Abstract
Chromosomal instability and the subsequent genetic mutations are considered to be critical factors in the development of the majority of solid tumors. Here, we describe how the nucleoside diphosphate kinase Nm23-H1, a protein with a known link to cancer progression, regulates a critical step during cytokinesis. Nm23-H1 acts to provide a local source of GTP for the GTPase dynamin. Loss of Nm23-H1 in diploid cells leads to cytokinetic furrow regression, followed by cytokinesis failure and generation of tetraploid cells. Loss of dynamin phenocopies loss of Nm23-H1, and ectopic overexpression of WT dynamin complements the loss of Nm23-H1. In the absence of p53 signaling, the tetraploid cells resulting from loss of Nm23-H1 continue cycling and develop classic hallmarks of tumor cells. We thus provide evidence that the loss of Nm23-H1, an event suspected to promote metastasis, may additionally function at an earlier stage of tumor development to drive the acquisition of chromosomal instability.
Keywords: aneuploidy, RNAi screen, cell cycle, tetraploidy, p53
Chromosomal instability, or the inability to segregate chromosomes equally during cell division, plays a critical role in the development of the majority of solid tumors. One mechanism by which chromosomal instability develops is through a transient tetraploidy state (1). In a companion article, we describe the execution of loss-of-function shRNA screens in untransformed diploid human cells to identify kinases whose loss leads to increased ploidy. We further show that sustained loss of one of the screening hits, NME1, leads to aneuploidy, chromosomal instability, loss of contact inhibition, and anchorage-independent growth.
NME1 is the genomic locus that encodes the nucleoside diphosphate kinase (NDK) Nm23-H1; for the sake of consistency, we will refer to the gene, mRNA, and protein as NME1 in this article. NME1 is a member of the NDK family, whose biochemical role is to use ATP to generate a different nucleoside triphosphate (2). Although initially thought of as a housekeeping gene that maintains cellular pools of nucleoside triphosphates, interest in NME1 was sparked by its identification as a potential metastasis suppressor by Steeg et al. (3). A comparison of expression levels across mouse melanoma cell lines revealed an inverse correlation between NME1 expression and metastasis (3). These expression data were supported by the observation that overexpression of NME1 in human carcinoma cells is sufficient to reduce tumor cell motility (4–6). Additional evidence points to a role for NME1 in the direct regulation of factors that may drive motility and invasion during metastasis (5–11).
Given its connection with tumor progression, we were intrigued to identify NME1 as a regulator of chromosomal stability. To clarify the link between NME1, chromosomal instability, and tumor development, we have carried out an extensive analysis of the mechanism by which NME1 loss leads to chromosomal instability and contributes to tumor development.
Results
NME1 Regulates Chromosomal Stability.
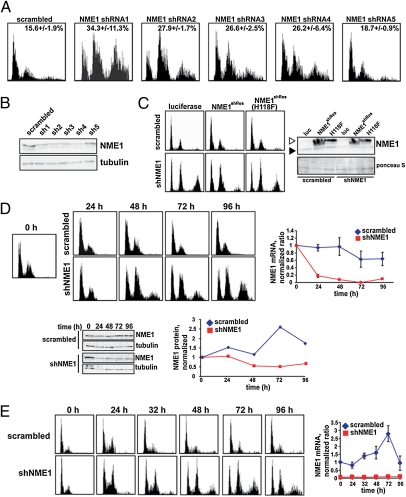
We first sought to link increased ploidy conclusively to a reduction in NME1 expression. First, we established that multiple shRNAs targeting different regions of the NME1 mRNA were capable of generating increased ploidy in HBE135 cells. As shown in Fig. 1A, four shRNAs targeting NME1 induced an increase in ploidy in HBE135 cells as measured by flow cytometry 4 d after the constructs were introduced. Also shown in Fig. 1A is the DNA content histogram of an additional shRNA targeting NME1 (shRNA 5), which did not induce a significant increase in ploidy. Consistent with the phenotypes of the five NME1 shRNAs, Western blot analysis showed that only those four shRNAs that reduced NME1 protein levels caused an increase in ploidy (Fig. 1B). Because the ploidy phenotype induced by shRNA 1 was slightly stronger than that of the other shRNAs, all experiments discussed below made use of NME1 shRNA 1 unless otherwise indicated.
Fig. 1.
NME1 regulates chromosomal stability. (A) Multiple shRNAs against NME1 cause an increase in ploidy. A scrambled control shRNA or five different shRNAs against NME1 were transduced into HBE135 cells. Cells were fixed for flow cytometric analysis at 4 d. (B) Protein knockdown for multiple NME1 shRNAs. Protein extracts were prepared in parallel when cells were fixed as in A. Normalized protein extracts were subjected to SDS/PAGE and Western blot analysis for the indicated proteins. (C) Overexpression of NME1 cDNA complements the NME1 knockdown phenotype. The indicated lentiviral shRNA and cDNA constructs were transduced into confluent HBE135 cells, and the cells were reseeded after 4 d and fixed for flow cytometric analysis after an additional 4 d. Protein extracts were also prepared and subjected to Western blot analysis to measure the level of endogenous (closed arrowhead) and Flag-tagged NME1 protein (open arrowhead). Ponceau S staining was used as a loading control. luc, luciferase; shNME1, short hairpin NME1. (D) Increase in tetraploid cells occurs 48 h following transduction of NME1 shRNA. Control or NME1 shRNAs were introduced into HBE135 cells, and cells were prepared for flow cytometric, mRNA, or Western blot analysis at the indicated time points. For quantification, data points were normalized to the mRNA or protein level at time 0. (E) Abortive mitosis occurs within 24 h of knockdown of NME1. HBE135 cells were infected at confluence with scrambled or NME1 shRNA virus in the LKO/H2BGFP vector. After 4 d, the infected cells were seeded in multiple wells, fixed for flow cytometric analysis, and lysed for mRNA analysis at the indicated time points after release from confluence. (Left) DNA content histograms are shown for the GFP-positive population only. (Right) First-strand cDNA was prepared and used for qPCR. The level of NME1 mRNA was normalized to the level of GFP mRNA in scrambled and shNME1 samples. The y axis represents the ratio of the normalized value at each time point and the normalized value of the scrambled control at the time of release (0 h).
