Abstract
In most vertebrate embryos and neonates studied to date unique antigen receptors (antibodies and T cell receptors) are expressed that possess a limited immune repertoire. We have isolated a subclass of IgM, IgM1gj, from the nurse shark Ginglymostoma cirratum that is preferentially expressed in neonates. The variable (V) region gene encoding the heavy (H) chain underwent V-D-J rearrangement in germ cells (“germline-joined”). Such H chain V genes were discovered over 10 years ago in sharks but until now were not shown to be expressed at appreciable levels; we find expression of H1gj in primary and secondary lymphoid tissues early in life, but in adults only in primary lymphoid tissue, which is identified in this work as the epigonal organ. H1gj chain associates covalently with light (L) chains and is most similar in sequence to IgM H chains, but like mammalian IgG has three rather than the four IgM constant domains; deletion of the ancestral IgM C2 domain thus defines both IgG and IgM1gj. Because sharks are the members of the oldest vertebrate class known to possess antibodies, unique or specialized antibodies expressed early in ontogeny in sharks and other vertebrates were likely present at the inception of the adaptive immune system.
In mammals, expressed Ig and T cell receptors (TCR) of fetuses and newborns are qualitatively different from those of adults. For Ig, a few particular variable (V) regions are expressed by an early subset of developing B cells (B1 cells) that are later supplanted by new precursors with a diverse repertoire (1). T cell precursors bearing γ/δ TCR with highly restricted repertoires develop in the fetal thymus and then are also replaced in this primary lymphoid tissue by conventional, major histocompatibility complex (MHC)-restricted α/β T cells (2). Lymphocytes bearing such “innate” receptors are produced early in ontogeny when little diversity is generated as a consequence of gene (V-D-J or V-J) rearrangement (3–5). In addition, innate cells can self-renew and are found in the peritoneal cavity (B1 cells in mice; ref. 6) and epithelial surfaces (γ/δ T cells; ref. 7), where they are presumed to serve specialized functions. The early appearance of innate lymphocytes, serving perhaps as a first line of defense, and the late appearance of “adaptive” B and T cells with great antigen receptor diversity and residing in secondary lymphoid tissues are the foundation of the “Layering Hypothesis” of the immune system proposed by the Herzenbergs (8).
Elasmobranchs (sharks, skate, and rays) are members of the oldest vertebrate taxon known to possess an adaptive immune system grounded on Ig, TCR, and the MHC (9). There are at least three Ig classes in elasmobranchs: IgM (10), IgNAR (11), and IgW (12–15). IgM is the orthologue of the IgM identified in all other vertebrates and is the best studied class in elasmobranchs; it is found in both monomeric (7S) and multimeric (19S) forms and comprises about half of the serum protein in adults (16). Neonatal nurse sharks (Ginglymostoma cirratum) were shown to have very low serum levels of IgM at birth and required over 1 month post partum to approach adult-like levels (17). Their total serum protein, however, was the same as in adults, indicating that other unidentified neonatal proteins were present. Nurse sharks are ovoviviparous, developing within an egg case in the uterus, so there is no maternal Ig transfer as in mammals although there might be transfer to embryos from the yolk (18).
All elasmobranch Ig heavy (H) and light (L) chain genes are in the “cluster” configuration, with each H chain cluster containing one variable (V), 2–3 diversity (D), one joining (J), and several constant (C) exons (19). There are estimated to be up to 200 IgM H chain clusters in the horn shark (Heterodontus francisci; refs. 20 and 21). Most expressed H chains are encoded by clusters that rearrange the V, D, and J genes in what is inferred to be the typical mammalian fashion; in contrast, up to 50% of the clusters have V, D, and J genes that are either partially or wholly pre-rearranged (“germline-joined”), and these genes are not expressed at appreciable levels, i.e., detected by Northern blotting or even as cDNAs from adult spleen libraries (22). However, such germline-joined clusters were theorized to be expressed early in ontogeny and/or in specialized tissues; it was also suggested that they could provide a selective advantage by perpetuating germline specificities and thus be used for specific functions (22–26).
Along with Ig, TCR, and MHC, cartilaginous fish are the oldest vertebrates to have both primary (hematopoietic cell-producing) and secondary (immune response-generating) lymphoid tissues (27, 28). Based upon morphological studies, the putative bone marrow equivalent is the epigonal organ, which is physically associated with gonadal tissue throughout life, or the Leydig organ, attached to the outer wall of the esophagus. Depending on the particular species examined, either one or both of these tissues is present and they have the same basic organization: >50% of the cells are myeloid but lymphoid and Ig-secretory cells have also been observed (29). The spleen is the only elasmobranch secondary lymphoid tissue with defined accumulations of mature lymphoid cells; it has distinct white and red pulp regions but no marginal zone separating the two. The white pulp areas contain predominantly lymphocytes of different sizes and a few mature and developing secretory B cells. The red pulp contains erythrocytes, leukocytes of many types (including secretory B cells), and venous sinuses (ref. 27; L.L.R. and M.F.F., unpublished results).
