Abstract
Changing environmental conditions and lessening fresh water supplies have sparked intense interest in understanding and manipulating abscisic acid signaling, which controls adaptive responses to drought and other abiotic stressors. We recently discovered a selective ABA agonist, pyrabactin, and used it to discover its primary target PYR1, the founding member of the PYR/PYL family of soluble ABA receptors. To understand pyrabactin's selectivity we have taken a combined structural, chemical and genetic approach. We show that subtle differences between receptor binding pockets control ligand orientation between productive and non-productive modes. Non-productive binding occurs without gate closure and prevents receptor activation. Observations in solution show that these orientations are in rapid equilibrium that can be shifted by mutations to control maximal agonist activity. Our results provide a robust framework for the design of new agonists and reveal a new mechanism for agonist selectivity.
Introduction
ABA biosynthesis is triggered in response to heat, cold, drought and other stressors1,2. Crop plants engineered to have increased ABA sensitivity show improved yield under conditions of moderate drought3 and the direct application of ABA or ABA analogs to plants in the field improves water use efficiency4. Synthetic ABA agonists that activate plant stress tolerance pathways therefore have potential to provide new options for managing stress tolerance and water use, which has important ramifications since ~70% of fresh water is utilized for agriculture.
We recently discovered a synthetic ABA agonist called pyrabactin (Fig. 1a)5. Unlike ABA, pyrabactin does not activate all ABA responses5. This unusual selectivity allowed the identification of receptor mutants, because unlike ABA, the effects of pyrabactin in seeds depend primarily on one protein, PYR1. While this selectivity was valuable for genetic dissection of ABA signaling, it limits the usefulness of pyrabactin for controlling ABA responses in vegetative tissues, which is an important new goal.
Figure 1.
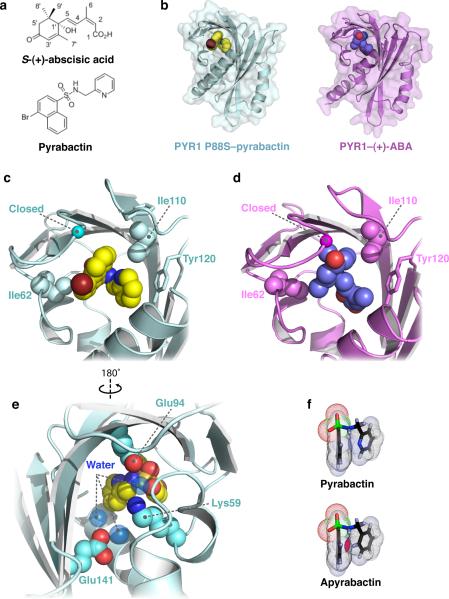
Pyrabactin binds the ABA-binding pocket of PYR1 and induces gate closure. (a) Chemical structures of abscisic acid and pyrabactin. (b) Structures of PYR1 P88S–pyrabactin (cyan) and PYR1–ABA10 (violet) contain ligands bound in the conserved START domain binding pocket. Close-up views in the same orientation as (a) and (b) illustrating the orientation of (c) pyrabactin and (d) ABA in the PYR1 binding pocket. The pyridyl ring packs against the aromatic side chain of Y120 and the naphthyl group of pyrabactin. Contacts with the bromine atom and napthyl group of pyrabactin stabilize the gate in the closed conformation, as indicated by the location of the conserved proline in the SerGlyLeuProAla gate. (e) The sulfonamide linkage forms polar contacts in the base of the binding pocket and functions analogously to ABA's carboxylate group. (f) Based on AM1 calculations, packing interactions between the pyridyl ring and naphthyl group in the conformation adopted by pyrabactin when bound to PYR1 would result in steric clash (magenta) in apyrabactin.
PYR1 is a member of the START-protein superfamily, a large and evolutionarily ancient family of hydrophobic ligand-binding proteins6. START-proteins share a conserved “helix-grip” fold that contains a central α-helix surrounded by a 7-stranded anti-parallel β sheet, which form a central hydrophobic ligand-binding pocket. PYR/PYL proteins bind directly to ABA5,7–9 and function as allosteric switches that control ABA signaling by inhibiting type 2C protein phosphatases (PP2Cs). Recent structural studies9–13 have revealed that ABA binds to PYR/PYL proteins within their conserved START-domain ligand-binding pockets, which are flanked by two loops that have been named “gate” and “latch”13. ABA binding induces closure of these loops, which seals ABA inside the ligand-binding pocket and creates an interaction surface that enhances PP2C binding. ABA-bound PYR/PYL proteins bind adjacent to the PP2C active site, which occludes substrate access and thus inhibits enzymatic activity11–13. Here we focus on features of the ligand binding pocket that allow pyrabactin to function selectively within the PYR/PYL family.
