Abstract
Fragile X syndrome is caused by the absence of functional fragile X mental retardation protein (FMRP), an RNA binding protein. The molecular mechanism of aberrant protein synthesis in fmr1 KO mice is closely associated with the role of FMRP in mRNA transport, delivery, and local protein synthesis. We show that GFP-labeled Fmr1 and CaMKIIα mRNAs undergo decelerated motion at 0–40 min after group I mGluR stimulation, and later recover at 40–60 min. Then we investigate targeting of mRNAs associated with FMRP after neuronal stimulation. We find that FMRP is synthesized closely adjacent to stimulated mGluR5 receptors. Moreover, in WT neurons, CaMKIIα mRNA can be delivered and translated in dendritic spines within 10 min in response to group I mGluR stimulation, whereas KO neurons fail to show this response. These data suggest that FMRP can mediate spatial mRNA delivery for local protein synthesis in response to synaptic stimulation.
Keywords: fragile X syndrome, dendritic mRNA targeting, local translation
Fragile X syndrome (FXS) is the most common form of inherited mental retardation and is caused by the loss of function of the FMR1 gene, which encodes fragile X mental retardation protein (FMRP) (1). FXS affects 1 in 4,000 males and 1 in 6,000 females on average and is characterized by hyperactivity, attention deficits, autistic-like behaviors, and seizures (2). Dendritic spine morphology in the cerebral cortex of FXS patients and in the fmr1 KO mouse model shows more immature long thin spines than mature stubby, mushroom-shaped spines (3). Furthermore, group I mGluR-dependent long-term depression in the hippocampus is exaggerated in the fmr1 KO model (4). These findings suggest that FMRP functions in synaptic development and plasticity.
Activity-dependent local translation is a fundamental mechanism underlying synaptic plasticity (5, 6). Inhibition of protein synthesis attenuates specific types of long-term plasticity (7, 8). Morphological changes in dendritic spines can be blocked by protein synthesis inhibitors (9). In the fmr1 KO model, it has been shown that aberrant synthesis of individual proteins such as CaMKIIα, PSD-95, and MAP1b, upon group I mGluR stimulation, is associated with defective long-term plasticity (10–12). Here we have studied specific molecular mechanisms to elucidate aberrant localized translation in the fmr1 KO model.
The molecular basis of FMRP's role in translation-dependent plasticity remains unclear despite extensive study. FMRP is a ribosome-associated RNA binding protein with selective affinity (13, 14). Upon neuronal stimulation, FMRP may regulate protein levels by mediating translational regulation and mRNA trafficking (11, 15). FMRP, mRNA, and other RNA binding proteins can form ribonucleoprotein (RNP) or granule structures and couple with motor proteins to be transported in dendrites (16–18). Dendritic transport of FMRP and associated mRNAs, such as Fmr1, CaMKIIα, and MAP1b, are regulated by group I mGluR signaling (15, 19). It is not yet fully understood how and when mRNA is delivered to the synapse and translated. Local delivery of mRNA to active synapses could provide a high degree of regulation and flexibility of protein synthesis (20–23).
To test the role of FMRP in local protein synthesis, we investigated the speed and directionality of mRNA movement upon group I mGluR stimulation using time-lapse imaging of primary WT and fmr1 KO neurons. We found that at 0–40 min after stimulation, the speed of mRNA-containing granules is reduced in WT but not KO dendrites, and at 40–60 min mRNAs resumed more directional motion. At 20 min after stimulation, FMRP was translated in regions closely adjacent to mGluR5. CaMKIIα mRNAs and protein synthesis were more enriched at dendritic spines in WT but not fmr1 KO neurons. This suggests lack of local translation-dependent plasticity in fmr1 KO neurons, originating from aberrant mRNA targeting function in the absence of FMRP.
Results
Study of mRNA Dynamic Motions in WT and fmr1 KO Hippocampal Neurons by Time-Lapse Imaging.
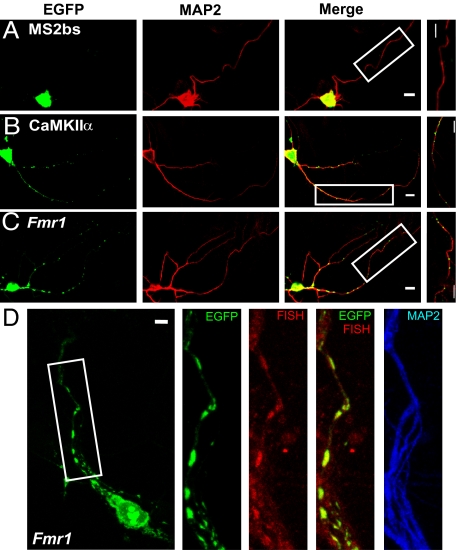
To test whether FMRP regulates the dynamics of dendritic mRNA movement, we used time-lapse imaging to investigate mRNA movement in primary cultures of WT and fmr1 KO hippocampal neurons. Two mRNAs, CaMKIIα and Fmr1, were indirectly labeled by GFP-MS2 (Fig. S1A) using the MS2 tethering method (24), and monitored by time-lapse imaging. CaMKIIα was used here because its translation is regulated by FMRP (10) and its dendritic trafficking was studied previously (24). Fmr1 was chosen because of its high-affinity association with FMRP (25). Because FMRP may associate with Fmr1 through its G quartet on the open reading frame (ORF) and/or the U-rich region on the 3′ untranslated region (UTR) (26, 27), we made a construct containing both the ORF and the 3′UTR of Fmr1 as the RNA of interest (Fig. S1A) to mimic endogenous Fmr1 mRNA. Fig. S1B shows that the ORF of the Fmr1 mRNA construct cannot be translated, consistent with its placement downstream of the LacZ gene stop codon. Fig. 1A shows that without a dendritic targeting signal, GFP-labeled MS2 binding site (MS2bs) cannot be transported to neuronal dendrites. Both MS2-GFP-labeled Fmr1 and CaMKIIα formed punctate mRNA granules in dendrites (Fig. 1 B and C). Moreover, Fmr1-containing GFP-labeled granules were colocalized with Fmr1 RNA signals as shown by fluorescence in situ hybridization (FISH) (Fig. 1D), confirming that GFP-labeled granules contained Fmr1 mRNA.
