Abstract
Retinitis pigmentosa (RP) is a phenotypically and genetically heterogeneous group of inherited retinal degenerations characterized clinically by night blindness, progressive constriction of the visual fields, and loss of vision, and pathologically by progressive loss of rod and then cone photoreceptors. Autosomal-recessive RP (arRP) in a consanguineous Pakistani family previously linked to chromosome 2p22.3-p24.1 is shown to result from a homozygous missense mutation (c.1015T>C [p.C339R]) in ZNF513, encoding a presumptive transcription factor. znf513 is expressed in the retina, especially in the outer nuclear layer, inner nuclear layer, and photoreceptors. Knockdown of znf513 in zebrafish reduces eye size, retinal thickness, and expression of rod and cone opsins and causes specific loss of photoreceptors. These effects are rescued by coinjection with wild-type (WT) but not p.C339R-znf513 mRNA. Both normal and p.C339R mutant ZNF513 proteins expressed in COS-7 cells localize to the nucleus. ChIP analysis shows that only the wild-type but not the mutant ZNF513 binds to the Pax6, Sp4, Arr3, Irbp, and photoreceptor opsin promoters. These results suggest that the ZNF513 p.C339R mutation is responsible for RP in this family and that ZNF513 plays a key role in the regulation of photoreceptor-specific genes in retinal development and photoreceptor maintenance.
Results and Discussion
Retinitis pigmentosa (RP [MIM 268000]) refers to a group of inherited retinal diseases with phenotypic and genetic heterogeneity. The estimated worldwide prevalence of RP is approximately 1 in 5000.
RP patients experience night blindness, followed by loss of peripheral visual field and later loss of central vision, often leading to complete blindness. RP initially affects the rod photoreceptors; the function of the cone photoreceptors is compromised as the disease progresses.1 Affected individuals often have severely abnormal or nondetectable rod responses in the electroretinogram (ERG) recordings even in the early stage of the disease.1 The classic symptom of patients with RP is the subjective complaint of nyctalopia, defined as difficulty adapting or seeing in a dimly lit environment. Nyctalopia is frequently present early in the disease course of arRP (median age of onset is 11 years) but occurs relatively later in association with autosomal-dominant RP (median age of onset is 24 years).2 Classically, RP has been classified based on the genetic inheritance pattern: autosomal-dominant (about 30%–40% of cases), autosomal-recessive (50%–60%), or X-linked (5%–15%).3–5
To date, more than 45 genes causing RP have been identified. These genes account for less than 40% of all patients; the remainder have defects in as yet unidentified genes. Many of these can be grouped by function, giving insights into the disease process. These include components of the phototransduction cascade, proteins involved in retinol metabolism, proteins involved in cell-cell interaction, photoreceptor structural proteins, transcription factors, intracellular transport proteins, and splicing factors.3,6–10
We previously mapped an arRP locus to a 12.31 cM (13.35 Mb) interval on chromosome 2p22.3-p24.1 flanked by D2S220 and D2S367 in a consanguineous Pakistani family (PKRP115, 61115).11 Here we identify a p.C339R mutation in ZNF513 as the cause of arRP in this family and show that knockdown of ZNF513 protein levels in zebrafish embryos causes defective retinal development and photoreceptor loss. In addition, normal but not p.C339R mutant ZNF513 binds to specific photoreceptor promoters, suggesting that it acts through changing the transcriptional control of genes involved in retinal development and maintenance.
Clinical details of the arRP family were previously described.11 IRB approval was obtained for this study from the Centre of Excellence in Molecular Biology (CEMB) (Lahore, Pakistan) and National Eye Institute (Bethesda, MD, USA). The participating subjects gave informed consent consistent with the tenets of the Declaration of Helsinki. Three markers (D2S2221, D2S2383, and D2S2347) were selected for fine mapping from the Marshfield map. Two-point linkage analyses were performed with the FASTLINK version of MLINK from the LINKAGE program package.12,13 arRP was analyzed as a fully penetrant trait with an affected allele frequency of 0.000001. Marker allele frequencies were estimated from 50 control chromosomes. Fine mapping (Table 1) confirms the 2p22.3-p24.1 locus but was unable to refine the critical interval further. Flanking recombinant events are seen at D2S367 distally in individual 8 and lack of homozygosity is seen at D2S220 proximally (Figure 1A).
Table 1.
Two-Point LOD Scores of 2p22.3-p24.1 Fine Mapping Markers
| Marker | cM | Mb | 0 | 0.001 | 0.01 | 0.05 | 0.1 | 0.2 | 0.3 | 0.4 | Zmax | θmax |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D2S149 | 34 | 14.31 | -∞ | −2.75 | −0.8 | 0.37 | 0.68 | 0.71 | 0.49 | 0.22 | 0.71 | 0.2 |
| D2S2150 | 40.5 | 20.39 | −2.43 | −1.72 | −0.84 | −0.3 | −0.2 | −0.1 | 0 | 0.03 | 0.03 | 0.4 |
| D2S220 | 42.7 | 20.95 | −0.85 | −0.13 | 0.73 | 1.2 | 1.21 | 0.92 | 0.52 | 0.17 | 1.23 | 0.08 |
| D2S2221 | 44.1 | 23.07 | 2.89 | 2.88 | 2.83 | 2.57 | 2.26 | 1.63 | 1 | 0.41 | 2.89 | 0 |
| D2S2247 | 46.9 | 27.15 | 3.24 | 3.23 | 3.18 | 2.92 | 2.59 | 1.91 | 1.24 | 0.59 | 3.24 | 0 |
| ZNF513(1015C>T) | 27.6 | 3.38 | 3.37 | 3.32 | 3.06 | 2.72 | 2.04 | 1.36 | 0.68 | 3.38 | 0 | |
| D2S223 | 45.8 | 28.19 | 2.89 | 2.88 | 2.83 | 2.58 | 2.26 | 1.61 | 0.98 | 0.38 | 2.89 | 0 |
| D2S165 | 47.1 | 28.45 | 3.15 | 3.14 | 3.09 | 2.85 | 2.54 | 1.89 | 1.23 | 0.58 | 3.15 | 0 |
| D2S2383 | 48.5 | 30.02 | 3.23 | 3.22 | 3.17 | 2.93 | 2.61 | 1.96 | 1.29 | 0.61 | 3.23 | 0 |
| D2S2347 | 50.6 | 33.23 | 3.09 | 3.08 | 3.03 | 2.79 | 2.48 | 1.85 | 1.19 | 0.55 | 3.09 | 0 |
| D2S367 | 55 | 34.29 | -∞ | −0.17 | 0.78 | 1.27 | 1.3 | 1.05 | 0.67 | 0.28 | 1.31 | 0.09 |
Figure 1.
