Abstract
A thorough understanding of the circadian clock requires qualitative evaluation of circadian clock gene expression. Thus far, no simple and effective method for detecting human clock gene expression has become available. This limitation has greatly hampered our understanding of human circadian rhythm. Here we report a convenient, reliable, and less invasive method for detecting human clock gene expression using biopsy samples of hair follicle cells from the head or chin. We show that the circadian phase of clock gene expression in hair follicle cells accurately reflects that of individual behavioral rhythms, demonstrating that this strategy is appropriate for evaluating the human peripheral circadian clock. Furthermore, using this method, we indicate that rotating shift workers suffer from a serious time lag between circadian gene expression rhythms and lifestyle. Qualitative evaluation of clock gene expression in hair follicle cells, therefore, may be an effective approach for studying the human circadian clock in the clinical setting.
Keywords: circadian rhythm, clock gene, shift work, jet lag, chronotherapy
Circadian behavioral and physiological rhythms are driven by autonomous oscillation of clock gene expression (1–3). Thus, a clear understanding of the circadian core clock requires a thorough examination of the rhythmic expression of clock genes. As evidenced by the correlation between circadian dysfunction and the incidence of pathological diseases such as sleep disorders, metabolic syndromes, and cancer (4–7), newer strategies for monitoring clock gene expression in humans are necessary for preventing circadian rhythm-related diseases. Furthermore, medical evaluation of circadian rhythm disorders, basic research on shift work and jet lag, and chronopharmacological applications also require effective strategies for studying human clock gene expression. However, currently there is no established simple and accurate method for detecting human circadian clock gene expression. This is underscored by the fact that despite the identification of major mammalian clock genes from 1997 to 1999 (8–14), there are only about 10 studies reporting the basic research and clinical application of human clock gene expression in vivo (15, 16). The lack of an established method for evaluating circadian clock gene expression has markedly impeded the progress for studying human circadian rhythms. Circadian clock gene expression is detectable not only in the central circadian pacemaker, the suprachiasmatic nucleus, but also in almost all peripheral tissues. The circadian clock is a cell-autonomous system, and numerous cell-autonomous peripheral clocks are synchronized and governed by the suprachiasmatic nucleus (17, 18). These findings suggest that it is possible to characterize the human circadian clock by determining clock gene mRNA expression in peripheral tissues. In previous reports, white blood cells or oral mucosa samples were used for the detection of human clock gene expression, although these methods have several potential issues and problems which may be avoidable with well-improved and sophisticated techniques. To avoid these potential pitfalls, we report that high-quality RNA can be obtained, less invasively, from head or chin hair follicle cells, and that circadian oscillation of clock gene expression can easily and reliably be detected with good sensitivity using this technique.
Results and Discussion
Method for Measuring Circadian Expression of Human Clock Genes Using Hair Follicle Cells.
