhcr24 and Gapdh. (D) RT-PCR analysis of RNA of J774 cells and mouse liver for detection of Dhcr24 cDNA fragments of increasing length and comprising regions expanding from exon 1 up to exon 7, as indicated. J774-D cells are J774 cells grown in DCCM-1 (see text). The upper panel indicates the position of the sense (no. 3 in Supplementary Table S1) and antisense (no. 4-8 in Supplementary Table S1) primers (filled and empty arrowheads, respectively) used for the amplifications and the resulting cDNA fragments (horizontal bars). stand., standard; bp, base pairs.
To explore the defect underlying the deficiency of DHCR24 activity, we first analyzed RNA extracts from J774 cells and mouse liver as control by Northern blot. As shown in Figure 1B, no Dhcr24 mRNA was detected in J774 cells. Then, the mRNA sequence corresponding to exons 8 and 9 of Dhcr24 was subjected to RT-PCR. Again, no cDNA was detected (Figure 1C). To further increase the sensitivity of the assay, quantitative real-time RT-PCR was performed to amplify different regions of Dhcr24, but no amplification product was obtained from J774 cells in any case (results not shown). Next, we tested for possible truncated mRNAs. Total RNA was reverse-transcribed and cDNA fragments of increasing length were amplified by combining a sense primer located in exon 1 of Dhcr24 mRNA with different antisense primers located in exons 2 to 7. In contrast to liver samples, only a faint band was detectable in J774 cells. This band comprised sequences up through exon 2. No detectable amplification product was obtained for any of the longer fragments studied (Figure 1D). These results demonstrate that J774 cells are defective in the production of a normal Dhcr24 transcript.
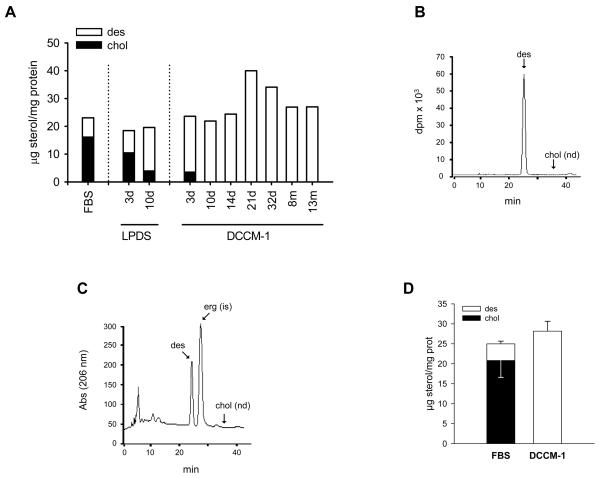
Next, we analyzed whether desmosterol is able to substitute for cholesterol for the survival and proliferation of J774 cells. Cholesterol was the predominant sterol in cells growing in 10% FBS. After 3 days growing in 10% LPDS, desmosterol became the major sterol (Figure 2A). Despite the inability of J774 cells to synthesize cholesterol, cholesterol levels were still approximately one-fifth of total sterol content after 10 days in 10% LPDS (Figure 2A). This observation may be explained by the low concentration of cholesterol usually present in 10% LPDS (approximately 0.2 μg/ml). To test whether biosynthetic desmosterol could completely replace cell cholesterol in viable J774 cells, the regular maintenance medium containing 10% FBS was replaced with a cholesterol-free medium (DCCM-1). As shown in Figure 2A, J774 cells have been growing in this medium, with passages every three days, for as long as 13 months, the time at which the culture was interrupted. We designated these J774-D cells. They were unable to express the normal Dhcr24 transcript (Figure 1D) or to synthesize cholesterol. Instead, they accumulated biosynthetic desmosterol, as measured by incubation with [2-14C]acetate for 72 h (Figure 2B). Consistently, cellular cholesterol was undetectable (less than 0.017 % of total sterols in J774 cells) from 10 days after the complete withdrawal of serum; desmosterol was the only quantifiable sterol (Figures 2A and 2C). Replacement of the regular maintenance medium was performed on multiple occasions and in each case the cells successfully adapted to growth in cholesterol-free medium, indicating that this was not the result of selecting a specific cell mutant. On average, cellular levels of desmosterol in cells growing in DCCM-1 were similar to the sum of cholesterol plus desmosterol levels in cells growing in 10% FBS (Figure 2D). Therefore, J774 cells were able to grow indefinitely after the virtually complete replacement of cholesterol by desmosterol. These cells were used for subsequent studies at least three weeks after complete withdrawal of FBS.
Figure 2.
Sterol content and biosynthesis in J774 cells adapted to grow in a cholesterol-free medium. (A) Cholesterol and desmosterol content of cells at different times of growth after the complete replacement of RPMI containing 10% FBS with either RPMI containing 10% LPDS or DCCM-1. (B and C) HPLC profiles of radioactivity (B) and absorbance at 206 nm (C) of sterols from cells at day 21 of growth in DCCM-1. DCCM-1 was supplemented with 40 μCi of [14C]acetate for 3 days and the cell extracts containing the nonsaponifiable lipid fraction were analyzed by reverse-phase HPLC and in-line radioactivity counting. The arrows indicate the elution times of desmosterol (des), cholesterol (chol) and ergosterol (erg). d, days; m, months; is, internal standard; nd, not detected. (D) Average cholesterol and desmosterol levels in cells maintained in RPMI containing 10% FBS or at days 21 to 32 of growth in DCCM-1. Bars show the mean and whiskers indicate the SEM of data from seven separate cell cultures.