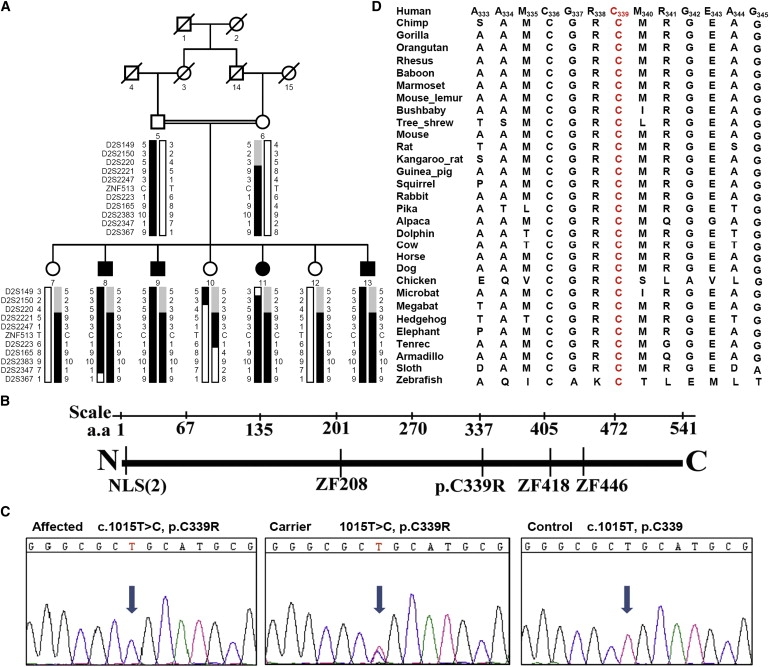
ZNF513 Region, Pedigree, Gene, and Sequence
(A) Haplotypes of the ZNF513 region of family 61115 showing the ZNF513 p.C339R mutation and surrounding microsatellite markers included in Table 1. The risk haplotype is shown in black.
(B) Diagram of the ZNF513 gene showing the two nuclear localization signals (NLS), the p.C339R mutation, and the three zinc finger binding domains (ZF) with their positions.
(C) Electropherograms show the affected sequence (left, individual 8), carrier sequence (middle, individual 12), and normal control sequence (right) surrounding the ZNF513 1015T>C mutation.
(D) Amino acid sequence alignment around the ZNF513 C339 amino acid (red) in 31 species ranging from human to zebrafish.
One hundred and forty seven genes in the linked region of chromosome 2p22.3-p24.1 were identified with the NCBI gene browser. Exons, including more than 100 bp of flanking sequence, were screened with an ABI PRISM 3130 automated sequencer (Applied Biosystems) and analyzed with Seqman software (Lasergene). Primer pairs for individual exons were designed with the primer3 program. A total of 33 genes in this region were sequenced, including the zinc finger 513 gene (ZNF513, AAH40650), which shows a homozygous missense mutation (c.1015T>C [p.C339R]) in exon 4 in all affected individuals and cosegregating with the disease (Figure 1C). This change is predicted to be damaging via Polyphen,14 which gives a position-specific independent count (PSIC) score difference of 3.17. The mutation is not seen in 242 ethnically matched control chromosomes, providing a power of more than 80% for detecting a polymorphism with a 1% frequency, and C339 is conserved in all species from Homo sapiens to Danio rerio as ascertained by a bioinformatic search of NCBI-Blast by means of human ZNF513 DNA and protein and MegAlign (DNASTAR Lasergene, Madison, WI). The entire region surrounding the mutation is relatively well conserved, especially among mammals (Figure 1D). No additional sequence changes were identified in ZNF513, and changes identified in other genes were either known SNPs or noncoding polymorphisms (data not shown).
Analysis with PSORT algorithms predicts the presence of two types of classical nuclear localization signals at the N-terminal of ZNF513 (Figure 1B): PRRK (pattern 4) and PRRKQSH (pattern 7).15 Further, PROSITE algorithms detect the presence of three Cys2His2-type zinc finger binding domain signatures starting at residue Cys208 (CPHCPFACSSLGNLRRHQRTH), Cys418 (CPLCPYACGNLANLKRHGRIH), and Cys446 (CSLCNYSCNQSMNLKRHMLRH). This bioinformatic analysis suggests that the p.C339R mutation in ZNF513 is located in a transcriptional regulation or effector domain.16 We hypothesized that the p.C339R mutation might modify the structure of the zinc finger and, consequently, the binding and activation of various target genes by ZNF513.
In order to check expression of Znf513 in various mouse eye tissues, quantitative (q) RT-PCR was performed according to previously published protocols,17,18 by means of the forward primer 5′-GCTCATCTGGATAACCTGAAACG-3′ and reverse primer 5′-GCTACACCGAAAAGGTTTGTCAC-3′. Although Znf513 is expressed in all tissues tested, ocular expression is greatest in the retina and least in the lens and cornea, in which expression also plateaus early (Figure 2A). Retinal expression of Znf513 increases progressively with age, beginning to level off between 180 and 300 days, the highest age tested (Figure 2A). This expression pattern suggests that Znf513 expression is important not only for eye and retinal development but also for maintenance of the adult retina.
Figure 2.
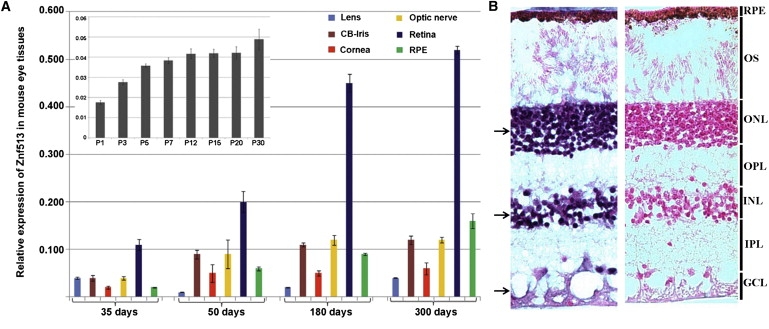
Relative Expression of Znf513 in Mouse Eye Tissues at Various Ages and Distribution of ZNF513 mRNA in the Human Adult Retina
(A) Expression of Znf513 (Zpf513) was measured in lens, ciliary body (CB), iris, cornea, optic nerve, retina, and RPE tissues by qRT-PCR at different time points during aging. Data represent the mean (±SD) on an arbitrary scale (y axis) representing expression relative to the housekeeping gene Rpl19 and were calculated from at least three independent experiments. The inset shows the expression pattern of Znf513 in the post natal-stage eye from P1 to P30.
