Abstract
The recent availability of genome-scale genotyping data has led to the identification of regions of the human genome that seem to have been targeted by selection. These findings have increased our understanding of the evolutionary forces that affect the human genome, have augmented our knowledge of gene function and promise to increase our understanding of the genetic basis of disease. However, inferences of selection are challenged by several confounding factors, especially the complex demographic history of human populations, and concordance between studies is variable. Although such studies will always be associated with some uncertainty, steps can be taken to minimize the effects of confounding factors and improve our interpretation of their findings.
The past few years have seen an explosion of studies using molecular data to detect Darwinian natural selection1–6. With the recent availability of large-scale genotyping data, genome-wide scans for genes or genomic regions that have been targeted by selection have become feasible. These studies have greatly advanced our understanding of human evolution and molecular evolution in general, but they have also sparked considerable controversy.
The interest in detecting selection is twofold. First, it stems from a natural curiosity about our evolutionary past and the basic mechanisms that govern molecular evolution. Much of the work in this field through the past four decades has focused on quantifying the relative importance of Darwinian selection and random genetic drift in determining levels of variability within species, as well as divergence between species (for example, REFS 7,8). However, as evidence accumulates for a strong role of selection, efforts are increasingly concentrating on identifying and characterizing particular instances of selection and adaptation at the molecular level. In humans, in particular, there has been a strong interest in identifying genes that have undergone recent selection relating to key human traits such as cognitive abilities4,9,10.
A second motivation for studying selection stems from the realization that inferences about selection can provide important functional information. For example, genes that are targeted by selection acting on segregating mutations are more likely to be associated with disease (for example, REF. 3). Even small fitness effects can, on an evolutionary timescale, leave a distinct pattern. Therefore, it might be possible to identify putative genetic disease factors by identifying regions of the human genome that currently are under selection3,11. In general, positions in the genome that are under selection must be of functional importance, otherwise selection could not be operating.
The aim of this Review is to discuss some of the major findings regarding selection in humans, and explain why the conclusions of these studies have at times been controversial with low levels of concordance among studies. We focus particularly on recent selection; that is, selection that might have affected current population genetic variation. We first address the question of the likely relative contributions of negative and positive selection to genetic variation in human populations, and explore how identifying these types of selection might contribute to our understanding of human evolution and gene function. We then discuss the different approaches that are taken to detecting the recent and ongoing positive selection that has affected the human genome, followed by a detailed discussion of recent genome-wide studies that have provided many new potentially selected genomic regions and individual genes. The key problems that face studies of selection are then addressed, along with a discussion of why low concordance has been seen among some of the studies that have been carried out so far. Finally, we bring together a discussion of studies that have provided insights into the patterns of selection that are likely to characterize Mendelian and complex human diseases, with the potential to aid the discovery of further disease-associated genes.
Positive and negative selection
Although it is clear that selection is pervasive in humans and other organisms, the relative importance of positive and negative selection is still debated. Much of the natural selection acting on genomes may be negative selection acting to remove new deleterious mutations. Most exons in protein-coding regions are highly conserved between species, because many potential mutations would disrupt protein function. Therefore, the conservation of genic regions provides evidence of past negative selection and provides an important route to genome annotation. Similar evidence for conservation and negative selection in non-coding regions provides the basis for an important approach for detecting functional elements, such as microRNAs (for example, REFS 12,13).
Eyre-Walker and Keightley14 estimated that at least 38% of all new amino-acid altering mutations in the human genome are being eliminated by negative selection, assuming that all mutations are either deleterious or neutral (that is, having no effect on organismal fitness). As noted by the authors of this study, this is probably an underestimate, and subsequent studies15–17 have suggested that as a much as 70–75% of amino-acid altering mutations are affected by moderate or strong negative selection. Importantly, however, much of this selection might act at the level of gametogenesis, on mature gametes or during early development. Mutations that are strongly deleterious will be quickly eliminated by natural selection, and only mutations that have, at worst, a mildly negative fitness effect will be observed as segregating in the population. A. R. Boyko et al. (unpublished observations) estimated that the proportion of amino-acid altering mutations in humans that have a negative fitness effect, but are so weakly selected that they might still be segregating in the population, is approximately 30–40%.
Positive selection occurs when a new (or previously rare) mutation confers a fitness advantage to the individuals carrying it. Much attention has focused on this type of selection because it provides the footprints of evolutionary adaptation at the molecular level. Identifying genomic regions that have been influenced by positive selection provides a key to understanding the processes that lead to differences among species and a subset of heritable phenotypic differences within species. For example, we can learn much about the biological basis of the evolution of modern humans by studying how positive selection has affected the human genome over the past few hundred thousand years. In general, positive selection is relevant when we seek to understand species-specific adaptations or processes that relate to dynamic interactions between the organism and its environment.
The signature of positive selection
Comparative studies
If positive selection acts on protein- coding genes, and if it occurs by repeated rounds of favouring multiple mutations in a gene (that is, selection is recurrent), positive selection can be detected as an increased rate of amino-acid substitution. Many studies have recently taken advantage of this fact to quantify positive selection acting in the genome on the human lineage leading from the ancestor of human and chimpanzees to modern humans18–20. In general, these studies have identified genes involved in immune-related functions, spermatogenesis, olfaction and sensory perception, and have highlighted several other functional gene categories with an increased likelihood of having experienced positive selection. Genes in these categories are likely to be involved in direct interactions with the environment, and will be under selective pressure in the face of environmental change. In particular, genes involved in dynamic competitive or co-evolutionary interactions are expected to experience more positive selection. A prime example of this is immunity and defence-related genes, which are involved in dynamic interactions with pathogens. As a category, these genes have experienced by far the most positive selection in humans and other organisms18–20.
There are several theories regarding selection acting on spermatogenesis, one being that most selection is related to post-mating competition between sperm from different males for fertilization21. In this case, the changing environment is the phenotype of sperm from other males. Alternative theories suggest that the selection is related to interactions between egg and sperm cells22,23, or that it is driven by selfish mutations causing segregating distortion20.
Several individual genes that might underlie human-specific adaptations have been highlighted in interspecies studies, including genes involved in speech and cognition, such as forkhead box P2 (FOXP2), genes associated with pregnancy, such as the progesterone receptor (PGR), genes associated with skeletal development, such as tolloid-like 2 (TLL2), and numerous other genes4,18. However, genomic comparisons between species alone do not inform us about ongoing and recent selection within species, and have little power if genes have been affected by only a single, recent selective event, even if the strength of selection acting on the mutation is strong. To detect such selection, population genetic data are needed. These data are informative about selection that occurs less than approximately 4Ne generations ago (where Ne is the effective population size).
Intraspecific studies
As a positively selected mutation increases in frequency in the population, it leaves a distinct mark on the pattern of genomic variation (FIG. 1). The pattern that is produced by such a ‘selective sweep’24 includes a reduction in the amount of variation, a temporary increase in the strength of linkage disequilibrium and a skew in the distribution of allele frequencies towards more alleles of low frequencies24–28 in the genomic region around the selected mutation. Because of recombination, the effect will be strongest in the immediate vicinity of the selected mutations, and will diminish with increasing genetic distance from them. When the selective sweep is ‘complete’, that is, when the favoured allele goes to fixation, all local variation is removed except that which has arisen by mutation and recombination during the sweep. If the sweep was rapid, then local variation will diminish to zero followed by a re-accumulation of variation through the combined processes of mutation and recombination, with a resulting site frequency spectrum that is skewed strongly towards rare alleles (FIG. 1).
Figure 1. Selective sweeps.
The lines indicate individual DNA sequences or haplotypes, and derived SNP alleles are depicted as stars. A new advantageous mutation (indicated by a red star) appears initially on one haplotype. In the absence of recombination, all neutral SNP alleles on the chromosome in which the advantageous mutation first occurs will also reach a frequency of 100% as the advantageous mutation become fixed in the population. Likewise, SNP-alleles that do not occur on this chromosome will be lost, so that all variability has been eliminated in the region in which the selective sweep occurred. However, new haplotypes can emerge through recombination, allowing some of the neutral mutations that are linked to the advantageous mutation to segregate after a completed selective sweep. As the rate of recombination depends on the physical distance among sites, the effect of a selective sweep on variation in the genomic regions around it diminishes with distance from the site that is under selection. Chromosomal segments that are linked to advantageous mutations through recombination during the selective sweep are coloured yellow. Data that are sampled during the selective sweep at a time point when the new mutation has not yet reached a frequency of 100% represent an incomplete selective sweep.
