Abstract
CD40 is a member of the tumor necrosis factor receptor superfamily. The interaction between CD40 and CD40 ligand (CD154) activates NF-κB, Jun N-terminal kinase, and Janus kinase/signal transducers and activators of transcription pathways and promotes B cell growth, differentiation, and survival as well as IL-12 production in macrophages and dendritic cells. We demonstrate here the existence of multiple isoforms of CD40 mRNA generated by alternative splicing and show that their expression is regulated differentially in activated macrophages and dendritic cells. Pre-CD40 RNA is spliced preferentially out to signal-transducible CD40 mRNA in the early stage of activation; half of the CD40 mRNA is replaced by the signal-nontransducible CD40 mRNAs in the later stages (24 h). Using IL-12 p40 gene expression as a reporter for CD40 signaling, we show that three of the alternative isoforms can disable signaling through CD40. The major alternative isoform lacks the membrane-associated endodomain and seems to reduce the amount of the signal-transducible form available on the cell surface. It would seem, therefore, that CD40 expression is controlled by posttranscriptional and posttranslational regulation through alternative splicing. Modulation of isoform expression may provide a mechanism by which cells regulate their susceptibility to CD40L signaling.
CD40 is a member of the tumor necrosis factor receptor (TNFR) superfamily and is expressed on a wide range of cell types including B cells, macrophages, and dendritic cells (1, 2). The interaction between CD40 and CD40 ligand (CD40L, CD154) promotes B cell growth, differentiation, and survival and also regulates IL-12 production and the overall activation state of dendritic cells (1–7). Functional CD40 presumably acts as a trimer receptor and activates NF-κB, Jun N-terminal kinase, and Janus kinase/signal transducers and activators of transcription (JAK/STAT) pathways (8–12). The intracellular domain contains the binding regions for several signal-transducible molecules including TNFR-associated factors family, JAK3, and Ku (8–11). The involvement of CD40–CD40L interactions in disease [e.g., X-linked hyper IgM syndrome (13), atherosclerosis (14), Hodgkin's disease (15), and Alzheimer's disease (16)] are now well established, but despite the important role that CD40 plays in the immune system, its gene expression is understood poorly.
We have made the surprising observation that CD40 expression is controlled by posttranscriptional regulation through alternative splicing. We have identified five CD40 isoforms generated by alternative splicing in the region between exon 5 and exon 9. In the major alternative-isoform mRNA, the sequence encoded in exon 6 is spliced out. The translated product from this isoform mRNA lacks the membrane-associated endodomain and seems to reduce the amount of the signal-transducible form available on the cell surface. These results suggest that CD40 expression is controlled by posttranscriptional and also posttranslational regulation through alternative splicing.
Materials and Methods
Reverse Transcriptase (RT)–PCR and Population Assay.
RT-PCR was performed as described in the text and previously (17). The primer sequences used were as follows: IL-12 and hypoxanthine phosphoribosyltransferase (HPRT) as described in ref. 17; CD40-P1, CTGCCCAGTCGGCTTCTTCTC; CD40-P2, CCTGTGTGACAGGCTGACAC; CD40-P3, GGCAAGCTTCCCTGCATGGTGTCTTTGCCTC; CD40-P4, GGCAGATCTCAAACTTCAAAGGTC; CD40-P5, GAACGAGTCAGACTAATGTCATC; CD40-P6, GCCGTCGAGCCGCAGGGGGTAA; CD40-P7, ATGGTGATGAGGATGCCCATC; suppressor of cytokine signaling-1 (SOCS-1) sense, GCGCGCTCCTGGACGCCTGCGGC; SOCS-1 antisense, CCGCACGCGGCGCTGGCGCAGC; type II transgene-sense, TCTTCCATTTCAGGTGTCGTG; type II transgene-antisense, CAATGTATCTTATCATGTCTG; human CD40-sense (H1), GCGAAGCTTGGTCCTGCCGCCTGGTCTCAC; and human CD40-antisense (H2), CCTCCTGGGTGACCGGTTGGC. For population assay, PCR primers P3 and P4 were used. To separate products from type I and IV mRNAs, cDNAs were amplified by using P6 and 32P-labeled P5, and the products were analyzed by 6% sequencing gel.
Immunoblotting, Immunoprecipitation, and Flow Cytometry.
Golgi/membrane-rich fractions were prepared by freezing/thawing. Burst cells (500 × g) and Golgi/membranes (which released to supernatants; 100,000 × g) were recovered by centrifugation. These pellets were combined and resuspended in lysis buffer (50 mM Tris⋅HCl/300 mM NaCl/0.5% Triton X-100). To prepare total-cell lysate, harvested cells were resuspended directly in lysis buffer. After removal of the insoluble fraction, protein concentration was measured. Then 100-μg or 5-μg protein samples were analyzed by immunoblotting. Anti-CD40 rabbit polyclonal IgG (L-17, Santa Cruz Biotechnology) and goat polyclonal IgG (T-20, Santa Cruz Biotechnology) were used. For immunoblotting, culture supernatants were concentrated (20-fold) by Centricon YM-10 (Amicon). FITC-conjugated 3/23 antibody (PharMingen) was used for flow cytometry. For immunoprecipitation, in vitro-translated products, T-20 antibody, and protein G conjugated agarose (Santa Cruz Biotechnology) were used. In vitro translation was performed by using 5 μg poly(A)+ RNA and rabbit reticulocyte lysate system (Amersham Pharmacia).
