Abstract
Toxoplasmosis causes significant morbidity and mortality and yet available medicines are limited by toxicities and hypersensitivity. Since improved medicines are needed urgently, rational approaches were used to identify novel lead compounds effective against Toxoplasma gondii enoyl reductase (TgENR), a type II fatty acid synthase enzyme essential in parasites but not present in animals. Fifty-three compounds, including three classes that inhibit ENRs, were tested. Six compounds have anti-parasite MIC90s ≤6μM without toxicity to host cells, three compounds have IC90s <45nM against recombinant TgENR and two protect mice. To further understand the mode of inhibition, the co-crystal structure of one of the most promising candidate compounds in complex with TgENR has been determined to 2.7Å. The crystal structure reveals that the aliphatic side chain of compound 19 occupies, as predicted, space made available by replacement of a bulky hydrophobic residue in homologous bacterial ENRs by Ala in TgENR. This provides a paradigm, conceptual foundation, reagents, and lead compounds for future rational development and discovery of improved inhibitors of T. gondii.
Introduction
Toxoplasmosis causes substantial morbidity and mortality, especially in persons who are congenitally infected or immune-compromised and this parasite is the most frequent infectious cause of uveitis.1–4 Toxoplasma gondii (T. gondii, Tga) is acquired as a sporozoite from oocysts formed in cats or bradyzoites from cysts in meat. In humans, this parasite has a simple life cycle consisting of two stages; tachyzoites and bradyzoites. The former are a rapidly growing, obligate intracellular forms of T. gondii present when parasites are first acquired in acute infections. T. gondii then develops into slowly growing, encysted, latent bradyzoites, sequestered within cysts inside cells, with a competent host immune response. When a cyst ruptures, stage transition from latent bradyzoites back to rapidly growing tachyzoites occurs, causing destruction of surrounding tissue.
Reasons for recrudescence of eye disease have not been completely defined but is a lifelong problem in individuals infected congenitally as well as some of those whose infection is acquired after birth.3,4 This is an especially pressing problem in Brazil, as 80% of the population is infected with particularly pathogenic parasite strains, with a high incidence beginning in childhood. In some regions of Brazil, 20% of these individuals and 50% of those over 50 years old have eye disease. In immunocompromised persons such as those with AIDS, disease due to recrudescence (especially in the brain) is frequent, occurring in 50% of those with AIDS whose HIV infection remains untreated. Life threatening toxoplasmosis occurs in those immunocompromised by malignancies, organ transplantations and autoimmune disease with associated treatments. Rarely, there is significant organ damage in those without known immune compromise. An epidemic of multivisceral, lethal disease caused by a hypervirulent strain of parasite was reported recently in Guyana, making this emerging infection potentially even more problematic with globalization of food supplies. This parasite can easily contaminate food supplies or the environment and is a potential bioterrorism pathogen. There have been several recent epidemics associated with contaminated water supplies.
Consequences of chronic infections present in ~30% of the population (~2 billion people) worldwide, throughout their lifetimes, are not thoroughly characterized. Recently, memory impairment was reported in healthy, young to middle aged professionals in association with this infection and presence of a susceptibility allele of a gene encoding an enzyme that degrades dopamine, catechol O-methyl transferase (COMT) (Yolken et al. 2009 Personal Communication). There is also a higher prevalence of antibody to T. gondii in those with cryptogenic epilepsy and schizophrenia, although cause and effect between T. gondii infection and these neurologic observations has not yet been proven.
There are only a few medicines that restrict growth of tachyzoites,1–4 and use of these medicines is associated with significant incidences of hypersensitivity (up to 25%) and toxicity.5 No medicines eliminate encysted, latent bradyzoites. Better approaches to treat this disease are greatly needed including medicines that eliminate active parasites causing disease and means to eliminate latent parasites.
Recent work by our group,6–15 and a recent report by others,16 provide the foundation for the present work to develop a new class of medicines to better treat toxoplasmosis. Specifically, the prokaryotic-like type II fatty acid biosynthetic (fas) pathway in T. gondii is a validated molecular target in tachyzoites it is essential for parasite survival in vitro and in vivo.16 In particular, the enoyl reductase (ENR) enzyme, which catalyses the last reductive step of the type II fatty acid synthesis pathway, is present in all T. gondii life cycle stages except microgametes11 and ENRs in other organisms have been shown to be the target for a wide range of potent inhibitors. Importantly, compounds which inhibit type II fatty acid synthesis (including triclosan and a number of newly designed and synthesized compounds) not only inhibit T. gondii tachyzoite growth but are effective against other apicomplexan parasites, such as the hepatic stage of Plasmodia,7,16–20 which causes malaria. However, while triclosan is a potent inhibitor of the enzyme, its poor solubility makes it unsuitable as a potential therapeutic. This prompted the investigation into different triclosan scaffolds as potential therapeutic agents for toxoplasmosis, with improved activity, as well as physicochemical properties and toxicity profiles.
This report details activity of a panel of 53 compounds (Figure 1) based on previously identified ENR inhibitors, with two of these compounds (compounds 2 and 19) showing low toxicity and activity in the low nM range. A promising adaptation to the triclosan scaffold is the addition of an n-propyl group at the 4-position, exploits an increase in space in the parasitic ENR binding pocket. In order to validate this, a co-crystal structure of one of the most promising inhibitors (compound 19) in complex with TgENR, was determined to 2.7Å. This structure revealed that these inhibitors utilize the extra space within the binding pocket of the parasitic ENR family. The data presented herein provides insights into lead compounds which form the basis for future development of highly active inhibitors of ENR with potential for progression to medicines that effectively treat toxoplasmosis and related apicomplexan diseases.
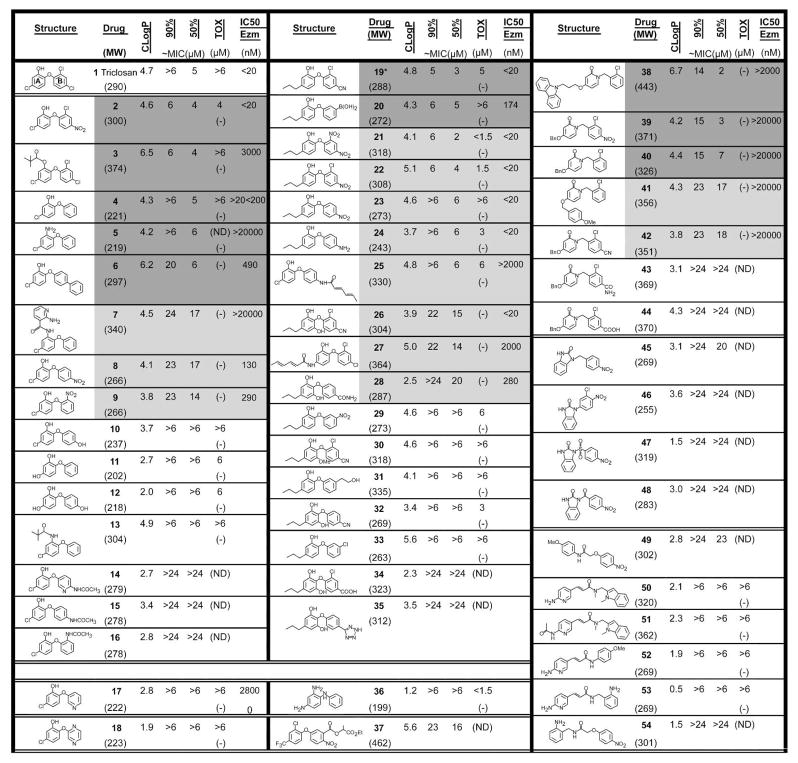
Figure 1.
Inhibitors of ENR: Structures of potential ENR inhibitors, ID #, MW, and ClogP of inhibitors (calculated by using ALOGPS 2.1 www.vcclab.org/lap/alogps/), inhibition of T. gondii in tissue culture and toxicity for HFF at highest concentration tested in simultaneous experiments; IC50 enzyme. * = co-crystal. MIC, Inhibition of T. gondii uptake of tritiated uracil; “Tox,” Effect of compound on nonconfluent fibroblast’s uptake of tritiated thymidine with visual analysis of normal confluent monolayer, (−) represents no toxicity at highest concentration tested. Dark grey shading indicates most promising inhibitors and light grey shading indicates next initially most promising inhibitors but with either greater toxicity to fibroblasts relative to effect on parasite or lower enzyme activity. Shaded compounds were prescreened in a T. gondii ENR enzyme assay at three concentrations: 0.2, 2, and 20μM. Compounds with significant inhibition at 2μM were further analyzed to determine IC50 values with all data collected in triplicate.
Results
Synthesis of Inhibitors
Synthesis of compounds tested is shown in Figure 2 (Synthetic Schemes). The numbers, molecular weights, ClogPs and structures of compounds are shown in Figures 1 and 2. The requisite diphenyl ethers were synthesized either by nucleophilic aromatic substitution (Method A) or through Cu catalyzed coupling reactions (Method B). The choice of the method was dependent on the product desired and the choice of the starting phenol. In each case, the methoxy diarylether precursors so obtained were demethylated to phenols using boron tribromide (Scheme 1). The aryl boronic acid 20 was prepared by lithiation of 1-(3-chlorophenoxy)-2-methoxy-4-propylbenzene followed by quenching with trimethyl borate followed by acidic hydrolysis. The diarylether derivatives 2, 4, 6, 8, 9–12, 14–19, 21–24, 29, 31,21 and the 2-pyridone derivatives 38–44 were previously published by us.22 Amides 7, 25 and 27 were prepared from their corresponding aniline precursors using standard amide bond forming procedures (Scheme 2). Pivaloyl ester 3 was prepared from triclosan (Scheme 2). Aminopyridines 52 and 53 were synthesized from 3-(6-aminopyridin-3-yl)-acrylic acid23 according to Scheme 3. Benzimidazolones 46–48 were prepared using standard procedures as depicted in Scheme 3. The HPLC and high resolution mass spectroscopy were employed to determine the purity of the tested compounds. Purity was between 96 and 100% as shown in Table S1.
Figure 2.
(a) Synthesis of compound in 3 schemes: Scheme 1. Synthesis of compounds: When X = F and R2 = Cl, R3 = H, R4 = CN; or when X = F and R2 = R3 = H, R4 = CN, Method A: K2CO3 or Cs2CO3, DMSO, 100 °C, 8–12 h. When X = I and R2 = R4 = H, R3 = Cl, Method B: KOtBu, DMF, (CuOTf)2·PhH, 140 °C, 16–20 h. a) Excess BBr3, CH2Cl2, −78 °C to rt, 2–6 h; b) n-BuLi, B(OMe)3, 2N HCl, −78 °C to rt, 18 h; c) 35% H2O2, 3N NaOH, EtOH, 30 °C, 18 h; d) 25% NaOH, EtOH, reflux, 20 h; e) NaN3, SiCl4, CH3CN, 90 °C, 6 h; (b) Scheme 2. Synthesis of compounds: a) Sorbic acid, EDAC?HCl, HOBt, Et3N, DMF, rt, 16 h; b) Excess BBr3, CH2Cl2, −78 °C to rt, 6 h; c) Pd/C, H2, 40 PSI, TEA, EtOH, rt, 16 h; d) Pivaloyl chloride, Et3N, THF, rt, 4 h; e) 2-Aminonicotinic acid, EDAC?HCl, HOBt, Et3N, DMF, rt, 16 h; f) Pivaloyl chloride, Et3N, DMAP, CH2Cl2, 0 °C to rt, 6 h; (c) Scheme 3. Synthesis of compounds: a) p-Anisidine, EDAC?HCl, HOBt, Et3N, DMF, rt, 20 h; b) 2-Aminobenzylamine, EDAC?HCl, HOBt, Et3N, DMF, rt, 16 h; c) 4-Fluoronitrobenzene, K2CO3, DMSO, 100 °C, 16 h; d) 4-Nitrobenzoyl chloride, pyridine, 0−80 °C, 4 h; e) 4-Nitrobenzyl bromide, K2CO3, MeOH, 100 °C, 4 h; f) 4-Nitrobenzenesufonyl chloride, Et3N, DMF, 0 °C–rt, 4 h.
Inhibition of T. gondii tachyzoites in vitro
A summary of ability of each compound to inhibit the parasite’s growth by 50% (MIC50) and 90% (MIC90) and effect on growth and survival of nonconfluent cultures of the same fibroblasts host cells in the parasite inhibition assay, as an indication of toxicity, are in Figure 1.
