Abstract
Non-alcoholic fatty liver disease (NAFLD), is the most common form of chronic liver disease in most western countries. Current NAFLD diagnosis methods (e.g. liver biopsy analysis or imaging techniques) are poorly suited as tests for such a prevalent condition, from both a clinical and financial point of view. The present work aims to demonstrate the potential utility of serum metabolic profiling in defining phenotypic biomarkers that could be useful in NAFLD management. A parallel animal model / human NAFLD exploratory metabolomics approach was employed, using ultra performance liquid chromatography-mass spectrometry (UPLC®-MS) to analyze 42 serum samples collected from non-diabetic, morbidly obese, biopsy-proven NAFLD patients, and 17 animals belonging to the glycine N-methyltransferase knockout (GNMT-KO) NAFLD mouse model. Multivariate statistical analysis of the data revealed a series of common biomarkers that were significantly altered in the NAFLD (GNMT-KO) subjects in comparison to their normal liver counterparts (WT). Many of the compounds observed could be associated with biochemical perturbations associated with liver dysfunction (e.g. reduced Creatine) and inflammation (e.g. eicosanoid signaling). This differential metabolic phenotyping approach may have a future role as a supplement for clinical decision making in NAFLD and in the adaption to more individualized treatment protocols.
Keywords: NAFLD, steatosis, NASH, metabolomics, biomarkers
Introduction
According to the World Health Organization, there are more than 1 billion overweight adults [body mass index (BMI) > 25 kg/m2], of which at least 300 million are obese (BMI > 30 kg/m2)1. Morbid obesity (BMI > 40 kg/m2) prevalence is also increasing rapidly worldwide.
Obesity poses a major risk factor for nonalcoholic fatty liver disease (NAFLD)2–7. NAFLD is a progressive disease, ranging from the simple accumulation of fat in the liver (steatosis) to the more severe necroinflammatory complication non-alcoholic steatohepatitis (NASH), and affecting up to 24% of the US population2–8. Fortunately, only a small fraction of NAFLD patients develop cirrhosis and hepatocellular carcinoma (HCC)9, 10, although rising obesity prevalence may result in a corresponding increase in these more severe diseases, representing a major health risk. Several molecular mechanisms have been proposed to explain how steatosis progresses to NASH, including free fatty acid-induced apoptosis, endoplasmic reticulum and oxidative stress, and altered methionine metabolism11–13. The contribution of molecules secreted by visceral adipose tissue depots, including inflammatory mediators, has also been underlined14.
Although there is currently no generally accepted medical therapy for NAFLD, weight loss, induced by caloric restriction diets, bariatric surgery or drug-induced fat mal-absorption, improves the condition in some cases15–17. Efficient diagnosis methods are needed for the facile identification of NAFLD patients, disease progression risk assessment, and monitoring the response to potential new treatment strategies.
NAFLD may be suspected in subjects with one or more components of the metabolic syndrome, especially obesity and type 2 diabetes, and elevated serum aminotransferase levels [alanine aminotransferase (ALT) and aspartate aminotransferase (AST)]18–20. Currently, the most reliable methods for NAFLD diagnosis include imaging techniques such as ultrasound and magnetic resonance imaging, and the histological examination of a liver biopsy specimen21, 22. However, imaging techniques are expensive and non-specific (they are unable to distinguish NASH from simple steatosis, or detect hepatic fibrosis), whilst liver biopsy is an expensive, invasive and subjective procedure, associated with potential complications and prone to sampling error23. Transient elastography or FibroScan has been proposed for the non-invasive diagnosis of liver fibrosis24. Its main application is to avoid liver biopsy in assessing disease progression in patients with chronic hepatitis C. Several predictive panels, based on the multivariate analysis of well-established clinical and laboratory variables (such as age, BMI, ALT, AST, glucose, insulin resistance, albumin)25 have been proposed as non-invasive markers for the quantitative assessment of fibrosis (FibroTest,)26, 27, steatosis (SteatoTest)28, NASH (NashTest)29, and fibrosis in patients with NAFLD (ELF test, NAFLD fibrosis score)30–32. However, these methods are not all validated in obese or morbidly obese patients.
Offering a physiologically holistic, non-invasive platform, the emergent field of metabolomics has the potential to provide new NAFLD diagnostic tools33, 34. Recent technological breakthroughs have provided researchers with the capacity to measure hundreds or even thousands of small-molecule metabolites in as little as a few minutes per sample, paving the way for hypothesis generation studies ideally suited to complex diseases such as NAFLD35, 36. Metabolomics is particularly suited to liver injury assessment applications, where serum or urine are the most common samples made available for laboratory tests; as opposed to other disease scenarios, such as cancer, where tissue is readily available for transcriptomic and proteomic analysis. Targeted metabolite analysis studies have already shown that alterations of critical hepatic metabolic pathways, such as methionine and phospholipid metabolism, are strongly associated with NAFLD development13, 37. Such changes are expected to be reflected in wider coverage metabolic profiles, which may in turn be explored as potential biomarkers for NAFLD assessment and treatment stratification.
One of the great promises of the metabolomics approach is the fact that groups of metabolite biomarkers are expected to be less species dependent than gene or protein markers, facilitating the direct comparison of animal models with human studies, which in turn improves the potential of the technique to rapidly convert laboratory based research into clinical practice38. Although the NAFLD condition is typically associated with key metabolic syndrome factors such as obesity, insulin resistance, diabetes, and hypertriglyceridemia, the mechanisms of disease pathogenesis and progression remain unclear. This has brought about the need for the development of animal models, which have provided further insight into the many complex processes which may occur as the liver progresses through different NAFLD stages39. Ideal animal models should resemble as closely as possible the disease pathological characteristics observed in humans. For the study of NAFLD, they should show, together with biochemical alterations, liver fat accumulation, progressing through hepatocyte degeneration and inflammation39, 40. All of these features are observed in the glycine N-methyltransferase knockout (GNMT-KO) mouse model, based on methionine metabolism perturbations, where animals have elevated serum ALT, AST and S-adenosylmethionine (SAMe) levels and develop liver steatosis, fibrosis, and hepatocellular carcinoma (HCC)41. Mammalians catabolize up to half of their daily methionine intake in the liver, via conversion to SAMe in a reaction catalyzed by methionine adenosyltransferase I/III (MATI/III)42. SAMe is involved in a number of different key metabolic pathways, amongst which transmethylation reactions involve the donation of a methyl group to a variety of acceptor molecules, catalyzed by methyltransferases43–45. In quantitative terms, the most important methlytransferase acting upon hepatic SAMe is GNMT, catalyzing its conversion to S-adenosylhomocysteine (SAH), a potent inhibitor of methylation reactions. The importance of the GNMT enzyme is therefore to maintain a constant SAMe/SAH ratio, thus avoiding aberrant methylation events46. This mechanism was recently exemplified by the finding that as well as provoking NAFLD, loss of GNMT induces aberrant methylation of DNA and histones, resulting in epigenetic modulation of critical carcinogenic pathways in mice41.
Further evidence supporting the suitability of the GNMT-KO model for comparison with human NAFLD includes the finding that several children with GNMT mutations had mild to moderate liver disease, and the report of a loss of heterozygosity of GNMT in around 40% of HCC patients, with GNMT being proposed as a tumor-susceptibility gene for liver cancer47, 48.
In this study we aimed to use the differential global serum metabolite profile of GNMT-KO animals, as compared to their wild type (WT) littermates to help define a NAFLD metabolic signature for comparison with that found in humans. Common biomarkers may provide further mechanistic insights, and have great potential for practical use in NAFLD management applications. A liquid chromatography-mass spectrometry (UPLC®-MS) platform-based metabolomics approach was used to explore the serum metabolic profiles of the GNMT-WT/KO animals and biopsy-proven22 human NAFLD patients. Multivariate statistical analysis of the UPLC®-MS data revealed some similarities in the GNMT-KO and human NAFLD patients’ relative serum metabolite levels, as compared to normal liver subjects. The results illustrate the potential of metabolite profiling to provide biomarkers for staging, prognosis and therapy selection in NAFLD management.
Materials and Methods
Clinical population and animal experiments
Human NAFLD patients
Serum samples were collected from a total of 42 morbidly obese (BMI > 40 kg/m2), non-diabetic subjects. The individuals were bariatric surgery candidates, all being weight stable before intervention. Oral glucose tolerance tests (OGGT) were performed to confirm the absence of diabetes. The clinicopathological characteristics of the patients are summarized in Table 1. All of the patients were of Caucasian origin; there were 41 females (98%) and 1 male (2%), with a mean age of 41 ± 2 years at the time of NAFLD diagnosis. For all patients the diagnosis of hepatic steatosis - grades S0 (normal liver), S1, S2, S3 (in increasing order of steatosis severity) - and NASH (grade 1) was established histologically in liver biopsy samples, in the absence of other (viral-, alcohol-, metabolic-, or drug-induced) causes of NAFLD22. The study was approved by the human research review committee of the two participating hospitals (Nice and Pitié-Salpêtrière Paris). No clinical differences were observed between patients recruited at the two sites.
Table 1.