To rule out an off-target effect conclusively, we carried out cDNA complementation experiments of the shRNA knockdown phenotype using NME1 cDNA in a lentiviral expression vector. Silent mutations were introduced into the coding sequence of the NME1 cDNA used for complementation [NME1 shRNA resistant (NME1shRes)] to prevent the resulting mRNA from being targeted by the shRNA. HBE135 cells were coinfected with lentiviruses expressing either scrambled or NME1 shRNA constructs and lentiviruses expressing luciferase or Flag-tagged NME1shRes cDNA overexpression constructs. The NME1 cDNA represented either the catalytically active (NME1shRes) or inactive [NME1shRes(H118F)] version (12). As shown in Fig. 1C Left, ectopic expression of WT but not catalytically inactive NME1shRes significantly abrogated the increase in ploidy generated by expression of the NME1 shRNA. Western blot analysis showed that rescue was not simply attributable to failure to eliminate the endogenous protein (Fig. 1C Right). Our validation experiments with NME1 both unequivocally eliminated off-target effects and showed that catalytic activity of NME1 is required for its function in regulating chromosomal stability.
Increased Ploidy Occurs Within 24 h of NME1 Knockdown.
We next sought to determine the kinetics of mRNA and protein knockdown and increased ploidy resulting from NME1 knockdown. Control or NME1 shRNAs were transduced into HBE135 cells. Cells were fixed for flow cytometry, and mRNA and protein extracts were prepared 24, 48, 72, and 96 h following infection. As shown in Fig. 1D, a substantial increase in the population of cells with greater than 4N DNA content was detected in cells containing the NME1 shRNA at 48 h. This increase occurred 24 h following the initial decrease in mRNA and was concurrent with the reduction in protein concentration.
The asynchronous nature of the cell population in Fig. 1D made it difficult to demonstrate conclusively that the appearance of a tetraploid population was a direct consequence of NME1 knockdown. To address this, we introduced the lentiviral shRNA constructs into confluent HBE135 cells, ensuring that knockdown would occur while the majority of cells were arrested in G0/G1. To mark infected cells, we made use of a modified lentiviral shRNA construct containing GFP-tagged histone H2B (H2BGFP) in place of the puromycin acetyltransferase gene. Cells were released from confluence 4 d after infection and were harvested for flow cytometry, immunofluorescence, and mRNA analysis at various time points.
In Fig. 1E, DNA content histograms are shown for only the GFP-positive (and thus infected) populations. At the time of release, both control and NME1 shRNA-treated cells were arrested primarily in G0/G1. The absence of any increased ploidy in the cells with low levels of NME1 that were arrested in G0/G1 suggested that the cells must pass through the cell cycle in order for the phenotype to be observed. In short hairpin (sh) NME1 cells, unlike scrambled controls, a significant and persistent 4-N peak (likely diploid cells in mitosis) was observed by 24 h and a population of cells with greater than 4N DNA content appeared by 48 h. The 4N and greater than 4N populations in the cells lacking NME1 were composed of tetraploid cells in G1 and S/G2 phases, respectively. These data were consistent with those shown in Fig. 1D and suggested that the increase in ploidy was the result of an error in the first mitosis after knockdown of NME1.
Knockdown of NME1 Results in Cytokinesis Failure.
To understand the mechanism by which loss of NME1 leads to tetraploidy, we visualized mitoses in cells depleted of NME1 using time-lapse video microscopy to follow infected cells through the cell cycle. For the time-lapse studies, we introduced scrambled or NME1 shRNA constructs in the H2BGFP vector into four different cell lines: HBE135; HeLa; and two variants of primary human foreskin keratinocytes (HFKs), one carrying an empty vector (HFK) and one expressing the HPV16 E6 oncoprotein (HFK/E6). The former two cell lines would allow us to compare the impact of NME1 reduction in near-diploid and aneuploid cells. Use of the HFK paired cell lines allowed us to determine the role of NME1 in primary cells and to elucidate the importance of p53 activity on the NME1 knockdown phenotype.
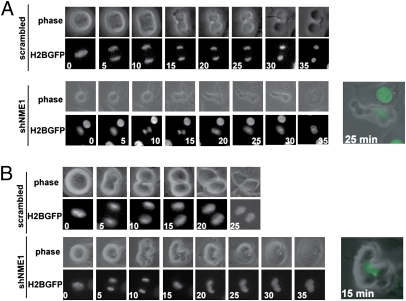
As shown in Fig. 2 and Figs. S1 and S2, in each cell line, knockdown of NME1 had the same profound effect on the completion of cell division. Although the appearance of the cells in metaphase was unremarkable (0-min time point), abnormalities began to appear on initiation of anaphase and cytokinesis. Equatorial furrow ingression was initiated; however, while the furrow paused for several minutes, the segregating daughter genomes collapsed and the chromosomes decondensed. Finally, the equatorial furrow regressed, giving rise to binucleate tetraploid cells. For each cell line expressing the NME1 shRNA, there was a substantial increase in the percentage of cells experiencing this cytokinesis failure. The phenotypic penetrance was highest in HFK/E6 (68%), followed by HBE135 (61%), HFK (40%), and HeLa (16%).