Analysis of the 53 compounds identified 6 compounds that robustly inhibited parasites (~60–95% inhibition of parasite growth) with less than 20% toxicity to host cells with MIC90s <6μM (Figure 1). An additional six compounds had modest inhibitory effect on parasites (~20% to 60% inhibition of parasite growth) and an additional three had ~20–40% inhibition of host cell uptake of tritiated thymidine (Figure 1). The next six compounds had less than 40% inhibitory effect on uptake of uracil by parasites or were very toxic to host cells precluding interpretation of parasite inhibition assays. The last compounds shown had minimal inhibitory effect and/or harmed host cell growth substantially (Figure 1). These data showing IC50 and IC90 are in Figure 1. To illustrate these results in specific experiments with more detail, a representative experiment with compounds 25, which was toxic for fibroblasts, and 19, which was efficacious and not toxic are shown in Figure 3. Of the initial 53 compounds tested, compounds that displayed the greatest effect and least toxicity were 2, 3, 19, and 39.
Figure 3.
(a,b) Representative experiments showing effects of compounds 25 and 19 on parasite uptake of tritiated uracil and on growth of nonconfluent host cell fibroblasts. (a) Inhibition of T. gondii uptake of tritiated uracil by compound 19 and lack of inhibition by compound 25. (b) Lack of toxicity to fibroblasts (uptake of 3H Thymidine by nonconfluent HFF) by compound 19. Toxicity to HFF from compound 25. (c,d,e) Representative experiments showing inhibition of T. gondii tachyzoites (top panels), lack of inhibition of fibroblast growth (middle panels) and inhibition of ENR enzyme activity (bottom panels) by compounds 2, 19, and 39. Error bars in panels A–E show the standard deviation between triplicate measurements. Compound 2 IC50 = 45nM and 19 = 20nM, and 39 less active (low micro molar), possibly off target.
Inhibition of TgENR activity
Compounds that inhibited the growth of T. gondii in culture (Figure 1, shaded regions) were initially tested for inhibition of TgENR enzymatic activity at three concentrations (0.2, 2, and 20μM). Compounds which displayed significant inhibitory activity at 2μM were assayed in triplicate at ten concentrations to determine IC50 values as summarized in Figure 1 and shown in detail in Figure 3. This assay uses 20nM TgENR, preventing the accurate measurement of IC50 values below this concentration. Seven compounds (including triclosan) are listed as having IC50 values below 20nM. Compound 39 turned out to be a poor inhibitor of TgENR and hence appears to have an off target effect on parasites and requires further investigation. This result indicates that this compound may be of interest for further development but does not target ENR specifically.
Co-crystallization and structure solution of TgENR in complex with NAD+ and compound 19
In order to gain insights into the mode of binding for compound 19 which is very active against the ENR enzyme (IC50 <20nM; Figure 3 bottom panel) and the parasite in tissue culture, structural studies were conducted. TgENR was co-crystallized in the presence of NAD+ and compound 19 with the subsequent crystals diffracting to beyond 2.7 Å (Data are in Table 1). The refined structure showed clear and continuous density for the bound inhibitor allowing for its unambiguous placement within the structure (Figure 5a). The superposition of TgENR in complex with compound 19 and triclosan shows that the mode of binding for both inhibitors is similar with the aromatic rings of both compounds adopting the same position, with ring A in both cases forming stacking interactions with the NAD+ co-factor. The additional bulk of the alkyl substituent of compound 19, when compared to triclosan, occupies the space made available by replacement of a bulky hydrophobic residue common to the bacterial enzyme family (Met206 in E. coli) by alanine (Ala247 in TgENR) which is conserved in the apicomplexan family (Figure 5c). Comparison of the triclosan and compound 19 TgENR complex also reveals that there is little change in the position of the mainchain atoms with an overall Cα r.m.s.d value of 0.3 Å (Figure 5b). However, the side chain of Phe 243 does move significantly by ~1.5Å about its Cβ atom to accommodate the extra bulk of compound 19 compared to triclosan, without significant change to its mainchain position (Figure 5b, c). As such elaborating the nature of the substituent on the phenoxy ring of the candidate compound exploits the increase in space which is a conserved feature of the apicomplexan ENR family, without perturbing the overall fold of the binding pocket.
Table 1.
| Data collection/processing | |
| Space group | P3221 |
| Resolution (Å) | 2.7 |
| Unique reflections | 16525 |
| Completeness (%) | 90.85 |
| I/σ(I) >3 (%) | 26 |
| Rmerge (%) | 0.378 |
| Refinement statistics | |
| Rcryst (%)a/Rfree (%)b | 0.26/0.32 |
| R.m.s.d. values | |
| Bond length (Å) | 0.0054 |
| Bond angle (deg.) | 1.00 |
| Ramachandran plotc | |
| Most favoured (%) | 93.0 |
| Additionally allowed (%) | 7.0 |
| Generously allowed (%) | 0.0 |
| Disallowed (%) | 0.0 |
| Protein/substrate atoms/waters | 4486/128/51 |
| Mean B-values (Å2) | |
| Protein | 38 |
| Co-factors | 38 (compound 19) 36 (NAD) |
| Water molecules | 43 |
Refinement statistics for TgENR/NAD+/compound 19 complex.
Rcryst = Σhkl(|Fobs| - |Fcalc|)/Σhkl |Fobs|.
Rfree was calculated on 5% of the data omitted randomly.
Percentage of residues in regions of the Ramachandran plot, according to PROCHECK.
Figure 5.
(a) 2Fobs – 1Fcalc electron density map contoured at 1σ for compound 19 in complex with TgENR/NAD+ after refinement in REFMAC5. (b) Superposition of TgENR in complex with triclosan (blue) and compound 19 (green) shown in cartoon format, to show the conservation of the overall fold between the two complexes. Both the NAD+ cofactor and compound 19 are displayed in stick format and coloured yellow, red, blue, green and purple for carbon, oxygen, nitrogen, chlorine and phosphorus, respectively. (c) Stereo diagram of the TgENR triclosan (green) and compound 19 (blue) complex structures superposed. The NAD+ cofactor and inhibitors are shown in stick format in addition to Phe243 which display the greatest change between the different complexes.
Inhibition of T. gondii tachyzoites in vivo
Effect of Compounds 2 and 19 on T. gondii in vivo are in Figure 4. Representative experiment of two replicate studies, indicate that compounds 2 and 19 can reduce parasite burden in mice (P<0.05).
Figure 4.
Reduction of parasite burden by 2 and 19 in mice.
Cytochrome P450 inhibition and human liver microsomal stability
2 and 19 were tested for in vitro inhibition of recombinant CYP450 isozymes 2C9, 2D6 and 3A4. At 10μM, 2 and 19 showed strong inhibition of CYP2C9 (both >85%) and relatively low inhibition of CYP2D6 and CYP3A4 (both <22%). Further, metabolic stabilities in pooled human liver microsomes (HLM) indicated that they were moderately metabolized in HLM, and remaining parent drugs after incubation for 60 minutes were 51.23% and 45.14%, respectively. Intrinsic clearances, metabolic rates, of 2 and 19 in HLM were slower than the control drug verapamil.
Discussion
Triclosan is a broad spectrum antibacterial agent incorporated into a wide range of consumer products.24–26 It inhibits the ENR (FabI) enzyme in a number of microorganisms like E. coli,27,28 P. aeruginosa29 and S. aureus.30 Significant variation in the degree of inhibition of different pathogens by triclosan has been observed28–30 suggesting that it should be possible to modify the compound to improve its efficacy against a particular organism through systematic structure-based as well as ligand-based drug design methods to improve properties such as potency, solubility and toxicity.
The crystal structure of T. gondii and P. falciparum ENR in complex with triclosan shows the binding geometry of triclosan to be similar to that seen with other organisms. Thus the phenoxy ‘ring A’ (Figure 1 and 2) of triclosan and the nicotinamide ring of NAD+ engage in π-stacking interactions, while the phenolic hydroxyl group on ring A (Figure 1) is involved in a hydrogen bond with a conserved Tyr (OH), and the ether linkage has two hydrogen bonds, one to the hydroxyl group of Tyr, and the other to the ribose unit of the NAD+. Encouraged by our earlier results on triclosan activity against TgENR,7 we tested a number of newly synthesized triclosan derivatives in addition to those available to us from our previous work.21,22 In addition, we designed and tested compounds based on diazaborine and aminopyridine inhibitor families shown to inhibit the enzyme with the goal of attaining improved inhibitors.
Several of the triclosan derivatives identified were active against parasites at low micromolar levels with sub-micromolar inhibitory activity toward TgENR (Figures 1 and 3). In general, presence of a strong electron withdrawing group on the B ring contributed to improved activity while the presence of an electron donating group reduced the activities of these compounds (compare compounds 2 vs 1, 8 vs 10, 4 vs 10. Freundlich and co-workers showed that compound 2 has an IC50 of 0.2μM against P. falciparum ENR.31 This compound is highly active against TgENR as well, which is not surprising as the active sites of these two enzymes share significant homology. Unlike the observation of Sivaraman et al,32,33 in the case of TgENR, deletion of chlorine atoms on the B ring of triclosan (as in compound 4) does not improve activity toward both TgENR and the parasite, suggesting that the presence of a more lipophilic group would favor the improvement in activity. However, modification of the B ring to a biphenyl to increase its hydrophobicity did not improve activity (compound 6). This is not surprising since this group faces the NAD+ co-factor and would cause a large degree of steric hindrance accounting for the large increase in IC50 value (>2000 nM). When the B ring of triclosan is replaced by a hetero-aromatic ring resulted in compounds 17 and 18 with compound 17, the pyridine analog of compound 4 showing some activity. However, compound 18, the corresponding pyrazine analog had no activity. Since compound 18 is smaller than triclosan this lack of activity is unlikely to be caused simply through steric hindrance within the binding pocket highlighting the importance of presentation of an appropriate electronic distribution in this ring to allow for optimal interaction with the active site. Moreover, the pyrazine may be attracted towards the NAD+ cofactor, destabilizing both the rings A and B, driven in part by the formation of a hydrogen bond between the backbone oxygen of NAD+ and nitrogen of compound 18. Further studies are required to test this hypothesis. Although of the 11 compounds reflecting changes focused on the B ring (2, 4, 8–10, 14–18) only compound 2 displays a comparable efficacy to triclosan. These observations, taken together open up additional avenues for further development of a new class of heteroaryl analogs of triclosan. However, caution must be taken to not disturb the important interaction the A ring makes with the NAD+ cofactor through changes in the B ring.
The phenolic hydroxyl group on ring A is considered to be critical to triclosan’s activity, as it is involved in hydrogen bonding to a conserved Tyr residue in the active site and forming packing interactions with the NAD+ cofactor. This has been observed by us in our earlier work,10 and is also evident by the inactivity of compound 5. The significant change in potency between compounds 4 (IC50 >20<200 nM) and 5 (IC50 >20000 nM), can only be attributed to a change of the OH group to NH2. In light of this observation, it is interesting to note that 3 shows improved activity on the parasites over triclosan itself. Since the pivalate group would produce severe steric clashes within the binding pocket (as highlighted by the increased IC50 value) we suspect that this heightened activity may be due to the enhanced ability of this pro-drug form to enter parasites, whereby the ester is hydrolyzed. Similarly, while the antiparasitic activities of compounds 4 and 13 were comparable, there is significant difference in their TgENR activity indicating that a tertiary butyl group is indeed too bulky at this position. The differences in the antiparasitic activity and TgENR inhibitory activity observed in some of the analogs is not surprising and could be attributed to off target effects and problems with delivery, particularly with delivery of an inhibitory compound into the apicoplast that has 4 surrounding membranes and in which ENR resides. We intend to investigate this finding further.
Diazaborines are known to inhibit ENR through covalent bond formation between the 2′-hydroxyl of the nicotinamide ribose unit of the NAD+ molecule and the boron atom of the drug.34 Consequently, we believed that it would be valuable to incorporate this type of functionality into our triclosan based ENR inhibitors, and thus we prepared the boronic acid analog 20. This compound also proved to be a very effective inhibitor of TgENR, although further studies are needed to determine whether a covalent bond has been formed with the nicotinamide ribose. Other attempts to vary the meta position of the B ring with H-bond donors and acceptors did not give encouraging results (compounds 29, 31, and 33).