Results
Pyrabactin binding closes the PYR1 gate and latch loops
To understand how pyrabactin functions as a PYR1 agonist, we compared ABA and pyrabactin receptor complexes using both NMR and X-ray crystallography. Both ligands perturb signals in the PYR1 1H-15N HSQC spectrum near the gate (85SerGlyLeuProAla89) suggesting that pyrabactin mimics ABA by stabilizing the closed-gate configuration observed previously (Supplementary Fig. 1). Because crystals of wild-type PYR1–pyrabactin complex failed to diffract well enough for structural studies, we used PYR1 P88S, which binds both ABA5 and pyrabactin (Supplementary Fig. 2), to solve the X-ray structure of PYR1 P88S–pyrabactin to a resolution of 2.5 Å (Table 1). Pyrabactin occupies the ABA binding site within the hydrophobic pocket of PYR1 (Fig. 1b) in a unique compact configuration that folds the pyridyl and naphthyl groups tightly, therefore allowing the gate to pack against the bromine atom (Fig. 1c). Comparison of the PYR1 P88S–pyrabactin structure with the PYR1–ABA structure (Fig. 1d)9 shows that the gate loop (85SerGlyLeuProAla89) adopts a closed arrangement in response to both ligands. Pyrabactin binding also positions the backbone of the latch loop similarly, but with greater variability in the side chain positions between the two PYR1 structures. As observed previously for ABA-bound receptor structures, a complex network of polar and hydrophobic interactions facilitates pyrabactin's binding to PYR1. Pyrabactin's sulfonamide is positioned similarly to ABA's carboxylate and hydrogen bonds to the amino group of K59. Both direct and water-mediated contacts coordinated by E94 and E141 explain the necessity of these residues for pyrabactin responsiveness in vivo as originally defined by mutational studies5 (Fig. 1e). These results show that ABA and pyrabactin exploit similar contacts with PYR1 to elicit the conformational changes in the gate and latch needed for receptor activation.
Table 1.
Data collection and refinement statistics
| PYR1P88S:pyrabactin (refinement) | PYL2:pyrabactin (refinement) | PYL2:pyrabactin (Anomalous Br data) | PYL2V1141:pyrabactin (refinement) | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P6122 | C2221 | C2221 | P6122 |
| Cell dimensions | ||||
| a,b,c (Å) | 60.3, 60.3, 527.6 | 62.5, 105.2, 187.2 | 62.4, 104.9, 186.3 | 61.8, 61.8, 220.0 |
| α,β,γ (°) | 90, 90, 120 | 90, 90, 90 | 90, 90, 90 | 90, 90, 120 |
| Resolution (Å) | 50-2.47(2.51−2.47) | 50-1.89(1.92−1.89) | 45-2.05(2.09−2.05) | 40-1.95(1.98−1.95) |
| R merge | 0.086(0.458) | 0.073(0.468) | 0.082(0.357) | 0.068(0.619) |
| I / σI | 13.6(2.4) | 23.8(2.0) | 34.9(5.1) | 41.5(1.7) |
| Completeness (%) | 95.3(93.1) | 97.1(71.4) | 100.0(99.7) | 98.6(82.9) |
| Redundancy | 5.4(4.4) | 7.2(3.6) | 14.2(9.8) | 16.3(6.0) |
| Refinement | ||||
| Resolution (Å) | 32.0-2.47(2.54−2.47) | 29.7-1.89(1.94−1.89) | - | 30.5-1.89(2.00−1.95) |
| No. reflections | 19,707 | 46,723 | - | 18,358 |
| Rwork / Rfree | 0.226/0.265 | 0.178/0.205 | - | 0.195/0.217 |
| No. atoms | 4,301 | 4,886 | - | 1,668 |
| Protein | 4,156 | 4,282 | - | 1,506 |
| Ligand/ion | 77 | 284 | - | 49 |
| Water | 68 | 320 | - | 113 |
| B-factors | ||||
| Protein | 55.2 | 39.1 | - | 39.3 |
| Ligand/ion | 59.3 | 59.9 | - | 38.8 |
| Water | 48.2 | 41.8 | - | 41.3 |
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.026 | 0.010 | - | 0.017 |
| Bond angles (°) | 1.57 | 1.18 | - | 1.26 |
Values in parentheses are for highest-resolution shell. A single crystal was used for each structure determination.