Fig. 2.
Knockdown of NME1 results in cytokinesis failure. (A) Examples of the late stages of mitosis in HBE135 cells expressing a scrambled control or NME1 shRNA. HBE135 cells were infected with shRNAs in the LKO/H2BGFP vector. After 24 h, cells were split into glass-bottom dishes. At 48 h postinfection, cells were imaged for 24 h for phase contrast and GFP. Images were collected every 5 min at 6 (scramble) or 15 [short hairpin NME1 (shNME1)] stage positions. Still images begin at the frame before the initiation of anaphase; numbers indicate time in minutes after anaphase initiation. (B) Examples of the late stages of mitosis in control or shNME1 primary HFKs. Cells were infected and analyzed as described in A. Images were captured at eight stage positions for each sample.
From these experiments, we concluded that NME1 loss leads to cytokinesis failure. This phenomenon appeared to be general because it was observed in primary, immortalized, and aneuploid tumor cell lines. Further, because identical phenotypes were observed in both HFK cells and HFK/E6 cells, we concluded that the presence of active p53 does not prevent tetraploid formation driven by NME1 loss. We will further address this conclusion below.
NME1 Localizes to the Cortex of Mitotic Cells.
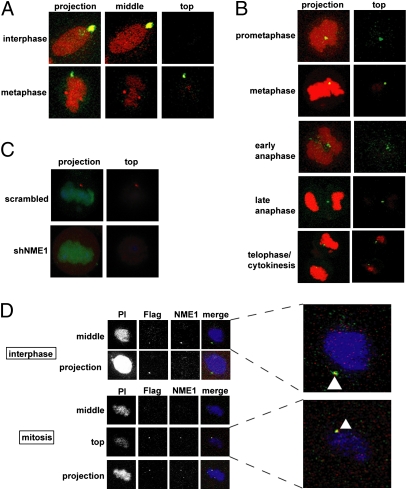
To understand how NME1 might regulate cytokinesis, we carried out experiments to determine its intracellular localization during the cell division process. Two groups have previously demonstrated that NME1 localizes to the centrosome (13, 14), which was difficult to reconcile with a function in cytokinesis. Consistent with this, we observed NME1 in the centrosome in interphase cells (Fig. 3A). However, during metaphase, NME1 was found to localize to the cortex along with a fraction of γ-tubulin (Fig. 3A) and was notably absent from the spindle poles. Analysis of other stages of mitosis revealed a cortical localization of NME1 during other stages of mitosis, with the localization being in the equatorial region during anaphase and telophase/cytokinesis (Fig. 3B).
Fig. 3.
NME1 localizes to the cortex of mitotic cells. (A) NME1 colocalizes with γ-tubulin in interphase and metaphase. HBE135 cells were grown on coverslips and fixed for immunofluorescence. Cells were stained with antibodies against NME1 (green) and γ-tubulin (red). DNA was stained with propidium iodide (red). Images of cells were captured as ≈50 0.37-μm confocal sections. middle, section through the middle of the cell; top, section at the top of the cell; projection, all sections projected onto a single image. (B) NME1 localizes to the cortex of mitotic cells. HBE135 cells were processed and stained with antibodies against NME1 (green). DNA was stained with propidium iodide (red). Confocal sections were obtained as in A. Images shown are merged images of a projection of all confocal sections and merged images of a section at the top of the cell. (C) NME1 cortical signal is absent in cells expressing the NME1 shRNA. HBE135 cells were transduced with lentiviral scrambled or NME1 shRNAs containing H2BGFP. After 32 h, cells were fixed and stained with anti-GFP and anti-NME1. Merged confocal projections are shown. shNME1, short hairpin NME1. (D) Flag-tagged NME1 localizes to the cortex. HBE135 cells were transduced with lentiviral Flag-tagged NME1. After 2 d, cells were fixed and stained with anti-Flag and anti-NME1 antibodies. Nuclei were stained with propidium iodide (PI). Confocal sections are as described in A. (Right) Zoomed images with arrowheads show colocalization (yellow) of the Flag (green) and NME1 (red) signals.
There are several lines of evidence that support this unexpected localization of NME1. First, we found that the fluorescence signal at the cortex disappeared in cells expressing NME1 shRNA (Fig. 3C). Second, the cortical localization of NME1 was observed using two independent antibody preparations: rabbit polyclonal anti-NME1 (Fig. 3 A and B, prometaphase and anaphase) and mouse monoclonal anti-NME1 (Fig. 3B, metaphase and cytokinesis). Third, similar cortical localization was observed with both paraformaldehyde/methanol fixation (Figs. 3A and 3B, prometaphase and anaphase) and trichloroacetic acid fixation (Fig. 3B, metaphase and cytokinesis). Fourth, ectopically expressed Flag-tagged NME1, which we established above, can functionally rescue NME1 knockdown (Fig. 1C) localized to the cortex when detected with an anti-Flag antibody (Fig. 3D). Finally, immunofluorescence experiments in primary HFK cells confirmed the cortical localization of NME1 (Fig. S3). Taken together, these data showed that NME1 localizes to the cortex in mitosis, and functional studies suggested that NME1 acts there to ensure completion of cytokinesis.
GTPase Dynamin Acts Downstream of NME1 During Cytokinesis.
Proteins whose activity is regulated by the binding and hydrolysis of GTP, or G proteins, play a significant role in a number of cellular signaling events. Because the NDK activity of NME1 is required to complement the NME1 knockdown phenotype (Fig. 1C), we imagined that the function of NME1 at the cortex may be to provide a local supply of GTP for one or more downstream GTPases that act during cytokinesis.