Although the indole aminopyridine compound 5023 was very effective against S. aureus and E. coli ENR, it shows only moderate activity toward TgENR. We synthesized several aminopyridine mimics of this compound, namely 50, 52 and 53, in order to determine whether this series had any special merits that might warrant further investigation. Unfortunately, none of these showed any promising activity, this may in part be due to the steric hindrance which can be brought about by Phe243 (TgENR numbering) in the binding pocket as previously reported.15
When the A ring was modified by replacing the chlorine atom at the para position by an hydrophilic OH group, the activity decreased (compounds 11 and 12). These changes on ring A may be brought about by the disturbance of the stacking interactions with the NAD+ cofactor which are critical for inhibitor binding. Furthermore, this inactivity could also be influenced by the lower lipophilicity of these compounds rather than the electron-donor effect of the hydroxyl group. To test this hypothesis, we attempted to append a long hydrophobic alkyl chain at the para position. This can be accommodated since the bacterial and plant ENR family contain a bulky hydrophobic residue (Met, Leu or Ile) close to the ring A of triclosan, whereas the ENR of apicomplexan species such as T. gondii and Plasmodium have a fully conserved alanine residue. This sequence change produces an increase in space at the base of the binding pocket and reduces the van der Waals packing interactions with the triclosan based inhibitors in comparison to other members of the ENR family.15 When we synthesized a number of triclosan derivatives bearing an n-propyl group at the 4-position which were predicted to be optimal length to fit within the binding site, these inhibitors showed, in general, better or similar activities than the triclosan scaffold (compounds 19–24), with no detrimental effect of this extension to their IC50 values with derivatives that bear electron withdrawing groups on the B ring (compounds 19, 21–24) producing among the most active compounds identified to date.
To further investigate the mode of binding of the most promising inhibitor which contains an n-propyl group at the 4 position, TgENR was co-crystallized in the presence of compound 19 and NAD+. The structure reveals that compound 19 binds in very similar mode to triclosan (Figure 5c).35–45 A structural comparison of the complex with that of the E. coli ENR/NAD+/triclosan enzyme reveals that the additional bulk of the alkyl substituent of compound 19 compared to triclosan does occupy the space made available by the replacement of a bulky hydrophobic residue of the E. coli enzyme Met by alanine in TgENR within the substrate binding pocket. Moreover, the structure shows that n-propyl is not the most optimal group as there is still some additional space at the base of the pocket which may be further utilized through further modifications. The only significant difference in side chain positions between the compound 19 and triclosan complex is a small change in the position of Phe243 to accommodate the larger bulk of compound 19 (Figure 5c). Presence of an n-propyl group on the 4-position clearly does not adversely affect the activity of these inhibitors (compare compounds 2 and 22), hence this side chain could be further modified to modulate both physicochemical properties and activity of these compounds. Moreover, despite triclosan and compound 19 showing similar solubility, as shown by their ClogP, ALOGpS, and tPSA values (Table 2), there is more scope for improvement through modifications of compound 19, (in particular the additional n-propyl group) when compared to triclosan. Since the ability to bind the triclosan analogues with the extension on the n-propyl group is only achieved within the parasitic ENR it may be possible to develop an inhibitor which is specific for the parasitic and not bacterial family. Although the addition of the n-propyl side chain maximizes packing interactions within the active site, these interactions are not fully exploited and could explain why the n-propyl group did not enhance activity. The additional n-propyl group does however produce a compound which is more tailored towards the apicomplexan and not bacterial ENR family. Moreover, further development of this inhibitor family is feasible particularly with the crystal structure showing there is still additional space at the base of the inhibitor binding pocket which could be further modified to improve binding.
Table 2.
 Triclosan |
 2 |
 19 |
 39 |
|
|---|---|---|---|---|
| ClogPa | 5.5 | 4.5 | 4.8 | 2.9 |
| ALOGpSb | −4.68 | −4.44 | −4.76 | −4.63 |
| tPSAc | 29.46 | 75.28 | 53.25 | 78.52 |
calculated with ChemDraw Ultra 7.0, CambridgeSoft;
Predicted water solubility calculated by using ALOGPS 2.1 (www.vcclab.org/lab/alogps/);
total Polar Surface Area calculated by using http://www.molinspiration.com/cgi-bin/properties.
Reference for vcclab
Tetko, I. V.; Gasteiger, J.; Todeschini, R.; Mauri, A.; Livingstone, D.; Ertl, P.; Palyulin, V. A.; Radchenko, E. V.; Zefirov, N. S.; Makarenko, A. S.; Tanchuk, V. Y.; Prokopenko, V. V. J. Comput. Aid. Mol. Des., 2005, 19, 453-63.
By maintaining the A ring of these compounds which is involved in packing interactions with the NAD+, and changing the B ring, it is clear that subtle variations in the MIC90 value can be achieved. Since these changes are small it may be that they affect the packing of the B ring within the active site or change its electrostatic properties without destabilizing the interaction with the A ring and NAD+ such that the mode of binding will be very similar to triclosan. Among these, inhibitor 19, bearing a 4-cyano group on the B ring seemed to be most promising with the least toxicity to host cells. Interestingly, compound 24 with a 4-amino functionality on the B ring also showed reasonable activity albeit lower than compound 19. This lower activity is caused through the removal of the chlorine and replacement of CN with NH2 on ring B, this again affects the properties of the B ring whilst allowing the A ring to stack onto the NAD+ cofactor such that the IC50 value is still comparable with triclosan. From a medicinal chemistry perspective, triclosan with two chlorine atoms at the 2′ and 4′ positions on the ring B presents little opportunity to incorporate any polar susbstitutions in order to reduce the lipophilicity of the drug. The rationale behind introduction of functionalities such as CN or NO2 on ring B is to create an opportunity to modify these groups in future rounds of ligand optimization to improve the physicochemical properties of these compounds. However, increased electron density on the A ring (a difference between compound 19 vs. 30, and 32) which enhances ability of compound 19 to π-stack with the nicotinamide ring of NAD+ did not improve activity.
As a final proof of concept for the promising activity of compounds 2 and 19, they were tested in an animal model for Toxoplasma infection with both protecting mice against parasite burden in vivo with an efficacy approximately 5 times higher than triclosan itself (Figure 4). In addition, a further study into the metabolism of these derivatives was performed. Both compounds 2 and 19 were tested for in vitro inhibition of recombinant CYP450 isozymes 2C9, 2D6 and 3A4. At 10μM, 2 and 19 showed strong inhibition of CYP2C9 (both >85%) and relatively low inhibition of CYP2D6 and CYP3A4 (both <22%). Further metabolic stabilities in pooled HLM indicated that they were moderately metabolized in HLM, and remaining parent drugs after incubation for 60 minutes were 51.23% and 45.14%, respectively. Intrinsic clearances and metabolic rates of 2 and 19 in HLM were slower than the control drug verapamil. Interestingly, when certain of these compounds recently were used to test their efficacy against Bacillus anthracis some were noted to be effective.21,22 The scavenging of fatty acids which abrogates the lethal effect of the elimination of Type 2 FAS with Staphlococus aureus in mice is not seen with T. gondii.46 Triclosan was noted to be safe in mice in earlier studies.10,47
Conclusions
In all a series of fifty-three compounds were tested for their activity against T. gondii, with six compounds having anti-parasite MIC90s ≤6μM without toxicity to host cells. Three compounds had IC90s <45nM against recombinant TgENR and of the 44 triclosan analogs tested, 6 showed better activity than triclosan. Moreover, those with the best toxicological profiles were tested in mice (compounds 2 and 19), and were found to be several times more active than triclosan itself in protecting mice. To investigate if, as predicted, the larger bulk of a subset of these compounds exploits the increase in space in the parasitic enzymes, a co-crystal structure of compound 19 in complex with TgENR, and NAD+ was solved. This structure clearly revealed that the compound’s aliphatic side chain does, as predicted, occupy space made available in the parasitic ENR family and reveals that further modifications may be possible. Moreover compound 19 with its adaptations on both the A and B ring is more amenable to chemical modifications which could improve both potency and solubility.
This work provides a foundation for discovery of medicines to treat toxoplasmosis by targeting ENR and for a program which includes a structure based program to develop new medicines against ENR. This work has produced new insights into the efficacy of a range of inhibitor scaffolds. The co-crystal structures proved useful in understanding structure activity relationships. In particular the co-crystal structure of compound 19 has revealed that a subset of these compounds exploit the increase in space within the parasitic ENR family which these inhibitors target. This strategy can now be utilized in future medicinal chemistry efforts to develop compounds that inhibit TgENR with increased avidity along with modifications which will increase bioavailability.
Experimental Section
Chemistry
Synthesis of Inhibitors
1H NMR and 13C NMR spectra were recorded on Bruker spectrometer with TMS as an internal standard. Standard abbreviation indicating multiplicity was used as follows: s = singlet, d = doublet, t = triplet, q = quadruplet, quin = quintuplet, m = multiplet and br = broad. HRMS experiment was performed on Q–TOF–2TM (Micromass). The progress of all reactions was monitored by TLC on precoated silica gel plates (Merck Silica Gel 60 F254). Preparative TLC was performed with Analtech 1000–mm silica gel GF plates. Column chromatography was performed using Merck silica gel (40–60 mesh).
Compounds triclosan, 5, 36, 49, 54 and 37 were commercially available and have been purchased. Compounds 50 and 51 were synthesized according to the reported procedures.23 The new diarylether analogs of triclosan were synthesized by Schemes 1 and 2.21 The synthesis of aminopyridine compounds 53 and 52, as well as benzimidazole-2-one derivatives 46, 47, and 48 were carried out according to the Scheme 3. All the compounds not illustrated in the synthetic schemes were reported earlier.21,22 Descriptions of synthetic experimental details and analytical data of compounds in Figure 1 is as follows:
General Methods
Method A
The aryl halide (1 mmol), phenol (1 mmol), and K2CO3 or Cs2CO3 (2–4 mmol) in DMSO (1.5 mL) were heated to 100 °C under nitrogen until the reaction was shown to be complete by TLC (8–12 h). After cooling to rt, the reaction mixture was diluted with ethyl acetate, and washed with 5% aqueous NaOH solution. The aqueous layer was further extracted with ethyl acetate, and the combined organic layers were washed with brine. The organic layer was dried over Na2SO4, and concentrated under vacuum to give the crude product, which was subsequently purified by flash chromatography on silica gel.
Method B
To the phenol (1 mmol) dissolved in DMF (1.75 mL) was added KOtBu (1.1 mmol) in one portion and the mixture was heated at 45 °C under mild vacuum. After 2 h, the reaction mixture was cooled to rt. Aryl halide (1 mmol) and (CuOTf)2. Ph (0.05 mmol) were added and the mixture was heated to reflux at 145 °C for 16–20 h. After cooling to rt, the reaction mixture was diluted with ethyl acetate, filtered over Celite, and extracted with ethyl acetate. The extract was washed with 5% aqueous NaOH solution. The aqueous layer was then extracted with ethyl acetate, and the combined organic layers were washed with brine. The organic layer was dried over Na2SO4, and concentrated under vacuum to give the crude product, which was subsequently purified by flash chromatography on silica gel.
Method C
In each case, the methoxy diarylether derivatives obtained by the above methods were converted to the corresponding phenols using boron tribromide according to the following procedure: A solution of boron tribromide (2–8 mmol of 1.0 M solution in dichloromethane) was added to a solution of the corresponding methoxy diphenylether analog (1 mmol) in dry dichloromethane (4 mL) maintained at −78 °C under nitrogen. The reaction mixture was stirred at the same temperature (1 h), and then at rt (3–8 h) while monitoring by TLC. Following completion, the reaction was quenched with methanol at −78 °C and concentrated under vacuum. The concentrate was further dissolved in ethyl acetate, washed with 10% aqueous sodium bicarbonate solution, and the organic layer was separated and washed with water and then with brine. The aqueous layer was extracted twice with ethyl acetate. The combined organic layers were then dried over Na2SO4, concentrated under vacuum, and the crude product was purified by flash chromatography on silica gel.