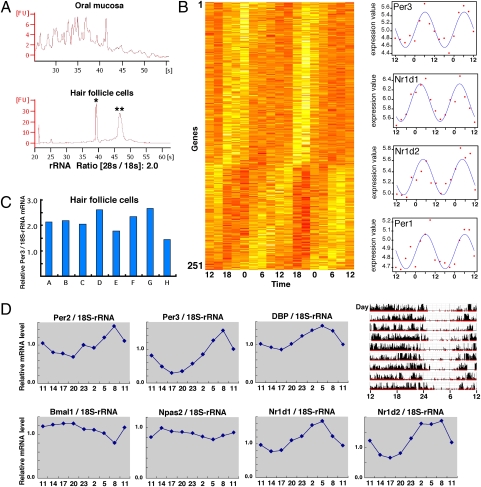
To determine the circadian expression of human clock genes, we examined mRNA levels of clock genes in white blood cells and oral mucosa, as previously reported (15, 16). In the case of white blood cells, as noted above, physical stimuli and time delays due to the processing of cell separation may affect levels of expression of clock genes. Consequently, we were unable to obtain reproducible results using this method, although some of the difficulties were overcome when blood samples were collected around the clock. As for the oral mucosa, total RNA samples extracted from oral mucosa were severely fragmented (Fig. 1A) and, therefore, we were unable to obtain reproducible results using this method. Because of the difficulties with these two previous approaches, we decided to focus our efforts on another source of peripheral tissue, the hair follicle cells, which remain attached to plucked hairs. The distinct advantages of using hair follicle cells are that they could be obtained relatively noninvasively and fresh naked cells could be obtained simply by plucking hairs without additional cell separation. Indeed, total RNA purified from scalp hair follicle cells exhibits clearly distinguishable peaks representing 18S- and 28S-rRNA signals (Fig. 1A), suggesting that these cells are suitable for isolation of high-quality total RNA. As a next step, we examined whether hair follicle cells have a functional cellular clock, because of reports that certain clocks may not be present in a few specific peripheral tissues (19). We therefore performed a comprehensive survey of genes expressed in a circadian fashion in human hair follicle cells by DNA microarray analysis (Fig. 1B). Genes with significant expression changes were extracted from microarray data with ANOVA. Fast Fourier transform (FFT) and cosine fitting correlation (CF) analysis were performed on the time-course expression data of these genes. Based on the 48-h diurnal profiles of gene expression, we found that mRNA levels of 220 (CF) and 251 (FFT) genes oscillated in circadian fashion with various phases (Fig. S1A; the names of these genes are listed in Dataset S1). Clock genes such as Period1 (Per1), Period3 (Per3), Nr1d1 (Rev-erbα), and Nr1d2 (Rev-erbβ) were highly ranked in both CF- and FFT-selected gene lists (Fig. 1B and Fig. S1B).
Fig. 1.
Biological rhythms exist in hair follicle cells of the scalp and are useful for measuring human clock gene expression rhythms. (A) Qualitative comparison of total RNA obtained from the oral mucosa or hair follicle cells of the scalp. Total RNA was collected from both samples and isolated by electrophoresis. Single and double asterisks indicate 18S and 28S rRNA, respectively. (B) DNA microarray analysis was performed to comprehensively measure rhythms of gene expression in hair follicle cells. Based on the 48-h diurnal expression profiles, genes with circadian expression were organized in a phase sequence (see SI Materials and Methods for details). Dataset S1 presents gene names in detail. The darker the color, the higher the gene expression, in each time zone. This gene group includes Per1, Per3, Nr1d1, and Nr1d2. The right figures show the actual values for these four clock genes (scatter diagram) and the fitted cosine curves. (C) Using total RNA collected from 10 scalp hairs, real-time PCR was performed to measure the expression of Per3. This was repeated eight times to assess fluctuation. Expressions relative to 18S rRNA are shown. (D) Scalp hair samples were collected every 3 h to ascertain rhythms of clock gene expression by real-time PCR. Expressions relative to 18S rRNA are shown. (Upper Right) Activity data over a period of about 9 d for the same individual. The final day in the actogram is the sampling day. The P values in cosine curve fitting are Per2 (0.0062), Per3 (0.0010), Dbp (0.00029), Bmal1 (0.016), Npas2 (0.030), Nr1d1 (0.00019), and Nr1d2 (0.00039).