We first hypothesized that knocking down the expression of a protein that is downstream of NME1 should phenocopy the knockdown of NME1 itself. We considered three GTPases known to have roles in cytokinesis: the small GTPases RhoA and Cdc42 and the large GTPase dynamin (15–18). After preparing shRNAs that effectively knocked down these genes (Fig. S4), we transferred the shRNAs into the H2BGFP shRNA vector described above and carried out time-lapse imaging experiments as described in Fig. 2. To eliminate potential premitosis phenotypes, HBE135 cells were virally transduced at confluence and were released after 3 d and imaged 24 h later. Although cells with reduced Cdc42 expression rarely reached cytokinesis and often died, we were able to obtain significant numbers of mitoses in cells with reduced RhoA or dynamin expression.
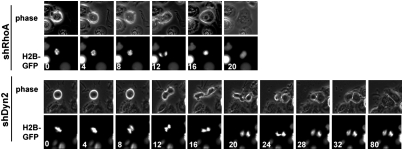
As shown in Fig. 4, knockdown of either RhoA or dynamin resulted in highly penetrant but phenotypically distinct cytokinesis failure phenotypes (Fig. 4). In mitotic cells with reduced RhoA expression, initiation of anaphase was followed by either minimal furrow ingression or no furrow ingression (Fig. 4 Upper, 12 min). In cells with reduced dynamin expression, equatorial furrow ingression occurred to near completion before pausing and regressing (Fig. 4 Lower, 16 min). Quantification of the time-lapse data revealed that in the majority of dynamin knockdown cells (but not RhoA knockdown cells), cytokinesis failure was preceded by regression of a nearly fully ingressed furrow (Fig. S5). This is consistent with work in Caenorhabditis elegans showing that expression of a mutant dynamin or knockdown of dynamin by RNAi causes cytokinesis failure following furrow regression (17). This furrow ingression followed by cytokinesis failure was reminiscent of the phenotype of knocking down NME1 and provided good evidence that dynamin might be a downstream mediator of NME1 during cytokinesis.
Fig. 4.
Dynamin knockdown phenocopies NME1 knockdown. Knockdown of RhoA (shRhoA) or dynamin-2 (shDyn2) results in cytokinesis failure. H2BGFP-containing shRNA vectors targeting RhoA or Dyn2 were introduced into HBE135 cells. Live cell imaging was carried out as in Fig. 2A.
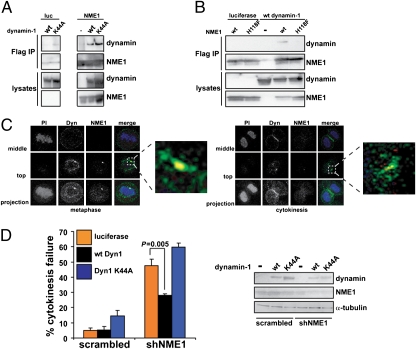
Consistent with a functional relationship for NME1 and dynamin, the two proteins have been previously shown to interact physically (19). To test whether GTP binding by dynamin is required for this interaction, Flag-tagged NME1 was transfected into 293T cells along with untagged WT dynamin-1 or dynamin-1K44A (see below). As shown in Fig. 5A, both variants of dynamin coimmunoprecipitated with Flag-tagged NME1. To determine whether the catalytic activity of NME1 was required for the interaction with dynamin, Flag-tagged constructs that synthesized either WT NME1 or NME1H118F were cotransfected into 293T cells along with WT dynamin-1. Anti-Flag immunoprecipitation followed by Western blotting revealed that only WT NME1 interacted with dynamin in cells, suggesting that the catalytic activity of NME1 is required to form a complex with dynamin (Fig. 5B).
Fig. 5.
NME1 and dynamin interact biochemically and genetically. (A) NME1 and dynamin physically interact. 293T cells were transfected with luciferase (luc) or Flag-tagged NME1 along with dynamin-1 WT (wt) or K44A. Cells were lysed and subjected to anti-Flag immunoprecipitation and anti-Flag or anti-dynamin1/2 Western blotting. (B) Catalytic activity of NME1 is required for the interaction with dynamin. Luciferase or Flag-tagged NME1 (wt or H118F) was transfected into 293T cells along with wt dynamin-1. Immunoprecipitation and Western blotting were carried out as in A. (C) NME1 and dynamin (Dyn) colocalize at the cortex. HBE135 cells were fixed and stained with anti-NME1 (red) and anti-dynamin1/2 (green). Nuclei were stained with propidium iodide (PI, blue). Confocal sections were obtained as in Fig. 3A. (Right) Zoomed images are of the merged top sections; arrowheads indicate colocalization (yellow) of NME1 and dynamin. (D) Ectopic expression of dynamin rescues the NME1 knockdown phenotype. Luciferase or dynamin-1 (Dyn1; WT or K44A) cDNA was transduced into HBE135 cells along with the H2BGFP-containing vector containing scrambled or NME1 shRNA. Live cell imaging was carried out as described in Fig. 2A 32 h after infection. The percentage of mitotic cells that showed cytokinesis failure is shown. Error bars represent SEM [n = 5 for scrambled/luciferase and short hairpin NME1 (shNME1)/luciferase and n = 3 for scrambled/dynamin and shNME1/dynamin]. A minimum of 50 mitoses was counted in each experiment. The P value was obtained using Student's t test. Western blot analysis was carried out to determine the levels of NME1, dynamin, and α-tubulin.