Much interest has focused on identifying incomplete selective sweeps, which are seen when positively selected mutations are currently on the rise in the human populations but have not yet reached a frequency of 100%. The pattern that is left by such mutations is distinctive, involving some locally identical haplotypes that segregate at moderate or high frequencies, whereas the remaining haplotypes show normal levels of variability (FIG. 1).
One of the most famous examples of an incomplete sweep is that at the lactase (LCT) locus in European populations. Variants in this gene influence whether the ability to produce lactase, which enables the digestion of milk, persists into adulthood. Lactase persistence is thought to have increased in frequency as a result of positive selection during the past 10,000 years after the emergence of dairy farming29–33. The striking pattern of genomic variability that is observed in this locus involves a long, high-frequency haplotype that contains an allele associated with lactase persistence34 (FIG. 2). The haplotypes that carry the allele are almost identical in regions close to the location of the causative SNP, whereas haplotypes that do not carry the allele show a normal level of variability. This is exactly the pattern we would expect to observe if the allele has recently increased in frequency as a result of positive selection. Even more striking is that some African populations that use dairy farming also carry a high-frequency, long-range haplotype associated with lactase persistence, but the mutation is distinct from the one observed in Europeans35. Another gene that shows almost as strong evidence for an incomplete selective sweep is the glucose-6-phosphate dehydrogenase gene (G6PD): deficiency alleles confer resistance to malaria and show a signature of positive selection36–38.
Figure 2. The signature of an incomplete selective sweep in the region containing the lactase (LCT) gene.
The LCT region shows a characteristic signature of an incomplete selective sweep. There is a haplotype of high frequency with strongly increased homozygosity as illustrated by the iHS (integrated haplotype score) statistic (data from REF. 40). There is a skew in the frequency spectrum as illustrated by the negative values of Tajima’s D (data from REF. 45), and a reduction in variability as shown by the estimate of the population genetic parameter θ (I. Hellmann, unpublished observations). Characteristically of many regions that show statistical evidence for an incomplete selective sweep, there is also a reduction in the local recombination rate (cM Mb−1; data from REF. 70). For the top two panels, the red lines represent the Asian and the blue lines represents the CEPH HapMap samples.
Genome-wide scans for sweeps in humans
The LCT and G6PD loci provide some of the most striking examples of ongoing selective sweeps in the human genome. These genes were identified a priori as candidate genes on the basis of functional information. Several recent papers have aimed at detecting loci under positive selection without such prior knowledge, based on genome-wide genotyping data. These methods can be used equally well to detect selection in non-coding and protein-coding regions, but the results are usually interpreted in terms of predictions for protein-coding regions, because most functional annotation is focused on genes. The following discussion will, therefore, also focus mostly on the results obtained for protein-coding regions.
Haplotype-based scans
The striking haplotype pattern that is observed at the LCT locus helped to motivate the development of the relative extended haplotype homozygosity (rEHH) and integrated haploytype score (iHS) tests for incomplete selective sweeps39,40 (BOX 1). These methods identify selection when a high-frequency haplotype with little intra-allelic variability is observed. The most comprehensive application of these methods made use of samples from the International HapMap Project, using some 800,000 SNPs in 89 Japanese and Chinese, 60 European and 60 Yoruban individuals40. Although there was significant overlap among populations, much of the evidence for selection was found to be specific to just one of them. This is not surprising as these methods have power primarily to detect incomplete selective sweeps, which might not have spread among different human populations. In addition to LCT and G6PD loci, Voight et al.40 also found signatures of selection for the 17q21 inversion in Europeans41, many cytochrome P450 genes, including CYP3A5 (REF. 42), the alcohol dehydrogenase (ADH) cluster in Asians43 and the olfactory-receptor clusters on chromosome 11 in Africans44. Cytochrome P450 genes, which are important in detoxification of plant secondary compounds, showed a signature of excess positive selection in all populations, with CYP3A5, CYP2E1 and CYP1A2 standing out. Finally, many genes involved in skin pigmentation showed signatures of positive selection outside of Africa, which would be consistent with the hypothesis that alleles that confer lighter skin colour have a selective advantage in regions that are further from the tropics. Interestingly, some of these same genes, and categories of genes, are also detected by comparative studies of selection based on human–chimpanzee divergence, including the olfactory-receptor clusters on chromosome 11 (REFS 18,20), suggesting that the selection acting on these genes occurred not only during recent human evolution, but also deeper in our ancestral past.
Box 1. Statistical methods for detecting selective sweeps.
Haplotype-based and linkage-disequilibrium-based methods
An influential approach for detecting recent and strong natural selection is the extended haplotype test91 and its derivatives40. The extended haplotype test relies on the linkage-disequilibrium structure of local regions of the genome. A haplotype at high frequency with high homozygosity that extends over large regions is a sign of an incomplete selective sweep. The method identifies tracts of homozygosity within a ‘core’ haplotype, using the ‘extended haplotype homozygosity’ (EHH) as a statistic. A relative EHH (rEHH) is calculated by comparing the EHH of the core haplotype to the EHH of all other haplotypes in the region. In the version by Voight et al.40, the EHH is summed over all sites away from a core SNP, and compared between the haplotypes that carry the ancestral and the derived allele in the SNP. The statistic (iHS — integrated haplotype score) is then normalized to have a mean of 0 and variance of 1. A related test was proposed by Wang et al.6, called the linkage-disequilibrium decay (LDD) test, which makes use of only homozygous SNP sites and therefore does not require separate phasing of haplotypes.
Site frequency spectrum (SFS)-based methods
Several classical methods for detecting selection are based on the distribution of allele frequencies in SNPs, or SFS. The SFS can be ‘unfolded’, in which case the spectrum tallies the counts in the sample of the derived (more recently arisen) allele. For a sample of k chromosomes, the unfolded SFS has the frequencies of chromosomes with 1, 2, 3, …, k–1 copies of the derived allele in the site. The ‘folded’ SFS is applied when one does not know which is the ancestral and which is the derived allele. In this case, the classes with j and k–j, where j < k/2, copies of the derived allele are not distinguishable, and so they are pooled. A selective sweep strongly affects the SFS, leading to a deficiency of alleles of intermediate frequency right after a selective sweep and an increase of such alleles during the selective sweep. Several methods for detecting selective sweeps take advantage of this fact46,92. Tajima’s D test46 detects an excess (indicated by negative values of D) or deficiency (indicated by positive values of D) of mutations of intermediate frequency relative to derived mutations that segregate at low or high frequencies. Fay and Wu’s47 test extends the Tajima’s D test by providing the power to detect an excess of high-frequency derived alleles, a clear signal of positive selection. The method of Kim and Stephan93 and its derivatives49,64 use the spatial pattern of the SFS to identify the location of a selective sweep.
Tests based on population subdivision
Locally increased levels of population subdivision can be caused by recent positive selection (for example, REF. 52). Several methods have been proposed for detecting selection based on this idea, for example, that of Akey et al.1, which identifies areas of increased FST, the traditional population genetic measure of population subdivision.
Tests based on levels of variability
A selective sweep causes a strong temporary reduction in the level of variability. The first and most well-known test of neutrality based on detecting regions is the HKA test94, which compares levels of diversity in different genes or genomic regions (calibrated by interspecific divergence rates) to test whether the rates are significantly increased or reduced in a particular region.
The length of the conserved haplotypes that can be detected by the extended haplotype-based tests depends on the timing and strength of selection. A strongly selected mutation, caught at a time when its frequency is around 0.5–0.7, will show the strongest signature. Two particular regions of the genomes that were highlighted in the Voight et al.40 study are noteworthy for the length of the haplotypes observed. Near the β-glucosidase gene (GBA), which is associated with Gaucher disease, a glycogen-storage disorder, East Asians have a common haplotype that extends 1.39 cM and well over 1 Mb in its physical span. Another gene that is associated with carbohydrate metabolism and blood-sugar regulation is NKX2-2, which has a 1.25-cM haplotype in the European population. It is tempting to speculate that whole-genome association studies will be able to detect differences in some physiological attributes between alternative genotypes in these regions, given the strength of selection that is implied by these large haplotypes.
