Abstract
We describe the integration of microfluidic selection with high-throughput DNA sequencing technology for rapid and efficient discovery of nucleic acid aptamers. The Quantitative Selection of Aptamers through Sequencing method tracks the copy number and enrichment-fold of more than 10 million individual sequences through multiple selection rounds, enabling the identification of high-affinity aptamers without the need for the pool to fully converge to a small number of sequences. Importantly, this method allows the discrimination of sequences that arise from experimental biases rather than true high-affinity target binding. As a demonstration, we have identified aptamers that specifically bind to PDGF-BB protein with Kd < 3 nM within 3 rounds. Furthermore, we show that the aptamers identified by Quantitative Selection of Aptamers through Sequencing have ∼3–8-fold higher affinity and ∼2–4-fold higher specificity relative to those discovered through conventional cloning methods. Given that many biocombinatorial libraries are encoded with nucleic acids, we extrapolate that our method may be extended to other types of libraries for a range of molecular functions.
Keywords: high-throughput DNA sequencing, PDGF-BB, Quantitative Selection of Aptamers through Sequencing, MicroMagnetic Separation device
Aptamers are nucleic acid-based affinity reagents (1, 2) that can be chemically synthesized and modified (3, 4). Aptamers have been generated against many classes of molecular targets, including small molecules (5, 6), proteins (7, 8), and cell-surface markers (9, 10), for a wide range of applications including diagnostics (11, 12), molecular imaging (13, 14), targeted therapeutics (15), gene delivery (16), and drug delivery (17). Systematic evolution of ligands by exponential enrichment (SELEX) offers an effective in vitro selection method for the isolation of aptamers from random libraries of nucleic acids. Unfortunately, SELEX requires a significant investment of time and resources, involving numerous rounds of selection (typically 8–15) to isolate molecules with sufficient binding affinity and specificity (8, 18). We recently described the use of microfluidics technology to accelerate the process of aptamer selection (M-SELEX); due to multiple advantages that occur at the microscale, specific aptamers that bind target proteins with nanomolar affinity can be generated by M-SELEX within 1–3 rounds (19, 20). Upon completion of selection, the enriched pool of nucleic acids is cloned, typically into Escherichia coli, to identify the sequences of individual aptamers. This entails PCR amplification of the isolated aptamer pool, followed by insertion of the purified PCR product into a pCR4-TOPO vector and transformation into competent bacterial cells (19–21). Typically, ∼100 colonies are randomly picked, sequenced, and aligned to identify consensus sequences. Finally, the aptamers are synthesized to measure target affinity via surface plasmon resonance (19, 22), radioactivity (23, 24), or fluorescence (20, 25).
It is hypothesized that, prior to cloning, the final selected pool contains a large number of unique aptamer sequences of varying affinity and specificity (26, 27). However, it is difficult to identify optimal sequences from this pool using traditional cloning and sequencing approaches, which only enable sampling of a small portion of the sequence space (19, 20). Furthermore, conventional approaches do not offer the capability to track the evolution of individual sequences across multiple rounds of selection, which would provide valuable information about the enrichment process.
Recently, the Schroeder group has shown that the combination of traditional genomic SELEX with high-throughput sequencing, yields a powerful method for the identification of genomic aptamers (28). As a further step toward rapid and efficient identification of high-affinity aptamer sequences, we have developed the Quantitative Selection of Aptamers through Sequencing (QSAS) method, which pairs M-SELEX with high-throughput DNA sequencing. Using platelet derived growth factor BB (PDGF-BB) protein (8)—a useful biomarker for various pathological states (29)—as a model target, we demonstrate the capability to track the evolution and enrichment of > 10 million individual aptamer sequences throughout the selection process, and thereby quantitatively identify high-affinity aptamer sequences within three rounds (Fig. 1). Our method improves the speed and outcome of selection, and by comparing the enrichment-fold of sequences between selection rounds, we were able to quantitatively identify sequences with binding affinities ∼3–8-fold higher than those obtained by traditional cloning approaches.
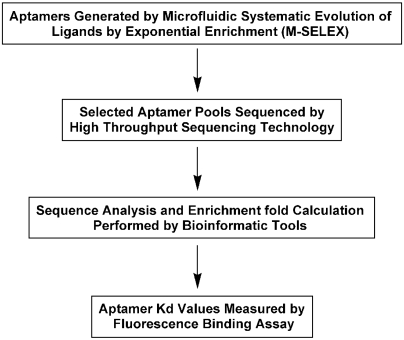
Fig. 1.
An overview of the QSAS method. The QSAS method begins with three rounds of M-SELEX against the target. The aptamer pools from each round are subjected to high-throughput sequencing, and the millions of resulting sequences are computationally filtered, aligned and analyzed. We ranked the 100 most highly enriched aptamer sequences against PDGF-BB between round 3 and round 2 or between round 3 and round 1, and then synthesized the top three sequences and measured their target affinity and specificity.
Results and Discussion
Microfluidic-SELEX.
We have described the M-SELEX experimental procedure in our previous work (20). Briefly, we immobilized PDGF-BB target molecules on the surface of micron-sized magnetic beads and incubated the coated beads with a ssDNA library (∼1 nanomole total). The DNA library design features a central 40-base random domain flanked by two 20-nucleotide PCR primer sites. We subjected the aptamer-bound, target-coated beads to high-stringency continuous washing at a high flow-rate (50 mL/hr) within the MicroMagnetic Separation device (MMS), which has been shown to effectively remove weakly and nonspecifically bound molecules (30). After the separation, the external magnets were removed and the beads carrying the selected aptamers were eluted from the device and PCR amplified; finally, we generated ssDNA from the amplicons for use in a subsequent round of selection.