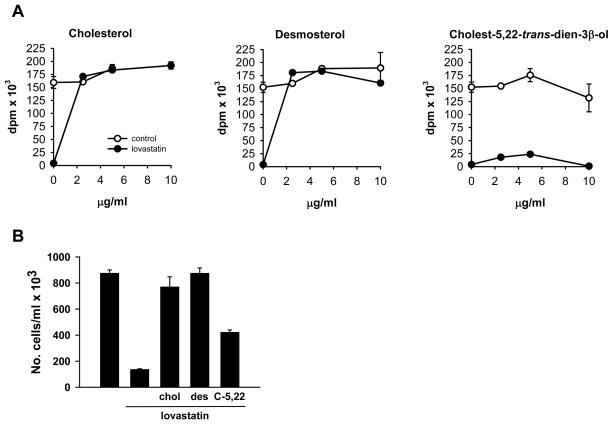
To assess the effect of the addition of cholesterol or desmosterol on the proliferation of J774-D cells, incorporation of [3H-methyl]thymidine was analyzed in DCCM-1 medium. The addition of either cholesterol or desmosterol up to a final concentration of 10 μg/ml slightly increased [3H-methyl]thymidine incorporation in the control condition (Figure 3A). Treatment with 0.5 μM lovastatin completely abolished cell proliferation (Figure 3A). However, a dose of 2.5 μg/ml of either cholesterol or desmosterol was sufficient to completely overcome the inhibitory effect of the drug (Figure 3A). Similar results were obtained by counting the number of viable cells (Figure 3B), indicating that the [3H-methyl]thymidine incorporation assay reflected actual effects on cell proliferation. These findings indicate that desmosterol can sustain J774 cell proliferation in the absence of cholesterol.
Figure 3.
Effect of cholesterol, desmosterol and cholest-5,22-trans-dien-3β-ol on the proliferation of lovastatin-treated J774-D cells. (A) [3H-methyl]thymidine incorporation into DNA of cells incubated in DCCM-1 in the absence or presence of 0.5 μM lovastatin alone or in combination with the indicated concentrations of cholesterol, desmosterol or cholest-5,22-trans-dien-3β-ol. Data correspond to the mean ± SEM of triplicates. (B) Cells were incubated in the presence or absence of 0.5 μM lovastatin alone or in combination with 2.5 μg/ml cholesterol, desmosterol or cholest-5,22-trans-dien-3β-ol. At 72 h of incubation, the cells were harvested for counting of viable cells. Data correspond to the mean ± SEM of duplicates. Data in A and B correspond to a representative experiment out of five. chol, cholesterol, des, desmosterol; C-5,22, cholest-5,22-trans-dien-3β-ol.
Cholest-5,22-trans-dien-3β-ol, a structural analog of desmosterol that is not converted into cholesterol [39], has also been shown to support the growth of S2 cells, a sterol auxotrophic cell line derived from L cells and which fails to demethylate lanosterol in the course of cholesterol biosynthesis [13]. Given the ability of J774-D cells to proliferate with only desmosterol, we tested the effect of cholest-5,22-trans-dien-3β-ol on proliferation of J774-D cells. As shown in Figure 3, this sterol partially prevented the inhibitory effect of lovastatin, but it was unable to restore the proliferation rate to control values. Thus, cholest-5,22-trans-dien-3β-ol was not equivalent to its analogue desmosterol in supporting J774-D cell proliferation.
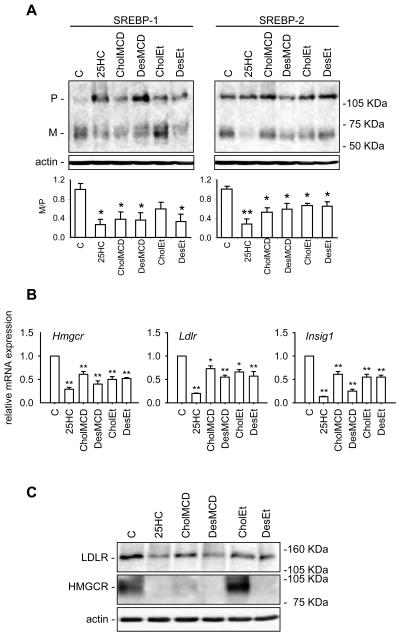
A functional SREBP pathway is required for cell survival and proliferation [9]. The SREBP family of transcription factors regulates the expression of many genes involved in lipid homeostasis in response to changes in cellular cholesterol content. Thus, we next analyzed the ability of desmosterol, cholesterol and 25HC to inhibit the SREBP pathway in J774-D cells. For this, we tested the short-term effect of these sterols on SREBP-1 and -2 processing and on SREBP-targeted gene expression when added to sterol-deprived J774-D cells. As shown in Figure 4A, 25HC markedly decreased the ratio between the mature (M) and the precursor (P) forms of both SREBP-1 and -2, indicating an inhibition of their processing as compared with the controls. Accordingly, the mRNA levels of HMG-CoA reductase (Hmgcr), the LDL receptor (Ldlr) and Insig1 were lower in 25HC-treated cells than in controls (Figure 4B). Such effects of 25HC on Hmgcr and Ldlr mRNAs were paralleled by reductions of HMG-CoA reductase and LDL receptor protein levels (Figure 4C). Cholesterol and desmosterol were added to the cells dissolved in ethanol, as was 25HC, or complexed with MCD to increase their solubility. Cholesterol, either dissolved in ethanol or complexed with MCD, inhibited SREBP-1 and SREBP-2 processing (Figure 4A) and decreased Hmgcr, Ldlr and Insig1 mRNA levels (Figure 4B), although less potently than 25HC. Cholesterol also caused a reduction of HMG-CoA reductase and LDL receptor protein levels, although the effect on the former protein was null when cholesterol was added in ethanol (Figure 4C). Desmosterol, added in any of the two vehicles, suppressed SREBP-1 and SREBP-2 processing and the expression of the above mentioned genes and proteins at least as efficiently as cholesterol did (Figure 4).
Figure 4.
