Abstract
Pre-mRNA processing is coupled with transcription. It is still unclear if the transcription machinery can also directly affect the cytoplasmic fate of a transcript, such as its intracellular localization. In yeast, the RNA-binding protein She2p binds several mRNAs and targets them for localization at the bud. Here we report that She2p is recruited cotranscriptionally to the nascent bud-localized ASH1, IST2, and EAR1 mRNA. She2p interacts in vivo with the elongating forms of RNA polymerase II (pol II) via the transcription elongation factor Spt4–Spt5. Mutations in either SPT4 or SPT5 reduce the cotranscriptional recruitment of She2p on the ASH1 gene, disrupt the proper localization of ASH1 mRNA at the bud tip, and affect Ash1p sorting to the daughter cell nucleus. We propose that She2p is recruited by the RNA pol II machinery prior to its transfer to nascent bud-localized mRNAs. Indeed, She2p is present with RNA pol II on genes coding for localized or nonlocalized transcripts, but is associated with nascent mRNA only on genes coding for bud-localized transcripts. Moreover, a She2p mutant defective in RNA binding still associates with RNA pol II transcribed genes. This study uncovers a novel mechanism for the cotranscriptional assembly of mRNP complexes primed for localization in the cytoplasm.
Keywords: mRNA, localization, yeast, She2p, Spt4p, Spt5p, cotranscriptional
Eukaryotes use cytoplasmic localization of mRNAs to control, in space and time, the expression of proteins regulating cell fate, cellular polarity, or asymmetric cell division (Du et al. 2007; Martin and Ephrussi 2009). With >30 transcripts localized asymmetrically at its bud tip, the budding yeast Saccharomyces cerevisiae is an excellent model organism for studying the molecular mechanisms behind mRNA transport and localization (Chartrand et al. 2001; Darzacq et al. 2003). Yeast bud localization depends on the RNA-binding protein She2p, as it binds to specific localization elements, or zipcodes, within bud-localized transcripts (Olivier et al. 2005). She2p promotes the formation of an mRNA localization complex, or “locasome,” by recruiting the type V myosin Myo4p via the bridging protein She3p (Bohl et al. 2000; Long et al. 2000; Takizawa and Vale 2000).
One of the localized transcripts is the ASH1 mRNA, which accumulates at the bud tip of daughter cells during anaphase (Long et al. 1997; Takizawa et al. 1997). This asymmetric localization of the transcript results in the specific sorting of the Ash1 protein to the nucleus of the daughter cell, where it controls mating type switching and cell size (Jansen et al. 1996; Sil and Herskowitz 1996; Di Talia et al. 2009). During its transport to the bud tip, translation of the ASH1 mRNA is repressed by the RNA-binding proteins Khd1p (Irie et al. 2002; Paquin et al. 2007) and Puf6p (Gu et al. 2004). The phosphorylation of these factors at the bud tip promotes the local synthesis of the Ash1 protein (Paquin et al. 2007; Deng et al. 2008). Recent evidence has shown that the proper localization and translational repression of the ASH1 transcript is dependent on some nuclear events. Indeed, She2p actively shuttles between the cytoplasm and the nucleus, and this nuclear transit is important for the recruitment of the translational repressor Puf6p on the ASH1 mRNA (Du et al. 2008; Shen et al. 2009). However, the mechanism by which She2p is recruited to the ASH1 mRNA in the nucleus in order to promote its cytoplasmic localization is still unknown.
To ensure bud tip localization, She2p may be recruited to localized mRNAs during their transcription. Indeed, RNA polymerase II (RNA pol II) transcription is already known to be coupled to several events, like capping, splicing, 3′ end processing, and nuclear export of the transcript (Moore and Proudfoot 2009). These steps in mRNP biogenesis are coordinated mainly via interactions between factors involved in these processes and the RNA pol II transcription elongation complex (TEC) (Perales and Bentley 2009). The large subunit of RNA pol II, Rpb1p, is one of the main actors in this coupling. The C-terminal domain (CTD) of Rpb1p is comprised of heptad repeats with the consensus sequence YS2PTS5PS7, which are differentially phosphorylated during transcription (Phatnani and Greenleaf 2006; Buratowski 2009). Phosphorylation of Ser5 of the heptad sequence by the TFIIH-associated kinase Cdk7 (Kin28 in yeast) occurs during transcription initiation (Komarnitsky et al. 2000). This promotes mRNA capping, as the capping enzyme and guanylyltransferase bind the Ser5-phosphorylated CTD (Ho and Shuman 1999). Ser2 phosphorylation by the kinase Cdk9/P-TEFb (Ctk1 and Bur1 in yeast) occurs later (Peterlin and Price 2006; Qiu et al. 2009). Factors involved in 3′ end processing, like the cleavage/polyadenylation factor Pcf11, bind the Ser2-phosphorylated CTD during transcription elongation (Licatalosi et al. 2002).
Another member of the TEC involved in cotranscriptional RNA processing is the Spt4–Spt5 transcription elongation factor, also known as DSIF (DRB [5,6-dichloro-1-b-d-ribofuranosylbenzimidazole] sensitivity-inducing factor) (Wada et al. 1998). Spt4 and Spt5 form a heterodimeric complex that regulates the processivity of RNA pol II (Hartzog et al. 1998). Beside its role in elongation, Spt5 is also implicated in mRNA capping (as it interacts and activates the cap guanylyltransferase) (Wen and Shatkin 1999) and in mRNA splicing (Lindstrom et al. 2003). The coupling between transcription and pre-mRNA processing involves steps in mRNP biogenesis that occur prior to nuclear export (from capping to mRNP export complex assembly). However, it is not clear if the cytoplasmic fate of an mRNA—like its translational regulation, cytoplasmic localization, or degradation—could also be modulated by the transcription machinery via interactions between the TEC and factors involved in these cytoplasmic processes.
In this study, we used chromatin immunoprecipitation (ChIP) to show that She2p interacts cotranscriptionally with the nascent ASH1 mRNA. She2p was found associated with the elongating form of RNA pol II in vivo, and this interaction depended on the transcription elongation factor Spt4–Spt5. Mutations in either SPT4 or SPT5 reduced the cotranscriptional recruitment of She2p on the ASH1 gene, and resulted in a decreased localization of both ASH1 mRNA and Ash1p. We hypothesize that She2p is recruited by the RNA pol II machinery prior to its transfer to nascent bud-localized mRNAs. Consistent with this model, ChIPs showed that She2p is associated with genes coding for both nonlocalized and bud-localized mRNAs, but this association is RNA-dependent only for genes coding for bud-localized transcripts. Altogether, these data reveal a mechanism for the cotranscriptional assembly of mRNPs competent for localization in the cytoplasm.
Results
She2p is recruited cotranscriptionally at the ASH1 and IST2 genes
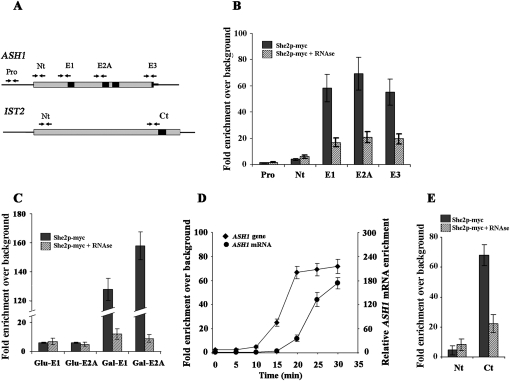
Previous results have shown that She2p actively shuttles between the nucleus and the cytoplasm, and that nuclear She2p promotes the binding of the nuclear proteins Loc1 and Puf6 to the ASH1 mRNA in vivo (Shen et al. 2009). These results led us to suppose that She2p might be recruited early to the nascent ASH1 mRNA, where it could promote the formation of the mRNP complex before nuclear export. To explore this possibility, we used ChIP to determine if She2p is associated cotranscriptionally with the ASH1 gene. A she2 yeast strain transformed with a plasmid expressing endogenous levels of She2p-myc was fixed with formaldehyde, the chromatin was extracted and fragmented, and an anti-Myc antibody was used to immunoprecipitate She2p-myc complexes. After ChIP, PCR amplification of specific regions of the endogenous ASH1 gene was performed and normalized to a nontranscribed intergenic region to determine if She2p-myc was enriched at the ASH1 locus in vivo (Fig. 1A). As shown in Figure 1B, PCR amplification of regions surrounding the localization elements E1, E2A, and E3 (which are bound by She2p) were enriched after ChIP, showing that She2p-myc is associated with the ASH1 gene in vivo. In contrast, amplicons from the ASH1 promoter (Pro) and the terminus (Nt) of the ASH1 coding sequence were poorly amplified. To determine if the recruitment of She2p to the ASH1 gene depends on the interaction between She2p and the nascent ASH1 transcript, the ChIP was repeated in the presence of RNase. A threefold reduction in chromatin enrichment was observed for the E1, E2A, and E3 amplicons after RNase treatment (Fig. 1B), suggesting that the interaction between She2p and the ASH1 gene is in part RNA-dependent.