Cell Culture and Transfection.
To prepare bone-marrow-derived dendritic cells (bmDC), bone marrow cells from CBA/Ca mice were cultured for 7 days with granulocyte macrophage colony-stimulating factor (5 ng/ml; ref. 18). A B cell enriched fraction was prepared from T cell-depleted CBA/Ca mice by passage of splenocytes over a Sephadex G-10 column (19). Peritoneal macrophages were obtained from CBA/Ca mice injected 5 days previously with thioglycollate.
Plasmid constructs for producing transfectants of the type I, II, III, and IV CD40 isoform (E-I to E-IV) were derived by cloning into an expression vector (pMTF) containing the elongation factor-1α promoter and a neomycin resistance gene. RAW 264 cells were transfected by using the resulting expression plasmids or the vector only (controls), and stable transfectants were selected by using G418 (1 mg/ml). For double transfectant C-I/E-II, type I cDNA was cloned into pcDNA3.1/Zeo (Invitrogen) and transfected into E-II transfectant, and stable lines were selected by using G418 and zeocin (0.5 mg/ml). For the double transfectants E-I/II-1 and E-I/II-2, type II cDNA was cloned into pcDNA3.1/Zeo and the resulting plasmid alone (for E-I/II-1) or with the type II/pMTF plasmid (for E-I/II-2) was transfected into the E-I transfectant. Stable transfectants were selected by using G418 and zeocin. Where appropriate, cells were activated by lipopolysaccharide (LPS; 20 μg/ml) or FGK-45 antibody (10 μg/ml) together with subactivating low concentration of LPS (10 ng/ml).
Determination of Human CD40 Gene Structure.
Human CD40 gene was amplified by using primers binding to the exon sequences. The amplified fragments were sequenced partially to determine exon–intron junction sequences.
Results
Identification of CD40 Isoforms Generated by Alternative Splicing.
We identified alternative CD40 isoforms in the course of analyzing expression levels of CD40 mRNA by RT-PCR. To distinguish PCR products from contaminating genomic DNA, the PCR primers (P1 and P2) were designed to bind to sequences encoded in exon 5 and exon 9 of mouse CD40 gene (Fig. 1A). To perform semiquantitative RT-PCR, the number of PCR cycles was kept low. To detect these products, Southern blot hybridization was performed. For RT-PCR, cDNAs were prepared by using an oligo-dT primer and cytoplasmic RNAs from the LPS-activated mouse B cell-line A20, the mouse macrophage cell lines J774 and RAW 264, the temperature-sensitive dendritic-cell line tsDC (20), and bmDC. Surprisingly, at least three different-sized PCR products between 0.27 and 0.46 kb were detected (Fig. 1B). Each fragment was isolated, cloned, and sequenced. Comparison of these sequences with CD40 cDNA (21) and genomic DNA (22) sequences shows that these PCR products were amplified from CD40 isoform mRNAs generated by alternative splicing (type I to type V). The published mouse CD40 (type I) cDNA was identified within the medium-sized PCR products (Fig. 1B, I + IV). The structures of CD40 isoform mRNAs generated by alternative splicing are shown in Fig. 1C.
Figure 1.
Identification and structures of CD40 isoform mRNAs. (A) CD40 mRNA comprises sequences included in exons 1–9 of this gene. The relative position of PCR primers used for RT-PCR and population assay are indicated by arrows (P1 to P7). Positions of the first ATG and the stop codon for translation of CD40 are indicated. (B) CD40 mRNA was analyzed by RT-PCR using P1 and P2 primers (as in A), and cytoplasmic RNAs from LPS-activated (18 to 24 h) cells. Three different-sized PCR products (III, I+IV, II+V) were detected by Southern blot hybridization using CD40 cDNA as a probe. (C) Structure of CD40 isoform (type I–V) mRNAs generated by alternative splicing. Positions of alternative splicing are indicated by thick solid lines. The alternative splice site in exon 8 (5 bp downstream of the 5′ end of the exon 8) is indicated by dotted lines. Position of stop codons for translation are indicated by arrow heads. (D) Population assay of CD40 isoform mRNAs. All PCR products amplified by using P3 and P4 primers (as in A) and cytoplasmic RNA from LPS-activated A20 cells were cloned without size fractionation. The number of each isoform is indicated with the percentage in brackets.