Clinicopathological characteristics of the human patients included in the study. NAFLD diagnoses were established histologically21.
| Group | N (males) | Age (years) | BMI (kg/m2) | AST (IU) | ALT (IU) | Glucose (mM) | Cholesterol (mM) | Triglycerides (mM) |
|---|---|---|---|---|---|---|---|---|
| S0 | 9 (0) | 35.0 ± 3.5 | 47.0 ± 1.9 | 23.3 ± 2.3 | 25.1 ± 3.2 | 5.0 ± 0.2 | 4.8 ± 0.2 | 1.2 ± 0.2 |
| S1 | 8 (0) | 43.8 ± 3.8 | 45.4 ± 1.7 | 21.8 ± 3.5 | 24.8 ± 3.1 | 5.0 ± 0.2 | 6.2 ± 0.6 | 1.9 ± 0.4 |
| S2 | 7 (0) | 41.2 ± 5.1 | 43.5 ± 2.0 | 24.9 ± 2.3 | 34.9 ± 3.2 | 5.2 ± 0.2 | 5.6 ± 0.7 | 1.6 ± 0.2 |
| S3 | 9 (0) | 39.9 ± 4.7 | 45.5 ± 2.7 | 27.8 ± 2.5 | 40.8 ± 7.2 | 5.3 ± 0.2 | 4.8 ± 0.4 | 1.4 ± 0.2 |
| S3 + NASH | 9 (1) | 44.6 ± 3.5 | 43.2 ± 1.5 | 32.8 ± 3.2 | 44.6 ± 5.6 | 5.5 ± 0.4 | 5.1 ± 0.3 | 1.4 ± 0.2 |
Values are given as mean ± 1 standard error of the mean. ALT, a known biomarker of liver damage, is the only parameter found significantly altered (p < 0.05) between the groups of patients under comparison.
Animal handling and sample collection
All animal experimentation was conducted in accordance with Spanish guidelines for the care and use of laboratory animals, and protocols approved by the CIC bioGUNE ethical review committee. The generation of GNMT-KO mice has been described previously49. All animals were supplied with a standard laboratory diet and water ad libitum. Male homozygous GNMT-KO mice were killed at 4 (n = 4) and 6.5 (n = 3), and their WT littermates at 4 (n = 6) and 6.5 (n = 4) months of age. Histological examination of the 4-month-old mutant mice showed steatosis and fibrosis, which was more prominent in the 6.5-month-old mice41. The WT mice histologies were normal at both the 4- and 6.5-month-old time points41. Serum samples were collected from the animals at the time of death, for determination of ALT, AST levels, and metabolic profiling experiments.
Metabolic profiling
A global metabolite profiling UPLC®-MS methodology was employed where all endogenous metabolite related features, characterized by their mass-to-charge ratio m/z and retention time Rt, are included in a subsequent multivariate analysis procedure used to study metabolic differences between the different groups of samples50–53. Where possible, Rt-m/z features corresponding to putative biomarkers were identified. The analytical methodology was designed to provide maximum coverage over classes of compounds involved in key hepatic metabolic pathways, such as major phospholipids, fatty acids, and organic acids, whilst offering relatively high-throughput with minimal injection-to-injection carryover effects.
Sample Preparation
Proteins were precipitated from the defrosted serum samples (50 μL) by adding four volumes of methanol in 1.5 mL microtubes at room temperature. After brief vortex mixing the samples were incubated overnight at −20 °C. Supernatants were collected after centrifugation at 13,000 rpm for 10 minutes, and transferred to vials for UPLC®-MS analysis.
Chromatography
Chromatography was performed on a 1 mm i.d. × 100 mm ACQUITY 1.7 μm C8 BEH column (Waters Corp., Milford, USA) using an ACQUITY UPLC® system (Waters Corp., Milford, USA). The column was maintained at 40 ºC and eluted with a 10 minute linear gradient. The mobile phase, at a flow rate of 140 μL/min, consisted of 100% solvent A (0.05% formic acid) for 1 minute followed by an incremental increase of solvent B (acetonitrile containing 0.05% formic acid) up to 50% over a further minute, increasing to 100% B over the next 6 minutes before returning to the initial composition in readiness for the subsequent injection which proceeded a 45 s system re-cycle time. The volume of sample injected onto the column was 1 μL.
Mass spectrometry
The eluent was introduced into the mass spectrometer (LCT PremierTM, Waters Corp., Milford, USA) by electrospray ionisation, with capillary and cone voltages set in the positive and negative ion modes to 3200 V and 30 V, and 2800 V and 50 V respectively. The nebulisation gas was set to 600 L/h at a temperature of 350 ºC. The cone gas was set to 50 L/h and the source temperature set to 150 ºC. Centroid data were acquired from m/z 50–1000 using an accumulation time of 0.2 s per spectrum. All spectra were mass corrected in real time by reference to leucine enkephalin, infused at 50 μL/min through an independent reference electrospray, sampled every 10 s. A test mixture of standard compounds (Acetaminophen, Sulfaguanidine, Sulfadimethoxine, Val-Tyr-Val, Terfenadine, Leucine-Enkephaline, Reserpine and Erythromicyn – all 5nM in water) was analyzed before and after the entire set of randomized, duplicated sample injections in order to examine the retention time stability (generally < 6 s variation, injection-to-injection), mass accuracy (generally < 3 ppm for m/z 400–1000, and < 1.2 mDa for m/z 50–400) and sensitivity of the system throughout the course of the run which lasted a maximum of 26 h per batch of samples injected. For each injection batch, the overall quality of the analysis procedure was monitored using five repeat extracts of a pooled serum sample. For all biomarker metabolites reported in this work, coefficients of variation, CV, between the repeat extracts were less than 25%.
Online tandem mass spectrometry (MS/MS) experiments for metabolite identification were performed on a Waters QTOF PremierTM (Waters Corp., Milford, USA) instrument operating in both the positive and negative ion electrospray modes; source parameters were identical to those employed in the profiling experiments, except for the cone voltage which was increased (30–70 V) when pseudo MS/MS/MS data was required. During retention time windows corresponding to the elution of the compounds under investigation the quadrupole was set to resolve and transmit ions with appropriate mass-to-charge values. The selected ions then traversed an argon-pressurized cell, with a collision energy voltage (typically between 5 and 50 V) applied in accordance with the extent of ion fragmentation required. Subsequent TOF analysis of the fragment ions generated accurate mass (generally < 3 ppm for m/z 400–1000, and < 1.2 mDa for m/z 50–400) MS/MS or pseudo MS/MS/MS spectra corrected in real time by reference to leucine enkephalin, infused at 50 μL/min through an independent reference electrospray, sampled every 10 s. Centroid data were acquired between m/z 50–1000 using an accumulation time of 0.2 s per spectrum.
Data processing
All data were processed using the MarkerLynx application manager for MassLynx 4.1 software (Waters Corp., Milford, USA). The LC/MS data are peak-detected and noise-reduced in both the LC and MS domains such that only true analytical peaks are further processed by the software (e.g. noise spikes are rejected). A list of intensities (chromatographic peak areas) of the peaks detected is then generated for the first chromatogram, using the Rt-m/z data pairs as identifiers. This process is repeated for each LC-MS analysis and the data sorted such that the correct peak intensity data for each Rt-m/z pair are aligned in the final data table. The ion intensities for each peak detected are then normalized, within each sample, to the sum of the peak intensities in that sample. There was no significant correlation (F < Fcrit) between the total intensities used for normalization and the sample groups being compared in the study. The resulting normalized peak intensities form a single matrix with Rt-m/z pairs for each file in the dataset. All processed data were mean centered and pareto scaled during multivariate data analysis54.
Multivariate data analysis
The first objective in the data analysis process is to reduce the dimensionality of the complex data set to enable easy visualization of any metabolic clustering of the different groups of samples. This has been achieved (Figure 1) by principal components analysis (PCA)55 where the data matrix is reduced to a series of principal components (PCs), each a linear combination of the original Rt-m/z pair peak areas. Each successive PC explains the maximum amount of variance possible, not accounted for by the previous PCs. Hence the scores plots shown in the figures – where the first two principal components, t[1] and t[2], are plotted – represent the most important metabolic variation in the samples captured by the analysis.
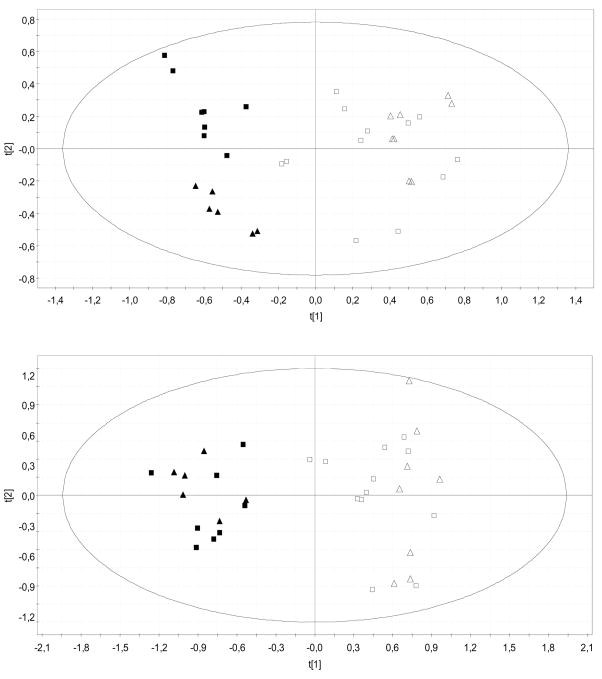
Figure 1.
PCA scores plots discriminating GNMT-KO mice from their WT littermates [Upper plot was obtained from negative ion UPLC™-MS data (t[1]: R2X = 0.28, Q2 = 0.20; t[2]: R2X = 0.09, Q2 = 0.03), lower plot from positive ion data (t[1]: R2X = 0.23, Q2 = 0.12; t[2]: R2X = 0.10, Q2 = 0.007) ]: 4 month old WT (n = 6), open squares; 6.5 month old WT (n = 4), open triangles; 4 month old GNMT-KO (n = 4), squares; 6.5 month old GNMT-KO (n = 3), triangles. Duplicate sample injection data are shown in the plots.