Figure 1.
She2p-myc interacts cotranscriptionally with bud-localized mRNAs. (A) Diagram of ASH1 and IST2 genes, and the relative position of the amplicons used for each gene. Dark boxes in ASH1 and IST2 correspond to the localization elements. In the ASH1 gene, amplicon Pro spans a region 500 base pairs (bp) upstream of the start codon, while the amplicon Nt is upstream of the start codon. Amplicons E1, E2A, and E3 overlap with the 5′ half of the corresponding localization element. (B) She2p-myc immunoprecipitates the endogenous ASH1 coding sequence near the localization elements in an RNA-dependent manner. She2p-myc ChIP was performed using an anti-myc antibody, followed by PCR amplification of amplicons specific to endogenous ASH1 (Pro, Nt, E1, E2A, and E3). Chromatin was treated (+RNase) or not with RNase A prior immunoprecipitation. (C) Transcription-dependent interaction between She2p-myc and a galactose-inducible ASH1 gene. She2p-myc ChIP was performed using an anti-myc antibody, followed by PCR amplification of amplicons specific to the galactose-induced (galactose) or repressed (glucose) ASH1 gene (E1 and E2A). Chromatin was treated (+RNase) or not with RNase A prior immunoprecipitation. (D) Kinetics of She2p-myc recruitment on the ASH1 gene, and interaction with ASH1 mRNA. Interaction of She2p-myc with the ASH1 gene was determined by ChIP, followed by qPCR amplification of amplicon E1. Interaction of She2p-myc with ASH1 mRNA was determined by immunoprecipitation followed by qRT–PCR. ASH1 mRNA enrichment was normalized to IST2 mRNA levels. (Left Y-axis) ASH1 amplicon enrichment by ChIP. (Right Y-axis) ASH1 mRNA enrichment by RT–PCR. (E) She2p-myc immunoprecipitates the IST2 coding sequence near the localization element. She2p-myc ChIP was performed using an anti-myc antibody, followed by PCR amplification of the amplicon specific to endogenous IST2 (Nt or Ct). Chromatin was treated (+RNase) or not with RNase A prior immunoprecipitation. The data are presented as the mean ± SEM (N ≥ 3).
To further confirm that the interaction between She2p-myc and the ASH1 gene depends on transcription, a galactose-inducible ASH1 gene was integrated in the genome of a yeast strain deleted of its endogenous ASH1 gene, and ChIP with She2p-myc was performed in the presence of glucose (low transcription) or galactose (active transcription). PCR amplification in regions of the ASH1 gene containing the localization elements E1 and E2A were positive in the presence of galactose, while lower PCR amplification was detected when yeasts were grown in glucose (Fig. 1C). In controls, no PCR amplification was detected from the ASH1 gene when IgG antibodies were used for the immunoprecipitation, or with anti-Myc antibody when She2p was expressed without a myc tag (data not shown). To confirm that the interaction between She2p-myc and the ASH1 gene is RNA-dependent, RNase treatment of the chromatin was performed before immunoprecipitation of She2p-myc. While, in galactose-induced cells, She2p-myc ChIP of the ASH1 gene was positive, pretreatment with RNase A strongly reduced this ChIP (Fig. 1C). Altogether, these results suggest that She2p is recruited at the ASH1 locus during the transcription of the nascent ASH1 mRNA.
Since the data presented above reflect a steady-state association of She2p with the ASH1 gene, it is unclear whether or not the recruitment of She2p to this gene precedes the interaction between She2p and the ASH1 transcript. To answer this question, a kinetic analysis of She2p recruitment to the ASH1 gene and transcript was performed using the galactose-inducible ASH1 gene. After addition of galactose to yeast culture, samples were taken every 5 min, fixed with formaldehyde, and split in two fractions. On one fraction, ChIP of the ASH1 gene was performed using She2p-myc, and on the other, immunoprecipitation of She2p-myc was followed by quantitative RT–PCR (IP + RT–qPCR) of ASH1 mRNA. This allowed quantitative measurement of She2p association with ASH1 chromatin (by ChIP) and ASH1 transcript (by IP + RT–qPCR) during induction of this gene. As shown in Figure 1D, the kinetics revealed that She2p associates first with the ASH1 gene, 10–15 min after induction. Interaction between She2p and ASH1 mRNA was detected later, 15–20 min after induction. These results show that the association of She2p with the ASH1 gene precedes its interaction with the ASH1 transcript.
She2p is known to be involved in the localization of >30 mRNAs to the bud of yeast cells (Shepard et al. 2003; Aronov et al. 2007). To determine if She2p was also associated cotranscriptionally with another bud-localized mRNA, PCR amplification of a region of the endogenous IST2 gene, near the localization element of the IST2 mRNA (Olivier et al. 2005), was performed after immunoprecipitation of She2p-myc. Indeed, an amplicon in the IST2 gene near the localization element was detected after ChIP (Fig. 1E). Pretreatment with RNase strongly reduced this PCR amplification, suggesting an RNA-dependent interaction. These results show that She2p is recruited cotranscriptionally to bud-localized mRNAs like ASH1 and IST2.
She2p interacts with the elongating form of RNA pol II in vivo
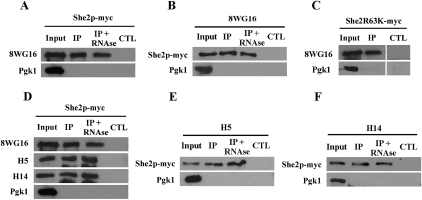
The cotranscriptional interaction between She2p and the ASH1 mRNA raised the question of how this RNA-binding protein gets recruited to the nascent transcript. One possibility is that She2p interacts with the RNA pol II transcription machinery. To test this hypothesis, coimmunoprecipitation (co-IP) between She2p-myc and Rpb1p was performed. Immunoprecipitation of She2p-myc expressed at endogenous levels resulted in the pull-down of Rpb1p (Fig. 2A). This interaction between She2p and Rpb1p was confirmed by performing reverse co-IP, in which Rpb1p was immunoprecipitated using the 8WG16 antibody (Fig. 2B). Interestingly, this interaction was independent of RNA, since RNase treatment did not affect the co-IP between She2p-myc and Rpb1p (Fig. 2A,B). To confirm that the interaction is RNA-independent, the co-IP was repeated using a mutant of She2p that cannot bind RNA, She2R63K (Gonsalvez et al. 2003). This confirmed that She2p is coimmunoprecipitated with RNA pol II via protein–protein interactions (Fig. 2C).
Figure 2.