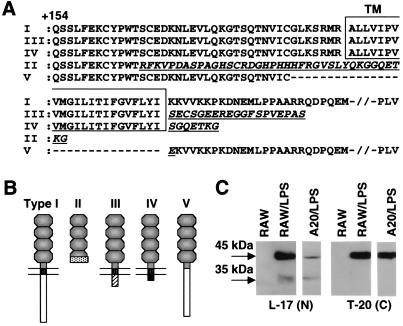
To exclude artifacts and identify any other isoforms, RT-PCR was performed by using a further set of primers that bind to sequences containing the first ATG encoded in the first exon and the 3′ untranslated region encoded in the last exon of the CD40 gene (Fig. 1A, P3 and P4) and cytoplasmic RNAs from LPS-activated A20 (Fig. 1D), RAW 264, and bmDC (Fig. 4C). Products were cloned without size fractionation and approximately 70 positive clones were selected randomly and sequenced (population assay). Additional isoforms lacking exons 2 or 3 also were identified, albeit at low abundance (Figs. 1D, Others and 4C, Others). Types I and II isoform mRNAs were the major species (Fig. 1D). The amino acid sequence between +1 (first Methionine) and +165 of all isoforms were identical. The amino acid sequences (from +154; Fig. 2A) and the proposed protein structures (Fig. 2B) are shown. Type I CD40 comprises an exodomain including four cysteine-rich repeats and a membrane-associated endodomain containing TNFR-associated factors (10, 11), JAK3 (8), and Ku (9) binding regions. Intracellular regions of type III and IV are replaced by distinct 19-aa (type III) and 7-aa (type IV) sequences, respectively (Fig. 2A). The type II isoform contains three cysteine-rich repeats, but the C-terminal 22-aa sequence in the fourth cysteine-rich repeat is replaced by a distinct 38-aa sequence (Fig. 2A). No signal-transducible endodomain would be present in this isoform. Type V isoform lacks the transmembrane region and the C-terminal 7-aa sequence in the fourth cysteine-rich repeat.
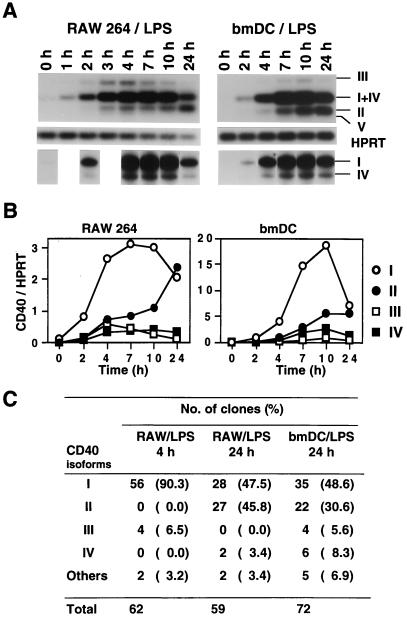
Figure 4.
Differential expression of CD40 isoform mRNAs. (A) CD40 isoforms type I–V (III, I+IV, II, and V) and HPRT were analyzed by RT-PCR using cytoplasmic RNAs from LPS-activated RAW 264 and bmDC. To separate PCR products from type I and IV (5 nt smaller than type I) RNAs, cDNAs were amplified by using P6 and 32P-labeled P5 primers (Fig. 1A). The PCR products then were analyzed on 6% sequence gel and are shown below the HPRT results. Cytoplasmic RNAs were isolated at different time points after LPS activation, as indicated. (B) RT-PCR shown in Fig. 4A was analyzed also by using a PhosphorImager. Intensity of type I (○), type II (●), type III (▫), and type IV (▪) bands are indicated. Type V was not detected in bmDC or RAW 264 cells (3 and 4 h LPS-activated RAW 264 cells might express very small amounts of type V). Population of type I and IV was calculated by using the results analyzed by sequence gel. Expression levels were normalized to HPRT levels. (C) Population assay (see text and Fig. 1D) of CD40 isoform mRNAs from indicated cells.
Figure 2.
Amino acid sequences and structures of CD40 isoforms. (A) Amino acid sequences of CD40 isoforms (I–V) from +154 are aligned. Transmembrane domains (TM) are boxed. Type II-, III-, IV-, and V-specific sequences are indicated by underlined, italicized letters. (B) Structures of CD40 isoforms presumed from amino acid sequences are indicated. (C) CD40 isoforms from indicated cells were analyzed by immunoblotting using antibodies binding to the N terminus (L-17, Santa Cruz Biotechnology) and C terminus (type I-specific, T-20, Santa Cruz Biotechnology).
The type II–IV isoforms are predicted to be 6- to 10-kDa smaller than type I (45 kDa). An approximately 35-kDa molecule was detected by immunoblotting by using an anti-CD40 antibody (L-17, binding to the N terminus) in the Golgi/membrane-rich fraction from LPS-activated RAW 264 and A20 cells but not by using a type I-specific antibody (T-20, binding to the C terminus; Fig. 2C).
Differential Expression of CD40 Isoform mRNAs.