Effect of 25HC, cholesterol and desmosterol on the SREBP pathway in sterol deprived J774-D cells. Cells were incubated in DCCM-1 supplemented with 1% HPCD, 2.5 μM lovastatin and 100 μM mevalonate for 1 h. Then, cells were switched to the same medium but without HPCD and containing or not (control, C) 0.5 μg/mL 25HC dissolved in ethanol, 11.6 μg/mL cholesterol (CholMCD) or desmosterol (DesMCD) complexed with MCD or 25 μg/mL cholesterol (CholEt) or desmosterol (DesEt) dissolved in ethanol, and incubated for additional 5 h. (A) Western blot analysis of SREBP-1 and SREBP-2 processing. Fifty μg of protein were subjected to SDS-PAGE electrophoresis. The precursor and mature form of SREBPs are denoted P and M, respectively. The results correspond to a representative experiment out of three. Lower panels represent the ratio between the respective optical densities of M and P bands; the ratio obtained for the control condition has been arbitrarily defined as 1. Bars show the mean and whiskers indicate the SEM of data from three experiments. *, P<0.05; and **, P<0.01 vs. the control condition by paired t-test. (B) Hmgcr, Ldlr and Insig1 mRNA levels. The results are expressed as the relative amount of mRNA compared to the level in control condition without sterol. Bars show the mean and whiskers indicate the SEM of data from three experiments. *, P<0.05; and **, P<0.01 vs. the control condition by paired t-test. (C) Western blot analysis of HMG-CoA reductase and LDLR protein levels. Fifty μg of protein were subjected to SDS-PAGE electrophoresis. The results correspond to a representative experiment out of three.
The inhibition of SREBP processing and targeted gene expression by desmosterol was also observed in sterol-deprived parental J774 cells (Supplementary Figure S1), indicating that such responses of J774-D cells to desmosterol were not a consequence of adapting the cells to growth in the cholesterol-free medium. We then questioned whether desmosterol addition would be able to regulate the SREBP pathway in L-cell mouse fibroblasts, another cell model defective in DHCR24 activity [17]. Similarly to J774-D and parental J774 cells, in sterol-deprived L cells, desmosterol was at least as efficient as cholesterol suppressing SREBP-1 and SREBP-2 processing as well as Hmgcr, Ldlr and Insig1 expression (Supplementary Figure S2). Together, these findings demonstrate that desmosterol inhibits SREBP processing and the expression of target genes, and that cholesterol-free J774 cells retain the ability to respond to sterols through the suppression of the SREBP pathway.
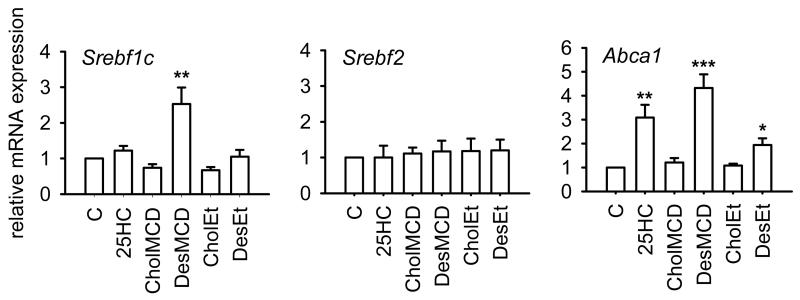
A noticeable finding arising from the analysis of SREBP processing in J774-D (Figure 4A), parental J774 (Supplementary Figure S1A) and L cells (Supplementary Figure S2A) is that the absolute amount of the precursor form of SREBP-1 (P-SREBP-1) was markedly increased by the addition of 25HC, cholesterol or desmosterol as compared with control cells, whereas the absolute amount of P-SREBP-2 was not changed appreciably by sterol addition. Such an accumulation of P-SREBP-1 may be a direct consequence of the sterol-induced inhibition of its cleavage. However, it may also result from an increased expression of the SREBP-1c isoform through the activation of the liver X receptor (LXR), which is known to stimulate Srebf1c transcription [40]. We then analyzed the effect of those sterols on the levels of Srebf1c mRNA and, for comparison, those of Srebf2 mRNA. As shown in Figure 5, when desmosterol was delivered complexed with MCD it significantly increased Srebf1c mRNA levels as compared with control cells, whereas no effect was produced by other additions. Neither addition influenced Srebf2 mRNA levels (Figure 5). The expression of another LXR-targeted gene, Abca1, was also analyzed. It was found that 25HC and desmosterol, when added either dissolved in ethanol or complexed with MCD, increased Abca1 mRNA levels (Figure 5). Therefore, desmosterol can stimulate the expression of LXR-targeted genes in J774-D cells.
Figure 5.
Effect of 25HC, cholesterol and desmosterol on Srebf1c, Srebf2 and Abca1 mRNA levels in sterol deprived J774-D cells. Cells were treated as described in Figure 4. The results are expressed as the relative amount of mRNA compared to the level in control condition without sterol (C). Bars show the mean and whiskers indicate the SEM of data from three experiments. *, P<0.05; **, P<0.01; and ***, P<0.001 vs. control condition by paired t-test.
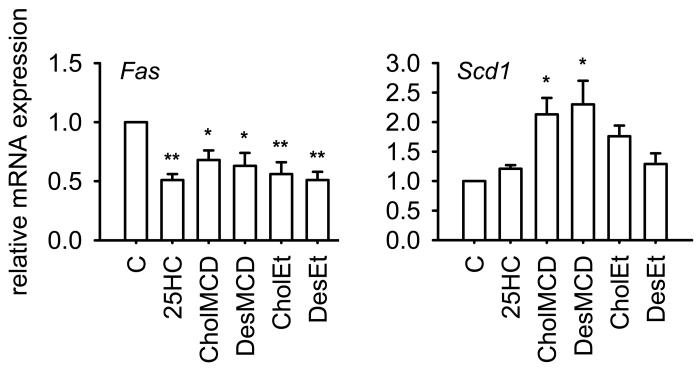
The fact that desmosterol increased Srebf1c expression while inhibiting SREBP-1 processing led us to ask what would be the net effect on the expression of genes required for fatty acid biosynthesis, which are characteristic SREBP-1c targets [23]. Thus, we addressed the effect of desmosterol addition on the expression of fatty acid synthase (Fas) and stearoyl-CoA desaturase-1 (Scd1) in J774-D cells. As shown in Figure 6, desmosterol, 25HC and cholesterol decreased Fas mRNA levels. By contrast, the different sterols did not decrease Scd1 mRNA levels, but, instead, desmosterol and cholesterol significantly enhanced it when added complexed with MCD (Figure 6).