2-(3-Chlorophenoxy)-5-propylphenol (33)
To the 2-methoxy-4-propyl-phenol (1 g, 6.0 mmol) dissolved in DMF (11 mL) was added KOtBu (0.81 g, 7.2 mmol) in one portion and the mixture was heated at 45 °C under vacuum. After 2 h, the reaction mixture was cooled to room temperature. 1-chloro-3-iodobenzene (1.72 g, 7.2 mmol), and (CuOTf)2. Ph (0.31 g, 0.6 mmol) were added, and the mixture was heated to reflux at 145 °C for 18 h (Method B above). Usual workup and purification by flash chromatography (SiO2, 10% EtOAc/Hexanes) gave analytically pure 1-(3-chlorophenoxy)-2-methoxy-4-propylbenzene as a colorless oil (62%). 1H NMR (400 MHz, CDCl3): δ = 1.00 (t, J = 7.3 Hz, 3H), 1.70 (quin, J = 7.5 Hz, 2H), 2.62 (t, J = 7.5 Hz, 2H), 3.83 (s, 3H), 6.79 (dd, J = 8.0, 1.7 Hz, 1H), 6.85–6.83 (m, 2H), 6.91 (t, J = 2.1 Hz, 1H), 6.95 (d, J = 8.0 Hz, 1H), 7.00 (dd, J = 7.9, 1.0 Hz, 1H), 7.21 ppm (t, J = 8.1 Hz, 2H); 13C NMR (100 MHz, CDCl3): δ = 13.5, 24.3, 37.6, 55.5, 112.8, 114.3, 116.3, 120.6, 121.4, 121.7, 129.8, 134.4, 140.3, 141.2, 151.0, 159.0 ppm.
A mixture of above intermediate 1-(3-chlorophenoxy)-2-methoxy-4-propylbenzene (0.22 g, 0.79 mmol) and BBr3 (1M, 2.5 mL, 2.5 mmol) were subjected to the general demethylation procedure outlined in method C above. The crude product was purified by flash chromatography (SiO2, 10% EtOAc/Hexanes) to give analytically pure 33 as a colorless oil (62%). 1H NMR (400 MHz, CDCl3): δ = 0.99 (t, J = 7.3 Hz, 3H), 1.67 (quin, J = 7.5 Hz, 2H), 2.58 (t, J = 7.5 Hz, 2H), 5.43 (br s, 1H), 6.72 (d, J = 8.1 Hz, 2H), 6.86 (d, J = 8.2 Hz, 2H), 6.92–6.89 (m, 2H), 7.02 (s, 1H), 7.09 (d, J = 8.0 Hz, 1H), 7.26 ppm (t, J = 8.1 Hz, 1H); 13C NMR (100 MHz, CDCl3): δ = 13.5, 24.1, 37.2, 115.1, 116.2, 117.3, 119.2, 120.5, 122.9, 130.2, 134.8, 139.9, 140.3, 146.9, 157.8 ppm. HRMS (ESI-positive): calcd for C15H15ClO2 ([M–H]+): 261.06878, found: 261.0693.
3-Chloro-4-(2-hydroxy-6-methoxy-4-propylphenoxy)benzonitrile (30)
2,6-Dimethoxy-4-propylphenol (1.0 g, 5.1 mmol), 3-chloro-4-fluoro-benzonitrile (0.79 g, 5.1 mmol) and K2CO3 (1.41 g, 10.2 mmol) in DMSO (8 mL) were heated to 100 °C under nitrogen for 12 h (Method A above). Usual workup followed by purification of the crude reaction mixture by column chromatography (SiO2, 10% EtOAc/Hexanes) afforded the compound 3-chloro-4-(2,6-dimethoxy-4-propylphenoxy)benzonitrile (1.57 g, 93%). 1H NMR (400 MHz, CDCl3): δ = 1.00 (t, J = 7.3 Hz, 3H), 1.70 (quin, J = 7.5 Hz, 2H), 2.62 (t, J = 7.5 Hz, 2H), 3.78 (s, 6H), 6.49 (s, 2H), 6.62 (d, J = 8.6 Hz, 1H), 7.38 (dd, J = 8.6, 1.8 Hz, 1H), 7.73 ppm (d, J = 1.8 Hz, 1H); 13C NMR (100 MHz, CDCl3): δ = 13.4, 24.2, 38.2, 55.8, 105.0, 114.5, 117.6, 122.8, 128.3, 131.5, 133.4, 141.4, 151.9, 157.6 ppm. MS (ESI) m/z: 354.1 [M + Na]+
A mixture of above intermediate 3-chloro-4-(2,6-dimethoxy-4-propylphenoxy)benzonitrile (0.29 g, 0.86 mmol) and BBr3 (1M, 3.4 mL, 3.4 mmol) were subjected to the general demethylation procedure outlined in method C above. The reaction resulted in two products (30, 20% yield) and (26, 60% yield) that were separated by flash chromatography (SiO2, 15% EtOAc/Hexanes) to give analytically pure samplesas white solids.
3-Chloro-4-(2-hydroxy-6-methoxy-4-propylphenoxy)benzonitrile (30)
1H NMR (400 MHz, CDCl3): δ = 0.99 (t, J = 7.3 Hz, 3H), 1.68 (quin, J = 7.5 Hz, 2H), 2.57 (t, J = 7.5 Hz, 2H), 3.73 (s, 3H), 5.67 (s, 1H), 6.40 (s, 1H), 6.54 (s, 1H), 6.73 (d, J = 8.6 Hz, 1H), 7.40 (d, J = 8.6 Hz, 1H), 7.70 ppm (s, 1H); 13C NMR (100 MHz, CDCl3): δ = 13.4, 23.9, 37.8, 55.6, 104.4, 105.8, 108.9, 115.0, 117.2, 123.0, 126.8, 131.6, 133.5, 141.8, 148.3, 151.2, 156.8 ppm. HRMS (ESI-positive): calcd for C17H16ClNO3 ([M–H]+): 316.0746, found: 316.0755.
3-Chloro-4-(2,6-dihydroxy-4-propylphenoxy)benzonitrile (26)
1H NMR (400 MHz, CDCl3): δ = 0.98 (t, J = 7.3 Hz, 3H), 1.66 (quin, J = 7.5 Hz, 2H), 2.53 (t, J = 7.5 Hz, 2H), 5.04 (s, 2H), 6.47 (s, 2H), 6.85 (d, J = 8.6 Hz, 1H), 7.40 (d, J = 8.6 Hz, 1H), 7.77 ppm (d, J = 1.9 Hz, 1H); 13C NMR (100 MHz, CDCl3): δ = 13.4, 23.7, 37.4, 106.7, 108.8, 115.1, 117.0, 123.2, 126.0, 131.9, 133.8, 142.3, 147.7, 156.1 ppm. HRMS (ESI-positive): calcd for C16H14ClO3 ([M–H]+): 302.05894, found: 302.0606.
4-(2,6-Dihydroxy-4-propylphenoxy)benzonitrile (32)
2,6-Dimethoxy-4-propylphenol (0.63 g, 3.2 mmol), 4-fluoro-benzonitrile (0.39 g, 3.2 mmol) and Cs2CO3 (2.1 g, 6.4 mmol) in DMSO (9 mL) were reacted according to the Method A above. Usual workup followed by purification of the crude reaction mixture by column chromatography (SiO2, 10% EtOAc/Hexanes) afforded the compound 4-(2,6-dimethoxy-4-propylphenoxy)benzonitrile (0.81 g, 86%). A mixture of this intermediate compound (0.20 g, 0.67 mmol) and BBr3 (1M, 3.4 mL, 3.4 mmol) were subjected to the general demethylation procedure outlined in Method C above. The crude product was purified by flash chromatography (SiO2, 10% EtOAc/Hexanes) to give analytically pure 32 as a colorless oil (74%). 1H NMR (400 MHz, (CD3)2SO): δ = 0.90 (t, J = 7.2 Hz, 3H), 1.55 (quin, J = 7.2 Hz, 2H), 2.40 (t, J = 7.2 Hz, 2H), 6.27 (s, 2H), 6.91 (d, J = 8.8 Hz, 2H), 7.73 (d, J = 8.8 Hz, 2H), 9.41 ppm (s, 2H); 13C NMR (100 MHz, (CD3)2SO): δ = 14.1, 24.2, 37.6, 103.7, 107.9, 116.3, 119.6, 127.0, 134.4, 140.5, 150.7, 162.3 ppm. HRMS (ESI-positive): calcd for C16H15NO3 ([M–H]+): 268.09792, found: 268.0988.
3-Chloro-4-(2,6-dihydroxy-4-propylphenoxy)benzoic acid (34)
A mixture of the nitrile 3-chloro-4-(2,6-dimethoxy-4-propylphenoxy)benzonitrile (0.60 g, 1.8 mmol) whose synthesis is described in the preparation of 30 above and 25% aqueous NaOH (0.9 mL) in ethanol (5 mL) was refluxed with stirring (20 h). After cooling, the reaction mixture was acidified with dilute hydrochloric acid. The precipitate was filtered, dried, and purified by column chromatography (SiO2, 6% MeOH/CHCl3) to yield the 3-chloro-4-(2,6-dimethoxy-4-propylphenoxy)benzoic acid (0.51 g, 80%) as a white solid. 1H NMR (400 MHz, (CD3)2SO): δ = 0.94 (t, J = 7.2 Hz, 3H), 1.66 (quin, J = 7.6 Hz, 2H), 2.58 (t, J = 7.6 Hz, 2H), 3.38 (br s, 1H), 3.71 (s, 6H), 6.53 (d, J = 8.8 Hz, 1H), 6.67 (s, 2H), 7.76 (d, J = 8.8 Hz, 1H), 7.97 ppm (s, 1H); 13C NMR (100 MHz, (CD3)2SO): δ = 14.2, 24.5, 38.2, 56.4, 105.9, 114.1, 121.0, 125.4, 128.3, 130.3, 131.6, 141.7, 152.3, 157.4, 166.3 ppm. MS (ESI) m/z: 372.1 [M + Na]+ A mixture of this intermediate (0.40 g, 1.14 mmol) and BBr3 (1M, 6.8 mL, 6.8 mmol) were subjected to the general demethylation procedure outlined in Method C above. The crude product was purified by flash chromatography (SiO2, 5% MeOH/Chloroform) to give analytically pure 34 as a white solid (76%). 1H NMR (400 MHz, (CD3)2SO): δ = 0.91 (t, J = 7.2 Hz, 3H), 1.56 (quin, J = 7.2 Hz, 2H), 2.41 (t, J = 7.6 Hz, 2H), 6.28 (s, 2H), 6.61 (d, J = 8.4 Hz, 1H), 7.77 (dd, J = 8.6, 1.6 Hz, 1H), 7.95 (d, J = 2.0 Hz, 1H), 9.45 ppm (br s, 1H); 13C NMR (100 MHz, CDCl3): δ = 14.1, 24.2, 37.6, 107.9, 114.6, 121.3, 125.0, 127.2, 130.1, 131.4, 140.6, 150.6, 157.8, 166.4 ppm. HRMS (ESI-positive): calcd for C16H15ClO5 ([M–H]+): 321.0535, found: 321.0545.
3-(2-Hydroxy-4-propylphenoxy)phenyl boronic acid (20)
1-(3-chlorophenoxy)-2-methoxy-4-propylbenzene (0.83 g, 3.0 mmol), whose synthesis is described in the preparation of 33, was dissolved in 20 mL of anhydrous THF and treated with n-BuLi (1.6 M in hexanes, 3.8 mL, 6 mmol) at −78 °C for 2.5 h under Ar. Trimethyl borate (3.1 g, 30 mmol) was added drop wise to this solution at −78 °C and the reaction mixture was stirred for 3 h. The reaction was quenched by careful addition of 14 mL of 2N HCl at −78 °C and stirred at room temperature for 18 h. The solvent was evaporated and the concentrate was further dissolved in ethyl acetate, washed with 10% aqueous sodium bicarbonate solution, and the organic layer was separated and washed with water and brine. The aqueous layer was extracted twice with ethyl acetate. The combined organic layers were then dried over Na2SO4, concentrated under vacuum, and the crude product was purified by flash chromatography on silica gel (20% EtOAc/Hexanes) to result in 3-(2-methoxy-4-propylphenoxy)phenyl boronic acid (0.65 g, 76%) as a grey solid. 1H NMR (400 MHz, (CD3)2SO): δ = 0.91 (t, J = 7.3 Hz, 3H), 1.62 (quin, J = 7.4 Hz, 2H), 2.56 (t, J = 7.4 Hz, 2H), 3.35 (s, 3H), 6.36 (d, J = 8.2 Hz, 1H), 6.77 (d, J = 8.0 Hz, 1H), 6.93 (d, J = 8.0 Hz, 1H), 7.00–6.98 (m, 2H), 7.16 (t, J = 8.0 Hz, 1H), 8.38 ppm (s, 2H); 13C NMR (100 MHz, (CD3)2SO): δ = 13.7, 24.2, 37.2, 55.7, 112.6, 120.8, 121.7, 121.8, 128.5, 130.1, 135.7, 139.9, 142.0, 151.2, 160.6 ppm. A mixture of this boronic acid intermediate (0.34 g, 1.2 mmol) and BBr3 (1M, 6.0 mL, 6.0 mmol) were subjected to the general demethylation procedure outlined in Method C above. The crude product was purified by flash chromatography (SiO2, 2% MeOH/chloroform) to give analytically pure 20 as a grey solid (72%). 1H NMR (400 MHz, (CD3)2SO): δ = 0.89 (t, J = 7.1 Hz, 3H), 1.57 (quin, J = 7.2 Hz, 2H), 2.48 (t, J = 7.1 Hz, 2H), 6.65 (d, J = 7.4 Hz, 1H), 6.82–6.78 (m, 3H), 6.91 (d, J = 8.0 Hz, 1H), 7.03 (d, J = 7.6 Hz, 1H), 7.29 ppm (t, J = 8.1 Hz, 1H); 13C NMR (100 MHz, (CD3)2SO): δ = 13.7, 24.0, 36.9, 114.5, 115.6, 117.3, 119.7, 121.5, 122.2, 130.9, 133.7, 139.4, 140.4, 149.1, 159.4 ppm.