To investigate the differential expression of CD40 isoforms, RNAs from different organs, peritoneal macrophages (Macrophage), a B cell-enriched fraction (B cell rich) from normal mice, and cytokine-treated macrophage cell lines and several transfectants were analyzed by RT-PCR (Fig. 3 A and B). Liver cells expressed mainly type I+IV CD40 mRNA. In lung, the type II mRNA was higher than that in other tissues. LPS-treated (activated) peritoneal macrophages expressed type I+IV, II, and V, but type III was minor. The B cell-enriched population expressed all isoform RNAs, although the type II population was increased slightly after activation for 72 h with IL-4 and the agonist CD40 antibody FGK-45 (23). Type V mRNA was not detected in bmDC (Fig. 4 A and C). None of a wide range of cytokines tested influenced differential gene expression in macrophage cell lines. However, a SOCS-1 (SSI-1 or JAB) transfectant of RAW 264, generated to investigate the impact of SOCS-1 on JAK/STAT signaling (24), failed to up-regulate CD40 mRNA expression on IFN-γ stimulation (Fig. 3B, IFN-γ) and selectively inhibited type II but not type I expression on LPS activation (Fig. 3B, LPS and LPS + IFN-γ).
Figure 3.
mRNAs of CD40 isoforms from indicated cells and organs were analyzed by RT-PCR using P1 and P2 primers (Fig. 1A). HPRT mRNA was analyzed also by RT-PCR as control. (A) RNA was isolated from organs and cells from normal mice. If required, cells were activated by phorbol 12-myristate 13-acetate, ConA, LPS, or IL-4 + anti-CD40 antibody FGK 45. (B) RNA was isolated from the SOCS-1 transfectant. If required, cells were activated by IFN-γ and LPS.
We found also that CD40 isoform mRNAs are regulated differentially in LPS-activated RAW 264 and bmDC. CD40 type I mRNA expression was up-regulated and reached maximum levels 7–10 h after LPS activation and then declined (Fig. 4 A and B). Type II mRNA expression, however, was maintained at low levels by 7 h after LPS activation and then up-regulated (Fig. 4A and B). It seems, therefore, that pre-CD40 RNA is spliced out preferentially to transducible CD40 type I mRNA at an early stage of LPS activation in RAW 264 and bmDC. Indeed, in 2-h activated cells, the type I/type II mRNA ratio was 5.7 (RAW 264) and 5.5 (bmDC). On the other hand, the type I/type II ratio declined to 0.9 (RAW 264) and 1.3 (bmDC) in 24-h activated cells. In these cells, half of the CD40 mRNA was replaced by the signal-nontransducible CD40 type II, III, and IV mRNAs through alternative splicing. Comparable data were obtained also when analysis was performed by using a population assay (Fig. 4C). More 24-h activated cells were not analyzed because of an increase of dead-cell population.
Alternative CD40 Isoforms Can Disable Signals Through CD40 Type I.
The type I CD40 isoform normally signals through its endodomain. What then might be the function of the type II, III, and IV isoforms that lack this domain? Because CD40 isoform expression in normal bmDC is similar to that of RAW 264 cells that have been used to define IL-12 p40 gene expression (25), we used IL-12 p40 mRNA expression as a reporter for CD40 signaling in the RAW 264 cells. Transfectants were generated by using type I, II, III, and IV cDNAs as transgenes. These transfectants expressed both endogenous and transfected CD40 isoforms. Negative controls and transfectants were activated with the agonist CD40 antibody FGK-45 (23), and the extent of activation was monitored by measurement of IL-12 p40 mRNA (Fig. 5A). Transfection with type I CD40 (E-I) resulted in enhanced expression of IL-12 p40 mRNA, whereas transfection of type II (E-II), III (E-III), and IV (E-IV) resulted in down-regulation of IL-12 p40 mRNA. As a positive control for IL-12 p40 induction, cells were activated with LPS (Fig. 5A), and IL-12 p40 mRNA was detected. Surprisingly, IL-12 p40 mRNA levels were increased in the type I transfectant and reduced in type II, III, and IV transfectants. This result suggests that type I CD40 is required also for high-level IL-12 p40 mRNA expression induced by LPS. Because CD40L also is expressed in the RAW 264 cells (data not shown) and DC (26), it may be that LPS promotes CD40L interactions with CD40 on the same cell.
Figure 5.