She2p-myc interacts with the elongating form of RNA pol II in vivo. (A) Rpb1p coimmunoprecipitates with She2p-myc. She2p-myc was immunoprecipitated with an anti-myc antibody (9E10), followed by detection of Rpb1p by Western blot using the 8WG16 antibody. (Input) Total yeast extract; (IP) immunoprecipitate; (IP + RNase) yeast extract treated with RNase prior to immunoprecipitation; (CTL) strain K699 with untagged She2p. Pgk1 was used as a negative control. (B) She2p-myc coimmunoprecipitates with Rpb1p. Rpb1p was immunoprecipitated with the 8WG16 antibody, followed by detection of She2p-myc by Western blot using the 9E10 antibody. (Input) Total yeast extract; (IP) immunoprecipitate; (IP + RNase) yeast extract treated with RNase prior to immunoprecipitation; (CTL) strain K699 with untagged She2p. Pgk1 was used as negative control. (C) Rpb1p interacts with She2R63K-myc, a mutant defective in RNA binding. She263K-myc was immunoprecipitated with an anti-myc antibody (9E10), followed by detection of Rpb1p by Western blot using the 8WG16 antibody. (Input) Total yeast extract; (IP) immunoprecipitate; (CTL) strain K699 with untagged She2R63K. Pgk1 was used as a negative control. (D) The elongating form of Rpb1p coimmunoprecipitates with She2p-myc. She2p-myc was immunoprecipitated with an anti-myc antibody (9E10), followed by detection of hypophosphorylated (8WG16), Ser5-phosphorylated (H5), or Ser-2-phosphorylated (H14) Rpb1p by Western blot. (Input) Total yeast extract; (IP) immunoprecipitate; (IP + RNase) yeast extract treated with RNase prior to immunoprecipitation; (CTL) strain K699 with untagged She2p. Pgk1 was used as negative control. (E) She2p-myc coimmunoprecipitates with Ser5-phosphorylated Rpb1p. Ser5-phosphorylated Rpb1p was immunoprecipitated with the H5 antibody, followed by detection of She2p-myc by Western blot using the 9E10 antibody. (Input) Total yeast extract; (IP) immunoprecipitate; (IP + RNase) yeast extract treated with RNase prior to immunoprecipitation; (CTL) strain K699 with untagged She2p. Pgk1 was used as negative control. (F) She2p-myc coimmunoprecipitates with Ser2-phosphorylated Rpb1p. Ser2-phosphorylated Rpb1p was immunoprecipitated with the H14 antibody, followed by detection of She2p-myc by Western blot using the 9E10 antibody. (Input) Total yeast extract; (IP) immunoprecipitate; (IP + RNase) yeast extract treated with RNase prior to immunoprecipitation; (CTL) strain K699 with untagged She2p. Pgk1 was used as a negative control.
RNA pol II is phosphorylated on the CTD of Rpb1p during transcription. Phosphorylation of Ser5 and Ser2 of the YSPTSPS heptad repeat in the CTD occurs during transcription initiation and elongation, respectively (Phatnani and Greenleaf 2006). Antibodies that recognize these phosphoresidues were used to determine if She2p is associated with the active, elongating form of RNA pol II. Immunoprecipitation of She2p-myc was followed by detection of hypophosphorylated Rpb1p (8WG16 antibody), Ser5-phosphorylated Rpb1p (H5 antibody), or Ser2-phosphorylated Rpb1p (H14 antibody) by Western blot. As shown in Figure 2D, all of the forms of Rpb1p interacted with She2p-myc. Treatment with RNase prior to immunoprecipitation had no effect on these interactions. The reverse co-IPs were also performed, using the anti-P-Ser5 (H5) or anti-P-Ser2 (H14) antibodies for immunoprecipitation of Rpb1p, followed by detection of She2p-myc by Western blot. For both antibodies, She2p-myc was detected in the coimmunoprecipitate, even after treatment with RNase (Fig. 2E,F). Altogether, these results show that She2p interacts with the active, elongating RNA pol II independently of its binding to RNA.
The interaction between She2p and RNA pol II occurs via the transcription elongation factor Spt4–Spt5
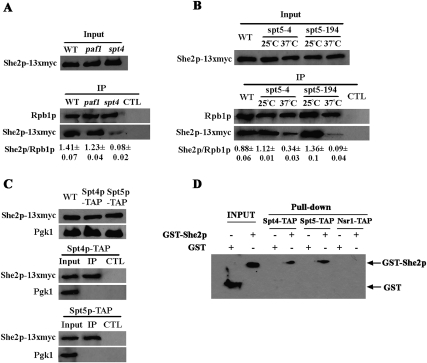
To better assess the biological relevance of the interaction between She2p and RNA pol II, we decided to identify the binding site of She2p on the RNA pol II machinery. Since the CTD of Rpb1p is a major binding site for RNA processing factors (Perales and Bentley 2009), a possible interaction between She2p and Pol II CTD was explored. However, no interaction between She2p-myc and the nonphosphorylated or phosphorylated CTD was detected using GST pull-down experiments (data not shown). Since She2p did not interact with the pol II CTD, we hypothesized that it may be associated indirectly with RNA pol II via interaction with transcription factors. Therefore, we decided to focus on transcription elongation factors, which are associated with the phosphorylated forms of RNA pol II, as is She2p. Using co-IP, we measured the interaction between She2p and Rpb1p in strains carrying mutations or deletions of genes coding for the transcription elongation factors (1) Rad6p and Bre1p, which are required for histone H2B ubiquitylation and H3 methylation; (2) Paf1p and Rtf1p, which are part of the PAF1 complex that promotes histone methylation; and (3) Spt4p and Spt5p, which regulate RNA pol II processivity. For each mutant strain and its isogenic wild-type strain, a 13xmyc tag was integrated at the C-terminal end of the coding sequence of the SHE2 gene, and co-IP of Rpb1p was performed. None of these mutants had an effect on She2p-myc expression levels (data not shown). Of the seven deletion mutants (rad6, bre1, paf1, spt4, and rtf1) or temperature-sensitive mutants (spt5-4 and spt5-194), only the SPT4 and SPT5 mutants had an impact on the interaction between She2p-myc and Rpb1p (Fig. 3A,B; data not shown). Indeed, deletion of SPT4 strongly reduced the interaction between She2p-myc and Rpb1p (Fig. 3A), while deletion of PAF1 had no effect. For SPT5, two temperature-sensitive mutants were tested: spt5-4 and spt5-194 (Hartzog et al. 1998). Both mutants displayed a reduced interaction between She2p-myc and Rpb1p when shifted to a nonpermissive temperature (Fig. 3B). RNase treatment prior to immunoprecipitation did not further reduce the interaction between She2p and Rpb1p in spt4 and spt5 mutants, confirming an RNA-independent interaction between these factors (Supplemental Fig. 1).
Figure 3.
She2p-myc interacts with RNA pol II via the transcription elongation factor Spt4–Spt5. (A) Deletion of SPT4 decreases the interaction between She2p-myc and Rpb1p in vivo. Rpb1p was immunoprecipitated with the 8WG16 antibody, followed by detection of She2p-myc by Western blot using the 9E10 antibody. (Input) Total yeast extract; (IP) immunoprecipitate; (CTL) yeast extract treated with beads only (no antibody). The ratio She2p/Rpb1p reflects the mean ± SEM (N = 3). (B) Temperature-sensitive mutations in SPT5 decrease the interaction between She2p-myc and Rpb1p in vivo. Rpb1p was immunoprecipitated with 8WG16 antibody, followed by detection of She2p-myc by Western blot using the 9E10 antibody. (Input) Total yeast extract; (IP) immunoprecipitate; (CTL) yeast extract treated with beads only (no antibody). The ratio She2p/Rpb1p reflects the mean ± SEM (N = 3). (C) She2p-myc coimmunoprecipitates with Spt4-TAP and Spt5-TAP. Spt4-TAP or Spt5-TAP was immunoprecipitated with IgG, followed by detection of She2p-myc by Western blot using the 9E10 antibody. (Input) Total yeast extract; (IP) immunoprecipitate; (CTL) yeast extract treated with beads only (no antibody). Pgk1 was used as a negative control. (D) Recombinant GST-She2p interacts with yeast-purified Spt4–Spt5 complex in vitro. The Spt4–Spt5 complex was purified from yeast either as a Spt4-TAP/Spt5 complex (Spt4-TAP) or Spt4/Spt5-TAP complex (Spt5-TAP). (Input) Purified recombinant proteins.
Spt4–Spt5 forms a complex called DSIF, which is highly conserved from yeast to humans (Wada et al. 1998). Mutations in both members of the DSIF complex affect the interaction between She2p and Rpb1p, which strongly suggests that Spt4–Spt5 participates in the recruitments of She2p to RNA pol II. To determine if Spt4 and Spt5 interact with She2p, pull-down experiments were performed using TAP-tagged Spt4 and Spt5 and myc-tagged She2p, all expressed from their chromosomal locus. As shown in Figure 3C, both Spt4-TAP and Spt5-TAP were able to pull down She2p-myc. To further confirm the interaction between She2p and the Spt4–Spt5 complex in vitro, recombinant GST-She2p was purified from bacteria, while the Spt4–Spt5 complex was purified from yeast using Spt4-TAP- or Spt5-TAP-tagged strains (Supplemental Fig. 2; Krogan et al. 2002). As shown in Figure 3D, the purified Spt4–Spt5 complex (containing either Spt4-TAP or Spt5-TAP bound to calmodulin beads) was able to pull down recombinant GST-She2p, but not GST alone. As a control, GST-She2p did not interact with yeast-purified Nsr1-TAP. Altogether, these results demonstrate that She2p interacts with the transcription elongation factor Spt4–Spt5 in vivo and in vitro, and that this interaction is important for the association between She2p and RNA pol II.