In subsequent experiments, we adopted a commonly used real-time PCR method to examine clock gene expression, because our aim was to establish a simple and reliable method for accurately assessing human circadian rhythm. As shown in Fig. 1C, we examined the number of hairs required for reproducible results by measuring levels of expression of Per3, and found that 10 head hairs were sufficient to reduce experimental error to less than 2-fold. However, the number of hairs required is strongly affected by individual variation and gender. In the case of thick hairs, 5 head hairs was sufficient for reproducible detection, whereas in other cases such as thin hairs, 20 hairs were required for reliable detection. In addition, results vary depending on experimental conditions, such as the method used to pluck hairs, the method of total RNA extraction, the efficiency of reverse transcription, and the real-time PCR protocol used. Almost all of the results shown in this report were obtained with general methods commonly used to determine mRNA levels (details are described in Materials and Methods). As shown in Fig. 1D, we collected head hair samples at 3-h intervals around the clock, and examined the circadian expression of seven clock genes. Sampling was begun after a period of maintenance of a fixed waking/meal/sleep schedule (the final day in the actogram is the sampling day). Although the expression of Per3, Nr1d1, and Nr1d2 genes exhibited circadian fluctuations, Per2 and Dbp genes oscillated with lower amplitudes and therefore were less useful for the detection of circadian properties. For Bmal1 and Npas2, only slight oscillations were detected. These results are consistent with our finding that only Per3, Nr1d1, and Nr1d2 genes were identified as rhythmically expressed clock genes in the DNA microarray analysis. Peak time intervals among these seven clock genes were conserved compared with their corresponding genes in mice (1). Although PER3, NR1D1, and NR1D2 are not components of the classical core negative feedback loop (20), these three genes meet the criteria for rhythm markers of the circadian clock.
Correlation Between Human Behavioral Rhythms and Circadian Clock Gene Expression in Hair Follicle Cells.
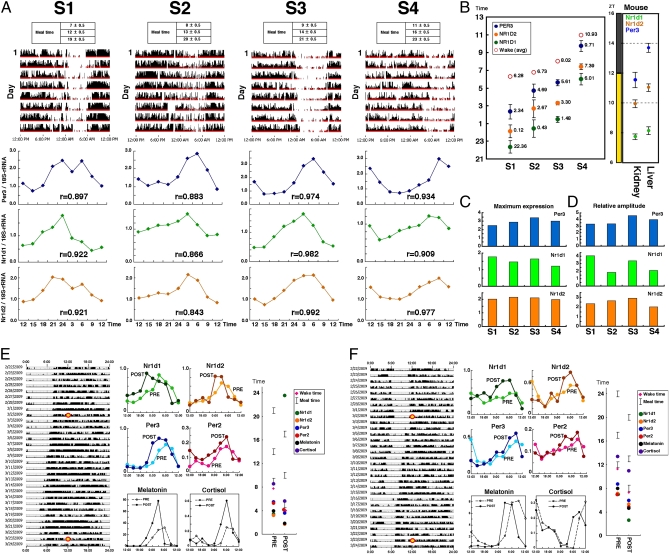
To demonstrate that circadian properties of clock gene expression in hair follicle cells can be used as markers for determination of the individual circadian clock pacemaker, we examined whether the circadian phase of clock gene expression in hair follicle cells reflects that of individual behavioral rhythms. We compared the circadian phases of Per3, Nr1d1, and Nr1d2 mRNA rhythms in head hair follicle cells obtained from four normal healthy subjects who maintained a regular lifestyle with a specific phase (Fig. 2A and Figs. S1–S4). Sampling was begun after a period of maintenance of a fixed waking/meal/sleep schedule based on the lifestyle of each individual (the final day in the actogram is the sampling day). The intervals between wakefulness, meals, and sleep were similar among these subjects. As shown in Fig. 2, we performed objective monitoring of behavioral rhythms with a wristwatch-type device, the Actiwatch (Mini Mitter). Clock gene expression in head hair follicle cells was thus measured under these conditions, and circadian fluctuation of Per3, Nr1d1, and Nr1d2 gene expression was detected in all subjects. These time-series data were fitted to a cosine curve with a high correlation coefficient (shown as “r”). As shown in Fig. 2B, the phase intervals among these three genes were strictly maintained in all subjects, and were almost the same as those in mouse peripheral tissues, indicating that the mechanism driving circadian gene expression is highly conserved across species. The peak time of expression of these genes also correlated with the average of wakefulness times over the 9-d period just before time-course sampling. For example, the earliest phase of circadian gene expression was observed in subject S1, who woke up earliest. Interestingly, in both species, Per3 expression peaked just around waking time (note that the onset of activity occurs in mice immediately after lights-off), although the phase was tissue-specific. Thus, the phase correlation between behavioral rhythms and circadian clock gene expression is conserved and is independent of whether the animals are diurnal or nocturnal. Furthermore, there were no remarkable individual differences in the maximum level or the circadian amplitude of clock gene expression in hair follicle cells (Fig. 2 C and D). These findings indicate that circadian properties other than phase time (such as phase correlation, absolute amount of expression, and amplitude) in circadian clock gene expression are not severely affected by differences in genomic background or lifestyles. Based upon these findings, we concluded that the circadian phase of clock gene expression in hair follicle cells accurately reflects that of individual behavioral rhythms, indicating that this method can be used to evaluate the human peripheral circadian clock.