We performed a total of three rounds of positive selection with PDGF-BB-coated beads with increasing selection stringency. The first two rounds only incorporated positive selection, with no negative selection. In the first round (R1), the molar ratio between ssDNA and target was kept at ∼10∶1 by using a controlled number of PDGF-BB-coated magnetic beads (2 × 107 total beads at ∼105 PDGF-BB molecules per bead). In the second round (R2), we used a higher molar ratio of ssDNA∶target (∼100∶1) to increase the selection pressure (20). Prior to the third round of positive selection, we subjected the selected R2 pool to two sets of negative selection using magnetic beads coated with PDGF-AA or Tris (tris(hydroxymethyl)aminomethane) molecules to increase the specificity of the selected aptamers. Tris was also used to block the remaining active group after N-ethyl-N′-(3-(dimethylamino)propyl)carbodiimide (EDC)/N-hydroxysuccinimide (NHS) immobilization of protein onto the magnetic beads. We used 2 × 107 total beads at ∼105 PDGF-AA molecules per bead or 2 × 107 total Tris-coated beads to efficiently deplete aptamers with affinity for PDGF-AA or Tris molecules from the R2 pool. We then used this depleted pool for a third round of positive selection (R3) with the same molar ratio as R2 to further increase aptamer affinity.
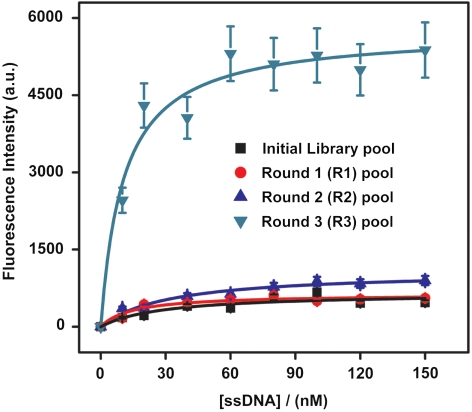
After the selection, we measured the bulk equilibrium dissociation constants (Kd) of the three aptamer pools (R1, R2, and R3) via a fluorescence assay described below (20, 25) (Fig. 2). As expected, the initial naïve library displayed negligible binding affinity to the target (Fig. 2). However, the binding affinity of the aptamer pools increased after each successive round of selection, and all three postselection pools showed nanomolar Kd values: Kd(R1) = 57.3 ± 12.0 nM, Kd(R2) = 32.1 ± 7.06 nM, and Kd(R3) = 10.5 ± 2.63 nM (Fig. 2). We believe that the significantly greater fluorescence intensity of the R3 pool relative to the other pools can be attributed to the higher affinity and greater target specificity of these aptamers. We postulate that the negative selection performed between R2 and R3, as well as the higher washing stringency used in R3, contributed to this large difference.
Fig. 2.
Fluorescence measurements were used to determine dissociation constants (Kd) of the selected aptamer pools after each round of selection. The naïve initial library pool exhibited negligible binding to the PDGF-BB target. After R1, the average Kd of the enriched pool was 57.3 ± 12.0 nM. Subsequently, R2 yielded a pool with an average Kd of 32.1 ± 7.06 nM. Following two sets of negative selection against Tris-coated beads or PDGF-AA-coated beads, R3 positive selection resulted in a pool with an average Kd of 10.5 ± 2.63 nM.
High-Throughput DNA Sequencing Technology.
We analyzed the DNA sequences contained in the R1, R2 and R3 pools by performing high-throughput DNA sequencing with the Genome Analyzer II (Illumina) (31, 32). To prepare the samples, we first PCR amplified the aptamer pools from each round, then purified the double-stranded DNA products and used ten nanograms of the purified products for 3′-end repair, adenosine tailing, and adapter ligation. Ligated fragments in the 150–200 bp size range were then isolated via gel extraction and amplified with a limited number of cycles of PCR (< 10 cycles). After we had obtained single-stranded, adapter-ligated molecules, we used a flow-cell cluster station to bridge amplify each individual molecule 35 times to generate cloned clusters. After each cluster was linearized, the free ends of the clusters were blocked and the sequencing primer was added and hybridized. Finally, we sequenced 75 bases on the Genome Analyzer II, and used Illumina Pipeline software to process the resulting data. We developed custom software to perform downstream sequence analysis (without the Eland alignment step), which we used to obtain 40 base aptamer sequences from the combination of forward and reverse complement tags and then counted the tag sequences from all samples.
Sequence Analysis.
We obtained > 1.7 × 107 raw sequences from each selection round. We subsequently filtered out sequences that were not 75 bases in length (i.e., 20-base 5′ primer +40-base aptamer +15-base 3′ primer) or did not contain the correct primer on each end, and then trimmed out the two PCR primer sites from the remaining sequences, yielding ∼2–9 × 106 unique aptamer sequences from each pool (Table 1). We observed that the number of unique sequences decreased and the percentage of duplicate sequences increased in later selection rounds, indicating the convergence of potential aptamers in the resulting pools. As additional evidence of this, we determined that the ten most highly represented sequences in each pool appeared with an average copy number of 594, 2,789 and 216,488 in the R1, R2, and R3 pools, respectively (Fig. S1).