4-(2,6-Dihydroxy-4-propylphenoxy)benzamide (28)
A mixture of the (2,6-dimethoxy-4-propylphenoxy)benzonitrile (0.60 g, 1.8 mmol) whose synthesis is described in the preparation of 32 above, H2O2 (38%, 1.5 mL, 12.4 mmol) and 3N NaOH (0.3 mL) in ethanol (10 mL) was refluxed with stirring (20 h). After cooling, the reaction mixture was acidified with dilute hydrochloric acid. The precipitate was filtered, dried to obtain 4-(2,6-dimethoxy-4-propylphenoxy)benzamide as a white solid. A mixture of this intermediate compound (0.15 g, 0.48 mmol) and BBr3 (1M, 3.4 mL, 3.4 mmol) were subjected to the general demethylation procedure outlined in Method C above. The crude product was purified by flash chromatography (SiO2, 10% MeOH/chloroform). The product was recrystallized from MeOH/chloroform to give analytically pure 28 as white prisms (74%). 1H NMR (400 MHz, (CD3)2SO): δ = 0.91 (t, J = 7.2 Hz, 3H), 1.55 (quin, J = 7.2 Hz, 2H), 2.40 (t, J = 7.2 Hz, 2H), 6.26 (s, 2H), 6.78 (d, J = 8.4 Hz, 2H), 7.18 (br s, 1H), 7.80–7.78 (m, 3H), 9.28 ppm (s, 2H); 13C NMR (100 MHz, (CD3)2SO): δ = 14.2, 24.3, 37.6, 107.9, 114.6, 127.5, 129.5, 140.0, 150.9, 161.1, 167.9 ppm.
5-Propyl-2-(4-(2H-tetrazol-5-yl)phenoxy)-3-hydroxyphenol (35)
A mixture of the 4-(2,6-dimethoxy-4-propylphenoxy)benzamide (0.18 g, 0.6 mmol) (from above), SiCl4 (0.20 g, 1.2 mmol) and NaN3 (0.15 g, 2.3 mmol) was refluxed at 90 °C in CH3CN (0.5 mL) for 6 h under Ar. After cooling, the reaction mixture was diluted with EtOAc, washed with saturated sodium bicarbonate solution, the organic layer was separated and washed with water followed by brine. The aqueous layer was extracted twice with ethyl acetate. The combined organic layers were then dried over Na2SO4, concentrated under vacuum, and the crude product was purified by flash chromatography on silica gel (8% MeOH/chloroform) to result in 5-(4-(2,6-dimethoxy-4-propylphenoxy)phenyl)-2H-tetrazole (0.18 g, 97%) as a grey solid. 1H NMR (400 MHz, CDCl3): δ = 0.96 (t, J = 7.2 Hz, 3H), 1.66 (quin, J = 7.4 Hz, 2H), 2.57 (t, J = 7.6 Hz, 2H), 3.72 (s, 6H), 6.45 (s, 2H), 6.88 (d, J = 8.1 Hz, 2H), 8.01 (d, J = 8.2 Hz, 2H), 12.56 ppm (br s, 1H); 13C NMR (100 MHz, (CD3)2SO): δ = 13.9, 24.2, 37.9, 55.9, 105.5, 114.9, 121.1, 128.2, 128.5, 140.6, 152.4 ppm. A mixture of this intermediate compound (0.17 g, 0.50 mmol) and BBr3 (1M, 4.0 mL, 4.0 mmol) were subjected to the general demethylation procedure outlined in Method C above. The crude product was purified by flash chromatography (SiO2, 10% MeOH/chloroform) to give analytically pure 35 as off-white solid (78%). 1H NMR (400 MHz, (CD3)2SO): δ = 0.91 (t, J = 7.2 Hz, 3H), 1.56 (quin, J = 7.2 Hz, 2H), 2.40 (t, J = 7.6 Hz, 2H), 6.28 (s, 2H), 6.97 (d, J = 8.0 Hz, 2H), 7.94 (d, J = 8.4 Hz, 2H), 9.34 ppm (s, 2H); 13C NMR (100 MHz, (CD3)2SO): δ = 14.2, 24.3, 37.6, 107.9, 116.1, 127.4, 128.9, 140.1, 150.9, 161.0 ppm. HRMS (ESI-positive): calcd for C16H16N4O3 ([M+H]+): 313.1295, found: 313.1296.
Hexa-2,4-dienoic acid (4-(4-chloro-2-hydroxyphenoxy)-phenyl)amide (25)
A mixture of 4-(4-chloro-2-methoxyphenoxy)phenylamine21 (0.19 g, 0.8 mmol), EDAC·HCl (0.15 g, 0.8 mmol) and HOBt (0.1 g, 0.8 mmol) in 8 mL DMF was stirred for 10 minutes at rt under Ar. To this, a mixture of sorbic acid (0.09 g, 0.8 mmol) and triethylamine (0.08 g, 0.8 mmol) in 2 mL of DMF was added at rt and the reaction mixture was stirred at rt. After 16 h, the reaction mixture was diluted with EtOAc, washed with saturated sodium bicarbonate solution, the organic layer was separated and washed with water followed by brine. The aqueous layer was extracted twice with ethyl acetate. The combined organic layers were then dried over Na2SO4, concentrated under vacuum, and the crude product was purified by flash chromatography on silica gel (3% MeOH/chloroform) to result in hexa-2,4-dienoic acid (4-(4-chloro-2-methoxyphenoxy)-phenyl)amide (0.18 g, 67%) as a grey solid. 1H NMR (300 MHz, CDCl3): δ = 1.87 (d, J = 7.1 Hz, 3H), 3.85 (s, 3H), 5.89 (d, J = 19.7 Hz, 1H), 6.20 (dd, J = 19.7, 6.5 Hz, 2H), 7.03–6.89 (m, 5H), 7.42–7.32 (m, 2H), 7.53–7.50 ppm (m, 1H). A mixture of this intermediate compound (0.18 g, 0.50 mmol) and BBr3 (1M, 1.5 mL, 1.5 mmol) were subjected to the general demethylation procedure outlined in Method C above. The crude product was purified by flash chromatography (SiO2, 2% MeOH/chloroform) to give analytically pure 25 as a brown gel (76%). 1H NMR (300 MHz, CDCl3): δ = 1.88 (d, J = 6.9 Hz, 3H), 6.18–5.74 (m, 2H), 6.22–6.16 (m, 2H), 6.80–6.77 (m, 2H), 7.07–6.98 (m, 3H), 7.54 ppm (d, J = 11.2 Hz, 2H); 13C NMR (100 MHz, CD3OD): δ = 17.2, 116.7, 117.1, 117.2, 119.3, 121.0, 121.4, 121.5, 121.6, 129.0, 129.7, 137.9, 141.7, 149.7, 154.0 ppm. HRMS (ESI-positive): calcd for C18H16ClNO3 ([M+H]+): 330.0891, found: 330.0883.
Hexa-2,4-dienoic acid (4-(2,4-dichlorophenoxy)-3-hydroxyphenyl)amide (27)
A mixture of 2,4-dichlorophenol (1.0 g, 6.3 mmol), KOtBu (0.83 g, 7.5 mmol), 2-iodo-5-nitroanisole (1.9 g, 6.9 mmol), and (CuOTf)2 PhCH3 (0.16 g, 0.3 mmol) in DMF (11 mL) were reacted according to the Method B above. Usual workup and purification by flash chromatography (SiO2, 5% EtOAc/Hexanes) gave analytically pure 4-(2,4-dichlorophenoxy)-3-methoxynitrobenzene as a brown oil (62%). 1H NMR (400 MHz, (CD3)2SO): δ = 4.14 (s, 3H), 6.36 (d, J = 4.8 Hz, 1H), 7.13 (d, J = 5.2 Hz, 1H), 7.46 (dd, J = 8.8, 2.4 Hz, 1H), 7.61 (dd, J = 4.2, 2.4 Hz, 1H), 7.69 (d, J = 2.4 Hz, 1H), 8.09 (d, J = 8.4 Hz, 2H); 13C NMR (100 MHz, (CD3)2SO): δ = 57.5, 105.9, 108.7, 117.3, 117.6, 118.1, 122.2, 125.7, 129.5, 129.7, 130.7, 140.2, 150.2 ppm. A suspension of the above compound (2.0 g, 6.5 mmol), and 10% Pd/C (0.32 g) in EtOH (25 mL) was stirred under hydrogen atmosphere at 40 PSI at room temperature for 16 h. The catalyst was removed by filtration through a pad of Celite, and the residue was thoroughly washed with EtOAc. The solvent was evaporated and purification by flash chromatography (SiO2, 20% EtOAc/Hexanes) gave the compound 4-(2,4-dichlorophenoxy)-3-methoxyaniline (0.7 g, 38%). 1H NMR (400 MHz, CDCl3): δ = 3.48 (br s, 2H), 3.69 (s, 3H), 6.27 (dd, J = 4.2, 2.4 Hz, 1H), 6.36 (d, J = 2.4 Hz, 1H), 6.61 (d, J = 8.8 Hz, 1H), 6.85 (d, J = 8.4 Hz, 1H), 7.06 (dd, J = 4.4, 2.3 Hz, 1H), 7.42 ppm (d, J = 2.4 Hz, 1H); 13C NMR (100 MHz, CDCl3): δ 55.4, 100.2, 106.7, 116.2, 122.4, 122.9, 126.1, 127.1, 129.5, 144.7, 151.7, 153.2 ppm. A mixture of this aniline intermediate (0.15 g, 0.5 mmol), EDAC·HCl (0.10 g, 0.5 mmol) and HOBt (0.07 g, 0.5 mmol) in 6 mL DMF was stirred for 10 minutes at rt under Ar. To this, a mixture of sorbic acid (0.06 g, 0.5 mmol) and triethylamine (0.08 g, 0.8 mmol) in 2 mL of DMF was added at rt and the reaction mixture was stirred at rt. After 16 h, the reaction mixture was diluted with EtOAc, washed with saturated sodium bicarbonate solution, the organic layer was separated and washed with water followed by brine. The aqueous layer was extracted twice with ethyl acetate. The combined organic layers were then dried over Na2SO4, concentrated under vacuum, and the crude product was purified by flash chromatography on silica gel (3% MeOH/chloroform) to result in hexa-2,4-dienoic acid (4-(2,4-dichlorophenoxy)-3-methoxyphenyl)amide (0.18 g, 93%) as brown liquid. 1H NMR (400 MHz, (CD3)2SO): δ = 1.84 (d, J = 6.2 Hz, 3H), 3.34 (s, 3H), 6.35–6.09 (m, 3H), 6.64 (d, J = 8.6 Hz, 1H), 7.04 (d, J = 8.6 Hz, 1H), 7.28–7.15 (m, 3H), 7.65 (d, J = 8.6 Hz, 2H), 10.15 ppm (s, 1H); 13C NMR (100 MHz, (CD3)2SO): δ = 18.8, 56.0, 105.1, 112.0, 117.5, 122.2, 122.9, 123.2, 126.4, 128.7, 130.1, 130.3, 138.3, 138.5, 141.4, 151.2, 153.2, 164.4 ppm. MS (ESI) m/z: 402.1 [M + H]+ A mixture of this intermediate compound (0.12 g, 0.33 mmol) and BBr3 (1M, 1.6 mL, 1.6 mmol) were subjected to the general demethylation procedure outlined in Method C above. The crude product was purified by flash chromatography (SiO2, 3% MeOH/chloroform) to give analytically pure 27 as a brown oil (76%). 1H NMR (400 MHz, (CD3)2SO): δ = 1.83 (d, J = 6.2 Hz, 3H), 6.33–6.09 (m, 2H), 6.64 (d, J = 8.6 Hz, 1H), 6.96 (d, J = 8.6 Hz, 1H), 7.06 (d, J = 8.4 Hz, 1H), 7.19–7.13 (m, 1H), 7.28 (d, J = 8.6 Hz, 1H), 7.55 (s, 1H), 7.65 (s, 1H), 10.02 ppm (s, 1H); 13C NMR (100 MHz, (CD3)2SO): δ = 18.8, 108.7, 111.0, 117.5, 122.4, 122.8, 123.4, 126.1, 128.6, 129.9, 130.3, 137.4, 137.9, 138.2, 141.2, 149.4, 153.3, 164.2 ppm.