Figure 6.
Effect of 25HC, cholesterol and desmosterol on Fas and Scd1 mRNA levels in sterol deprived J774-D cells. Cells were treated as described in Figure 4. The results are expressed as the relative amount of mRNA compared to the level in control condition without sterol (C). Bars show the mean and whiskers indicate the SEM of data from three experiments. *, P<0.05; and **, P<0.01 vs. control condition by paired t-test.
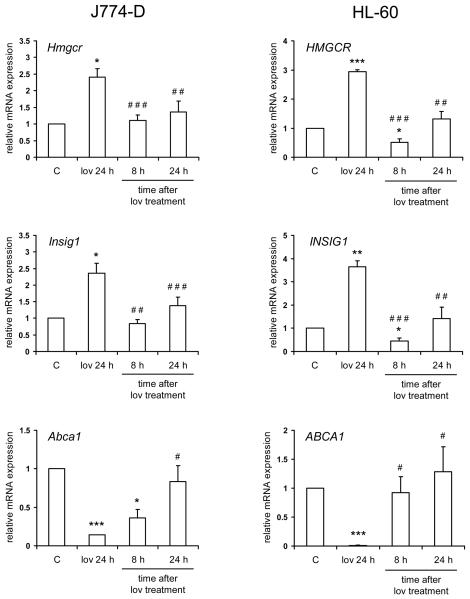
Finally, we tested the ability of endogenously produced desmosterol to regulate the expression of SREBP and LXR gene targets. For this, J774-D and HL-60 cells, as a control, were treated with 0.25 μM lovastatin for 24 h, which was then removed to promote a burst of de novo sterol biosynthesis [41]. The expression of Hmgcr, Insig1 and Abca1 was studied up to 24 h after the removal of the drug. The results are shown in Figure 7. In J774-D cells, lovastatin increased the mRNA levels of Hmgcr and Insig1, and both returned to control levels by 8 h after lovastatin removal. As regards Abca1, lovastatin decreased its mRNA levels, which increased up to control levels after lovastatin removal. Qualitatively similar results were obtained in HL-60 cells for HMGCR, INSIG1 and ABCA1, respectively (Figure 7). These findings suggest that endogenously produced desmosterol is able to regulate the expression of SREBP and LXR gene targets in a similar manner to that of endogenously produced cholesterol.
Figure 7.
Effect of endogenously produced desmosterol on the expression of SREBP and LXR gene targets. J774-D and HL-60 cells (5×105 cells/ml) were preincubated in the absence (control, C) or presence of 0.25 μM lovastatin for 24 h. Media with lovastatin (lov) were then removed and the cells were washed with PBS and incubated in fresh media without lovastatin for additional 8 or 24 h. The levels of Hmgcr, Insig1 and Abca1 mRNA were measured. The results are expressed as the relative amount of mRNA compared to the level in control condition. Bars show the mean and whiskers indicate the SEM of data from four experiments. *, P<0.05; **, P<0.01 and ***, P<0.001 vs. control; and #, P<0.05; ##, P<0.01 and ###, P<0.001 vs. lovastatin pretreatment by paired t-test.
DISCUSSION
J774 cells are unable to synthesize cholesterol and instead accumulate desmosterol. Present results show that the lack of cholesterol biosynthesis in J774 cells results from their inability to produce a normal Dhcr24 transcript. This leads to the complete absence of enzyme, and is consistent with the virtually total replacement of cholesterol by desmosterol in J774-D cells when growing in a cholesterol-free medium. Using this cell model we addressed the capacity of desmosterol to substitute for cholesterol in two essential and tightly linked functions in mammalian cells, namely, the maintenance of cell growth and the control of regulatory responses involved in the maintenance of lipid homeostasis.
The ability of J774-D cells to grow indefinitely in a cholesterol-free medium demonstrates that desmosterol is sufficient to support cell survival and proliferation. Therefore, desmosterol can perform the dual role attributed to cholesterol in mammalian cell growth, namely, the formation of membranes, an ability shared by different sterols, and the highly specific regulatory function, which is fulfilled by small amounts of the species' selected sterol, cholesterol in mammalian cells [3, 4, 12, 13]. This finding extends previous results obtained in L-cell mouse fibroblasts [13, 17] and indicates that the ability of desmosterol to substitute for cholesterol is not a peculiarity of fibroblasts. The fact that the addition of either desmosterol or cholesterol overcomes the inhibitory effect of lovastatin on proliferation of J774-D cells further supports the suggestion that both sterols can be used interchangeably for cell growth in J774 cells.
As far as we know, desmosterol is the only sterol proven to be able to completely substitute for cholesterol in mammalian cells for an indefinite period of time. Although our observations in J774 cells and previous findings with L cells are consistent with the obvious proliferation that takes place in the Dhcr24−/− mice and in human subjects with desmosterolosis, the Dhcr24−/− mice actually represents a low-cholesterol, high-desmosterol model [29, 32], and there is no evidence that subjects with desmosterolosis were cholesterol-free during development [30, 31]. It is conceivable that desmosterol could replace cholesterol for cell proliferation in vivo, but the severe abnormalities associated with desmosterolosis indicate that cholesterol has a unique role in embryogenesis. Another process in which cholesterol and desmosterol appear not to be equivalent is in the lipid raft/caveolae function, since exchanging the bulk of cell cholesterol for desmosterol increases caveolin-1 phosphorylation [42] and impairs insulin signaling [43].