(B) In situ hybridization of ZNF513 probes in the retina of human eyes at 55°C–69°C. Hybridization with the antisense probe (left) shows signal (black arrows) in the ONL, INL, and GCL when contrasted to hybridization with the sense probe (right). Counter stain is nuclear fast red. RPE, retinal pigmented epithelia; OS, outer segment; ONL, outer nuclear layer; INL, inner nuclear layer; OPL, outer plexiform layer; IPL inner plexiform layer; GCL, ganglion cell layer.
In situ hybridization analysis was used to localize expression of ZNF513 mRNA in the human retina. ZNF513 RNA probes were generated from a ZNF513 cDNA clone in the pOTB7 vector. The pOTB7-ZNF513 construct was linearized with ClaI. SP6 and T7 RNA polymerases were used to create sense and antisense digoxigenin (DIG)-labeled RNA probes, respectively, from the linearized DNA. In situ hybridization was performed on 7 μm cryosections from the posterior segment of human eyes according to previously published protocols.19,20 In situ hybridization of ZNF513 shows expression in human retinal layers including the outer nuclear layer (ONL), inner nuclear layer (INL), and ganglion cell layer (GCL) (Figure 2B), consistent with a potential role for the mutant in retinal degeneration.
znf513 mRNA is expressed in early developing zebrafish embryo brain and eyes according to the zebrafish model database, suggesting that zebrafish might be a reasonable model system to evaluate the role of znf513 in eye development and function. An antisense morpholino oligonucleotide (znf513-MO, 5′-GATTCTGCTGTTTTCTTCTTGGCAT-3′) targeting the znf513 translation initiation site and a mismatch control morpholino (MM-MO, 5′-GATTGTCCTCTTTTCTTGTTCGCAT-3′) were dissolved in distilled water, mixed with 2X injection buffer, and injected into egg yolks of 1- to 4-cell stage zebrafish embryos in order to investigate wild-type and p.C339R znf513 expression and function. Wild-type zebrafish were maintained as described by Westerfield,21 and embryos were produced by natural matings. All experiments with animals were approved by the NEI Animal Use and Care Committee.
Morpholino-sensitive EGFP was constructed by adding the sequence 5′-ATGCCAAGAAGAAAACAGCAGAATC-3′ to the 5′-end of EGFP cDNA and cloning into the pCS2+ vector, as was unmodified EGFP cDNA. To test morpholino inhibitory activity, EGFP mRNA fused to the morpholino-sensitive sequence at the 5′-end (5′-modified EGFP) was injected prior to morpholino injections. High-intensity EGFP fluorescence is observed in 5′-modified EGFP mRNA-injected embryos and eliminated by znf513-MO injection (Figures 3A and 3B). znf513-MO injection does not reduce expression of native EGFP without the morpholino-sensitive sequence, and MM-MO injection has no effect on expression of 5′-modified EGFP (Figures 3C and 3D), consistent with the ability of the morpholino to inhibit expression of znf513 specifically.
Figure 3.
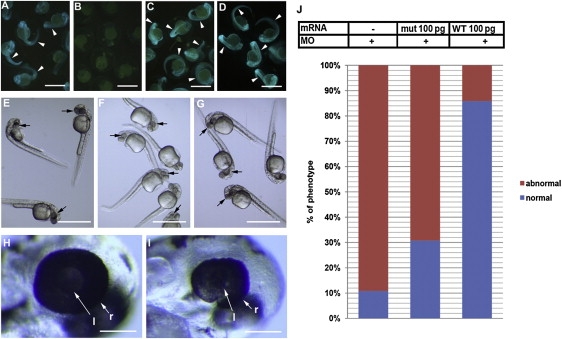
Zebrafish Eye Development Disturbed by Knockdown of znf513 Expression and Rescue of znf513 Morphants
Morpholino-sensitive 5′-modified EGFP mRNA (A, B, C) or native unmodified EGFP (D) was injected to single cell stage embryos. znf513-MO (B, D, F, I), MM-MO (C, G, H), or injection buffer (E) was injected into embryos.
(A–D) Validation of the activity of znf513-MO on the expression of znf513. znf513-MO injection eliminated the fluorescence signal (green on the embryo body, arrowheads) from coinjected morpholino-sensitive 5′-modified EGFP mRNA (B), and it did not reduce the expression of native unmodified EGFP without the znf513 sequence (D). MM-MO had no effect on morpholino-sensitive GFP expression (C). Note that all embryos contain faint yellowish autofluorescence in the egg yolk not derived from EGFP.
(E–G) 0.5 ng Znf513-MO (F) dramatically reduced eye size (black arrows) compared to MM-MO-injected (G) and buffer-injected (E) embryos.
(H and I) Higher magnification of the heads from embryos shown in (G) and (F) showing reduced size of the retina in a znf513-MO-injected embryo relative to a MM-MO-injected embryo (l, lens; r, retina, white arrows).
(J) Graph depicting proportions of embryos with abnormal phenotype (eye size) associated with znf513-MO injection and rescue by coinjected znf513 mut and WT mRNA.
(A–D) 24 hpf, (E–I) 37 hpf. Scale bars represent 1 mm in (A)–(G) and 100 μm in (H) and (I).