Fig. 1.
Labeling of CaMKIIα and Fmr1 mRNA in primary hippocampal neurons. (A) A neuron transfected with GFP-MS2-nls (nuclear localization signal) and MS2bs showed that GFP signals stay in soma. (B and C) The neuron transfected with GFP-MS2-nls and MS2bs-CaMKIIα or MS2bs-Fmr1 showed that mRNA puncta distribute in dendrites. Higher magnification of the boxed images shows GFP-labeled granules in dendrites. (D) Fmr1-containing GFP-labeled granules (green) in dendrites colocalized with Fmr1 mRNA detected by FISH (red). (Scale bars, 10 μm.)
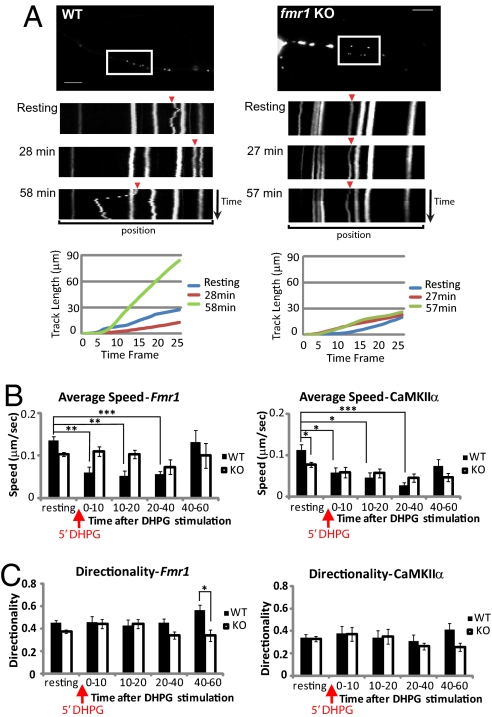
Next, to compare the dynamic movement of mRNA in WT and fmr1 KO hippocampal neurons, either CaMKIIα- or Fmr1-labeling constructs were transfected into WT and fmr1 KO neurons. mRNA granules in single dendrites were imaged 5 s per frame for 25 frames (2 min in total) before and after stimulation by the group I mGluR agonist (S)-3,5-dihydroxyphenylglycine (DHPG) (Fig. 2A and Movies S1, S2, S3, S4, S5, and S6). We found that the majority of granules was stationary, as reported before (24). Therefore, we measured the trafficking pattern of granules which are motile in at least two time series. The movement dynamics of CaMKIIα or Fmr1 granules were measured as average speed and as directionality, which is a measurement of the degree to which the granule travels in a single direction. In WT neurons, the average speed of Fmr1 granules (Fig. 2BLeft and Movies S1, S2, S3, S4, S5, and S6) was decreased between 0 and 40 min after DHPG stimulation compared with the speed before stimulation. However, in fmr1 KO neurons, average speed did not change significantly after stimulation. The average speed of CaMKIIα granules showed similar changes (Fig. 2B Right); in WT, but not KO neurons, CaMKIIα mRNA significantly slowed during 0–40 min poststimulation. This suggests FMRP may act to decelerate mRNA movement at the early phase (0–40 min) after group I mGluR treatment. The average speed of CaMKIIα (82.96 nm/s) was comparable to a previous report (24). Interestingly, the average speed before stimulation was significantly lower in fmr1 KO neurons compared with WT neurons.
Fig. 2.
CaMKIIα and Fmr1 mRNA dynamic motions in WT and fmr1 KO hippocampal neurons. (A) Representation of Fmr1 mRNA movement in WT and fmr1 KO neurons by time-lapse imaging. Kymograph (upper) shows granule motion in the boxed region of a WT or KO neuron transfected with GFP-MS2-nls and MS2bs-Fmr1. The time point of the image taken is labeled next to each kymograph, which represents a 2-min series of images at 5-s intervals (25 frames in total). (Scale bars, 20 μm.) Track length (lower) of each quantified Fmr1 granule (arrowhead in kymograph) was represented. Track length is the total length of displacements within the track. (B) Average speed of GFP-labeled Fmr1 or CaMKIIα mRNA calculated for WT or fmr1 KO neurons shows that particle movement was retarded in WT from 0 to 40 min after stimulation. (C) Directionality of GFP-labeled Fmr1 or CaMKIIα in WT or fmr1 KO neurons was calculated, showing increased unidirectional movement of Fmr1 mRNA in WT neurons. Bar graph represents data from three experiments, total of at least 20 mRNA particles in each group. Experiments and definition of average speed and directionality were as described in Materials and Methods. Statistical analysis by two-way ANOVA with Tukey's HSD posttest. Error bars denote SEM.