When bound to PYR1, the pyridyl and naphthyl rings of pyrabactin are packed against each other at a ~60° angle. In order to satisfy this conformation, the pyridyl nitrogen must pack close to the naphthyl ring (Fig. 1f), as its rotation would cause steric clash. In a previous analysis of pyrabactin structure-activity relationships, any substitution involving the pyridyl nitrogen led to a complete loss of activity in germination and hypocotyl growth assays, while changes to the naphthyl group had only a modest effect5. Interestingly, the PYR1-bound conformation of pyrabactin is energetically unfavorable for the inactive analog apyrabactin, in which the replacement of the pyridyl N with a C–H is predicted by semi-empirical quantum mechanical calculations to cause steric clash with the naphthyl group (Fig. 1f). We suggest that this clash prevents apyrabactin from binding deeply enough in PYR1's pocket to allow gate closure. Thus, differences in the accessible conformations of these two ligands are sufficient to explain their differential effects on PYR1.
Pyrabactin binds to PYL2 in a non-productive mode
Our structure showed that pyrabactin, like ABA, triggers gate closure, but suggested no obvious residues that might explain pyrabactin's selectivity. We reasoned that pyrabactin's selectivity could reflect a lack of binding to non-responsive family members. To test this, we used PYL2 as a representative model since it does not show strong pyrabactin responsiveness in comparison to PYR15. In contrast to this expectation, NMR titration data show that pyrabactin binds PYL2 (Fig. 2a) at comparable stoichiometry to PYR1. This in turn suggested that a non-productive binding interaction could explain the differential effects of pyrabactin on PYR1 and PYL2. To investigate this, we solved the crystal structure of the PYL2–pyrabactin complex to 1.9 Å resolution (Table 1). As observed with PYR1, pyrabactin's pyridyl ring packs against the aromatic ring of Tyr124, but in PYL2 it slides ~2 Å away from the long C-terminal helix allowing the naphthyl group to rotate ~90° into the space vacated by the pyridyl ring (Fig. 2b). Pyrabactin positions PYL2's latch in a similar conformation to that observed in PYR1 (Fig. 2c), though weak or missing electron density for latch residues 117GEH119 suggests that it may be partially disordered. Notably, the gate remains open, with the central proline (Pro92) shifted 9 Å away from its closed position relative to the PYL2–ABA and PYR1–pyrabactin complexes. Thus, pyrabactin binds within the ABA-binding pocket of both PYR1 and PYL2, but does not elicit gate closure in PYL2 due to non-productive, incompletely buried ligand orientation (Supplementary Fig. 3).
Figure 2.
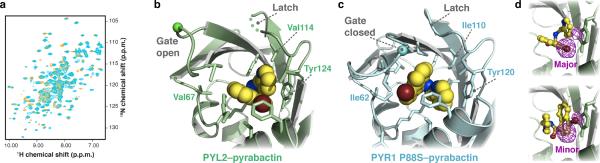
Crystal structure of the PYL2–pyrabactin complex. (a) 1H-15N HSQC spectra of PYL2 (500 μM) in the absence (cyan) and presence (orange) of pyrabactin (750 μM) demonstrates that pyrabactin is a PYL2 ligand. Pyrabactin saturation was observed at 1:1 stoichiometry demonstrating that PYL2 does not have a defect in pyrabactin binding. (b) Pyrabactin occupies the PYL2 binding pocket in different orientation than observed in (c) the PYR1 P88S–pyrabactin structure. While the latch conformation is similar, the gate loop remains in an open conformation due to the loss of stabilizing contacts with the pyrabactin naphthyl group and bromine atom. The position of the conserved proline in the SerGlyLeuProAla gate is indicated by a green (PYL2) or blue (PYR1 P88S) sphere. (d) Electron density from anomalous bromine scattering (magenta) identifies two binding modes for pyrabactin in a 2:1 ratio. The sidechain for Tyr124 is shown.
During refinement of our PYL2–pyrabactin model, analysis of the electron density in PYL2's ligand-binding pocket suggested that multiple pyrabactin conformations were present. To probe these binding orientations more directly, we exploited the anomalous X-ray scattering generated by pyrabactin's bromine atom to resolve multiple ligand conformations (Table 1). The anomalous density map reveals two signals corresponding to the major orientation described above and a minor species representing a second unproductive ligand orientation in a ~2:1 ratio. Relative to the major PYL2 binding mode, the naphthyl ring revolves ~90° around the bromine-sulfur axis to pack against the aromatic ring of Tyr124 while the pyridyl ring rotates ~180° to pack against the naphthyl ring (Fig. 2d). This rearrangement moves the pyridyl nitrogen the greatest distance, ~5.5 Å, when the conformations are compared. Thus, the conclusion from these three X-ray data sets is that PYR1 enables a specific and productive pyrabactin binding orientation, while PYL2 accommodates multiple non-productive orientations.