The second stage of the data analysis process concerns the identification of metabolites contributing to the clustering observed in the PCA plots. This information was obtained either by studying the corresponding PCA loadings plot, where the projections of the model variables onto the principal components (p[1], p[2] etc.) are represented, or more specifically using orthogonal partial least-squares to latent structures discriminant analysis (OPLS-DA)56, 57. The latter method is a supervised approach, allowing pre-defined inter-class variance to be captured within a single predictive component. The performance of the OPLS-DA models were evaluated using the Q2 parameter (a Q2 score between 0.7 – 1.0 is indicative of a reliable classifier), calculated by iteratively leaving out samples from the model and predicting their group classification. Paired sample injections were randomly distributed over the cross-validation groups (7–fold), minimizing the risk of calculating falsely predictive components. Appropriate filtration of the loading profile associated with the OPLS-DA predictive components resulted in a set of candidate biomarkers that were further evaluated by calculating group percentage changes and unpaired Student’s t-test p-values.
Metabolite biomarker identification
Exact molecular mass data from redundant m/z peaks corresponding to the formation of different parent (e.g. cations in the positive ion mode, anions in the negative ion mode, adducts, multiple charges) and product (formed by spontaneous “in-source” CID) ions were first used to help confirm the metabolite molecular mass. This information was then submitted for database searching, either in-house or using the online ChemSpider database (www.chemspider.com) where the Kegg, Human Metabolome Database and Lipid Maps data source options were selected. MS/MS data analysis highlights neutral losses or product ions, which are characteristic of metabolite groups and can serve to discriminate between database hits. Specific metabolite groups were characterized as follows: (1) Glycerophospholipids: All species showed clear, diagnostic headgroup ions corresponding to the different lipid sub-classes – m/z = 184.0739 (phosphoryl choline, observed in the MS/MS spectra of [M+H]+ monoacyl, monoalkenyl, monoalkyl, diacyl, and 1-alkenyl,2-acyl glycerophosphocholine) in the positive ion mode, and m/z = 168.0426 (demethylated phosphoryl choline observed in MS/MS spectra of [M-CH3]− monoacyl, monoalkenyl, monoalkyl, diacyl, and 1-alkenyl,2-acyl glycerophosphocholine), 196.0380 (dilyso phosphoryl ethanolamine, observed in MS/MS spectra of [M-H] − monoalkenylglycerophosphoethanolamine) in the negative ion mode58. The two ether subclasses of lysoglycerophosphocholine could be readily distinguished from the monoacyl species by analysis of the MS/MS behaviour of their [M+H]+ and [M-CH3]− ions in the positive and negative modes respectively59. Positive ion mode MS/MS spectra of the [M+H]+ ions showed strong water loss for the acyl species, which was barely observed in the ether subclass spectra. This was consistent with previous reports that rationalized this specificity in fragmentation behavior in terms of different protonation sites for the ether molecular ions that did not eliminate water59. Negative ion mode MS/MS spectra of the [M-CH3]− ions corresponding to these species also showed highly specific fragmentation behavior of the acyl species, which display clear elimination of a highly stable carboxylate ion, whilst the spectra corresponding to the ether species showed strong loss of dimethylaminoethylene followed by dehydration. Monoacylglycerophosphocholine regiochemistry was determined by the fragmentation behavior of the corresponding [M+Na]+ ions, as observed in their positive ion mode MS/MS spectra. This followed previous reports that have shown that spectra of sn-1 monoacyl species show a high m/z = 104.1070 (choline) / 146.9818 (sodiated cyclic ethylene phosphate) product ion ratio from the [M+Na]+ ion60. The converse situation is observed in the case of the sn-2 monoacyl species60. The vinyl ether subclass was distinguished from the alkyl ether species by observation of the m/z = 224.0688 ion in the MS/MS spectra of the [M-CH3]− ions, formed by a charge remote fragmentation mechanism facilitated by the vinyl ether double bond, resulting in the loss of the vinyl ether moiety – no similar ion was observed in the corresponding alkyl ether subclass spectra59. The only corresponding glycerophosphoethanolamine species putative biomarker [PE(P-16:0/0:0)] described in this work showed cleavage of the glycerol backbone in both the positive (m/z = 198.0531) and negative (m/z = 239.2375) ion mode MS/MS spectra of the [M+H]+ and [M-H]− ions respectively. Diacylglycerophosphocholine species were readily identified by the facile loss of the sn-1 and sn-2 carboxylate ions (the sn-2 ion was taken to be the most intense carboxylate moiety – taking into account the formation of second generation fragments, in accordance with previous reports58). Composite nomenclature, referred to as the sum of fatty acid pairs was used where evidence was found for the contribution of multiple species to a single chromatographic peak [e.g. PC 36:5 = PC(16:0/20:5) + PC(16:1/20:4)]. The only corresponding ether subclass described as a putative biomarker in this work [PC(P-18:0/20:4)] showed strong agreement with published (www.lipidmaps.org) MS/MS spectra of the compound in both the positive [[M+H]+; showing loss of trimethylammonia (m/z = 735.5329) and cleavage of the vinyl ether backbone (m/z = 526.3298)] and negative [[M-CH3]− showing loss of the arachidonic acid moiety as a neutral ketene (m/z = 492.3454) and as a carboxylate ion (m/z = 303.2324)] ion modes. (2) Phosphosphingolipids: The putative biomarkers discussed in the current work also showed clear, diagnostic headgroup ions – m/z = 184.0739 (phosphoryl choline, observed in the MS/MS spectra of [M+H]− ions) in the positive ion mode, and m/z = 168.0426 (demethylated phosphoryl choline observed in MS/MS spectra of [M-CH3] − ions) in the negative ion mode. These compounds could be easily distinguished from glycerophospholipids by their odd m/z values (nitrogen rule) and highly stable [M-CH3]− fragment ions61. Specific phosphosphingolipids were further identified by high collision energy CID of the [M-CH3] − moiety in the negative ion mode. Loss of the N-fatty acyl chain in the corresponding MS/MS spectra allowed identification of the sphingoid bases d18:0 (m/z = 451.3301), d18:1 (m/z = 449.3144) or d18:2 (m/z = 447.2988)61. Composite nomenclature, referred to as the sum of sphingoid base and N-linked fatty acid was used where evidence was found for the contribution of multiple sphingoid base lipids to a single chromatographic peak [e.g. SM 34:2 = SM(d18:1/16:1) + SM(d18:2/16:0)].
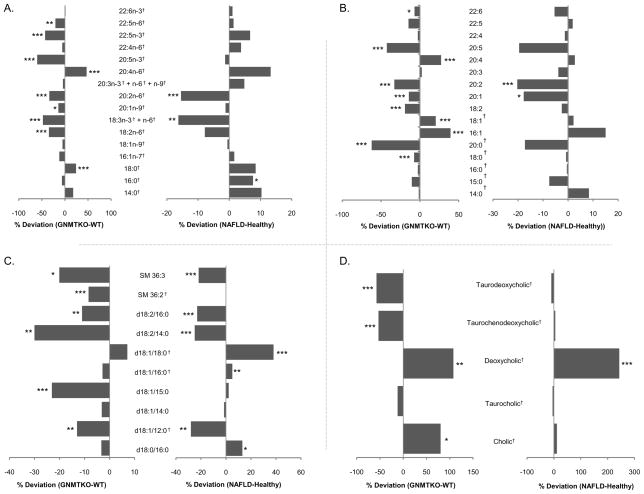
The MassFragment™ application manager (Waters MassLynx v4.1, Waters corp., Milford, USA) was used to facilitate the MS/MS fragment ion analysis process by way of chemically intelligent peak-matching algorithms. The identities of free fatty acids (Figure 2A), bile acids (Figure 2D), and other selected metabolites (all indicated by † in the tables and figures) were confirmed by comparison of their mass spectra and chromatographic retention times with those obtained using commercially available reference standards (Sigma Aldrich, Avanti Polar Lipids Inc.). A full spectral library, containing MS/MS data obtained in the positive and negative ion modes, for all metabolites reported in this work is available on request from the authors.
Figure 2.
Mean percent changes of (a) free fatty acids, (b) sn-1 monoacylglycerophosphocholine, (c) phosphosphingolipids, (d) bile acids in human NAFLD (S0 vs. S1, S2, S3, S3+NASH - right) and GNMT mice (GNMT-WT vs. GNMT-KO - left) sera. Positive and negative percentages indicate higher levels of metabolites in NAFLD (GNMT-KO) and healthy (GNMT-WT) sera, respectively. Unpaired Student’s t-test p-values are indicated where appropriate: *p < 0.15, **p < 0.1, ***p < 0.05. †Metabolite identifications performed by comparison of mass spectra and chromatographic retention times with those obtained using commercially available standards. All other identifications were performed by accurate mass database searching with fragment ion analysis. Lipid nomenclature follows the LIPID MAPS convention (www.lipidmaps.org). Raw data mean values and standard deviations within the different subgroups are detailed in supplementary tables 1 and 2.
Results
GNMT WT/KO Metabolic Profiles
In total, sera taken from 10 WT and 7 GNMT-KO mice were analyzed by UPLC®-MS. PCA was used to produce a two-dimensional visual summary of the observed variation in the serum metabolic profiles of these samples (Figure 1). The results reveal clear metabolic differentiation between the WT and KO animals, with KO samples having a more negative score in the first principal component, t[1]. Additional separation, between the KO mice at 4 (fibrosis + steatosis) and 6.5 (fibrosis + NASH) months is also apparent in the second principal component, t[2], in the negative ion mode – KO mice at 6.5 months of age have a more negative score. The top three variable loadings – increased (negative loading, p[1]) and decreased (positive loading, p[1]) in the GNMT-KO mice with respect to their WT littermates - corresponding to the PCA model in Figure 1 are displayed in Table 2.
Table 2.