Zebrafish embryos treated with the znf513-MO show normal development until 24 hr postfertilization (hpf). After 37 hpf, znf513-MO treatment (Figures 3F and 3I) causes a reduction in eye size not seen in larvae treated with MM-MO (Figures 3G and 3H) or buffer (Figure 3E). The eye size phenotype induced by znf513-MO is partially rescued with coinjection of WT but not p.C339R zebrafish znf513 mRNA. Injection of the znf513-MO results in 89% of fish with an abnormal phenotype and only 11% with a normal phenotype. For rescue, the full-length cDNA (Open Biosystems) was subcloned into the pCS2+ vector. znf513 RNA was synthesized with a mMESSAGE mMACHINE kit (Ambion). Synthesized RNAs were injected into 1-cell stage embryos with/without indicated MOs. Rescue of the normal phenotype is almost complete with coinjected WT mRNA, giving 86% of fish with a normal phenotype as compared to coinjection with p.C339R znf513 mRNA, which gave only 31% of fish with a normal phenotype. This suggests that the effect is specifically caused by knockdown of endogenous znf513 protein (Figure 3J) and that the p.C339R mutation inhibits znf513 protein function.
Immunofluorescence studies show that znf513-MO injection preferentially affects specific retinal layers. Fixed zebrafish embryos were equilibrated in 20% sucrose and embedded in OCT (Electron Microscopy Sciences). Frozen sections (10 μm) were produced with a cryostat (CM3050, Leica Microsystems). Sections were blocked with 1% normal goat serum/1% BSA and incubated with anti-PKCβ1 (1:300, Santa Cruz), Zpr-1 (1:200, Zebrafish International Resource Center), 1D1 (1:50),22 1D4 (1:1000), and rabbit anti-opsin (anti-blue, 1:250; anti-green, 1:500; anti-red, 1:500; anti-UV, 1:1000).23 Slides were washed in PBS and incubated with Alexa488- or Alexa595-conjugated goat anti-mouse or anti-rabbit IgG (1:400, Invitrogen) and with DAPI for nuclear staining. Images were collected with Axioplan 2 and LSM 700 confocal microscope (Carl Zeiss). The thickness of retinal layers and the number of cells were measured with ImageJ software. The inner plexiform layer (IPL), INL, GCL, and ONL of znf513-MO-treated larvae (Figures 4A and 4B versus Figures 4C and 4D) are much thinner in the znf513-MO-treated than MM-MO-treated larvae (see also Figure 4M, p < 0.05). The znf513-MO-treated larvae also show reduced staining for rod photoreceptors markers. By 5 dpf, rod photoreceptor cells are almost completely absent from the retina and are observed only at the retinal margin, a region of persistent neurogenesis in teleost fish (Figure 4D). Even at the margin, rod cells often have an abnormal morphology or are pyknotic, suggesting that these rods might be directed to cell death. Cone opsin staining in znf513-MO-treated larvae (Figures 4F, 4H, and 4L) decreased compared to MM-MO-treated larvae (Figures 4E, 4G, and 4K), while rhodopsin (Figures 4F, 4H, 4J, and 4L) and red opsin (Figure 4J) signals are completely absent from znf513-MO-treated larvae. Reduction of the green opsin and elimination of the red opsin in MO-treated retina suggests that remaining green/red cones are dysfunctional cone bodies (Figure 4C).
Figure 4.
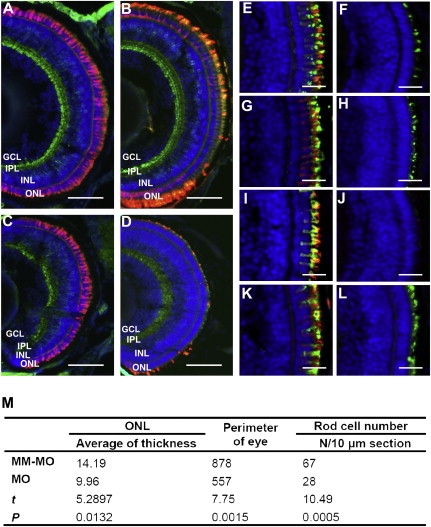
Inhibition of Cone and Rod Photoreceptor Development by Knockdown of znf513 Gene Expression
(A–D) Merged photos of frozen sections from MM-MO-injected (A, B) and znf513-MO-injected (C, D) zebrafish embryos stained for PKCβ1 (bipolar cells, green), Zpr-1 (cone photoreceptors, red, A and C), 1D1 (rod receptors, red, B and D), and DAPI (nuclei, blue). znf513-MO-injected embryos show decreased thickness of all layers except the OPL.
(E–L) Staining of znf513-MO- and MM-MO-injected embryos for specific opsins. Frozen sections from MM-MO-injected (E, G, I, K) and znf513-MO-injected (F, H, J, L) embryos were stained with anti-blue opsin (E, F, green), anti-green opsin (G, H, green), anti-red opsin (I, J, green), anti-UV opsin (K, L, green), 1D4 (all, Rhodopsin, red), and DAPI (all, nuclei, blue). There is a decrease in each opsin in znf513-MO-injected embryos, with rhodopsin and red opsin being essentially absent.
(M) Comparison of retina layer thicknesses in μm, eye perimeter, and cell number between the znf513-MO-treated (n = 5) and MM-MO-treated (n = 7) larvae. t is the unpaired Student's t test statistic. IN, inner nuclear layer cells. p < 0.05 was considered statistically significant.
(A–L) 5 dpf. Scale bars represent 50 μm in (A)–(D) and 15 μm in (E)–(L). ONL, outer nuclear layer; INL, inner nuclear layer; IPL, inner plexiform layer; GCL ganglion cell layer.
Wild-type and mutant human GFP-ZNF513 fusion proteins were analyzed by western blotting after transient transfection of COS-7 cells with wild-type and mutant GFP-ZNF513 constructs. The cells were harvested 48 hr after transfection, and western blot analysis was performed with 30 μg reduced proteins separated on 10% Bis-Tris gels (Invitrogen). Wild-type and mutant GFP-ZNF513 proteins are expressed with an approximate molecular weight of 82 kDa (Figure 5A), consistent with the expected molecular mass.
Figure 5.
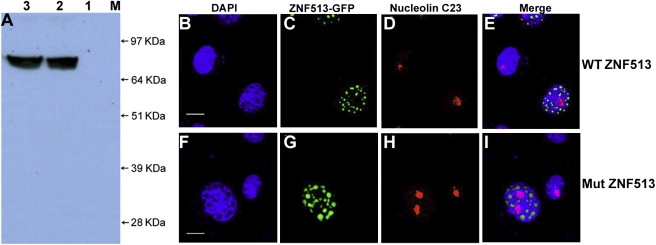
Characterization of Mutant and Wild-Type GFP-ZNF513 Proteins by Western Blot Analysis and Localization of ZNF513 in COS-7 Cells by Immunofluorescence
(A) Blot of COS-7 cell lysates probed with GFP antibodies. Lane 1, untransfected lysate; lane 2, transfected wild-type ZNF513-GFP lysate; lane 3, lysate transfected with mutant ZNF513-GFP; lane M, SeeBlue2 Plus molecular weight marker. Both WT and mutant proteins migrate at the predicted MW of 82 kDa.