The ability of desmosterol to substitute for cholesterol in maintaining membrane integrity may not be surprising, since it differs from cholesterol only in the presence of a double bond at C24, and it otherwise satisfies the structural features required to support cell growth [10, 11]. Interestingly, cholest-5,22-trans-dien-3β-ol, which only differs from desmosterol by the position of the double bond (C22 vs. C24) in the side chain, was not able to effectively support J774-D cell proliferation, thus demonstrating the specificity of this effect. The partial prevention of the inhibitory effect of lovastatin by cholest-5,22-trans-dien-3β-ol suggests that this compound has some capacity to cooperate with desmosterol in a process that is similar to the synergism between sterols described previously [3, 12-14]. The discrepancy between the effects of desmosterol and cholest-5,22-trans-dien-3β-ol on cell proliferation may reflect a distinct requirement for accompanying membrane lipids imposed by the distance of the side chain double bond to the steroid nucleus. Our findings contrast with those obtained by Rujanavech and Silbert in S2 cells, an L cell-derived sterol auxotrophic cell line, which grow normally on either desmosterol or cholest-5,22-trans-dien-3β-ol [13]. The reason why J774-D cells discriminate between these two sterols whereas S2 cells do not is unknown, but it may be due to a differing ability to undergo accompanying compensatory changes in membrane lipids.
In response to changes in cholesterol levels, SREBP-2 controls the cholesterol biosynthesis pathway and lipoprotein uptake, whereas SREBP-1, and especially the SREBP-1c isoform, regulates the biosynthesis of free fatty acids and acylglycerols, also required for membrane biosynthesis. It was predictable that J774-D cells would be able to control lipid homeostasis as to preserve cell viability and proliferation. We were, therefore, interested in studying the ability of desmosterol to substitute for cholesterol in the regulation of the SREBP pathway. We found that desmosterol is able to inhibit SREBP-1 and SREBP-2 processing, as observed both in J774 and L cells. Accordingly, in these cells the expression of Hmgcr and Ldlr, two preferential SREBP-2 target genes [23], was suppressed by desmosterol. Similarly, desmosterol reduced the expression of Insig1, whose expression can be stimulated by SREBP-1c [18]. Addition of desmosterol to J774-D cells also decreased Fas expression, which is regulated principally by SREBP-1c [23]. Consistently with these studies, the treatment of J774-D cells with lovastatin produced an up-regulation of SREBP-targeted genes, whereas after lovastatin removal the expression of these genes returned to control levels. This suggests that, like cholesterol, endogenously produced desmosterol can regulate the SREBP pathway. However, contributions to these effects by other endogenously produced sterols, notably 24(S),25-epoxycholesterol, cannot be excluded [41]. Such effects of desmosterol are in keeping with the finding that desmosterol, like cholesterol, can induce a conformational change in Scap when assayed in cell membrane suspensions [19]. This conformational change causes Scap to bind to Insig, thereby retaining the Scap/SREBP complex in the ER and precluding SREBP processing [8, 18]. Consistent with this, desmosterol treatment of CHO cells has recently been shown to reduce SREBP-2 processing [20, 44], although the possibility existed that its conversion into cholesterol mediated such an effect. Our findings unambiguously show that desmosterol inhibits SREBP processing in intact cells that cannot convert it to cholesterol at least as efficiently as does cholesterol added with the same vehicle.
Current results demonstrate for the first time that cells virtually free of cholesterol retain the capacity to respond to sterols such as desmosterol and cholesterol through the inhibition of the SREBP pathway. They also respond to 25HC, whose potent inhibitory effect is exerted by binding to Insig [20, 22], but not to Scap [19, 21]. Therefore, the replacement of cholesterol with desmosterol in cell membranes is compatible with the functioning of the SREBP pathway, sterols being able to suppress SREBP processing by binding to either one of the two sterol sensors, Scap or Insig.
Increased expression of both Abca1 and Srebf1c, two well known LXR targets, following addition of desmosterol to J774-D cells suggests activation of this nuclear receptor. Consistently, we also found that Abca1 is up-regulated in response to increased production of endogenous desmosterol after lovastatin pretreatment. However, the involvement of other sterols, such as 24(S),25-epoxycholesterol, in the effect of lovastatin removal cannot be excluded [41]. It has recently been demonstrated that desmosterol is an LXR ligand [44]. Moreover, all these observations are in agreement with the up-regulation of hepatic Srebf1c, Cyp7a1, Abcg5 and Abcg8, all of which are LXR targets, in the Dhcr24−/− mouse [45]. Surprisingly, hepatic Abca1 expression was not enhanced in this animal model, which may reflect a differential regulation of this gene by LXR in hepatocytes and monocyte/macrophages like J774 cells. Desmosterol-induced stimulation of Abca1 expression suggests that desmosterol itself might be released from cells through the ABCA1 transporter, thus contributing to the reported high efficiency of the efflux of biosynthetic desmosterol [46, 47].
Addition of 25HC also raised Abca1 expression, although, in contrast to desmosterol, it failed to affect Srebf1c expression. The latter result is consistent with the notion that 25HC is a weak activator of LXR [48], but other mechanisms may be involved. Besides being stimulated by LXR, Srebf1c transcription is also up-regulated by SREBPs, including SREBP-1c [49, 50]. Therefore, the lack of effect of 25HC on Srebf1c expression may be the consequence of the counterbalance between two opposite mechanisms: the activation of the LXR pathway and the suppression of the SREBP pathway. Relative to 25HC and cholesterol, excess desmosterol appears to be more effective activating the LXR pathway, thus markedly increasing the expression of both Abca1 and Srebf1c.