Finally, Wang et al. Conducted a similar study using another haplotype-based test, the linkage-disequilibrium decay (LDD) test6 (Box 1) , to scan the genome for selection based on the HapMap data. They argued that 1,800 genes, or 1.6% of the genes in the genome, are currently undergoing selective sweeps. In contrast to Voight et al.40, they find that most of these regions are not specific to one population, but are shared among at least two. The categories of genes that show excess evidence for selection in this study included pathogen response, cell cycle, neuronal function, reproduction, DNA metabolism and protein metabolism.
Tests based on site frequency spectrum (SFS)
These tests use the allele frequencies in individual segregating nucleotide sites to detect selection (BOX 1). As previously mentioned, a selective sweep causes a skew in the distribution of allele frequencies towards more alleles of low frequencies. Carlson et al.45 used Tajima’s D46 and Fay and Wu’s H tests47 (BOX 1) to scan for distortions from the expected neutral-site frequencies in the human genome using the Perlegen data48, which was supported by additional sequencing of candidate genes. They identified 7, 23 and 29 aberrant regions for populations of African, European and Chinese descent, respectively. One region contained CYP3A4 and CYP3A5, which have a central role in the metabolism of some prescribed drugs. Another region contained vitamin K epoxide reductase complex, subunit 1 (VKORC1), which has been linked to human warfarin dosing. Carlson et al. Proposed that regions with extreme frequency spectra may provide important targets for genotype–phenotype studies45. However, they did not provide an analysis of the correlation of these regions with specific functional or biological categories of genes.
Williamson et al.5 used a composite-likelihood approach49 to detect selection, also based on the Perlegen SNP set48. This method compares models that are fitted to the data with and without a selective sweep to quantify the evidence in favour of such a sweep having occurred in a particular region of the genome. In contrast to haplotype-based methods, this method primarily detects recently completed selective sweeps, and is likely to have little or reduced power to detect incomplete sweeps. This is illustrated by the fact that the prime example of an incomplete sweep, the LCT locus, was not identified in this study. Williamson et al.5 found 101 genes for which there was strong evidence for a selective sweep having occurred (at a significance level of P < 10−5) and, as in Voight et al.40, there was wide variation among populations in locations of these regions. As in previous studies, genes that showed evidence of a selective sweep included those for olfactory receptors, as well as genes related to the nervous system, pigmentation and immunity. In addition, the method found increased signatures of selective sweeps around centromeres. There are several possible explanations for this pattern, one of which is increased selection associated with genetic elements that are responsible for meiotic drive in these regions50.
Evidence based on population subdivision
Another approach for detecting ongoing selection is to study between-population differences in allele frequencies. Positive selection may increase levels of genetic differentiation among populations for two reasons. First, selection might act locally and be related to adaptations to the local environment. An example might be genes relating to skin pigmentation in humans where selection is possibly related to adaptation to the local climate. Second, selective sweeps acting on mutations that arise in specific geographical regions might cause increased levels of population subdivision during the period of time in which the mutation is still increasing in frequency51,52. Even if the mutation is beneficial in all environments, the fact that the mutation arose in a particular geographical location might temporarily increase levels of population differentiation in the genomic region that is affected by selection. For example, allele frequencies for the LCT locus differ dramatically among European populations and between Europe and other continents34. The genetic differentiation in this locus among geographical regions may not be primarily caused by differences in the strength or direction of selection among regions, but rather by a historical contingency. The beneficial mutation that confers lactose tolerance in adults might have arisen in Europe first, and might therefore have so far reached the highest frequency in Europe as opposed to other continents.
Although the first attempts to use population subdivision to identify selection dates back to 1973 (REF. 53), the methodology has been reviewed in the context of genome-wide studies1,54,55. In a study of 26,530 SNPs from African Americans, European Americans and East Asians, Akey et al.1 identified several candidate genes including the CFTR gene, the gene that is famously associated with cystic fibrosis (BOX 1). Weir et al.54 examined the HapMap data using a similar approach, and also identified several regions that showed a drastically increased level of population subdivision, including the LAC locus. The HapMap phase I publication56 also included the results of scanning genome-wide SNP data for evidence for selective sweeps based on haplo-type structure, population differentiation and SFS. They identified a number of candidate regions that harbour genes with extreme differences in allele frequencies among populations, such as ALMS1, which is associated with Alström syndrome, a rare hereditary disease with clinical features including hyperinsulinaemia, visual impairment and obesity.
Problems in identifying selection
Inferences about human adaptation are often controversial; this is particularly clear from the debate surrounding studies of the FOXP2, asp (abnormal spindle) homolog, microcephaly-associated (ASPM) and microcephalin (MCPH1) genes, for which claims of human-specific adaptation have been made (BOX 2). Unfortunately, there is little consensus in the field of population genetics about what should be considered convincing evidence of selection. In the following, we will discuss some of the statistical issues that plague inferences of selection.
Box 2. Adaptation in humans.
Much of the positive selection in humans, as well as in other organisms, is related to immune-defence functions and other processes that are unrelated to human-specific evolution. A list of some of the genes that show evidence for positive selection is given in REF. 4. However, there has recently been much interest in identifying loci that are involved in human-specific adaptation, particularly adaptation of cognitive skills. A prime example is the forkhead box P2 (FOXP2) gene, mutations in which cause deficiencies in language skills including grammatical competence, and are additionally associated with the motor-control of craniofacial muscles95–97. Only four FOXP2 mutations occur in the evolutionary tree of mice, macaques, orangutans, gorillas, chimpanzees and humans, two of which occur in the evolutionary lineage leading to humans. This relative speed-up in the evolution of this gene in humans is highly suggestive of positive selection1,10,98. One possible interpretation is that this selection introduced a change in the FOXP2 gene that was a necessary step to the development of speech. However, the selected phenotype could also have been unrelated to speech. FOXP2 is fairly ubiquitously expressed and also has an essential role in lung development. In addition, one of the mutations seems to have occurred independently on the carnivore lineage61, suggesting that this substitution might have been selected for reasons other than language development. Nonetheless, the gene shows a very negative value of Tajima’s D (–2.2) and other evidence for a recent selective sweep that might have coincided temporally with the emergence of anatomically modern humans10.
Other examples of genes for which there is evidence of potential involvement in human-specific adaptation include the asp (abnormal spindle) homologue, microcephaly-associated gene (ASPM) and the microcephalin gene (MCPH1). These genes are cell-cycle regulators, and loss-of-function mutations in these genes are known to cause several deleterious phenotypic effects, including reduced brain size. These genes have been proposed to have unusually long haplotypes and extensive geographical variation, which are both typical signs of recent ongoing selection9,99; the authors of these studies suggested that these patterns arose as a result of positive selection relating to increased brain size. However, Currat et al. 100 re-analysed their data and concluded that human demographic models that include population structure followed by population growth can explain the patterns observed for ASPM and MCPH1 without invoking selection. Furthermore, Yu et al.101 demonstrated that ASPM is not unusual compared with other anonymously selected regions in the genome, with respect to tests for selection based on population subdivision, haplotype structure and frequency spectrum, and argued that recent positive selection at this locus is unlikely. In addition, subsequent tests for an association between IQ and specific variants of ASPM and MCPH1 proved to be negative102.
Demography
Methods for detecting selection have historically been challenged by the confounding effects of demography57–62. For example, it is well known57,60 that Tajima’s D will falsely reject neutrality in the presence of population bottlenecks or certain types of population structure (FIG. 3). For example, a recent bottleneck tends to mimic the effects of a selective sweep in several ways63,27. In light of this concern, there has been a considerable effort to develop more robust methods of detecting recent positive selection, and some recently described methods are highly robust under various demographic models that are relevant for human populations20,64.
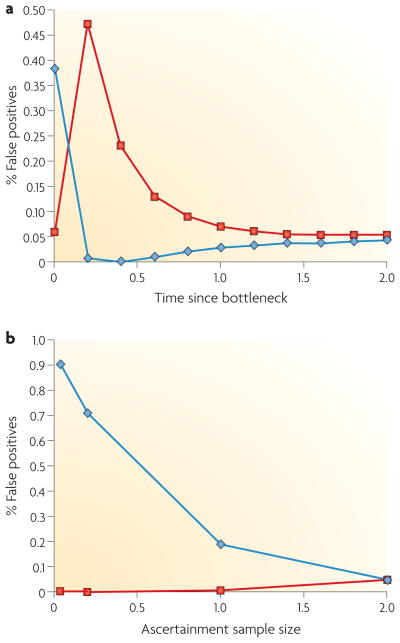
Figure 3. Effects of demography and ascertainment bias on tests of selection.
a. The false positive rate of Tajima’s D46 in the presence of a population bottleneck. A sample of 50 chromosomes was simulated using coalescent simulations105, and the time since a population bottleneck that reduced the population size tenfold was varied. The duration of the bottleneck was 0.1 × 2Ne generations, and time in the figure is measured in 2Ne generations, where Ne is the effective population size of a diploid population. Each simulated data set had 20 segregating sites. The proportion of time the test rejects at the 5% significance level in a one-sided test based on negative values (shown in red) and positive values (shown in blue) is shown. B. The false-positive rate of Tajima’s D46 in the presence of an ascertainment bias. Simulation conditions are as described in panel a, but the size of the ascertainment sample (expressed as a proportion of the final sample) used for SNP discovery is varied.