Mutations in SPT4–SPT5 disrupt the cotranscriptional recruitment of She2p to the ASH1 gene, and affect ASH1 mRNA localization
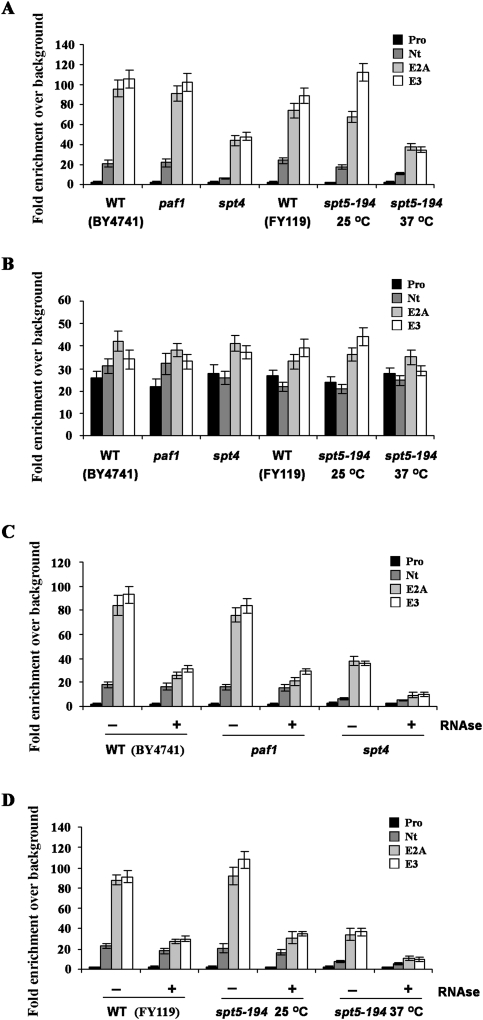
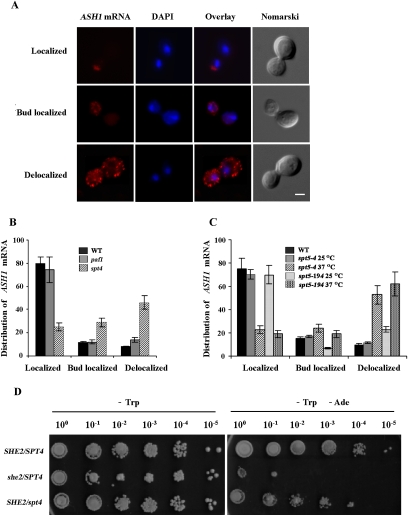
With the identification of mutants that affect the interaction between She2p and Rpb1p, we could explore the role of the Spt4–Spt5 complex in the cotranscriptional recruitment of She2p to the ASH1 gene. We repeated the ChIP of She2p-myc in the spt4, spt5-4, and spt5-194 strains and measured the enrichment of the ASH1 gene using qPCR. To control for the possibility that the disruption of an elongation factor could affect the transcription level of the ASH1 gene, we tested another elongation factor, Paf1p, whose function depends on the Spt4–Spt5 complex. Paf1p is part of the PAF1 complex (PAF1C) TEC, and depends on Spt4p for its association with RNA pol II (Qiu et al. 2006). Moreover, phosphorylation of the CTD of Spt5 by the Bur1 kinase stimulates the recruitment of PAF1C to RNA pol II (Liu et al. 2009; Zhou et al. 2009). Also, ASH1 mRNA expression levels were measured in all mutant strains using RT–qPCR, and revealed no significant decrease in the spt4, paf1, and spt5-4 or spt5-194 strains at a nonpermissive temperature (Supplemental Fig. 3).
ChIP was performed using She2p-myc, and enrichment of the Pro, Nt, E2A, and E3 amplicons of the ASH1 gene was determined by qPCR. As shown in Figure 4A, deletion of SPT4 resulted in more than twofold reduction of the ASH1 Nt, E2A, and E3 amplicons compared with the isogenic wild-type or paf1 strains. Similar results were observed in the spt5-4 and spt5-194 strains at a nonpermissive temperature, where a twofold to 2.5-fold decrease in the enrichment of ASH1 E2A and E3 amplicons was also measured (Fig. 4A; Supplemental Fig. 4). Spt4–Spt5 is known to increase the level of elongating RNA pol II on genes, as a spt4 mutant reduces RNA pol II levels at the 3′ end of a number of specific genes in yeast (Rodriguez-Gil et al. 2010). Therefore, it is possible that the defective recruitment of She2p to the ASH1 gene in the spt4 and spt5 mutant strains may be caused by a reduction in elongating RNA pol II levels on this gene. To eliminate this possibility, ChIP using Rpb1p was performed in parallel with She2p-myc ChIP in wild-type, paf1, spt4, and spt5-194 strains. This experiment revealed that there was no difference in enrichment of the Pro, Nt, E2A, and E3 amplicons after Rpb1p ChIP between wild-type, paf1, and spt4 strains, or in the spt5-194 strain at permissive and restrictive temperatures (Fig. 4B). Only the ChIP on the amplicon E3 was slightly reduced (∼30%) in the spt5-194 strain at restrictive versus permissive temperatures (Fig. 4B). However, this small effect cannot explain the nearly threefold reduction in She2p-myc ChIP on the amplicon E3 observed in this mutant (Fig. 4A).
Figure 4.
Cotranscriptional interaction of She2p-myc with ASH1 depends on the transcription elongation factor Spt4–Spt5. (A) Mutation in SPT4 or SPT5 decreases She2p-myc ChIP of the endogenous ASH1 gene. She2p-myc ChIP was performed using an anti-myc antibody, followed by qPCR amplification of amplicons specific to endogenous ASH1 (Pro, Nt, E2A, and E3) in isogenic wild-type (WT; BY4741 SHE2-MYC), paf1, and spt4 strains, or in isogenic wild-type (WT; FY119 SHE2-MYC) and spt5-194 mutant strains at 25°C or 37°C. (B) Mutations in SPT4 and SPT5 do not affect Rpb1p ChIP on the ASH1 gene. Experiments were performed as in A, with the exception that the 8WG16 antibody was used for ChIP. (C) RNase treatment reduces She2p-myc ChIP in the spt4 strain. ChIP was performed as in A, with the addition (+) or not (−) of RNase A prior to immunoprecipitation. (D) RNase treatment reduces She2p-myc ChIP in the spt5-194 mutant strain. ChIP was performed as in A, with the addition (+) or not (−) of RNase A prior to immunoprecipitation. The data are presented as the mean ± SEM (N = 3).
Intriguingly, while the spt4 and spt5 mutants reduced the interaction between She2p and Rpb1p by fourfold to 12-fold depending on the mutant (Fig. 3A,B), ChIP of She2p-myc was reduced by only twofold to threefold in the same mutant strains (Fig. 4A). This discrepancy could be due to the presence of Spt4–Spt5-independent interactions between She2p and the ASH1 gene, possibly via the nascent ASH1 transcript. Indeed, treatment with RNase prior to ChIP using She2p-myc further reduced the residual enrichment of the E2A and E3 amplicons to near background levels in the spt4 strain (Fig. 4C) and in the spt5-194 strain at a nonpermissive temperature (Fig. 4D). This result shows that She2p can be recruited to the nascent ASH1 transcript via its E2A and E3 zipcodes independently of the presence of Spt4–Spt5, although this recruitment is less efficient. Altogether, these results suggest that the Spt4–Spt5 complex promotes the cotranscriptional recruitment of She2p to the ASH1 gene.