Apart from influencing the expression of genes involved in cholesterol homeostasis, desmosterol altered the expression of genes for fatty acid biosynthesis, such as Fas and Scd1, as well as their upstream regulator Srebf1c. Similarly to cholesterol and 25HC, desmosterol had a differential effect on Fas and Scd1 expression. While lowering Fas mRNA levels, in correspondence to the sterol-induced inhibition of SREBP-1 processing, Scd1 mRNA levels were increased (desmosterol and cholesterol) or unchanged (25HC). A possible involvement of an LXR-mediated up regulation of Scd1 can not be ruled out [51]. Similarly to our findings, it has been observed previously that feeding mice a cholesterol-rich diet can produce different responses of hepatic expression of Fas and Scd1 [40]. Considered together, these results suggest a differential regulation of both genes by sterols, in such a way that the expression of Fas is more responsive than that of Scd1 to the sterol-induced inhibition of SREBP-1 cleavage.
Current results show that desmosterol can replace cholesterol in sustaining J774 cell proliferation. These cells have a functional SREBP pathway, which, by regulating lipid homeostasis, presumably contributes to cell viability and growth. Desmosterol, like cholesterol and 25HC, is able to suppress SREBP processing and targeted gene expression. On the other hand, in comparison with cholesterol and 25HC, desmosterol is especially effective activating LXR-targeted genes. Such effects of desmosterol on SREBP and LXR pathways may be relevant for lipid homeostasis in cells and tissues with high desmosterol content, such as spermatozoa [26], astrocytes [27] and the developing brain [28].
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Angela Murúa and Lorena Crespo for excellent technical support and to Dr. Iain Patten for editorial advice on a version of the manuscript.
FUNDING
This study was supported by grants from Fondo de Investigación Sanitaria (00/0229), Ministerio de Ciencia y Tecnología (BMC2002-01262) and Ministerio de Educación y Ciencia (SAF2005-07038). S.R.A. was a recipient of a fellowship “Beca de Formación en Investigación” from the Instituto de Salud Carlos III. J.M.B. is an investigator of FIBio-HRC, supported by the Comunidad Autónoma de Madrid. The “CIBER de Fisiopatología de la Obesidad y Nutrición” is an initiative of the Instituto de Salud Carlos III.
Abbreviations used
- Abca1
ATP-binding cassette, subfamily A, member 1
- Cypb
cyclophilin B
- DHCR24
sterol-Δ24-reductase
- ER
endoplasmic reticulum
- Fas
fatty acid synthase
- FBS
fetal bovine serum
- Gapdh
glyceraldehyde-3-phosphate dehydrogenase
- 25HC
25-hydroxycholesterol
- HMG-CoA
3-hydroxy-3-methylglutaryl coenzyme A
- HPCD
hydroxypropyl-β-cyclodextrin
- Insig
insulin-induced gene
- ldlr
low density lipoprotein receptor
- LPDS
lipoprotein deficient serum
- LXR
liver X receptor
- MCD
methyl-β-cyclodextrin
- Scap
SREBP cleavage-activating protein
- Scd1
stearoyl-CoA desaturase-1
- SRE
sterol regulatory element
- SREBP
sterol regulatory element-binging protein
- Srebf1c
sterol regulatory element-binding factor 1c
- rebf2
sterol regulatory element-binding factor 2
REFERENCES
- 1.Martinez-Botas J, Suarez Y, Ferruelo AJ, Gomez-Coronado D, Lasuncion MA. Cholesterol starvation decreases p34(cdc2) kinase activity and arrests the cell cycle at G2. Faseb J. 1999;13:1359–1370. doi: 10.1096/fasebj.13.11.1359. [DOI] [PubMed] [Google Scholar]
- 2.Martinez-Botas J, Ferruelo AJ, Suarez Y, Fernandez C, Gomez-Coronado D, Lasuncion MA. Dose-dependent effects of lovastatin on cell cycle progression. Distinct requirement of cholesterol and non-sterol mevalonate derivatives. Biochim Biophys Acta. 2001;1532:185–194. doi: 10.1016/s1388-1981(01)00125-1. [DOI] [PubMed] [Google Scholar]
- 3.Suarez Y, Fernandez C, Ledo B, Ferruelo AJ, Martin M, Vega MA, Gomez-Coronado D, Lasuncion MA. Differential effects of ergosterol and cholesterol on Cdk1 activation and SRE-driven transcription. Eur J Biochem. 2002;269:1761–1771. doi: 10.1046/j.1432-1327.2002.02822.x. [DOI] [PubMed] [Google Scholar]
- 4.Fernandez C, Lobo Md Mdel V, Gomez-Coronado D, Lasuncion MA. Cholesterol is essential for mitosis progression and its deficiency induces polyploid cell formation. Exp Cell Res. 2004;300:109–120. doi: 10.1016/j.yexcr.2004.06.029. [DOI] [PubMed] [Google Scholar]
- 5.Tabas I. Consequences of cellular cholesterol accumulation: basic concepts and physiological implications. J Clin Invest. 2002;110:905–911. doi: 10.1172/JCI16452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schoonjans K, Brendel C, Mangelsdorf D, Auwerx J. Sterols and gene expression: control of affluence. Biochim Biophys Acta. 2000;1529:114–125. doi: 10.1016/s1388-1981(00)00141-4. [DOI] [PubMed] [Google Scholar]
- 7.Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 8.Bengoechea-Alonso MT, Ericsson J. SREBP in signal transduction: cholesterol metabolism and beyond. Curr Opin Cell Biol. 2007;19:215–222. doi: 10.1016/j.ceb.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 9.Goldstein JL, Rawson RB, Brown MS. Mutant mammalian cells as tools to delineate the sterol regulatory element-binding protein pathway for feedback regulation of lipid synthesis. Arch Biochem Biophys. 2002;397:139–148. doi: 10.1006/abbi.2001.2615. [DOI] [PubMed] [Google Scholar]