(B–I) COS-7 cells were transfected with pEGFPC1 fused in-frame with wild-type (B–E) or p.C339R mutant (F–I) ZNF513. Cells were immunostained with C23 antibody (red, nucleolus) or DAPI (4′,6-diamidino-2-phenylindole, blue, nucleus). Overlays of images from the first three columns are shown in (E) and (I). Scale bars represent 10 μm. Both wild-type (WT) and mutant (Mut) proteins localize to the nucleus in a speckled pattern.
In order to localize the human ZNF513 protein within the cell, COS-7 cells were transfected with expression constructs via Fugene 6 HD transfection reagent (Roche). Fixed cells were incubated with primary antibody: C23 (H-6) mouse monoclonal antibody (1:1000 dilution, Santa Cruz) and GFP mouse monoclonal antibody (Santa Cruz) followed by washing with PBS and incubation with the secondary antibody. Subsequently, washed cells were mounted with Vectashield anti-fade mounting medium containing DAPI.24 Immunofluorescence was visualized with a Zeiss LSM 510 laser scanning confocal microscope. Both the wild-type and p.C339R mutant GFP-ZNF513 proteins localize indistinguishably as dispersed foci in the nucleus but not with the nucleolus as indicated by lack of overlap with the nucleolin fluorescence (Figures 5B–5I). This is consistent with a possible role for ZNF513 as a transcriptional regulator and suggests that the p.C339R mutation does not affect nuclear localization and of the ZNF513 protein.
The mechanism through which the p.C339R ZNF513 mutant acts was further investigated by expression and chromatin immunoprecipitation (ChIP) studies. ChIP assay was performed as per the manufacturer's instructions (Active Motif) with modifications in washing conditions to minimize high background and nonspecific reactions. NIH 3T3 cells were transfected with the wild-type human GFP-ZNF513 and mutant GFP-ZNF513 plasmids with Fugene HD transfection reagent (Roche) as described above. The cells grown in a 10 cm plate were treated with formaldehyde at a final concentration of 1% for 10 min at room temperature (RT). The reaction was quenched with the addition of glycine to a final concentration of 125 mM. 1 ml of cold 1× PBS supplemented with PMSF was added to each plate and the cells were scrapped with a rubber policeman. The cell pellet was collected by centrifugation and resuspended in 1 ml of ice cold 1× PBS supplemented with protease inhibitor cocktail (PIC) (Sigma) and incubated on ice for 30 min. To aid nuclei release, the cell suspension was stroked for at least 10 times with a Dounce homogenizer and then centrifuged at 5000 rpm for 10 min to pellet the nuclei. The nuclei pellet was resuspended in digestion buffer and enzymatic shearing cocktail and incubated at 37°C for 10 min with intermittent vortexing to ensure that the chromatin was evenly sheared. The reaction was stopped with ice-cold 0.5 M EDTA and chilled on ice for 10 min. The sheared chromatin was centrifuged at 15,000 rpm at 4°C for 10 min, and the supernatant containing the sheared chromatin was collected. Agarose gel electrophoresis via the chromatin from NIH 3T3 cells transfected with wild-type GFP-ZNF513 is shown in Figure 6A. Lane 1 shows the pulldown chromatin fraction bound to ZNF513 by the GFP antibody as a faint smear, which is absent in lane 2 that shows the fraction immunoprecipitated with normal serum IgG. The input DNA is shown in lane 3 as a dense smear. These data suggest that a small subset of sheared DNA is bound by the ZNF513 protein, consistent with its putative role as a transcriptional regulator.
Figure 6.
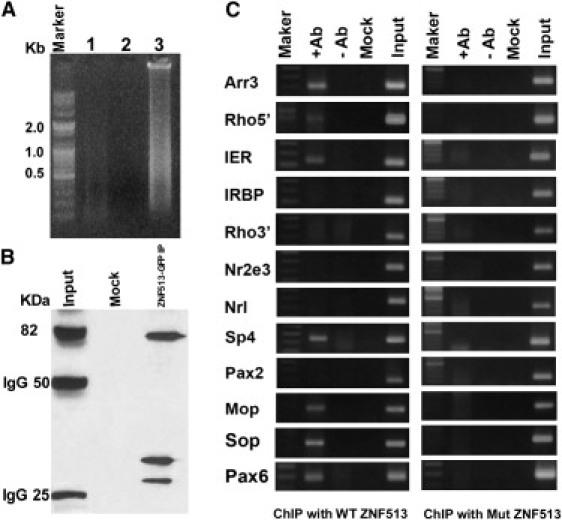
Chromatin Binding and ChIP Analysis of ZNF513
(A) Agarose gel electrophoresis with the chromatin from NIH 3T3 cells transfected with wild-type GFP-ZNF513.
(B) The DNA binding proteins present in the immunoprecipitated chromatin analyzed by western blotting via GFP antibody.
(C) ChIP assays with NIH 3T3 cells transfected with wild-type GFP-ZNF513 (left) or p.C339R mutant GFP-ZNF513 (right) with GFP monoclonal antibody (+Ab). Normal rabbit IgG (–Ab) and no chromatin (Mock) samples serve as negative controls, whereas input (chromatin samples without IP) serve as positive controls. Arr3, cone arrestin; Rho 5′, rhodopsin 5′ regulatory region; IER, enhancer of interphotoreceptor retinoid binding protein (IRBP); Rho3′, exonic 3′ region of Rhodopsin; Nr2e3, nuclear receptor subfamily 2, group E, member 3; Nrl, neural retina leucine zipper; Sp4, Sp4 transcription factor; Pax2, paired box gene 2; Mop, M-cone opsin; Sop, S-cone opsin; Pax6, paired box gene 6. Specific bands are seen for Arr3, Rho5′, IER, Sp4, Mop, Sop, and Pax6 with the wild-type but not the p.C339R mutant GFP-ZNF513.