The effect of the spt4 and spt5 mutations on ASH1 mRNA localization was then determined using fluorescent in situ hybridization (FISH) against the ASH1 transcript. Wild-type and mutant strains were scored for either bud tip localization of ASH1 mRNA (localized), full bud localization (bud-localized), or delocalization of this mRNA (Fig. 5A). While 80% of wild-type and paf1 late-anaphase cells displayed localization of the ASH1 mRNA at their bud tips, this percentage dropped to 20% in spt4 cells (Fig. 5B). A similar effect was observed in spt5-4 and spt5-194 strains when they were shifted to a nonpermissive temperature, as >50% of late-anaphase cells had delocalized ASH1 mRNA (Fig. 5C). Mutations in both spt4 and spt5 had little additive effect on ASH1 mRNA localization compared with each single mutant (Supplemental Fig. 5). The disruptive effect of spt4 and spt5 mutants on ASH1 mRNA localization was not as penetrant as in a she2 strain, in which only 1% of the cells maintain localization of this transcript (Long et al. 1997). The effect of the disruption of the Spt4–Spt5 complex on the asymmetric distribution of the Ash1 protein was also determined using a genetic assay in which the Ash1p-repressed promoter from the HO gene controls the expression of the ADE2 gene (Jansen et al. 1996). In this assay, asymmetric distribution of Ash1p maintains expression of the HO-ADE2 gene in the mother cell and growth on plates lacking adenine (−Ade), while symmetric Ash1p represses HO-ADE2 expression and growth of both mother and daughter cells in this medium. Deletion of SHE2 (and disruption of ASH1 mRNA localization and Ash1p sorting to the daughter cell nucleus) resulted in a growth defect on −Ade plates (Fig. 5D). Deletion of SPT4 in this genetic background led to a partial growth defect on −Ade plates (Fig. 5D), suggesting that the defect in ASH1 mRNA localization observed in this mutant resulted in an incomplete sorting of the Ash1 protein. Altogether, these results provide evidence that the cotranscriptional recruitment of She2p on the ASH1 gene via the Spt4–Spt5 elongation complex is important for cytoplasmic ASH1 mRNA localization.
Figure 5.
Spt4–Spt5 is required for proper ASH1 mRNA localization and Ash1p sorting. (A) Phenotypes of ASH1 mRNA localization as detected by FISH. (Localized) ASH1 mRNA localized at the bud tip; (Bud-localized) ASH1 mRNA filling the entire bud. (Delocalized) ASH1 mRNA in both the bud and the mother cell. Bar, 2 μm. (B) Scores on ASH1 mRNA localization phenotypes from wild-type (WT), paf1, and spt4 strains. (C) Scores on ASH1 mRNA localization phenotypes from wild-type (WT) and spt5-4 and spt5-194 mutant strains at 25°C or 37°C. (D) Yeast genetic assay for Ash1p asymmetric distribution. Tenfold dilutions of exponentially growing K5547 (HO-ADE2, she2) transformed with either YCP22-She2-myc (SHE2/SPT4) or YCP22 empty (she2/SPT4), or strain K5547 spt4∷KAN transformed with YCP22-She2-myc (SHE2/spt4) were spotted on plates lacking tryptophan (−Trp) or lacking tryptophan and adenine (−Trp −Ade), and were incubated at 30°C.
She2p is associated with genes coding for both bud-localized and nonlocalized mRNAs
The results presented above show that She2p interacts with Rpb1p via the Spt4–Spt5 TEC, and this interaction is independent of the RNA-binding property of She2p. From these results, a possible model for the recruitment of She2p on bud-localized mRNAs could be proposed: She2p interacts first with the TEC via the Spt4–Spt5 complex, and is then transferred to the nascent mRNA when a localization element RNA motif emerges from the elongating RNA polymerase. This model predicts that She2p would be associated with RNA pol II transcribed genes coding for both bud-localized and nonlocalized mRNAs via its interaction with the TEC. However, this association would be in part RNA-dependent only for genes coding for bud-localized mRNAs. Indeed, the ChIP data in Figure 1 show that, even after RNase treatment, there was still a significant association of She2p-myc with the ASH1 and IST2 genes. Therefore, for these two genes, part of the She2p-myc ChIP may reflect the interaction between She2p and the nascent mRNA (which is RNase-sensitive), and some of the association may reflect the direct interaction between She2p and the TEC (which is RNase-insensitive).
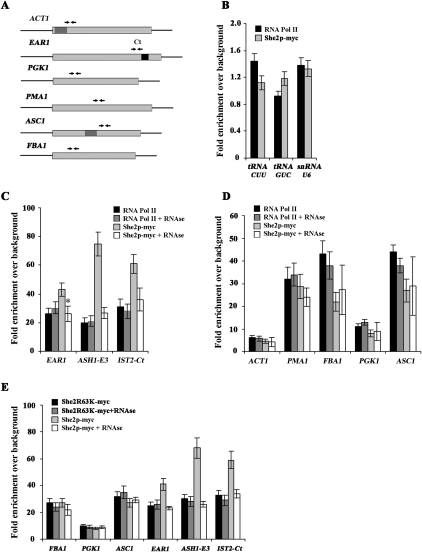
To test this hypothesis, we repeated the She2p-myc ChIP on genes whose mRNAs are not bound by She2p (Shepard et al. 2003; Oeffinger et al. 2007). In parallel, we performed ChIP on the same genes using Rpb1p to estimate their level of transcription. Both She2p-myc and Rpb1p ChIPs were treated or not with RNase to determine if the association was RNA-dependent. We focused on five genes—ACT1, PMA1, FBA1, PGK1, and ASC1—that have been characterized previously by ChIP (Abruzzi et al. 2004) and do not encode for known bud-localized/She2p-associated mRNAs. These five genes were compared with ASH1, IST2, and EAR1, three genes coding for bud-localized mRNAs. As negative controls, we used genes transcribed by RNA pol III, coding for tRNACUU, tRNAGUC, and U6 snRNA. As anticipated, the RNA pol III genes showed no association with either Rpb1p or She2p-myc by ChIP (Fig. 6B). The ASH1, IST2, and EAR1 genes, however, were enriched after Rpb1p and She2p-myc ChIP (Fig. 6C). While treatment of chromatin with RNase prior to immunoprecipitation had no effect on the Rpb1p ChIP, it resulted in a decreased level of She2p-myc ChIP for all three genes, suggesting that part of the association of She2p-myc with these genes is RNA-dependent (Fig. 6C).
Figure 6.
She2p-myc interacts cotranscriptionally with genes coding for both nonlocalized and bud-localized mRNAs. (A) Diagram of EAR1, PGK1, PMA1, ACT1, ASC1, and FBA1 genes, and the relative position of the amplicons used for each gene. Dark boxes in EAR1 correspond to localization elements. The gray box in ACT1 and ASC1 corresponds to their introns. For all ChIP experiments, RNA pol II (Rpb1p) and She2p-myc ChIPs were performed using 8WG16 and anti-myc antibodies, respectively. qPCR amplification of amplicons specific to endogenous genes was performed on immunoprecipitated chromatin. Chromatin was treated (+RNase) or not with RNase A prior to immunoprecipitation. All ChIPs were performed using strain W303-SHE2-MYC, with the exception of E. (B) She2p-myc is not associated with genes coding for tRNACUU, tRNACUG, and U6 snRNA, which are transcribed by RNA pol III. (C) She2p-myc immunoprecipitates the ASH1, IST2, and EAR1 genes, which encode bud-localized mRNAs. (*) P < 0.001. (D) She2p-myc immunoprecipitates the ACT1, PMA1, FBA1, PGK1, and ASC1 genes, which do not encode for bud-localized/She2p-associated mRNAs. (E) An RNA-binding-defective mutant of She2p, She2R63K, still associates with pol II transcribed genes by ChIP. Experiments were performed using strain K699 she2 strain transformed with YCP22-She2-myc or YCP22-She2R63K-myc. The data are presented as the mean ± SEM (N = 3).
Interestingly, She2p-myc ChIP revealed that the ACT1, PMA1, FBA1, PGK1, and ASC1 genes were all associated with She2p (Fig. 6D). Their enrichment in She2p-myc ChIP followed the same pattern as for the Rpb1p ChIP (low for ACT1 and PGK1; high for PMA1, FBA1, and ASC1), suggesting a relationship between She2p interaction and the transcription level of these genes. As predicted in our hypothesis, RNase treatment had no effect on the association of She2p-myc with the ACT1, PMA1, FBA1, PGK1, and ASC1 by ChIP (Fig. 6D), suggesting an RNA-independent interaction. To further confirm these results, we performed ChIP using the RNA-binding mutant She2R63K, and measured the effect of this mutation on the association between She2p-myc and the above-mentioned genes. As shown in Figure 6E, while the R63K mutation reduced (but did not abolish) She2p-myc ChIP on the bud-localized EAR1, ASH1, and IST2 genes, it had no effect on the RNase-insensitive occupancy of She2p on these genes. Moreover, the R63K mutation had no effect on the association between She2p-myc and the FBA1, PGK1, and ASC1 genes. Altogether, these results show that She2p is associated with genes transcribed by RNA pol II independently of their nascent mRNAs. However, RNA-dependent interaction of She2p occurs only on genes whose mRNAs contain zipcodes.