5-Chloro-2-phenoxy-N-pivaloyl aniline (13)
A suspension of commercially available 5-chloro-2-phenoxyaniline (0.13 g, 0.61 mmol) and Et3N (0.15 g, 1.5 mmol) in 2 mL methylene chloride at 0 °C was treated with pivaloyl chloride (0.07 g, 0.61 mmol), and the reaction mixture was stirred for 3h. The reaction mixture was diluted with ethyl acetate, extracted and the combined organic layers were washed with brine. Purification by flash chromatography (SiO2, 5% EtOAc/Hexanes) gave analytically product. 1H NMR (400 MHz, CD3OD): δ = 1.14 (s, 9H), 6.97 (t, J = 8.5 Hz, 3H), 7.12 (t, J = 7.4 Hz, 3H), 7.18 (dd, J = 4.4, 2.4 Hz, 3H), 7.35 ppm (t, J = 8.2 Hz, 3H); 13C NMR (100 MHz, CD3OD): δ = 25.7, 38.8, 99.6, 116.5, 120.3, 122.9, 123.6, 124.8, 128.4, 129.3, 130.4, 145.9, 156.4, 177.7 ppm. HRMS (ESI-positive): calcd for C17H18ClNO2 ([M+H]+): 304.1099, found: 304.1098.
2-Amino-N-(5-chloro-2-phenoxyphenyl)nicotinamide (7)
A mixture of 2-aminonicotinic acid (0.27 g, 1.9 mmol), EDAC·HCl (0.38 g, 1.9 mmol) and HOBt (0.26 g, 1.9 mmol) in 15 mL DMF was stirred for 10 minutes at rt under Ar. To this, a mixture of commercially available 5-chloro-2-phenoxyaniline (0.43 g, 1.9 mmol) and triethylamine (0.2 g, 1.9 mmol) in 5 mL of DMF was added at rt and the reaction mixture was stirred at rt. After 16 h, DMF was evaporated, the reaction mixture was diluted with EtOAc, washed with saturated sodium bicarbonate solution. The organic layer was separated and washed with water followed by brine. The aqueous layer was extracted twice with ethyl acetate. The combined organic layers were then dried over Na2SO4, concentrated under vacuum, and the crude product was purified by flash chromatography on silica gel (1% MeOH/chloroform) to result in 7. 1H NMR (400 MHz, CDCl3): δ = 6.46 (br s, 2H), 6.63–6.59 (m, 1H), 6.83 (d, J = 8.7 Hz, 1H), 7.08–7.02 (m, 3H), 7.21 (t, J = 7.4 Hz, 1H), 7.41 (t, J = 8.2 Hz, 2H), 7.59–7.57 (m, 1H), 8.21–8.13 (m, 1H), 8.40 (br s, 1H), 8.61 ppm (d, J = 2.2 Hz, 1H); 13C NMR (100 MHz, (CD3)2SO): δ = 109.7, 111.8, 118.6, 121.1, 124.0, 126.4, 126.5, 127.5, 130.4, 131.0, 137.7 ppm. HRMS (ESI-positive): calcd for C18H14ClN3O2 ([M+H]+): 340.0847, found: 340.0851.
5-chloro-2-(2,4-dichlorophenoxy)phenyl pivaloate (3)
To a mixture of 1 (0.26 g, 0.9 mmol) and DMAP (0.02 g, 0.2 mmol) in 8 mL of methylene chloride at 0 °C was added Et3N (0.10 g, 1.0 mmol) and stirred for 10 minutes. To this, pivaloyl chloride (0.12 g, 1.0 mmol) was added at 0 °C and the reaction mixture was stirred for 1h. The reaction mixture was stirred for an additional 2 h at rt. After evaporation of methylene chloride, the reaction mixture was diluted with ethyl acetate, washed with water followed by brine. The combined organic layers were extracted by ethyl acetate, dried over Na2SO4 and concentrated under vacuum. Purification by flash chromatography (SiO2, 3% EtOAc/Hexanes) gave analytically product 3 (85%). 1H NMR (400 MHz, CDCl3): δ = 1.25 (s, 9H), 6.81 (d, J = 8.8 Hz, 1H), 7.21–7.18 (m, 3H), 7.46 ppm (d, J = 2.4 Hz, 3H); 13C NMR (100 MHz, CDCl3): δ = 26.5, 38.7, 118.7, 120.5, 124.1, 124.8, 126.4, 127.6, 128.5, 129.3, 129.9, 142.2, 145.6, 150.9, 175.6 ppm.
3-(6-Aminopyridin-3-yl)-N-(4-methoxyphenyl)acrylamide (52)
A mixture of 3-(6-aminopyridin-3-yl)-acrylic acid23 (0.12 g, 0.7 mmol), EDAC·HCl (0.13 g, 0.7 mmol) and HOBt (0.10 g, 0.7 mmol) in 15 mL DMF was stirred for 10 minutes at rt under Ar. To this, a mixture of p-anisidine (0.09 g, 0.7 mmol) and triethylamine (0.07 g, 0.7 mmol) in 5 mL of DMF was added at rt and the reaction mixture was stirred at rt. After 16 h, DMF was evaporated, the reaction mixture was diluted with EtOAc, washed with saturated sodium bicarbonate solution. The organic layer was separated and washed with water followed by brine. The aqueous layer was extracted twice with ethyl acetate. The combined organic layers were then dried over Na2SO4, concentrated under vacuum, and the crude product was purified by flash chromatography on silica gel (5% MeOH/chloroform) to result in 52. 1H NMR (400 MHz, CD3OD): δ = 3.79 (s, 3H), 6.56 (d, J = 15.6 Hz, 1H), 6.62 (d, J = 15.6 Hz, 1H), 6.90 (d, J = 9.0 Hz, 1H), 7.50 (s, 1H), 7.55 (d, J = 9.1 Hz, 2H), 7.75 (dd, J = 4.4, 2.2 Hz, 1H), 8.08 ppm (d, J = 1.8 Hz, 1H); 13C NMR (100 MHz, CD3OD): δ = 54.1, 108.6, 113.2, 115.7, 119.7, 121.0, 131.3, 135.1, 137.6, 148.0, 156.1, 159.9, 164.9 ppm. HRMS (ESI-positive): calcd for C15H15N3O2 ([M+H]+): 270.1237, found: 270.1239.
N-(2-Aminobenzyl)-3-(6-aminopyridin-3-yl)acrylamide (53)
A mixture of 3-(6-aminopyridin-3-yl)-acrylic acid23 (0.13 g, 0.8 mmol), EDAC·HCl (0.15 g, 0.8 mmol), HOBt (0.10 g, 0.8 mmol), 2-aminobenzylamine (0.09 g, 0.8 mmol) and triethylamine (0.08 g, 0.8 mmol) in 15 mL DMF were reacted according to the procedure described above for 52. 1H NMR (400 MHz, CD3OD): δ = 4.42 (s, 2H), 6.41 (d, J = 15.6 Hz, 1H), 6.62 (d, J = 15.7 Hz, 1H), 6.60 (d, J = 8.8 Hz, 1H), 6.68 (t, J = 7.3 Hz, 1H), 6.75 (d, J = 7.9 Hz, 1H), 7.06 (t, J = 7.5 Hz, 1H), 7.11 (d, J = 7.4 Hz, 1H), 7.45 (d, J = 15.7 Hz, 1H), 7.72 (d, J = 8.6 Hz, 1H), 8.04 ppm (s, 1H); 13C NMR (100 MHz, CD3OD): δ = 39.4, 108.7, 115.4, 116.1, 117.2, 119.7, 122.2, 127.9, 129.3, 135.2, 137.2, 145.1, 147.7, 159.8, 167.1 ppm. HRMS (ESI-positive): calcd for C15H16N4O ([M+H]+): 269.1397, found: 269.1402.
1-(4-Nitrophenyl)-1,3-dihydrobenzimidazol-2-one (46)
1,3-Dihydrobenzimidazol-2-one (0.50 g, 3.7 mmol), 1-fluoro-4-nitrophenol (0.53 g, 3.7 mmol) and K2CO3 (1.00 g, 7.5 mmol) in DMSO (8 mL) were heated to 100 °C under nitrogen for 12 h (Method A above). The reaction mixture was cooled to rt and filtered. The yellow solid was repeatedly washed with water (3 × 5 mL), EtOAc (3 × 2 mL), ether (3 × 2 mL) and finally with hexanes (3 × 2 mL). The solid was dried in vacuum to obtain analytically pure 46 (0.70 g, 74%). 1H NMR (400 MHz, (CD3)2SO): δ = 6.92 (s, 2H), 7.26 (br s, 1H), 7.34 (br s, 1H), 8.00 (d, J = 8.5 Hz, 2H), 8.48 ppm (d, J = 8.7 Hz, 1H).
1-(4-Nitrobenzoyl)-1,3-dihydrobenzimidazol-2-one (48)
To a mixture of 1,3-dihydrobenzimidazol-2-one (0.51 g, 3.8 mmol) in 9 mL of anhydrous pyridine at 0 °C was added 4-nitrobenzoyl chloride (1.00 g, 5.6 mmol) portion wise and the reaction mixture was stirred for 15 minutes. The contents were then refluxed at 80 °C for 4 h. The reaction mixture solidified when cooled to rt, to which, 10 mL water and 20 mL of EtOAc were added and stirred for 10 minutes. The organic layer was separated and washed with 1N HCl followed by water and brine. The aqueous layer was extracted twice with ethyl acetate. The combined organic layers were then dried over Na2SO4, concentrated under vacuum to result in 48 (1.50 g, 98%). 1H NMR (400 MHz, (CD3)2SO): δ = 6.91 (s, 2H), 8.16 (d, J = 8.5 Hz, 2H), 8.31 (d, J = 8.6 Hz, 1H), 10.59 ppm (s 1H); 13C NMR (100 MHz, CDCl3): δ = 108.9, 120.8, 124.1, 130.1, 131.1, 136.9, 150.0, 150.4, 155.7, 166.3 ppm.
1-(4-Nitrobenzyl)-1,3-dihydrobenzimidazol-2-one (45)
1,3-Dihydrobenzimidazol-2-one (0.20 g, 1.5 mmol), 4-nitrobenzyl bromide(NOTE: strong lachrymator) (0.32 g, 1.5 mmol) and K2CO3 (0.31 g, 2.2 mmol) in MeOH (6 mL) were refluxed at 100 °C under nitrogen for 4 h. The reaction mixture was cooled to rt and MeOH was evaporated, 10 mL water and 20 mL of EtOAc were added and stirred for 10 minutes. The organic layer was separated and washed with water followed by brine. The aqueous layer was extracted twice with ethyl acetate. The combined organic layers were then dried over Na2SO4, concentrated under vacuum and the crude product was purified by flash chromatography on silica gel (15% EtOAc/Hexanes) to result in 45. 1H NMR (400 MHz, (CD3)2SO): δ = 4.98 (s, 2H), 6.98 (d, J = 8.5 Hz, 2H), 7.15 (d, J = 8.5 Hz, 2H), 8.28 (d, J = 8.6 Hz, 2H), 11.38 ppm (s, 1H); 13C NMR (100 MHz, CDCl3): δ = 43.8, 108.4, 108.9, 109.5, 120.8, 121.1, 121.7, 122.0, 154.7 ppm. HRMS (ESI-positive): calcd for C14H11N3O3 ([M+H]+): 270.0873, found: 270.0875.