The presence of ZNF513 protein in both the input and the immunoprecipitated fractions was tested by western blot analysis with GFP monoclonal antibody. An immunoprecipitation reaction was set up with 25 μl protein G magnetic beads, 20 μl ChIP buffer 1 (a proprietary component of the Active Motif ChIP-IT Express magnetic chromatin immunoprecipitation kit, Carlsbad, CA), 100 μl sheared chromatin, 1 μl PIC, dH2O, 2 μg of monoclonal GFP antibody (Clontech, Mountain View, CA) and incubated overnight at 4°C. The magnetic beads were pelleted on a magnetic stand and the supernatant was discarded. The beads were washed with three volumes of ChIP Buffer 1, two volumes with high salt buffer (20 mM Tris-Cl, 1 mM EDTA, 0.1% SDS, 1% Triton X-100, 500 mM NaCl [pH 7.4]), and three volumes of ChIP Buffer 2 (a proprietary component of the Actve Motif ChIP-IT Express magnetic chromatin immunoprecipitation kit, Carlsbad, CA). The bound chromatin was eluted from the beads by incubating the beads in 50 μl of elution buffer AM2 for 20 min at RT on an end to end rocker. Reverse cross-linking buffer was added to eluted chromatin and the magnetic beads were pelleted with a magnetic stand. The supernatant containing the chromatin bound to the antibody was incubated at 65°C for 3 hr and treated with RNAase A and Proteinase K before precipitating it with 3M sodium acetate (pH 5.2) and ethanol. Western blot analysis reveals the presence of GFP-ZNF513 protein in the total chromatin fraction (Input) and in the pulldown fraction with GFP antibody (ZNF513-GFP, 82 KDa, Figure 6B). No bands are seen after immunoprecipitation with normal IgG (Mock). Both immunoprecipitation and western analysis indicate the ability of ZNF513 to bind DNA.
Interaction of ZNF513 with the promoters of other transcription factors (TFs) known to play a role in the development and maintenance of photoreceptor genes and their targets was investigated by examining ZNF513 binding to chromatin isolated from NIH 3T3 cells transfected with either wild-type GFP-ZNF513 or mutant GFP-ZNF513 plasmids. Chromatin immunoprecipitated with antibodies to GFP was tested for the presence of known photoreceptor gene promoters by PCR analysis with specific primers via 1.5% agarose gels.25,26 The Pax6 (MIM 607108), Pax2 (MIM 167409), and Sp4 (MIM 600540) promoters and those of their targets including Arr3 (MIM 301770, cone arrestin), Rhodopsin (MIM 180380, 5′ regulatory region and 3′ exon region), IRBP (MIM 180290, interphotoreceptor Retinoid Binding Protein), IER (IRBP enhancer region), Nr2e3 (MIM 604485, nuclear receptor subfamily 2, group E, member 3), Nrl (MIM 162080, neural retina leucine zipper), Mop (Opn1m2, MIM 303800, M-cone opsin), and Sop (OPN1SW, MIM 190900, S-cone opsin) were tested. Only wild-type ZNF513 is found to be associated with the proximal promoter regions of Pax6 and Sp4 (Figure 6C) and their known targets including Arr3, Rhodopsin, Mop, Sop, and IER. PCR amplification for the minus antibody or minus input DNA controls is negative as are negative controls with normal rabbit IgG. The mutant ZNF513 shows no interaction with the above genes, indicating that the p.C339R mutation disrupts the binding site in ZNF513 necessary for interaction with the specific targets mentioned above (Figure 6C).
Here, we have identified a homozygous missense mutation in the ZNF513 gene in affected members of a Pakistani family with arRP. These data strongly implicate the p.C339R mutation in ZNF513 with RP. ZNF513 is highly expressed in the retina, and both the failure of the p.C339R mutant znf513 to rescue the zebrafish phenotype and the failure of the mutant protein to bind to the promoters of retinal-specific genes argue for its pathogenicity. Knockdown of znf513 in zebrafish causes abnormal retinal development with major effects on photoreceptors, including retinal thinning with photoreceptor degeneration and decreased numbers. That ZNF513 binds to the promoters of a variety of retinal transcription factors and their known targets suggest that it acts through transcriptional regulation. Taken together, these results suggest that ZNF513 has an important role in the early retinal development and later in maintenance. They also show that inhibition of ZNF513 expression causes structural and functional defects in the retina, consistent with the clinical and pathologic findings in human eyes with RP.
Although little is currently understood about the physiological functions of ZNF513, it is a widely expressed zinc finger DNA binding protein with a potential role in regulating the transcription of specific genes in the retina. Rod and cone photoreceptors in vertebrate retina are regulated by a network of photoreceptor transcription factors. Some of these factors regulate expression of themselves by binding to their own promoters, or regulate other factors acting in parallel or downstream. A few of these factors also act synergistically.27 ChIP analysis reveals that ZNF513 interacts with the Sp4, IER, Rho, Arr3, M-cone opsin, S-cone Opsin, and Pax628,29 promoters whereas no interaction is observed with transcription factors Nrl, Nr2e3, and Pax2. Both Nrl30 and especially Nr2e3 can repress the transcription of cone-specific genes and activate rod-specific genes. Crx, a key regulator of photoreceptor transcription, regulates rod opsin expression synergistically with Nrl. In addition, Sp4, a widely expressed protein, also activates transcription from the rod opsin promoter synergistically with Crx. Sp4 could have a more universal role in cell type-specific expression of certain genes in rods and possibly other retinal cell populations by interacting with different arrays of transcription factors.31 Although ZNF513 does not interact directly with Nrl and Nr2e3, which are primarily involved in the regulation of rod cell development, it shows strong interaction with the promoter sequences of rhodopsin, IER, and Sp4.
Regulation of cone photoreceptor development is much more complex than that of rods. Extrinsic factors such as retinoic acid, thyroid hormone, and other positive and negative regulators of cone-specific gene expression have been implicated in development and maintenance of cones.27 These regulators bind to cone-specific gene promoter sequences including the human locus control region (LCR) of red and green opsin genes and participate in regulation of expression of these genes, cone cell development, and distribution. ChIP assays show that ZNF513 can directly bind to the promoters of cone genes, including cone arrestin (Arr3), IRBP, Mop, and Sop. Structurally, the putative ZNF513 protein contains a nuclear localization signal in addition to zinc finger domains. Transient expression of ZNF513 shows localization of this protein to the nucleus. Therefore, the nuclear protein ZNF513, which binds to Sop and Mop promoter sequences, could be a new member of the factors that regulate the expression of opsins and participate in regulation of cone cell development and function. These observations in combination with retinal abnormalities observed in zebrafish knockdown model suggest a role for ZNF513 in regulation of photoreceptor gene expression in the retina.