The directionality of Fmr1 granules in WT neurons was significantly higher during 40 and 60 min after DHPG treatment (Fig. 2C Left). There was a similar trend (not significant) for CaMKIIα granules in WT neurons compared with fmr1 KO neurons (Fig. 2C Right). The apparent increase in unidirectional Fmr1 mRNA movement might indicate a regulatory interaction between FMRP and motor proteins and mRNA in response to stimulation (17–19). It is important to note that total granule number and the percentage of motile granules were not significantly different between WT and fmr1 KO neurons. However, the brightness of Fmr1 granules in WT was significantly higher after DHPG treatment, but not in fmr1 KO neurons (Fig. S2). This suggests that mRNA, previously below detection and measurement levels, may be incorporated into granule structures by DHPG stimulation in WT but not KO neurons. We hypothesized that shortly after group I mGluR treatment, FMRP could facilitate mRNA deceleration and docking to specific synaptic targets for a subsequent translation process.
FMRP Was Translated near Group I mGluRs.
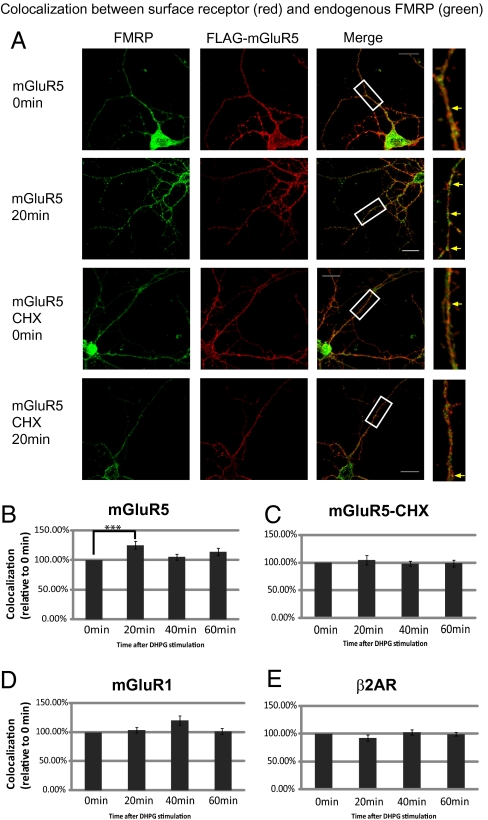
Next, we used double-label immunofluorescence to examine translation of FMRP in regions near group I mGluR. We chose the cellular microdomain of group I mGluR because it is linked to signal pathways: mGluR1a-mediated ERK phosphorylation is enriched after stimulation in the membrane fraction (28), and the ERK pathway is crucial for group I mGluR-dependent plasticity (29). N-terminal FLAG-tagged mGlu1a and mGluR5, two members of the group I mGluR family, were individually expressed in primary hippocampal neurons to label surface receptor regions. Surface mGluR1a and mGluR5 were recognized by anti-FLAG under nonpermeabilized conditions (Fig. S3A). Colocalization between endogenous FMRP and the surface receptor was measured at different time points after DHPG treatment (Fig. 3A). Our results show that colocalization between FMRP and surface mGluR5 was significantly elevated at 20 min after DHPG treatment (Fig. 3B). Although treated with a protein synthesis blocker, cycloheximide (CHX), there was not a greater colocalization (Fig. 3C). This suggests that FMRP was newly synthesized in regions close to surface mGluR5 in response to group I mGluR stimulation, although some FMRP transport may still occur. There was only a slight increase in colocalization between FMRP and surface mGluR1a, and this change did not occur until 40 min (Fig. 3D). The difference between mGluR5 and mGluR1a may have been caused by different representation of surface receptor constructs because surface mGluR5 staining showed better representation of endogenous mGluR5 (Fig. S3B). Last, surface β2 adrenergic receptor (β2AR), another G-protein-coupled receptor, was used as a negative control because it cannot be stimulated by group I mGluR agonist. DHPG stimulation caused no change in colocalization between FMRP and β2AR (Fig. 3E). This suggests translation of FMRP could be enriched temporally within active receptor regions.
Fig. 3.
Colocalization between surface group I mGluRs and FMRP after DHPG treatment. (A) Representative deconvolved image (Z projection) detected surface mGluR5 (red) and endogenous FMRP (green). Higher magnification of the boxed images shows that more FMRP colocalized with surface mGluR5 at 20 min after DHPG (yellow arrow), but not in the presence of CHX. (Scale bars, 20 μm.) (B–E) The increased colocalization between FMRP and surface mGluR5 at 20 min was measured as Manders's coefficient. The time points indicate the time post-washout after 5-min DHPG incubation. Data were analyzed from at least 15 dendrites in each group from three independent experiments. Statistical analysis by one-way ANOVA with Dunnett's posttest. Error bars denote SEM.
FMRP Targets Translation of CaMKIIα to Dendritic Spines.