The issue of sensitivity to demographic factors for different types of test for selection has not been fully explored. However, some methods, such as the composite-likelihood ratio test, which is based on comparing allele frequencies in different parts of the genome, have been shown directly, using simulations, to be highly robust towards perturbations of the demographic model49. Other methods for detecting selective sweeps that are based on haplotypes are possibly also more robust than methods based on population subdivision or allele frequencies, because they compare variation between different allelic classes. Although the overall pattern of genetic variation can be strongly influenced by demographics, this influence might be smaller for the relative variability in different allelic classes. For example, under the simulation conditions of FIG. 3, we have found that the rEHH test never reveals more than 5% significant results at the 5% level when applied to a random haplotype. Although the test is unlikely to be applied to a random haplo-type in real-life applications, this finding suggests some robustness to assumptions about demography. Nonetheless, no method can claim to be 100% robust to such assumptions.
A particularly worrying effect that was recently discovered is the phenomenon of ‘allelic surfing’65,66. As a population expands geographically, rare alleles can, by random genetic drift, be caught on the haplotype with the favoured allele, and ride the wave in expansion of the favoured allele, increasing in frequency in restricted geographical areas. The resulting pattern includes large extended haplotypes, and may mimic selective sweeps. Allelic surfing may provide a new challenge that many current methods do not address, and it is currently unknown whether it will be possible to develop methods that can distinguish between allelic surfing and positive selection with great accuracy.
Ascertainment bias
Another fundamental problem is that ascertainment bias confounds much of the large-scale human genomic data, affecting allele frequencies, levels of population subdivision and patterns of linkage disequilibrium (BOX 3; FIG. 3b). This ascertainment bias arises when the data are obtained not through direct sequencing but by genotyping SNPs that have been discovered in another sample. Patterns of allele frequencies, linkage disequilibrium, population differentiation and so on that are observed in the data depend on the exact procedure that is used to discover SNPs for inclusion in the genotyping effort. The effects of this bias on neutrality tests can be particularly worrying when the SNP-discovery protocol differs among different genomic regions. Regions in which many sequences were used for ascertainment might appear to have more SNP alleles segregating at low frequencies and a more defined haplotype structure — that is, more haplotypes are represented in the data. The observed levels of population subdivision will also depend on the ethnic make-up of the SNP-discovery panel in the local genomic region. In such cases, apparent evidence for selection might simply be caused by variation in the discovery protocol among different genomic regions. Only when the SNP-discovery protocol is well defined can this effect can be controlled for statistically5,67,68. However, in cases in which the SNP-discovery protocol is not known, it might not be possible to directly control for this effect. In most studies, little or no attempt has been made to correct for ascertainment bias, and the effect of this bias on these studies is currently unknown. However, several studies, such as the study by Carlson et al.45, used only the Perlegen data48, and not the full HapMap data, because it is known that ascertainment does not vary among regions in the former data set. In the Voight et al.40 study, a simulation procedure was used to generate data with the same allele frequencies as in the HapMap data, in order to at least partially control for the effects of ascertainment biases.
Box 3. Ascertainment biases.
Almost all genome-wide human population genetic data sets are based on SNP genotyping data (so far, the prime exception is that of Bustmante et al.3, which is based on resequencing of coding genes). This type of data is obtained through a process in which the SNPs are first identified by direct sequencing of a small panel of individuals, and then subsequently genotyped in a larger panel. This two-stage process introduces biases in allele frequencies, patterns of linkage disequilibrium, overall levels of variability, and so on67,68,103,104. For example, because alleles that segregate at low frequencies are unlikely to be detected in the small panel that is initially sequenced, the genotyped data will be deficient in low-frequency alleles. The effect on the statistical methods for detecting selection can be severe. An example is shown in FIG. 3b, using the Tajima’s D46 statistic (BOX 1). When the ascertainment sample size is small, Tajima’s D49 is biased towards large values, with a decreased rate of false positives from below (shown in red in FIG. 3a) and an increased rate of false positives from above (shown in blue in FIG. 3a).
Although it is clear that some methods, for example methods that gain much of their information from the distribution of allele frequencies, such as Tajima’s D test46, will be largely invalidated by the ascertainment bias (FIG. 3b), the effect on other methods, such as methods based on haplotype patterns, is largely unexplored. The effect on patterns of linkage disequilibrium can be severe67, but the effect on many of the statistics that are used to detect selection is unclear. Tests based on haplotype structure, such as extended haplotype homozygosity (EHH) and integrated haplotype score (iHS), are commonly thought to be less affected by ascertainment bias because, even when there is a strong bias towards alleles of high frequency, haplotype structure might not be strongly affected as long as the selected SNPs adequately tag the haplotypes. For example, for a simple ascertainment scheme such as the one in FIG. 3b, with constant ascertainment based on the same chromosomes for all SNPs, the distribution of the relative EHH (rEHH) statistics for a randomly chosen haplotype is largely unaffected, with 6% false positives at the 5% significance level for an ascertainment sample size of n = 2.
Most methods can be corrected for ascertainment bias problems if the SNP-discovery protocol is well documented67. Unfortunately, such information is not available for much of the genome-wide data that have been generated for humans.
Recombination rate
Tests for selection can also be highly sensitive to assumptions about recombination rate. Regions of low recombination may produce an upward bias in detection of selection, because the variance in most statistics is increased in regions of low recombination. Distinct from the issue of bias, the statistical power of all of the tests will be a function of the recombination rate, owing to the fact that a selective sweep will leave a much stronger signal in regions of low recombination (FIG. 1). For example, one of the reasons the signal seems to be so strong for LCT might be that this gene occurs in a region of low recombination (FIG. 2).
We examined the average recombination rate for genes or regions that have been determined to be under selection in various studies (TABLE 1), based on the linkage maps of Kong et al.69 and the linkage-disequilibrium map of Meyers et al.70. The regions identified by some of the studies show a drastically reduced level of recombination. For example, the regions in the Wang et al.6 study show an average recombination rate of 0.49 cM Mb−1 and 0.26 cM Mb−1, according to the Kong et al.69 and Meyers et al.70 estimates, respectively. This should be compared with an average recombination rate in the genome of 1.29 cM Mb−1 and 1.33 cM Mb−1 from the two genetic maps, respectively. This does not directly show that the conclusions from the Wang et al.6 study, or any other study, are invalid. It is entirely possible that the low level of recombination in the regions identified in the Wang et al.6 study is caused by an increase in statistical power to detect selection in regions of low recombination. Nonetheless, the results of these studies should be interpreted in the context of the fact that they primarily identify regions of low recombination. An obvious solution to this problem in future studies is to use critical values that are specific to the local recombination rate, and/or to primarily compare regions with similar recombination rates71. It might also be preferable to use methods with low sensitivity to the local recombination rate. However, the relative robustness of different methods to assumptions about recombination rates has not been systematically explored.
Table 1.
Average recombination rates for human genes and regions identified as being under selection
| Study | Units | cM Mb−1 (from Kong et al.69) | cM Mb−1 (from Myers et al.70) | Number of regions or genes |
|---|---|---|---|---|
| Genome-wide average | N/A | 1.29 | 1.33 | N/A |
| Williamson et al.5 | Location of peak | 1.27 | 1.20 | 179 |
| Voight et al.40 | 100-kb windows | 0.92 | 0.76 | 713 |
| Carlson et al.45 | ~400-kb windows | 0.69 | 0.69 | 59 |
| Wang et al. (genes overlapping between all three populations)6 | Genes | 0.49 | 0.26 | 90 |
| Bustamante et al. (PS p < 0.025)3 | Genes | 1.35 | 1.26 | 301 |
| Bustamante et al. (NS p > 0.975)3 | Genes | 1.40 | 1.38 | 802 |
PS, positive selection; NS, negative selection; N/A, not available.