While carrying out studies of the developmental regulation of IgM and IgNAR expression, we identified serendipitously a subclass of IgM expressed during the early life of nurse sharks. As reported here, its expression and gene structure conform to speculations tendered above for the function of cartilaginous fish germline-joined Ig genes, and reveal that antigen receptors expressed early in development are different from those in adult life in a wide range of vertebrates. Furthermore, its expression led us to reexamine, with molecular means, the hypothesis that the epigonal organ serves as a primary lymphoid tissue in elasmobranchs.
Materials and Methods
Animals.
Nurse shark (G. cirratum) pups were delivered by Caesarian section from three females as described (30). Because there is no table of nurse shark development, we do not know the precise ages of these animals, but offspring were collected in October/November in South Florida when females drop their pups (mating in June/July). The pups were maintained in aerated seawater and fed seafood twice a week. Blood was obtained (16) and plasma was collected and either used immediately for biochemical studies or frozen in aliquots at −20°C.
Pup Spleen and Epigonal Organ cDNA Libraries and Genomic Cosmid Library.
Two newborn nurse shark pups were anesthetized in MS-222. Tissues were collected, and total RNA was prepared per the TRIzol RNA isolation protocol (GIBCO/BRL). mRNA was prepared per Ambion (Austin, TX) Poly(A) Pure kit. The cDNA libraries were constructed per Stratagene ZAP-cDNA library kit using 5 μg mRNA pooled from each tissue. The cosmid library was constructed commercially by using erythrocyte genomic DNA from one adult nurse shark per Stratagene SuperCos 1 Cosmid library. Whole blood was centrifuged, and the peripheral blood lymphocyte buffy coat was discarded. The erythrocyte pellet was subjected to two rounds of Ficoll gradient centrifugation to rigorously deplete all lymphocytes. The spleen and epigonal libraries were screened under both high and low stringency conditions as described (31) with the conventional IgM VH probe (32).
Probes.
Splenic or thymic cDNA or plasmid DNA containing cloned cDNA inserts were used as templates for PCR amplification of all probes as described (30). Oligo(dT)-primed cDNA was made from 5 μg nurse shark total RNA thymus and spleen by using GIBCO/BRL Superscript First Strand cDNA Synthesis kit. The probes were the following: conventional IgM V (no. M92851, nucleotides 1–477); IgM1gj V and CH1 (no. AF327520, see Fig. 2A for priming sites); recombination-activating gene 1 (RAG1) (no. U13982, nucleotides 6–309); terminal deoxynucleotidyl transferase (TdT) (no. AF327519) (S.B., M.D., and M.F.F., unpublished results); the nucleotide diphosphate kinase (NDPK) probe was a gift from M. Kasahara (33). Twenty-five nanograms of probe was labeled with 32P[dCTP] with the Random Primed Labeling kit (Boehringer). Typical counts were 1 × 106 cpm/μl.
Figure 2.
IgM1gj is germline joined, and the CH2 domain exon has been deleted from the H1gj gene cluster. (A) Alignment of IgM1gj with conventional nurse shark IgM (Accession no. M92851). The CDRs in the VH domain are boxed. Note that the short CDR3 uses only one D gene (22). The CH1 cys used for L chain association in IgM is substituted by asn (N) in H1gj (bolded, asterisk). The ancestral IgM CH2 domain is missing. Putative CH glycosylation sites are boxed–an additional glycosylation site occurs in CH3 (H1gjCH2) that is not found in conventional IgM. The arrowheads at the beginning of the V and CH1 domains and at the end of the J and CH1 regions denote the primers used for amplification of exon-specific probes. (B) Cartoon comparing the V and C domains of IgM and IgM1gj H chains. The ancestral IgM CH2 domain is missing in H1gj. Putative glycosylation sites (black lollipops) and interchain disulfide bonds (—) are noted. (C) IgM1gj germline V-D-J join. The VDJ junction nucleotide sequence aligned to a fully sequenced horn shark Ig cluster (ref. 22, see Fig. 2D) suggests the presence of only one D gene, D2, in the join. D2 nucleotides matching to the horn shark D2 segment have one underline (see Fig. 2D); D2 nucleotides possibly derived from a P-nucleotide addition are double underlined (we propose that the GA dinucleotide was part of the original D2 region.) (D) Coding ends and RSS (heptamers and nonamers bolded) of rearranging segments (V, D2, and J) of a representative horned shark variable region gene (ref. 22; accession no. X13447). If a similar gene gave rise to the IgM1gj join (Fig. 2C), a cleavage would have occurred between the last base of the V and D2 3′ coding ends and the heptamers (bold), and two bases (italics) would have been trimmed from the J coding end. In the C legend, we describe a potential scenario for processing of the IgM1gj 5′ D2 junction.
Southern and Northern Blots.
Southern blots were done under low or high stringency conditions (34). Five micrograms of nurse shark pup or adult genomic DNA was digested for 1.5 days with various enzymes. For Northerns (31), gels were loaded with 20 μg total RNA or 4 μg mRNA per lane. The labeled probes were hybridized with filters for a minimum of 20 h at 42°C and then washed under high or low stringency conditions (34).