Principal variable loadings p[1] in the GNMTWT/KO PCA model (negative ion data).
| Metabolite | p[1] (p[1]cvSE) |
|---|---|
| PC (20:4/0:0) | −0.25 (0.03) |
| PC (18:1/0:0)† | −0.18 (0.03) |
| PC (16:0/20:4)† | −0.16 (0.03) |
| PC (18:2/0:0) | 0.26 (0.06) |
| PC (20:0/0:0)† | 0.14 (0.01) |
| PC (18:2/18:2)† | 0.14 (0.03) |
The standard error of the loading p[1]cvSE generated from the cross-validation rounds is shown in parenthesis.
Metabolite identifications performed by comparison of mass spectra and chromatographic retention times with those obtained using commercially available standards. All other identifications were performed by accurate mass database searching with fragment ion analysis (all mass spectra are available on request). Lipid nomenclature follows the LIPID MAPS convention (www.lipidmaps.org).
Common biomarkers – GNMT-KO / Human NAFLD
Having established that the animal model metabolic profiles recorded by UPLC®-MS were correlated with NAFLD progression, the proceeding analysis was focused towards the identification of biomarkers with similar trends in the human NAFLD samples. OPLS-DA models of the animal data were created, comparing the WT animals with their KO littermates. The model diagnostics, R2(Y) and Q2(Y) were 0.99 and 0.93, and 0.99 and 0.91, in the negative and positive modes respectively, indicating a robust trend, as was suggested by the PCA result. The loadings profiles associated with the OPLS-DA predictive component were filtered according to the ratio of the loadings p[1] to the standard error of the loadings, pcvSE[1], as obtained from the cross-validation rounds. Variables having this ratio less than 2 were eliminated from further investigation, and those remaining ordered by their p[1] value. These data were then compared with percentage changes and unpaired Student’s t-test p-values obtained from the human sample UPLC®-MS data, comparing subjects with a normal liver biopsy to those with NAFLD. Significant overlap was found between the two data sets, with common perturbed groups of compounds including free fatty acids (FFAs), lysophosphatidylcholine (LPC), bile acids (BAs), and sphingomyelin (SM). Detected compounds belonging to these metabolite classes, showing percentage changes of the metabolites in NAFLD (S1, S2, S3, S3+NASH for humans; GNMT-KO for animals) compared to normal liver (S0 for humans, WT for animals) samples, and unpaired Student’s t-test p-values are represented in Figure 2. Two further common biomarkers were also observed: Creatine [−28% (NAFLD-normal liver), p-value 0.063; −24% (GNMT-KO/WT), p-value 0.011] and PE(P-16:0/0:0) [+28% (NAFLD-normal liver), p-value 0.063; +17% (GNMT-KO/WT), p-value 0.13]. In all cases, negative ion biomarker data are shown, for the most intense ionic species observed. Percentage changes for other Rt-m/z pairs, corresponding to the formation of different parent (e.g. cations in the positive ion mode, adducts, multiple charges) and product (formed by spontaneous “in-source” CID) ions were broadly consistent with these data, further validating the structural assignment of the biomarkers.
Metabolite biomarkers discriminating between human steatosis and NASH
Since NASH is considered to be a significant NAFLD development, corresponding to a more aggressive condition that may progress to cirrhosis2, the human samples were further subjected to univariate statistical analyses, focusing on metabolite biomarkers differentiating between the S3 (severe steatosis) and S3 + NASH (severe steatosis + NASH) sample groups. No corresponding supervised analysis was performed for the animal model samples, since the number of samples in the corresponding groups (4 month-old GNMT-KO, n = 4; 6.5 month old GNMT-KO, n = 3) was deemed to be too low to provide statistically significant results. A list of significant (p < 0.1) biomarkers, discriminating between the human S3 and S3 + NASH sample groups is provided in Table 3. Amongst compound classes found significantly altered between the S3 + NASH and S3 sample groups were antioxidative ether glycerophospholipids, sn-2 arachidonly diacylglycerophosphocholine, and free arachidonic acid, the latter two species being involved in eicosanoid signaling pathways.
Table 3.
Biomarker metabolites found in human sera. Mean percentage changes are provided, comparing the S3 + NASH and S3 sample groups.
| Metabolite | % Change (NASH – S3) | p-value (NASH – S3) |
|---|---|---|
| PC (14:0/20:4) | 73.6 | 0.033 |
| PC (16:0/20:3) | 14.4 | 0.086 |
| PC (18:1/0:0)† | 60.8 | 0.028 |
| PC (P-18:0/20:4) | 24.7 | 0.068 |
| PC (P-24:0/0:0) | −38.9 | 0.049 |
| PC (P-22:0/0:0) | −48.0 | 0.097 |
| PC (O-20:0/0:0) | −47.5 | 0.051 |
| Arachidonic acid† | −32.5 | 0.083 |
| Glutamic acid† | −32.1 | 0.061 |
Positive and negative percentages indicate higher levels of metabolites in S3 + NASH and S3 sera, respectively. Statistical p-value calculated using the unpaired Student’s t-test.
Metabolite identifications performed by comparison of mass spectra and chromatographic retention times with those obtained using commercially available standards. All other identifications were performed by accurate mass database searching with fragment ion analysis (all mass spectra are available on request). Lipid nomenclature follows the LIPID MAPS convention (www.lipidmaps.org).
Discussion
Targeted metabolite / lipid profiling studies have already indicated that alterations of critical hepatic metabolic pathways, such as methionine and phospholipid metabolism are strongly associated with NAFLD progression; this information has been collected in independent animal and human studies13, 62–65. To the best of our knowledge, this is the first serum global metabolite profiling study correlating with biopsy proven NAFLD histology in a BMI matched, non-diabetic subject population. Highly controlled subject populations are of key importance in the study of metabolic syndrome related disease where associated factors such as obesity or diabetes can have a large effect on the interpretation of results.
The results from parallel metabolic profiling experiments in the current work, in human NAFLD and in the GNMT-KO NAFLD mouse model, show evidence for strong metabolic correlation with progression of the disease. A series of common, putative biomarker metabolites were observed in the GNMT-WT/KO animal model and human NAFLD patients. Since the pathological characteristics of the GNMT-KO animals closely resemble those observed in human patients across the whole NAFLD spectrum, it is reasonable to presume that similar mechanisms may be responsible for the common metabolic alterations observed. Additionally, the fact that the metabolites are found in both species provides extra confidence for their selection as candidate biomarkers for targeted validation studies. Metabolite markers forwarded from global profiling experiments to such validation studies are often discarded at an early stage, mainly due to the large natural variation that is found in human metabolism, as well as sample handling effects.
Whether the metabolite biomarkers play a role in promoting NAFLD progression or are due to secondary phenomena will require further investigation. However, the results demonstrate the potential of the mass spectrometric based metabolomics approach to facilitate physiologically holistic inter-species studies via the rapid identification and quantification of common metabolites.
The groups of metabolites altered in NAFLD with respect to normal liver subjects include organic acids, free fatty acids, phosphatidylcholine (PC), lysophosphatidylcholine (LPC), bile acids (BAs) and sphingomyelin (SM). All these compound classes are involved in key hepatic metabolic pathways. Creatine, found to be decreased in both the animal (−20%, p-value 0.09) and human (−24%, p-value 0.01) NAFLD samples with respect to their normal liver counterparts, has long been used to assess possible liver dysfunction - lower levels indicating that metabolic reactions in the liver are not occurring to their full capacity66–68. The potential role of fatty acid composition in NAFLD progression has been studied previously37, 69–72, and most recently in the work of Puri et.al. where fatty acid compositions of plasma lipids were studied in lean controls versus obese NAFLD and NASH patients37. In the current work, the overall fatty acid profile observed in the human NAFLD patients bares a strong resemblance to that found in the GNMT-KO/WT animals (Figure 2a). Data collected in various experimental models have suggested that lipid-induced cell toxicity and apoptosis are specific to or made more severe by saturated fatty acids (SFA) when compared to their monounsaturated fatty acid derivatives (MUFA)69. Consistent with these reports, in this work SFA were generally increased in the NAFLD animals and human patients whilst MUFA were either decreased or remained constant, when compared with their normal liver counterparts. Increased Δ6-desaturase activity upon essential fatty acids has recently been associated with NAFLD progression71. The current study also shows evidence for Δ6-desaturase activity modulation, in both the animal model GNMT-KO samples and human NAFLD patients, where decreased levels of circulating linoleic (18:2n-6) and α-linolenic (18:3n-3) building blocks are observed. One of the downstream products along this pathway, arachidonic acid, is observed in higher quantities in the NAFLD samples. Arachidonic acid may be converted to prostaglandins, leukotrienes and lipoxins, of which prostaglandins and leukotrienes are potent pro-inflammatory lipid mediators. Endocannabinoids are also metabolites of arachidonic acid, which have been recently linked with the development of hepatic steatosis73. The lipoxygenase (LOX) pro-inflammatory metabolites of arachidonic acid, also very recently described as correlating with human NAFLD37, were found to be positively expressed in the GNMT-KO mice [12-HETE (+96.0%, p-value 3.4E-05), 15-HETE (+151.7%, p-value 4.2E-06)] with respect to their WT littermates; unfortunately the levels of these compounds were too low in the human NAFLD patients to be measured using the current global metabolite profiling methodology.