Ile110 and Ile62 position pyrabactin for agonist activity
Since the gate and latch loops are highly conserved and the PYR1 and PYL2 structures are very similar, we reasoned that subtle sequence differences between the pockets of PYR1 and PYL2 influence pyrabactin's binding orientations. To define the specific residues underlying this behavior, we screened for PYR1 mutants that respond to ABA but not pyrabactin using a mutagenized PYR1 library in a yeast based receptor assay5. Sequence analysis of mutants that selectively block pyrabactin response identified a number of residues (Supplementary Table 1 online). Some play no obvious role in pyrabactin selectivity since they are conserved in responsive and non-responsive family members or are located far from the ligand binding pocket (e.g. Lys170). We focused on the specific mutations I110V and I62V because they correspond to natural substitutions found in family members such as PYL2, PYL3 and PYL4 that are non-responsive to pyrabactin. The side chains of both Ile110 and Ile62 project into the ligand–binding pockets of PYR/PYL proteins and make close contact to pyrabactin. These observations suggested that natural variation at positions homologous to Ile110 and Ile62 contribute to pyrabactin's selectivity. To test this, we constructed and analyzed PYR1 I62V, PYR1 I110V and PYR1 I110V I62V mutants; all mutants generated retain ABA responsiveness (Fig. 3a), but achieve only partial PP2C interaction (Fig. 3b) or inhibition (Fig. 3c) in response to saturating levels of pyrabactin. These results define Ile110 and Ile62 as key determinants of pyrabactin response and demonstrate that subtle changes in pocket residues control agonist activity.
Figure 3.
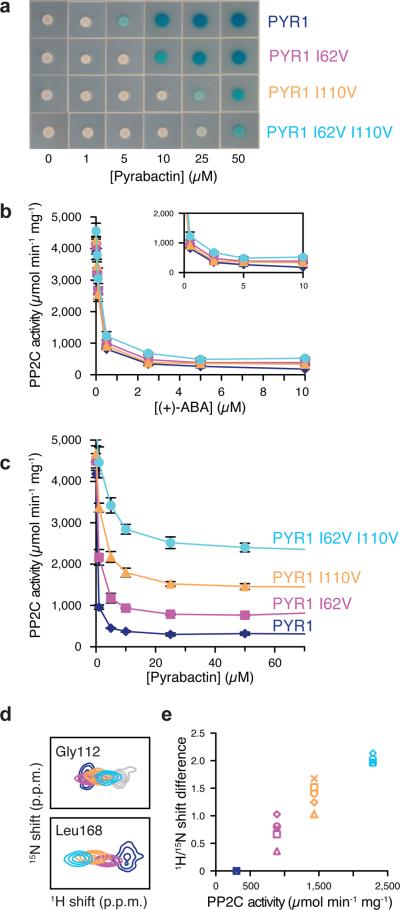
Genetic Identification of Pyrabactin Selectivity Determinants. (a) Yeast two hybrid screening for PYR1 mutants that become unresponsive to pyrabactin but retain ABA receptor activity identified I62V and I110V. Both substitutions, which correspond to amino acids in the pyrabactin-insensitive PYL2 receptor (Val67 and Val114), reduce pyrabactin-dependent PYR1-HAB1 binding and the PYR1 I62V I110V double mutant reflects the additive nature of the individual mutations. (b) In vitro PP2C assays conducted using ABA as the agonist show no adverse effects from the mutations on receptor activity. (c) Receptor mediated PP2C inhibition assays were used to further characterize effects of the swap mutations on pyrabactin responsiveness. Error bars in panels (b) and (c) represents standard deviations. (d) HSQC peak positions for pyrabactin-saturated PYR1 L166R (blue), PYR1 I62V L166R (magenta), PYR1 I110V L166R (orange), or PYR1 I62V I110V L166R (cyan) show a linear progression of shifts toward the ligand-free spectrum (gray), consistent with mutations shifting a two-state conformational equilibrium toward the inactive conformation. (e) Shifts in the HSQC peak positions for residues around the ligand binding pocket and PP2C binding loops vary linearly with the change in maximal PP2C inhibition at saturating concentrations of pyrabactin. The HSQC experiments in panels (d) and (e) utilize PYR1 L166R as a reference molecule; this is a functional monomeric PYR1 variant that was created for NMR analysis to provide more complete residue coverage in comparison to wild type (dimeric) PYR1.