- 10.Esfahani M, Scerbo L, Devlin TM. A requirement for cholesterol and its structural features for a human macrophage-like cell line. J Cell Biochem. 1984;25:87–97. doi: 10.1002/jcb.240250204. [DOI] [PubMed] [Google Scholar]
- 11.Suarez Y, Fernandez C, Ledo B, Martin M, Gomez-Coronado D, Lasuncion MA. Sterol stringency of proliferation and cell cycle progression in human cells. Biochim Biophys Acta. 2005;1734:203–213. doi: 10.1016/j.bbalip.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 12.Xu F, Rychnovsky SD, Belani JD, Hobbs HH, Cohen JC, Rawson RB. Dual roles for cholesterol in mammalian cells. Proc Natl Acad Sci U S A. 2005;102:14551–14556. doi: 10.1073/pnas.0503590102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rujanavech C, Silbert DF. LM cell growth and membrane lipid adaptation to sterol structure. J Biol Chem. 1986;261:7196–7203. [PubMed] [Google Scholar]
- 14.Fernandez C, Martin M, Gomez-Coronado D, Lasuncion MA. Effects of distal cholesterol biosynthesis inhibitors on cell proliferation and cell cycle progression. J Lipid Res. 2005;46:920–929. doi: 10.1194/jlr.M400407-JLR200. [DOI] [PubMed] [Google Scholar]
- 15.Dahl JS, Dahl CE, Bloch K. Effect of cholesterol on macromolecular synthesis and fatty acid uptake by Mycoplasma capricolum. J Biol Chem. 1981;256:87–91. [PubMed] [Google Scholar]
- 16.Ramgopal M, Bloch K. Sterol synergism in yeast. Proc Natl Acad Sci U S A. 1983;80:712–715. doi: 10.1073/pnas.80.3.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rothblat GH, Burns CH, Conner RL, Landrey JR. Desmosterol as the major sterol in L-cell mouse fibroblasts grown in sterol-free culture medium. Science. 1970;169:880–882. doi: 10.1126/science.169.3948.880. [DOI] [PubMed] [Google Scholar]
- 18.Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 19.Brown AJ, Sun L, Feramisco JD, Brown MS, Goldstein JL. Cholesterol addition to ER membranes alters conformation of SCAP, the SREBP escort protein that regulates cholesterol metabolism. Mol Cell. 2002;10:237–245. doi: 10.1016/s1097-2765(02)00591-9. [DOI] [PubMed] [Google Scholar]
- 20.Radhakrishnan A, Ikeda Y, Kwon HJ, Brown MS, Goldstein JL. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: oxysterols block transport by binding to Insig. Proc Natl Acad Sci U S A. 2007;104:6511–6518. doi: 10.1073/pnas.0700899104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adams CM, Reitz J, De Brabander JK, Feramisco JD, Li L, Brown MS, Goldstein JL. Cholesterol and 25-hydroxycholesterol inhibit activation of SREBPs by different mechanisms, both involving SCAP and Insigs. J Biol Chem. 2004;279:52772–52780. doi: 10.1074/jbc.M410302200. [DOI] [PubMed] [Google Scholar]
- 22.Sun LP, Seemann J, Goldstein JL, Brown MS. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: Insig renders sorting signal in Scap inaccessible to COPII proteins. Proc Natl Acad Sci U S A. 2007;104:6519–6526. doi: 10.1073/pnas.0700907104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Brown MS, Goldstein JL. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci U S A. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greeve I, Hermans-Borgmeyer I, Brellinger C, Kasper D, Gomez-Isla T, Behl C, Levkau B, Nitsch RM. The human DIMINUTO/DWARF1 homolog seladin-1 confers resistance to Alzheimer's disease-associated neurodegeneration and oxidative stress. J Neurosci. 2000;20:7345–7352. doi: 10.1523/JNEUROSCI.20-19-07345.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin DS, Connor WE, Wolf DP, Neuringer M, Hachey DL. Unique lipids of primate spermatozoa: desmosterol and docosahexaenoic acid. J Lipid Res. 1993;34:491–499. [PubMed] [Google Scholar]
- 27.Mutka AL, Lusa S, Linder MD, Jokitalo E, Kopra O, Jauhiainen M, Ikonen E. Secretion of sterols and the NPC2 protein from primary astrocytes. J Biol Chem. 2004;279:48654–48662. doi: 10.1074/jbc.M405345200. [DOI] [PubMed] [Google Scholar]
- 28.Fumagalli R, Paoletti R. The identification and significance of desmosterol in the developing human and animal brain. Life Sci. 1963;5:291–295. doi: 10.1016/0024-3205(63)90159-0. [DOI] [PubMed] [Google Scholar]
- 29.Wechsler A, Brafman A, Shafir M, Heverin M, Gottlieb H, Damari G, Gozlan-Kelner S, Spivak I, Moshkin O, Fridman E, Becker Y, Skaliter R, Einat P, Faerman A, Bjorkhem I, Feinstein E. Generation of viable cholesterol-free mice. Science. 2003;302:2087. doi: 10.1126/science.1090776. [DOI] [PubMed] [Google Scholar]
- 30.Andersson HC, Kratz L, Kelley R. Desmosterolosis presenting with multiple congenital anomalies and profound developmental delay. Am J Med Genet. 2002;113:315–319. doi: 10.1002/ajmg.b.10873. [DOI] [PubMed] [Google Scholar]
- 31.FitzPatrick DR, Keeling JW, Evans MJ, Kan AE, Bell JE, Porteous ME, Mills K, Winter RM, Clayton PT. Clinical phenotype of desmosterolosis. Am J Med Genet. 1998;75:145–152. [PubMed] [Google Scholar]
- 32.Brown AJ. Of cholesterol-free mice and men. Curr Opin Lipidol. 2004;15:373–375. doi: 10.1097/00041433-200406000-00021. [DOI] [PubMed] [Google Scholar]
