Abstract
Retinal stem cells (RSCs) are present in the ciliary margin of the adult human eye and can give rise to all retinal cell types. Here we show that modulation of retinal transcription factor gene expression in human RSCs greatly enriches photoreceptor progeny, and that strong enrichment was obtained with the combined transduction of OTX2 and CRX together with the modulation of CHX10. When these genetically modified human RSC progeny are transplanted into mouse eyes, their retinal integration and differentiation is superior to unmodified RSC progeny. Moreover, electrophysiologic and behavioral tests show that these transplanted cells promote functional recovery in transducin mutant mice. This study suggests that gene modulation in human RSCs may provide a source of photoreceptor cells for the treatment of photoreceptor disease.
Keywords: Retinal stem cells, Photoreceptor, Regeneration
Introduction
During the development of the mammalian retina, retinal precursor cells give rise to all of the morphologically and functionally distinct retinal cell types perinatally [1, 2]. However, in adult mammals, there is little evidence of further retinal growth or regeneration. Nevertheless, in vitro studies have shown that the ciliary margin of the adult rodent and adult human eyes contains retinal stem cells (RSCs) that can self-renew and give rise to all retinal cell types including photoreceptors [3-5]. These findings suggest that human retinal stem cells (hRSCs) could provide a source of retinal cells for regenerative therapy of blindness. Moreover, autologous RSC transplantation following expansion in culture would avoid immune rejection. The principal cell type that must be replaced in individuals with retinal disease are the photoreceptors. Photoreceptors are light detectors that transfer visual signals through other retinal neurons to the brain. Photoreceptors become compromised in retinal diseases such as retinitis pigmentosa [6], retinal detachment [7], and age-related macular degeneration [8]. At present, there is no proven therapy available to rescue the blindness caused by photoreceptor diseases.
A recent report suggested that RSCs do not exist, and posited instead that all ciliary epithelial cells have the ability to transdifferentiate to neural cells [9]. However, we suggest that the prospective in vitro isolation of a specific rare population of RSCs from the ciliary margin using high Pax6 expression [10] and high pigmentation [3] speaks strongly in favor of the stem cell hypothesis.
A major limitation in using the progeny of RSCs to replace photoreceptors is that these cells are only a minority of the progeny differentiated from RSCs in vitro. To address this problem, we manipulated, in RSCs, the expression of genes known to influence photoreceptor development using a lentiviral mediated gene system [11, 12]. Recent studies have demonstrated that certain combinations of retinal transcription factors contribute to the development of multiple retinal cell types in cultured retinal systems [13, 14]. Initially, we focused on the CHX10 gene, which is required for retinal progenitor proliferation [15] and for promoting bipolar cell development at the expense of rods [16] and works as a transcriptional repressor [17]. We asked whether converting CHX10 to an activator would increase photoreceptor progeny. To reverse CHX10 activity to an activating form, CHX10VP16 was engineered [18] by fusing CHX10 to the VP16 activator domain, which works to convert the construct to a transcriptional activator by promoting the assembly of a transcription activation complex [19]. Then, we examined the key regulator genes of photoreceptor formation such as OTX2 [20] and CRX [21]. We hypothesized that modulating the expression of these genes that are important during normal eye development would increase the number of photoreceptor progeny of hRSCs. To assess the efficiency of photoreceptor induction, these hRSCs were subjected to in vitro differentiation and transplanted in vivo into mouse eyes. We demonstrate that coexpression of CHX10VP16, OTX2, and CRX enhances photoreceptor differentiation from hRSCs. After transplantation to immunosuppressed wild-type mice, these genetically modified progeny of hRSCs produce progeny that survive and differentiate into photoreceptors in vivo at a higher frequency than unmanipulated hRSCs. Furthermore, transplantation of RSCs into the eyes of transducin mutant mice, which lack functional rod photoreceptors, can significantly improve visual function as measured by electrophysiologic and behavioral methods.
Materials and Methods
Human Retinal Stem Cells Isolation and Culture In Vivo and Sphere Passaging
We performed hRSC isolation using human eyes from the Eye Bank of Canada within 24 hours postmortem as previously described [4]. RSC-derived sphere passaging was performed as previously described [4].