Fig. 2.
Clock gene expression in hair follicle cells of the scalp reflects behavioral rhythms of each individual. (A) The upper and lower figures show activity data over a period of about 8 d and clock gene expression (Per3, Nr1d1, and Nr1d2) for four healthy individuals (S1–S4). The subjects were asked to rise and have meals at set times based on normal daily routines. Scalp hair samples were collected every 3 h to ascertain rhythms of clock gene expression by real-time PCR. Expressions relative to 18S rRNA are shown. r, correlation coefficient calculated by cosine curve fitting. (B) Cosine curve fitting was applied to clock gene expression rhythm data for each subject, and peak times were calculated. The peak times of these clock genes in the mouse liver and kidney are shown at right for interspecies comparison. (C and D) To compare individual differences in clock gene expression, maximum levels of expression (peak values) (C) and relative amplitudes (D) of rhythms of clock gene expression were calculated for each subject. (E and F) The effect of a 4-h phase advance of lifestyle rhythms on clock gene expression rhythms in hair follicle cells. (Left) The subjects were asked to rise and have meals at set times based on their normal daily routines for more than 1 wk before a scheduled phase shift. The lifestyle schedule (wakefulness/meals/sleep) was phase-advanced by 1 h per 5 d. Scalp hair samples were collected every 3 h from the time points marked with a yellow circle. (Upper Center) Expression rhythms of the four clock genes were examined. PRE, data before the phase shift; POST, data after the 4-h phase advance of lifestyle rhythms. (Lower Center) Salivary melatonin and cortisol concentrations were measured with ELISA. (Right) Cosine curve fitting was applied to clock gene expression and endocrine rhythm data for each subject, and the peak times were calculated. The average waking time, meal times, and peak time of clock gene expression and endocrine rhythms are shown. The P values of the phase difference between PRE and POST are Nr1d1 (E: 0.00015; F: 0.0018), Nr1d2 (E: 0.093; F: 0.038), Per3 (E: 0.019; F: 0.013), Per2 (E: 0.18; F: 0.022), melatonin (E: 0.034; F: 0.0019), and cortisol (E: 0.0018; F: 0.0060).
Next, we examined a phase shift of clock gene expression rhythms caused by a phase advance of lifestyle. The lifestyle (waking/meal/sleep schedule) of normal healthy subjects was gradually phase-advanced by 4 h over 3 wk (−1 h/5 d; Fig. 2 E and F). To support the phase advance, the subjects were required to be exposed to ∼10,000-lux light for 30 min just after waking. The average phase advances of clock gene expression rhythms in hair follicle cells were about 2.1 h (Fig. 2E) and 2.8 h (Fig. 2F). In melatonin and cortisol rhythms, about 2.5 h (Fig. 2E) and 2.9 h (Fig. 2F) phase advances were observed. These results reemphasize that our hair follicle-based strategy is powerful for evaluating the human peripheral circadian clock, and indicate that the 3-wk adaptation period is not enough to advance the phase of the molecular clock by 4 h.