Assigning statistical significance
Because of the statistical uncertainties in detecting selection that are discussed above, several studies have not assigned p values to specific genes, but have simply relied on detecting outliers in the genome (for example, REF. 40). We stress that this is not the equivalent of a determination of statistical significance. Teshima et al. evaluated the accuracy of such outlier approaches using simulations and concluded that, although these approaches can identify many genes under selection, they may tend to give a biased view of which genes are under selection, depending on the specific assumptions that are used to define outliers71. Application of an outlier approach does not circumvent the problem of assigning statistical confidence. It is obviously preferable to have an accurately assigned p value which can be used to measure confidence in inferences about selection.
Concordance between results
One potentially worrying issue is that many of the studies described above have identified different sets of genes as having been subject to recent or ongoing selection. For example, only three regions have been identified as being under selection in each of the Williamson et al.5, Voight et al.40 and Carlson et al.45 studies. A summary of the overlap among gene sets is provided in TABLE 2. The relatively low rate of overlap is not surprising for comparisons of some of these studies, such as those of Williamson et al.5 and Voight et al.40, as these studies aimed to detect incomplete and completed sweeps, respectively, and are therefore expected to pick up different genes. The power of each test depends on the strength of selection, the time that has elapsed since the mutation arose, the dominance of the selected mutations and so on4,71,72.
Table 2.
Concordance between genome-wide studies of natural selection in the human genome
| Williamson et al.5 | Voight et al.40 | Carlson et al.45 | Wang et al.6 | Bustamante et al.3 (PS p < 0.025) | Bustamante et al.3 (NS p > 0.975) | |
|---|---|---|---|---|---|---|
| Williamson et al.5* | 179 | 12 | 20 | 0 | 0 | 4 |
| Voight et al.40* | 13 | 713 | 6 | 7 | 22 | 32 |
| carlson et al.45* | 23 | 7 | 59 | 5 | 3 | 10 |
| Wang et al.6* | 0 | 7 | 3 | 90 | 3 | 1 |
| Bustamante et al.3 (PS p < 0.025)‡ | 0 | 22 | 3 | 3 | 301 | # |
| Bustamante et al.3 (NS p > 0.975)‡ | 3 | 30 | 10 | 2 | # | 802 |
For each pairwise combination of studies, the table shows the number of genes that were identified as being under positive selection in both studies. The data from each study were, as far as possible, mapped to the University of California Santa Cruz (UCSC) hg16 genome assembly, using either the liftOver tool for genome coordinate data (*) or the NCBI Reference Sequence (RefSeq) (‡) and known gene tables for gene data. Values shown in the cells that run diagonally from top left to bottom right are the total count of genes that were found to be under selection in each study. PS, positive selection; NS, negative selection.
More worrying, however, is that only 7 of the 90 genes flagged by the Wang et al.6 study are also found among the 713 candidate genes of the Voight et al.40 study. Both aim at detecting incomplete selective sweeps based on extended haplotype methods. Although this might lead to some natural concern about the validity of the conclusions in genome-wide scans for selection, there are several reasons why we believe that the results of most of the studies discussed are reliable, if they are interpreted correctly. First, several of the methods have been evaluated extensively in simulation studies or by theory, and have been shown to be highly robust to the underlying assumptions3,5,49. Second, several of the methods find their strongest signal in regions that have been independently identified as being under selection (for example, REF. 40) such as the LCT region. And finally, we know from theory and simulations that all of the methods are sufficiently statistically sensitive to identify regions of strong selection. So, in the presence of selection, they will pick up selected loci. However, genome-wide studies should be interpreted in light of the fact that the false-negative and false-positive rates are often effectively unknown owing to statistical uncertainties, and that any set of candidate genes, except the most restrictive ones, can contain some false positives. Inferences of positive selection from population genetic data may always be associated with some uncertainty. However, as we learn more about the robustness of individual tests, the demographic history of human populations and the recombination landscape, this level of uncertainty is reduced.
Functional verification
The uncertainty regarding inferences of natural selection has led to calls for functional studies to verify claims of natural selection73. One example of such a functional study is that from Shu et al.74 showing that FOXP2 knockout mice are unable to produce certain vocalizations. However, it is important to realize that much of the selection that is occurring might be acting without any detectable phenotypic effects — it might be acting only under certain conditions (for example, in the presence of certain pathogens), and the selective effects might be so subtle that they are not easily measurable. Additionally, whereas functional effects are often caused by selection, functional differences alone do not demonstrate the past or present action of selection75. For example, the fact that humans from northern Europe have light-coloured hair does not in itself show that there has been selection for light hair colour in these populations. In theory, the evolutionary change in hair colour could have been caused by genetic drift as selection to maintain a dark hair colour has been relaxed, or it could have been caused by indirect effects due to selection relating to skin colour. Almost 30 years ago, Gould and lewontin76 launched a crusade against the adaptive paradigm that functional differences must be adaptive, that is, caused by natural selection. Although their arguments were controversial at the time, they have been highly influential on the community of evolutionary biologists. As a new generation of biologists with a background in genomics, molecular biology or bioinformatics has taken leadership in the field of genomic evolutionary biology, the old lessons from Gould and lewontin76 seem to have been forgotten. It is a desirable addition to a story of selection to identify possible functional reasons why selection might be acting, but it will never be a method for identifying or verifying selection.
Selection and disease
Genetic factors underlying both complex and Mendelian diseases should, under most circumstances, be affected by selection, because most diseases will have an effect on organismal survival or reproduction. One exception might be diseases with late onset under the assumption that older individuals do not contribute to the fitness of their offspring. Examples of negative selection acting on disease-causing mutations include mutations in GBA causing Gaucher disease77, mutations in nucleotide-binding oligomerization-domain-containing 2 gene (NOD2; also known as CARD15) causing Crohn disease78, mutations in CMH1, CMH2, CMH3, and CMH4 causing familial hypertrophic cardiomyopathy79, and a host of other genetic disorders that are caused by de novo mutations or segregating recessive mutations. If negative selection alone is operating, the disease mutations are expected to segregate at low frequencies, and to be predominantly recessive. However, there are rare examples of partially dominant disease mutations segregating, such as in some forms of familial hypertrophic cardiomyopathy79, in cases in which the fitness effects of the mutations are relatively low.
In some cases, disease-causing mutations can segregate at relatively high frequencies (for example, diabetes32,80–82), which is not easily explainable if only negative selection has been acting on the disease mutations. Possible explanations for this include balancing selection, for example, in the case of mutations in the G6PD locus or in the α-globin gene, which cause G6PD enzyme deficiency and sickle-cell anaemia, respectively, in the homozygous state, but can confer partial protection against malaria in the heterozygous state83,84. Another example is provided by mutations in the CFTR locus, which causes cystic fibrosis in the homozygous state but protects against asthma in the heterozygous state85.
Another possible explanation for the segregation of disease alleles at moderate or high frequencies is that genetic drift has acted on mutations that have only moderate fitness effects, possibly exacerbated by bottlenecks in the population size, as suggested for Gaucher disease in Askhenazi Jews77. Yet another explanation is that there might have been a recent change in the direction of selection. For example, according to the popular thrifty-genotype hypothesis86, selection has originally worked to maximize metabolic efficiency, especially in population groups that often encountered a scarcity of food. With (evolutionarily) recent dietary changes, the direction of selection might have been reversed, causing many common alleles that are now related to metabolic diseases and/or diabetes to be selected against. There are also other reasons why disease genes might be associated with positive selection; for example, an increased frequency of moderately deleterious mutations due to genetic hitch-hiking25 during a selective sweep.
Empirical studies
There is considerable interest in further elucidating the relationship between heritable diseases and selection. Bustamante et al.3 compared human genetic variation across more than 11,000 protein-coding genes that were re-sequenced in 39 individuals (19 African Americans and 20 European Americans). Comparing polymorphism and divergence between humans and chimpanzees at synonymous versus non-synonymous sites, they quantified the amount of positive or negative selection acting on each gene, including both current selection and that which has occurred during the shared evolutionary history of humans and chimpanzees. The study showed that mutations in evolutionarily constrained genes are disproportionately associated with heritable disorders. Specifically, although less than 12% of known genes have been associated with a Mendelian disorder, genes that show plentiful amino-acid variation in human populations (at least four amino-acid- replacement SNPs), but no divergence between humans and chimpanzees, have a 50% chance of causing at least one Mendelian disease. This is the expected pattern in genes if negative selection is acting on new mutations.