Lentivirus Construct
Replication-defective, self-inactivating lentiviral vectors [11, 12] with EF1α as an internal promoter (pCSII-EF was a gift from Dr. H. Miyoshi) containing an internal ribosome entry site (IRES)-EGFP (CSEIE), a phosphoglycerate kinase (PGK) promoter-EGFP (CSEPE), or a PGK promoter-neomycin resistance gene (CSEPneo) were prepared. Each cDNA was cloned into CSEIE or CSEPneo, which directs the expression of the cloned genes together with EGFP from the internal promoter. For CSEPE-OTX2/CRX, CSEPE-CHX10VP16/OTX2, and CSEPE-CHX10VP16/OTX2/CRX, IRES-OTX2 and IRES-CRX followed OTX2 or CHX10VP16. Overxpression or coexpression of all these genes in cells was confirmed by immunocytochemistry or polymerase chain reaction (PCR). The lentiviral vectors were produced by cotransfecting 293T cells with the lentiviral expression vector and pLP/VSVG (encoding the VSV-G envelope protein), along with the packaging constructs pLP1 and pLP2 (Invitrogen, Carlsbad, CA, http://www.invitrogen.com). High-titer viral vector stocks were prepared for transfection by ultracentrifugation for transfection (1.0 × 109 transduction units (TU)/ml).
Lentiviral Transfection of Stem Cells
Adult hRSC-derived spheres were dissociated into single cells and the cells were seeded at 1.0 × 105 cells per well in 1 ml of serum-free media (SFM). The cells were transfected at the multiplicity of infection (MOI) 10 for 12 hours at 37°C in 5% CO2. After infection, the cells were harvested, washed twice, and then plated at 10 cells/μl in SFM containing fibroblast growth factor-2 and heparin. The cells then were allowed to proliferate for 7 to 14 days. The spheres that arose were visualized using a fluorescent microscope, and only green spheres were harvested for the differentiation or transplantation assays.
In Vitro Differentiation Assay
To assay the differentiation potential of the RSC progeny transfected with different genes, single clonally derived hRSC spheres were selected 7 to 14 days after lentivirus infection and plated as whole spheres [4]. Each experiment was repeated at least five times.
Immunochemical Analysis
Immunofluorescent staining was performed using antibodies directed to specific markers: human-specific Nestin (Chemicon, Temecula, CA, http://www.chemicon.com), Pax6 (Chemicon), Chx10 (from lab of R. McInnes), Brn3B (Santa Cruz Biotechnology Inc., Santa Cruz, CA, http://www.scbt.com), NF-M (Chemicon), Rho1D4 (Abcam, Cambridge, U.K., http://www.abcam.-com), Rho4D2 (gift of Dr. R. Molday), Rom1 (R. McInnes), human cone arrestin (from lab of C. Craft), 10E4 (Cedarlane, Hornby, ON, Canada, http://www.cedarlanelabs.com), HPC-1 (Sigma-Aldrich, St. Louis, MO, http://sigmaaldrich.com), calbindin (Chemicon), RPE65 (Chemicon), bestrophin (Abcam), PKCα (Abcam), active Caspase-3 (Promega, Madison, WI, http://www.promega.com), Ki-67 (BD Biosciences, San Diego, CA, http://www.bdbiosciences.com), desmin (Chemicon), cytokeratin-17 (Abcam), human nuclei antigen (Chemicon), and green fluorescent protein (GFP) (Chemicon). Antigens were visualized using appropriate fluorescent secondary antibodies.
CAT Assay
NG108 cells were maintained and transfected as previously described [22] with the following plasmids: HD4-pG5EC (chloramphenicol acetyl transferase [CAT] reporter containing four homeodomain binding sites and 5 GAL4 DNA binding sites), GAL4-HSF1 (HSF1 activator), pMXIE, pMXIE-CHX10, and pMXIE-CHX10VP16. For the CAT assay, briefly, NG108 cells were cotransfected with equimolar amounts of control effector plasmid or pMXIE-CHX10 or pMXIE-CHX10VP16 along with GAL4-HSF1 activator and HD4-pG5EC CAT reporter. One hundred percent CAT activity is taken as that obtained in the presence of control effector plasmid. CAT activity was corrected for transfection efficiency using a β-galactosidase internal control.
Luciferase Assay
Luciferase assay was carried out as previously described [17]. Luciferase reporters (pGL3-Basic, Promega) under the control of the β-actin promoter together with the OTX2 5′ genomic region (0.5 μg) were transfected into hRSCs, which were plated in the differentiation condition described above, with or without CHX10-expression vectors (CSEIE-CHX10, 1 μg). The vector for renilla luciferase gene under the control of the SV40 promoter (pRL-SV40, 6 ng) was cotransfected as an internal standard to normalize the transfection efficiency. After 40 to 48 hours, the cells were harvested and luciferase activity was measured.