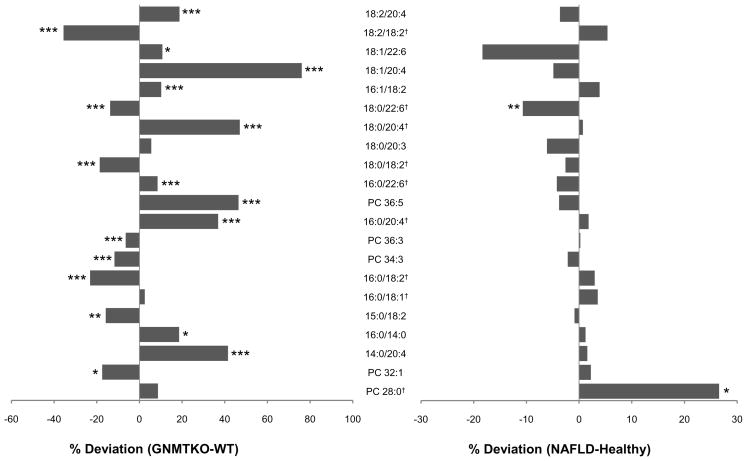
Interestingly, two of the three top variable loadings found in the animal model PCA model (Table 2), correlating positively in GNMT-KO with respect to WT, correspond to phospholipids which are involved in key arachidonic acid metabolic pathways. Indeed, all six sn-2 arachidonoyl diacylglycerophosphocholine compounds [PC(18:2/20:4), PC(18:1/20:4), PC(18:0/20:4), PC(16:0/20:4), PC(14:0/20:4), and PC(36:5) – containing PC(16:1/20:4)] detected were significantly (p<0.05) increased in GNMT-KO with respect to WT (Figure 3). Although this finding was not repeated in the human NAFLD patients as compared to normal liver controls, similar lipids [PC(14:0/20:4) and PC(P-18:0/20:4)] were found significantly increased in the S3+NASH group with respect to the S3 patients (Table 3). Phospholipases A1 (PLA1) and A2 (PLA2) (much more abundant) constitute a large family of enzymes that catalyze the hydrolysis of phospholipids at the sn-1 and sn-2 positions respectively, producing free fatty acid and lysophospholipid74, 75. The ability of PLA2s to selectively mobilize arachidonic acid from endogenous phospholipid storage depots has served as an important and defining characteristic in identifying enzymes contributing to eicosanoid-mediated signaling processes. It is well established that cPLA2α possesses high hydrolytic selectivity towards lipids containing arachidonic acid at the sn-2 position76–79, making it an important enzymatic candidate for intracellular eicosanoid signalling studies80, 81. Modulation of PLA2 activity has been associated with NAFLD in the past70; the observation that GNMT-KO and human NASH patients have elevated sn-2 arachidonoyl diacylglycerophosphocholine may also reflect phospholipase enzymatic activity changes, perturbing key phospholipid deacylation/reacylation reactions involved in eicosanoid signalling.
Figure 3.
Mean percent changes of diacylglycerophosphocholine in human NAFLD (S0 vs. S1, S2, S3, S3+NASH -right) and GNMT mice (GNMT-WT vs. GNMT-KO - left) sera. Positive and negative percentages indicate higher levels of metabolites in NAFLD (GNMT-KO) and healthy (GNMT-WT) sera, respectively. Unpaired Student’s t-test p-values are indicated where appropriate: *p < 0.15, **p < 0.1, ***p < 0.05. †Metabolite identifications performed by comparison of mass spectra and chromatographic retention times with those obtained using commercially available standards. All other identifications were performed by accurate mass database searching with fragment ion analysis. Lipid nomenclature follows the LIPID MAPS convention (www.lipidmaps.org). Raw data mean values and standard deviations within the different subgroups are detailed in supplementary tables 1 and 2.
Strong overlap of the animal model samples and human NAFLD patients is also observed in the sn-1 monoacylglycerophosphocholine profile (Figure 2b). Lysophospholipids are biologically active lipids that are involved in a variety of important processes, including cell proliferation, cell migration, angiogenesis, and inflammation82. Inflammatory effects promoted by LPC include expression of endothelial cell adhesion molecules, growth factors, chemotaxis, and activation of monocytes / macrophages83.
A significant increase of bile acids was also found (Figure 2c); deoxycholic acid was found significantly higher in both the animal model samples and in the human NAFLD patients. Elevated serum bile acids have been strongly related to liver disease in a number of recent studies84–88.
Seven sphingomyelin type lipids: (SM 36:3), (d18:2/16:0), (d18:2/14:0), (d18:1/18:0), (d18:1/16:0), (d18:1/12:0), and (d18:0/16:0) were found to be significantly altered in the human NAFLD patients compared to normal liver subjects; similar tendencies were also found in the animal model samples (Figure 2C). Sphingolipids have been previously associated with stress and death ligand-induced hepatocellular death, which contributes to the progression of several liver diseases including steatohepatitis, ischaemia-reperfusion liver injury or hepatocarcinogenesis89–91.
A further series of metabolites were found to significantly discriminate between the human S3 and S3+NASH sample groups (Table 3). The potential role of the sn-2 arachidonoyl phospholipids [PC(14:0/20:4) and PC(P-18:1/20:4)] has already been discussed in terms of arachidonic acid storage / mobilization. Also associated with these pathways, free arachidonic acid (20:4n-6) was decreased in the S3+NASH group, with respect to S3 patients. This finding may reflect the increased utilization of arachidonic acid by the NASH patients in eicosanoid synthesis and / or its reduced mobilization from phospholipids, as suggested by the increased sn-2 arachidonoyl species. Two lyso plasmalogen species [PC(P-24:0/0:0) and PC(P-22:0/0:0)] were significantly decreased in the S3+NASH patients, as compared to the S3 group. Previous evidence has shown that plasmalogens, characterized by a vinyl-ether bond at the sn-1 position, have anti-oxidant properties92, 93. Glutamic acid was also found to be reduced in the NASH patients, as has been previously found in other non-alcoholic liver diseases94. A similar decrease of glutamic acid has also been reported in high-fat diet rat livers95.
Collectively, the results from this study identify a series of putative biomarkers that correlate with NAFLD progression. The biological implications of the changes that have been observed are likely to be complex and difficult to predict based on global metabolite profiling data alone. Nonetheless, the current data have allowed the identification of a number of altered metabolic pathways potentially involved in NAFLD, using comparisons with previously published information and drawing confidence from the fact that very similar changes have been observed in human patients and animal model samples.
From a practical point of view, one of the most significant obstacles facing the introduction of metabolomics technology into routine clinical practice is the inability to produce simple-to-use kits of the type that are the product of sister omics technologies such as transcriptomics or proteomics. Since it is highly unlikely that sufficiently specific antibodies, or other chemical detection technologies, will be developed for metabolites in the near future, clinical applications will have to rely on scaled-down versions of existing technology such as that used in the current work. In this regard, mass spectrometric metabolomics analysis has the potential to offer a number of significant practical advantages over rival technologies. The analytical turn-around time is short (~ 10 min / sample in this study) and specific, with little sample pre-treatment necessary, allowing for the possibility of high-throughput studies that are required, for example, in clinical trials used in drug development. Reduction of the wide-coverage analytical methodology presented here to a targeted, quantitative or semi-quantitative platform is the next step in the validation procedure for the putative biomarkers described. Longitudinal studies will need to be performed, where the levels of the discriminating metabolites are studied during NAFLD progression / regression and in response to treatment. Moreover, it will be necessary to study much larger patient cohorts, in particular those belonging to different age and ethnic groups. If successful, such a procedure may be optimized to provide a robust, fast-turnaround analytical platform that could be operated in a clinical setting for efficient, non-invasive NAFLD management.
In conclusion, UPLC®/MS metabolic profiling was found to be a suitable platform for the study of NAFLD. The metabolite profiles obtained revealed NAFLD perturbations that may be further exploited for future research in disease pathogenesis and development, or harnessed for use in diagnosis, monitoring, and treatment development applications.
Supplementary Material
Acknowledgments
This work is supported by grants from SAF 2008-04800 and ETORTEK-2008 (J.M.M. and M.L.M.-C.), NIH AT-1576 (S.C.L., M.L.M-C. and J.M.M.), INTEK 06-20, 07-29 and FIT-06-101 (JB), FIS PI060085 (J.C.), HEPADIP-EULSHM-CT-205 (J.M.M., M.L.M.-C., J.B., Y.L.M.B., P.G., K.C., J.T. and N.V.), the FLIP UP consortium (K.C., J.T., N.V.) the Institut National de la Santé et de la Recherche Médicale (France), the University of Nice, the Programme Hospitalier de Recherche Clinique (CHU of Nice), Assistance-Publique Hôpitaux de Paris, Hospitalier de Recherche Clinique, Paris region lle de France, and charities (ALFEDIAM and AFEF/Schering-Plough to P.G.). Y.L.M.B. and P.G. are the recipients of an Interface Grant from CHU of Nice. N.V. is supported by Fondation pour la Recherche Médicale (FRM). Ciberehd is funded by the Instituto de Salud Carlos III.
The contribution to this work from the technicians Ziortza Ispizua, Jessica Arribas, Mónica Martínez and Stephanie Bounnafous is gratefully acknowledged.