To explore the possibilities of optimizing the body area for sampling and the measurement method for mRNA, hair follicle cells were collected from the chin, and gene expression was detected using a branched-DNA (bDNA)-based assay, an experimental system completely different from the real-time PCR assay (Fig. S4). The advantages of using this experimental system include the following. (i) More reproducible results can be obtained with the collection of smaller numbers of chin hairs than of head hairs. In the 11 samples containing 3 beard hairs, reproducible results were obtained for levels of expression of Per3, Nr1d1, and Nr1d2 (Fig. S4A). (ii) The bDNA-based system does not require RNA purification and reverse transcription, and thus reduces both acquisition time and handling errors. (iii) High-throughput detection is possible with automation (see Fig. S3). Using this experimental procedure, we were able to measure a circadian pattern of clock gene expression in a healthy subject (Fig. S4B). The time-series data fit a cosine curve well, and the phase relationship between Per3 and Nr1d2 was reproducible.
Practical Application: Circadian Clock Gene Expression in Rotating Shift Workers.
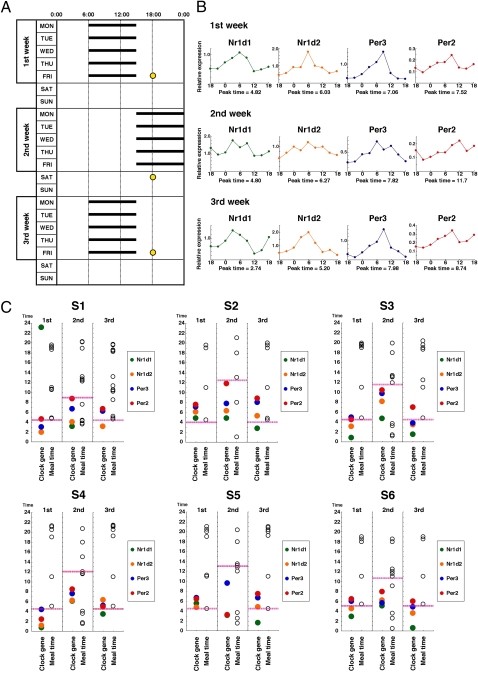
To confirm the usability of hair follicle cells, we tried applying this method to some field investigations. Physiological and epidemiological studies show that shift work causes various circadian rhythm disorders (21). Using our method, we characterized the circadian pacemaker of rotating shift workers by investigating circadian properties of clock gene expression in head hair follicle cells (Fig. 3). As shown in the pattern diagram of a shift-work schedule during 3 wk, an early shift (0600–1500 hours) and a late shift (1500–0000 hours) were alternately repeated (Fig. 3A). As noted below, an interesting pattern of circadian gene expression was observed in all of the six rotating shift workers (Fig. 3C; a representative raw dataset is shown in Fig. 3B). The phases of clock gene expression rhythms were not drastically shifted, compared with the phase shift of their lifestyle. The lifestyle was phase-delayed by about 7 h from the first to the second week, whereas the phase of circadian gene expression was delayed only by about 2 h (Fig. S5). A similar result was obtained in the case of the phase advance from the second to the third week. In healthy individuals, Per3 expression peaked more than 1 h before waking time (Fig. 2B). However, in rotating shift workers, it often peaked after waking time during the period of early shifts, indicating that late shift schedules started before the entrainment of the clock to early shifts was completed. The 1-wk adaptation was thus not sufficient for completing the entrainment. As shown in Fig. 2 E and F, these results are well-consistent with the fact that the 3-wk adaptation period is not enough for the 4-h phase advance of the molecular clock. However, the result would be dependent on the work schedule (Fig. S6). Previous studies assumed that about 10% of the genes expressed in peripheral tissues are regulated by the circadian clock pacemaker (22), suggesting that abnormal patterns of clock gene expression in rotating shift workers spread over the expression of downstream genes that are involved in various physiological processes. This propagation should markedly contribute not only to the development of circadian rhythm disorders but also to increased risk of heart attack, stroke, and sudden death in rotating shift workers. Thus, our hair cell-based strategy for the human circadian clock could be used to evaluate working conditions that do not disturb clock function.
Fig. 3.
