Abstract
Understanding how the information is conveyed from outside to inside the cell is a critical challenge for all biologists involved in signal transduction. The flow of information initiated by cell-cell and cell-extracellular matrix contacts is mediated by the formation of adhesion complexes involving multiple proteins. Inside adhesion complexes, connective membrane skeleton (CMS) proteins are signal transducers that bind to adhesion molecules, organize the cytoskeleton, and initiate biochemical cascades. Adhesion complex-mediated signal transduction ultimately directs the formation of supramolecular structures in the cell nucleus, as illustrated by the establishment of multi complexes of DNA-bound transcription factors, and the redistribution of nuclear structural proteins to form nuclear subdomains. Recently, several CMS proteins have been observed to travel to the cell nucleus, suggesting a distinctive role for these proteins in signal transduction. This review focuses on the nuclear translocation of structural signal transducers of the membrane skeleton and also extends our analysis to possible translocation of resident nuclear proteins to the membrane skeleton. This leads us to envision the communication between spatially distant cellular compartments (i.e., membrane skeleton and cell nucleus) as a bidirectional flow of information (a dynamic reciprocity) based on subtle multilevel structural and biochemical equilibria. At one level, it is mediated by the interaction between structural signal transducers and their binding partners, at another level it may be mediated by the balance and integration of signal transducers in different cellular compartments.
Keywords: signal transduction, cell adhesion complexes, membrane skeleton, nucleo-cytoplasmic translocation
The communication between the cellular microenvironment and the cell nucleus is critical to understanding the essence of cell behavior and tissue development. Signals to be transduced to the cell's interior upon attachment to extracellular matrix (ECM) components, or contact with another cell, are mediated by a variety of adhesion molecules. Using cells cultured in the presence of a reconstituted basement membrane, investigators at a number of laboratories have been able to show that the interaction between adhesion molecules of epithelial cells and basement membrane components induces a differentiation program that leads to the formation of functional tissue-like structures [Bissell et al., 1987; Li et al., 1987; Barcellos-Hoff et al., 1989; Caron, 1990; Petersen et al., 1992; Matter and Laurie, 1994; Hoffman et al., 1995]. Similarly, the interaction between adhesion molecules of osteoblasts and fibronectin promotes the production of osteopontin, one of the predominant proteins of the bone tissue [Carvalho et al.,1998]. ECM-mediated regulation of the expression of a defined repertoire of genes is conveyed, at the molecular level, by a modification of DNA-protein interactions [Owen et al., 1990] and the activation of ECM-response elements located in the promoters of some of the expressed genes [Schmidhauser et al., 1990, 1992; Liu et al., 1991]. Cell-cell interaction also induces the expression of specific genes that regulate tissue differentiation and morphogenesis [Takeichi, 1995; Gumbiner, 1996; Redfield et al., 1997]. Moreover, the interplay between the formation and loss of adhesion complexes and the correct balance of different kinds of adhesion molecules is essential for tissue development and maintenance of differentiation [Martins-Green and Bissell, 1995; Herminston et al., 1996], and the alteration of this equilibrium can lead to extreme behavior such as apoptosis [Tenniswood et al., 1992; Sympson et al., 1994; Frisch and Francis, 1994; Boudreau et al., 1995] and tumor formation [Gamallo et al., 1993; Sympson et al., 1995; Lochter and Bissell, 1995; Perl et al., 1998; Hagios et al., 1998].
Whereas it is now well established that cell-ECM and cell-cell interactions can regulate gene expression and cell behavior, the way information is transduced from the cell membrane to the nucleus remains an exciting challenge for biologists to solve at the dawn of the third millennium. Explored mechanisms of cell adhesion-mediated signal transduction encompass the induction of biochemical cascades that ultimately regulate the activity of transcription factors [Baichwal et al., 1991; Juliano and Haskill, 1993; O'Neill et al., 1994], alterations of histone acetylation [Loidl, 1994; Alberts et al., 1998; Myers et al., 1998], and remodeling of supramolecular organization of nuclear matrix proteins, defined by the redistribution of these components to distinct nuclear sites [Lelièvre et al., 1998]. In addition, the existence of a structural continuity from the cell membrane and cytoplasm to the nuclear matrix and chromatin [Capco et al., 1984] has been postulated to also participate in the rapid transmission of information within the cell [Bissell et al.,1982; Ingber, 1997]. Elements of proof for this concept have been brought to our attention during the past decade [Pienta and Coffey, 1992; Sims et al, 1992; Maniotis et al., 1997; Lelièvre et al., 1998].
Cell adhesion-mediated signal transduction is initiated by the supramolecular organization of adhesion molecules and proteins localized in the inner part of the cell membrane, referred to as the membrane skeleton [Luna and Hitt, 1992]. By acting both as inducers of biochemical cascades and as organizers of cytoskeletal fibers, proteins of the membrane skeleton bring together the chemical and mechanical aspects of intracellular signaling. Recently, their role in signal transduction has been underscored by their capacity to travel to the nucleus. The nuclear translocation of membrane skeleton proteins and, inversely, the possibility that proteins usually involved in supramolecular structures of the nucleus may translocate to the cell membrane, raise new and important mechanistic issues for signal transduction, and add a new dimension to the concept of dynamic reciprocity proposed almost two decades ago [Bissell et al., 1982].
Structural Signal Transducers of the Membrane Skeleton
Communication between spatially separated elements such as the cell membrane and the chromatin requires intracellular mediators, referred to as structural and biochemical signal transducers. Our general understanding is that a signal transducer can receive a signal and transfer the information to the next component of the signaling cascade by altering its molecular state and modifying its binding to other cellular components. Typically, signals initiated at cell adhesion sites, by cell-ECM or cell-cell contacts, are transduced by membrane skeleton proteins, also referred to as junctional plaque proteins [Kartenbeck et al., 1982; Ben-Ze'ev, 1997]. Although these structural signal transducers share common characteristics such as involvement in phosphorylation/dephosphorylation cascades and the capacity to induce cytoskeletal reorganization [for review, see Longhurst and Jennings, 1998], they are specific for each type of adhesion complex. Focal adhesions (FAs), a class of cell-ECM adhesion complexes formed by the interaction of various types of integrin heterodimers with specific ECM molecules, contain a large number of connective membrane skeleton (CMS) proteins (e.g., α-actinin, talin, tensin, vinculin, Cas, moesin, fimbrin, paxillin, and zyxin) [Weisberg et al., 1997; Brugge, 1998] that interact with a broad range of kinases and phosphatases, and are implicated in the control of actin and myosin filament assembly (Fig.1) [Schlaepfer and Hunter, 1996; Golsteyn et al., 1997; Helmke et al., 1998]. Similarly, hemidesmosomes are formed by the interaction between the ECM component laminin and α6-β4 integrin, and the recruitment of CMS proteins, plectin and bullous pemphigoid antigen 230, to form the hemidesmosomal plaque. Hemidesmosome formation directs the organization of intermediate filament type proteins and initiates phosphorylation cascades [Wiche et al., 1993; Giancotti, 1996; Jones et al., 1998; Rezniczek et al., 1998; Schaapveld et al., 1998] (Fig. 1). Cell-cell adhesion complexes (i.e., tight junctions, adherens junctions and desmosomes) have been characterized both microscopically [Farquhar et al., 1963] and by their composition. Their localization in a tissue is highly ordered. Tight junctions or zonula occludens are located at the outermost edge of the intercellular space (i.e., at the apical part of epithelial glandular cell assemblies) and are believed to participate in maintaining cell polarity [Cereijido et al., 1998] along with other cell-cell adhesion complexes [Nathke et al., 1994]. Several tight junction-associated CMS proteins have been identified, including ZO-1, ZO-2 and ZO-3, members of the membrane-associated guanylate kinase family (MAGUK), as well as 7H6, cingulin, and symplekin (Fig.1) [Citi, 1993; Haskins et al., 1998; Mitic and Anderson, 1998; Balda and Matter, 1998]. Tight junctions and adherens junctions together form apical junctional complexes. Adherens junction-associated CMS proteins encompass symplekin, plakoglobin, α-catenin, β-catenin, vinculin, and undoubtably another host of known and unknown proteins, that interact with actin, as is the case also in tight junctions [Weiss et al., 1998] (Fig.1). Desmosomes constitute a third type of cell-cell junction, where adhesion is mediated through the desmosomal cadherins desmocollin and desmoglein and the CMS proteins desmoplakin and plakoglobin that are connected to intermediate filament type proteins [Green et al., 1998; Smith and Fuchs, 1998] (Fig. 1). As with cell-ECM adhesion complexes, cell-cell adhesion complexes interact with various kinases and phosphatases.
Fig. 1.