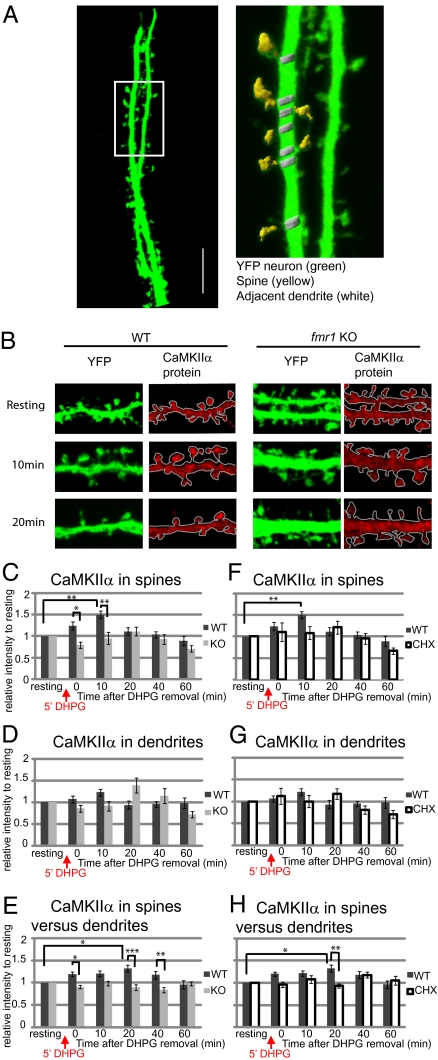
To investigate localization of both mRNA and newly translated protein at excitatory synaptic sites in the presence of FMRP, we looked at levels of CaMKIIα mRNA and protein in dendritic spines. We examined the change in staining intensity of CaMKIIα protein at spines and an adjacent area of dendrites in WT and fmr1 KO neurons that endogenously expressed YFP (Fig. 4 A and B). Based on our previous results, we defined five time points from the washout after 5 min of treatment with DHPG. At 10 min in WT spines, the level of CaMKIIα peaks and is significantly higher than at the pre-DHPG resting state. In KO spines, on the other hand, there is a delayed, nonsignificant increase above baseline at 20 min after DHPG treatment (Fig. 4C). Dendrites showed the same temporal pattern of protein translation as spines, although the changes were smaller, and nonsignificant in WT (Fig. 4D).
Fig. 4.
The differential distribution of CaMKIIα protein in neuronal spines and dendrites in response to group I mGluR stimulation. Cycloheximide blocks local translation of CaMKIIα in spines of WT neurons. (A) YFP-expressing hippocampal neurons (green) were used to outline neuronal dendrites and spines. A higher magnification of the boxed region shows that spine (yellow) and neighboring dendrite (white) regions could be selected based on YFP staining threshold. (Scale bar, 10 μm.) (B) Representative figures of CaMKIIα immunostaining (red) in WT or fmr1 KO YFP hippocampal neurons show relative CaMKIIα distribution in spines or dendrites in response to group I mGluR stimulation (DHPG). (C and D) The average intensity of CaMKIIα protein in WT or fmr1 KO YFP spines (C) or adjacent dendrites (D) was measured and compared with CaMKIIα level before stimulation. (E) Enrichment of CaMKIIα mRNA in spine relative to adjacent dendrite was calculated and normalized to the level before DHPG treatment. (F and G) In the presence or absence of 60 μM cycloheximide, a protein synthesis blocker, throughout the experiment, the level of CaMKIIα in spines (F) or neighboring dendrites (G) of WT neurons was measured. The relative level of CaMKIIα was standardized to the level before DHPG treatment. (H) In the presence or absence of 60 μM cycloheximide, the level of CaMKIIα in WT spines versus dendrites was calculated as the level of CaMKIIα in one spine divided by the level in the neighboring dendrite. Data were analyzed from at least 18 dendrites in each group from three independent experiments. Statistical analysis by two-way ANOVA with Tukey's HSD posttest. Error bars denote SEM.
Next, we compared the levels of enrichment of CaMKIIα at individual spines with the adjacent dendrite after DHPG stimulation in WT and fmr1 KO neurons (Fig. 4E). The ratios were also standardized to the level before DHPG. In WT neurons, the spine-to-dendrite ratio of CaMKIIα was significantly enriched at 20 min after DHPG removal compared with prestimulation. Compared with KO, WT neurons showed higher spine-to-dendrite enrichment of CaMKIIα at 0, 20, and 40 min following stimulation. Finally, we asked whether the elevated level of CaMKIIα is caused by de novo protein synthesis after group I mGluR stimulation. After neurons were treated with cycloheximide, the increase in CaMKIIα levels seen at 10 min in WT spines (Fig. 4F), as well as the spine-to-dendrite enrichment ratio at 20 min, were no longer apparent (Fig. 4H). These findings suggest that, in the presence of FMRP, CaMKIIα is translated and enriched at individual spines shortly after the cessation of stimulation.
Local Targeting of CaMKIIα mRNA to WT Spines.