These results have been extended in a recent analysis by R. M. Bleckham et al. (unpublished observations) aimed at identifying differences in selective constraint among Mendelian disease genes, genes that contribute to complex disease and genes that are not associated with a disease. This study used the same data as in the Bustamante et al.3 study, in addition to divergence data from Human–Macaque comparisons to quantify selective constraints. They correlated their findings with a hand-curated version of the Mendelian Inheritance in Man database (OMIM), and concluded that Mendelian disease genes tend to be more constrained than those that contribute to non-Mendelian disease, with stronger purifying selection acting on genes with dominant rather than recessive disease mutations. As previously demonstrated by Thomas et al.87, they also found that genes that are implicated in complex diseases tend to be under less purifying selection than either Mendelian disease genes or non-disease genes, with some showing evidence of recent positive selection as reflected by high values of Tajima’s D. This might be taken as support for the thrifty-genotype hypothesis, but could also be consistent with balancing selection acting on these genes. A recent survey by Zlotogora88 discusses 14 common autosomal recessive diseases that show genetic heterogeneity, even in isolated populations. They argue that the pattern observed of multiple mutations segregating at high frequency can be adequately explained only if selection is, or has been, acting in favour of the mutations.
Whatever the role of positive and negative selection in explaining the presence of disease mutations, it is clear that there is a well-established relationship between disease status and selection that can be exploited when searching for the genetic causes for heritable diseases. This can be done at two levels: by selecting candidate loci using bioinformatical methods for detecting selection, or by prioritizing candidate SNPs or haplotypes by ranking them according to the magnitude of their inferred fitness effects. The latter methodology is already well-established in the use of computational methods for determining levels of conservation, such as SIFT89 or PolyPhen90.
Future perspectives
Although the field of evolutionary genetics has been challenged by difficulties in separating the effects of demographic history and natural selection, the availability of improved statistical methods has helped to improve the accuracy of predictions of natural selection. With the emerging full-scale re-sequencing of human genomes, other challenges relating to the currently available SNP genotyping data will be eliminated. The re-sequencing of large, diverse panels of human populations will help to settle many of the outstanding questions regarding selection in humans. In addition, it will provide a vehicle for exploiting the link between selection and disease, which will inform many studies on heritable diseases. However, even in the presence of large data sets based on full re-sequencing, there may be debates about historical evolutionary events that cannot be settled. Evolutionary biology is by and large a historical science, and practitioners in this field will have to learn to live with the uncertainty that arises from making inferences about an experiment of nature that cannot be repeated.
Acknowledgments
We would like to thank D. Reich, M. Przeworski and two anonymous reviewers for their helpful comments on earlier versions of this manuscript. This work was supported by Danmarks Grundforskningsfond and the US National Insitutes of Health grants R01HG003229 and U01HL084706.
- Genetic drift
The stochastic change in population frequency of a mutation due to the sampling process that is inherent in reproduction
- Adaptation
Heritable changes in genotype or phenotype that result in increased fitness
- Fitness
A measure of the capacity of an organism to survive and reproduce
- Effective population size (Ne)
The size of a population measured by the expected effect (through genetic drift) of the population size on genetic variablity. is typically much Ne lower than the actual population size (N)
- Selective sweep
The process by which new favourable mutations become fixed so quickly that physically linked alleles also become either fixed or lost depending on the phase of the linkage
- Linkage disequilibrium
A measure of genetic associations between alleles at different loci, which indicates whether allelic or marker associations on the same chromosome are more common than expected
- Fixation
Describes the situation in which a mutation has achieved a frequency of 100% in a natural population
- Site frequency spectrum
The distribution of allele frequencies in a single site of a DNA sequence averaged over multiple sites
- Haplotype
Allelic composition over a contiguous chromosome stretch
- Whole-genome association studies
Also known as genome-wide association studies. Genetic variants across the whole genome (or markers linked to these variants) are genotyped in a population for which phenotypic information is available (such as disease occurrence, or a range of different trait values). If a correlation is observed between genotype and phenotype, there is said to be an association between the variant and the disease or trait
- Meiotic drive
Any process that causes some alleles to be overrepresented in gametes formed during meiosis
- Population bottleneck
A marked reduction in population size followed by the survival and expansion of a small random sample of the original population
- Population structure
A departure from random mating that is typically caused by geographical subdivision
- Balancing selection
A selection regime that results in the maintenance of two or more alleles at a single locus in a population
- Genetic hitch-hiking
The increase if frequency of a selective neutral or weakly selected mutation due to linkage with a positively selected mutation
Footnotes
DATABASES
Entrez Gene: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene
ALMS1 | ASPM | CFTR | CYP1A2 | CYP2E1 | CYP3A4 | CYP3A5 | FOXP2 | GBA | G6PD | LCT | MCPH1 | NKX2-2 | NOD2 | PGR | TLL2 | VKORC1
OMIM: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=OMIM
Alström syndrome | Crohn disease | cystic fibrosis | familial hypertrophic cardiomyopathy
References
- 1.Akey JM, Zhang G, Zhang K, Jin L, Shriver MD. Interrogating a high-density SNP map for signatures of natural selection. Genome Res. 2002;12:1805–1814. doi: 10.1101/gr.631202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andolfatto P. Adaptive evolution of non-coding DNA in Drosophila. Nature. 2005;437:1149–1152. doi: 10.1038/nature04107. [DOI] [PubMed] [Google Scholar]
- 3.Bustamante CD, et al. Natural selection on protein-coding genes in the human genome. Nature. 2005;437:1153–1157. doi: 10.1038/nature04240. This paper reports a genome-wide scan for selection in humans based on a derivative of the MacDonald–Kreitman test. [DOI] [PubMed] [Google Scholar]
- 4.Sabeti PC, et al. Positive natural selection in the human lineage. Science. 2006;312:1614–1620. doi: 10.1126/science.1124309. This review contains a comprehensive list of genes that are thought to be under selection in humans. [DOI] [PubMed] [Google Scholar]
- 5.Williamson SH, et al. Localizing recent adaptive evolution in the human genome. PLoS Genet. 2007;3:e90. doi: 10.1371/journal.pgen.0030090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang ET, Kodama G, Baldi P, Moyzis RK. Global landscape of recent inferred Darwinian selection for Homo sapiens. Proc Natl Acad Sci USA. 2006;103:135–140. doi: 10.1073/pnas.0509691102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge Univ. Press; New York: 1983. [Google Scholar]
- 8.Gillespie JH. The Causes of Molecular Evolution. Oxford Univ. Press; New York: 1991. [Google Scholar]
- 9.Evans PD, et al. Microcephalin, a gene regulating brain size, continues to evolve adaptively in humans. Science. 2005;309:1717–1720. doi: 10.1126/science.1113722. [DOI] [PubMed] [Google Scholar]
- 10.Enard W, et al. Molecular evolution of FOXP2, a gene involved in speech and language. Nature. 2002;418:869–872. doi: 10.1038/nature01025. [DOI] [PubMed] [Google Scholar]
- 11.Bamshad M, Wooding SP. Signatures of natural selection in the human genome. Nature Rev Genet. 2003;4:99A–111A. doi: 10.1038/nrg999. [DOI] [PubMed] [Google Scholar]
- 12.Blanchette M, Tompa M. Discovery of regulatory elements by a computational method for phylogenetic footprinting. Genome Res. 2002;12:739–748. doi: 10.1101/gr.6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eyre-Walker A, Keightley PD. High genomic deleterious mutation rates in hominids. Nature. 1999;397:344–347. doi: 10.1038/16915. [DOI] [PubMed] [Google Scholar]
- 15.Eyre-Walker A, Keightley PD, Smith NGC, Gaffney D. Quantifying the slightly deleterious model of molecular evolution. Mol Biol Evol. 2002;19:2142–2149. doi: 10.1093/oxfordjournals.molbev.a004039. [DOI] [PubMed] [Google Scholar]