ChIP Assay
Chromatin immunoprecipitation (ChIP) analysis was carried out as previously described [23]. The hRSC-derived spheres were cross-linked with 1% formaldehyde, sonicated, and incubated with anti-Chx10 antibody (Chemicon) or normal sheep serum (Sigma) for 12 hours. Immune complexes were incubated with protein A Sepharose beads (Upstate, Charlottesville, VA, http://www.upstate.com), which were then washed six times and incubated with 100 μg/ml proteinase K for DNA extraction. DNA was analyzed by PCR using 5′-TCTGCCATGGAAAGGCAACAGTCT-3′ and 5′-CGTGCCTTCAAATGCACACATTGC-3′ for CHX10BA, and 5′-ACTGGGCTGGACATTCCAGTTT-3′ and 5′-GGTGTTTGGTTGCACATGGCTAGA-3′ for the 3′UTR of the OTX2 genomic region.
Reverse transcription polymerase chain reaction and Real-Time PCR
Total RNA was extracted, and reverse-transcription reactions were performed using the Superscript-II enzyme (Invitrogen). Quantitative detection of specific mRNA transcripts was carried out by conventional reverse transcription polymerase chain reaction (RT-PCR) using Advantage-GC2 PCR Taq (BD Biosciences) or real-time PCR using SYBR Green PCR Master Mix (Applied BioSystems, Foster City, CA, http://www.appliedbiosystems.-com). Relative amounts of mRNA were determined by normalizing to GAPDH mRNA for each sample. To detect the corresponding gene expression, we used the following primers:
OTX2, 5′- ATCTGCCAAATCCAGGAA-3′ and 5′-TGCACT
GAAACTTTACGACA-3′; CHX10, 5′-TGGAGCACCGGGTGG
GCTCT-3′ and 5′-CCAGTCTCTCACCTCTGCCCT-3′; CRX, 5′-
TATTCTGTCAACGCCTTGGCCCTA-3′ and 5′-AACCCTGGAC
TCAGGCAGATTGAT-3′;
GAPDH, 5′-CTACTGGCGCTGCCAAGGCTGT-3′ and 5′-
GCCATGAGGTCCACCACCCTG-3′;
NESTIN, 5′-AGAGGGGAATTCCTGGAG-3′ and 5′-CTGA
GGACCAGGACTCTCTA-3′;
PAX6, 5′-CGGTGTGGTGGGTTGTGGAAT-3′ and 5′-ATG
GTTTTCTAATCGAAGGG-3′;
GATA-1, 5′-CCATTGCTCAACTGTATGGAGGG-3′ and 5′-
ACTATTGGGGACAGGGAGTGATG-3′;
BRACHYURY, 5′-TAAGGTGGATCTTCAGGTAGC-3′ and
5′-CATCTCATTGGTGAGCTCCCT-3′; AND GATA-4, 5′-TCC
CTCTTCCCTCCTCAAAT-3′ and 5′-TCAGCGTGTAAAGGC
ATCTG-3′.
Transplantation
We performed transplantation of hRSCs into mouse eyes as previously described [4, 24].
Control (GFP alone) or CHX10VP16/OTX2/CRX-transduced hRSC progeny were transplanted into the vitreous cavity of postnatal day 1 CD1 mice, as previously described. To suppress tissue rejection, cyclosporine was administered intraperitoneally (i.p.) to animals every day beginning just before the transplantation surgery, and continuing until the hosts were killed. Host mice were killed at 1, 3, or 5 weeks after transplantation, and the number of surviving hRSC progeny were counted at each time point. In Figure 4A, multiple comparison tests revealed that the group that did not receive cyclosporine showed a significant decrease in the numbers of cells surviving between 1 and 5 weeks after transplantation (post hoc Dunn’s correction, p < .05). However, the control vector and the CHX10VP16/OTX2/CRX-transduced groups treated with cyclosporine did not show significant differences in surviving cell numbers between week 1 and week 5 after transplantation (p > .05). Indeed, at 1 week after transplantation, similar numbers of human cells were seen in the host mouse eyes in the noncyclosporine-treated and cyclosporine-treated control vector groups (p > .05), but at 5 weeks after transplantation into the mouse eye, the transplanted hRSC progeny were integrated significantly better with than without i.p. cyclosporin treatment (p < .05). Integration and differentiation of transplanted hRSCs into photoreceptors (with either control or CHX10VP16/OTX2/CRX transfection) into the transducin mutant mice retinas were similar to that of the same cells transplanted into control CD1 retinas. The numbers of human cells in mouse eye sections were determined using Abercrombie’s correction. All experimental protocols were approved by the Animal Care Committee guidelines of the University of Toronto and the Government of Canada.