- 33.Tabas I, Feinmark SJ, Beatini N. The reactivity of desmosterol and other shellfish- and xanthomatosis-associated sterols in the macrophage sterol esterification reaction. J Clin Invest. 1989;84:1713–1721. doi: 10.1172/JCI114354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 35.Russell DW, Yamamoto T, Schneider WJ, Slaughter CJ, Brown MS, Goldstein JL. cDNA cloning of the bovine low density lipoprotein receptor: feedback regulation of a receptor mRNA. Proc Natl Acad Sci U S A. 1983;80:7501–7505. doi: 10.1073/pnas.80.24.7501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sato R, Goldstein JL, Brown MS. Replacement of serine-871 of hamster 3-hydroxy-3-methylglutaryl-CoA reductase prevents phosphorylation by AMP-activated kinase and blocks inhibition of sterol synthesis induced by ATP depletion. Proc Natl Acad Sci U S A. 1993;90:9261–9265. doi: 10.1073/pnas.90.20.9261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimano H, Horton JD, Hammer RE, Shimomura I, Brown MS, Goldstein JL. Overproduction of cholesterol and fatty acids causes massive liver enlargement in transgenic mice expressing truncated SREBP-1a. J Clin Invest. 1996;98:1575–1584. doi: 10.1172/JCI118951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shimano H, Shimomura I, Hammer RE, Herz J, Goldstein JL, Brown MS, Horton JD. Elevated levels of SREBP-2 and cholesterol synthesis in livers of mice homozygous for a targeted disruption of the SREBP-1 gene. J Clin Invest. 1997;100:2115–2124. doi: 10.1172/JCI119746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koroly MJ, Dempsey ME. Synthesis of delta 5,22-cholestadien-3 beta-ol from delta 5,7,22-cholestatrien-3 beta-ol by a liver enzyme. Lipids. 1981;16:755–758. doi: 10.1007/BF02535344. [DOI] [PubMed] [Google Scholar]
- 40.Repa JJ, Liang G, Ou J, Bashmakov Y, Lobaccaro JM, Shimomura I, Shan B, Brown MS, Goldstein JL, Mangelsdorf DJ. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev. 2000;14:2819–2830. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong J, Quinn CM, Gelissen IC, Brown AJ. Endogenous 24(S),25-epoxycholesterol fine-tunes acute control of cellular cholesterol homeostasis. J Biol Chem. 2008;283:700–707. doi: 10.1074/jbc.M706416200. [DOI] [PubMed] [Google Scholar]
- 42.Jansen M, Pietiainen VM, Polonen H, Rasilainen L, Koivusalo M, Ruotsalainen U, Jokitalo E, Ikonen E. Cholesterol substitution increases the structural heterogeneity of caveolae. J Biol Chem. 2008;283:14610–14618. doi: 10.1074/jbc.M710355200. [DOI] [PubMed] [Google Scholar]
- 43.Vainio S, Jansen M, Koivusalo M, Rog T, Karttunen M, Vattulainen I, Ikonen E. Significance of sterol structural specificity. Desmosterol cannot replace cholesterol in lipid rafts. J Biol Chem. 2006;281:348–355. doi: 10.1074/jbc.M509530200. [DOI] [PubMed] [Google Scholar]
- 44.Yang C, McDonald JG, Patel A, Zhang Y, Umetani M, Xu F, Westover EJ, Covey DF, Mangelsdorf DJ, Cohen JC, Hobbs HH. Sterol intermediates from cholesterol biosynthetic pathway as liver X receptor ligands. J Biol Chem. 2006;281:27816–27826. doi: 10.1074/jbc.M603781200. [DOI] [PubMed] [Google Scholar]
- 45.Heverin M, Meaney S, Brafman A, Shafir M, Olin M, Shafaati M, von Bahr S, Larsson L, Lovgren-Sandblom A, Diczfalusy U, Parini P, Feinstein E, Bjorkhem I. Studies on the cholesterol-free mouse: strong activation of LXR-regulated hepatic genes when replacing cholesterol with desmosterol. Arterioscler Thromb Vasc Biol. 2007;27:2191–2197. doi: 10.1161/ATVBAHA.107.149823. [DOI] [PubMed] [Google Scholar]
- 46.Aviram M, Bierman EL, Oram JF. High density lipoprotein stimulates sterol translocation between intracellular and plasma membrane pools in human monocyte-derived macrophages. J Lipid Res. 1989;30:65–76. [PubMed] [Google Scholar]
- 47.Johnson WJ, Fischer RT, Phillips MC, Rothblat GH. Efflux of newly synthesized cholesterol and biosynthetic sterol intermediates from cells. Dependence on acceptor type and on enrichment of cells with cholesterol. J Biol Chem. 1995;270:25037–25046. doi: 10.1074/jbc.270.42.25037. [DOI] [PubMed] [Google Scholar]
- 48.Janowski BA, Willy PJ, Devi TR, Falck JR, Mangelsdorf DJ. An oxysterol signalling pathway mediated by the nuclear receptor LXR alpha. Nature. 1996;383:728–731. doi: 10.1038/383728a0. [DOI] [PubMed] [Google Scholar]
- 49.Amemiya-Kudo M, Shimano H, Yoshikawa T, Yahagi N, Hasty AH, Okazaki H, Tamura Y, Shionoiri F, Iizuka Y, Ohashi K, Osuga J, Harada K, Gotoda T, Sato R, Kimura S, Ishibashi S, Yamada N. Promoter analysis of the mouse sterol regulatory element-binding protein-1c gene. J Biol Chem. 2000;275:31078–31085. doi: 10.1074/jbc.M005353200. [DOI] [PubMed] [Google Scholar]
- 50.Cagen LM, Deng X, Wilcox HG, Park EA, Raghow R, Elam MB. Insulin activates the rat sterol-regulatory-element-binding protein 1c (SREBP-1c) promoter through the combinatorial actions of SREBP, LXR, Sp-1 and NF-Y cis-acting elements. Biochem J. 2005;385:207–216. doi: 10.1042/BJ20040162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schultz JR, Tu H, Luk A, Repa JJ, Medina JC, Li L, Schwendner S, Wang S, Thoolen M, Mangelsdorf DJ, Lustig KD, Shan B. Role of LXRs in control of lipogenesis. Genes Dev. 2000;14:2831–2838. doi: 10.1101/gad.850400. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.