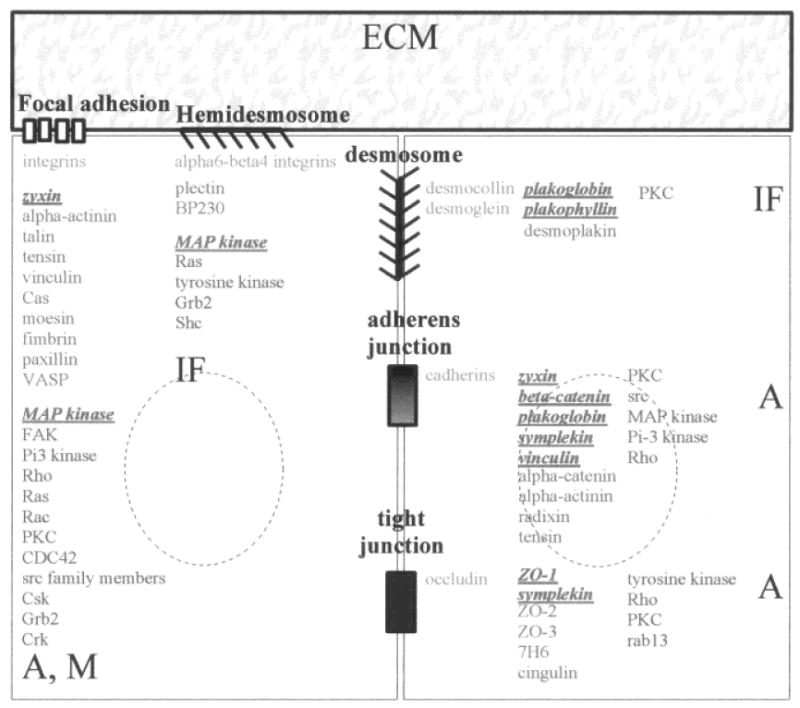
Organization of cell-cell adhesion complexes (tight junction, adherens junction, desmosome) and cell-ECM adhesion complexes (focal adhesion, hemidesmosome). The formation of cell-cell and cell-ECM interactions involves specific cell membrane adhesion molecules (blue), that interact with connective membrane skeleton (CMS) proteins (green). CMS proteins themselves interact with other cytoskeletal proteins to organize cytoskeleton networks (A: actin microfilaments; IF: intermediate filaments; M: myosin filaments). Moreover, biochemical signal transducers (purple) are found at the cell membrane or in its vicinity and participate in the regulation of cell adhesion complexes and/or in the transfer of signals initiated at cell adhesion complexes. Some CMS proteins (red), as well as biochemical signal transducers (red), have been observed in the nucleus under specific circumstances. The list of cell adhesion complex components is nonexhaustive.
Although adhesion complexes are built with different components, they are connected to the same network of cytoskeletal filaments, their integrity depends on tyrosine kinase activity, and they share similar biochemical signal transducers [Yamada and Geiger, 1997]. This indicates that from one complex to another, the organization of cell adhesion complexes follows the same linear path including adhesion molecules linked to complexes of CMS proteins and kinases/phosphatases that regulate the induction of biochemical cascades and the organization of the cytoskeleton (Fig. 2), but it does not mean that these structures behave similarly. For instance, treatment with protein phosphatases disrupts FAs and the underlying cytoskeleton [Schneider et al., 1998], while increased tyrosine phosphorylation induces the redistribution of adherens and tight junction proteins [Collares-Buzato et al., 1998]. More specifically, adherens junction disassembly is due to increased phosphorylation mediated through MAP kinase and PI 3-kinase pathways [Potempa and Ridley, 1998], whereas desmosome disassembly appears to be regulated by activation of PKC [Amar et al., 1998].
Fig. 2.
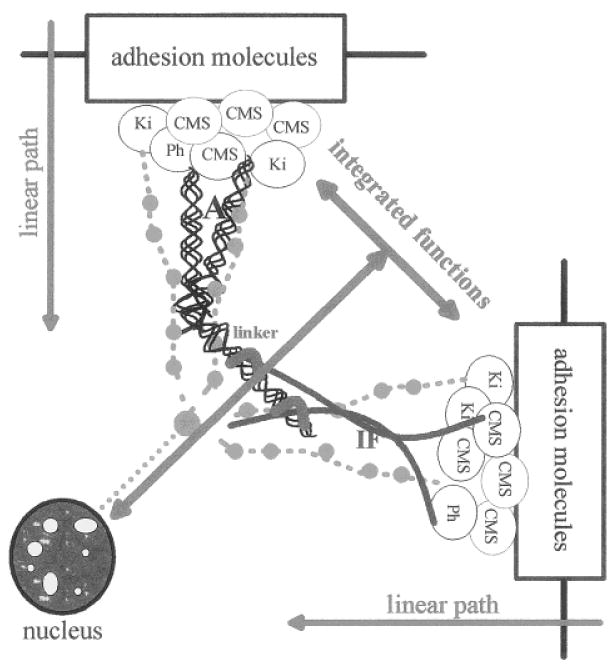
Linear path and integrated functions of cell adhesion complexes. All cell adhesion complexes follow the same organizational path, which involves the recruitment of connective membrane skeleton (CMS) proteins upon cell-cell and cell-ECM interaction, and the induction of the formation of multiprotein complexes at the inner part of the cell membrane. These complexes include biochemical signal transducers such as kinases (Ki) and phosphatases (Ph) and cytoskeletal proteins and lead to the initiation of biochemical cascades and the reorganization of cytoskeletal fibers (A, actin; IF, intermediate filaments). This defines the linear path of cell adhesion complexes. The integrative function results from the influence of the various cell adhesion complexes on each other through the physical interconnection of cytoskeletal networks (linker) and the interactions between biochemical signaling pathways. The integrative function is ultimately responsible for the regulation of cytoplasmic and nuclear activities that determine the cell and tissue phenotype.
Cell adhesion complexes participate in the coordinated regulation of cell division, survival, and differentiation [Weaver et al., 1997; Bailey et al., 1998; Balda and Matter, 1998; Bissell, 1998; Clark et al., 1998; Perl et al., 1998; Sharma, 1998; Short et al., 1998]. This coordinated regulation of cell behavior is mediated by the integration of the linear paths of cell adhesion complexes through interconnection with other signal transduction cascades [Wang et al., 1998], and the link between actin and intermediate filament networks [Yang et al., 1996; Yamada and Geiger, 1997; Fuchs and Cleveland, 1998] (Fig. 2). This defines an integrated function for cell adhesion complexes, which is ruled by the equilibrium between the different adhesion structures. In addition, alternate construction and deconstruction of cell adhesion complexes has been shown to be critical for developmental programs and cell migration. In this case, modulation of cellular behavior is due to the switch of dominant signaling pathways that results from equilibrium shifts and temporary delocalization of adhesion proteins and membrane skeleton proteins. However, if there is sustained imbalance, the equilibrium shift may also lead to the stimulation of tumor development [Sympson et al., 1995 and in preparation; Weaver et al., 1997; Thomasset et al., 1998; Efstathiou et al., 1998; Tselepis et al., 1998; Shinohara et al., 1998].
Connective Membrane Skeleton Proteins also Reside in the Nucleus
The CMS proteins, β-catenin, plakoglobin, plakophilin 2, symplekin, ZO-1, and zyxin have all been observed in the nucleus. Logically, only the CMS proteins free from their interaction with adhesion complexes will translocate into the nucleus. The constitution of a free pool of CMS proteins could result from the expression of these proteins above the level necessary for the formation of adhesion complexes, as was the case after overexpression of exogenous plakoglobin in transfection experiments [Karnovsky and Klymkowsky, 1995]. However, CMS proteins are more likely to be observed in the nucleus when the formation of adhesion complexes is impaired, as it has been described in naturally occurring situations. Endogenous β-catenin has been found in tumor cell nuclei in which cell adhesion complexes were altered [Bailey et al., 1998]. Endogenous symplekin and plakophilin 2 were observed in the nucleus of cells that usually do not form adherens junctions or desmosomes [Mertens et al., 1996; Keon et al., 1996]. The presence of endogenous ZO-1 in the nucleus of epithelial cells was inversely correlated with the extent or maturity of tight junctions [Gottardi et al., 1996], and apical polarity in mammary acini (Lelièvre and Bissell, unpublished observations). The localization of these CMS proteins in cell adhesion complexes and in the nucleus is not mutually exclusive, as both locations have been simultaneously observed in many cases. This finding suggests that there may be an equilibrium between membrane skeleton and nuclear localization of the CMS proteins.
The creation of a free pool of CMS proteins via their release from existing cell adhesion complexes or through other mechanisms is not sufficient to explain how these proteins can enter the nucleus. The study of shuttling proteins has shown that the mechanisms of nuclear translocation are highly regulated and it is conceivable that translocating CMS proteins may conform to the same mechanisms. It is known that proteins of >40 kD actively enter the nucleus by binding to the nucleopore proteins importins through a nuclear localization signal (NLS) and by translocating through an energy-dependent mechanism [Görlich and Mattaj, 1996]. While a putative NLS has been identified in the sequence of both ZO-1 and symplekin proteins [Gottardi et al,. 1996; Keon et al., 1996], the evidence that these NLS are functional is still lacking. If indeed they are functional, it will be worthwhile to analyze the possible nuclear localization of other NLS bearing CMS proteins (e.g., plectin) [Nikolic et al., 1996]. The presence of a functional NLS may not always be necessary for the nuclear translocation of CMS proteins. These proteins could “piggy-back” with other NLS-bearing molecules, as proposed for β-catenin, which travels as a complex with LEF-1 [Funayama et al., 1995; Behrens et al., 1996; Simcha et al., 1998]. NLS-free CMS proteins could also bind directly to the nuclear pore and translocate into the nucleus, as demonstrated recently for β-catenin [Fagotto et al., 1998]. It will be important to clearly classify those proteins that “piggy-back” from self-translocating CMS proteins. In the former case, the nuclear translocation is dependent not only on a free pool of CMS proteins, but also on the availability of their carrier.