Function of CD40 isoforms. (A) RAW 264 stable transfectants were generated by using type I, II, III, and IV CD40 isoform expression plasmids carrying the cDNAs downstream of the elongation factor (EF)-1α promoter. Negative control cells were generated by transfection using vector only. Control (Con), type I (E-I), type II (E-II), type III (E-III), and type IV (E-IV) transfectants were activated by agonist CD40 antibody FGK-45 or LPS. IL-12 p40 and HPRT (as control) mRNAs were analyzed by RT-PCR using cytoplasmic RNAs from indicated transfectants. (B) Expression levels of endogenous CD40 mRNA were analyzed by RT-PCR using P1 and P2 (endogenous CD40-specific) primers (Fig. 1A) and cytoplasmic RNAs from nonactivated (N) and LPS-activated (L) control cells (Con) and type II (E-II) transfectants. (C) Expression levels of CD40, IL-12 p40, and HPRT mRNAs were analyzed by RT-PCR using cytoplasmic RNAs from negative control (Con), type I (C-I), and type II (E-II) transfectants and a type I+II (C-I/E-II) double transfectant. Expression of type I mRNA was under-controlled by the cytomegalovirus promoter in C-I and C-I/E-II, and expression of type II mRNA was under-controlled by the EF-1α promoter in E-II and C-1/E-II. Endogenous (Endo.) plus transfected (T) type I and endogenous type II mRNAs were amplified by using P1 and P2 primers (Fig. 1A). Transfected type II mRNA was amplified by using primers binding to 5′ and 3′ untranslated regions encoded in the expression vector. (D) Protein (100 μg) in membrane-rich fractions from nonactivated (N) and LPS-activated (L) control (Con) cells and C-I/E-II cells were analyzed by immunoblotting using L-17 (binding to N terminus of CD40) and T-20 (binding to C terminus of CD40, type I-specific) antibodies. (E) Nonactivated control cells (Con/Non, dotted line), LPS-activated control cells (Con/LPS, solid line), and LPS-activated C-I/E-II cells (C-I/E-II/LPS, filled with gray) were analyzed by flow cytometry using anti-CD40 antibody (FITC-conjugated 3/23, PharMingen).
Although type III and IV isoforms lack the signaling endodomain of CD40, they contain a transmembrane region. Therefore, these isoforms can and are expressed on the cell surface (confirmed by flow cytometry, data not shown). It is conceivable that type III and IV isoforms might operate as dominant negative inhibitors by competing for CD40L. Alternatively, they might prevent proper receptor function by production of disabled hetero-oligomers at the cell surface as the Neu/erbB does for the epidermal growth-factor receptor (27).
Reduction of the Signal-Transducible Type I CD40 on the Cell Surface by Expression of Type II Isoform.
Although type II can disable signaling through type I CD40, this isoform lacks the transmembrane domain as well as the cytoplasmic region. Indeed, no CD40 type II was detected on the transfectant cell surface by flow cytometry (a small amount of type II protein was detected only in the highly concentrated culture supernatant with degraded type II molecules, data not shown). How might this isoform disable signaling through type I CD40? We investigated further the effects of transfected type II CD40 on expression of endogenous CD40 mRNA (Fig. 5B). As the transfected type II mRNA did not contain the primer P2-binding region (Fig. 1A), we could measure the endogenous CD40 mRNA products selectively by using P1 and P2 primers. Although LPS activation increased endogenous CD40 mRNA in control cells, it failed to do so in type II transfectants (Fig. 5B).
Given that endogenous type I CD40 was not up-regulated in the type II transfectant, we investigated the effects of the type II isoform on type I by generating a double transfectant (C-I/E-II) using the type II transfectant (E-II, Fig. 5A) as parent and type I cDNA as transgene. LPS activation of control cells and type I transfectants (Fig. 5C, C-I) resulted in significant up-regulation of IL-12 p40 mRNA. This up-regulation was not observed in the double transfectants (Fig. 5C ,C-I/E-II) even though they expressed large amounts of type I CD40 mRNA. This result suggests that the type II isoform inhibits the up-regulation of CD40 and consequently IL-12 p40 expression.
Functional CD40 may need to act as a trimer. Type II CD40 might interact with type I and regulate CD40 signaling. We therefore analyzed CD40 protein expression. Although the amount of the type I CD40 mRNA was greater in the LPS-activated transfectants (C-I/E-II) than in LPS-activated control cells (Fig. 5C, Con), the level of serologically detectable CD40 protein in the membrane-rich fraction from these transfectants was much lower (Fig. 5D). This result was consistent with flow-cytometry data (Fig. 5E). Type II protein seems to reduce signal-transducible type I on the cell surface.
To study the effect of the type II isoform at levels normally expressed in the cells examined, we made two additional double transfectants expressing type II mRNA [15% (E-I/II-1) and 40% (E-I/II-2) relative to the type I mRNA] using the type I transfectant (Fig. 5A, E-I) as the parent. All transfectants expressed comparable levels of type I mRNA (Fig. 6 A and B). However, the level of type I protein compared with control cells (E-I/Con) was reduced to 75% in E-I/II-1 and to 22% in the E-I/II-2 (Fig. 6C, Non). In LPS-activated cells, it was reduced to 45 and 16%, respectively (Fig. 6C, LPS). The level of expression of type I protein therefore seems to be inversely related to expression of type II mRNA.
Figure 6.