Monoclonal Antibodies, Immunoprecipitation, and Metabolic Labeling.
Monoclonal antibodies (mAbs) specific for nurse shark IgM were made as described (11, 35, 36). CB5, CB16, and LK45 recognize both the 19S and 7S forms of IgM, GA15 prefers 7S to 19S, and GA16 prefers 19S. LK14 recognizes both 19S and 7S IgM via an L chain epitope. NART1 is specific for the IgNAR C terminus. NK11 binds a shark cell surface protein not present in plasma and was used as a negative control (E.C.M. and M.F.F., unpublished results). JC4 is specific for shark J chains (V. S. Hohman, A. S. Greenberg, L.L.R., D.A., M.F.F., and L. A. Steiner, unpublished results). Nurse shark plasma proteins were labeled with 125I by using the chloramine T method (37). Radio-iodinated plasma (1–4 μl containing 106 cpm) was incubated with 100 μl neat mAb supernatants overnight at 4°C, and the complexes were captured with protein-G bead adsorbents (11). SDS/PAGE on 5% (nonreducing) or 9% (reducing) gels was done as described (11, 14). Biomax MR film (Kodak) was used for autoradiography. For metabolic labeling, nurse sharks were euthanized, and single cell suspensions of spleen and epigonal organ were prepared. Cells were labeled overnight with 35S Trans (radiolabeled methionine and cysteine, ICN) as described (14). Supernatants containing labeled secreted Igs were collected and immunoprecipitated with mAbs as described above for plasma.
Protein Sequencing.
IgM1gj was immunopurified on a protein-G-mAb GA15 affinity column as described (36), separated on a 13% SDS/PAGE gel under reducing conditions, and electroblotted to poly(vinylidene difluoride) membrane (Immobilon-Psq, Millipore) for 2 h at 250 mA constant current in 10 mM CAPS (3-cyclohexylamino-1-propanesulfonic acid), pH 11/10% methanol. Proteins were stained with 0.1% (wt/vol) Amido Black 1oB (Bio-Rad) in methanol/acetic acid/water (40:10:50) and then destained with methanol/acetic acid (50:10). Protein bands at 55.7 kDa and 53 kDa (see Fig. 1) were excised with a razor for Edman sequencing in a Biosystems 494 (Foster City, CA) Protein Sequencer (11).
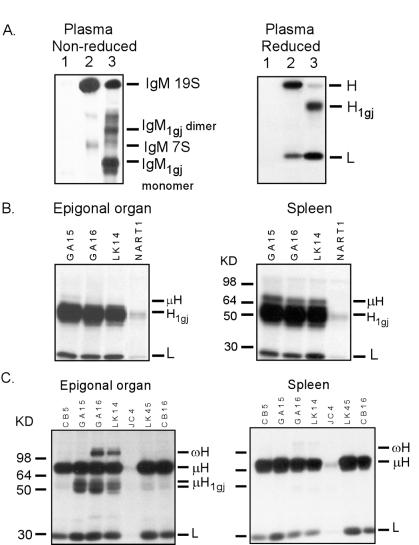
Figure 1.
IgM1gj immunoprecipitated from neonatal plasma, neonatal spleen and epigonal organ, and adult epigonal organ has a structure similar to mammalian IgG. (A) Radio-iodinated IgM1gj in plasma exists as monomers (lower IgM1gj band in lane 3) and dimers (upper IgM1gj band in lane 3) in nonreduced 5% PAGE. Under reducing conditions in 9% PAGE, the H1gj Mr is ≈55 kDa, associated covalently with an L chain (see text and supplementary Fig. 5). The 80-kDa H chain is derived from 19S and 7S IgM. Lanes 1, NK11 mAb (negative control), lanes 2, CB16 mAb (IgM-specific but noncrossreactive on IgM1gj), lanes 3, GA15 (IgM-specific and crossreactive on IgM1gj). (B) Neonatal spleen and epigonal organ cells preferentially secrete IgM1gj. Cells were metabolically labeled with 35S met/cys, and secreted IgM and IgM1gj were immunoprecipitated with the cross-reactive GA15, GA16, and LK14 mAbs and run on reducing 9% reducing PAGE. (C) Adult epigonal organ and spleen cells were metabolically labeled with 35S met/cys, and secreted IgM1gj was immunoprecipitated and run in 9% reducing PAGE. Relative to IgM, IgM1gj is maintained at high levels in the epigonal organ. Note that GA15, GA16, and LK14 mAbs recognize both IgM1gj and IgM, whereas the other IgM-specific mAbs do not cross-react. Additionally, putative IgW was immunoprecipitated from the epigonal organ supernatants by using the LK14 and GA16 mAbs (14).
Results
IgM1gj Predominates During Early Development.