Rhythms of clock gene expression in rotating shift workers. (A) The work shift table is shown for six rotating shift workers (S1–S6). At time points indicated with a yellow circle, scalp hair samples were collected every 3 h to ascertain rhythms of clock gene expression. (B) Clock gene expression rhythms in an individual are shown as representative of the six workers. Expression data relative to 18S rRNA are shown. Cosine curve fitting was applied to clock gene expression data, and the peak times were calculated. (C) In S1–S6, the average waking time (pink dashed line), meal times (open circles), and peak time of clock gene expression (colored circles) are plotted against time.
With Three-Point Sampling, It Is Possible to Predict the Phase of Clock Gene Expression Rhythms.
In the clinical setting, it may not be feasible to perform 24-h sampling every 3–4 h. Thus, biological clock phases will need to be predicted with less sampling. This is difficult, if not impossible, using experimental methods with inherently large fluctuations or errors. However, it may be possible with a stable measurement system using hair follicle cells. To use the phase interval value for phase prediction, we focused on Per3 and Nr1d2, whose expression rhythm data exhibited small individual differences in phase interval (Fig. 2B). Whereas marked individual differences were observed in the phases of rhythm of expression, the phase interval appeared stable for Per3 and Nr1d2 (2.2 ± 0.097 h; see details in SI Text, Result 1). When predicting rhythmic variations of gene expression using three-point data, accuracy of prediction is dependent on sampling interval. Although our mathematical analysis indicated high accuracy of prediction for many sampling time intervals (e.g., 6 h-6 h and 3 h-3 h intervals; see details in SI Text, Result 2), three-point prediction was not successful with some sampling time intervals (e.g., 9 h-15 h interval).
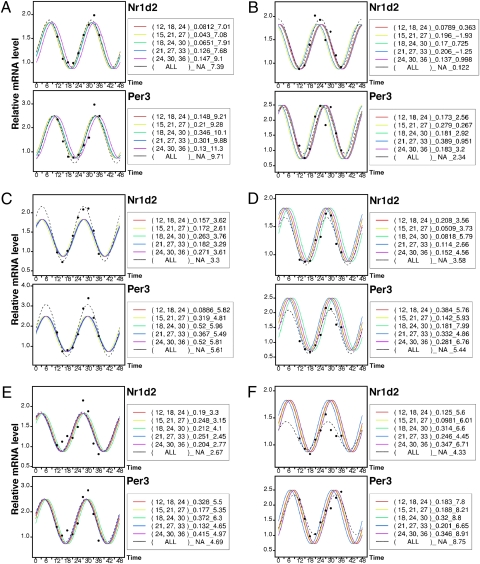
Next, based on these results, we attempted three-point phase prediction by the conjugate gradient method using actual data. Actual measurements of clock gene expression can deviate from expected values for various reasons. Phase prediction was performed with the above-noted optimal sampling interval, but three-point prediction including irregular values, which appear occasionally, resulted in poor prediction. With this method, although prediction was favorable in many cases, we sought a technique with more tolerance for fluctuations in data and errors. Previous findings for healthy individuals (Fig. 2 C and D) suggested that amplitude and level of clock gene expression rhythm remain within a certain range for all individuals. A cosine curve with the average amplitude and level of expression for healthy individuals was therefore defined as the standard curve, and the rhythm of expression in all healthy individuals was assumed to follow the standard curve, based on the results shown in Fig. 2. In other words, we assumed that no individual differences would be observed in wave elements, except for phases. The standard curve was obtained taking into account the phase interval in Per3 and Nr1d2 shown in SI Text, Result 1. In three-point phase prediction, phases were determined by the least-squares method to fit the standard curve as closely as possible. Fig. 4 shows the results of three-point phase prediction with a 6 h-6 h sampling interval (five three-point combinations). Unlike the conjugate gradient method, marked errors in prediction due to irregular data were not observed for six subjects (A–F), the maximum error in relation to nine-point calculated curves was about 2.5 h, and errors were within 1.5 h in most cases. To evaluate the validity of the standard-curve method, a 6-fold cross-validation was performed. The results showed that the method has enough accuracy for practical use: a maximum and average phase estimation error of 2.7 and 0.7 h, respectively. Furthermore, to examine the limitation of the standard-curve-based prediction method, with computer simulation, three point values (120,000 combinations) were selected from cosine curves with random amplitude, phase, and oscillation offset, and then phase estimations were performed. The phases predicted were plotted against each parameter (Fig. S8), which revealed that estimation errors fell within 1 h if the Per3-Nr1d2 phase difference (experimental average = ∼2.12 h) was between 1.2 and 3.2 h, regardless of amplitude and oscillation offset. Thus, the tolerance against data fluctuation is very high. In actual clinical settings, positioning of three sampling time points as close as possible is desirable. For example, in the case of 3 h-3 h intervals, the results also showed enough accuracy for practical use (SI Text, Result 3). We moreover succeeded in single-point phase prediction using expression data for nine clock genes obtained from mouse peripheral tissues; this suggests that even one-point phase prediction may be possible for human data with our standard-curve method, using more genes whose rhythms of expression are consistently detectable. There is a report on a single-point phase prediction method using expression profiles of more than 100 circadian output genes in mouse liver (23).