Once CMS proteins are trapped in the nucleus, they may stay there until a signal induces their release from nuclear complexes and initiates their degradation. De novo expression of proteins would re-create the pool of CMS proteins in the cytoplasm. However, since the control of the nuclear translocation of CMS proteins is achieved by their release from interactions with cell membrane and cytoskeletal components, it is similarly possible that their release from interactions with nuclear partners would lead to their return to the cytoplasm. The study of protein shuttling has shown that proteins can slowly diffuse out of the nucleus without any specific signal [Schmidt-Zachmann et al., 1993], while a fast re-entry into the cytoplasm is regulated by pathways distinct from nuclear import mechanisms [Moroainu, 1998]. The rapid transit of nuclear proteins to the cytoplasm is mediated by nuclear export sequences (NES) via energy-dependent extrusion mechanisms [Wen et al., 1995; Richards et al., 1996]. Thus far, only the CMS protein zyxin has been shown to possess a functional NES, and its traveling into and out of the nucleus has been observed in the course of antibody-injection experiments [Nix and Beckerle, 1997]. However, as with nuclear protein import, we could imagine that CMS proteins could also leave the nucleus by “piggy-backing.” Bidirectional transit of zyxin could be regulated by temporarily masking the NES, and/or by the presence of a nuclear retention signal or cytoplasmic retention signal that would be responsible for the binding of the protein to nuclear or cytoplasmic components, as has been proposed for shuttling proteins [Wen et al., 1995; Nakielny and Dreyfus, 1996; Richards et al.,1996]. The participation of CMS proteins in the formation of adhesion complexes is an example of cytoplasmic retention.
Do Nuclear CMS proteins act as signal transducers?
The release of CMS proteins from cell adhesion complexes and their nuclear translocation are both highly regulated; thus, we can anticipate that the journey of CMS proteins to the nucleus is another important facet of signal transduction. Most of the CMS proteins bear sequences that link them to families of proteins known to participate in the mediation of signal transduction events, such as the armadillo family for β-catenin, plakoglobin, and plakophilin 2 [Peifer et al., 1994], and the MAGUK family for ZO-1 [Willot et al., 1993]. Zyxin harbors LIM domains known to participate in protein-protein interactions and that might interact with nucleic acids [Schmeichel and Beckerle, 1994, 1997; Nix and Beckerle, 1997; Beckerle, 1997]. The role of the nuclear translocation of CMS proteins in signal transduction is further supported by the fact that some CMS proteins are found in the nucleus of cell types that do not use these CMS proteins in the formation of their adhesion complexes [Mertens et al., 1996; Keon et al., 1996]. Interestingly, MAP kinases (erk1, erk2, and Nlk) known to be associated with cell adhesion complexes-mediated signaling have also been shown to travel to the nucleus. Their role as nuclear signal transducers is easier to grasp compared with CMS proteins because MAP kinases could drive the continuity of biochemical transduction cascades by exerting their kinase activity both in cytoplasmic and nucleoplasmic compartments [Sanghera et al., 1992; Chen et al., 1992; Khokhlatchev et al., 1998; Brott et al., 1998]. Their translocation is regulated as shown for erk2, the nuclear import of which is promoted by phosphorylation-directed homodimerization [Khokhlatchev et al., 1998]. Another type of regulation of translocation has been described for MAPKAP kinase2, which is exported to the cytoplasm upon stress induction and also harbors an NLS [Engel et al., 1998]. Potential nuclear targets of MAP kinases include Rb [Taieb et al., 1998], histone H3, c-Fos, c-Jun, and transcription factors phosphorylated in response to growth stimuli [Chen et al., 1992]. Nuclear MAP kinases have also been proposed to participate in the regulation of insulin gene transcription [Benes et al.,1998].
The best evidence for a meaningful function for nuclear CMS proteins as signal transducers would require the characterization of the cellular behaviors associated with their presence in the cell nucleus and the identification of their nuclear binding partners. This goal has been achieved partly for nuclear β-catenin, a component of the Wnt signaling pathway, which has been implicated in the induction of embryonic axis in Xenopus and the regulation of gene expression in higher organisms via the formation of a complex with transcription factor LEF-1 and DNA [McCrea et al., 1993; Funayama et al., 1995; Gumbiner, 1995; Behrens et al., 1996; Papkoff et al., 1996; Brannon et al., 1997; Larabell et al., 1997; Miller and Moon, 1997]. More specifically, LEF-1-β catenin complexes have been shown to bind to the 5′ end of the E-cadherin gene [Huber et al., 1996]. This demonstrate that nuclear CMS proteins may act as repressors or activators of transcription.
Obviously, at this point there are more questions than answers for the mode of action of CMS proteins. Before answers are forthcoming, a number of technical points need to be clarified. The choice of immunostaining conditions appears to be critical for observing CMS proteins in the nucleus. Thus, caution has been advised in the use of permeabilization agents, and in some cases, a special protection buffer had to be used [Keon et al, 1996; Gottardi et al.,1996]. A careful monitoring of immunostaining conditions may allow the observation of other types of CMS proteins in the nucleus. The fact that, in a number of cases, CMS proteins located in the nucleus have been shown to be sensitive to the use of permeabilization agents indicates that although these proteins are associated with the cytoskeleton in the cytoplasm, they are not likely to be associated with the nucleoskeleton or nuclear matrix in the nucleus. The only CMS protein so far described to be a nuclear matrix protein, because it stays associated with cell remnants following DNA removal under high salt conditions, is protein band 4.1. This protein is associated with the membrane skeleton in red blood cells [Matsuzaki et al.,1985] and other cells [Granger and Lazarides, 1984; Leto et al., 1986], and has been reported to be present in the nuclei of numerous cell types [Coreas, 1991; de Carcer et al., 1995; Krauss et al., 1997; Lallena and Coreas, 1997]. However, because some of the isoforms found in the nucleus have been shown to be the product of alternative splicing, this undermines a possible role of protein 4.1 in direct signal transduction between the cell membrane and the nucleus [Tang et al., 1988; Luque et al., 1998; Lallena et al., 1998]. A demonstration that the same isoform of protein 4.1 shuttles between the cytoplasm and nucleus requires further experiments and proof.
A Current Perspective on the Mechanisms of Communication
The dual function of the CMS proteins located at the cell membrane that act both as initiators of biochemical cascades and organizers of the cytoskeleton, plays a pivotal role in cell adhesion-mediated extracellular information. Once released from adhesion complexes, CMS proteins are available for interaction with other cellular components and for possible nuclear translocation. Thus, they become powerful signal transducers by directly transferring information from the source, a specific type of cell adhesion complex formed as a response to microenvironmental cues, to nuclear targets. Whether additional intermediate steps are needed is an open question. An interesting hypothesis for a new level of regulation of directed-protein localization has been proposed. In this model, proteins would reach specific cell membrane or nuclear substructures via a defined targeting sequence [Wu et al., 1998; Cardoso and Leonhardt, 1998]. This model seems especially tempting in the case of shuttling proteins that may bear targeting sequences for substructures in both cytoplasmic and nuclear compartments. Specific sequences of β-catenin are responsible for its binding to E-cadherin and α-catenin in the cytoplasm and although its arm sequence has been shown to be necessary for nuclear targeting, its nuclear substructural destination is not yet known [Behrens et al.,1996].
CMS proteins can be considered as active mediators of the dynamic reciprocity between the microenvironment and the cells. We suggest that there is a bidirectional flow of information between the microenvironment and the nucleus, part of which depends on a molecular equilibrium defined by the binding of CMS proteins to their various partners. This concept is well illustrated by the data generated for β-catenin (Fig. 3). Free cytoplasmic β-catenin, the presence of which depends on its association with other adherens junction molecules and cell membrane receptors, has to override the APC-regulated degradation mechanism [Munemitsu et al.,1995] before going to the nucleus. The number of free β-catenin molecules also has to exceed the amount required to form complexes with free ZO-1 that were shown to participate in the formation of tight junctions [Rajasekaran et al., 1996].
Fig. 3.