While examining the ontogeny of expression of known secreted nurse shark Ig classes, we immunoprecipitated a new form of Ig, IgM1gj,** from radio-iodinated plasma proteins of neonates with a subset of our mAbs specific for conventional IgM (e.g., lane(s) 3 in Fig. 1A). The protein recognized by the crossreacting mAbs (GA15, GA16, and LK14) had the same biochemical properties under reducing and nonreducing conditions in each neonate examined (16 newborn pups from 3 families), by using fresh or frozen plasma. The H1gj chain Mr is ≈55 kDa, suggested to be smaller by one constant domain from conventional IgM H chains, but the same size as mammalian IgG H chains (38). H1gj associates covalently with IgL chains, determined by two-dimensional gels (first dimension under nonreducing conditions and the second under reducing conditions; see Fig. 5, which is published as supplemental data on the PNAS web site, www.pnas.org). The mature IgM1gj forms dimers and monomers under nonreducing conditions (Fig. 1A); the quantity of each form varies among individuals, which seems related to the sharks' age (not shown). Immunoprecipitations of IgM1gj from metabolically labeled spleen and epigonal organ cells from 1-week-old sharks demonstrated that it is made de novo and indeed is the major form of Ig produced by the pups (Fig. 1B, and see Fig. 4 for mRNA expression).
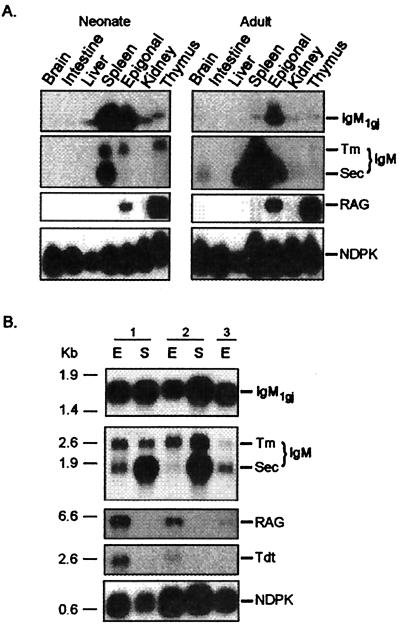
Figure 4.
IgM1gj is expressed in neonatal spleen and epigonal organ and perpetuated in adult epigonal organ. (A) Northern blot with neonatal and adult tissue RNA showing relative expression of IgM1gj, bona fide IgM, RAG1, and positive control nucleotide diphosphate kinase (NDPK). IgM1gj is highly expressed in the spleen and epigonal organ of newborn pups and maintained in adult epigonal organ. Canonical IgM RNA in the newborn spleen (exposed 80 h) is at much lower levels than in the adult (exposed 17 h) and is enriched for transmembrane (Tm) over secretory (Sec) Ig. RAG1 is expressed in the thymus and epigonal organ throughout life. (B) Northern blot by using mRNA from 1-day-old (1), 1-week-old (2), and 41-day-old (3) pups. The same probes in Fig. 4A were used, except TdT was included.
IgM1gj protein is expressed in adults, but attempts to isolate it from adult plasma were unsuccessful because of the large amounts of conventional IgM present at maturity (16). However, metabolic labeling of cells from adult epigonal organ and spleen clearly showed IgM1gj protein produced at appreciable levels relative to conventional IgM only in the epigonal organ (Fig. 1C). Note that the two closely spaced H1gj bands under reducing conditions may be the result of differential glycosylation (there being five putative N-linked glycosylation sites, Fig. 2 A and B).
The H1gj V Gene Is Germline Joined.
IgM1gj was affinity purified with the GA15 mAb from neonatal plasma, and H1gj was sequenced. Both closely spaced bands described above at ≈56 and 53 kDa (Fig. 1) yielded the same N-terminal amino acid sequence, DIMLTQPEAETSIXGGXLKL. To isolate cDNAs encoding H1gj, two cDNA libraries were constructed with mRNA from neonatal spleen and epigonal tissue, respectively, and screened under high and low stringency conditions with a conventional IgM V probe. Five clones were isolated that hybridized only under low stringency conditions and had deduced N-terminal sequences that strictly matched the H1gj N-terminal protein sequence (Fig. 2A). The V regions of the five clones were also identical in sequence, with no replacements in the complementarity-determining regions (CDRs), particularly in CDR3, the site of greatest diversity because of somatic rearrangement of V-D-J genes (38). Note that conventional IgM cDNAs were also obtained under high stringency conditions in this library screen, with extensive CDR3 diversity generated via addition of nontemplated nucleotides (not shown). These results suggested that a germline-joined gene encoded H1gj; furthermore, unlike other vertebrates [frog (4, 5) and mouse (3)], TdT is expressed by neonatal sharks and diversifies rearranging junctions with template-independent nucleotide additions (also see Fig. 4 and M.D. and M.F.F., unpublished results).
To prove that H1gj V region was germline joined, we screened a nurse shark genomic cosmid library, prepared from erythrocyte DNA of one shark from which all lymphocytes were rigorously depleted, with the H1gj V and CH1 cDNA probes under high stringency conditions. Isolated clones were sequenced and revealed the following: (i) the leader exon is split by an intron that is found in all IgV genes (39), indicating that H1gj is encoded by a typical V gene (not shown), and (ii) the VH1gj gene is germline joined with a V-D-J join that precisely matches the sequences of the cDNA clones (Fig. 2C). Note that we further confirmed these features of the H1gj gene by PCR amplification of the region encoding CDR3 from erythrocyte genomic DNA of two other sharks (not shown).