Abbreviations
- ALT
Alanine aminotransferase
- AST
aspartate aminotransferase
- BMI
body mass index
- NAFLD
non-alcoholic fatty liver disease
- NASH
non-alcoholic steatohepatitis
- HCC
hepatocellular carcinoma
- GNMT
glycine N-methyltransferase
- SAMe
S-adenosylmethionine
- SAH
S-adenosylhomocysteine
- FFAs
free fatty acids
- LPC
lysophosphatidylcholine
- BAs
bile acids
- SM
sphingomyelin
- PC
phosphatidylcholine
- MUFA
monounsaturated fatty acid derivatives
- SFA
saturated fatty acids
- PCA
principal component analysis
- OPLS-DA
orthogonal partial least-squares to latent structures discriminant analysis
- UPLC®-MS
ultra performance liquid chromatography-mass spectrometry
Footnotes
Supporting Information Available
Raw data mean values and standard deviations within the different subgroups shown in Figures 2 and 3 are included in supplementary Tables 1 and 2. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.World Health Organization. Obesity and Overweight. Geneva: 2006. Factsheet No. 3011. [Google Scholar]
- 2.Reid AE. Nonalcoholic steatohepatitis. Gastroenterology. 2001;121(3):710–23. doi: 10.1053/gast.2001.27126. [DOI] [PubMed] [Google Scholar]
- 3.Clark JM, Diehl AM. Defining nonalcoholic fatty liver disease: implications for epidemiologic studies. Gastroenterology. 2003;124(1):248–50. doi: 10.1053/gast.2003.50032. [DOI] [PubMed] [Google Scholar]
- 4.Marchesini G, Bugianesi E, Forlani G, Cerrelli F, Lenzi M, Manini R, Natale S, Vanni E, Villanova N, Melchionda N, Rizzetto M. Nonalcoholic fatty liver, steatohepatitis, and the metabolic syndrome. Hepatology. 2003;37(4):917–23. doi: 10.1053/jhep.2003.50161. [DOI] [PubMed] [Google Scholar]
- 5.Adams LA, Lindor KD. Nonalcoholic fatty liver disease. Ann Epidemiol. 2007;17(11):863–9. doi: 10.1016/j.annepidem.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 6.Angulo P. Obesity and nonalcoholic fatty liver disease. Nutr Rev. 2007;65(6 Pt 2):S57–63. doi: 10.1111/j.1753-4887.2007.tb00329.x. [DOI] [PubMed] [Google Scholar]
- 7.Parekh S, Anania FA. Abnormal lipid and glucose metabolism in obesity: implications for nonalcoholic fatty liver disease. Gastroenterology. 2007;132(6):2191–207. doi: 10.1053/j.gastro.2007.03.055. [DOI] [PubMed] [Google Scholar]
- 8.Matteoni CA, Younossi ZM, Gramlich T, Boparai N, Liu YC, McCullough AJ. Nonalcoholic fatty liver disease: a spectrum of clinical and pathological severity. Gastroenterology. 1999;116(6):1413–9. doi: 10.1016/s0016-5085(99)70506-8. [DOI] [PubMed] [Google Scholar]
- 9.Caldwell SH, Crespo DM, Kang HS, Al-Osaimi AM. Obesity and hepatocellular carcinoma. Gastroenterology. 2004;127(5 Suppl 1):S97–103. doi: 10.1053/j.gastro.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 10.El-Serag HB. Hepatocellular carcinoma: recent trends in the United States. Gastroenterology. 2004;127(5 Suppl 1):S27–34. doi: 10.1053/j.gastro.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 11.Malhi H, Gores GJ. Molecular mechanisms of lipotoxicity in nonalcoholic fatty liver disease. Semin Liver Dis. 2008;28(4):360–9. doi: 10.1055/s-0028-1091980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mato JM, Lu SC. Role of S-adenosyl-L-methionine in liver health and injury. Hepatology. 2007;45(5):1306–12. doi: 10.1002/hep.21650. [DOI] [PubMed] [Google Scholar]
- 13.Mato JM, Martinez-Chantar ML, Lu SC. Methionine metabolism and liver disease. Annu Rev Nutr. 2008;28:273–93. doi: 10.1146/annurev.nutr.28.061807.155438. [DOI] [PubMed] [Google Scholar]
- 14.Tordjman J, Poitou C, Hugol D, Bouillot JL, Basdevant A, Bedossa P, Guerre-Millo M, Clement K. Association between omental adipose tissue macrophages and liver histopathology in morbid obesity: influence of glycemic status. J Hepatol. 2009;51(2):354–62. doi: 10.1016/j.jhep.2009.02.031. [DOI] [PubMed] [Google Scholar]
- 15.Farrell GC, Larter CZ. Nonalcoholic fatty liver disease: from steatosis to cirrhosis. Hepatology. 2006;43(2 Suppl 1):S99–S112. doi: 10.1002/hep.20973. [DOI] [PubMed] [Google Scholar]
- 16.Dixon JB, Bhathal PS, Hughes NR, O'Brien PE. Nonalcoholic fatty liver disease: Improvement in liver histological analysis with weight loss. Hepatology. 2004;39(6):1647–54. doi: 10.1002/hep.20251. [DOI] [PubMed] [Google Scholar]
- 17.Klein S, Mittendorfer B, Eagon JC, Patterson B, Grant L, Feirt N, Seki E, Brenner D, Korenblat K, McCrea J. Gastric bypass surgery improves metabolic and hepatic abnormalities associated with nonalcoholic fatty liver disease. Gastroenterology. 2006;130(6):1564–72. doi: 10.1053/j.gastro.2006.01.042. [DOI] [PubMed] [Google Scholar]
- 18.Wanless IR, Lentz JS. Fatty liver hepatitis (steatohepatitis) and obesity: an autopsy study with analysis of risk factors. Hepatology. 1990;12(5):1106–10. doi: 10.1002/hep.1840120505. [DOI] [PubMed] [Google Scholar]
- 19.Marchesini G, Brizi M, Morselli-Labate AM, Bianchi G, Bugianesi E, McCullough AJ, Forlani G, Melchionda N. Association of nonalcoholic fatty liver disease with insulin resistance. Am J Med. 1999;107(5):450–5. doi: 10.1016/s0002-9343(99)00271-5. [DOI] [PubMed] [Google Scholar]
- 20.Marra F, Gastaldelli A, Svegliati Baroni G, Tell G, Tiribelli C. Molecular basis and mechanisms of progression of non-alcoholic steatohepatitis. Trends Mol Med. 2008;14(2):72–81. doi: 10.1016/j.molmed.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 21.Oh MK, Winn J, Poordad F. Review article: diagnosis and treatment of non-alcoholic fatty liver disease. Aliment Pharmacol Ther. 2008;28(5):503–22. doi: 10.1111/j.1365-2036.2008.03752.x. [DOI] [PubMed] [Google Scholar]
- 22.Kleiner DE, Brunt EM, Van Natta M, Behling C, Contos MJ, Cummings OW, Ferrell LD, Liu YC, Torbenson MS, Unalp-Arida A, Yeh M, McCullough AJ, Sanyal AJ. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41(6):1313–21. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 23.Ratziu V, Charlotte F, Heurtier A, Gombert S, Giral P, Bruckert E, Grimaldi A, Capron F, Poynard T. Sampling variability of liver biopsy in nonalcoholic fatty liver disease. Gastroenterology. 2005;128(7):1898–906. doi: 10.1053/j.gastro.2005.03.084. [DOI] [PubMed] [Google Scholar]
- 24.Castera L, Foucher J, Bertet J, Couzigou P, de Ledinghen V. FibroScan and FibroTest to assess liver fibrosis in HCV with normal aminotransferases. Hepatology. 2006;43(2):373–4. doi: 10.1002/hep.21019. author reply 375–6. [DOI] [PubMed] [Google Scholar]
- 25.Kotronen A, Peltonen M, Hakkarainen A, Sevastianova K, Bergholm R, Johansson LM, Lundbom N, Rissanen A, Ridderstrale M, Groop L, Orho-Melander M, Yki-Jarvinen H. Prediction of non-alcoholic fatty liver disease and liver fat using metabolic and genetic factors. Gastroenterology. 2009;137(3):865–72. doi: 10.1053/j.gastro.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 26.Poynard T, IBF, Munteanu M, Messous D, Myers RP, Thabut D, Ratziu V, Mercadier A, Benhamou Y, Hainque B. Overview of the diagnostic value of biochemical markers of liver fibrosis (FibroTest, HCV FibroSure) and necrosis (ActiTest) in patients with chronic hepatitis C. Comp Hepatol. 2004;4:10–23. doi: 10.1186/1476-5926-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imbert-Bismut F, Messous D, Thibault V, Myers RB, Piton A, Thabut D, Devers L, Hainque B, Mercadier A, Poynard T. Intra-laboratory analytical variability of biochemical markers of fibrosis (Fibrotest) and activity (Actitest) and reference ranges in healthy blood donors. Clin Chem Lab Med. 2004;42(3):323–33. doi: 10.1515/CCLM.2004.058. [DOI] [PubMed] [Google Scholar]
- 28.Poynard T, Ratziu V, Naveau S, Thabut D, Charlotte F, Messous D, Capron D, Abella A, Massard J, Ngo Y, Munteanu M, Mercadier A, Manns M, Albrecht J. The diagnostic value of biomarkers (SteatoTest) for the prediction of liver steatosis. Comp Hepatol. 2005;4:10. doi: 10.1186/1476-5926-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poynard T, Ratziu V, Charlotte F, Messous D, Munteanu M, Imbert-Bismut F, Massard J, Bonyhay L, Tahiri M, Thabut D, Cadranel JF, Le Bail B, de Ledinghen V. Diagnostic value of biochemical markers (NashTest) for the prediction of non alcoholo steato hepatitis in patients with non-alcoholic fatty liver disease. BMC Gastroenterol. 2006;6:34. doi: 10.1186/1471-230X-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morra R, Munteanu M, Imbert-Bismut F, Messous D, Ratziu V, Poynard T. FibroMAX: towards a new universal biomarker of liver disease? Expert Rev Mol Diagn. 2007;7(5):481–90. doi: 10.1586/14737159.7.5.481. [DOI] [PubMed] [Google Scholar]