We next investigated the localization of endogenous NME1 and dynamin in HBE135 cells during multiple stages of cell division (Fig. 5C). In prometaphase (Fig. S6A), metaphase (Fig. 5C Left), and anaphase (Fig. S6A), NME1 colocalized with a fraction of the cellular pool of dynamin in cortical dots. During cytokinesis, we observed that dynamin localized not only to the midzone region, as expected from published data (17), but to the equatorial cortex, where it colocalized with NME1 (Fig. 5C Right). The cortical localization of dynamin did not require the presence of NME1, because we observed dynamin at the cortex of mitotic cells expressing the NME1 shRNA (Fig. S6B). These data demonstrated that a significant fraction of cellular dynamin colocalized with NME1 at the cortex before and following furrow ingression. Taken together, the coimmunoprecipitation and colocalization experiments suggested that dynamin could be a downstream mediator of NME1 during cytokinesis.
In the presence of GTP, dynamin organizes into oligomeric rings and is competent to interact with downstream mediators (20, 21). Because dynamin oligomerization is cooperative, if the role of NME1 is to provide a local source of GTP that drives dynamin oligomerization, ectopic expression of dynamin may be sufficient to drive dynamin oligomerization in the presence of reduced NME1 (and thus GTP) concentrations (22). To test this hypothesis, HBE135 cells were transduced with scrambled or NME1 shRNAs in the H2BGFP vector along with lentiviral cDNA constructs expressing either luciferase or one of two variants of dynamin-1: WT (Dyn1wt) or a GTP-binding mutant (Dyn1K44A) (23). Cells were followed by time-lapse microscopy 48 h after infection to assess the effects of ectopic dynamin expression on cytokinesis failure induced by NME1 knockdown. As shown in Fig. 5D, knockdown of NME1 in the presence of luciferase gave rise to the expected level of cytokinesis failure in cells that entered mitosis. Ectopic expression of WT dynamin-1 significantly reduced the percentage of mitotic cells that experienced cytokinesis failure in the presence of reduced NME1. Rescue of the cytokinesis failure phenotype was not observed on expression of the GTP-binding mutant of dynamin-1, suggesting that GTP binding was still necessary for dynamin function and that the reduced levels of GTP present in cells with decreased NME1 were sufficient when dynamin was overexpressed. Taken together with the interaction and colocalization data, the rescue of the NME1 knockdown phenotype by expression of dynamin strongly suggests that dynamin functions downstream of NME1 on the same pathway during cytokinesis.
NME1 Loss Leads to Senescence in Primary Cells.
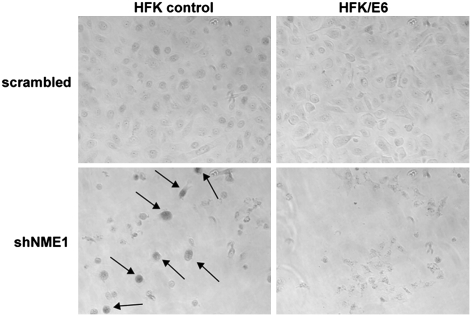
To order NME1 knockdown and subsequent chromosomal instability relative to another key step in tumor progression, we considered the relationship between NME1 knockdown and the loss of the p53 tumor suppressor (24). As shown in Fig. 2B and Fig. S1, the knockdown of NME1 resulted in cytokinesis failure and tetraploidy regardless of the status of p53 signaling. However, we noticed that primary cells with reduced NME1 expression stopped proliferating and showed a large flat morphology that was observed in both diploid (mononucleate) and tetraploid (binucleate) cells (Fig. S7). Further, primary HFK cells in which NME1 was knocked down stained positive for acidic β-galactosidase, an established marker of replicative senescence (25) (Fig. 6). Cell cycle arrest of both diploids and tetraploids can also be observed in DNA content histograms of HFK cells in which NME1 was knocked down (Fig. S8). These phenotypes were dependent on p53 because they were not observed in cells expressing the E6 oncoprotein. These data argue that loss of NME1 leads to tetraploidy in all contexts but that tumorigenic phenotypes driven by downstream chromosomal instability may only occur with the prior loss of p53 signaling. This places the loss of NME1 after the loss of p53 on the timeline of tumor development and implies that chromosomal instability, at least of the type induced by loss of NME1, may not itself be sufficient to initiate tumor formation.
Fig. 6.
Knockdown of NME1 results in senescence in the presence of p53. Loss of NME1 results in senescence in primary cells. Cells were fixed for acidic β-galactosidase staining. Cells positive for β-galactosidase are indicated by arrows in the black and white images. shNME1, short hairpin NME1.
Discussion
NME1, Cytokinesis Failure, and Dynamin.
We carried out multiple experiments to elucidate the mechanism by which NME1 regulates diploid chromosome content. Using time-lapse imaging, we found that NME1 loss results in cytokinesis failure and tetraploidy. In contrast to the cytokinesis failure associated with the loss of RhoA, abortive cytokinesis associated with NME1 loss occurs after nearly complete furrow ingression. Some clues as to the nature of this abortive cytokinesis may come from more detailed examination of the cells while the furrow is paused. We consistently observed multiple nonequatorial blebs and projections of the dividing cell in multiple directions away from the equator and division plane. These observations suggested that NME1 may be involved in regulating some aspect of the cellular architecture that maintains the bipolar morphology of a dividing cell.
Two possibilities for targets of NME1 during cytokinesis are the actin cytoskeleton and plasma membrane trafficking, both of which play critical roles during cytokinesis (26, 27). Consistent with a function downstream of NME1 during cytokinesis, dynamin has a well-established role in the establishment of clathrin-coated vesicles during endocytosis (28), and a role for dynamin in the regulation of actin cytoskeleton has also been established (29). Further, dynamin may function to anchor actin filaments to membrane vesicles (30). NME1 may thus function through dynamin to regulate some combination of endocytic processes, actin organization, or actin–membrane interactions. The colocalization of only a fraction of dynamin with NME1 suggests that some of the activities of dynamin during cytokinesis are regulated in an NME1-independent manner.