In conventional IgM of the horn shark and ratfish (Hydrolagus collei), the rearrangement join generally includes two D segments, designated D1 and D2; VH1gj CDR3 is short, in part because it appears to use only one D gene (D2, deduced from horn shark genomic sequences; ref. 22; Fig. 2 A, C, and D). It is possible that the dinucleotide TC found in the V-D junction is the result of a P-nucleotide addition (ref. 39; double-underlined in Fig. 2C). This mature V-D-J junction appears to have been formed neither with extensive trimming nor nucleotide addition (see Discussion and Fig. 2D legend).
The V domain framework regions (FR) 1–3 are most similar to horn shark IgM VH [65% amino acid (aa) identity] but only 55% identical to any particular nurse shark IgM VH. Phylogenetic analysis of VH1gj indicates that it is a new nurse shark V family having only 70% nucleotide identity and less than 70% aa identity with conventional IgM VH (38 and see below). Besides the short CDR3, the other conspicuous feature is the paucity of serines usually found in a high percentage of CDR1 and CDR2 aa residues in conventional IgM; there is an average of six serines in CDR1 and CDR2 from horn shark (22), sandbar shark (Carcharhinus plumbeus, ref. 40), and nurse shark (32) H chain sequences (Fig. 2A).
Features of the H1gj C Domains, Especially the Lack of a CH2 Domain Exon in the Gene Clusters.
The CH1 domain of IgM1gj lacks the cysteine (cys) commonly used for L chain association (38) and is substituted by asparagine (bold, asterisk in Fig. 2A). Because biochemical evidence showed that H1gj is disulfide bonded to L chains, we predict that another cys, perhaps at the beginning of the CH2 domain (asterisk in Fig. 2A), forms this bridge. The biochemical analysis above predicted only three C domains for H1gj, which was confirmed by the cDNA sequence. Homology searches suggested that CH1-1gj correlates to IgM CH1 (58% aa identity), CH2-1gj to IgM CH3 (51% identity), and CH3-1gj to IgM CH4 (77% identity, Fig. 2A, supplementary Table 1, and supplementary Fig. 6). Thus, like mammalian IgG (38), the ancestral IgM CH2 is “missing” in H1gj. Sequencing of the 1750-bp intron between the CH1 and CH2 domain exons of the H1gj genomic clone revealed no remnant of an exon, showing that the ancestral IgM CH2 exon was lost via deletion and not alternative splicing.
IgM1gj Is a Single Copy Gene.
Southern blot analysis was done to determine the number of VH-1gj and CH1-1gj genes. V1gj and CH1-1gj exons apparently hybridize to only one cluster under high stringency conditions, indicating that there is only one member in this new family (Fig. 3); longer exposures did not reveal other hybridizing genes, nor did the genomic library screen uncover related H1gj clusters. A conventional IgM VH probe cross-hybridized to IgM1gj under low, but not high, stringency conditions. As expected from studies in the horn shark (20, 21) and skate (Raja erinacea; ref. 41), the conventional IgM VH probe hybridized to many genes under both high and low stringency conditions; however, IgM1gj VH was detected with this probe only under low stringency conditions (diamonds, Fig. 3), consistent with the nucleotide similarity analyses described above.
Figure 3.
The IgM1gj gene cluster is single copy. Southern blot analysis of VH1gj and CH1-1gj genes under high stringency conditions reveals only one band with four different enzymes. A canonical IgM VH probe hybridizes to the VH-1gj gene only under low stringency conditions. The white diamond shapes show the positions of VH-1gj in the three VH blots. B, BamHI; E1, EcoRI; EV, EcoRV; H, HindIII.
Newborn Expression and Maintenance in Adults.
Because IgM1gj protein was shown to be secreted from neonatal spleen and from both neonatal and adult epigonal organs, a Northern blot analysis was done to ascertain which pup and adult tissues expressed IgM1gj. The relative mRNA expression of IgM1gj, IgM, RAG1, TdT, and the positive control nucleoside diphosphate kinase (NDPK) is displayed in Fig. 4. IgM1gj RNA is produced at highest levels in neonatal spleen and epigonal organ and much lower levels in the liver, kidney, and thymus. Expression is sustained in the spleen for at least the first few weeks after birth (Fig. 4B; unfortunately, the spleen RNA sample was degraded in the oldest animal, but serum protein levels have apparently decreased by this time, not shown). In adults, IgM1gj RNA expression is found only in epigonal organ, consistent with the previous immunoprecipitation analyses (compare Figs. 1B and 4A). There are higher amounts of IgM1gj protein in newborn plasma relative to conventional IgM most likely because cells in both the epigonal organ and spleen produce IgM1gj (the exposure for IgM expression in the neonate was 80 h and the adult only 17 h; for all other probes exposure times were the same). A transmembrane form of IgM1gj has not been identified from cDNA clones and may not exist.
RAG1 expression is continuous in the thymus and epigonal organ throughout life (Fig. 4 and data not shown). This molecular evidence fits with the hypothesis, based on morphological grounds, that the epigonal organ (and by inference the Leydig organ) functions as a primary lymphoid tissue in elasmobranchs.