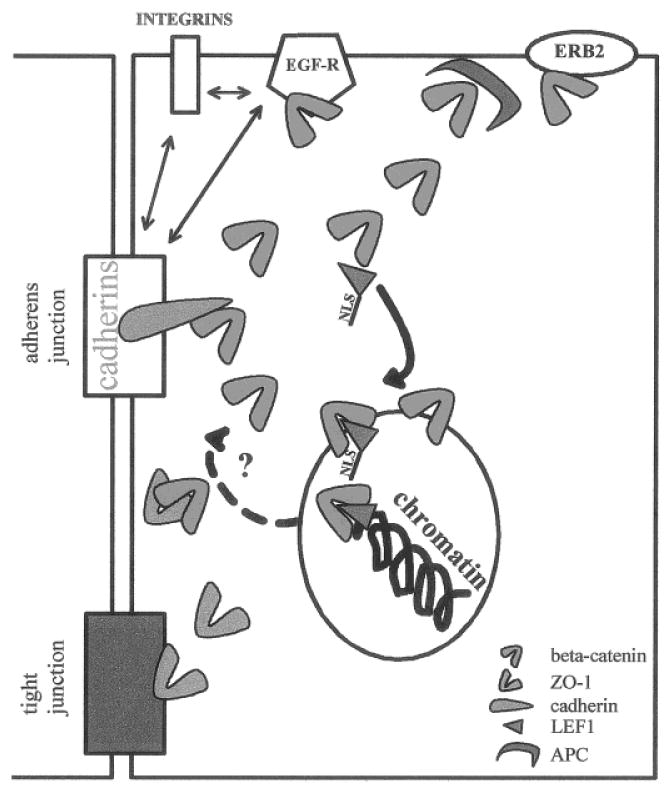
Molecular equilibrium. CMS proteins are engaged in a number of interactions with different cellular components. The relative abundance of the binding partners of CMS proteins regulates the molecular equilibrium, which in turn affects cellular function. For example, β-catenin can interact with cadherins (light blue), ZO-1 (green), APC (dark blue), EGF receptor (EGF-R), and erb-2 at the cell membrane. Moreover, cadherin/catenin, EGF-R, β-1-integrin and α6-β4 integrins interact with each other. β-Catenin binding with transcription factor LEF-1(brown) and its translocation to the nucleus, depends on the existence of a free pool of β-catenin; it may also be influenced by the amount of LEF-1 available. Other factors may contribute to the shift of the molecular equilibrium, like the interaction of plakoglobin with β-catenin partners, and Wnt-1 expression which influences the degradation process associated with the formation of APC/β-catenin complexes. The presence GSK3 β and Axin that interact with APC/β-catenin complexes may also influence the molecular equilibrium. The return of β-catenin from the nucleus (dotted arrow) to interact with cell membrane binding partners, upon reception of specific signals, remains to be demonstrated.
Displacement of individual molecular equilibria resulting in the accumulation of CMS proteins in the nucleus, subsequently influences another level of equilibrium that exists between distant cellular compartments (e.g., membrane skeleton and nucleus) (Fig. 4). A prolonged shift in the compartmental equilibrium may ultimately lead to the development of aberrant cellular behavior, including malignancy. We have recently shown that it is possible to reestablish the equilibrium and to revert the malignant phenotype by correcting the levels and the signaling of cell adhesion components [Weaver et al., 1997], as well as other cell surface receptors [Wang et al., 1998]. Thus, an “oncogene” [β-catenin—Peifer, 1997] or tumor suppressor [ZO-1-Willot et al., 1993] function could be linked to the relative localization of CMS proteins, depending on whether they are in the nuclear or in the membrane skeleton compartment. The cytoplasmic localization of c-abl kinase has been associated with the expression of the malignant phenotype [Van Etten et al., 1989; Sawyers 1992], whereas in nontumor cells, its nuclear localization has been demonstrated to be essential for the inhibition of cell growth [Sawyers et al., 1994]. Interestingly, c-abl is primarily considered a nuclear protein [Van Etten et al., 1989]. It binds to DNA and RNA polymerase II [Kipreos and Wang, 1992; Rajasekaran et al., 1996]. Moreover, Rb protein has been reported to bind c-abl during G1 phase and to inhibit its kinase activity [Welch and Wang, 1993]. Nevertheless, c-abl does not require Rb to exert its growth suppressive activity [Sawyers et al., 1994]. c-abl harbors NLSs and a functional NES and has been shown to shuttle between the nucleus and the cytoplasm [Lewis et al., 1996; Taagepera et al., 1998]. The rate of nuclear import and export of c-abl is regulated by adhesion to the ECM, which also regulates c-abl tyrosine kinase activity [Lewis et al., 1996]. Since c-abl activation requires cell adhesion, and adhesion to ECM recruits c-abl to early focal adhesions coincident with export of the protein from the nucleus, and finally the active nuclear c-abl seems to originate from the cytoplasmic pool activated by adhesion, it has been proposed that c-abl is an important mediator of integrin signals to the nucleus [Lewis et al., 1996]. It has also been proposed recently that CMS or adhesion plaque proteins (e.g., plakophilins, β-catenin, plakoglobin, symplekin) may be genuine nuclear proteins that could be recruited to the membrane skeleton to participate in the assembly of cell adhesion complexes during cell differentiation [Keon et al., 1996]. This seductive hypothesis will require more evidence starting with the elucidation of the role played by CMS proteins in the nucleus. CMS proteins that translocate to the nucleus may also be shuttling proteins, as suggested for zyxin [Nix and Beckerle, 1997].
Fig. 4.
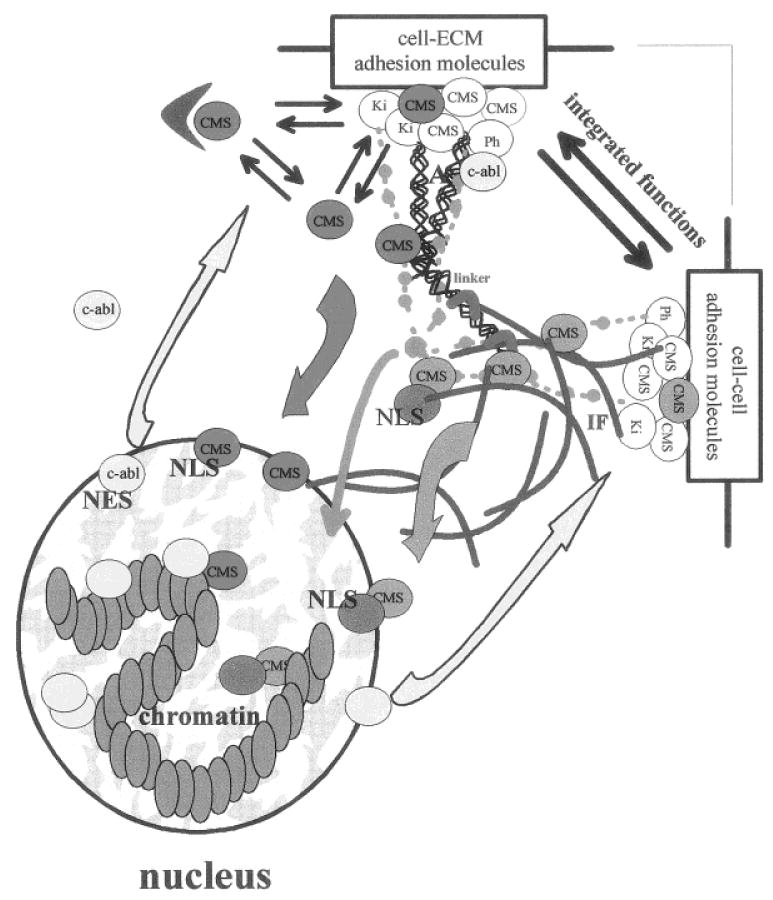
Compartmental equilibrium and dynamic reciprocity. Nuclear translocation of CMS proteins (red and orange) modifies the balance of these proteins between the membrane skeleton compartment and the nuclear compartment and shifts their molecular equilibrium toward an increase of interactions with nuclear targets. Translocating CMS proteins can be considered as structural signal transducers that act as mediators of cell-cell and cell-ECM signaling, along with biochemical cascades (light green) possibly superimposed on signaling via tension generated through actin (A) and intermediate filament (IF) networks. CMS proteins could translocate into the nucleus on their own (red) and bind to nuclear proteins (yellow), or they could travel with a carrier (dark green). The transfer of information from the nucleus to the cell membrane includes the synthesis of membrane skeleton, cell membrane, and ECM components, it may also involve feedback reactions to generated tensional force (not represented). The translocation of nuclear CMS proteins back to the membrane skeleton may also participate in inside/out signaling. Similarly, proteins primarily located in the nucleus could travel to the cell membrane (yellow), as shown with c-abl. We propose that nuclear structural proteins involved in supramolecular organization of the nucleus also may travel to the cell membrane. The balance between these interactive signaling pathways represents the dynamic reciprocity that governs cellular and tissue behavior.
Shuttling proteins are defined as proteins that continuously travel from the cytoplasm to the nucleus [Schmidt-Zachmann et al., 1993]. Although not always demonstrated, it is understood that shuttling must be associated with the transport of information. If primary localization sites of shuttling proteins are in the cytoplasm, as determined by the visualization of supramolecular organizations due to the cytoplasmic concentration of shuttling protein-binding partners or the masking/unmasking of specific targeting sequences, a very rapid translocation in and out of the nucleus will result in immunostaining seen almost exclusively in the cytoplasm. The same line of reasoning is true for a primary localization site in the nucleus. Many types of supramolecular organization have been described in the nucleus, including transcription sites, speckles, coiled bodies, and PML bodies [Nickerson et al., 1995]. Specific types of supramolecular organization in the nucleus have also been described to occur in association with tissue-like morphogenesis [Lelièvre et al., 1998]. The shuttling of components found primarily in the nucleus has been described for snRNAs and RNA-binding proteins, as well as for proteins participating in nuclear import, heat shock proteins, and nucleolar proteins [Schmidt-Zachmann et al., 1993; Görlich and Mattaj, 1996], and the shuttling of molecules from sites predominantly located in the nucleus to the cytoplasm has been suggested to participate in the regulation of nuclear functions.
Sooner or later, we must put a plan together as to how parts are integrated to bring about homeostasis. Recent data demonstrating the coupling of integrins and EGF receptor pathways in epithelial cells cultured three-dimensionally in the presence of a reconstituted basement membrane, as opposed to monolayer culture [Wang et al., 1998], as well as the demonstration of movement of specific mRNAs to focal adhesion complexes [Chicurel et al., 1998], point to the intimate relationship between positional and functional information. To complete the dynamic reciprocity scheme, we anticipate that, as with the behavior of CMS proteins, more resident structural components of the nucleus may travel in the opposite direction to the cell membrane and hence act as nuclear signal transducers.