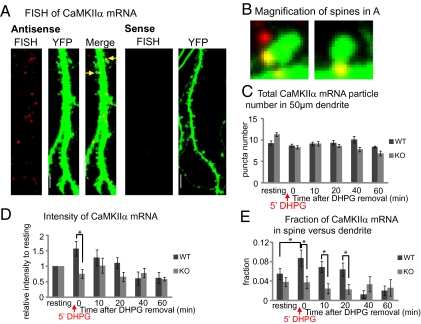
To test targeting of CaMKIIα mRNA at dendrites or spines after group I mGluR stimulation, we examined endogenous CaMKIIα mRNA localization in YFP-labeled WT and fmr1 KO neurons by FISH (Fig. 5 A and B). The total number and average intensity of RNA particles (larger than 0.3 μm), as well as the ratio of CaMKIIα mRNA localized in spines to total CaMKIIα mRNA, were calculated for each 50-μm segment of dendrite. The total number of CaMKIIα mRNA particles inside 50 μm of dendrite did not change over time after stimulation in either WT or fmr1 KO dendrites (Fig. 5C). The intensity of total CaMKIIα mRNA was rapidly and significantly elevated in WT immediately after DHPG washout (0 min), compared with fmr1 KO neurons (Fig. 5D). This suggests that the elevated CaMKIIα mRNA intensity could be due to loading of mRNAs previously under FISH detection level, which could be regulated by FMRP-dependent dendritic transport in response to group I mGluR stimulation. Interestingly, the fraction of CaMKIIα mRNA at WT spines compared with total dendritic puncta was significantly higher at 0 min compared with baseline (Fig. 5E). As a control for mRNA binding specificity of FMRP, we further showed that the level of polyA mRNA does not change over time after stimulation (Fig. S4). These data suggest that in response to group I mGluR stimulation, mRNA granules/mRNPs can be loaded and delivered to excitatory synapses for a period following stimulation, and that this mechanism requires FMRP.
Fig. 5.
The differential distribution of endogenous CaMKIIα mRNA in neuronal spines and dendrites of WT or fmr1 KO neurons in response to group I mGluR stimulation. (A) FISH-detected CaMKIIα mRNA (red) in dendrite of YFP hippocampal neurons (left). Hybridization with a sense probe showed no detectable labeling in dendrite (right). (Scale bars, 10 μm.) (B) Two magnified figures of spines (arrows in A) show that CaMKIIα mRNA could be localized in spine head, adjacent dendrite region (left), or spine base (right). (C) The number of CaMKIIα mRNA particles in 50-μm dendrite segments was calculated in WT and fmr1 KO neurons before or at different time points after 5 min DHPG treatment. (D) The average intensity of CaMKIIα mRNA was measured in each 50-μm dendrite segment. The level of average intensity was compared with the level before DHPG stimulation in WT or fmr1 KO neurons. (E) The ratio of the number of CaMKIIα mRNA localized in spines versus total number of CaMKIIα in each 50-μm dendrite segment was calculated, showing a peak ratio immediately after DHPG stimulation in WT but not KO. Data were analyzed from at least 20 dendrites in each group from three independent experiments. In C and D, data were analyzed by two-way ANOVA with Tukey's HSD posttest. In E, the value of the ratio was transformed to meet the normality requirement and then analyzed by two-way ANOVA with Tukey's HSD. Error bars denote SEM.
Discussion
We have presented evidence that FMRP can target mRNAs toward specialized locations for de novo protein synthesis. First, using time-lapse imaging, we showed that Fmr1 and CaMKIIα RNA particles decelerated their motion during 0–40 min after group I mGluR stimulation and returned to their basal level of movement during a later stage. Second, we showed that translation of FMRP occurs locally adjacent to mGluR5-rich regions. Last, our experiments using YFP-labeled spines revealed that CaMKIIα mRNAs and protein synthesis of CaMKIIα are enriched at spines compared with neighboring dendritic regions only in the presence of FMRP. These data strongly corroborate our hypothesis whereby FMRP enhances RNA targeting to specialized regions for local translation in response to neuronal stimulation.
Using time-lapse imaging, we observed that during the period 0–40 min after neurotransmitter treatment, mRNA granules exhibited slower motion than before stimulation. We speculate that this stage might represent the docking of mRNA granules. First, it has been shown that the “hotspots” of dendritic translation are spatially stable after stimulation and colocalized with ribosomes (21, 22). Second, the selective association of FMRP between microtubules or polyribosomes may provide translation initiation control (30). Third, it has been demonstrated that FMRP-mRNP complexes relocate into dendritic spines after stimulation (31). Last, Myosin Va associates with another RNA binding protein, TLS, to localize mRNA into dendritic spines (32–34). We have now shown quantitative data comparing the temporal and spatial distribution of CaMKIIα mRNA and CaMKIIα protein in WT and fmr1 KO neurons upon group I mGluR stimulation. Our data suggest that although dendritic translation events are spatially static, spines could be the targets for FMRP-dependent mRNA docking and protein synthesis in response to neuronal stimulation.
FMRP can also facilitate directional movement of Fmr1 mRNA granules/mRNPs (Fig. 2C) at 40–60 min after group I mGluR stimulation. This agrees with a previous finding that in dfmr mutant neurons, CG9293 mRNA exhibits less directional movement (35). CaMKIIα has been shown to have other associated mRNPs, possibly modifying directionality (36). The heightened unidirectional movement at 40–60 min in the presence of FMRP may be association with motor proteins. Translation-primed mRNAs may be transported to active synaptic regions and be available there for the next translation event to induce plasticity, including morphological and physiological changes in dendritic spines that may strengthen or weaken the synapse as necessary (37).
In an earlier study, Dictenberg et al. (19) compared movement of labeled CaMKIIα granules in WT and fmr1 KO dendrites. Under conditions of chronic (15-min) DHPG treatment, they found faster movement of granules in WT than KO. We also observed faster movement in WT dendrites under basal conditions. We then compared this basal movement to movement after acute (5-min) DHPG stimulation to imitate more closely a natural stimulation event. In this case, both CaMKIIα- and Fmr1-bearing granules exhibited an initial decrease in movement, recovering to basal level 40 min poststimulation. In addition, we observed that the large motile particles become brighter after stimulation (Fig. S2 and Fig. 5D); we attribute this to aggregation with smaller subthreshold particles. This would be in agreement with studies of mRNA transport dynamics, based on fluorescence recovery after photobleaching (15), that showed rapid recovery of average fluorescence intensity in a photobleached dendritic segment after stimulation.