The localization of NME1 and dynamin to cortical dots was unexpected but not unreasonable. Both proteins have been localized to the centrosome, and there is evidence that centrosomal proteins are translocated to the cortex during mitosis to define the location of cytokinetic furrow ingression (31, 32). Interestingly, we observed NME1 in the centrosome during interphase and at the cortex during mitosis. It will be interesting to determine whether there is transport of NME1 and other proteins to the cortex during mitosis and to identify the factors responsible for this transport.
Because the catalytic activity of NME1 is required to complement NME1 knockdown (Fig. 2C), we propose that it acts to provide a local source of GTP for dynamin at the cortex during cytokinesis. A link between NME1 and dynamin is not unprecedented, although it has not been observed in the context of cytokinesis. As mentioned above, dynamin and NME1 have been shown to interact physically both in vitro and in mammalian cells. In C. elegans, homologs of NME1 and dynamin have been proposed to interact (33). In Drosophila, dynamin and the homolog of NME1 (AWD) have been shown to interact genetically in multiple biological contexts (34, 35). Finally, the localized production of GTP by NME1 regulates dynamin during adherens junction disassembly (36). It will be interesting to learn what classes of GTPase rely on this type of localized GTP generation for activation and how this regulation is established and maintained.
NME, p53, and Tumorigenesis.
The majority of the experiments in this and the companion article were carried out using immortalized cell lines lacking p53 signaling. To determine the impact of the signaling context on the NME1 knockdown phenotype, we investigated the effect of knocking down NME1 in primary human cells. We noted that NME1 knockdown resulted in cytokinesis failure regardless of cellular context but that continued cycling of tetraploid cells required dysfunctional p53 signaling. Although this would seem to support the existence of a “tetraploidy checkpoint,” (37) our observation of a similar p53-dependent arrest in diploid cells argues that, at least in this case, the arrest is not specific to tetraploids. Rather, it is possible that NME1 knockdown in diploids before S phase triggers a similar p53-dependent arrest to that triggered by tetraploid cells that inappropriately exit mitosis without dividing. This would be consistent with the theory that observed tetraploidy checkpoints are, in fact, responses to cellular stress to which, in certain contexts, tetraploids are more sensitive (38, 39).
Over the past 20 y, evidence has pointed to a metastasis suppressor activity for NME1. Based on our data, we propose that the role for NME1 in tumor progression may be more complicated than previously thought and that it may additionally act early in tumor development. Following the loss of p53, NME1 loss would lead to chromosomal instability and the acquisition of mutations favorable for tumor progression. Later in the life of the tumor, NME1 loss would drive metastasis through its activity on proteins that regulate invasion and motility. These two functions may occur within the development of a single tumor, because NME1 loss in cancers is primarily attributable to potentially reversible down-regulation of gene expression and not mutation (40). A role for NME1 in early tumor development is not inconsistent with its identification as a metastasis suppressor, which was based on a comparison of expression levels between primary and metastatic tumors. One might imagine that the development of chromosomal instability requires only transient loss of NME1 and that higher expression is again required during the rapid proliferation of a primary tumor. Alternatively, given the fact that we observed the cytokinesis defects with only 50% protein knockdown, it is possible that NME1 is haploinsufficient in regard to cytokinesis regulation. This would suggest that NME1 might be gradually lost over the course of tumor development (i.e., one allele is silenced to lead to primary tumor formation, and loss of the second allele promotes metastasis).
NME1 is a known metastasis suppressor that we now show to have an additional role earlier in tumor development. Our data suggest that the loss of NME1 may be one method by which a nascent tumor cell acquires chromosomal instability. We anticipate that the remaining hits from our screen will provide fertile ground for the identification of additional mechanisms by which diploid cells acquire chromosomal instability on the way to becoming aneuploid tumors.
Materials and Methods
Cell Lines and Antibodies.
HBE135 E6/E7 and 293T cells were obtained from the American Type Culture Collection and cultured according to the supplier's instructions. Primary HFKs and a variant expressing HPV 16 E6 (HFK/E6) were the kind gifts of A. Baldwin and K. Münger (Brigham and Women’s Hospital and Harvard Medical School, Boston, MA) and were cultured in Keratinocyte Serum Free Media (Invitrogen).
Primary antibodies used were anti-NME1 (rabbit polyclonal sc-343 or mouse monoclonal sc-465; Santa Cruz), anti-γ-tubulin (goat polyclonal sc-7396; Santa Cruz), anti-β-actin (Cell Signaling Technology), anti-Flag (mouse monoclonal M2; Sigma), or antidynamin 1/2 (Hudy1; Upstate). Secondary antibodies for immunofluorescence were obtained from Invitrogen and used at a ratio of 1:400. Secondary antibodies for Western blotting were obtained from Cell Signaling and used at a ratio of 1:1,000–1:100,000.
Time-Lapse Video Microscopy.
For imaging of nuclei, a lentiviral shRNA vector containing H2BGFP was constructed by standard PCR subcloning in which the puromycin acetyltransferase gene of pLKO.1 was swapped for H2BGFP from the vector pLE-H2BGFP (a kind gift of R. King, Harvard Medical School). Virus was prepared from this vector containing scrambled, NME1 (shRNA 1), dynamin-2, Cdc42, or RhoA shRNAs and was used to infect HBE135, HFK, HFK/E6, or HeLa cells in a six-well plate. After 24 h, cells were split to glass-bottom tissue culture dishes (MatTek). Alternatively, cells were infected at confluence and reseeded 4 d postinfection in glass-bottom dishes. After an additional 24 h, time-lapse data were collected using a Nikon TE2000E Automated Inverted Microscope and a 20× objective at the Nikon Imaging Center, Harvard Medical School. Cells were maintained at 37 °C and 5% (vol/vol) CO2, and phase-contrast and GFP images were obtained every 4–5 min in multiple fields for 24–48 h as indicated.
cDNA Complementation.