Fig. 4.
Prediction of expression rhythm phase by three-point data. Three-point phase prediction using actual data. Using the clock gene expression data obtained from scalp hair follicle cells of six healthy volunteers (A–F), phase prediction was performed using the standard curve for three-point assays with sampling time intervals of 6 h-6 h. The standard curve refers to the average cosine curve prepared using actual data. Factors other than phase time, such as period, amplitude, levels of expression, and the phase interval between Per3 and Nr1d2, were fixed. Black dots indicate actual measurements, the broken black line indicates a curve calculated based on all nine-point data (ALL), and colored lines indicate three-point predicted curves. Numbers in parentheses beside the figures indicate the time points of the three samplings used for prediction (actual data were obtained at nine points every 3 h, and three of the nine points were selected), and numbers at the right indicate root-mean-square errors of the three-point prediction curves and three measurements (the smaller the value, the more accurate the prediction). Additionally, numbers on the right show predicted peak times. The closer the nine-point calculated phase is to the three-point predicted phase, the more accurate the prediction.
The clock gene expression profiling data detailed in this report reveal phenotypes resulting from both endogenous and environmental factors. When a patient is diagnosed with a circadian rhythm disorder by clock gene expression profiling, it will be necessary to determine whether the disorder is due to lifestyle habits (environmental factors) or to autonomous clock dysfunction (endogenous factors). We therefore propose two-step diagnosis: first, clock gene expression profiling performed by our hair-based detection method (including both endogenous and environmental factors), and then tissue culture-based diagnosis for autonomous clock function (basically including only endogenous factors). As reported by Brown et al., the second step can be performed by monitoring the activity of a clock-regulated reporter gene transfected into primary-cultured fibroblasts obtained from the skin (24). By using this method, they have already succeeded in characterizing human chronotypes (25). However, because skin biopsy is burdensome to patients, another similar method may need to be developed without this type of biopsy. Brown et al. also demonstrated that on rare occasions, they succeeded in monitoring a clock-regulated gene activity by cultivating human primary hair root keratinocytes.
Although 10 y have passed since the first mammalian clock gene was identified, there are still only a handful of reports on in vivo observation of circadian expression of human clock genes. Researchers in the field of circadian rhythms can easily adopt our experimental strategy using hair follicle cells without specialized experimental techniques. Furthermore, if the technical improvements mentioned above are achieved (see details in SI Discussion), our approach could be used in the clinical setting. Because circadian clock dysfunction causes various human disorders, characterization of the circadian pacemaker would be of great value in predicting and preventing these diseases. Perhaps it could also be useful for evaluating therapeutic efficacy. In chronopharmacological treatment, estimation of internal time is required for objective determination of timing of administration, and this type of estimation could be carried out through the hair-based assay of clock gene expression. Finally, our method for assessing the circadian clock could also be used to optimize the schedule for shift work, thereby minimizing perturbations of endogenous clock function.