Interestingly, the PP2C-inhibition curves for these PYR1 selectivity mutants achieve incomplete inhibition in response to saturating concentrations of pyrabactin (Fig. 3c), suggesting that a mixture of active and inactive receptor populations is formed. To interrogate the stability of these complexes in solution, we performed a comparative NMR analysis of a monomeric PYR1 mutant (PYR1 L166R) and the equivalent set of pyrabactin responsiveness mutants (PYR1 L166R I62V, PYR1 L166R I110V, and PYR1 L166R I62V I110V) in the presence of saturating pyrabactin concentrations. NMR titration experiments indicated that PYR1 L166R binds ABA and pyrabactin with higher affinity than wild-type PYR1, suggesting that ligand binding may compete with PYR1 self-association. PYR1 L166R also retains ABA-dependent PP2C binding in a yeast two-hybrid assay, indicating that receptor function is not dependent on the dimer interface being intact.
An HSQC overlay of all four pyrabactin-saturated PYR1 L166R proteins shows a linear pattern of 1H/15N shift perturbations for multiple residues (Fig. 3d), which is a hallmark of an equilibrium in which two conformations interconvert on microsecond-to-millisecond time scales14. The progression of HSQC signals for each PYR1–pyrabactin complex has the appearance of a titration experiment, as if incorporation of each inactivating mutations increases the relative abundance of a new conformational state. Since the alternative binding mode observed in the PYL2–pyrabactin complex correlates with an `open' gate-latch arrangement, we speculated that the loss of sidechain methyl groups at positions 62 and 110 in the PYR1 binding pocket would permit pyrabactin to sample similar non-productive orientations. In the case of Gly112, a latch residue that does not contact pyrabactin, the selectivity mutations progressively shift its HSQC signal toward the ligand-free `open' chemical shift despite the presence of pyrabactin in the binding pocket. Strikingly, the magnitude of the chemical shift for each mutant relative to wild-type PYR1 is directly proportional to the residual PP2C activity observed at saturating pyrabactin concentrations (Fig. 3e). Because residues in the pocket, gate and latch respond coordinately, we conclude that the Ile62V and Ile110V mutations shift the ratio between the productive ligand orientation observed in PYR1 P88S–pyrabactin and a non-productive orientation and that these states interconvert rapidly in solution.
Pyrabactin shifts orientation in reciprocal PYL2 mutants
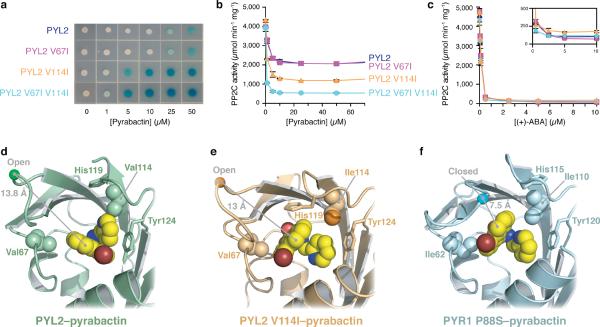
Since removal of a delta methyl group at either Ile62 or Ile110 promotes non-productive pyrabactin binding in PYR1, we reasoned that reciprocal mutations in PYL2 would have the opposite effect. Indeed, when compared to wild-type PYL2, the PYL2 V67I, PYL2 V114I and PYL2 V67I V114I mutants are increasingly pyrabactin-sensitive (Fig. 4a). Likewise, maximal PP2C inhibition in response to pyrabactin levels rises significantly with the addition of each delta methyl group into the binding pocket (Fig. 4b), but ABA responses are not changed (Fig. 4c). To investigate the structural basis for this, we solved the PYL2 V114I–pyrabactin complex to 2.0 Å resolution (Table 1). Relative to the ligand orientation in the wild-type PYL2–pyrabactin structure (Fig. 4d), pyrabactin binds PYL2 V114I in a single well-defined position resembling a productive ligand orientation in which the pyridyl ring is stacked between the aromatic ring of Tyr124 and the naphthyl ring (Fig. 4e). However, the gate remains in an open conformation with the central proline (Pro92) ~11 Å away from its closed position observed in the PYR1 P88S–pyrabactin structure (Fig. 4f). PYL2 V114I enables strong pyrabactin agonist activity, but like the reciprocal selectivity mutants in PYR1, it does not enable complete PP2C inhibition in response to pyrabactin, which implies the formation of a mixed population of open and closed gate conformations. We therefore suggest that the PYL2 V114I crystal structure captures a non-productive state in which pyrabactin sits ~1.5 Å deeper in the PYL2 binding pocket than that observed in the PYR1–pyrabactin structure. This subtle difference allows His119 to pack inside PYL2's pocket and block gate closure. Irrespective of the explanation for its open gate, this structure clearly reveals that the addition of a single delta methyl carbon in PYL2 V114I is sufficient to shift pyrabactin's binding orientation. The inverse trends in PP2C binding and inhibition for the PYR1 and PYL2 reciprocal mutants, the linear pattern of 1H/15N shift perturbations observed for the PYR1 mutants, and the changes in pyrabactin orientation observed crystallographically in PYL2 and PYL2 V114I bolster our conclusion that removal of the delta methyls from Ile110 and Ile62 alters the pyrabactin orientation in PYR1.