- 16.Eyre-Walker A, Woolfit M, Phelps T. The distribution of fitness effects of new deleterious amino acid mutations in humans. Genetics. 2006;173:891–900. doi: 10.1534/genetics.106.057570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kryukov GV, Pennacchio LA, Sunyaev SR. Most rare missense alleles are deleterious in humans: implications for complex disease and association studies. Am J Hum Genet. 2007;80:727–739. doi: 10.1086/513473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clark AG, et al. Inferring nonneutral evolution from human-chimp-mouse orthologous gene trios. Science. 2003;302:1960–1963. doi: 10.1126/science.1088821. This paper provides a list of genes under positive selection in the human evolutionary lineage based on the ratio of non-synonymous to synonymous mutations. [DOI] [PubMed] [Google Scholar]
- 19.Consortium TCS. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 20.Nielsen R, et al. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 2005;3:e170. doi: 10.1371/journal.pbio.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wyckoff GJ, Wang W, Wu CI. Rapid evolution of male reproductive genes in the descent of man. Nature. 2000;403:304–309. doi: 10.1038/35002070. [DOI] [PubMed] [Google Scholar]
- 22.Swanson WJ, Nielsen R, Yang Q. Pervasive adaptive evolution in mammalian fertilization proteins. Mol Biol Evol. 2003;20:18–20. doi: 10.1093/oxfordjournals.molbev.a004233. [DOI] [PubMed] [Google Scholar]
- 23.Gavrilets S. Rapid evolution of reproductive barriers driven by sexual conflict. Nature. 2000;403:886–889. doi: 10.1038/35002564. [DOI] [PubMed] [Google Scholar]
- 24.Kaplan NL, Hudson RR, Langley CH. The hitchhiking effect revisited. Genetics. 1989;123:887–899. doi: 10.1093/genetics/123.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maynard Smith J, Haigh J. The hitch-hiking effect of a favorable gene. Genet Res. 1974;23:23–35. The original paper describing the effect of a selective sweep in a population. [PubMed] [Google Scholar]
- 26.Braverman JM, Hudson RR, Kaplan NL, Langley CH, Stephan W. The hitchhiking effect on the site frequency spectrum of DNA polymorphisms. Genetics. 1995;140:783–796. doi: 10.1093/genetics/140.2.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barton N. The effect of hitch-hiking on neutral genealogies. Genet Res. 1998;72:123–133. [Google Scholar]
- 28.Stephan W, Song YS, Langley CH. The hitchhiking effect on linkage disequilibrium between linked neutral loci. Genetics. 2006;172:2647–2663. doi: 10.1534/genetics.105.050179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simoons FJ. Primary adult lactose intolerance and milking habit — a problem in biologic and cultural interrelations.2 A culture historical hypothesis. Am J Dig Dis. 1970;15:695–710. doi: 10.1007/BF02235991. [DOI] [PubMed] [Google Scholar]
- 30.Cavalli-Sforza L. Analytic review: some current problems of population genetics. Am J Hum Genet. 1973;25:82–104. [PMC free article] [PubMed] [Google Scholar]
- 31.Beja-Pereira A, et al. Gene–culture coevolution between cattle milk protein genes and human lactase genes. Nature Genet. 2003;35:311–313. doi: 10.1038/ng1263. [DOI] [PubMed] [Google Scholar]
- 32.Bersaglieri T, et al. Genetic signatures of strong recent positive selection at the lactase gene. Am J Hum Genet. 2004;74:1111–1120. doi: 10.1086/421051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burger J, Kirchner M, Bramanti B, Haak W, Thomas MG. Absence of the lactase-persistence-associated allele in early Neolithic Europeans. Proc Natl Acad Sci USA. 2007;104:3736–3741. doi: 10.1073/pnas.0607187104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bersaglieri T, et al. Genetic signatures of strong recent positive selection at the lactase gene. Am J Hum Genet. 2004;74:1111–1120. doi: 10.1086/421051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tishkoff SA, et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nature Genet. 2007;39:31–40. doi: 10.1038/ng1946. A recent paper that demonstrates positive selection acting independently on different lactase alleles that confer lactose tolerance in adults, in African and European populations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tishkoff SA, et al. Haplotype diversity and linkage disequilibrium at human G6PD: recent origin of alleles that confer malarial resistance. Science. 2001;293:455–462. doi: 10.1126/science.1061573. [DOI] [PubMed] [Google Scholar]
- 37.Verrelli BC, Argyropoulos G, Destro-Bisol G, Williams SM, Tishkoff SA. Signature of selection at the G6PD locus inferred from patterns of nucleotide variation and linkage disequilibrium in Africans. Am J Hum Genet. 2001;69:395–395. [Google Scholar]
- 38.Saunders MA, Hammer MF, Nachman MW. Nucleotide variability at G6PD and the signature of malarial selection in humans. Genetics. 2002;162:1849–1861. doi: 10.1093/genetics/162.4.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sabeti PC, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 40.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. This paper provides the result of a genome-wide scan for selective sweeps based on haplotype structure information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stefansson H, et al. A common inversion under selection in Europeans. Nature Genet. 2005;37:129–137. doi: 10.1038/ng1508. [DOI] [PubMed] [Google Scholar]
- 42.Thompson EE, et al. CYP3A variation and the evolution of salt-sensitivity variants. Am J Hum Genet. 2004;75:1059–1069. doi: 10.1086/426406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Osier MV, et al. A global perspective on genetic variation at the ADH genes reveals unusual patterns of linkage disequilibrium and diversity. Am J Hum Genet. 2002;1:84–99. doi: 10.1086/341290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gilad Y, Bustamante CD, Lancet D, Paabo S. Natural selection on the olfactory receptor gene family in humans and chimpanzees. Am J Hum Genet. 2003;73:489–501. doi: 10.1086/378132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carlson CS, et al. Genomic regions exhibiting positive selection identified from dense genotype data. Genome Res. 2005;15:1553–1565. doi: 10.1101/gr.4326505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. This paper describes the most common methods for detecting selection based on population genetic data. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fay JC, Wu CI. Hitchhiking under positive Darwinian selection. Genetics. 2000;155:1405–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hinds DA, et al. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- 49.Nielsen R, et al. Genomic scans for selective sweeps using SNP data. Genome Res. 2005;15:1566–1575. doi: 10.1101/gr.4252305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Henikoff S, Malik HS. Selfish drivers. Nature. 2002;417:227–227. doi: 10.1038/417227a. [DOI] [PubMed] [Google Scholar]
- 51.Charlesworth B, Nordborg M, Charlesworth D. The effects of local selection, balanced polymorphism and background selection on equilibrium patterns of genetic diversity in subdivided populations. Genet Res. 1997;70:155–174. doi: 10.1017/s0016672397002954. [DOI] [PubMed] [Google Scholar]
- 52.Slatkin M, Wiehe T. Genetic hitch-hiking in a subdivided population. Genet Res. 1998;71:155–160. doi: 10.1017/s001667239800319x. [DOI] [PubMed] [Google Scholar]
- 53.Lewontin RC, Krakauer J. Distribution of gene frequency as a test of theory of selective neutrality of polymorphisms. Genetics. 1973;74:175–195. doi: 10.1093/genetics/74.1.175. The original paper discussing the effect of selection on measures of population subdivision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weir BS, Cardon LR, Anderson AD, Nielsen DM, Hill WG. Measures of human population structure show heterogeneity among genomic regions. Genome Res. 2005;15:1468–1476. doi: 10.1101/gr.4398405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kayser M, Brauer S, Stoneking M. A genome scan to detect candidate regions influenced by local natural selection in human populations. Mol Biol Evol. 2003;20:893–900. doi: 10.1093/molbev/msg092. [DOI] [PubMed] [Google Scholar]
- 56.The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simonsen KL, Churchill GA, Aquadro CF. Properties of statistical tests of neutrality for DNA polymorphism data. Genetics. 1995;141:413–429. doi: 10.1093/genetics/141.1.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Andolfatto P, Przeworski M. A genome-wide departure from the standard neutral model in natural populations of Drosophila. Genetics. 2000;156:257–268. doi: 10.1093/genetics/156.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Przeworski M, Hudson RR, Di Rienzo A. Adjusting the focus on human variation. Trends Genet. 2000;16:296–302. doi: 10.1016/s0168-9525(00)02030-8. [DOI] [PubMed] [Google Scholar]