FMRP-mediated translation-dependent synaptic plasticity may be regulated at several levels. First, dendritic transport and synaptic docking of mRNA could regulate translation initiation by mediating the availability of specific mRNAs for local protein synthesis (15, 19). We have shown that FMRP can facilitate the localization of CaMKIIα mRNA (one of several mRNAs associated with FMRP) at dendritic spines for subsequent translation, lending support to the role of FMRP in regulatory synaptic delivery of specific mRNA. Second, the phosphorylation state of FMRP can govern translation state during the elongation process (38). Third, the restriction of protein distribution by proteasome degradation could be important for synaptic function as well (10). Last, the involvement of microRNAs associated with FMRP in synaptic protein expression is also emerging (39, 40). FMRP is critically involved in several levels of regulation of protein synthesis for synaptic plasticity, and the current work suggests a dynamic role of FMRP in transport, spine localization, and rapid translational control.
Materials and Methods
Primary Hippocampal Neuron Culture and Transfection.
Primary neurons were prepared from hippocampi of WT or fmr1 KO C57BL/6 mice at postnatal day 1–2 and maintained in Neurobasal medium supplemented with B27 and glutamine (Invitrogen). Neurons were transfected using Lipofectamine LTX (Invitrogen). All studies were performed in compliance with the Institutional Animal Care and Use Committee of the University of Illinois at Urbana–Champaign.
Time-Lapse Imaging.
Primary WT or fmr1 KO hippocampal neurons were transfected and imaged within 24 h posttransfection. Neurons were maintained in Liebovitz's L-15 supplemented with B27 at 37 °C in a 5% CO2 live-cell incubation chamber and imaged using the 40× objective (NA 1.4) on a Zeiss Axiovert 200M microscope, before and after exposure to 50 μM (S)-3,5-dihydroxyphenylglycine (DHPG; Tocris), a group I mGluR agonist, for 5 min. Images were taken every 5 s for 25 frames.
Immunocytochemistry.
Primary hippocampal cells on coverslips were fixed using 4% paraformaldehyde in PBS and permeabilized with methanol. Neurons were incubated in primary antibody (diluted in 1% normal donkey serum) at 4 °C overnight. This was followed by incubation with species-appropriate secondary antibodies. For surface receptor staining, neurons were incubated with rabbit anti-FLAG (Sigma) at room temperature for 5 min to allow labeling of N-terminal FLAG-tagged receptor. In cycloheximide-treatment groups, 60 μM cycloheximide was included in medium 30 min before and during experiment periods. After stimulation, neurons were permeabilized and examined by regular immunocytochemistry procedures. (For further details, see SI Materials and Methods.)
Fluorescence In Situ Hybridization.
Digoxigenin (DIG)-labeled riboprobes were generated from plasmids with T3 or T7 RNA polymerase sites. Primary neurons were fixed with 4% paraformaldehyde, permeabilized with methanol, and prehybridized with hybridization buffer. Then neurons were incubated with probes in hybridization buffer overnight at 55 °C for CaMKIIα probes or at 42 °C for Fmr1 probes or 2 h at 37 °C for poly-dT oligos. After hybridization, cells were washed in 0.5× SSC with 50% formamide, 0.5× formamide, and PBS. Cells were incubated with an HRP-linked DIG antibody (Roche) and the signal was amplified by a Cy3 TSA-Plus system (PerkinElmer). (For further details, see SI Materials and Methods.)
CaMKIIα Protein and mRNA Localization in YFP Spines.
WT or fmr1 KO neurons containing Thy1-YFP (yellow fluorescent protein) derived from B6.Cg-Tg(Thy1-YFPH)2Jrs/J mice (Jax Mice) were cultured to days in vitro (DIV)18–21, stimulated with 50 μM DHPG for 5 min, and left for the indicated period after DHPG was removed. After fixation, neurons were subjected to immunostaining or in situ hybridization as described above. Images were taken by a Zeiss LSM710 with a 63× (NA 1.4) objective as Z stacks with a 0.3-μm interval. All images in a single time-series group were taken under the same acquisition parameters for relative comparisons. Because imaging of six time points within each sample group required 7∼8 h, it was not practicable to image and compare WT and KO samples.
Imaging Analysis.
For time-lapse imaging, granules consistently motile during at least two time series were analyzed. Time-lapse imaging series were analyzed by ImarisTrack software (Bitplane). The track displacement is the distance between the first and last position. The track length is the total length of displacements within the track. Total trafficking length of motile particles was divided by time as average speed. The directionality, calculated by track displacement divided by track length, is the measurement of unidirectional movement. Colocalization between surface receptors and FMRP of primary dendrites, selected 20 μm away from soma, in deconvolved 3D rendering images was quantified by Manders's coefficient of FMRP staining (41), which is detailed in SI Materials and Methods. Three-dimensional reconstruction and surface rendering were applied to YFP neuron images by using the Surface function of Imaris. Spine and dendrite regions of interest were defined by YFP signals without visualizing other immunofluorescence channels. Intensity of CaMKIIα protein or mRNA was calculated as absolute intensity (pixel) per volume unit (voxel) and standardized by the value before DHPG in each group.
Statistical Analysis.