Figure 4.
Ligand orientation polymorphisms underlie selectivity. (a) Yeast two hybrid measurement of PYL2 responsiveness to pyrabactin using wild type PYL2, or the selectivity mutants shown. PP2C enzymatic activity assays conducted using PYL2 or the mutants shown in response to (b) pyrabactin or (c) ABA. Enzymatic assays were conducted using 600 nM receptor and PP2C (HAB1), as described in the experimental methods. Error bars in panels (b) and (c) represents standard deviations. Orientations of bound pyrabactin relative to the selectivity determining residues are shown for (d) PYL2, (e) PYL2 V114I and (f) PYR1 P88S. The conformation of pyrabactin is different in all three states, as is the response of the critical protein loops and their resulting ability to inhibit the PPC2 phosphatase.
Discussion
Our structural comparisons and mutational analysis of pyrabactin binding to the abscisic acid receptors PYR1 and PYL2 has shown that subtle changes in the binding pockets of PYR/PYL proteins control ligand orientation inside the pocket, and this is sufficient to explain whether pyrabactin acts as an effective receptor agonist. One question this raises is whether the natural variation in pocket-lining residues has functional relevance in vivo. One possibility is that plants may contain endogenous signals that exploit this variability to achieve selective receptor activation, a hypothesis we are currently investigating. We initiated our studies anticipating that pocket variation would control pyrabactin binding, however this is not the case. Our results are consistent with frequently observed promiscuity of ligand-binding to START proteins and show that a key feature of agonist activity is not receptor binding per se, but stabilization of gate closure. The implications of our work for agonist design are important and we now have several tools with which to measure and monitor effective gate closure. Additionally, we suggest that the chemical modification of pyrabactin at sites designed to alter ligand orientation will enable agonist design for other PYR/PYL receptors.
Methods
Protein expression and purification
PYR1 P88S, PYL2 and PYL2 V114I were expressed in E. coli and purified as previously described for PYR15. Briefly, proteins were expressed in E. coli BL21(DE3), lysed using a French press, and purified by IMAC chromatography. Since PYR/PYL proteins are dimers, they pose inherent problems for NMR and initial analyses, and we have previously shown that residues at the dimer interface are undetectable due to monomer-dimer exchange13. We therefore tested multiple mutations that we predicted would destabilize the dimer interface. The most promising variant created, PYR1 L166R, elutes as a monomer in gel filtration experiments and yields NMR spectra in which backbone chemical shifts for >90% of the residues were assigned. Affinity tags were proteolytically removed and the proteins were further purified by gel filtration chromatography. Proteins were concentrated to 20–30 mg mL−1 in preparation for crystallization.
Crystallization
Conditions for PYR1 P88S crystallization in the presence of a 2-fold molar excess of pyrabactin were identified using the UW192 screen developed by the Center for Eukaryotic Structural Genomics, and the Hampton Index HT screen (Hampton Research, Aliso Viejo, CA) at 4° and 20° C. Diffraction quality crystals were grown at 20° C in a sitting drop vapor diffusion experiment from 2 M ammonium sulfate and 100 mM BisTris, pH 5.5. Crystals were transferred to Fomblin 06/6 and excess crystallization solution was mechanically removed from the crystals. Crystals were frozen directly in liquid nitrogen.
PYL2 crystallization conditions in the presence of a 2.5-fold molar excess of pyrabactin were identified using screens described above at 20° C. Diffraction quality crystals were grown at 20° C in a hanging drop vapor diffusion experiment from 180 mM magnesium acetate, 16% (w/v) PEG-8000, and 100 mM sodium cacodylate, pH 6.5. PYL2 V114I crystals were grown at 20° C in a hanging drop vapor diffusion experiment from 220 mM ammonium citrate, and 19.5% (v/v) PEG-3350. Crystals were transferred to well solution containing 20% (v/v) glycerol prior to freezing in liquid nitrogen.