Table 1.
Analysis of selected sequences after each round
| Selection rounds | Raw tags | Filtered (by primers) | Combined + merged (unique sequences) | Duplicate % |
| Round 1 (R1) | 17,319,764 | 13,496,744 | 8,943,339 | 33.7370628 |
| Round 2 (R2) | 18,991,417 | 14,722,556 | 7,994,342 | 45.7000401 |
| Round 3 (R3) | 17,052,851 | 12,512,480 | 2,027,839 | 83.7934686 |
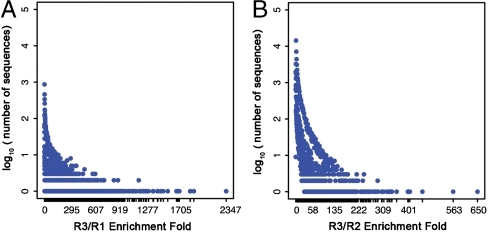
To identify high affinity sequences, we calculated and ranked the enrichment-fold (i.e., ratio of copy numbers between two selection rounds) for every sequence. The top 36 sequences are shown in Table 2 for R3/R2 and in Table S1 for R3/R1. We then plotted the number sequences with a particular enrichment-fold as a function of its enrichment-fold from R1 to R3 (Fig. 3A) or R2 to R3 (Fig. 3B). It should be noted that highly represented sequences did not necessarily exhibit high enrichment between rounds, and there is a large population of sequences with high copy numbers but with minimal enrichment-fold (Fig. 3, Upper Left). Interestingly, the same ten sequences were identified with the highest copy numbers in all three pools (R1, R2, and R3), but none of them displayed significant enrichment-fold. We suspect that these sequences are overrepresented as a result of biases during library synthesis or PCR (26, 33). On the other hand, the sequences with highest enrichment-fold (e.g., 2,347-fold for R3/R1 and 650-fold for R3/R2) appear infrequently in earlier rounds (Fig. 3, Lower Right), and we hypothesize that these represent true high-affinity aptamers. In this way, QSAS offers the capability to distinguish artifactual sequences from those that have become enriched through target binding.
Table 2.
Top 36 enriched aptamer sequences from R2 to R3
| Aptamer ID | Sequences of selected aptamers (5′ → 3′) | Enrichment fold (R3/R2) |
| 1 | ATAAGCTGAGCATCTTAGATCCCCGTCAAGGGCAGCGTAA | 650 |
| 2 | GATACTGAGCATCGTACATGATCCCGCAACGGGCAGTATT | 563 |
| 3 | AGTTGAATGGTGTGGTCACTTCCAGTCCCGCAGGGCACAC | 453 |
| 4 | CCGTTACAGTCTAAGACTCCTATCAACATAGGCGGAAGTA | 408 |
| 5 | TGTGGGTATGGTCCTAATTTTTTAGGCACGGAGGTACCAT | 403 |
| 6 | CCGTTACAGTCTAAGACTTCTATGAACATAGGCGGAAGTA | 401 |
| 7 | TACGAGTTTGATCCTTTCCATTAGGCGTACAGCTCATCAA | 361 |
| 8 | TTGGGGCGGTCCTGTGAAAGGCAAAAATCTATTATACCGC | 344 |
| 9 | ATAAGCTGAGCATCTTAGATCCCTGTTAAGGGCAGCGTAA | 343 |
| 10 | GCTGAGTTAGATCCCTTTCGTAAGGGCAGCCGGGCATCGA | 337 |
| 11 | CTGAGCATCACGAAGTGATCCTGTCGGGGACGGGCAGTTA | 333 |
| 12 | CCGGGAACATTGTCTAAACAATGTCATTTATGTCGGAAGC | 327 |
| 13 | ATAAGCTGAGCACCTTAGATCCCTGTCAAGGGCAGCGTAA | 315 |
| 14 | TAAGGGCACTATTGCATGGTGAGCATCCTTGGATCCCGCT | 309 |
| 15 | TACGAGGTTGATCCTTTTTATTAGGCGTACAGCTCATCAA | 307 |
| 16 | CCGTTACAGTCTAAGACTCCCATGAACATAGGCGGAAGTA | 294 |
| 17 | GCTGAGTTAGATCCATTTCGTAAGGGCAGCCGGGCATCTA | 293 |
| 18 | AATGGATGGGCACCGCTATAGTTGGTCCCGAAGGGCATGC | 293 |
| 19 | TACGAGTTTGATCCTTTTTATTATGCGTACAGCTCATCAA | 287 |
| 20 | AGTTGAATAGTGTGGTCACTTCCAGTCCCGCGGGGCACAC | 284 |
| 21 | ATAAGCTGAGCATCTTAGATCCCTGTCAAGGGCAGTGTAA | 277 |
| 22 | GATACTGAGCATCGTACATGATCCCGCAACGAGCAGTATC | 268 |
| 23 | GATACTGAGAATCGTACATGATCCCGCAACGGGCAGTATC | 267 |
| 24 | AGGTGAGCATCTTTAGATCCCATTTGGCGCCTTTTCTTTC | 263 |
| 25 | CGGGGCAACAACGTTGAGCATCATTCATGATCGCCGCTAT | 260 |
| 26 | GCTCCGTACCATTCTTATGGTCCCGGCACGGGCGCAAACG | 259 |
| 27 | CTTATTGTGCGGGCACCTCAGGTCCTAAAGTTAGGCGCAC | 258 |
| 28 | TTTATTGTGCGGGCACCTCAGTTCCTAAAGTTAGGCGCAC | 253 |
| 29 | CATACTGAGCATCGTACATGATCCCGCAACGGGCAGTATC | 251 |
| 30 | CCGTTACAGTCTAAGACTCCTATGAACATAGGCGGAACTA | 249 |
| 31 | GCTGAGTTAGATCCCTTTCGTAAGGGCAGCCGGGTATCTA | 244 |
| 32 | CTGAGCATCACGAAGTGGTCCTGTCGAGGACGGGCAGTTA | 244 |
| 33 | CGAATGCAAGTCCTGACGGAGGCCAGGCGCTGAAACTTGC | 244 |
| 34 | CTGTTACAGTCTAAGACTCCTATGAACATAGGCGGAAGTA | 243 |
| 35 | TAAGGTGGGGATAAGGACGTCGGGGATCGGGGGGGGGGAT | 243 |
| 36 | GAGGGCTGATGCAATCCTAGTATAGGCAGCATAACATTGC | 242 |
Fig. 3.