Acknowledgments
We thank Dr. C. Hagios and R. Boudreau for critical reading of the manuscript, and Drs. C. Larabell and K. Schmeichel for helpful comments. This work was supported by the U.S. Department of Energy, Office of Biological and Environmental Research, contract DE-AC03–76SF00098; by the National Institutes of Health, grants CA64786 and CA57621; and by a Department of Defense/Breast Cancer Research Program fellowship (to S.A.L.).
Contract grant sponsor: U.S. Department of Energy, Office of Biological and Environmental Research; Contract grant number: DE-AC03–76SF00098; Contract grant sponsor: National Institutes of Health, Contract grant number: CA64786; Contract grant number: CA57621; Contract grant sponsor: Department of Defense/Breast Cancer Research Program.
Abbreviations
- CMS protein
connective membrane skeleton protein
- FA
focal adhesion
- NLS
nuclear localization signal
- NES
nuclear export signal
References
- Alberts AS, Geneste O, Treisman R. Activation of SRF-regulated chromosomal templates by Rho-family GTPases requires a signal that also induces H4 hyperacetylation. Cell. 1998;92:475–487. doi: 10.1016/s0092-8674(00)80941-1. [DOI] [PubMed] [Google Scholar]
- Amar LS, Shabana AH, Oboeuf M, Martin N, Forest N. Desmosomes are regulated by protein kinase C in primary rat epithelial cells. Cell Adhes Commun. 1998;5:1–12. doi: 10.3109/15419069809005594. [DOI] [PubMed] [Google Scholar]
- Baichwal VR, Park A, Tjian R. v-Src and EJ Ras alleviate repression of c-Jun by cell-specific inhibitor. Nature. 1991;352:164–168. doi: 10.1038/352165a0. [DOI] [PubMed] [Google Scholar]
- Bailey T, Biddlestone L, Shepherd N, Barr H, Warner P, Jankowski J. Altered cadherin and catenin complexes in the Barrett's eosophagus-dysplasia-adenocarcinoma sequence: Correlation with disease progression and dedifferentiation. Am J Pathol. 1998;152:135–144. [PMC free article] [PubMed] [Google Scholar]
- Balda MS, Matter K. Tight junctions. J Cell Sci. 1998;111:541–547. doi: 10.1242/jcs.111.5.541. [DOI] [PubMed] [Google Scholar]
- Barcellos-Hoff MH, Aggeler J, Ram TJ, Bissell MJ. Functional differentiation and alveolar morphogenesis of primary mammary cultures on reconstituted basement membrane. Development. 1989;105:223–235. doi: 10.1242/dev.105.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckerle MC. Zyxin: Zinc fingers at sites of cell adhesion. BioEssays. 1997;19:949–957. doi: 10.1002/bies.950191104. [DOI] [PubMed] [Google Scholar]
- Behrens J, von Kries JP, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- Benes C, Roisin MP, Van Tan H, Creuzet C, Miyazaki J, Fagard R. Rapid activation and nuclear translocation of mitogen-activated protein kinases in response to physiological concentration of glucose in the MIN6 pancreatic beta cell line. J Biol Chem. 1998;273:15507–15513. doi: 10.1074/jbc.273.25.15507. [DOI] [PubMed] [Google Scholar]
- Ben-Ze'ev A. Cytoskeletal and adhesion proteins as tumor suppressors. Curr Opin Cell Biol. 1997;9:99–108. doi: 10.1016/s0955-0674(97)80158-5. [DOI] [PubMed] [Google Scholar]
- Bissell MJ. Glandular structure and gene expression. Lessons from the mammary gland. Ann NY Acad Sci. 1998;842:1–6. doi: 10.1111/j.1749-6632.1998.tb09625.x. [DOI] [PubMed] [Google Scholar]
- Bissell DM, Arenson DM, Maher JJ, Roll RF. Support of cultured hepatocytes by a laminin-rich gel. Evidence for a functionally significant subendothelial matrix in normal rat liver. J Clin Invest. 1987;79:801–812. doi: 10.1172/JCI112887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissell MJ, Hall HG, Parry G. How does the extracellular matrix direct gene expression? J Theor Biol. 1982;99:31–68. doi: 10.1016/0022-5193(82)90388-5. [DOI] [PubMed] [Google Scholar]
- Boudreau N, Sympson CJ, Werb Z, Bissell MJ. Suppression of ICE and apoptosis in mammary epithelial cells by extracellular matrix. Science. 1995;267:891–893. doi: 10.1126/science.7531366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannon M, Gomperts M, Sumoy L, Moon R, Kimelman D. A beta-catenin/XTcf-3 complex binds to the siamois promoter to regulate dorsal axis specification in Xenopus. Genes Dev. 1997;11:2359–2370. doi: 10.1101/gad.11.18.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brott BK, Pinsky BA, Erikson RL. Nlk is a murine kinase related to Erk/MAP kinases and localized in the nucleus. Proc Natl Acad Sci USA. 1998;95:963–969. doi: 10.1073/pnas.95.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugge JS. Casting light on focal adhesions. Nature Genet. 1998;19:309–311. doi: 10.1038/1189. [DOI] [PubMed] [Google Scholar]
- Capco DG, Kromalnic G, Penman S. A new method for embedment-free sections for transmission electron microscopy: Applications to the cytoskeletal framework and other three-dimensional networks. J Cell Biol. 1984;98:1878–1885. doi: 10.1083/jcb.98.5.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso C, Leonhardt H. Protein targeting to subnuclear higher order structures: A new level of regulation and coordination of nuclear processes. J Cell Biochem. 1998;70:222–230. [PubMed] [Google Scholar]
- Caron J. Induction of albumin gene transcription in hepatocytes by extracellular matrix. Mol Cell Biol. 1990;10:1239–1243. doi: 10.1128/mcb.10.3.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho RS, Schaffer JL, Gerstenfeld LC. Osteoblasts induce osteopontin expression in response to attachment on fibronectin: demonstration of a common role for integrin receptors in the signal transduction processes of cell attachment and mechanical stimulation. J Cell Biochem. 1998;70:376–390. doi: 10.1002/(sici)1097-4644(19980901)70:3<376::aid-jcb11>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Cereijido M, Valdes J, Shoshani L, Contreras RG. Role of tight junctions in establishing and maintaining cell polarity. Annu Rev Physiol. 1998;60:161–177. doi: 10.1146/annurev.physiol.60.1.161. [DOI] [PubMed] [Google Scholar]
- Chen RH, Sarnecki C, Blenis J. Nuclear localization and regulation of erk- and rsk-encoded protein kinases. Mol Cell Biol. 1992;12:915–927. doi: 10.1128/mcb.12.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicurel ME, Singer RH, Meyer CJ, Ingber DE. Integrin binding and mechanical tension induce movement of mRNA and ribosomes to focal adhesions. Nature. 1998;392:730–733. doi: 10.1038/33719. [DOI] [PubMed] [Google Scholar]
- Citi S. The molecular organization of tight junctions. J Cell Biol. 1993;121:485–489. doi: 10.1083/jcb.121.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark EA, King KW, Brugge JS, Symons M, Hynes RO. Integrin-mediated signals regulated by members of the rho family of GTPases. J Cell Biol. 1998;142:573–586. doi: 10.1083/jcb.142.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collares-Buzato CB, Jepson MA, Simmons NL, Hirst BH. Increased tyrosine phosphorylation causes redistribution of adherens junction and tight junction proteins and perturbs paracellular barrier function in MDCK epithelia. Eur J Cell Biol. 1998;76:85–92. doi: 10.1016/S0171-9335(98)80020-4. [DOI] [PubMed] [Google Scholar]
- Correas I. Characterization of isoforms of protein 4.1 present in the nucleus. Biochem J. 1991;279:581–585. doi: 10.1042/bj2790581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carcer G, Lallena MJ, Correas I. Protein 4.1 is a component of the nuclear matrix of mammalian cells. Biochem J. 1995;312:871–877. doi: 10.1042/bj3120871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efstathiou JA, Noda M, Rowan A, Dixon C, Chinery R, Jawhari A, Hattori T, Wright NA, Bodmer WF, Pignatelli M. Intestinal trefoil factor controls the expression of the adenomatous polyposis coli-catenin and the E-cadherin-catenin complexes in human colon carcinoma cells. Proc Natl Acad Sci USA. 1998;95:3122–3127. doi: 10.1073/pnas.95.6.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel K, Kotlyarov A, Gaestel M. Leptomycin B-sensitive nuclear export of MAPKAP kinase 2 is regulated by phosphorylation. EMBO J. 1998;17:3363–3371. doi: 10.1093/emboj/17.12.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagotto F, Gluck U, Gumbiner BM. Nuclear localization signal-independent and importin/karyopherin-independent nuclear import of beta-catenin. Curr Biol. 1998;8:181–190. doi: 10.1016/s0960-9822(98)70082-x. [DOI] [PubMed] [Google Scholar]