Reduction of the amount of signal-transducible type I CD40 by expression of type II isoform. (A) mRNA levels of CD40 type I and II were analyzed by RT-PCR using primers P1 and P7 (Fig. 1A) and cytoplasmic RNAs from control cells (Con; transfected by using vector only), E-I/Con, E-I/II-1, and E-I/II-2. These transfectants were generated by using E-I (Fig. 5A) as a parent and the type II expression plasmids or the vector only. Low level type II mRNA expression was shown by using a longer-exposed film (L). (B) RT-PCR shown in A was analyzed also by using a PhosphorImager. Intensity of CD40 type I and II bands were normalized to HPRT levels and compared with that of type I in E-I (100%). (C) Proteins (5 μg) in membrane-rich fraction from nonactivated (Non) and LPS-activated (LPS) indicated cells were analyzed by immunoblotting using T-20 antibody (type I-specific). The intensities of these bands were compared with those of type I in nonactivated or LPS-activated E-I cells (shown below the blots). (D) In vitro translation was performed by using [35S]Cys and 5 μg poly(A)+ RNA from the indicated cells or in vitro-synthesized CD40 RNA from Sp6 promoter (CD40 RNA). Immunoprecipitation was performed by using type I-specific T-20 antibody. Type I is indicated by an arrow. (E) Type I protein was detected in total-cell lysates (Total) and culture supernatants (Sup) from indicated cells by immunoblotting using T-20 antibody. The 45- and 27-kDa molecules were detected in type I transfectants. The 27-kDa molecules in total lysates from indicated cells (27 kDa) were detected by immunoblotting using T-20.
To investigate whether the reduction of type I is controlled by posttranslational regulation, we quantitated the amount of immunoprecipitable CD40 type I protein generated by an in vitro-translated system using poly(A)+ RNAs from the type I transfectant (E-I/Con) and type I+II transfectant (E-I/II-2). We observed similar amounts of type I protein in both (the normalized ratio of type I in E-I/Con to type I in E-I/II-2 was 1.01; Fig. 6D). This result suggests that any variation in the level of type I must be controlled by posttranslational regulation. Also, we could not detect accumulation of type I in the supernatant or total-cell lysate from E-I/II-2 cells (Fig. 6E), excluding all but the remaining explanation that type I in E-I/II-2 may be degraded rapidly. We sought evidence of such degradation and have found a 27-kDa fragment of CD40 degraded protein by using T-20 antibody (binding to C terminus) in both transfectants (Fig. 6E). Because this molecule can be detected in independent type I transfectants including a HeLa cell transfectant, it cannot be an artifact from recombination of the CD40 transgene. The 27-kDa molecule seems to contain a transmembrane domain and lack the 18-kDa of the N terminus region. The cleavage site seems to be located in the exodomain. Although this level of 27-kDa protein in E-I/Con was slightly greater than that in E-I/II-2, the 27-kDa/45-kDa ratio was 0.74 in E-I/Con but 1.08 in E-I/II-2 (Fig. 6E). This observation suggests that the 27-kDa molecule is produced more efficiently in E-I/II-2. We therefore conclude that degradation to the 27-kDa molecule by cleavage of type I may be enhanced by type II.
Identification of Human Type II CD40.
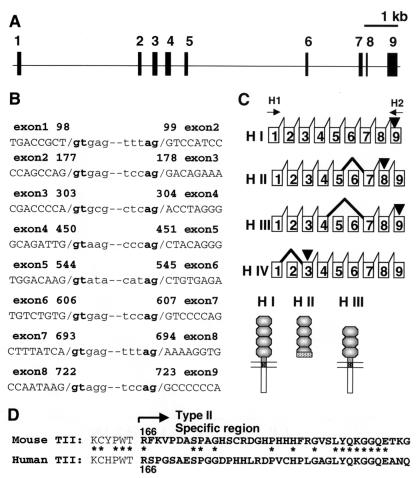
We have identified the major type II CD40 isoform in the mouse. Here we show that this CD40 isoform is conserved also in humans. We have determined the gene structure of human CD40 as well as the exon–intron junction sequences (Fig. 7 A and B). Human CD40 isoform mRNA was analyzed by RT-PCR using primers H1 and H2 (Fig. 7C) and RNA from human B cell line Raji cells. All amplified fragments were cloned, and DNA sequences of 62 randomly selected clones were determined. Of the 62, 24 clones carried type I CD40 cDNA. We also found human type II isoform (13 clones) that was also a major isoform in Raji cells. Additional isoform mRNAs lacking exon 2 (H-IV, 9 clones) and exon 5 + 6 (H-III, 6 clones) also were identified (Fig. 7C). H-III isoform lacks the fourth cysteine-rich region. Only a 21-aa peptide from the first Methionine would be translated from H-IV isoform mRNA. Partial amino acid sequences of human and mouse type II are aligned in Fig. 7D. Type II CD40 isoforms seem to be conserved in evolutionary history.
Figure 7.
Identification of human CD40 type II. (A) Gene structure of human CD40. Exons are indicated by boxes. (B)Exon–intron junction sequences are indicated with the nucleotide number of human CD40 cDNA. (C) Structure of human CD40 isoform RNA. Position of alternative splicing and stop codons are indicated by thick solid lines and arrow heads, respectively. Predicted protein structures also are indicated. (D) Partial amino acid sequences of human and mouse CD40 type II are aligned. Type II-specific sequences start from 166 in both. Similarities are indicated by asterisks.