- 31.Guha IN, Parkes J, Roderick P, Chattopadhyay D, Cross R, Harris S, Kaye P, Burt AD, Ryder SD, Aithal GP, Day CP, Rosenberg WM. Noninvasive markers of fibrosis in nonalcoholic fatty liver disease: Validating the European Liver Fibrosis Panel and exploring simple markers. Hepatology. 2008;47(2):455–60. doi: 10.1002/hep.21984. [DOI] [PubMed] [Google Scholar]
- 32.Guha IN, Rosenberg WM. Noninvasive assessment of liver fibrosis: serum markers, imaging, and other modalities. Clin Liver Dis. 2008;12(4):883–900. x. doi: 10.1016/j.cld.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 33.Nicholson JK, Connelly J, Lindon JC, Holmes E. Metabonomics: a platform for studying drug toxicity and gene function. Nat Rev Drug Discov. 2002;1(2):153–61. doi: 10.1038/nrd728. [DOI] [PubMed] [Google Scholar]
- 34.Nicholson JK, Lindon JC, Holmes E. 'Metabonomics': understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29(11):1181–9. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 35.Gowda GA, Zhang S, Gu H, Asiago V, Shanaiah N, Raftery D. Metabolomics-based methods for early disease diagnostics. Expert Rev Mol Diagn. 2008;8(5):617–33. doi: 10.1586/14737159.8.5.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaddurah-Daouk R, Kristal BS, Weinshilboum RM. Metabolomics: a global biochemical approach to drug response and disease. Annu Rev Pharmacol Toxicol. 2008;48:653–83. doi: 10.1146/annurev.pharmtox.48.113006.094715. [DOI] [PubMed] [Google Scholar]
- 37.Puri P, Wiest MM, Cheung O, Mirshahi F, Sargeant C, Min HK, Contos MJ, Sterling RK, Fuchs M, Zhou H, Watkins SM, Sanyal AJ. The plasma lipidomic signature of nonalcoholic steatohepatitis. Hepatology. 2009;50(6):1827–38. doi: 10.1002/hep.23229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heijne WH, Kienhuis AS, van Ommen B, Stierum RH, Groten JP. Systems toxicology: applications of toxicogenomics, transcriptomics, proteomics and metabolomics in toxicology. Expert Rev Proteomics. 2005;2(5):767–80. doi: 10.1586/14789450.2.5.767. [DOI] [PubMed] [Google Scholar]
- 39.Varela-Rey M, Embade N, Ariz U, Lu SC, Mato JM, Martinez-Chantar ML. Non-alcoholic steatohepatitis and animal models: understanding the human disease. Int J Biochem Cell Biol. 2009;41(5):969–76. doi: 10.1016/j.biocel.2008.10.027. [DOI] [PubMed] [Google Scholar]
- 40.Ariz U, Mato JM, Lu SC, Martinez Chantar ML. Nonalcoholic steatohepatitis, animal models, and biomarkers: what is new? Methods Mol Biol. 593:109–36. doi: 10.1007/978-1-60327-194-3_6. [DOI] [PubMed] [Google Scholar]
- 41.Martinez-Chantar ML, Vazquez-Chantada M, Ariz U, Martinez N, Varela M, Luka Z, Capdevila A, Rodriguez J, Aransay AM, Matthiesen R, Yang H, Calvisi DF, Esteller M, Fraga M, Lu SC, Wagner C, Mato JM. Loss of the glycine N-methyltransferase gene leads to steatosis and hepatocellular carcinoma in mice. Hepatology. 2008;47(4):1191–9. doi: 10.1002/hep.22159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kotb M, Mudd SH, Mato JM, Geller AM, Kredich NM, Chou JY, Cantoni GL. Consensus nomenclature for the mammalian methionine adenosyltransferase genes and gene products. Trends Genet. 1997;13(2):51–2. doi: 10.1016/s0168-9525(97)01013-5. [DOI] [PubMed] [Google Scholar]
- 43.Avila MA, Garcia-Trevijano ER, Martinez-Chantar ML, Latasa MU, Perez- Mato I, Martinez-Cruz LA, del Pino MM, Corrales FJ, Mato JM. S-Adenosylmethionine revisited: its essential role in the regulation of liver function. Alcohol. 2002;27(3):163–7. doi: 10.1016/s0741-8329(02)00228-8. [DOI] [PubMed] [Google Scholar]
- 44.Corrales FJ, Perez-Mato I, Sanchez Del Pino MM, Ruiz F, Castro C, Garcia-Trevijano ER, Latasa U, Martinez-Chantar ML, Martinez-Cruz A, Avila MA, Mato JM. Regulation of mammalian liver methionine adenosyltransferase. J Nutr. 2002;132(8 Suppl):2377S–2381S. doi: 10.1093/jn/132.8.2377S. [DOI] [PubMed] [Google Scholar]
- 45.Martinez-Chantar ML, Garcia-Trevijano ER, Latasa MU, Perez-Mato I, Sanchez del Pino MM, Corrales FJ, Avila MA, Mato JM. Importance of a deficiency in S-adenosyl- L-methionine synthesis in the pathogenesis of liver injury. Am J Clin Nutr. 2002;76(5):1177S–82S. doi: 10.1093/ajcn/76/5.1177S. [DOI] [PubMed] [Google Scholar]
- 46.Finkelstein JD. Metabolic regulatory properties of S-adenosylmethionine and S-adenosylhomocysteine. Clin Chem Lab Med. 2007;45(12):1694–9. doi: 10.1515/CCLM.2007.341. [DOI] [PubMed] [Google Scholar]
- 47.Heady JE, Kerr SJ. Alteration of glycine N-methyltransferase activity in fetal, adult, and tumor tissues. Cancer Res. 1975;35(3):640–3. [PubMed] [Google Scholar]
- 48.Chen SY, Lin JR, Darbha R, Lin P, Liu TY, Chen YM. Glycine Nmethyltransferase tumor susceptibility gene in the benzo(a)pyrene-detoxification pathway. Cancer Res. 2004;64(10):3617–23. doi: 10.1158/0008-5472.CAN-03-3726. [DOI] [PubMed] [Google Scholar]
- 49.Luka Z, Capdevila A, Mato JM, Wagner C. A glycine N-methyltransferase knockout mouse model for humans with deficiency of this enzyme. Transgenic Res. 2006;15(3):393–7. doi: 10.1007/s11248-006-0008-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Griffiths WJ, Karu K, Hornshaw M, Woffendin G, Wang Y. Metabolomics and metabolite profiling: past heroes and future developments. Eur J Mass Spectrom (Chichester, Eng) 2007;13(1):45–50. doi: 10.1255/ejms.850. [DOI] [PubMed] [Google Scholar]
- 51.Burton L, Ivosev G, Tate S, Impey G, Wingate J, Bonner R. Instrumental and experimental effects in LC-MS-based metabolomics. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;871(2):227–35. doi: 10.1016/j.jchromb.2008.04.044. [DOI] [PubMed] [Google Scholar]
- 52.Theodoridis GGHG, Wilson ID. LC-MS-based methodology for global metabolite profiling in metabonomics/metabolomics. Trend in analytical chemistry. 2008;27(3):238–250. [Google Scholar]
- 53.Bedair MSLW. Current and emerging mass-spectrometry technologies for metabolomics. Trends in analytical chemistry. 2008;27(3):251–260. [Google Scholar]
- 54.Wold SJE, Cocchi M. 3D-QSAR in drug design, theory, methods, and applications. ESCOM Science; Lieden: 1993. [Google Scholar]
- 55.Jolliffe IT. Principal component analysis. Springer; New York: 2002. [Google Scholar]
- 56.Byleso M. OPLS discriminant analysis: combining the stregths of PLS-DA and SIMCA classification. J Chemometrics. 2006;20:341–351. [Google Scholar]
- 57.Wiklund S, Johansson E, Sjostrom L, Mellerowicz EJ, Edlund U, Shockcor JP, Gottfries J, Moritz T, Trygg J. Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Anal Chem. 2008;80(1):115–22. doi: 10.1021/ac0713510. [DOI] [PubMed] [Google Scholar]
- 58.Brugger B, Erben G, Sandhoff R, Wieland FT, Lehmann WD. Quantitative analysis of biological membrane lipids at the low picomole level by nano-electrospray ionization tandem mass spectrometry. Proc Natl Acad Sci U S A. 1997;94(6):2339–44. doi: 10.1073/pnas.94.6.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Khaselev N, Murphy RC. Electrospray ionization mass spectrometry of lysoglycerophosphocholine lipid subclasses. J Am Soc Mass Spectrom. 2000;11(4):283–91. doi: 10.1016/s1044-0305(99)00158-0. [DOI] [PubMed] [Google Scholar]
- 60.Han XGRW. Structural Determination of Lysophospholipid Regioisomers by Electrospray Ionization Tandem Mass Spectrometry. Journal of the american chemical society. 1996;118:451–457. [Google Scholar]
- 61.Houjou T, Yamatani K, Nakanishi H, Imagawa M, Shimizu T, Taguchi R. Rapid and selective identification of molecular species in phosphatidylcholine and sphingomyelin by conditional neutral loss scanning and MS3. Rapid Commun Mass Spectrom. 2004;18(24):3123–30. doi: 10.1002/rcm.1737. [DOI] [PubMed] [Google Scholar]
- 62.Duce AM, Ortiz P, Cabrero C, Mato JM. S-adenosyl-L-methionine synthetase and phospholipid methyltransferase are inhibited in human cirrhosis. Hepatology. 1988;8(1):65–8. doi: 10.1002/hep.1840080113. [DOI] [PubMed] [Google Scholar]