A lentiviral cDNA expression vector was constructed using the pLKO.1 backbone. Gene expression is under the control of a CMV promoter, and the blasticidin resistance gene downstream of the gene of interest is translated using an internal ribosome entry site. Luciferase, NME1 (WT and mutant), or dynamin-1 (WT or mutant) was inserted into the vector by standard PCR subcloning.
For complementation experiments with NME1 cDNA, shRNA lentivirus and cDNA lentivirus were added to confluent HBE135 cells. After 8 h, virus was removed and fresh media were added. After 4 d, the cells were reseeded in media containing puromycin (0.5 μg/mL) and blasticidin (7.5 μg/mL). After an additional 4 d, half of the cells were lysed for protein extract preparation and half were fixed for flow cytometry analysis. For experiments with dynamin-1 cDNA, HBE135 cells were infected with H2BGFP shRNA and cDNA lentivirus and imaged by time-lapse video microscopy.
Supplementary Material
Acknowledgments
We acknowledge J. Walter for insight into a potential connection with dynamin. We thank J. Sawyer for preparation of the kinase shRNA virus, J. Doench for construction of the lentiviral cDNA expression vector, and W. Endege for construction of the Flag-tagged lentiviral cDNA expression vector. We are also indebted to A. L. Conery and A. Rolfes for critical reading of the manuscript and members of the Harlow laboratory for helpful discussions. A.R.C. was supported by Postdoctoral Fellowship PF-07-030-01-CCG from the American Cancer Society.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1010633107/-/DCSupplemental.
References
- 1.Ganem NJ, Storchova Z, Pellman D. Tetraploidy, aneuploidy and cancer. Curr Opin Genet Dev. 2007;17:157–162. doi: 10.1016/j.gde.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 2.Lascu I, Gonin P. The catalytic mechanism of nucleoside diphosphate kinases. J Bioenerg Biomembr. 2000;32:237–246. doi: 10.1023/a:1005532912212. [DOI] [PubMed] [Google Scholar]
- 3.Steeg PS, et al. Evidence for a novel gene associated with low tumor metastatic potential. J Natl Cancer Inst. 1988;80:200–204. doi: 10.1093/jnci/80.3.200. [DOI] [PubMed] [Google Scholar]
- 4.Hartsough MT, Steeg PS. Nm23/nucleoside diphosphate kinase in human cancers. J Bioenerg Biomembr. 2000;32:301–308. doi: 10.1023/a:1005597231776. [DOI] [PubMed] [Google Scholar]
- 5.Leone A, et al. Reduced tumor incidence, metastatic potential, and cytokine responsiveness of nm23-transfected melanoma cells. Cell. 1991;65:25–35. doi: 10.1016/0092-8674(91)90404-m. [DOI] [PubMed] [Google Scholar]
- 6.Kantor JD, McCormick B, Steeg PS, Zetter BR. Inhibition of cell motility after nm23 transfection of human and murine tumor cells. Cancer Res. 1993;53:1971–1973. [PubMed] [Google Scholar]
- 7.Murakami M, et al. Nm23-H1 modulates the activity of the guanine exchange factor Dbl-1. Int J Cancer. 2008;123:500–510. doi: 10.1002/ijc.23568. [DOI] [PubMed] [Google Scholar]
- 8.Murakami M, Meneses PI, Lan K, Robertson ES. The suppressor of metastasis Nm23-H1 interacts with the Cdc42 Rho family member and the pleckstrin homology domain of oncoprotein Dbl-1 to suppress cell migration. Cancer Biol Ther. 2008;7:677–688. doi: 10.4161/cbt.7.5.5665. [DOI] [PubMed] [Google Scholar]
- 9.Otsuki Y, et al. Tumor metastasis suppressor nm23H1 regulates Rac1 GTPase by interaction with Tiam1. Proc Natl Acad Sci USA. 2001;98:4385–4390. doi: 10.1073/pnas.071411598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu J, et al. Interaction of the Ras-related protein associated with diabetes rad and the putative tumor metastasis suppressor NM23 provides a novel mechanism of GTPase regulation. Proc Natl Acad Sci USA. 1999;96:14911–14918. doi: 10.1073/pnas.96.26.14911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nallamothu G, Woolworth JA, Dammai V, Hsu T. Awd, the homolog of metastasis suppressor gene Nm23, regulates Drosophila epithelial cell invasion. Mol Cell Biol. 2008;28:1964–1973. doi: 10.1128/MCB.01743-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilles AM, Presecan E, Vonica A, Lascu I. Nucleoside diphosphate kinase from human erythrocytes. Structural characterization of the two polypeptide chains responsible for heterogeneity of the hexameric enzyme. J Biol Chem. 1991;266:8784–8789. [PubMed] [Google Scholar]
- 13.Roymans D, et al. Identification of the tumor metastasis suppressor Nm23-H1/Nm23-R1 as a constituent of the centrosome. Exp Cell Res. 2001;262:145–153. doi: 10.1006/excr.2000.5087. [DOI] [PubMed] [Google Scholar]
- 14.Du J, Hannon GJ. The centrosomal kinase Aurora-A/STK15 interacts with a putative tumor suppressor NM23-H1. Nucleic Acids Res. 2002;30:5465–5475. doi: 10.1093/nar/gkf678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rincon S, Coll PM, Perez P. Spatial regulation of Cdc42 during cytokinesis. Cell Cycle. 2007;6:1687–1691. doi: 10.4161/cc.6.14.4481. [DOI] [PubMed] [Google Scholar]