1-(4-Nitrobenzenesulfonyl)-1,3-dihydrobenzimidazol-2-one (47)
To a mixture of 1,3-dihydrobenzimidazol-2-one (0.50 g, 3.7 mmol) in 8 mL of methylene chloride and 2 mL of DMF at 0 °C was added Et3N (0.42 g, 4.1 mmol) and stirred for 10 minutes. To this, 4-nitrobenzenesulfonyl chloride (0.83 g, 3.73 mmol) was added at 0 °C and the reaction mixture was stirred for 15 minutes. The reaction mixture was stirred for an additional 2 h at rt. The reaction mixture was quenched by adding crushed ice. The precipitated yellow solid was repeatedly washed with water (3 × 5 mL), EtOAc (3 × 2 mL), ether (3 × 2 mL) and finally with hexanes (3 × 2 mL). The solid was dried in vacuum to obtain analytically pure 46 (0.70 g, 74%). 1H NMR (400 MHz, (CD3)2SO): δ = 6.91 (s, 2H), 7.85 (d, J = 8.4 Hz, 2H), 8.20 (d, J = 8.4 Hz, 2H), 10.56 ppm (s, 1H); 13C NMR (100 MHz, CDCl3): δ = 108.6, 120.5, 123.4, 127.0, 129.7, 155.3 ppm.
Biology
Testing of inhibitors in vitro against T. gondii Tachyzoites
Four-day old confluent cultures of human foreskin fibroblasts (HFF) were infected with 103 tachyzoites of RH strain of T. gondii and cultured for 1 hour to allow parasite invasion, inhibitors added and cells cultured for 3 days, supplemented with 3H uracil and incubated for a further day, whereupon uracil incorporation into cells and thus parasite growth were assessed by liquid scintillation counting.47 Lack of toxicity for mammalian host cells was demonstrated first by visual inspection of monolayers and in separate 3H thymidine incorporation assays using non-confluent cell monolayers.
Cloning, Sequencing, Overexpression, and Purification of TgENR
TgENR inhibitor assay
Activity of TgENR was assayed by monitoring consumption of NADH (ε340=6220M−1cm−1) with a scanning spectrophotometer. ENR enzymes catalyze the conversion of trans-2-acyl-ACP to acyl-ACP, however, they are typically assayed with a surrogate substrate, trans-2-butyryl-Coenzyme A (crotonyl-CoA).18 We used a reaction mixture which would contain 20nM TgENR, 100mM Na/K phosphate buffer pH7.5, 150mM NaCl, and 100μM NADH (Sigma) when diluted to a final volume of 50μL. Compounds dissolved in DMSO were added to this mixture (2% final concentration of DMSO) and reactions were initiated at 25°C by addition of 100μM crotonyl-CoA (Sigma). This assay had also previously been used to monitor TgENR inhibition by an octaarginine tagged triclosan compound over a 24-hour period. Hydrolysis of the octaarginine tag released triclosan, leading to decreasing IC50 values (11nM at 24 hours) over the time course of the experiment10. Nonlinear regression analysis was performed using GraphPad Prism software
Co-crystallization and structure determination of TgENR in complex with NAD+ and compound 19. Preparation of protein crystals
Crystals of TgENR in complex with NAD+ and compound 19 were grown using the hanging drop vapor diffusion technique at 290K and by screening around the conditions which had previously been shown to be appropriate for crystallizing the TgENR/NAD+/triclosan complex (0.1M Tris-HCL pH9.0 and 6% PEG 8,000).14 In order to co-crystallize the TgENR with compound 19 the poor solubility was overcome by dissolving the inhibitor in DMSO and then diluting with the appropriate buffer for co-crystallization experiments.
Data collection and image processing
Crystals of both the TgENR/NAD+/compound 19 complex were flash frozen in 20% glycerol and data were collected at 100K to 2.7Å at the Daresbury SRS. The data were subsequently processed using the DENZO/SCALEPACK package (Table 1).49 The structure of the complex was determined by molecular replacement using the program Amore and the TgENR/NAD+/triclosan structure as a search model from which the triclosan and NAD+ coordinates were omitted. A clear solution was obtained in Amore and the model was subjected to rigid-body refinement and was subsequently rebuilt and refined in an iterative process using REFMAC550 and WinCoot. Although omitted in the search model, clear and continuous density could be seen for both the NAD+ cofactor and compound 19 allowing for their unambiguous fitting. Data collection and model refinement statistics can be seen in Table 1.
Testing of inhibitors in vivo
Compounds effective in enzyme assays and then in vitro (IC50 <10μM) were tested in an acute parenteral infection.10 CD1 mice were infected intraperitoneally with 2000 T. gondii (RH strain, Type 1). This inoculation dose is capable of killing one hundred percent of mice by day 10 post-infection. Mice were administered inhibitors intraperitoneally at 10mg/kg doses. Protection was measured by determining parasite burdens in the peritoneal cavity by microscopy at day 5 post-infection and survival up to 5 days post infection.
ADMET assays
Cyp450 inhibition and human liver microsomal stability assays were performed as is standard.
Statistical Analyses
Sample size and number of experiments. There were 6 replicate samples per group for in vitro experiments and 6 mice for each experimental group. All experiments were performed with sufficient sample sizes to have an 80% power to detect differences at the 5% level of significance. Groups included untreated or controls. Results were compared using students T test, Chi square analysis or Fisher’s exact test.
Supplementary Material
Acknowledgments
Supported by NIAID U01 AI082180-01. We gratefully acknowledge support of this work by gifts from the Finley John Gubbins Samuel Special Needs Trust, R Blackfoot, R Thewind, A Akfortseven, S Gemma, S Jackson, AK Bump, S. Pettican, the Rooney Aldens, the Dominique Cornwell and Peter Mann Family Foundation, the Morel, Rosenstein, Kapnick and Kiewit families, Intervet/Schering Plough, Toxoplasmosis Research Institute, and The Research to Prevent Blindness Foundation. This work was also supported by the Johns Hopkins Malaria Research Institute and the Bloomberg Family Foundation (JLZ and STP). We thank J. McCammon and Dr. Marco Pieroni for their assistance in preparation of this manuscript.
Footnotes
Abbreviations: T. gondii and Tg, Toxoplasma gondii; ENR, enoyl reductase; COMT, catechol O-methyl transferase; HLM, Human liver microsomes.
The PDB ID for TgENR co-crystallized with compound 19 is 3NJ8.
Supporting Information Available
Table S1 contains HPLC purity analysis data of the tested TgENR inhibitors.
References
- 1.McLeod R, Boyer K, Karrison T, Kasza K, Swisher C, Roizen N, Jalbrzikowski J, Remington J, Heydemann P, Noble AG, Mets M, Holfels E, Withers S, Latkany P, Meier P. Outcome of treatment for congenital toxoplasmosis, 1981–2004: the National Collaborative Chicago-Based, Congenital Toxoplasmosis Study. Clin Infect Dis. 2006;42:1383–1394. doi: 10.1086/501360. [DOI] [PubMed] [Google Scholar]
- 2.Roizen N, Kasza K, Karrison T, Mets M, Noble AG, Boyer K, Swisher C, Meier P, Remington J, Jalbrzikowski J, McLeod R, Kipp M, Rabiah P, Chamot D, Estes R, Cezar S, Mack D, Pfiffner L, Stein M, Danis B, Patel D, Hopkins J, Holfels E, Stein L, Withers S, Cameron A, Perkins J, Heydemann P. Impact of visual impairment on measures of cognitive function for children with congenital toxoplasmosis: implications for compensatory intervention strategies. Pediatrics. 2006;118:e379–390. doi: 10.1542/peds.2005-1530. [DOI] [PubMed] [Google Scholar]
- 3.Glasner PD, Silveira C, Kruszon-Moran D, Martins MC, Burnier Junior M, Silveira S, Camargo ME, Nussenblatt RB, Kaslow RA, Belfort Junior R. An unusually high prevalence of ocular toxoplasmosis in southern Brazil. Am J Ophthalmol. 1992;114:136–144. doi: 10.1016/s0002-9394(14)73976-5. [DOI] [PubMed] [Google Scholar]
- 4.Roberts F, Mets MB, Ferguson DJ, O’Grady R, O’Grady C, Thulliez P, Brezin AP, McLeod R. Histopathological features of ocular toxoplasmosis in the fetus and infant. Arch Ophthalmol. 2001;119:51–58. [PubMed] [Google Scholar]
- 5.McLeod R, Khan AR, Noble GA, Latkany P, Jalbrzikowski J, Boyer K. Severe sulfadiazine hypersensitivity in a child with reactivated congenital toxoplasmic chorioretinitis. Pediatr Infect Dis J. 2006;25:270–272. doi: 10.1097/01.inf.0000202070.59190.9a. [DOI] [PubMed] [Google Scholar]
- 6.Zuther E, Johnson JJ, Haselkorn R, McLeod R, Gornicki P. Growth of Toxoplasma gondii is inhibited by aryloxyphenoxypropionate herbicides targeting acetyl-CoA carboxylase. Proc Natl Acad Sci U S A. 1999;96:13387–13392. doi: 10.1073/pnas.96.23.13387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McLeod R, Muench SP, Rafferty JB, Kyle DE, Mui EJ, Kirisits MJ, Mack DG, Roberts CW, Samuel BU, Lyons RE, Dorris M, Milhous WK, Rice DW. Triclosan inhibits the growth of Plasmodium falciparum and Toxoplasma gondii by inhibition of apicomplexan Fab I. Int J Parasitol. 2001;31:109–113. doi: 10.1016/s0020-7519(01)00111-4. [DOI] [PubMed] [Google Scholar]
- 8.Muench SP, Rafferty JB, McLeod R, Rice DW, Prigge ST. Expression, purification and crystallization of the Plasmodium falciparum enoyl reductase. Acta Crystallogr D Biol Crystallogr. 2003;59:1246–1248. doi: 10.1107/s0907444903008813. [DOI] [PubMed] [Google Scholar]
- 9.Roberts CW, McLeod R, Rice DW, Ginger M, Chance ML, Goad LJ. Fatty acid and sterol metabolism: potential antimicrobial targets in apicomplexan and trypanosomatid parasitic protozoa. Mol Biochem Parasitol. 2003;126:129–142. doi: 10.1016/s0166-6851(02)00280-3. [DOI] [PubMed] [Google Scholar]
- 10.Samuel BU, Hearn B, Mack D, Wender P, Rothbard J, Kirisits MJ, Mui E, Wernimont S, Roberts CW, Muench SP, Rice DW, Prigge ST, Law AB, McLeod R. Delivery of antimicrobials into parasites. Proc Natl Acad Sci U S A. 2003;100:14281–14286. doi: 10.1073/pnas.2436169100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferguson DJ, Henriquez FL, Kirisits MJ, Muench SP, Prigge ST, Rice DW, Roberts CW, McLeod RL. Maternal inheritance and stage-specific variation of the apicoplast in Toxoplasma gondii during development in the intermediate and definitive host. Eukaryot Cell. 2005;4:814–826. doi: 10.1128/EC.4.4.814-826.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McLeod R, Roberts CW. Apicomplexan parasites: fascinating mosaics provide possible, novel antimicrobial targets. Leadership Medica. 2006:1. [Google Scholar]