Discussion
We have identified CD40 isoforms generated by alternative splicing. The amount of signal-transducible CD40 (type I) mRNA is reduced to half in 24-h LPS-activated bmDC and RAW 264 cells. Furthermore, three of the isoforms can disable signals through signal-transducible CD40 type I. Isoforms generated by alternative splicing also are observed among the other TNFR superfamily members such as soluble CD95 (Fas, Apo-1; ref. 28) and LARD (29). Signals through other members of the TNFR family also may be regulated by alternative splicing.
We have found that type II mRNA was not produced in the RAW 264 transfectant constitutively expressing SOCS-1, which was identified originally as an inhibitor of the JAK/STAT pathway (24). We generated the SOCS-1 transfectant to investigate the involvement of the JAK/STAT pathway in CD40 expression. CD40 expression is up-regulated in IFN-γ stimulated RAW 264 cells but not in the SOCS-1 transfectant. SOCS-1 seems also to block the JAK/STAT signaling through the IFN-γ receptor. Interestingly, type II mRNA expression was blocked selectively in the LPS-activated SOCS-1 transfectant cells but not in LPS-activated nontransfectant cells. The mechanisms by which SOCS-1 could regulate alternative splicing is not clear, because its primary role is considered to be inhibition of STAT phosphorylation by binding to JAKs. Perhaps its impact on alternative splicing reflects additional functions for this molecule such as regulation of alternative splicing factor expression or regulation of alternative splicing by inhibiting phosphorylation of splicing factors such as SR proteins.
We have shown that type I protein levels are reduced by type II. How might the type II isoform do this? We propose that the type II isoform interacts with the type I to form a hetero-oligomer. This hetero-oligomer formation in turn may induce a structural change in the type I molecule to expose the cleavage site, consequently resulting in enhancement of type I degradation to the shorter 27-kDa molecule.
Alternatively, degradation of newly synthesized type I may be enhanced by type II. As type II CD40 lacks structurally important two Cysteine residues but contains one distinct Cysteine (Fig. 2A), very likely it would misfold and be degraded in the endoplasmic reticulum. Interaction between type I and II precursors may cause misfolding of CD40 and rapid degradation. More information is needed now to establish the mechanisms of degradation in more detail.
Based on our results, we suggest the following sequence of events for signaling through CD40: (i) nonactivated cells constitutively express low levels of CD40 mRNA, (ii) stimuli such as LPS up-regulate pre-CD40 mRNA, which is spliced out preferentially to the CD40 type I isoform, (iii) SOCS-1 may act at this level, which increases the level of signal-transducible CD40 on the cell surface and consequently enhances activation through the functional endodomain, and (iv) eventually, much of the pre-CD40 mRNA is spliced to type II mRNA, thus reducing the amount of signal through type I by posttranscriptional and posttranslational regulation.
We conclude that signaling through CD40 is regulated by negative feedback that depends on alternative splicing and its products. As the activation status of many antigen-presenting cells is determined through the NF-κB pathway, this form of regulation may provide a means by which antigen-presenting cells vary their activation thresholds thus determining the degree of responsiveness to antigens, be they self or foreign.
Acknowledgments
We thank A. N. Barclay and G. G. MacPherson for helpful advice. This work was supported by the Medical Research Council (United Kingdom).
Abbreviations
- TNFR
tumor necrosis factor receptor
- JAK/STAT
Janus kinase/signal transducers and activators of transcription
- RT
reverse transcriptase
- HPRT
hypoxanthine phosphoribosyltransferase
- SOCS-1
suppressor of cytokine signaling-1
- bmDC
bone-marrow-derived dendritic cells
- LPS
lipopolysaccharide
Footnotes
References
- 1.Grewal I S, Flavell R A. Annu Rev Immunol. 1998;16:111–135. doi: 10.1146/annurev.immunol.16.1.111. [DOI] [PubMed] [Google Scholar]