Supplementary Material
Acknowledgments
We thank Noriko Ezaki, Rie Ichiyama, Tomoko Tsuboi, Masami Haraguchi, Sakae Toshimitsu, and Rumiko Imaizumi for their expert technical assistance. We also thank Kazuhiro Nakagawa, Tomohiro Hayakawa, Fan Wang, Noriyuki Kishii, Tetsuaki Hirase, and Teruo Inoue for general support. We deeply appreciate Joseph Takahashi for his critical reading of the manuscript. This study was supported by a grant from the Mitsubishi Pharma Research Foundation and fellowships from the Japan Society for the Promotion of Science.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1003878107/-/DCSupplemental.
References
- 1.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 2.Lowrey PL, Takahashi JS. Mammalian circadian biology: Elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: Implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toh KL, et al. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- 5.Turek FW, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- 7.Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Young MW, Kay SA. Time zones: A comparative genetics of circadian clocks. Nat Rev Genet. 2001;2:702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- 9.King DP, et al. Positional cloning of the mouse circadian clock gene. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tei H, et al. Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature. 1997;389:512–516. doi: 10.1038/39086. [DOI] [PubMed] [Google Scholar]
- 11.Sun ZS, et al. RIGUI, a putative mammalian ortholog of the Drosophila period gene. Cell. 1997;90:1003–1011. doi: 10.1016/s0092-8674(00)80366-9. [DOI] [PubMed] [Google Scholar]
- 12.Gekakis N, et al. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 13.Kume K, et al. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 14.Lowrey PL, et al. Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science. 2000;288:483–492. doi: 10.1126/science.288.5465.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boivin DB, et al. Circadian clock genes oscillate in human peripheral blood mononuclear cells. Blood. 2003;102:4143–4145. doi: 10.1182/blood-2003-03-0779. [DOI] [PubMed] [Google Scholar]
- 16.Cajochen C, et al. Evening exposure to blue light stimulates the expression of the clock gene PER2 in humans. Eur J Neurosci. 2006;23:1082–1086. doi: 10.1111/j.1460-9568.2006.04613.x. [DOI] [PubMed] [Google Scholar]
- 17.Schibler U, Sassone-Corsi P. A web of circadian pacemakers. Cell. 2002;111:919–922. doi: 10.1016/s0092-8674(02)01225-4. [DOI] [PubMed] [Google Scholar]
- 18.Hastings MH, Reddy AB, Maywood ES. A clockwork web: Circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci. 2003;4:649–661. doi: 10.1038/nrn1177. [DOI] [PubMed] [Google Scholar]
- 19.Alvarez JD, Sehgal A. The thymus is similar to the testis in its pattern of circadian clock gene expression. J Biol Rhythms. 2005;20:111–121. doi: 10.1177/0748730404274078. [DOI] [PubMed] [Google Scholar]
- 20.Shearman LP, Jin X, Lee C, Reppert SM, Weaver DR. Targeted disruption of the mPer3 gene: Subtle effects on circadian clock function. Mol Cell Biol. 2000;20:6269–6275. doi: 10.1128/mcb.20.17.6269-6275.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knutsson A. Health disorders of shift workers. Occup Med (Lond) 2003;53:103–108. doi: 10.1093/occmed/kqg048. [DOI] [PubMed] [Google Scholar]
- 22.Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 23.Ueda HR, et al. Molecular-timetable methods for detection of body time and rhythm disorders from single-time-point genome-wide expression profiles. Proc Natl Acad Sci USA. 2004;101:11227–11232. doi: 10.1073/pnas.0401882101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown SA, et al. The period length of fibroblast circadian gene expression varies widely among human individuals. PLoS Biol. 2005;3:e338. doi: 10.1371/journal.pbio.0030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown SA, et al. Molecular insights into human daily behavior. Proc Natl Acad Sci USA. 2008;105:1602–1607. doi: 10.1073/pnas.0707772105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.