In contrast to the wild-type protein, the mutant ZNF513 protein does not bind the Sp4, Arr3, Irbp, Rho, Mop, or Sop promoters, suggesting that p.C339 is located in a domain essential for recognition of specific sites for these genes. This is consistent with a role for ZNF513 as a common regulator of photoreceptor-specific gene expression within the combinatorial model of transcriptional regulation of cell-specific gene expression.
During early development of the eye, Pax6 plays a critical role in retinal cell fate and oculogenesis. Defects in Pax6 expression are involved in several abnormalities, including aniridia and Peter's anomaly in humans and the small eye syndrome in mice.32 Photoreceptors may initially require expression of Pax6 to regulate their proliferation, and disappearance of this expression precedes the onset of their differentiation.33 Lack of interaction of the mutant ZNF513 with the Pax6 promoter sequence and smaller size of the eyes in morpholino-injected zebrafish suggests a role for ZNF513 in eye development that is potentially mediated through its interaction with the Pax6 promoter.
Although ZNF513 is expressed broadly throughout the body including white blood cells, bladder, brain, parathyroid gland, ovary, and testis, affected individuals in this family have isolated RP without any systemic signs or symptoms. The reason for the retinal specificity of the phenotype is not clear, but suggests that the p.C339R mutation might possibly retain some activity for genes important in nonocular tissues. The retinal phenotype of patients from the Pakistani family with the p.C339R mutation in ZNF513 include significantly diminished rod and cone responses, macular atrophy with pigment clumping, and peripheral retinal abnormalities, suggesting the involvement of both rod and cone photoreceptors in the disease pathology.11 It is unclear at present why individuals with this mutation do not have a more severe or early-onset retinal disease, although it is possible that this might also relate to some residual activity in the p.C339R mutant protein. ZNF513 may influence the terminal differentiation and maintenance of photoreceptors through interactions with both a network of retinal transcription factors and retinal-specific functional genes. These observations strongly suggest that the p.C339R mutation interferes with retinal development and photoreceptor survival through disruption of the specific binding of ZNF513 to a subset of these genes. Interaction of ZNF513 with members of the photoreceptor transcriptional network may involve protein-protein or protein-promoter interactions or both. Understanding these interactions and the role of ZNF513 in the retina may assist in unraveling the mechanism underlying retinal degeneration because of mutant ZNF513.
Acknowledgments
We thank Dr. David Hyde and Dr. James Fadool for the generous gifts of zebrafish cone opsin and 1D1 monoclonal antibodies, respectively. Supported by National Eye Institute Grant EY13198 (R.A.); Foundation Fighting Blindness (R.A.); and Research to Prevent Blindness, Inc. (R.A.).
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
Primer3 program, http://primer3.sourceforge.net/
PROSITE, http://www.expasy.ch/prosite/
Zebrafish Model Organism Database, http://zfin.org/cgi-bin/webdriver?MIval=aa-fxfigureview.apg&OID=ZDB-FIG-050630-2875
References
- 1.Bird A.C. Retinal photoreceptor dystrophies LI. Edward Jackson Memorial Lecture. Am. J. Ophthalmol. 1995;119:543–562. doi: 10.1016/s0002-9394(14)70212-0. [DOI] [PubMed] [Google Scholar]
- 2.Bhatti M.T. Retinitis pigmentosa, pigmentary retinopathies, and neurologic diseases. Curr. Neurol. Neurosci. Rep. 2006;6:403–413. doi: 10.1007/s11910-996-0021-z. [DOI] [PubMed] [Google Scholar]
- 3.Bunker C.H., Berson E.L., Bromley W.C., Hayes R.P., Roderick T.H. Prevalence of retinitis pigmentosa in Maine. Am. J. Ophthalmol. 1984;97:357–365. doi: 10.1016/0002-9394(84)90636-6. [DOI] [PubMed] [Google Scholar]
- 4.Rivolta C., Sharon D., DeAngelis M.M., Dryja T.P. Retinitis pigmentosa and allied diseases: numerous diseases, genes, and inheritance patterns. Hum. Mol. Genet. 2002;11:1219–1227. doi: 10.1093/hmg/11.10.1219. [DOI] [PubMed] [Google Scholar]
- 5.Grondahl J. Estimation of prognosis and prevalence of retinitis-pigmentosa and Usher Syndrome in Norway. Clin. Genet. 1987;31:255–264. doi: 10.1111/j.1399-0004.1987.tb02804.x. [DOI] [PubMed] [Google Scholar]
- 6.Boughman J.A., Conneally P.M., Nance W. Population genetic studies of retinitis pigmentosa. Am. J. Hum. Genet. 1980;32:223–225. [PMC free article] [PubMed] [Google Scholar]
- 7.Boughman J.A., Caldwell R.J. Genetic and clinical characterization of a survey population with retinitis pigmentosa. In: Daentl D.L., editor. Clinical, Structural, and Biochemical Advances in Hereditary Eye Disorders. Alan R. Liss Inc; New York: 1982. pp. 147–166. [PubMed] [Google Scholar]
- 8.Dryja T.P., Li T. Molecular genetics of retinitis pigmentosa. Hum. Mol. Genet. 1995;4(Spec No):1739–1743. doi: 10.1093/hmg/4.suppl_1.1739. [DOI] [PubMed] [Google Scholar]
- 9.Inglehearn C.F. Molecular genetics of human retinal dystrophies. Eye. 1998;12(Pt 3b):571–579. doi: 10.1038/eye.1998.147. [DOI] [PubMed] [Google Scholar]
- 10.Hims M.M., Daiger S.P., Inglehearn C.F. Retinitis pigmentosa: Genes, proteins and prospects. Dev. Ophthalmol. 2003;37:109–125. doi: 10.1159/000072042. [DOI] [PubMed] [Google Scholar]