The plot of log 10 (number of sequences) as a function of enrichment-fold from R1 to R3 (A) and from R2 to R3 (B).
Dissociation Constant (Kd) Measurement via Fluorescence Assay.
To test this hypothesis, we measured the equilibrium dissociation constants (Kd) and binding specificity for the three sequences with the highest enrichment-fold based on both R3/R2 and R3/R1 metrics (Table 2 and Table S1). Only the variable sequence (40-mer) is shown in the tables, but affinity and specificity measurements were performed with full-length (80-mer) molecules containing the flanking PCR primer binding sites. We measured Kd values via a standard fluorescence assay (20, 25), in which we labeled the aptamers with an Alexa 488 fluorophore-modified primer by PCR amplification. The labeled aptamer was then diluted to several different concentrations between 0 and 200 nM in 100 μL of 1 × binding buffer [0.1 mM Na2HPO4, 1.8 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl, and 1 mM MgCl2 (PBSM), pH 7.4]. The aptamer dilutions were denatured at 95 °C for 10 min, rapidly cooled to 0 °C in an ice bath, and then incubated for another 10 min at room temperature before use. After adding 7 × 105 PDGF-BB-coated beads into the denatured aptamer solution, we incubated the mixtures in the dark at room temperature for 2 h, after which unbound aptamers were removed by washing four times with 1 × binding buffer. Finally, we eluted the bound aptamers into 100 μL of 1 × binding buffer by heating the beads at 95 °C for 10 min while shaking, and quantified the eluted aptamers by fluorescence measurement, with the dissociation constant Kd calculated by fitting to a kinetic model, assuming 1∶1 Langmuir binding.
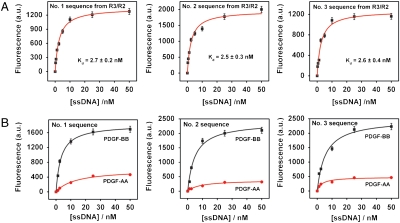
The three sequences with the highest R3/R2 enrichment-fold showed high-affinity binding to the PDGF-BB protein with low nM Kd values (Fig. 4), which is ∼4-fold higher than that shown by the R3 pool overall (Fig. 2). The measured Kd values for these top three sequences were 2.7 ± 0.2 nM, 2.5 ± 0.3 nM, and 2.6 ± 0.4 nM, respectively (Fig. 4A). By way of comparison, we performed a control experiment to measure the affinity of the sequence with the highest copy number in all three pools (Table S2 and Fig. S2). This sequence exhibited a Kd of 47.6 ± 16.1 nM—significantly lower than aptamers selected based on R3/R2 enrichment.
Fig. 4.
The three most highly enriched sequences from QSAS R3/R2 were tested for their affinity (A) and specificity (B) for the PDGF-BB target via a fluorescence binding assay.
Furthermore, the binding of these sequences was specific to PDGF-BB, with affinity for PDGF-AA that was 4.0-, 6.8-, and 4.9-fold lower, respectively (Fig. 4B). We performed a similar analysis using the three sequences with the highest R3/R1 enrichment-fold. These three aptamers demonstrated Kd values of 6.8 ± 1.3 nM, 8.2 ± 1.0 nM, and 2.7 ± 0.6 nM, respectively (Fig. S3A). Interestingly, these three displayed significantly less specificity for PDGF-BB in comparison to the sequences selected from R3/R2, with 2.0- (#1), 2.2- (#2), and 1.9-fold (#3) higher binding to PDGF-BB target relative to PDGF-AA protein (Fig. S3B). We conclude that the sequences with the highest R3/R2 enrichment-fold demonstrated better affinity and specificity than those with the highest R3/R1 enrichment-fold because the R2 pool is more specific to the target than the R1 pool (26). We also performed a nitrocellulose filter binding assay to confirm the specific binding of the most highly enriched sequence from R3/R2 relative to the initial library. The results show negligible binding (< 0.1%) of PDGF-BB protein by the unselected library, whereas the top R3/R2 sequence showed approximately 47% binding.