Data collection and structure determination
X-ray diffraction data were gathered for single crystals at the 21-ID-D and 21-ID-G beamlines of the Advanced Photon Source at Argonne National Laboratories. Data collection wavelengths for pyrabactin complexes with PYR1 P88S and the PYL2's were 1.0781 and 0.97856, respectively. The anomalous Br data for each of the PYL2 complexes was collected at 0.92017 Å. All data was collected at 100 K. The PYR1 P88S -pyrabactin, PYL2-pyrabactin and PYL2 V114I -pyrabactin complexes diffracted to 2.47, 1.89, and 1.95Å, respectively. The observed reflections were indexed, integrated and internally scaled using HKL200015. Molecular replacement was utilized to evaluate the initial phases using PYL1 (PDB code 3JRS, subunit A11) and PYL2 (PDB code 3KB013) as the search models for PYR1 P88S -pyrabactin and PYL2-pyrabactin, respectively. Our completed PYL2-pyrabactin structure served as the search model for the PYL2 V114I -pyrabactin complex. Phenix.AutoMR16–18 solved the initial phases and automatically built in the majority of the amino acid residues for each of the complexes. Models were completed through successive rounds of manual model building with Coot19 and refinement with phenix.refine using TLS and individual atomic displacement parameters. Selection of TLS groups was facilitated using the TLSMD web server (http://skuld.bmsc.washington.edu/~tlsmd/)20,21. Pyrabactin was modeled using Monomer Library Sketcher from the CCP4 software package. Geometry of the final structural model was validated with Molprobity22 and Procheck23. The steric clashes within pyrabactin and apyrabactin in the bound state were examined using AM1 within the phenix.elbow package24. Ramachandran statistics were 93.1 and 6.9, 97.5 and 2.5, and 97.2 and 2.8 for the favored and additionally allowed regions of the Ramachandran plot for the PYR1 P88S –pyrabactin, PYL2-pyrabactin, and PYL2 V114I -pyrabactin complexes, respectively. PyMOL was used to generate all structure images.
NMR spectroscopy
PYR1, PYR1 L166R, PYR1 I62V L166R, PYR1 I110V L166R, PYR1 I62V I110V L166R and PYL2 proteins for NMR were expressed in E. coli and purified as described previously5. PYR1–pyrabactin complexes were obtained by addition of a two-fold excess of pyrabactin to the purified protein. NMR experiments were performed at 37 °C on a Bruker Avance III 500MHz or Avance 600MHz spectrometer equipped with a 5 mm TCI CryoProbe. Backbone 1H, 15N and 13C chemical shifts of PYR1 L166R–ABA were obtained using automated assignment software as described previously5 and transferred to the pyrabactin complexes by inspection.
Isolation of Pyrabactin Non-Responsive mutants
To identify PYR1 residues selectively involved in pyrabactin responsiveness, the coding sequence for PYR1 was mutated by error-prone PCR and cloned into the yeast two hybrid vector pBD, yielding a library of ~70,000 mutants. This library was amplified in E. coli and then transformed into S. cerevisiae strain MAV99 containing pAD-HAB1. MAV9925 is a reverse two hybrid strain that contains a gal4∷URA3 reporter; growth of this strain in the absence of added uracil enables positive selection for protein-protein interactions, whereas growth on FOA (which is metabolized by URA3 to a toxic metabolite) allows selection against interactions. A pool of pyrabactin non-responsive (PyrA−) PYR1 mutants was identified by growth of the mutant library on 0.15% FOA and 10 μM pyrabactin. To subsequently remove non-functional mutants, the pool of PyrA− mutants were grown on media containing 5μM ABA, but lacking added uracil. The coding sequences for 49 ABA+, PyrA− colonies were determined using PCR products. To confirm the importance of specific residues identified by this analysis, defined mutations were introduced into PYR1 or PYL2 using the QuickChange site directed mutagenesis protocol, using pBD-PYR1 or pBD-PYL2 as templates. The mutated clones were sequence validated and transformed into a pAD-HAB1 expressing yeast two hybrid reporter, as previously described5. To characterize the mutant receptor proteins in in vitro PP2C assays, the mutants were cloned into pET28 to create 6X-HIS recombinant proteins, which were expressed and purified as described previously. PP2C assays were run using pNPP as the phosphatase substrate and 600 nM HAB1 and the appropriate PYR/PYL protein, as described previously13.