- Farquhar MG, Palade GE. Junctional complexes in various epithelia. J Cell Biol. 1963;17:375–412. doi: 10.1083/jcb.17.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch SM, Francis H. Disruption of epithelial cell-matrix interactions induces apoptosis. J Cell Biol. 1994;124:619–626. doi: 10.1083/jcb.124.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E, Cleveland DE. A structural scaffolding of intermediate filaments in health and disease. Science. 1998;279:514–519. doi: 10.1126/science.279.5350.514. [DOI] [PubMed] [Google Scholar]
- Funayama N, Fagotto F, McCrea P, Gumbiner BM. Embryonic axis induction by the armadillo repeat domain of beta-catenin. J Cell Biol. 1995;128:959–968. doi: 10.1083/jcb.128.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamallo C, Palacios J, Suarez A, Pizarro A, Navarro P, Quintanilla M, Cano A. Correlation of E-cadherin expression with differentiation grade and histological type in breast carcinoma. Am J Pathol. 1993;142:987–993. [PMC free article] [PubMed] [Google Scholar]
- Giancotti FG. Signal transduction by the alpha6-beta4 integrin: Charting the path between laminin binding and nuclear events. J Cell Sci. 1996;109:1165–1172. doi: 10.1242/jcs.109.6.1165. [DOI] [PubMed] [Google Scholar]
- Goldsteyn RM, Beckerle MC, Koay T, Friederich E. Structural and functional similarities between the human cytoskeletal protein zyxin and the ActA protein of Listeria monocytogenes. J Cell Sci. 1997;110:1893–1906. doi: 10.1242/jcs.110.16.1893. [DOI] [PubMed] [Google Scholar]
- Görlich D, Mattaj LW. Nucleocytoplasmic transport. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- Gottardi CJ, Arpin M, Fanning AS, Louvard D. The junction-associated protein zonula occludens-1, localizes to the nucleus before the maturation and during the remodeling of cell-cell contacts. Proc Natl Acad Sci USA. 1996;93:10779–10784. doi: 10.1073/pnas.93.20.10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger BL, Lazarides E. Membrane skeletal protein 4.1 of avian erythrocytes is composed of multiple variants that exhibit tissue-specific expression. Cell. 1984;37:595–607. doi: 10.1016/0092-8674(84)90390-8. [DOI] [PubMed] [Google Scholar]
- Green KJ, Kowalczyk AP, Bornslaeger EA, Palka HL, Norvell SM. Desmosomes: integrators of mechanical integrity in tissues. Biol Bull. 1998;194:374–376. doi: 10.2307/1543117. [DOI] [PubMed] [Google Scholar]
- Gumbiner BM. Signal transduction of beta-catenin. Curr Opin Cell Biol. 1995;7:634–640. doi: 10.1016/0955-0674(95)80104-9. [DOI] [PubMed] [Google Scholar]
- Gumbiner BM. Cell adhesion: The molecular basis of tissue architecture and morphogenesis. Cell. 1996;84:345–357. doi: 10.1016/s0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- Hagios C, Lochter A, Bissell MJ. Tissue architecture: the ultimate regulator of epithelial function? Philos Trans R Soc Lond. 1998;353:857–870. doi: 10.1098/rstb.1998.0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haskins J, Gu L, Wittchen ES, Hibbard J, Stevenson BR. ZO-3, a novel member of the MAGUK protein family found at the tight junction, interacts with ZO-1 and occludin. J Cell Biol. 1998;141:199–208. doi: 10.1083/jcb.141.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmke S, Lohse K, Mikule K, Wood MR, Pfenninger KH. SRC binding to the cytoskeleton, triggered by growth cone attachment to laminin, is protein tyrosine phosphatase-dependent. J Cell Sci. 1998;111:2465–2475. doi: 10.1242/jcs.111.16.2465. [DOI] [PubMed] [Google Scholar]
- Herminston ML, Wong MH, Gordon JI. Forced expression of E-cadherin in the mouse intestinal epithelium slows cell migration and provides evidence for non autonomous regulation of cell fate in a self-renewing system. Genes Dev. 1996;10:985–996. doi: 10.1101/gad.10.8.985. [DOI] [PubMed] [Google Scholar]
- Hoffman M, Kibbey M, Nomizu M, Kleinman HK. Laminin peptides promote acinar-like development of a human submandibular gland cell line (HSG) in vitro. Mol Biol Cell. 1995;6(suppl):169a. [Google Scholar]
- Huber O, Korn R, McLaughlin J, Oshugi M, Herrmann BG. Nuclear localization of beta-catenin by interaction with transcription factor LEF-1. Mech Dev. 1996;59:3–11. doi: 10.1016/0925-4773(96)00597-7. [DOI] [PubMed] [Google Scholar]
- Ingber DE. Tensegrity: the architectural basis of cellular mechano-transduction. Annu Rev Physiol. 1997;59:575–599. doi: 10.1146/annurev.physiol.59.1.575. [DOI] [PubMed] [Google Scholar]
- Jones JC, Hopkinson SB, Goldfinger LE. Structure and assembly of hemidesmosomes. BioEssays. 1998;20:488–494. doi: 10.1002/(SICI)1521-1878(199806)20:6<488::AID-BIES7>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Juliano RL, Haskill S. Signal transduction from the extracellular matrix. J Cell Biol. 1993;120:577–585. doi: 10.1083/jcb.120.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnowsky A, Klymkowsky MW. Anterior axis duplication in Xenopus induced by the over-expression of the cadherin-binding protein plakoglobin. Proc Natl Acad Sci USA. 1995;92:4522–4526. doi: 10.1073/pnas.92.10.4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartenbeck J, Schmid E, Franke WW, Geiger B. Different modes of internalization of proteins associated with adhaerens junctions and desmosomes: experimental separation of lateral contacts induces endocytosis of desmosomal plaque material. EMBO J. 1982;1:725–732. doi: 10.1002/j.1460-2075.1982.tb01237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keon BH, Schäfer S, Kuhn C, Grund C, Franke WW. Symplekin, a novel type of tight junction plaque protein. J Cell Biol. 1996;134:1003–1018. doi: 10.1083/jcb.134.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khokhlatchev AV, Canagarajah B, Wilsbacher J, Robinson M, Atkinson M, Goldsmith E, Cobb M. Phosphorylation of the MAP kinase ERK2 promotes its homodimerization and nuclear translocation. Cell. 1998;93:605–615. doi: 10.1016/s0092-8674(00)81189-7. [DOI] [PubMed] [Google Scholar]
- Kipreos ET, Wang JY. Cell cycle-regulated binding of c-Abl tyrosine kinase to DNA. Science. 1992;256:382–385. doi: 10.1126/science.256.5055.382. [DOI] [PubMed] [Google Scholar]
- Krauss SW, Larabell CA, Lockett S, Gascard P, Penman S, Mohandas N, Chasis JA. Structural protein 4.1 in the nucleus of human cells: Dynamic rearrangements during cell division. J Cell Biol. 1997;137:275–289. doi: 10.1083/jcb.137.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallena MJ, Correas I. Transcription-dependent redistribution of nuclear protein 4.1 to SC35-enriched nuclear domains. J Cell Sci. 1997;110:239–247. doi: 10.1242/jcs.110.2.239. [DOI] [PubMed] [Google Scholar]
- Lallena MJ, Martinez C, Varcarcel J, Correas I. Functional association of nuclear protein 4.1 with pre-mRNA splicing factors. J Cell Sci. 1998;111:1963–1971. doi: 10.1242/jcs.111.14.1963. [DOI] [PubMed] [Google Scholar]
- Larabell CA, Torres M, Rowning BA, Yost C, Miller JR, Wu M, Kimelman D, Moon R. Establishment of the dorso-ventral axis in Xenopus embryos is presaged by early asymetries in beta-catenin that are modulated by the Wnt signaling pathway. J Cell Biol. 1997;136:1123–1136. doi: 10.1083/jcb.136.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelièvre SA, Weaver VM, Nickerson JA, Larabell CA, Bhaumik A, Petersen OW, Bissell MJ. Tissue phenotype depends on reciprocal interactions between the extracellular matrix and the structural organization of the nucleus. Proc Natl Acad Sci USA. 1998 doi: 10.1073/pnas.95.25.14711. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leto TL, Pratt BM, Madri JA. Mechanisms of cytoskeletal regulation: modulation of aortic endothelial cell protein band 4.1 by the extracellular matrix. J Cell Physiol. 1986;127:423–431. doi: 10.1002/jcp.1041270311. [DOI] [PubMed] [Google Scholar]
- Lewis JM, Baskaran R, Taagepera S, Schwartz MA, Wang JY. Integrin regulation of c-Abl tyrosine kinase activity ad cytoplasmic-nuclear transport. Proc Natl Acad Sci USA. 1996;93:15174–15179. doi: 10.1073/pnas.93.26.15174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ML, Aggeler J, Farson DA, Hatier C, Hassell J, Bissell MJ. Influence of a reconstituted basement membrane and its components on casein gene expression and secretion in mouse mammary epithelial cells. Proc Natl Acad Sci USA. 1987;84:136–140. doi: 10.1073/pnas.84.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JK, Di Persio CM, Zaret KS. Extracellular signals that regulate liver transcription factors during hepatic differentiation in vitro. Mol Cell Biol. 1991;11:773–784. doi: 10.1128/mcb.11.2.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochter A, Bissell MJ. Involvement of extracellular matrix constituents in breast cancer. Semin Cancer Biol. 1995;6:165–173. doi: 10.1006/scbi.1995.0017. [DOI] [PubMed] [Google Scholar]