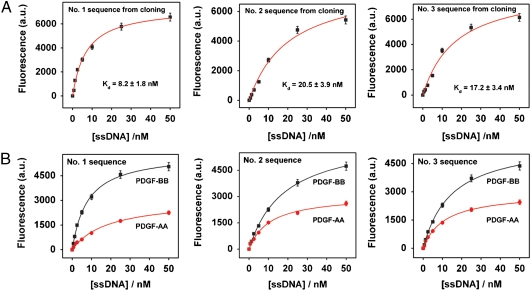
To compare the performance of aptamers obtained by QSAS to those obtained via standard cloning methods (19–21), we cloned the R3 aptamer pool into E. coli and randomly picked 98 individual clones. Three dominant consensus sequences emerged, which constituted ∼40% of the selected population; Table S3 shows a representative subset of 36 of these cloned sequences. We synthesized the sequences that appeared most frequently from each group, and measured their binding affinity and specificity for PDGF-BB. The selected group I, II, and III aptamers exhibited affinities of Kd = 8.2 ± 1.8 nM, 20.5 ± 3.9 nM, and 17.2 ± 3.5 nM (Fig. 5A), respectively, although we note that they also exhibited significant binding to PDGF-AA in comparison to the QSAS R3/R2 enriched pool, with 2.2- (#1), 1.82- (#2), and 1.8-fold (#3) higher binding to PDGF-BB target relative to PDGF-AA (Fig. 5B). Interestingly, all three sequences exhibited Kd values that were similar to the bulk Kd of the R3 pool (10.5 ± 2.63 nM), and therefore higher (i.e., lower affinity) than those obtained from QSAS analysis. When we aligned the top 36 sequences obtained based on R3/R2 and R3/R1 metrics (Table S4 and Table S5) and compared these against aptamer sequences derived via cloning, the only apparent similarity that we observed was between group 1 from the QSAS R3/R1 set and group 1 from the cloning assay.
Fig. 5.
The three most highly enriched sequences among 98 clones derived from the R3 pool were tested for their affinity (A) and specificity (B) for PDGF-BB target via a fluorescence assay.
Comparison of Aptamer Secondary Structures.
To understand the structural differences among the aptamers obtained by QSAS R3/R2, QSAS R3/R1 and standard cloning methods, we modeled the secondary structures of each of the top three enriched sequences using mfold software (34). We found that all nine sequences contained significant secondary structure, including protruding loops and stems (Figs. S4, S5, and S6). In comparing the structures, we observed that sequence #1 from the QSAS R3/R2 (Fig. S7A) differs from sequence #3 obtained by standard cloning methods (Fig. S7B) by only a single nucleotide. However, it is remarkable that their binding affinities varied by almost an order of magnitude, with significant differences in specificity. We have noted a number of examples wherein a single nucleotide difference at the loop region of an aptamer structure may play a critical role in binding affinity and specificity (35), and large differences in binding affinity of aptamers resulting from single-nucleotide changes within secondary structure elements have been reported previously (36–38). In general, it is difficult to analytically estimate the effects of a single nucleotide substitution on the binding affinity, and such analyses often require significant experimental efforts (36) including site-directed mutagenesis at or near the target-binding site (37, 38).
Conclusion
In this work, we report the use of M-SELEX with high-throughput sequencing for the rapid discovery of aptamers. The QSAS method, which combines microfluidic aptamer selection with high-throughput sequencing, displays a number of advantages over standard SELEX methods. First, the selection can be performed rapidly—We demonstrate the isolation of specific aptamers with Kd < 3 nM for PDGF-BB within 3 rounds. Second, our method enables the quantitative measurement of enrichment-fold of an individual sequence as a function of selection rounds. This allows for the identification of high-affinity sequences without the need for the pool to fully converge to a small number of sequences, while also discriminating those sequences that arise from experimental biases rather than true target binding. Though not demonstrated here, this capability to track the population of sequences may also be used to tune the selection stringency. Thirdly, the aptamers isolated by QSAS show better affinity and specificity compared to those obtained with standard cloning and sequencing methods; the aptamers identified using the R3/R2 enrichment parameters exhibited ∼3–8-fold higher affinity and ∼2–4-fold higher specificity compared to those found through random cloning of nearly 100 sequences. Finally, given that many biocombinatorial libraries are encoded with nucleic acids, we believe that our method may be generalized for the quantitative selection of other types of libraries, including tagged small molecules, phage display, cell surface display, as well as ribosome and mRNA displays (39–41), and we extrapolate that the method may ultimately be extended for the isolation of molecules with useful functions beyond binding, such as cooperative assembly (42), enzymatic activity (43) and binding-induced folding (44).
Materials and Methods
1. Materials and Instruments.
Recombinant human PDGF-BB and PDGF-AA were purchased from R&D Systems. Random DNA library, unlabeled primers, and modified primers (Alexa 488-modified forward and biotinylated reverse primer) were synthesized and PAGE-purified by Integrated DNA Technologies. N-hydroxysuccinimide (NHS), 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC), and all other chemicals were purchased from Sigma–Aldrich, Inc. Fluorescence measurements were performed in black 96-well microplates (Microfluor 2, Thermo Scientific) using the Infinite® M1000 (TECAN).