Discussion
We have discovered an IgM subclass in the nurse shark, IgM1gj, which is expressed predominantly in neonatal sharks. The mAbs crossreacting with IgM1gj also recognize 7S and/or 19S IgM; however, neonatal IgM levels are much lower than in adults, which “allowed” the identification of IgM1gj. As pups mature, large amounts of IgM are produced that dilute the IgM1gj, so serum levels of the latter appear to decrease. It is possible that, like innate lineages of mammalian lymphocytes, the IgM1gj cell lineage develops at the inception of the immune system and undergoes self-renewal in the epigonal organ throughout life. Another, equally likely possibility is that IgM1gj cells are continually produced from a small number of stable precursors in the epigonal organ.
Previous studies in the horn shark reported a very large number of germline-joined IgH genes, but apparently no expression of them (22, 23). Ratfish germline-joined H chain clusters were also isolated, and their expression could be detected only by a sensitive reverse transcription-PCR procedure (42). One hypothesis to account for such low-to-undetectable expression in adult spleen was that some of these prejoined clusters encoding proteins with specific functions might be expressed in a tissue-specific and/or developmentally controlled manner; this hypothesis is fulfilled from our study of IgM1gj, at least for the restricted tissue expression and ontogenetic appearance. We have detected a similar mode of expression of an IgNAR partially germline-joined cluster (M.D. and M.F.F., unpublished results) and L chain clusters (in collaboration with E. Hsu, ref. 25), and we predict this expression of such clusters will be the rule in all elasmobranchs when there is a mixture of joined and unjoined genes of particular antigen receptor families. It may be that the germline-joined clusters have a transcriptional advantage early in development, and that some clusters have been selected for particular roles before development of the adaptive response (25, 26). It must also be noted that, in certain L chain families, the clusters are entirely germline joined in some elasmobranch species and are expressed throughout life (43–45).
It is very likely that the H1gj gene was derived from a RAG-mediated rearrangement event in the germline, an activity that was inferred from many previous studies (reviewed in ref. 23), and recently proven for shark L chains (25). Proposed precise cleavages at two heptamer/coding end junctions, and the potential P nucleotide addition strongly suggest that the rearrangement was RAG mediated (Fig. 2D). It is not known whether a germline-joined variable (V-J) gene also encodes the covalently associated L chain. If IgM1gj indeed acts as an antibody, its function may be to recognize a conserved epitope on a marine pathogen common to nurse sharks. Alternatively, it may have a function that is not immune related at all, e.g., acting to clear waste products from the body, especially during development (46).
IgM1gj is similar to mammalian IgG by having the domain homologous to IgM CH2 deleted over evolutionary time in both molecules (38). Such deletions must have occurred independently in genes ancestral to shark IgM1gj and to mammalian IgG C region genes; based on sequence similarity to other Igs, the IgM1gj V family seems to have emerged some time near the divergence of nurse and horn sharks 120–160 million years ago (see supplemental data). Its smaller size may allow for easier passage into tissue spaces compared with monomeric IgM; furthermore, like a mutant mammalian IgG that lacks a hinge region (47), IgM1gj may be unable to interact with complement, thus limiting inflammatory responses in developing sharks. Like human IgG1 (48), the disulfide bridge bonding H and L chains has been altered but in a way difficult to predict, which might induce an atypical conformation of the VH/VL pair.
Much work is still needed as well to establish the epigonal organ as the elasmobranch bone marrow equivalent, but the new evidence of RAG and TdT expression only in this tissue and thymus throughout life is compelling. Such expression of enzymes involved in rearrangement, morphological studies demonstrating lymphoid aggregates (29), and the fact that all species of cartilaginous fish have similar tissues regardless of their location within the body (28, 29), strongly suggest that epigonal/Leydig organ is a primary lymphoid tissue. Furthermore, the data in Fig. 4 show that, early in life, cells in the epigonal predominantly express mRNAs encoding the transmembrane form of conventional IgM and the secreted form of IgM1gj, whereas later in life high levels of secreted IgM and IgM1gj (as well as IgW and IgNAR, not shown for RNA but protein levels of IgW shown in Fig. 1) are found. Such data suggest that, like mammalian bone marrow, the epigonal organ is both a primary lymphoid tissue and a reservoir for secretory B cells.
The discovery of Igs expressed early in development derived from germline-joined genes fits well with Herzenberg's Layering Hypothesis (8). The dichotomy between antigen receptors expressed early and late in development has now been described in mammals, frogs (49), and cartilaginous fish, and this basic truth should herald a rich area of future research (50). In particular, the elasmobranchs, with their germline-joined gene clusters, may provide a wealth of information on this topic; the fact that we and our colleagues have detected at least one joined cluster expressed early in development for each antibody class tested (and indeed only one for the IgM class in nurse sharks, Fig. 3) strongly suggests an important function for such gene products.