For mean comparisons, paired t test or one- or two-way ANOVA were performed. Tukey's honestly significant difference (HSD) or Dunnett's was carried out as post hoc analysis as mentioned in the figure legends. In all figures, data were presented as mean ± SEM, and *P < 0.05, **P < 0.01, ***P < 0.001.
Supplementary Material
Acknowledgments
We thank Dr. Kenneth S. Kosik (University of California, Santa Barbara, CA) for GFP-MS2-nls (nuclear localization signal) and MS2 binding site-CaMKIIα 3′UTR constructs and Dr. Stephan Ferguson (Robarts Research Institute, Canada) for FLAG-tagged mGluR1a, mGluR5, and β2AR constructs. We are grateful for pBS-CaMKIIα provided by Dr. Oswald Steward (University of California, Irvine, CA) and pCRFMR1e1 provided by Dr. Jim Eberwine (University of Pennsylvania) to generate cRNA probes. 1C3 is a kind gift of Dr. Jean-Louis Mandel (Collège de France, France). We thank Drs. Mayandi Sivaguru and Duohai Pan for assistance with imaging acquisition and analysis. We also thank W.T.G. laboratory members for helpful discussions. This work is funded in part by National Institutes of Health Grants MH35321 and HD002274-40S1, and the Spastic Paralysis Research Foundation of the Illinois-Eastern Iowa District of Kiwanis International.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1010564107/-/DCSupplemental.
References
- 1.Sutcliffe JS, et al. DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum Mol Genet. 1992;1:397–400. doi: 10.1093/hmg/1.6.397. [DOI] [PubMed] [Google Scholar]
- 2.Hagerman RJ. Lessons from fragile X regarding neurobiology, autism, and neurodegeneration. J Dev Behav Pediatr. 2006;27:63–74. doi: 10.1097/00004703-200602000-00012. [DOI] [PubMed] [Google Scholar]
- 3.Irwin SA, Galvez R, Greenough WT. Dendritic spine structural anomalies in fragile-X mental retardation syndrome. Cereb Cortex. 2000;10:1038–1044. doi: 10.1093/cercor/10.10.1038. [DOI] [PubMed] [Google Scholar]
- 4.Huber KM, Gallagher SM, Warren ST, Bear MF. Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proc Natl Acad Sci USA. 2002;99:7746–7750. doi: 10.1073/pnas.122205699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steward O, Schuman EM. Protein synthesis at synaptic sites on dendrites. Annu Rev Neurosci. 2001;24:299–325. doi: 10.1146/annurev.neuro.24.1.299. [DOI] [PubMed] [Google Scholar]
- 6.Weiler IJ, et al. Fragile X mental retardation protein is translated near synapses in response to neurotransmitter activation. Proc Natl Acad Sci USA. 1997;94:5395–5400. doi: 10.1073/pnas.94.10.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang H, Schuman EM. A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science. 1996;273:1402–1406. doi: 10.1126/science.273.5280.1402. [DOI] [PubMed] [Google Scholar]
- 8.Nosyreva ED, Huber KM. Metabotropic receptor-dependent long-term depression persists in the absence of protein synthesis in the mouse model of fragile X syndrome. J Neurophysiol. 2006;95:3291–3295. doi: 10.1152/jn.01316.2005. [DOI] [PubMed] [Google Scholar]
- 9.Vanderklish PW, Edelman GM. Dendritic spines elongate after stimulation of group 1 metabotropic glutamate receptors in cultured hippocampal neurons. Proc Natl Acad Sci USA. 2002;99:1639–1644. doi: 10.1073/pnas.032681099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hou L, et al. Dynamic translational and proteasomal regulation of fragile X mental retardation protein controls mGluR-dependent long-term depression. Neuron. 2006;51:441–454. doi: 10.1016/j.neuron.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 11.Todd PK, Mack KJ, Malter JS. The fragile X mental retardation protein is required for type-I metabotropic glutamate receptor-dependent translation of PSD-95. Proc Natl Acad Sci USA. 2003;100:14374–14378. doi: 10.1073/pnas.2336265100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu R, et al. The fragile X protein controls microtubule-associated protein 1B translation and microtubule stability in brain neuron development. Proc Natl Acad Sci USA. 2004;101:15201–15206. doi: 10.1073/pnas.0404995101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyashiro KY, et al. RNA cargoes associating with FMRP reveal deficits in cellular functioning in Fmr1 null mice. Neuron. 2003;37:417–431. doi: 10.1016/s0896-6273(03)00034-5. [DOI] [PubMed] [Google Scholar]
- 14.Brown V, et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- 15.Antar LN, Afroz R, Dictenberg JB, Carroll RC, Bassell GJ. Metabotropic glutamate receptor activation regulates fragile X mental retardation protein and FMR1 mRNA localization differentially in dendrites and at synapses. J Neurosci. 2004;24:2648–2655. doi: 10.1523/JNEUROSCI.0099-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: Isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 17.Ling SC, Fahrner PS, Greenough WT, Gelfand VI. Transport of Drosophila fragile X mental retardation protein-containing ribonucleoprotein granules by kinesin-1 and cytoplasmic dynein. Proc Natl Acad Sci USA. 2004;101:17428–17433. doi: 10.1073/pnas.0408114101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davidovic L, et al. The fragile X mental retardation protein is a molecular adaptor between the neurospecific KIF3C kinesin and dendritic RNA granules. Hum Mol Genet. 2007;16:3047–3058. doi: 10.1093/hmg/ddm263. [DOI] [PubMed] [Google Scholar]