- 2.van Kooten C, Banchereau J. Adv Immunol. 1996;61:1–77. doi: 10.1016/s0065-2776(08)60865-2. [DOI] [PubMed] [Google Scholar]
- 3.Scott D W, Grdina T, Shi Y. J Immunol. 1996;156:2352–2356. [PubMed] [Google Scholar]
- 4.Trinchieri G. Adv Immunol. 1998;70:83–243. doi: 10.1016/s0065-2776(08)60387-9. [DOI] [PubMed] [Google Scholar]
- 5.Ridge J P, Rosa F D, Matzinger P. Nature (London) 1998;393:474–478. doi: 10.1038/30989. [DOI] [PubMed] [Google Scholar]
- 6.Bennett S R M, Carbone F R, Karamalis F, Flavell R A, Miller J F A P, Heath W R. Nature (London) 1998;393:478–480. doi: 10.1038/30996. [DOI] [PubMed] [Google Scholar]
- 7.Schoenberger S P, Toes R E M, van-der-Voort E I H, Offringa R, Melief C J M. Nature (London) 1998;393:480–483. doi: 10.1038/31002. [DOI] [PubMed] [Google Scholar]
- 8.Hanissian S H, Geha R S. Immunity. 1997;6:379–387. doi: 10.1016/s1074-7613(00)80281-2. [DOI] [PubMed] [Google Scholar]
- 9.Morio T, Hanissian S H, Bacharier L B, Teraoka H, Nonoyama S, Seki M, Kondo J, Nakano H, Lee S-K, Geha R S, Yata J. Immunity. 1999;11:339–348. doi: 10.1016/s1074-7613(00)80109-0. [DOI] [PubMed] [Google Scholar]
- 10.Tsukamoto N, Kobayashi N, Azuma S, Yamamoto T, Inoue J. Proc Natl Acad Sci USA. 1999;96:1234–1239. doi: 10.1073/pnas.96.4.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pullen S S, Dang T T A, Crute J J, Kehry M R. J Biol Chem. 1999;274:14246–14254. doi: 10.1074/jbc.274.20.14246. [DOI] [PubMed] [Google Scholar]
- 12.Leo E, Welsh K, Matuzawa S, Zapata J M, Kitada S, Mitchell R S, Ely K R, Reed J C. J Biol Chem. 1999;274:22414–22422. doi: 10.1074/jbc.274.32.22414. [DOI] [PubMed] [Google Scholar]
- 13.Allen R C, Armitage R J, Conley M E, Rosenblatt H, Jenkins N A, Copeland N G, Bedell M A, Edelhoff S, Disteche C M, Simoneaux D K, et al. Science. 1993;259:990–993. doi: 10.1126/science.7679801. [DOI] [PubMed] [Google Scholar]
- 14.Mach F, Schonbeck U, Sukhova G K, Atkinson E, Libby P. Nature (London) 1998;394:200–203. doi: 10.1038/28204. [DOI] [PubMed] [Google Scholar]
- 15.Costello R T, Gastaut J-A, Olive D. Immunol Today. 1999;20:488–493. doi: 10.1016/s0167-5699(99)01507-8. [DOI] [PubMed] [Google Scholar]
- 16.Tan J, Town T, Paris D, Mori T, Suo Z, Crawford F, Mattson M P, Flavell R A, Mullan M. Science. 1999;286:2352–2355. doi: 10.1126/science.286.5448.2352. [DOI] [PubMed] [Google Scholar]
- 17.Tone M, Thompson S A J, Tone Y, Fairchild P J, Waldmann H. J Immunol. 1997;159:6156–6163. [PubMed] [Google Scholar]
- 18.Inaba K, Inaba M, Romani N, Aya H, Deguchi M, Ikehara S, Muramatsu S, Steinman R M. J Exp Med. 1992;176:1693–1702. doi: 10.1084/jem.176.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ly I A, Mishell R I. J Immunol Methods. 1974;5:239–247. doi: 10.1016/0022-1759(74)90108-2. [DOI] [PubMed] [Google Scholar]
- 20.Volkmann A, Neefjes J, Stockinger B. Eur J Immunol. 1996;26:2565–2572. doi: 10.1002/eji.1830261105. [DOI] [PubMed] [Google Scholar]
- 21.Torres R M, Clark E A. J Immunol. 1992;148:620–626. [PubMed] [Google Scholar]
- 22.Grimaldi J C, Torres R, Kozak C A, Chang R, Clark E A, Howard M, Cockayne D A. J Immunol. 1992;149:3921–3926. [PubMed] [Google Scholar]
- 23.Rolink A, Melchers F, Andersson J. Immunity. 1996;5:319–330. doi: 10.1016/s1074-7613(00)80258-7. [DOI] [PubMed] [Google Scholar]
- 24.Chen X P, Losman J A, Rothman P. Immunity. 2000;13:287–290. doi: 10.1016/s1074-7613(00)00028-5. [DOI] [PubMed] [Google Scholar]
- 25.Ma X, Chow J M, Gri G, Carra G, Gerosa F, Wolf S F, Dzialo R, Trinchieri G. J Exp Med. 1996;183:147–157. doi: 10.1084/jem.183.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pinchuk L M, Klaus S J, Magaletti D M, Pinchuk G V, Norsen J P, Clark E A. J Immunol. 1996;157:4363–4370. [PubMed] [Google Scholar]
- 27.O'Rourke D M, Qian X, Zhang H-T, Davis J G, Nute E, Meinkoth J, Greene M I. Proc Natl Acad Sci USA. 1997;94:3250–3255. doi: 10.1073/pnas.94.7.3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Papoff G, Cascino I, Eramo A, Starace G, Lynch D H, Ruberti G. J Immunol. 1996;156:4622–4630. [PubMed] [Google Scholar]
- 29.Screaton G R, Xu X-N, Olsen A L, Cowper A E, Tan R, McMichael A J, Bell J I. Proc Natl Acad Sci USA. 1997;94:4615–4619. doi: 10.1073/pnas.94.9.4615. [DOI] [PMC free article] [PubMed] [Google Scholar]