Supplementary Material
Acknowledgments
We thank Louis Du Pasquier and Ellen Hsu for general discussion, Becky Lohr for technical support, David Nemazee and Ferenc Livak for critical reading of the manuscript, an anonymous reviewer for suggested modifications, and Robyn Stanfield and Ian Wilson for discussion of the IgM1gj structure. The work was supported by National Institutes of Health Grant RR06603.
Abbreviations
- TCR
T cell receptor
- MHC
major histocompatibility complex
- CDR
complementarity-determining region
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AF327520 (IgM1gj) and AF327519 (TdT)].
IgM1gj is so-named because it is related to conventional IgM; 1 refers to the 1st IgM gene expressed in ontogeny and expression in primary lymphoid tissue; and gj signifies that the variable gene is germline joined.
References
- 1.Perlmutter R M, Kearney J F, Chang S P, Hood L E. Science. 1985;227:1597–1601. doi: 10.1126/science.3975629. [DOI] [PubMed] [Google Scholar]
- 2.Itohara S, Nakanishi N, Kanagawa O, Kubo R, Tonegawa S. Proc Natl Acad Sci USA. 1989;86:5094–5098. doi: 10.1073/pnas.86.13.5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feeney A J. J Exp Med. 1990;172:1377–1390. doi: 10.1084/jem.172.5.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwager J, Buerckert N, Courtet M, Du Pasquier L. EMBO J. 1991;10:2461–2470. doi: 10.1002/j.1460-2075.1991.tb07785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee A, Hsu E. J Immunol. 1994;152:4500–4507. [PubMed] [Google Scholar]
- 6.Herzenberg L A, Kantor A B, Herzenberg L A. Ann NY Acad Sci. 1992;651:1–9. doi: 10.1111/j.1749-6632.1992.tb24588.x. [DOI] [PubMed] [Google Scholar]
- 7.Chien Y-H, Jores R, Crowley M P. Annu Rev Immunol. 1996;14:511–532. doi: 10.1146/annurev.immunol.14.1.511. [DOI] [PubMed] [Google Scholar]
- 8.Herzenberg L A, Herzenberg L A. Cell. 1989;59:953–954. doi: 10.1016/0092-8674(89)90748-4. [DOI] [PubMed] [Google Scholar]
- 9.Flajnik M F, Rumfelt L L. Curr Top Microbiol Immunol. 2000;248:249–270. doi: 10.1007/978-3-642-59674-2_11. [DOI] [PubMed] [Google Scholar]
- 10.Marchalonis J, Edelman G M. J Exp Med. 1965;122:601–618. doi: 10.1084/jem.122.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greenberg A S, Avila D, Hughes M, Hughes A, McKinney E C, Flajnik M F. Nature (London) 1995;374:168–173. doi: 10.1038/374168a0. [DOI] [PubMed] [Google Scholar]
- 12.Harding F A, Amemiya C T, Litman R T, Cohen N, Litman G W. Nucleic Acids Res. 1990a;18:6369–6376. doi: 10.1093/nar/18.21.6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernstein R M, Schluter S F, Shen S, Marchalonis J J. Proc Natl Acad Sci USA. 1996;93:3289–3293. doi: 10.1073/pnas.93.8.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenberg A S, Hughes A L, Guo J, Avila D, McKinney E C, Flajnik M F. Eur J Immunol. 1996;26:1123–1129. doi: 10.1002/eji.1830260525. [DOI] [PubMed] [Google Scholar]
- 15.Anderson M K, Strong S J, Litman R T, Luer C A, Amemiya C T, Rast J P, Litman G W. Immunogenetics. 1999;49:56–67. doi: 10.1007/s002510050463. [DOI] [PubMed] [Google Scholar]
- 16.Clem L W, DeBoutaud F, Sigel M M. J Immunol. 1967;99:1226–1235. [PubMed] [Google Scholar]
- 17.Fidler J E, Clem L W, Small P A. Comp Biochem Physiol. 1969;31:365–371. doi: 10.1016/0010-406x(69)91660-0. [DOI] [PubMed] [Google Scholar]
- 18.Hamlett W C, Koob T J. In: Sharks, Skates, and Rays: The Biology of Elasmobranch Fishes. Hamlett W C, editor. Baltimore: Johns Hopkins Univ. Press; 1999. pp. 398–443. [Google Scholar]
- 19.Hinds K R, Litman G W. Nature (London) 1986;320:546–549. doi: 10.1038/320546a0. [DOI] [PubMed] [Google Scholar]
- 20.Kokubu F, Hinds K, Litman R, Shamblott M J, Litman G W. Proc Natl Acad Sci USA. 1987;84:5868–5872. doi: 10.1073/pnas.84.16.5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kokubu F, Hinds K, Litman R, Shamblott M J, Litman G W. EMBO J. 1988a;7:1979–1988. doi: 10.1002/j.1460-2075.1988.tb03036.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kokubu F, Litman R T, Shamblott M J, Hinds K, Litman G W. EMBO J. 1988b;7:3413–3422. doi: 10.1002/j.1460-2075.1988.tb03215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoder J A, Litman G W. Curr Top Microbiol Immunol. 2000;248:271–282. doi: 10.1007/978-3-642-59674-2_12. [DOI] [PubMed] [Google Scholar]