- Loidl P. Histone acetylation: Facts and questions. Chromosoma. 1994;103:441–449. doi: 10.1007/BF00337382. [DOI] [PubMed] [Google Scholar]
- Longhurst CM, Jennings LK. Integrin-mediated signal transduction. Cell Mol Life Sci. 1998;54:514–526. doi: 10.1007/s000180050180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna EJ, Hitt AL. Cytoskeleton-plasma membrane interactions. Science. 1992;258:955–964. doi: 10.1126/science.1439807. [DOI] [PubMed] [Google Scholar]
- Luque CM, Lallena MJ, Alonso MA, Correas I. An alternative domain determines nuclear localization in multifunctional protein 4.1. J Biol Chem. 1998;273:11643–11649. doi: 10.1074/jbc.273.19.11643. [DOI] [PubMed] [Google Scholar]
- Maniotis AJ, Chen CS, Ingber DE. Demonstration of mechanical connections between integrins, cytoskeletal filaments and nucleoplasm that stabilize nuclear structure. Proc Natl Acad Sci USA. 1997;94:849–854. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins-Green M, Bissell MJ. Cell-ECM interactions in development. Semin Dev Biol. 1995;6:149–159. [Google Scholar]
- Matsuzaki F, Sutoh K, Ikai A. Structural unit of the erythrocyte cytoskeleton. Isolation and electron microscopic examination. Eur J Cell Biol. 1985;39:153–160. [PubMed] [Google Scholar]
- Matter ML, Laurie G. A novel laminin E8 cell adhesion site required for lung alveolar formation in vitro. J Cell Biol. 1994;124:1083–1090. doi: 10.1083/jcb.124.6.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCrea PD, Brieher WM, Gumbiner B. Induction of a secondary body axis in Xenopus by antibodies to beta-catenin. J Cell Biol. 1993;123:477–484. doi: 10.1083/jcb.123.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens C, Kuhn C, Franke WW. Plakophilins 2a and 2b: Constitutive proteins of dual location in the karyoplasm and the desmosomal plaque. J Cell Biol. 1996;135:1009–1025. doi: 10.1083/jcb.135.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JR, Moon RT. Analysis of the signaling of localization of mutants of beta-catenin during axis specification in Xenopus. J Cell Biol. 1997;139:229–243. doi: 10.1083/jcb.139.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitic LL, Anderson JM. Molecular architecture of tight junctions. Annu Rev Physiol. 1998;60:121–142. doi: 10.1146/annurev.physiol.60.1.121. [DOI] [PubMed] [Google Scholar]
- Moroianu J. Distinct nuclear import and export pathways mediated by members of the karyopherin beta family. J Cell Biochem. 1998;70:231–239. [PubMed] [Google Scholar]
- Munemitsu S, Albert I, Souza B, Rubinfeld B, Polakis P. Regulation of intracellular beta-catenin levels by the adenomatous polyposis coli (APC) tumor-suppressor protein. Proc Natl Acad Sci USA. 1995;92:3046–3050. doi: 10.1073/pnas.92.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers CA, Schmidhauser C, Mellentin-Michelotti J, Fragoso G, Roskelley CD, Casperson G, Mossi R, Pujuguet P, Hager G, Bissell MJ. Characterization of BCE-1, a transcriptional enhancer regulated by prolactin and extracellular matrix and modulated by the state of histone acetylation. Mol Cell Biol. 1998;18:2184–2195. doi: 10.1128/mcb.18.4.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny S, Dreyfuss G. The hnRNP C proteins contain a nuclear retention sequence that can override nuclear export signal. J Cell Biol. 1996;134:1365–1373. doi: 10.1083/jcb.134.6.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathke IS, Hinck L, Swedlow JR, Papkoff J, Nelson WJ. Defining interactions and distributions of cadherin and catenin complexes in polarized epithelial cells. J Cell Biol. 1994;125:1341–1352. doi: 10.1083/jcb.125.6.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson JA, Blencowe BJ, Penman S. The architectural organization of nuclear metabolism. Int Rev Cytol. 1995;162A:67–123. doi: 10.1016/s0074-7696(08)61229-2. [DOI] [PubMed] [Google Scholar]
- Nikolic B, Mac Nulty E, Mir B, Wiche G. Basic amino acid residue cluster within nuclear targeting sequence motif is essential for cytoplasmic plectin-vimentin network junctions. J Cell Biol. 1996;134:1455–1467. doi: 10.1083/jcb.134.6.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nix DA, Beckerle MC. Nuclear-cytoplasmic shuttling of the focal contact protein, zyxin: a potential mechanism for communication between sites of cell adhesion and the nucleus. J Cell Biol. 1997;138:1139–1147. doi: 10.1083/jcb.138.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill EM, Rebay I, Tjian R, Rubin GM. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell. 1994;78:137–147. doi: 10.1016/0092-8674(94)90580-0. [DOI] [PubMed] [Google Scholar]
- Owen TA, Holthuis J, Markose E, van Wijnen AJ, Wolfe SA, Grimes SR, Lian JB, Stein GS. Modifications of protein-DNA interactions in the proximal promoter of a cell growth-regulated histone gene during onset and progression of osteoblast differentiation. Proc Natl Acad Sci USA. 1990;87:5129–5133. doi: 10.1073/pnas.87.13.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papkoff J, Rubinfeld B, Schryver B, Polakis P. Wnt-1 regulates free pools of catenins and stabilizes APC-catenin complexes. Mol Cell Biol. 1996;16:2128–2134. doi: 10.1128/mcb.16.5.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M. beta-catenin as oncogene: the smoking gun. Science. 1997;275:1752–1753. doi: 10.1126/science.275.5307.1752. [DOI] [PubMed] [Google Scholar]
- Peifer M, Berg S, Reynolds AB. A repeating amino acid motif shared by proteins with diverse cellular roles. Cell. 1994;76:789–791. doi: 10.1016/0092-8674(94)90353-0. [DOI] [PubMed] [Google Scholar]
- Perl AK, Wilgenbus P, Dahl U, Semb H, Christofori G. A causal role for E-cadherin in the transition from adenoma to carcinoma. Nature. 1998;392:190–192. doi: 10.1038/32433. [DOI] [PubMed] [Google Scholar]
- Petersen OW, Ronnov-Jessen L, Howlett AR, Bissell MJ. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc Natl Acad Sci USA. 1992;89:9064–9068. doi: 10.1073/pnas.89.19.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pienta KJ, Coffey DS. Nuclear-cytoskeletal interactions: Evidence for physical connections between the nucleus and cell periphery and their alteration by transformation. J Cell Biochem. 1992;49:357–365. doi: 10.1002/jcb.240490406. [DOI] [PubMed] [Google Scholar]
- Potempa S, Ridley AJ. Activation of both MAP kinase and phosphatidylinositide 3-kinase by ras is required for hepatocyte growth factor/scatter factor-induced adherens junction disassembly. Mol Biol Cell. 1998;9:2185–2200. doi: 10.1091/mbc.9.8.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasekaran AK, Hojo M, Huima T, Rodriguez-Boulan E. Catenins and zonula occludens-1 form a complex during early stages in the assembly of tight junctions. J Cell Biol. 1996;132:451–463. doi: 10.1083/jcb.132.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redfield A, Nieman MT, Knudsen KA. Cadherins promote skeletal muscle differentiation in three-dimensional cultures. J Cell Biol. 1997;138:1323–1331. doi: 10.1083/jcb.138.6.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezniczek GA, de Pereda JM, Reipert S, Wiche G. Linking integrin alpha6beta4-based cell adhesion to the intermediate filament cytoskeleton: direct interaction between the beta4 subunit and plectin at multiple molecular sites. J Cell Biol. 1998;141:209–225. doi: 10.1083/jcb.141.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards SA, Lousbury KM, Carey KL, Macara IG. A nuclear export signal is essential for the cytosolic localization of the ran binding protein, RanBP1. J Cell Biol. 1996;134:1157–1168. doi: 10.1083/jcb.134.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanghera JS, Peter M, Nigg EA, Pelech SL. Immunological characterization of avian MAP kinases: Evidence for nuclear localization. Mol Biol Cell. 1992;3:775–787. doi: 10.1091/mbc.3.7.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyers CL. The bcr-abl gene in chronic myelogenous leukemia. Cancer Surveys. 1992;15:37–51. [PubMed] [Google Scholar]
- Sawyers CL, McLaughlin J, Goga A, Havlik M, Witte O. The nuclear tyrosine kinase c-Abl negatively regulates cell growth. Cell. 1994;77:121–131. doi: 10.1016/0092-8674(94)90240-2. [DOI] [PubMed] [Google Scholar]