Supplementary Material
Acknowledgements
This work was supported by the NIGMS Protein Structure Initiative (U54 GM074901 and NSF (IOS-003725-002). We acknowledge LS-CAT at sector 21 at the Advanced Photon Source at Argonne National Laboratory for X-ray beamline access. SRC thanks Jeffrey Bachant (UC Riverside) for sharing yeast strains.
Footnotes
Database accession numbers. Crystal structures of PYR1 P88S–pyrabactin, PYL2–pyrabactin, and PYL2 V114I–pyrabactin were deposited in the RCSB under accession numbers 3NJO, 3NJ0 and 3NJ1 respectively.
Supplementary Information is linked to the online version of the paper
References
- 1.Nambara E, Marion-Poll A. ABSCISIC ACID BIOSYNTHESIS AND CATABOLISM. Annual Review of Plant Biology. 2005;56:165–185. doi: 10.1146/annurev.arplant.56.032604.144046. [DOI] [PubMed] [Google Scholar]
- 2.Cutler S, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic Acid: Emergence of a Core Signaling Network. Annual Review of Plant Biology. 2009 doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y, et al. Molecular tailoring of farnesylation for plant drought tolerance and yield protection. Plant J. 2005;43:413–424. doi: 10.1111/j.1365-313X.2005.02463.x. [DOI] [PubMed] [Google Scholar]
- 4.Hawkins AF, Stead AD, Pinfield NJ. Plant Growth Regulators for Agricultural and Amenity Use. British Crop Protection Council; 1987. [ED: This is a book - ISBN 0-948404-02-7] [Google Scholar]
- 5.Park SY, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iyer LM, Koonin EV, Aravind L. Adaptations of the helix-grip fold for ligand binding and catalysis in the START domain superfamily. Proteins. 2001;43:134–144. doi: 10.1002/1097-0134(20010501)43:2<134::aid-prot1025>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 7.Szostkiewicz I, et al. Closely related receptor complexes differ in their ABA selectivity and sensitivity. Plant J. 2009;61:25–35. doi: 10.1111/j.1365-313X.2009.04025.x. [DOI] [PubMed] [Google Scholar]
- 8.Ma Y, et al. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 9.Santiago J, et al. The abscisic acid receptor PYR1 in complex with abscisic acid. Nature. 2009;462:665–668. doi: 10.1038/nature08591. [DOI] [PubMed] [Google Scholar]
- 10.Nishimura N, et al. Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science. 2009;326:1373–1379. doi: 10.1126/science.1181829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyazono K, et al. Structural basis of abscisic acid signalling. Nature. 2009;462:609–614. doi: 10.1038/nature08583. [DOI] [PubMed] [Google Scholar]
- 12.Yin P, et al. Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nat Struct Mol Biol. 2009;16:1230–1236. doi: 10.1038/nsmb.1730. [DOI] [PubMed] [Google Scholar]
- 13.Melcher K, et al. A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature. 2009;462:602–608. doi: 10.1038/nature08613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Volkman BF, Lipson D, Wemmer DE, Kern D. Two-state allosteric behavior in a single-domain signaling protein. Science. 2001;291:2429–2433. doi: 10.1126/science.291.5512.2429. [DOI] [PubMed] [Google Scholar]
- 15.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 16.Frederick RO, et al. Small-scale, semi-automated purification of eukaryotic proteins for structure determination. J Struct Funct Genomics. 2007;8:153–166. doi: 10.1007/s10969-007-9032-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 18.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr D Biol Crystallogr. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 19.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 20.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 21.Painter J, Merritt EA. TLSMD web server for the generation of multi-group TLS models. J Appl Crystallogr. 2006;39:109–111. [Google Scholar]
- 22.Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laskowski RA, MacArthur MW, Moos DS, Thornton JM. PROCHECK - a program to check the stereochemical quality of protein structures. Journal of Applied Crystallography. 1993;26:283–291. [Google Scholar]
- 24.Moriarty NW, Grosse-Kunstleve RW, Adams PD. electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr D Biol Crystallogr. 2009;65:1074–1080. doi: 10.1107/S0907444909029436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vidal M, Brachmann RK, Fattaey A, Harlow E, Boeke JD. Reverse two-hybrid and one-hybrid systems to detect dissociation of protein-protein and DNA-protein interactions. Proc Natl Acad Sci U S A. 1996;93:10315–10320. doi: 10.1073/pnas.93.19.10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.