- 24.Lohr R L. Thesis. Wilmington: University of North Carolina; 1998. [Google Scholar]
- 25.Lee S S, Fitch D, Flajnik M F, Hsu E. J Exp Med. 2000;191:1637–1648. doi: 10.1084/jem.191.10.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis S M, Wu G E. J Exp Med. 2000;191:1631–1635. doi: 10.1084/jem.191.10.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zapata A G, Torroba M, Sacedón R, Varas A, Vicente A. J Exp Zool. 1996;275:125–143. [Google Scholar]
- 28.Matthews L H. Philos Trans R Soc London. 1950;234:248–316. doi: 10.1098/rstb.1950.0003. [DOI] [PubMed] [Google Scholar]
- 29.Fange R, Pulsford A. Cell Tissue Res. 1983;230:337–351. doi: 10.1007/BF00213808. [DOI] [PubMed] [Google Scholar]
- 30.Ohta Y, Okamura K, McKinney E C, Bartl S, Hashimoto K, Flajnik M F. Proc Natl Acad Sci USA. 2000;97:4712–4717. doi: 10.1073/pnas.97.9.4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bartl S, Baish M A, Flajnik M F, Ohta Y. J Immunol. 1997;159:6097–6104. [PubMed] [Google Scholar]
- 32.Vazquez M, Mizuki N, Flajnik M F, McKinney E C, Kasahara M. Mol Immunol. 1992;29:1157–1158. doi: 10.1016/0161-5890(92)90050-8. [DOI] [PubMed] [Google Scholar]
- 33.Kasahara M, Canel C, McKinney E C, Flajnik M F. In: NATO ASI Series: Molecular Evolution of the Major Histocompatibility Complex. Klein J, Klein D, editors. H59, Berlin: Springer; 1991. pp. 491–499. [Google Scholar]
- 34.Flajnik M F, Kasahara M, Shum B P, Salter-Cid L, Taylor E, Du Pasquier L. EMBO J. 1993;12:4385–4396. doi: 10.1002/j.1460-2075.1993.tb06123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kearney J F, Radbruch A, Liesegang B, Rajewsky K. J Immunol. 1979;123:1548–1550. [PubMed] [Google Scholar]
- 36.Roux K H, Greenberg A S, Greene L, Strelets L, Avila D, McKinney E C, Flajnik M F. Proc Natl Acad Sci USA. 1998;95:11804–11809. doi: 10.1073/pnas.95.20.11804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsu E, Du Pasquier L. Mol Immunol. 1984;21:257–270. doi: 10.1016/0161-5890(84)90096-8. [DOI] [PubMed] [Google Scholar]
- 38.Frazer J K, Capra J D. In: Fundamental Immunology. 4th Ed. Paul W E, editor. Philadelphia: Lippincott; 1999. pp. pp.37–74. [Google Scholar]
- 39.Max E E. In: Fundamental Immunology. 4th Ed. Paul W E, editor. Philadelphia: Lippincott; 1999. pp. 111–182. [Google Scholar]
- 40.Marchalonis J J, Schluter S F, Bernstein R M, Shen S, Edmundson A B. Adv Immunol. 1998;70:417–506. doi: 10.1016/s0065-2776(08)60392-2. [DOI] [PubMed] [Google Scholar]
- 41.Harding F A, Cohen N, Litman G W. Nucleic Acids Res. 1990;18:1015–1020. doi: 10.1093/nar/18.4.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rast J P, Amemiya C T, Litman R T, Strong S J, Litman G W. Immunogenetics. 1998;47:234–245. doi: 10.1007/s002510050353. [DOI] [PubMed] [Google Scholar]
- 43.Hohman V S, Schuchman D B, Schluter S F, Marchalonis J J. Proc Natl Acad Sci USA. 1993;90:9882–9886. doi: 10.1073/pnas.90.21.9882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anderson M K, Shamblott M J, Litman R T, Litman G W. J Exp Med. 1995;182:109–119. doi: 10.1084/jem.182.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee S S, Greenberg A S, Hsu E. Curr Topics Microbiol Immunol. 2000;248:285–300. doi: 10.1007/978-3-642-59674-2_13. [DOI] [PubMed] [Google Scholar]
- 46.Alving C R, Wassef N M. Immunol Today. 1999;20:362–366. doi: 10.1016/s0167-5699(99)01496-6. [DOI] [PubMed] [Google Scholar]
- 47.Guddat L W, Herron J N, Edmundson A B. Proc Natl Acad Sci USA. 1993;90:4271–4275. doi: 10.1073/pnas.90.9.4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poljak R J. Adv Immunol. 1975;21:1–33. [PubMed] [Google Scholar]
- 49.Mussmann R, Courtet M, Du Pasquier L. Eur J Immunol. 1998;8:2989–3001. doi: 10.1002/(SICI)1521-4141(199809)28:09<2947::AID-IMMU2947>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 50.Mouthon L, Lacroix-Desmazes S, Nobrega A, Barreau C, Coutinho A, Kazatchkine M D. Scand J Immunol. 1996;44:243–251. doi: 10.1046/j.1365-3083.1996.d01-306.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.