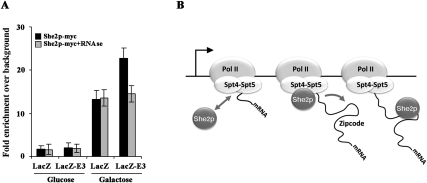
To further confirm the hypothesis that the presence of a zipcode in a gene confers RNA-dependent recruitment of She2p, the ASH1 E3 zipcode was inserted in the nonlocalized LacZ mRNA, and recruitment of She2p to the LacZ gene was determined by ChIP. While She2p-myc ChIP on the LacZ gene was insensitive to RNase treatment, the presence of a zipcode in the LacZ-E3 gene resulted in an RNase-sensitive recruitment of She2p (Fig. 7A). These results support the model that zipcodes mediate the RNA-dependent recruitment of She2p on RNA pol II transcribed genes.
Figure 7.
RNA-dependent recruitment of She2p on a gene requires a zipcode. (A) The presence of a zipcode in the LacZ gene results in RNase-sensitive association of She2p-myc by ChIP. Yeast strains with integrated galactose-inducible LacZ or LacZ-E3 genes were grown under repressed (glucose) or activated (galactose) transcription. Chromatin was treated (+RNase) or not with RNaseA prior to She2p-myc ChIP. The data are presented as the mean ± SEM (N = 3). (B) Model for cotranscriptional recruitment of She2p to bud-localized mRNAs. (Left) She2p interacts with RNA pol II via the Spt4–Spt5 transcription elongation factor. Association of She2p with the TEC (middle) allows the transfer of She2p to a zipcode in the nascent mRNA as it emerges from the RNA polymerase (right).
Discussion
Recent literature shows that most processes related to mRNA maturation (capping, splicing, and processing) and mRNA export are initiated on nascent transcripts (Komili and Silver 2008). Cotranscriptional recruitment of mRNA processing, splicing, and export factors has been well documented, and many of these factors interact specifically with the TEC (Perales and Bentley 2009). Since several RNA-binding proteins involved in cytoplasmic mRNA localization are known to be exclusive residents of the nucleus or to shuttle between the cytoplasm and the nucleus (Farina and Singer 2002), it is possible that some of these factors are also recruited cotranscriptionally to nascent localized mRNAs.
In this study, we show that the mRNA localization factor She2p interacts cotranscriptionally with the nascent ASH1, IST2, and EAR1 mRNAs. While She2p was already known to be associated with mRNAs in the nucleus (Kruse et al. 2002), the mechanism for its recruitment to bud-localized transcripts was not clear. ChIP revealed that She2p is associated with the ASH1, IST2, and EAR1 genes, and that this association is in part RNA-dependent and occurs only during transcription. These data are supported by evidence from metazoans that a factor involved in mRNA localization can associate with the gene of the localized transcript. In fibroblasts, the RNA-binding protein ZBP1, which is involved in β-actin mRNA localization at the leading edge, is recruited on nascent β-actin mRNA (Oleynikov and Singer 2003; Pan et al. 2007), but it is still unclear if this recruitment is directly coupled to the transcription machinery.
In this study, we found that She2p is associated with the phosphorylated, elongating form of RNA pol II in vivo via an interaction with the transcription elongation factor Spt4–Spt5. Mutations in either SPT4 or SPT5 reduced the cotranscriptional recruitment of She2p on the ASH1 gene, pointing toward a role of the TEC in the recruitment of She2p on the nascent ASH1 transcript. These results reveal a novel function for the Spt4–Spt5 complex, beside its roles in RNA pol II elongation, mRNA capping, and splicing (Hartzog et al. 1998; Wen and Shatkin 1999; Lindstrom et al. 2003). Spt4–Spt5 interacts with RNA pol II throughout most of the transcription process, as it gets recruited early on initiating pol II and stays during elongation (Pokholok et al. 2002). Since most of the known RNA localization motifs in yeast have been found within the coding sequence of localized mRNAs—as close as 600 nucleotides from the start codon in the case of the element E1 in ASH1 mRNA (Chartrand et al. 1999; Jambhekar et al. 2005; Olivier et al. 2005)—the early recruitment of She2p on the TEC via Spt4–Spt5 may ensure that She2p is present when a localization element emerges from the RNA polymerase.
More importantly, mutations in SPT4 and SPT5 significantly reduced the localization of the ASH1 mRNA at the bud tip, and disrupted the asymmetric sorting of Ash1p. None of the spt4 and spt5 mutations used had any effect on ASH1 mRNA expression or RNA pol II association with the ASH1 gene, suggesting a post-transcriptional role of Spt4–Spt5 on ASH1 mRNA localization. Altogether, these results suggest that the cotranscriptional recruitment of She2p on the ASH1 gene is important for the proper cytoplasmic localization of this mRNA. By binding directly on the nascent mRNA, She2p makes this mRNP competent for localization, and defines the fate of the transcript before its export to the cytoplasm. Moreover, She2p has to compete with more abundant nonspecific nuclear RNA-binding proteins for interaction with the nascent transcript—like mRNA export factors, for instance. Since several of these mRNA export factors are known to interact with the TEC and are recruited cotranscriptionally to mRNAs (Perales and Bentley 2009), interaction between She2p and the TEC may enable She2p to compete effectively with these RNA-binding proteins. In the spt4 and spt5 mutants, She2p still interacts with the nascent ASH1 mRNA, but it does so less efficiently.
Since the Spt4–Spt5 complex, also known as DSIF in metazoans, is highly conserved from yeast to humans (Wada et al. 1998), it may play a similar role in promoting the assembly of mRNA localization complexes in metazoans. Indeed, Spt5 regulates the transcription of the pair rule mRNAs hairy, runt, and even-skipped, which are localized to the apical cytoplasm of the Drosophila blastoderm embryo (Jennings et al. 2004). Also, in Drosophila, the hsp83 mRNA, which is localized to the posterior pole plasm by a protection/degradation mechanism, is regulated by Spt5 (Andrulis et al. 2000). Finally, ChIP in the zebrafish embryo showed that Spt5 is associated with the β-actin gene, whose transcript localizes to the neurite outgrowth and fibroblast leading edge in vertebrates (Krishnan et al. 2008).
Based on our observation that She2p interacted with RNA pol II independently of its RNA-binding function, we hypothesize that She2p is recruited by the TEC prior to its transfer to nascent bud-localized mRNAs. Consistent with this model, ChIPs show that She2p is associated with pol II transcribed genes coding for both nonlocalized and bud-localized mRNAs, but this association is in part RNA-dependent only for localized transcripts. Altogether, these data outline a mechanism for the cotranscriptional assembly of mRNPs competent for localization in the cytoplasm (Fig. 7B). This model suggests that the TEC-associated She2p may “sample” nascent transcripts on pol II transcribed genes, and bind only nascent mRNAs destined for localization at the bud. Finally, one could speculate that such a model may even apply to other mechanisms regulating the cytoplasmic fate of a transcript, like its translational control or decay, which may also be primed cotranscriptionally.
Materials and methods
Growth media and yeast strains
Yeast cells were grown in either synthetic growth media lacking the nutrients indicated or rich media (Rose et al. 1990). Transformation was performed according to the protocol of Schiestl and Gietz (1989). A yeast gene disruption cassette was created by PCR amplification of the loxP-KAN-loxP construct in plasmid pUG6 and primers specifics for the gene of interest (Guldener et al. 1996). Specific disruption was confirmed by PCR analysis of genomic DNA. Yeasts strains and plasmids used in this study are described in Supplemental Tables 1 and 2, respectively.