- 16.Piekny A, Werner M, Glotzer M. Cytokinesis: Welcome to the Rho zone. Trends Cell Biol. 2005;15:651–658. doi: 10.1016/j.tcb.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 17.Thompson HM, Skop AR, Euteneuer U, Meyer BJ, McNiven MA. The large GTPase dynamin associates with the spindle midzone and is required for cytokinesis. Curr Biol. 2002;12:2111–2117. doi: 10.1016/s0960-9822(02)01390-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Konopka CA, Schleede JB, Skop AR, Bednarek SY. Dynamin and cytokinesis. Traffic. 2006;7:239–247. doi: 10.1111/j.1600-0854.2006.00385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baillat G, Gaillard S, Castets F, Monneron A. Interactions of phocein with nucleoside-diphosphate kinase, Eps15, and Dynamin I. J Biol Chem. 2002;277:18961–18966. doi: 10.1074/jbc.M108818200. [DOI] [PubMed] [Google Scholar]
- 20.Sever S, et al. Physical and functional connection between auxilin and dynamin during endocytosis. EMBO J. 2006;25:4163–4174. doi: 10.1038/sj.emboj.7601298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sever S. Dynamin and endocytosis. Curr Opin Cell Biol. 2002;14:463–467. doi: 10.1016/s0955-0674(02)00347-2. [DOI] [PubMed] [Google Scholar]
- 22.Warnock DE, Hinshaw JE, Schmid SL. Dynamin self-assembly stimulates its GTPase activity. J Biol Chem. 1996;271:22310–22314. doi: 10.1074/jbc.271.37.22310. [DOI] [PubMed] [Google Scholar]
- 23.Damke H, Baba T, Warnock DE, Schmid SL. Induction of mutant dynamin specifically blocks endocytic coated vesicle formation. J Cell Biol. 1994;127:915–934. doi: 10.1083/jcb.127.4.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levine AJ, Oren M. The first 30 years of p53: Growing ever more complex. Nat Rev Cancer. 2009;9:749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dimri GP, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boucrot E, Kirchhausen T. Endosomal recycling controls plasma membrane area during mitosis. Proc Natl Acad Sci USA. 2007;104:7939–7944. doi: 10.1073/pnas.0702511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eggert US, Mitchison TJ, Field CM. Animal cytokinesis: From parts list to mechanisms. Annu Rev Biochem. 2006;75:543–566. doi: 10.1146/annurev.biochem.74.082803.133425. [DOI] [PubMed] [Google Scholar]
- 28.Mettlen M, Pucadyil T, Ramachandran R, Schmid SL. Dissecting dynamin's role in clathrin-mediated endocytosis. Biochem Soc Trans. 2009;37:1022–1026. doi: 10.1042/BST0371022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schafer DA. Regulating actin dynamics at membranes: A focus on dynamin. Traffic. 2004;5:463–469. doi: 10.1111/j.1600-0854.2004.00199.x. [DOI] [PubMed] [Google Scholar]
- 30.Orth JD, McNiven MA. Dynamin at the actin-membrane interface. Curr Opin Cell Biol. 2003;15:31–39. doi: 10.1016/s0955-0674(02)00010-8. [DOI] [PubMed] [Google Scholar]
- 31.Rusan NM, Wadsworth P. Centrosome fragments and microtubules are transported asymmetrically away from division plane in anaphase. J Cell Biol. 2005;168:21–28. doi: 10.1083/jcb.200409153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson HM, Cao H, Chen J, Euteneuer U, McNiven MA. Dynamin 2 binds gamma-tubulin and participates in centrosome cohesion. Nat Cell Biol. 2004;6:335–342. doi: 10.1038/ncb1112. [DOI] [PubMed] [Google Scholar]
- 33.Li S, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krishnan KS, et al. Nucleoside diphosphate kinase, a source of GTP, is required for dynamin-dependent synaptic vesicle recycling. Neuron. 2001;30:197–210. doi: 10.1016/s0896-6273(01)00273-2. [DOI] [PubMed] [Google Scholar]
- 35.Dammai V, Adryan B, Lavenburg KR, Hsu T. Drosophila awd, the homolog of human nm23, regulates FGF receptor levels and functions synergistically with shi/dynamin during tracheal development. Genes Dev. 2003;17:2812–2824. doi: 10.1101/gad.1096903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palacios F, Schweitzer JK, Boshans RL, D'Souza-Schorey C. ARF6-GTP recruits Nm23-H1 to facilitate dynamin-mediated endocytosis during adherens junctions disassembly. Nat Cell Biol. 2002;4:929–936. doi: 10.1038/ncb881. [DOI] [PubMed] [Google Scholar]
- 37.Margolis RL, Lohez OD, Andreassen PR. G1 tetraploidy checkpoint and the suppression of tumorigenesis. J Cell Biochem. 2003;88:673–683. doi: 10.1002/jcb.10411. [DOI] [PubMed] [Google Scholar]
- 38.Uetake Y, Sluder G. Cell cycle progression after cleavage failure: Mammalian somatic cells do not possess a “tetraploidy checkpoint”. J Cell Biol. 2004;165:609–615. doi: 10.1083/jcb.200403014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wong C, Stearns T. Mammalian cells lack checkpoints for tetraploidy, aberrant centrosome number, and cytokinesis failure. BMC Cell Biol. 2005;6:6–17. doi: 10.1186/1471-2121-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sager R. Expression genetics in cancer: Shifting the focus from DNA to RNA. Proc Natl Acad Sci USA. 1997;94:952–955. doi: 10.1073/pnas.94.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.