2. Microfluidic-SELEX.
M-270 Carboxylic Acid Dynabeads (Invitrogen) were used to immobilize PDGF-BB and PDGF-AA. The magnetic beads were activated with EDC and NHS and target proteins were immobilized after activation, following a manufacturer’s procedure and a previously reported procedure with minor modifications (19). The immobilized proteins were quantified using the NanoOrange® protein quantitation kit (Invitrogen). Each member of the ssDNA random library included 40 randomized nucleotides flanked by two 20-base primer binding sequences for PCR (5′-TCCCACGCATTCTCCACATC-[40N]-CCTTTCTGTCCTTCCGTCAC-3′). PDGF-BB-coated magnetic beads were washed three times with 100 μL of PBSM buffer (10.1 mM Na2HPO4, 1.8 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl, and 1 mM MgCl2, pH 7.4) before each selection. 2 × 107 PDGF-BB-coated beads were used in the first round of positive selection, and 2 × 106 PDGF-BB-coated beads were used for the next two rounds to maintain highly stringent selection conditions. The ssDNA library (∼1014 molecules) was denatured by heating at 95 °C for 10 min, and then cooled down to room temperature. This ssDNA library was incubated with PDGF-BB-coated beads in PBSM buffer for 2 h at room temperature. After incubation, the beads were trapped by MMS chip, which was fabricated and characterized as described previously (20). The trapped beads were washed with PBSM buffer at flow rates of 3 mL/h (sample) and 0.8 mL/h (buffer). Stringent washing was subsequently performed at a flow rate of 50 mL/h to continuously remove unbound and weakly bound DNAs. The aptamer-bound beads were collected in a final volume of 1.5 mL at the end of each round of selection. The eluted ssDNA aptamers were amplified by PCR using unmodified forward and biotinylated reverse primers to generate ssDNAs for next-round selection and high-throughput sequencing. To eliminate nonspecific aptamers, counter selections were also performed after the second round using 2 × 107 PDGF-AA- and Tris-coated beads.
3. High-Throughput Sequencing Technology.
To prepare samples for sequencing, the 600 μL of ssDNA obtained in each round were amplified via PCR at the optimized cycle number determined by a pilot PCR using unmodified forward and reverse primers, and then purified with the MinElute® PCR Purification Kit (Qiagen). The amplified PCR products at each round totaled ∼10 μg. The purity of the amplified DNA was examined on a 10% PAGE gel. For the unselected library control, 25 picomoles of library pool were amplified by PCR to obtain 6 μg of product, which was also purified using the PCR purification kit. Illumina’s single-read ChIP-Seq library preparation kit was used to prepare the double-stranded aptamers for sequencing on the Genome Analyzer II. 10 ng of sample were initially subjected to preparation for sequencing, which entails end repair, addition of adenosine to 3′ ends, adapter ligation, fragment size selection by gel extraction, and PCR following manufacturer’s instructions with modifications described below. After each step, samples were cleaned with DNA Clean & Concentrator columns from Zymo Research. For the adaptor ligation step, the adaptor mix was diluted 1∶20 to prevent an abundance of adaptors from being sequenced. Following adapter ligation, we ran a 2% agarose gel to select the specific size range of molecules needed for proper cluster formation on the cluster station. The size range from 200–300 bp was cut from the gel and cleaned using the Gel Purification Mini kit from Qiagen, based on the manufacturer’s protocol. We then performed 14 cycles of PCR to amplify selected fragments using Illumina-supplied PCR primers and the manufacturer’s suggested PCR recipe. The forward and reverse primers were diluted 1∶2 prior to being added to the sample. Finally, the sample was quantified with the Invitrogen Qubit fluorometer, after which the preparation was ready to load on the Illumina cluster station.
The cluster station is used to hybridize samples to a flow cell, which is then placed in the Genome Analyzer II to obtain the sequence data. The cluster station bridge amplifies each single molecule 35 times to make cloned bundles or clusters, which are large enough for individual nucleotides to be visualized by the optics of the Genome Analyzer II. Samples were loaded into the flow cell cluster station at a concentration of 4 pM. Once flow cell amplification was complete, each cluster was linearized, a blocking group was attached to the free end of the cluster and the sequencing primer was hybridized. The flow cell was subsequently loaded on the Genome Analyzer II, which was run for 75 cycles (equivalent to sequencing 75 bases). After the sequencing was complete, the data were processed using Illumina Pipeline software, which performs base calling and quality filtering and generates a lane-based fastaq format file. We performed downstream analysis using software developed internally. A correct sequence read should include 75 bp of sequence, including 20 bp 5′ primer +40 bp aptamer +15 bp 3′ primer; any sequences not matching this pattern were filtered out. The primer sequences were trimmed out after filtering, leaving only 40 bp aptamer sequences for downstream analysis. Reverse complement tags were combined with the sequenced forward tags, and tag count was done at this stage, followed by enrichment analysis.