ChIP
ChIPs were performed essentially as described (Ezhkova and Tansey 2006; Gilbert and Svejstrup 2006). Briefly, 100 mL of cells was grown to early log phase (OD600 ∼1) at 30°C in YPD or the appropriate medium. For galactose induction, galactose was added to cultures to a final concentration of 3% or 2% glucose for the negative control. To increase the number of cells with ASH1 transcription, yeasts were treated 2 h with nocodazole (at a final concentration of 15 μg/mL), then washed once with YPD. Formaldehyde was added to a final concentration of 1%, and cells were incubated for 20 min at room temperature. The cross-linking time was reduced from 20 min to 5 min when RNase treatment was performed. Glycine was added to a final concentration of 300 mM. Cells were washed twice in 1× PBS and lysed with glass beads in FA lysis buffer (50 mM HEPES/KOH at pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton, 0.1% sodium deoxycholate, 40 U RNasin/mL [Promega], 88 μg/mL PMSF, 10 μg/mL leupeptin, 10 μg/mL pepstatin, 5 μg/mL aprotinin). Cells were lysed by vortexing five times for 30 sec, with a pause of l min on ice between each vortex. A hole was punctured at the bottom of the tube with a hot 21-G needle, and cross-linked chromatin was collected by centrifugation for 5 min at top speed in a benchtop centrifuge. Pellets were washed twice in FA buffer and resuspended in 600 μL of FA buffer. Samples were sonicated for three cycles of a 20-sec pulse followed by a 60-sec pause on ice using a 100 Sonic Dismembranator (Fisher Scientific). Chromatin was sonicated to yield an average size of 300- to 500-base-pair (bp) DNA fragments.
Immunoprecipitations were performed as follows. Purified 9E10 antibody (Oncogene Science) against myc or monoclonal antibodies 8WG16 against total RNA pol II (Abcam) was coupled to protein A/G-sepharose beads. After 3 h incubation at 4°C on a rotator, beads were washed successively with 0.7 mL of FA lysis buffer, then 1 mL of FA 500 buffer (50 mM HEPES/KOH at pH 7.5, 500 mM NaCl, 1 mM EDTA, 1% Triton, 0.1% sodium deoxycholate, 40 U RNasin/mL), 0.7 mL of LiCl wash (10 mM Tris-Cl at pH 8.0, 250 mM LiCl, 0.5% [v/v] NP-40, 0.1% sodium deoxycholate, 1 mM EDTA), and, finally, 0.7 mL of TE and 100 mM NaCl. When an RNase treatment step was added to the ChIP protocol, chromatin from the same experiment was treated with four Kunitz units of RNase A (Sigma). After incubating for 45 min at room temperature, immunoprecipitations were performed as described above. Immunoprecipitated material was eluted from the beads by heating for 10 min at 65°C in 400 μL of 25 mM Tris-HCl (pH 7.5), 10 mM EDTA, and 0.5% SDS. To reverse cross-links, samples were treated with 1 mg/mL Proteinase K for 2 h at 37°C and overnight at 65°C. After extraction with phenol/chloroform, DNA was ethanol-precipitated 3 h in a dry-ice ethanol bath in the presence of 20 μg of glycogen, and resuspended in 40 μL of TE buffer.
The amount of DNA in the “input” and “IP” samples was quantified using real-time PCR (LightCycler 480, Caliper Life Sciences and Roche Applied Science). Real-time PCR reaction (20 μL) contained 2 μL of sample DNA, 10 μL of SYBR green I master mix (Roche), and 0.25 mM each primer. PCR was performed under the following conditions: preincubation for 5 min at 95°C, followed by 45 cycles of 10 sec at 94°C, 15 sec at 55°C, and 10 sec at 72°C. Each “input” and “IP” sample was analyzed in triplicate using real-time PCR, and the resulting values were averaged to determine the concentration of each sample. The primers used in this study are described in Supplemental Table 3. The PMA1, FBA1, PGK1, ASC1, tRNAGUC, tRNACUU, and intergenic primers (IntergenicV-1 and IntergenicV-2) have been described previously (Zenklusen et al. 2002; Abruzzi et al. 2004). The calculations and normalizations for each ChIP were carried out as follows. Briefly, after completing PCR cycles, the cycle threshold (Ct) was determined for each reaction. We calculated the average of the cycle threshold for each triplicate set of reactions. The enrichment of the protein of interest (i.e., She2p or Rpb1p) above background was calculated by dividing the relative immunopreciptiation signal for the gene-specific primer pair by the the relative input signal for the gene-specific primer pair. The “intergenic” primer pair is in a nontranscribed region of chromosome IV, and accounts for nonspecific DNA binding. A fold enrichment of 1 indicates that the amount of DNA immunoprecipitated by the protein of interest is the same as from nonspecific DNA binding.
Acknowledgments
We thank Drs. Grant Hartzog and Jackie Vogel for reagents and strains. We thank Amélie Forget for the artwork. We also thank Emmanuelle Querido and Daniel Zenklusen for critical reading of the manuscript. This work was supported by grant MOP43855 from the Canadian Institutes for Health Research (CIHR). P.C is a Senior Scholar from the Fond de Recherche en Santé du Québec (FRSQ).
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1937510.
Supplemental material is available at http://www.genesdev.org.
References
- Abruzzi KC, Lacadie S, Rosbash M 2004. Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes. EMBO J 23: 2620–2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrulis ED, Guzman E, Döring P, Werner J, Lis JT 2000. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: Roles in promoter proximal pausing and transcription elongation. Genes Dev 14: 2635–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronov S, Gelin-Licht R, Zipor G, Haim L, Safran E, Gerst JE 2007. mRNAs encoding polarity and exocytosis factors are cotransported with the cortical endoplasmic reticulum to the incipient bud in Saccharomyces cerevisiae. Mol Cell Biol 27: 3441–3455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohl F, Kruse C, Frank A, Ferring D, Jansen R-P 2000. She2p, a novel RNA-binding protein tethers ASH1 mRNA to the Myo4p myosin motor via She3p. EMBO J 19: 5514–5524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S 2009. Progression through the RNA polymerase II CTD cycle. Mol Cell 36: 541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartrand P, Meng X-H, Singer RH, Long RM 1999. Structural elements required for the localization of ASH1 mRNA and of a green fluorescent protein reporter particle in vivo. Curr Biol 9: 333–336 [DOI] [PubMed] [Google Scholar]
- Chartrand P, Singer RH, Long RM 2001. RNP localization and transport in yeast. Annu Rev Cell Dev Biol 17: 297–310 [DOI] [PubMed] [Google Scholar]
- Darzacq X, Powrie E, Gu W, Singer RH, Zenklusen D 2003. RNA asymmetric distribution and daughter/mother differentiation in yeast. Curr Opin Microbiol 6: 614–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Singer RH, Gu W 2008. Translation of ASH1 mRNA is repressed by Puf6p–Fun12p/eIF5B interaction and released by CK2 phosphorylation. Genes Dev 22: 1037–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Talia S, Wang H, Skotheim JM, Rosebrock AP, Futcher B, Cross FR 2009. Daughter-specific transcription factors regulate cell size control in budding yeast. PLoS Biol 7: e1000221 doi: 10.1371/journal.pbio.1000221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du T-G, Schmid M, Jansen R-P 2007. Why cells move messages: The biological functions of mRNA localization. Semin Cell Dev Biol 18: 171–177 [DOI] [PubMed] [Google Scholar]
- Du TG, Jellbauer S, Müller M, Schmid M, Niessing D, Jansen RP 2008. Nuclear transit of the RNA-binding protein She2 is required for translational control of localized ASH1 mRNA. EMBO Rep 9: 781–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E, Tansey WP 2006. Chromatin immunoprecipitation to study protein–DNA interactions in budding yeast. Methods Mol Biol 313: 225–244 [DOI] [PubMed] [Google Scholar]
- Farina KL, Singer RH 2002. The nuclear connection in RNA transport and localization. Trends Cell Biol 12: 466–472 [DOI] [PubMed] [Google Scholar]
- Gilbert C, Svejstrup JQ 2006. RNA immunoprecipitation for determining RNA–protein associations in vivo. Curr Protocols Mol Biol 75: 27.4.1–27.4.11 [DOI] [PubMed] [Google Scholar]