- Schaapveld RQ, Borradori L, Geerts D, van Leusden MR, Kuikman I, Nievers MG, Niessen CM, Steenbergen RD, Snijders PJ, Sonnenberg A. Hemidesmosome formation is initiated by the beta4 integrin subunit, requires complex formation of beta4 and HD1/plectin, and involves a direct interaction between beta4 and the bullous pemphigoid antigen 180. J Cell Biol. 1998;142:271–284. doi: 10.1083/jcb.142.1.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaepfer DD, Hunter T. Signal transduction from the extracellular matrix-A role for the focal adhesion protein-tyrosine kinase FAK. Cell Struct Funct. 1996;21:445–450. doi: 10.1247/csf.21.445. [DOI] [PubMed] [Google Scholar]
- Schmeichel KL, Beckerle MC. The LIM domain is a modular protein-binding interface. Cell. 1994;79:211–219. doi: 10.1016/0092-8674(94)90191-0. [DOI] [PubMed] [Google Scholar]
- Schmeichel KL, Beckerle MC. Molecular dissection of a LIM domain. Mol Biol Cell. 1997;8:219–230. doi: 10.1091/mbc.8.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidhauser C, Bissell MJ, Myers CA, Casperson GF. Extracellular matrix and hormones transcriptionally regulate bovine beta-casein 5′ sequences in stably transfected mouse mammary cells. Proc Natl Acad Sci USA. 1990;87:9118–9122. doi: 10.1073/pnas.87.23.9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidhauser C, Casperson GF, Myers CA, Sanzo KT, Bolten S, Bissell MJ. A novel transcriptional enhancer is involved in the prolactin and ECM-dependent regulation of beta-casein gene expression. Mol Biol Cell. 1992;3:699–709. doi: 10.1091/mbc.3.6.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Zachmann MS, Dargemont C, Kühn LC, Nigg EA. Nuclear export of proteins: the role of nuclear retention. Cell. 1993;74:493–504. doi: 10.1016/0092-8674(93)80051-f. [DOI] [PubMed] [Google Scholar]
- Schneider GB, Gilmore AP, Lohse DL, Pomer LH, Burridge K. Microinjection of protein tyrosine phosphatases into fibroblasts disrupts focal adhesions and stress fibers. Cell Adhes Commun. 1998;5:207–219. doi: 10.3109/15419069809040292. [DOI] [PubMed] [Google Scholar]
- Sharma SV. Rapid recruitment of p120RasGAP and its associated protein p190RhoGAP, to the cytoskeleton during integrin mediated cell-substrate interaction. Oncogene. 1998;17:271–281. doi: 10.1038/sj.onc.1201921. [DOI] [PubMed] [Google Scholar]
- Shinohara M, Hiraki A, Ikebe T, Nakamura S, Kurahara S, Shirasuma K, Garrod DR. Immunohistochemical study of desmosomes in oral squamous cell carcinoma: correlation with cytokeratin and E-cadherin staining, and with tumor behavior. J Pathol. 1998;184:369–381. doi: 10.1002/(SICI)1096-9896(199804)184:4<369::AID-PATH1236>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Short SM, Talbot GA, Juliano RL. Integrin-mediated signaling events in human endothelial cells. Mol Biol Cell. 1998;9:1969–1980. doi: 10.1091/mbc.9.8.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simcha I, Shtutman M, Salomon D, Zhurinsky J, Sadot E, Geiger N, Ben-Ze'ev A. Differential nuclear translocation and transactivation potential of beta-catenin and plakoglobin. J Cell Biol. 1998;141:1433–1448. doi: 10.1083/jcb.141.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims JR, Karp S, Ingber DE. Altering the cellular mechanical force balance results in integrated changes in cell, cytoskeletal and nuclear shape. J Cell Sci. 1992;303:1215–1222. doi: 10.1242/jcs.103.4.1215. [DOI] [PubMed] [Google Scholar]
- Smith EA, Fuchs E. Defining the interactions between intermediate filaments and desmosomes. J Cell Biol. 1998;141:1229–1241. doi: 10.1083/jcb.141.5.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sympson CJ, Bissell MJ, Werb Z. Mammary gland tumor formation in transgenic mice overexpressing stromelysin-1. Semin Cancer Biol. 1995;6:159–163. doi: 10.1006/scbi.1995.0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sympson CJ, Talhouk RS, Alexander CM, Chin JR, Clift SM, Bissell MJ, Werb Z. Targeted expression of stromelysin-1 in mammary gland provides evidence for a role of proteinases in branching morphogenesis and the requirement for an intact basement membrane for tissue-specific gene expression. J Cell Biol. 1994;125:681–693. doi: 10.1083/jcb.125.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taagepera S, McDonald D, Loeb JE, Whitazer LL, McElroy AK, Wang JYJ, Hope TJ. Nuclear-cytoplasmic shuttling of C-ABL tyrosine kinase. Proc Natl Acad Sci USA. 1998;95:7457–7462. doi: 10.1073/pnas.95.13.7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taieb F, Karaiskou A, Rime H, Jessus C. Human retinoblastoma protein (Rb) is phosphorylated by cdc2 kinase and MAP kinase in Xenopus maturing oocytes. FEBS Lett. 1998;425:465–471. doi: 10.1016/s0014-5793(98)00291-9. [DOI] [PubMed] [Google Scholar]
- Takeichi M. Morphogenetic roles of classical cadherins. Curr Opin Cell Biol. 1995;7:619–627. doi: 10.1016/0955-0674(95)80102-2. [DOI] [PubMed] [Google Scholar]
- Tang TK, Leto TL, Marchesi VT, Benz EJJ. Expression of specific isoforms of protein 4.1 in erythroid and non-erythroid tissues. Adv Exp Med Biol. 1988;241:81–95. doi: 10.1007/978-1-4684-5571-7_12. [DOI] [PubMed] [Google Scholar]
- Tenniswood MP, Guenette RS, Lakins J, Mooibroek M, Wong P, Welsh JE. Active cell death in hormone-dependent tissues. Cancer Metast Rev. 1992;11:197–220. doi: 10.1007/BF00048064. [DOI] [PubMed] [Google Scholar]
- Thomasset N, Lochter A, Sympson CJ, Lund LR, Williams DR, Behrendtsen O, Werb Z, Bissell MJ. Expression of autoactivated stromelysin-1 in mammary glands of transgenic mice leads to reactive stroma during early development. Am J Pathol. 1998;153:457–467. doi: 10.1016/S0002-9440(10)65589-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tselepis C, Chidgey M, North A, Garrod D. Desmosomal adhesion inhibits invasive behavior. Proc Natl Acad Sci USA. 1998;95:8064–8069. doi: 10.1073/pnas.95.14.8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Etten RA, Jackson P, Baltimore D. The mouse type c-abl gene product is a nuclear protein, and activation of transforming ability is associated with cytoplasmic localization. Cell. 1989;58:669–678. doi: 10.1016/0092-8674(89)90102-5. [DOI] [PubMed] [Google Scholar]
- Wang F, Weaver VM, Petersen OW, Larabell CA, Dedhar S, Briand P, Lupu R, Bissell MJ. Reciprocal interactions between beta1-integrin and EGF-R in three dimensional cultures: A new perspective in normal and malignant breast epithelial biology. Proc Natl Acad Sci (USA) 1998;95 doi: 10.1073/pnas.95.25.14821. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver VM, Petersen OW, Wang F, Larabell CA, Briand P, Damsky C, Bissell MJ. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J Cell Biol. 1997;137:231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg E, Sattler M, Ewaniuk DS, Salgia R. Role of focal adhesion proteins in signal transduction and oncogenesis. Crit Rev Oncogene. 1997;8:343–358. doi: 10.1615/critrevoncog.v8.i4.40. [DOI] [PubMed] [Google Scholar]
- Weiss EE, Kroember M, Rudiger AH, Jockush BM, Rudiger M. Vinculin is part of the cadherin-catenin junctional complex: Complex formation between alpha-catenin and vinculin. J Cell Biol. 1998;141:755–764. doi: 10.1083/jcb.141.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch PJ, Wang JY. A C-terminal protein-binding domain in the retinoblastoma protein regulates nuclear c-abl tyrosine kinase in the cell cycle. Cell. 1993;75:779–790. doi: 10.1016/0092-8674(93)90497-e. [DOI] [PubMed] [Google Scholar]
- Wen W, Meinkoth JL, Tsien RY, Taylor SS. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- Wiche G, Gromov D, Donovan A, Castanon MJ, Fuchs E. Expression of plectin mutant cDNA in cultured cells indicates a role of COOH-terminal domain in intermediate filament association. J Cell Biology. 1993;121:607–619. doi: 10.1083/jcb.121.3.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willot E, Balda MS, Fanning AS, Jameson B. The tight junction protein ZO-1 is homologous to the Drosophila discs-large tumor suppressor protein of septate junctions. Proc Natl Acad Sci USA. 1993;90:7834–7838. doi: 10.1073/pnas.90.16.7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Reuver SM, Kuhlendahl S, Chung WJ, Garner CC. Subcellular targeting and cytoskeletal attachment of SAP97 to the epithelial lateral membrane. J Cell Sci. 1998;111:2365–2376. doi: 10.1242/jcs.111.16.2365. [DOI] [PubMed] [Google Scholar]
- Yamada KM, Geiger B. Molecular interaction in cell adhesion complexes. Curr Opin Cell Biol. 1997;9:76–85. doi: 10.1016/s0955-0674(97)80155-x. [DOI] [PubMed] [Google Scholar]
- Yang Y, Dowling J, Yu QC, Kouklis P, Cleveland DW, Fuchs E. An essential cytoskeletal linker protein connecting actin microfilaments to intermediate filaments. Cell. 1996;86:655–665. doi: 10.1016/s0092-8674(00)80138-5. [DOI] [PubMed] [Google Scholar]