- 63.Kharbanda KK, Mailliard ME, Baldwin CR, Beckenhauer HC, Sorrell MF, Tuma DJ. Betaine attenuates alcoholic steatosis by restoring phosphatidylcholine generation via the phosphatidylethanolamine methyltransferase pathway. J Hepatol. 2007;46(2):314–21. doi: 10.1016/j.jhep.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 64.Ikura Y, Ohsawa M, Suekane T, Fukushima H, Itabe H, Jomura H, Nishiguchi S, Inoue T, Naruko T, Ehara S, Kawada N, Arakawa T, Ueda M. Localization of oxidized phosphatidylcholine in nonalcoholic fatty liver disease: impact on disease progression. Hepatology. 2006;43(3):506–14. doi: 10.1002/hep.21070. [DOI] [PubMed] [Google Scholar]
- 65.Li Z, Agellon LB, Allen TM, Umeda M, Jewell L, Mason A, Vance DE. The ratio of phosphatidylcholine to phosphatidylethanolamine influences membrane integrity and steatohepatitis. Cell Metab. 2006;3(5):321–31. doi: 10.1016/j.cmet.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 66.Nanji AA, Blank D. Low serum creatine kinase activity in patients with alcoholic liver disease. Clin Chem. 1981;27(11):1954. [PubMed] [Google Scholar]
- 67.Malnick SD, Bass DD, Kaye AM. Creatine kinase BB: a response marker in liver and other organs. Hepatology. 1994;19(1):261. [PubMed] [Google Scholar]
- 68.Vaubourdolle M, Chazouilleres O, Poupon R, Ballet F, Braunwald J, Legendre C, Baudin B, Kirn A, Giboudeau J. Creatine kinase-BB: a marker of liver sinusoidal damage in ischemia-reperfusion. Hepatology. 1993;17(3):423–8. [PubMed] [Google Scholar]
- 69.Gentile CL, Pagliassotti MJ. The role of fatty acids in the development and progression of nonalcoholic fatty liver disease. J Nutr Biochem. 2008;19(9):567–76. doi: 10.1016/j.jnutbio.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Puri P, Baillie RA, Wiest MM, Mirshahi F, Choudhury J, Cheung O, Sargeant C, Contos MJ, Sanyal AJ. A lipidomic analysis of nonalcoholic fatty liver disease. Hepatology. 2007;46(4):1081–90. doi: 10.1002/hep.21763. [DOI] [PubMed] [Google Scholar]
- 71.Kotronen A, Seppanen-Laakso T, Westerbacka J, Kiviluoto T, Arola J, Ruskeepaa AL, Oresic M, Yki-Jarvinen H. Hepatic stearoyl-CoA desaturase (SCD)-1 activity and diacylglycerol but not ceramide concentrations are increased in the nonalcoholic human fatty liver. Diabetes. 2009;58(1):203–8. doi: 10.2337/db08-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bass NM. Lipidomic dissection of nonalcoholic steatohepatitis: moving beyond foie gras to fat traffic. Hepatology. 51(1):4–7. doi: 10.1002/hep.23458. [DOI] [PubMed] [Google Scholar]
- 73.Kunos G, Osei-Hyiaman D. Endocannabinoids and liver disease IV. Endocannabinoid involvement in obesity and hepatic steatosis. Am J Physiol Gastrointest Liver Physiol. 2008;294(5):G1101–4. doi: 10.1152/ajpgi.00057.2008. [DOI] [PubMed] [Google Scholar]
- 74.Six DA, Dennis EA. The expanding superfamily of phospholipase A(2) enzymes: classification and characterization. Biochim Biophys Acta. 2000;1488(1–2):1–19. doi: 10.1016/s1388-1981(00)00105-0. [DOI] [PubMed] [Google Scholar]
- 75.Kudo I, Murakami M. Phospholipase A2 enzymes. Prostaglandins Other Lipid Mediat. 2002;68–69:3–58. doi: 10.1016/s0090-6980(02)00020-5. [DOI] [PubMed] [Google Scholar]
- 76.Leslie CC, Voelker DR, Channon JY, Wall MM, Zelarney PT. Properties and purification of an arachidonoyl-hydrolyzing phospholipase A2 from a macrophage cell line, RAW 264.7. Biochim Biophys Acta. 1988;963(3):476–92. doi: 10.1016/0005-2760(88)90316-5. [DOI] [PubMed] [Google Scholar]
- 77.Clark JD, Lin LL, Kriz RW, Ramesha CS, Sultzman LA, Lin AY, Milona N, Knopf JL. A novel arachidonic acid-selective cytosolic PLA2 contains a Ca(2+)–dependent translocation domain with homology to PKC and GAP. Cell. 1991;65(6):1043–51. doi: 10.1016/0092-8674(91)90556-e. [DOI] [PubMed] [Google Scholar]
- 78.Schievella AR, Regier MK, Smith WL, Lin LL. Calcium-mediated translocation of cytosolic phospholipase A2 to the nuclear envelope and endoplasmic reticulum. J Biol Chem. 1995;270(51):30749–54. doi: 10.1074/jbc.270.51.30749. [DOI] [PubMed] [Google Scholar]
- 79.Pawliczak R, Logun C, Madara P, Lawrence M, Woszczek G, Ptasinska A, Kowalski ML, Wu T, Shelhamer JH. Cytosolic phospholipase A2 Group IValpha but not secreted phospholipase A2 Group IIA, V, or X induces interleukin-8 and cyclooxygenase-2 gene and protein expression through peroxisome proliferator-activated receptors gamma 1 and 2 in human lung cells. J Biol Chem. 2004;279(47):48550–61. doi: 10.1074/jbc.M408926200. [DOI] [PubMed] [Google Scholar]
- 80.Leslie CC. Regulation of the specific release of arachidonic acid by cytosolic phospholipase A2. Prostaglandins Leukot Essent Fatty Acids. 2004;70(4):373–6. doi: 10.1016/j.plefa.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 81.Hirabayashi T, Murayama T, Shimizu T. Regulatory mechanism and physiological role of cytosolic phospholipase A2. Biol Pharm Bull. 2004;27(8):1168–73. doi: 10.1248/bpb.27.1168. [DOI] [PubMed] [Google Scholar]
- 82.Goetzl EJ. Pleiotypic mechanisms of cellular responses to biologically active lysophospholipids. Prostaglandins Other Lipid Mediat. 2001;64(1–4):11–20. doi: 10.1016/s0090-6980(01)00104-6. [DOI] [PubMed] [Google Scholar]
- 83.Kita TKN, Ishii K, Horiuchi H, Arai H, Yokode M. Oxidized LDL and expression of monocyte adhesion molecules. Diabetes Research and clinical practise. 1999;45(2–3):123–126. doi: 10.1016/s0168-8227(99)00041-8. [DOI] [PubMed] [Google Scholar]
- 84.Yang L, Xiong A, He Y, Wang Z, Wang C, Li W, Hu Z. Bile acids metabonomic study on the CCl4- and alpha-naphthylisothiocyanate-induced animal models: quantitative analysis of 22 bile acids by ultraperformance liquid chromatography-mass spectrometry. Chem Res Toxicol. 2008;21(12):2280–8. doi: 10.1021/tx800225q. [DOI] [PubMed] [Google Scholar]
- 85.Monte MJ, Marin JJ, Antelo A, Vazquez-Tato J. Bile acids: chemistry, physiology, and pathophysiology. World J Gastroenterol. 2009;15(7):804–16. doi: 10.3748/wjg.15.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Perez MJ, Briz O. Bile-acid-induced cell injury and protection. World J Gastroenterol. 2009;15(14):1677–89. doi: 10.3748/wjg.15.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gilat T, Leikin-Frenkel A, Goldiner I, Juhel C, Lafont H, Gobbi D, Konikoff FM. Prevention of diet-induced fatty liver in experimental animals by the oral administration of a fatty acid bile acid conjugate (FABAC) Hepatology. 2003;38(2):436–42. doi: 10.1053/jhep.2003.50348. [DOI] [PubMed] [Google Scholar]
- 88.Kobayashi N, Katsumata H, Uto Y, Goto J, Niwa T, Kobayashi K, Mizuuchi Y. A monoclonal antibody-based enzyme-linked immunosorbent assay of glycolithocholic acid sulfate in human urine for liver function test. Steroids. 2002;67(10):827–33. doi: 10.1016/s0039-128x(02)00036-3. [DOI] [PubMed] [Google Scholar]
- 89.Mari M, Colell A, Morales A, Paneda C, Varela-Nieto I, Garcia-Ruiz C, Fernandez-Checa JC. Acidic sphingomyelinase downregulates the liver-specific methionine adenosyltransferase 1A, contributing to tumor necrosis factor-induced lethal hepatitis. J Clin Invest. 2004;113(6):895–904. doi: 10.1172/JCI19852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mari M, Fernandez-Checa JC. Sphingolipid signalling and liver diseases. Liver Int. 2007;27(4):440–50. doi: 10.1111/j.1478-3231.2007.01475.x. [DOI] [PubMed] [Google Scholar]
- 91.Morales A, Lee H, Goni FM, Kolesnick R, Fernandez-Checa JC. Sphingolipids and cell death. Apoptosis. 2007;12(5):923–39. doi: 10.1007/s10495-007-0721-0. [DOI] [PubMed] [Google Scholar]
- 92.Engelmann B. Plasmalogens: targets for oxidants and major lipophilic antioxidants. Biochem Soc Trans. 2004;32(Pt 1):147–50. doi: 10.1042/bst0320147. [DOI] [PubMed] [Google Scholar]
- 93.Zoeller RA, Lake AC, Nagan N, Gaposchkin DP, Legner MA, Lieberthal W. Plasmalogens as endogenous antioxidants: somatic cell mutants reveal the importance of the vinyl ether. Biochem J. 1999;338 ( Pt 3):769–76. [PMC free article] [PubMed] [Google Scholar]
- 94.Tominaga T, Suzuki H, Mizuno H, Kouno M, Suzuki M, Kato Y, Sato A, Okabe K, Miyashita M. Clinical significance of measuring plasma concentrations of glutamine and glutamate in alcoholic liver diseases. Alcohol Alcohol Suppl. 1993;1A:103–9. doi: 10.1093/alcalc/28.supplement_1a.103. [DOI] [PubMed] [Google Scholar]
- 95.Xie Z, Li H, Wang K, Lin J, Wang Q, Zhao G, Jia W, Zhang Q. Analysis of transcriptome and metabolome profiles alterations in fatty liver induced by high-fat diet in rat. Metabolism. 2009 doi: 10.1016/j.metabol.2009.08.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.