- 19.Dictenberg JB, Swanger SA, Antar LN, Singer RH, Bassell GJ. A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Dev Cell. 2008;14:926–939. doi: 10.1016/j.devcel.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greenough WT, et al. Synaptic regulation of protein synthesis and the fragile X protein. Proc Natl Acad Sci USA. 2001;98:7101–7106. doi: 10.1073/pnas.141145998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aakalu G, Smith WB, Nguyen N, Jiang C, Schuman EM. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron. 2001;30:489–502. doi: 10.1016/s0896-6273(01)00295-1. [DOI] [PubMed] [Google Scholar]
- 22.Job C, Eberwine J. Identification of sites for exponential translation in living dendrites. Proc Natl Acad Sci USA. 2001;98:13037–13042. doi: 10.1073/pnas.231485698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steward O, Wallace CS, Lyford GL, Worley PF. Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron. 1998;21:741–751. doi: 10.1016/s0896-6273(00)80591-7. [DOI] [PubMed] [Google Scholar]
- 24.Rook MS, Lu M, Kosik KS. CaMKIIα 3′ untranslated region-directed mRNA translocation in living neurons: Visualization by GFP linkage. J Neurosci. 2000;20:6385–6393. doi: 10.1523/JNEUROSCI.20-17-06385.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ashley CT, Jr, Wilkinson KD, Reines D, Warren ST. FMR1 protein: Conserved RNP family domains and selective RNA binding. Science. 1993;262:563–566. doi: 10.1126/science.7692601. [DOI] [PubMed] [Google Scholar]
- 26.Schaeffer C, et al. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J. 2001;20:4803–4813. doi: 10.1093/emboj/20.17.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dolzhanskaya N, Sung YJ, Conti J, Currie JR, Denman RB. The fragile X mental retardation protein interacts with U-rich RNAs in a yeast three-hybrid system. Biochem Biophys Res Commun. 2003;305:434–441. doi: 10.1016/s0006-291x(03)00766-6. [DOI] [PubMed] [Google Scholar]
- 28.Burgueño J, et al. Metabotropic glutamate type 1α receptor localizes in low-density caveolin-rich plasma membrane fractions. J Neurochem. 2003;86:785–791. doi: 10.1046/j.1471-4159.2003.01842.x. [DOI] [PubMed] [Google Scholar]
- 29.Gallagher SM, Daly CA, Bear MF, Huber KM. Extracellular signal-regulated protein kinase activation is required for metabotropic glutamate receptor-dependent long-term depression in hippocampal area CA1. J Neurosci. 2004;24:4859–4864. doi: 10.1523/JNEUROSCI.5407-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang H, et al. Dynamic association of the fragile X mental retardation protein as a messenger ribonucleoprotein between microtubules and polyribosomes. Mol Biol Cell. 2008;19:105–114. doi: 10.1091/mbc.E07-06-0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferrari F, et al. The fragile X mental retardation protein-RNP granules show an mGluR-dependent localization in the post-synaptic spines. Mol Cell Neurosci. 2007;34:343–354. doi: 10.1016/j.mcn.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 32.Yoshimura A, et al. Myosin-Va facilitates the accumulation of mRNA/protein complex in dendritic spines. Curr Biol. 2006;16:2345–2351. doi: 10.1016/j.cub.2006.10.024. [DOI] [PubMed] [Google Scholar]
- 33.Fujii R, et al. The RNA binding protein TLS is translocated to dendritic spines by mGluR5 activation and regulates spine morphology. Curr Biol. 2005;15:587–593. doi: 10.1016/j.cub.2005.01.058. [DOI] [PubMed] [Google Scholar]
- 34.Ohashi S, et al. Identification of mRNA/protein (mRNP) complexes containing Purα, mStaufen, fragile X protein, and myosin Va and their association with rough endoplasmic reticulum equipped with a kinesin motor. J Biol Chem. 2002;277:37804–37810. doi: 10.1074/jbc.M203608200. [DOI] [PubMed] [Google Scholar]
- 35.Estes PS, O'Shea M, Clasen S, Zarnescu DC. Fragile X protein controls the efficacy of mRNA transport in Drosophila neurons. Mol Cell Neurosci. 2008;39:170–179. doi: 10.1016/j.mcn.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 36.Bramham CR, Wells DG. Dendritic mRNA: Transport, translation and function. Nat Rev Neurosci. 2007;8:776–789. doi: 10.1038/nrn2150. [DOI] [PubMed] [Google Scholar]
- 37.Grossman AW, Aldridge GM, Weiler IJ, Greenough WT. Local protein synthesis and spine morphogenesis: Fragile X syndrome and beyond. J Neurosci. 2006;26:7151–7155. doi: 10.1523/JNEUROSCI.1790-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ceman S, et al. Phosphorylation influences the translation state of FMRP-associated polyribosomes. Hum Mol Genet. 2003;12:3295–3305. doi: 10.1093/hmg/ddg350. [DOI] [PubMed] [Google Scholar]
- 39.Cheever A, Ceman S. Phosphorylation of FMRP inhibits association with Dicer. RNA. 2009;15:362–366. doi: 10.1261/rna.1500809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edbauer D, et al. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65:373–384. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manders EEM, Verbeek FJ, Aten JA. Measurement of co-localisation of objects in dual-colour confocal images. J Microsc. 1993;169:375–382. doi: 10.1111/j.1365-2818.1993.tb03313.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.