- Gonsalvez GB, Lehmann KA, Ho DK, Stanitsa ES, Williamson JR, Long RM 2003. RNA–protein interactions promote asymmetric sorting of the ASH1 mRNA ribonucleoprotein complex. RNA 9: 1383–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Deng Y, Zenklusen D, Singer RH 2004. A new yeast PUF family protein, Puf6p, represses ASH1 mRNA translation and is required for its localization. Genes Dev 18: 1452–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guldener U, Heck S, Fielder T, Beinhauer J, Hegemann J 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res 24: 2519–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzog GA, Wada T, Handa H, Winston F 1998. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev 12: 357–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CK, Shuman S 1999. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol Cell 3: 405–411 [DOI] [PubMed] [Google Scholar]
- Irie K, Tadauchi T, Takizawa PA, Vale RD, Matsumoto K, Herskowitz I 2002. The Khd1 protein, which has three KH RNA-binding motifs, is required for proper localization of ASH1 mRNA in yeast. EMBO J 21: 1158–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambhekar A, McDermott K, Sorber K, Shepard KA, Vale RD, Takizawa PA, DeRisi JL 2005. Unbiased selection of localization elements reveals cis-acting determinants of mRNA bud localization in Saccharomyces cerevisiae. Proc Natl Acad Sci 102: 18005–18010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RP, Dowzer C, Michaelis C, Galova M, Nasmyth K 1996. Mother cell-specific HO expression in budding yeast depends on the unconventional myosin Myo4p and other cytoplasmic proteins. Cell 84: 687–697 [DOI] [PubMed] [Google Scholar]
- Jennings BH, Shah S, Yamaguchi Y, Seki M, Phillips RG, Handa H, Ish-Horowicz D 2004. Locus-specific requirements for Spt5 in transcriptional activation and repression in Drosophila. Curr Biol 14: 1680–1684 [DOI] [PubMed] [Google Scholar]
- Komarnitsky P, Cho E-J, Buratowski S 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. GenesDev 14: 2452–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komili S, Silver PA 2008. Coupling and coordination in gene expression processes: A systems biology view. Nat Rev Genet 9: 38–48 [DOI] [PubMed] [Google Scholar]
- Krishnan K, Salomonis N, Guo S 2008. Identification of Spt5 target genes in zebrafish development reveals its dual activity in vivo. PLoS ONE 3: e3621 doi: 10.1371/journal.pone.0003621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF 2002. RNA polymerase II elongation factors of Saccharomyces cerevisiae: A targeted proteomics approach. Mol Cell Biol 22: 6979–6992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse C, Jaedicke A, Beaudouin J, Bohl F, Ferring D, Guttler T, Ellenberg J, Jansen RP 2002. Ribonucleoprotein-dependent localization of the yeast class V myosin Myo4p. J Cell Biol 159: 971–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Geiger G, Minet M, Schroeder S, Cilli K, McNeil JB, Bentley DL 2002. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol Cell 9: 1101–1111 [DOI] [PubMed] [Google Scholar]
- Lindstrom DL, Squazzo SL, Muster N, Burckin TA, Wachter KC, Emigh CA, McCleery JA, Yates JR III, Hartzog GA 2003. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol Cell Biol 23: 1368–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Warfield L, Zhang C, Luo J, Allen J, Lang WH, Ranish J, Shokat KM, Hahn S 2009. Phosphorylation of the transcription elongation factor Spt5 by yeast Bur1 kinase stimulates recruitment of the PAF complex. Mol Cell Biol 29: 4852–4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long RM, Singer RH, Meng X, Gonzalez I, Nasmyth K, Jansen R-P 1997. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science 277: 383–387 [DOI] [PubMed] [Google Scholar]
- Long RM, Gu W, Lorimer E, Singer RH, Chartrand P 2000. She2p is a novel RNA-binding protein that recruits the Myo4p–She3p complex to ASH1 mRNA. EMBO J 19: 6592–6601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin KC, Ephrussi A 2009. mRNA localization: Gene expression in the spatial dimension. Cell 136: 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Proudfoot NJ 2009. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136: 688–700 [DOI] [PubMed] [Google Scholar]
- Oeffinger M, Wei KE, Rogers R, DeGrasse JA, Chait BT, Aitchison JD, Rout MP 2007. Comprehensive analysis of diverse ribonucleoprotein complexes. Nat Methods 4: 951–956 [DOI] [PubMed] [Google Scholar]
- Oleynikov Y, Singer RH 2003. Real-time visualization of ZBP1 association with β-actin mRNA during transcription and localization. Curr Biol 13: 199–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier C, Poirier G, Gendron P, Boisgontier A, Major F, Chartrand P 2005. Identification of a conserved RNA motif essential for She2p recognition and mRNA localization to the yeast bud. Mol Cell Biol 25: 4752–4766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F, Huttelmaier S, Singer RH, Gu W 2007. ZBP2 facilitates binding of ZBP1 to β-actin mRNA during transcription. Mol Cell Biol 27: 8340–8351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquin N, Ménade M, Poirier G, Donato D, Drouet E, Chartrand P 2007. Local activation of yeast ASH1 mRNA translation through phosphorylation of Khd1p by the casein kinase Yck1p. Mol Cell 26: 795–809 [DOI] [PubMed] [Google Scholar]
- Perales R, Bentley D 2009. Cotranscriptionality: The transcription elongation complex as a nexus for nuclear transactions. Mol Cell 36: 178–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterlin BM, Price DH 2006. Controlling the elongation phase of transcription with P-TEFb. Mol Cell 23: 297–305 [DOI] [PubMed] [Google Scholar]
- Phatnani PH, Greenleaf AL 2006. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev 20: 2922–2936 [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Hannett NM, Young RA 2002. Exchange of RNA polymerase II initiation and elongation factors during gene expression. In Vivo 9: 799–809 [DOI] [PubMed] [Google Scholar]
- Qiu H, Hu C, Wong C-M, Hinnebusch AG 2006. The Spt4p subunit of yeast DSIF stimulates association of the Paf1 complex with elongating RNA polymerase II. Mol Cell Biol 26: 3135–3148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H, Hu C, Hinnebusch AG 2009. Phosphorylation of the Pol II CTD by KIN28 enhances BUR1/BUR2 recruitment and Ser2 CTD phosphorylation near promoters. Mol Cell 33: 752–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Gil A, Garcia-Martinez J, Pelechano V, Munoz-Centeno MC, Geli V, Perez-Ortin JE, Chavez S 2010. The distribution of active RNA polymerase II along the transcribed region is gene-specific and controlled by elongation factors. Nucleic Acids Res. doi: 10.1093/nar/gkq215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P 1990. Methods in yeast genetics. A laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Schiestl R, Gietz RD 1989. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet 16: 339–346 [DOI] [PubMed] [Google Scholar]
- Shen Z, Paquin N, Forget A, Chartrand P 2009. Nuclear shuttling of She2p couples ASH1 mRNA localization to its translational repression by recruiting Loc1p and Puf6p. Mol Biol Cell 20: 2265–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard KA, Gerber AP, Jambhekar A, Takizawa PA, Brown PO, Herschlag D, DeRisi JL, Vale RD 2003. Widespread cytoplasmic mRNA transport in yeast: Identification of 22 bud-localized transcripts using DNA microarray analysis. Proc Natl Acad Sci 100: 11429–11434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sil A, Herskowitz I 1996. Identification of asymmetrically localized determinant, Ash1p, required for lineage-specific transcription of the yeast HO gene. Cell 84: 711–722 [DOI] [PubMed] [Google Scholar]
- Takizawa PA, Vale RD 2000. The myosin motor, Myo4p, binds Ash1 mRNA via the adapter protein, She3p. Proc Natl Acad Sci 97: 5273–5278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa PA, Sil A, Swedlow J, Herskowitz I, Vale R 1997. Actin-dependent localization of an mRNA encoding a cell-fate determinant in yeast. Nature 389: 90–93 [DOI] [PubMed] [Google Scholar]
- Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, et al. 1998. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev 12: 343–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y, Shatkin AJ 1999. Transcription elongation factor hSPT5 stimulates mRNA capping. Genes Dev 13: 1774–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D, Vinciguerra P, Wyss J-C, Stutz F 2002. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol Cell Biol 22: 8241–8253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K, Kuo WHW, Fillingham J, Greenblatt JF 2009. Control of transcriptional elongation and cotranscriptional histone modification by the yeast BUR kinase substrate Spt5. Proc Natl Acad Sci 106: 6956–6961 [DOI] [PMC free article] [PubMed] [Google Scholar]