- 11.Naz S., Riazuddin S.A., Li L., Shahid M., Kousar S., Sieving P.A., Hejtmancik J.F., Riazuddin S. A novel locus for autosomal recessive retinitis pigmentosa in a consanguineous Pakistani family maps to chromosome 2p. Am. J. Ophthalmol. 2010;149:861–866. doi: 10.1016/j.ajo.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lathrop G.M., Lalouel J.M. Easy calculations of Lod scores and genetic risks on small computers. Am. J. Hum. Genet. 1984;36:460–465. [PMC free article] [PubMed] [Google Scholar]
- 13.Schaffer A.A., Gupta S.K., Shriram K., Cottingham R.W. Avoiding recomputation in linkage analysis. Hum. Hered. 1994;44:225–237. doi: 10.1159/000154222. [DOI] [PubMed] [Google Scholar]
- 14.Sunyaev S., Ramensky V., Koch I., Lathe W., Kondrashov A.S., Bork P. Prediction of deleterious human alleles. Hum. Mol. Genet. 2001;10:591–597. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- 15.Hicks G.R., Raikhel N.V. Protein import into the nucleus: An integrated view. Annu. Rev. Cell Dev. Biol. 1995;11:155–188. doi: 10.1146/annurev.cb.11.110195.001103. [DOI] [PubMed] [Google Scholar]
- 16.Sera T. Zinc-finger-based artificial transcription factors and their applications. Adv. Drug Deliv. Rev. 2009;61:513–526. doi: 10.1016/j.addr.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 17.Ayyagari R., Mandal M.N.A., Karoukis A.J., Chen L.C., McLaren N.C., Lichter M., Wong D.T., Hitchcock P.F., Caruso R.C., Moroi S.E. Late-onset macular degeneration and long anterior lens zonules result from a CTRP5 gene mutation. Invest. Ophthalmol. Vis. Sci. 2005;46:3363–3371. doi: 10.1167/iovs.05-0159. [DOI] [PubMed] [Google Scholar]
- 18.Ambasudhan R., Wang X.F., Jablonski M.M., Thompson D.A., Lagali P.S., Wong P.W., Sieving P.A., Ayyagari R. Atrophic macular degeneration mutations in ELOVL4 result in the intracellular misrouting of the protein. Genomics. 2004;83:615–625. doi: 10.1016/j.ygeno.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 19.Nieto M.A., Patel K., Wilkinson D.G. In situ hybridization analysis of chick embryos in whole mount and tissue sections. Methods Cell Biol. 1996;51:219–235. doi: 10.1016/s0091-679x(08)60630-5. [DOI] [PubMed] [Google Scholar]
- 20.Wilkinson D.G. Oxford University Press; New York: 1998. In Situ Hybridization: A Practical Approach. [Google Scholar]
- 21.Westerfield M. University of Oregon Press; Eugene, OR: 2000. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio rerio) [Google Scholar]
- 22.Morris A.C., Scholz T.L., Brockerhoff S.E., Fadool J.M. Genetic dissection reveals two separate pathways for rod and cone regeneration in the teleost retina. Dev. Neurobiol. 2008;68:605–619. doi: 10.1002/dneu.20610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vihtelic T.S., Hyde D.R. Light-induced rod and cone cell death and regeneration the adult albino zebrafish (Danio rerio) retina. J. Neurobiol. 2000;44:289–307. doi: 10.1002/1097-4695(20000905)44:3<289::aid-neu1>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 24.Vasireddy V., Vijayasarathy C., Huang J.B., Wang X.F.F., Jablonski M.M., Petty H.R., Sieving P.A., Ayyagari R. Stargardt-like macular dystrophy protein ELOVL4 exerts a dominant negative effect by recruiting wild-type protein into aggresomes. Mol. Vis. 2005;11:665–676. [PubMed] [Google Scholar]
- 25.Hirsch N., Harris W.A. Xenopus Pax-6 and retinal development. J. Neurobiol. 1997;32:45–61. [PubMed] [Google Scholar]
- 26.Peng G.H., Chen S.M. Chromatin immunoprecipitation identifies photoreceptor transcription factor targets in mouse models of retinal degeneration: New findings and challenges. Vis. Neurosci. 2005;22:575–586. doi: 10.1017/S0952523805225063. [DOI] [PubMed] [Google Scholar]
- 27.Hennig A.K., Peng G.H., Chen S. Regulation of photoreceptor gene expression by Crx-associated transcription factor network. Brain Res. 2008;1192:114–133. doi: 10.1016/j.brainres.2007.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lerner L.E., Peng G.H., Gribanova Y.E., Chen S.M., Farber D.B. Sp4 is expressed in retinal neurons, activates transcription of photoreceptor-specific genes, and synergizes with Crx. J. Biol. Chem. 2005;280:20642–20650. doi: 10.1074/jbc.M500957200. [DOI] [PubMed] [Google Scholar]
- 29.Hill R.E., Favor J., Hogan B.L.M., Ton C.C.T., Saunders G.F., Hanson I.M., Prosser J., Jordan T., Hastie N.D., Vanheyningen V. Mouse small eye results from mutations in a paired-like Homeobox-containing gene. Nature. 1991;354:522–525. doi: 10.1038/354522a0. [DOI] [PubMed] [Google Scholar]
- 30.Mears A.J., Kondo M., Swain P.K., Takada Y., Bush R.A., Saunders T.L., Sieving P.A., Swaroop A. Nrl is required for rod photoreceptor development. Nat. Genet. 2001;29:447–452. doi: 10.1038/ng774. [DOI] [PubMed] [Google Scholar]
- 31.Lerner L.E., Gribanova Y.E., Whitaker L., Knox B.E., Farber D.B. The rod cGMP-phosphodiesterase beta-subunit promoter is a specific target for Sp4 and is not activated by other Sp proteins or CRX. J. Biol. Chem. 2002;277:25877–25883. doi: 10.1074/jbc.M201407200. [DOI] [PubMed] [Google Scholar]
- 32.Hanson I.M., Fletcher J.M., Jordan T., Brown A., Taylor D., Adams R.J., Punnett H.H., Vanheyningen V. Mutations at the Pax6 locus are found in heterogeneous anterior segment malformations including Peters Anomaly. Nat. Genet. 1994;6:168–173. doi: 10.1038/ng0294-168. [DOI] [PubMed] [Google Scholar]
- 33.Garelli A., Rotstein N.P., Politi L.E. Docosahexaenoic acid promotes photoreceptor differentiation without altering Crx expression. Invest. Ophthalmol. Vis. Sci. 2006;47:3017–3027. doi: 10.1167/iovs.05-1659. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