- 60.Nielsen R. Statistical tests of selective neutrality in the age of genomics. Heredity. 2001;86:641–647. doi: 10.1046/j.1365-2540.2001.00895.x. [DOI] [PubMed] [Google Scholar]
- 61.Stajich JE, Hahn MW. Disentangling the effects of demography and selection in human history. Mol Biol Evol. 2005;22:63–73. doi: 10.1093/molbev/msh252. [DOI] [PubMed] [Google Scholar]
- 62.Wall JD, Andolfatto P, Przeworski M. Testing models of selection and demography in Drosophila simulans. Genetics. 2002;162:203–216. doi: 10.1093/genetics/162.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Galtier N, Depaulis F, Barton NH. Detecting bottlenecks and selective sweeps from DNA sequence polymorphism. Genetics. 2000;155:981–987. doi: 10.1093/genetics/155.2.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jensen JD, Kim Y, DuMont VB, Aquadro CF, Bustamante CD. Distinguishing between selective sweeps and demography using DNA polymorphism data. Genetics. 2005;170:1401–1410. doi: 10.1534/genetics.104.038224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Edmonds CA, Lillie AS, Cavalli-Sforza LL. Mutations arising in the wave front of an expanding population. Proc Natl Acad Sci USA. 2004;101:975–979. doi: 10.1073/pnas.0308064100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Klopfstein S, Currat M, Excoffier L. The fate of mutations surfing on the wave of a range expansion. Mol Biol Evol. 2006;23:482–490. doi: 10.1093/molbev/msj057. [DOI] [PubMed] [Google Scholar]
- 67.Nielsen R, Signorovitch J. Correcting for ascertainment biases when analyzing SNP data: applications to the estimation of linkage disequilibrium. Theor Popul Biol. 2003;63:245–255. doi: 10.1016/s0040-5809(03)00005-4. [DOI] [PubMed] [Google Scholar]
- 68.Nielsen R, Hubisz MJ, Clark AG. Reconstituting the frequency spectrum of ascertained single-nucleotide polymorphism data. Genetics. 2004;168:2373–2382. doi: 10.1534/genetics.104.031039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kong A, et al. A high-resolution recombination map of the human genome. Nature Genet. 2002;10:10. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- 70.Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- 71.Teshima KM, Coop G, Przeworski M. How reliable are empirical genomic scans for selective sweeps? Genome Res. 2006;16:702–712. doi: 10.1101/gr.5105206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Teshima KM, Przeworski M. Directional positive selection on an allele of arbitrary dominance. Genetics. 2006;172:713–718. doi: 10.1534/genetics.105.044065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.MacCallum C, Hill E. Being positive about selection. PLoS Biol. 2006;4:293–295. doi: 10.1371/journal.pbio.0040087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shu W, et al. Altered ultrasonic vocalization in mice with a disruption in the Foxp2 gene. Proc Natl Acad Sci USA. 2005;102:9643–9648. doi: 10.1073/pnas.0503739102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Williams GC. Adaptation and Natural Selection: A Critique of Some Current Evolutionary Thought. Princeton Univ. Press; Princeton: 1966. [Google Scholar]
- 76.Gould SJ, Lewontin RC. Spandrels of San-Marco and the Panglossian paradigm — a critique of the adaptationist program. Proc R Soc London Series B Biol Sci. 1979;205:581–598. doi: 10.1098/rspb.1979.0086. [DOI] [PubMed] [Google Scholar]
- 77.Diaz GA, et al. Gaucher disease: The origins of the Ashkenazi Jewish N370S and 84GG acid β-glucosidase mutations. Am J Hum Genet. 2000;66:1821–1832. doi: 10.1086/302946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hugot JP, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 79.Schwartz K, Carrier L, Guicheney P, Komajda M. Molecular-basis of familial cardiomyopathies. Circulation. 1995;91:532–540. doi: 10.1161/01.cir.91.2.532. [DOI] [PubMed] [Google Scholar]
- 80.Saxena R, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 81.Steinthorsdottir V, et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nature Genet. 2007;39:770–775. doi: 10.1038/ng2043. [DOI] [PubMed] [Google Scholar]
- 82.Zeggini E, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Verrelli BC, et al. Evidence for balancing selection from nucleotide sequence analyses of human G6PD. Am J Hum Genet. 2002;71:1112–1128. doi: 10.1086/344345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Allen SJ, et al. α+-thalassemia protects children against disease caused by other infections as well as malaria. Proc Natl Acad Sci USA. 1997;94:14736–14741. doi: 10.1073/pnas.94.26.14736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schroeder SA, Gaughan DM, Swift M. Protection against bronchial-asthma by Cftr δ-f508 mutation — a heterozygote advantage in cystic-fibrosis. Nature Med. 1995;1:703–705. doi: 10.1038/nm0795-703. [DOI] [PubMed] [Google Scholar]
- 86.Neel JV. Diabetes mellitus: a ‘thrifty’ genotype rendered detrimental by ‘progress’? Am J Hum Genet. 1962;14:353–362. [PMC free article] [PubMed] [Google Scholar]
- 87.Thomas PD, Kejariwal A. Coding single-nucleotide polymorphisms associated with complex vs. Mendelian disease: evolutionary evidence for differences in molecular effects. Proc Natl Acad Sci USA. 2004;101:15398–15403. doi: 10.1073/pnas.0404380101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zlotogora J. Multiple mutations responsible for frequent genetic diseases in isolated populations. Eur J Hum Genet. 2007;15:272–278. doi: 10.1038/sj.ejhg.5201760. [DOI] [PubMed] [Google Scholar]
- 89.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sunyaev S, et al. Prediction of deleterious human alleles. Hum Mol Genet. 2001;10:591–597. doi: 10.1093/hmg/10.6.591. This paper describes the most popular bioinformatical method for predicting disease mutations without phenotypic data. [DOI] [PubMed] [Google Scholar]
- 91.Sabeti PC, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 92.Fan Y, Linardopoulou E, Friedman C, Williams E, Trask BJ. Genomic structure and evolution of the ancestral chromosome fusion site in 2q13–12q14.1 and paralogous regions on other human chromosomes. Genome Res. 2002;12:1651–1662. doi: 10.1101/gr.337602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kim Y, Stephan W. Detecting a local signature of genetic hitchhiking along a recombining chromosome. Genetics. 2002;160:765–777. doi: 10.1093/genetics/160.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hudson RR, Kreitman M, Aguade M. A test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. The original paper describing the combined use of divergence and diversity data to detect selection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fisher SE, Vargha-Khadem F, Watkins KE, Monaco AP, Pembrey ME. Localisation of a gene implicated in a severe speech and language disorder. Nature Genet. 1998;18:168–170. doi: 10.1038/ng0298-168. [DOI] [PubMed] [Google Scholar]
- 96.Lai CS, et al. The SPCH1 region on human 7q31: genomic characterization of the critical interval and localization of translocations associated with speech and language disorder. Am J Hum Genet. 2000;67:357–368. doi: 10.1086/303011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature. 2001;413:519–523. doi: 10.1038/35097076. [DOI] [PubMed] [Google Scholar]
- 98.Zhang J, Webb DM, Podlaha O. Accelerated protein evolution and origins of human-specific features: Foxp2 as an example. Genetics. 2002;162:1825–1835. doi: 10.1093/genetics/162.4.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mekel-Bobrov N, et al. Ongoing adaptive evolution of ASPM, a brain size determinant in Homo sapiens. Science. 2005;309:1720–1722. doi: 10.1126/science.1116815. [DOI] [PubMed] [Google Scholar]
- 100.Currat M, et al. Comment on ‘Ongoing adaptive evolution of ASPM, a brain size determinant in Homo sapiens’ and ‘Microcephalin, a gene regulating brain size, continues to evolve adaptively in humans’. Science. 2006;313:172. doi: 10.1126/science.1122822. [DOI] [PubMed] [Google Scholar]
- 101.Yu FL, et al. Comment on ‘Ongoing adaptive evolution of ASPM, a brain size determinant in Homo sapiens’. Science. 2007;316:367. doi: 10.1126/science.316.5823.370a. [DOI] [PubMed] [Google Scholar]
- 102.Mekel-Bobrov N, et al. The ongoing adaptive evolution of ASPM and microcephalin is not explained by increased intelligence. Hum Mol Genet. 2007;16:600–608. doi: 10.1093/hmg/ddl487. [DOI] [PubMed] [Google Scholar]
- 103.Nielsen R. Estimation of population parameters and recombination rates from single nucleotide polymorphisms. Genetics. 2000;154:931–942. doi: 10.1093/genetics/154.2.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Clark AG, Hubisz MJ, Bustamante CD, Williamson SH, Nielsen R. Ascertainment bias in studies of human genome-wide polymorphism. Genome Res. 2005;15:1496–1502. doi: 10.1101/gr.4107905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hudson RR. Generating samples under a Wright–Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]