- 13.Ferguson DJ, Campbell SA, Henriquez FL, Phan L, Mui E, Richards TA, Muench SP, Allary M, Lu JZ, Prigge ST, Tomley F, Shirley MW, Rice DW, McLeod R, Roberts CW. Enzymes of type II fatty acid synthesis and apicoplast differentiation and division in Eimeria tenella. Int J Parasitol. 2007;37:33–51. doi: 10.1016/j.ijpara.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muench SP, Prigge ST, Zhu L, Kirisits MJ, Roberts CW, Wernimont S, McLeod R, Rice DW. Expression, purification and preliminary crystallographic analysis of the Toxoplasma gondii enoyl reductase. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006;62:604–606. doi: 10.1107/S1744309106018112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muench SP, Prigge ST, McLeod R, Rafferty JB, Kirisits MJ, Roberts CW, Mui EJ, Rice DW. Studies of Toxoplasma gondii and Plasmodium falciparum enoyl acyl carrier protein reductase and implications for the development of antiparasitic agents. Acta Crystallogr D Biol Crystallogr. 2007;63:328–338. doi: 10.1107/S0907444906053625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mazumdar J, EHW, Masek K, CAH, Striepen B. Apicoplast fatty acid synthesis is essential for organelle biogenesis and parasite survival in Toxoplasma gondii. Proc Natl Acad Sci U S A. 2006;103:13192–13197. doi: 10.1073/pnas.0603391103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baldock C, Rafferty JB, Sedelnikova SE, Baker PJ, Stuitje AR, Slabas AR, Hawkes TR, Rice DW. A mechanism of drug action revealed by structural studies of enoyl reductase. Science. 1996;274:2107–2110. doi: 10.1126/science.274.5295.2107. [DOI] [PubMed] [Google Scholar]
- 18.Surolia N, Surolia A. Triclosan offers protection against blood stages of malaria by inhibiting enoyl-ACP reductase of Plasmodium falciparum. Nat Med. 2001;7:167–173. doi: 10.1038/84612. [DOI] [PubMed] [Google Scholar]
- 19.Pidugu LS, Kapoor M, Surolia N, Surolia A, Suguna K. Structural basis for the variation in triclosan affinity to enoyl reductases. J Mol Biol. 2004;343:147–155. doi: 10.1016/j.jmb.2004.08.033. [DOI] [PubMed] [Google Scholar]
- 20.Perozzo R, Kuo M, Sidhu AS, Valiyaveettil JT, Bittman R, Jacobs WR, Jr, Fidock DA, Sacchettini JC. Structural elucidation of the specificity of the antibacterial agent triclosan for malarial enoyl acyl carrier protein reductase. J Biol Chem. 2002;277:13106–13114. doi: 10.1074/jbc.M112000200. [DOI] [PubMed] [Google Scholar]
- 21.Tipparaju SK, Mulhearn DC, Klein GM, Chen Y, Tapadar S, Bishop MH, Yang S, Chen J, Ghassemi M, Santarsiero BD, Cook JL, Johlfs M, Mesecar AD, Johnson ME, Kozikowski AP. Design and synthesis of aryl ether inhibitors of the Bacillus anthracis enoyl-ACP reductase. ChemMedChem. 2008;3:1250–1268. doi: 10.1002/cmdc.200800047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tipparaju SK, Joyasawal S, Forrester S, Mulhearn DC, Pegan S, Johnson ME, Mesecar AD, Kozikowski AP. Design and synthesis of 2-pyridones as novel inhibitors of the Bacillus anthracis enoyl-ACP reductase. Bioorg Med Chem Lett. 2008;18:3565–3569. doi: 10.1016/j.bmcl.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seefeld MA, Miller WH, Newlander KA, Burgess WJ, DeWolf WE, Jr, Elkins PA, Head MS, Jakas DR, Janson CA, Keller PM, Manley PJ, Moore TD, Payne DJ, Pearson S, Polizzi BJ, Qiu X, Rittenhouse SF, Uzinskas IN, Wallis NG, Huffman WF. Indole naphthyridinones as inhibitors of bacterial enoyl-ACP reductases FabI and FabK. J Med Chem. 2003;46:1627–1635. doi: 10.1021/jm0204035. [DOI] [PubMed] [Google Scholar]
- 24.White SW, Zheng J, Zhang YM, Rock The structural biology of type II fatty acid biosynthesis. Annu Rev Biochem. 2005;74:791–831. doi: 10.1146/annurev.biochem.74.082803.133524. [DOI] [PubMed] [Google Scholar]
- 25.Zhang YM, Lu YJ, Rock CO. The reductase steps of the type II fatty acid synthase as antimicrobial targets. Lipids. 2004;39:1055–1060. doi: 10.1007/s11745-004-1330-3. [DOI] [PubMed] [Google Scholar]
- 26.Rock CO, Jackowski S. Forty years of bacterial fatty acid synthesis. Biochem Biophys Res Commun. 2002;292:1155–1166. doi: 10.1006/bbrc.2001.2022. [DOI] [PubMed] [Google Scholar]
- 27.McMurry LM, Oethinger M, Levy SB. Triclosan targets lipid synthesis. Nature. 1998;394:531–532. doi: 10.1038/28970. [DOI] [PubMed] [Google Scholar]
- 28.Heath RJ, Rubin JR, Holland DR, Zhang E, Snow ME, Rock CO. Mechanism of triclosan inhibition of bacterial fatty acid synthesis. J Biol Chem. 1999;274:11110–11114. doi: 10.1074/jbc.274.16.11110. [DOI] [PubMed] [Google Scholar]
- 29.Escalada MG, Harwood JL, Maillard JY, Ochs D. Triclosan inhibition of fatty acid synthesis and its effect on growth of Escherichia coli and Pseudomonas aeruginosa. J Antimicrob Chemother. 2005;55:879–882. doi: 10.1093/jac/dki123. [DOI] [PubMed] [Google Scholar]
- 30.Heath RJ, White SW, Rock CO. Lipid biosynthesis as a target for antibacterial agents. Prog Lipid Res. 2001;40:467–497. doi: 10.1016/s0163-7827(01)00012-1. [DOI] [PubMed] [Google Scholar]
- 31.Freundlich JS, Anderson JW, Sarantakis D, Shieh HM, Yu M, Valderramos JC, Lucumi E, Kuo M, Jacobs WR, Jr, Fidock DA, Schiehser GA, Jacobus DP, Sacchettini JC. Synthesis, biological activity, and X-ray crystal structural analysis of diaryl ether inhibitors of malarial enoyl acyl carrier protein reductase. Part 1: 4′-substituted triclosan derivatives. Bioorg Med Chem Lett. 2005;15:5247–5252. doi: 10.1016/j.bmcl.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 32.Sivaraman S, Zwahlen J, Bell AF, Hedstrom L, Tonge PJ. Structure-activity studies of the inhibition of FabI, the enoyl reductase from Escherichia coli, by triclosan: kinetic analysis of mutant FabIs. Biochemistry (Mosc) 2003;42:4406–4413. doi: 10.1021/bi0300229. [DOI] [PubMed] [Google Scholar]
- 33.Sivaraman S, Sullivan TJ, Johnson F, Novichenok P, Cui G, Simmerling C, Tonge PJ. Inhibition of the bacterial enoyl reductase FabI by triclosan: a structure-reactivity analysis of FabI inhibition by triclosan analogues. J Med Chem. 2004;47:509–518. doi: 10.1021/jm030182i. [DOI] [PubMed] [Google Scholar]
- 34.Levy CW, Baldock C, Wallace AJ, Sedelnikova S, Viner RC, Clough JM, Stuitje AR, Slabas AR, Rice DW, Rafferty JB. A study of the structure-activity relationship for diazaborine inhibition of Escherichia coli enoyl-ACP reductase. J Mol Biol. 2001;309:171–180. doi: 10.1006/jmbi.2001.4643. [DOI] [PubMed] [Google Scholar]
- 35.Heerding DA, Chan G, DeWolf WE, Fosberry AP, Janson CA, Jaworski DD, McManus E, Miller WH, Moore TD, Payne DJ, Qiu X, Rittenhouse SF, Slater-Radosti C, Smith W, Takata DT, Vaidya KS, Yuan CC, Huffman WF. 1,4-Disubstituted imidazoles are potential antibacterial agents functioning as inhibitors of enoyl acyl carrier protein reductase (FabI) Bioorg Med Chem Lett. 2001;11:2061–2065. doi: 10.1016/s0960-894x(01)00404-8. [DOI] [PubMed] [Google Scholar]
- 36.Qiu X, Janson CA, Court RI, Smyth MG, Payne DJ, Abdel-Meguid SS. Molecular basis for triclosan activity involves a flipping loop in the active site. Protein Sci. 1999;8:2529–2532. doi: 10.1110/ps.8.11.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roujeinikova A, Sedelnikova S, de Boer GJ, Stuitje AR, Slabas AR, Rafferty JB, Rice DW. Inhibitor binding studies on enoyl reductase reveal conformational changes related to substrate recognition. J Biol Chem. 1999;274:30811–30817. doi: 10.1074/jbc.274.43.30811. [DOI] [PubMed] [Google Scholar]
- 38.Rozwarski DA, Vilcheze C, Sugantino M, Bittman R, Sacchettini JC. Crystal structure of the Mycobacterium tuberculosis enoyl-ACP reductase, InhA, in complex with NAD+ and a C16 fatty acyl substrate. J Biol Chem. 1999;274:15582–15589. doi: 10.1074/jbc.274.22.15582. [DOI] [PubMed] [Google Scholar]
- 39.Levy CW, Roujeinikova A, Sedelnikova S, Baker PJ, Stuitje AR, Slabas AR, Rice DW, Rafferty JB. Molecular basis of triclosan activity. Nature. 1999;398:383–384. doi: 10.1038/18803. [DOI] [PubMed] [Google Scholar]
- 40.Miller WH, Seefeld MA, Newlander KA, Uzinskas IN, Burgess WJ, Heerding DA, Yuan CC, Head MS, Payne DJ, Rittenhouse SF, Moore TD, Pearson SC, Berry V, DeWolf WE, Jr, Keller PM, Polizzi BJ, Qiu X, Janson CA, Huffman WF. Discovery of aminopyridine-based inhibitors of bacterial enoyl-ACP reductase (FabI) J Med Chem. 2002;45:3246–3256. doi: 10.1021/jm020050+. [DOI] [PubMed] [Google Scholar]
- 41.Payne DJ, Miller WH, Berry V, Brosky J, Burgess WJ, Chen E, DeWolf WE, Jr, Fosberry AP, Greenwood R, Head MS, Heerding DA, Janson CA, Jaworski DD, Keller PM, Manley PJ, Moore TD, Newlander KA, Pearson S, Polizzi BJ, Qiu X, Rittenhouse SF, Slater-Radosti C, Salyers KL, Seefeld MA, Smyth MG, Takata DT, Uzinskas IN, Vaidya K, Wallis NG, Winram SB, Yuan CC, Huffman WF. Discovery of a novel and potent class of FabI-directed antibacterial agents. Antimicrob Agents Chemother. 2002;46:3118–3124. doi: 10.1128/AAC.46.10.3118-3124.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kapoor M, Gopalakrishnapai J, Surolia N, Surolia A. Mutational analysis of the triclosan-binding region of enoyl-ACP (acyl-carrier protein) reductase from Plasmodium falciparum. Biochem J. 2004;381:735–741. doi: 10.1042/BJ20040302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rafferty JB, Simon JW, Baldock C, Artymiuk PJ, Baker PJ, Stuitje AR, Slabas AR, Rice DW. Common themes in redox chemistry emerge from the X-ray structure of oilseed rape (Brassica napus) enoyl acyl carrier protein reductase. Structure. 1995;3:927–938. doi: 10.1016/S0969-2126(01)00227-1. [DOI] [PubMed] [Google Scholar]
- 44.Baldock C, Rafferty JB, Stuitje AR, Slabas AR, Rice DW. The X-ray structure of Escherichia coli enoyl reductase with bound NAD+ at 2.1 A resolution. J Mol Biol. 1998;284:1529–1546. doi: 10.1006/jmbi.1998.2271. [DOI] [PubMed] [Google Scholar]
- 45.Kapoor M, Dar MJ, Surolia A, Surolia N. Kinetic determinants of the interaction of enoyl-ACP reductase from Plasmodium falciparum with its substrates and inhibitors. Biochem Biophys Res Commun. 2001;289:832–837. doi: 10.1006/bbrc.2001.6061. [DOI] [PubMed] [Google Scholar]
- 46.Brinster S, Lamberet G, Staels B, Trieu-Cuot P, Gruss A, Poyart C. Type II fatty acid synthesis is not a suitable antibiotic target for Gram-positive pathogens. Nature. 2009;458:83–86. doi: 10.1038/nature07772. [DOI] [PubMed] [Google Scholar]
- 47.Mui EJ, Jacobus D, Milhous WK, Schiehser G, Hsu H, Roberts CW, Kirisits MJ, McLeod R. Triazine Inhibits Toxoplasma gondii tachyzoites in vitro and in vivo. Antimicrob Agents Chemother. 2005;49:3463–3467. doi: 10.1128/AAC.49.8.3463-3467.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kapust RB, Waugh DS. Controlled intracellular processing of fusion proteins by TEV protease. Protein Expr Purif. 2000;19:312–318. doi: 10.1006/prep.2000.1251. [DOI] [PubMed] [Google Scholar]
- 49.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 50.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.