4. Determination of Dissociation Constants (Kd) and Specificities of Selected Aptamers.
Selected pools and individual ssDNA aptamers from those pools were tested for their binding affinity to PDGF-BB using a standard fluorescence binding assay. Various concentrations of Alexa 488-modified aptamers, generated via PCR with fluorescently labeled forward primers, were heated at 95 °C for 10 min in 93 μL of PBSM buffer, rapidly cooled down in an ice bath and then incubated for 10 min at room temperature. 7 × 105 PDGF-BB-coated beads were washed 3 times in PBSM buffer and incubated with the aptamers at room temperature for 2 h. After the binding reaction, the unbound aptamers were removed by washing 4 times with PBSM buffer. The bound aptamers were then eluted from the beads by heating at 95 °C for 10 min. The amount of bead-bound aptamers was measured by fluorescence measurement (excitation 495 nm, emission 519 nm) and the dissociation constant was calculated by nonlinear fitting analysis. For specificity tests, Alexa 488-modified aptamers (0–50 nM) were incubated with PDGF-BB-coated beads and PDGF-AA-coated beads, followed by washing steps as described above. The amount of aptamer in each elution fraction was determined by fluorescence measurement and calculation using a fitting curve as described above.
Supplementary Material
Acknowledgments.
We are grateful for the financial support of the Army Research Office (ARO) Institute for Collaborative Biotechnologies, Office of Naval Research, Charlotte Geyer Foundation, Midwest Progenitor Cell Consortium, and National Institutes of Health. Financial support for M.C. was partially provided by Korea Research Foundation Grant KRF-2008-357-C00086.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009331107/-/DCSupplemental.
References
- 1.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 2.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 3.Jayasena SD. Aptamers: An emerging class of molecules that rival antibodies in diagnostics. Clin Chem. 1999;45:1628–2650. [PubMed] [Google Scholar]
- 4.Mairal T, et al. Aptamers: molecular tools for analytical applications. Anal Bioanal Chem. 2008;390:989–1007. doi: 10.1007/s00216-007-1346-4. [DOI] [PubMed] [Google Scholar]
- 5.Huizenga DE, Szostak JW. A DNA aptamer that binds adenosine and ATP. Biochemistry. 1995;34:656–665. doi: 10.1021/bi00002a033. [DOI] [PubMed] [Google Scholar]
- 6.Mann D, Reinemann C, Stoltenburg R, Strehlitz B. In vitro selection of DNA aptamers binding ethanolamine. Biochem Biophys Res Commun. 2005;338:1928–1934. doi: 10.1016/j.bbrc.2005.10.172. [DOI] [PubMed] [Google Scholar]
- 7.Bock LC, Griffin LC, Latham JA, Vermass EH, Toole JJ. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 8.Green LS, et al. Inhibitory DNA ligands to platelet-derived growth factor B-chain. Biochemistry. 1996;35:14413–14424. doi: 10.1021/bi961544+. [DOI] [PubMed] [Google Scholar]
- 9.Lupold SE, Hicke BJ, Lin Y, Coffey DS. Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Cancer Res. 2002;62:4029–4033. [PubMed] [Google Scholar]
- 10.Mallikaratchy P, et al. Aptamer directly evolved from live cells recognizes membrane bound immunoglobin heavy mu chain in Burkitt’s lymphoma cells. Mol Cell Proteomics. 2007;6:2230–2238. doi: 10.1074/mcp.M700026-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Xiao Y, Lubin AA, Heeger AJ, Plaxco KW. Label-free electronic detection of thrombin in blood serum by using an aptamer-based sensor. Angew Chem Int Edit. 2005;44:5456–5459. doi: 10.1002/anie.200500989. [DOI] [PubMed] [Google Scholar]
- 12.Swensen JS, et al. Continuous, real-time monitoring of cocaine in undiluted blood serum via a microfluidic, electrochemical aptamer-based sensor. J Am Chem Soc. 2009;131:4262–4266. doi: 10.1021/ja806531z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen HW, et al. Molecular recognition of small-cell lung cancer cells using aptamers. ChemMedChem. 2008;3:991–1001. doi: 10.1002/cmdc.200800030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li W, et al. Real-time imaging of protein internalization using aptamer conjugates. Anal Chem. 2008;80:5002–5008. doi: 10.1021/ac800930q. [DOI] [PubMed] [Google Scholar]
- 15.McNamara JO, et al. Multivalent 4-1BB binding aptamers costimulate CD8 + T cells and inhibit tumor growth in mice. J Clin Invest. 2008;118:376–386. doi: 10.1172/JCI33365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou J, Li H, Li S, Zaia J, Rossi JJ. Novel dual inhibitory function aptamer-siRNA delivery system for HIV-1 therapy. Mol Ther. 2008;16:1481–1489. doi: 10.1038/mt.2008.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tong GJ, Hsiao SC, Carrico ZM, Francis MB. Viral capsid DNA aptamer conjugates as multivalent cell-targeting vehicles. J Am Chem Soc. 2009;131:11174–11178. doi: 10.1021/ja903857f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tasset DM, Kubik MF, Steiner W. Oligonucleotide inhibitors of human thrombin that bind distinct epitopes. J Mol Biol. 1997;272:688–698. doi: 10.1006/jmbi.1997.1275. [DOI] [PubMed] [Google Scholar]
- 19.Lou X, et al. Micromagnetic selection of aptamers in microfluidic channels. Proc Natl Acad Sci USA. 2009;106:2989–2994. doi: 10.1073/pnas.0813135106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qian J, Lou X, Zhang Y, Xiao Y, Soh HT. Generation of highly specific aptamers via micromagnetic selection. Anal Chem. 2009;81:5490–5495. doi: 10.1021/ac900759k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tok JB, Fischer NO. Single microbead SELEX for efficient ssDNA aptamer generation against botulinum neurotoxin. Chem Commun. 2008:1883–1885. doi: 10.1039/b717936g. [DOI] [PubMed] [Google Scholar]
- 22.Wang J, Lv R, Xu D, Chen H. Characterizing the interaction between aptamer and human IgE by use of surface plasmon resonance. Anal Bioanal Chem. 2008;390:1059–1065. doi: 10.1007/s00216-007-1697-x. [DOI] [PubMed] [Google Scholar]
- 23.Chen CH, Chernis GA, Hoang VQ, Landgraf R. Inhibition of heregulin signaling by an aptamer that preferentially binds to the oligomeric form of human epidermal growth factor receptor-3. Proc Natl Acad Sci USA. 2003;100:9226–9231. doi: 10.1073/pnas.1332660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee JH, et al. A therapeutic aptamer inhibits angiogenesis by specifically targeting the heparin binding domain of VEGF165. Proc Natl Acad Sci USA. 2005;102:18902–18907. doi: 10.1073/pnas.0509069102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stoltenburg R, Reinemann C, Strehlitz B. FluMag-SELEX as an advantageous method for DNA aptamer selection. Anal Bioanal Chem. 2005;383:83–91. doi: 10.1007/s00216-005-3388-9. [DOI] [PubMed] [Google Scholar]
- 26.Djordjevic M. SELEX experiments: new prospects, applications and data analysis in inferring regulatory pathways. Biomol Eng. 2007;24:179–189. doi: 10.1016/j.bioeng.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 27.Stoltenburg R, Reinemann C, Strehlitz B. SELEX-A (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol Eng. 2007;24:381–403. doi: 10.1016/j.bioeng.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 28.Zimmermann B, Gesell T, Chen D, Lorenz C, Schroeder R. Monitoring genomic sequences during SELEX using high-throughput sequencing: Neutral SELEX. PLoS One. 2010;5:e9169. doi: 10.1371/journal.pone.0009169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alvarez RH, Kantarjian HM, Cortes JE. Biology of platelet-derived growth factor and its involvement in disease. Mayo Clin Proc. 2006;81:1241–1257. doi: 10.4065/81.9.1241. [DOI] [PubMed] [Google Scholar]
- 30.Liu Y, et al. Controlling the selection stringency of phage display using a microfluidic device. Lab Chip. 2009;9:1033–1036. doi: 10.1039/b820985e. [DOI] [PubMed] [Google Scholar]
- 31.Morozova O, Marra MA. Applications of next-generation sequencing technologies in functional genomics. Genomics. 2008;92:255–264. doi: 10.1016/j.ygeno.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 32.Mardis ER. Next-generation DNA sequencing methods. Annu Rev Genom Hum G. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- 33.Acinas SG, Sarma-Rupavtarm R, Klepac-Ceraj V, Polz MF. PCR-induced sequence artifacts and bias: Insights from comparison of two 16S rRNA clone libraries constructed from the same sample. Appl Environ Microbiol. 2005;71:8966–8969. doi: 10.1128/AEM.71.12.8966-8969.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luchansky SJ, Nolan SJ, Baranger AM. Contribution of RNA conformation to the stability of a high-affinity RNA–protein complex. J Am Chem Soc. 2000;122:7130–7131. [Google Scholar]
- 36.Katilius E, Flores C, Woodbury NW. Exploring the sequence space of a DNA aptamer using microarrays. Nucleic Acids Res. 2007;35:7626–7635. doi: 10.1093/nar/gkm922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Green L, et al. Comprehensive chemical modification interference and nucleotide substitution analysis of an RNA pseudoknot inhibitor to HIV-1 reverse transcriptase. J Mol Biol. 1995;247:60–68. doi: 10.1006/jmbi.1994.0122. [DOI] [PubMed] [Google Scholar]
- 38.Burgstaller P, Kochoyan M, Famulok M. Structural probing and damage selection of citrulline and arginine-specific RNA aptamers identify base positions required for binding. Nucleic Acids Res. 1995;23:4769–4776. doi: 10.1093/nar/23.23.4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoogenboom HR. Selecting and screening recombinant antibody libraries. Nat Biotechnol. 2005;23:1105–1116. doi: 10.1038/nbt1126. [DOI] [PubMed] [Google Scholar]
- 40.Aina OH, et al. From combinatorial chemistry to cancer-targeting peptides. Mol Pharm. 2007;4:631–651. doi: 10.1021/mp700073y. [DOI] [PubMed] [Google Scholar]
- 41.Lee JS, Kim YK, Vendrell M, Chang YT. Diversity-oriented fluorescence library approach for the discovery of sensors and probes. Mol Biosyst. 2009;5:411–421. doi: 10.1039/b821766c. [DOI] [PubMed] [Google Scholar]
- 42.Silverman J, et al. Multivalent avimer proteins evolved by exon shuffling of a family of human receptor domains. Nat Biotechnol. 2005;23:1556–1561. doi: 10.1038/nbt1166. [DOI] [PubMed] [Google Scholar]
- 43.Puthenveetil S, et al. Yeast display evolution of a kinetically efficient 13-amino acid substrate for lipoic acid ligase. J Am Chem Soc. 2009;131:16430–16438. doi: 10.1021/ja904596f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nutiu R, Li YF. In vitro selection of structure-switching signaling aptamers. Angew Chem Int Edit. 2005;44:1061–1065. doi: 10.1002/anie.200461848. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.