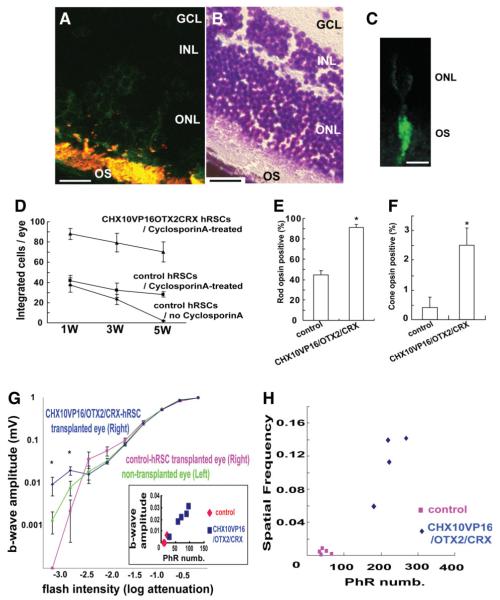
Figure 4.
In vivo human retinal stem cell (hRSC) transplantation into mouse eye. (A): Human retinal stem cell progeny (green fluorescent protein [GFP] positive) are immunostained with a photoreceptor marker Rom1 (red), which marks a outer segment protein in transplanted human (double labeled with two markers shown as yellow) and host (red) CD1 retinal cells (scale bar: 100 μm). GCL; retinal ganglion layer, INL; inner nuclear layer, ONL; outer nuclear layer, OS; outer segment. (B): Mouse retina section (adjacent to the one shown in A) stained with cresyl violette to show the retinal location of the transplanted human cells shown in (A) (scale bar: 100 μm). (C): High-power image of a single hRSC-derived phtotoreceptor (GFP positive) integrated into the host retina. The human donor cell shows the morphology of a photoreceptor (scale bar: 20 μm). (D): Transplanted hRSC progeny transfected with CHX10VP16/OTX2/CRX show improved integration and survival compared with control transfection at 1, 3, and 5 weeks after transplantation. (E,F): Improved photoreceptor differentiation in hRSC progeny transfected with CHX10VP16/OTX2/CRX compared with control transfected cells at 5 weeks after transplantation (rods (E) and cones (F)) (analysis of variance [ANOVA] and Dunnette’s multiple comparison test, *p < .05). Rho1D4 was used as a rod photoreceptor marker and human cone arrestin as a cone photoreceptor marker. The y-axis indicates the percentage of photoreceptor marker and GFP coexpressing cell number/GFP expressing cell number. (G): At the lowest flash intensities (−3.2 and −2.8) the CHX10VP16/OTX2/CRX group shows a higher response than the nontransplanted and GFP-only vector-treated groups (ANOVA and a Dunnette’s multiple comparison test, *p < .05). Inset shows that a significant correlation of maximal b wave response and surviving human photoreceptor cell number (PhR numb) was seen. For reasons of space within this inset, the two data points for the control animals represent the data from four animals. (H): As a within-animal control in the transplantation model, the differences in the minimal spatial frequency detected between the transplanted eye (right) and nontransplanted eye (left) in each individual mice were estimated. Abbreviations: GCL, retinal ganglion layer; hRSCs, human retinal stem cells; INL, inner nuclear layer; ONL, outer nuclear layer; OS, outer segment.
Electroretinogram
Electroretinogram (ERG) recordings were performed as previously described [25]. Briefly, mice were dark adapted for more than 12 hours, and pupils were fully dilated. ERGs were recorded from the corneal surface of one eye using a silver-impregnated nylon fiber. Electrodes were connected to a differential amplifier and the signal amplified 10,000-fold with an opened bandwidth of 3-1.000 Hz. A scotopic bright flash response with a well delineated a- and b-wave was obtained with the flash stimuli. The b-wave was measured from the a-wave trough to the maximum positive peak.
Behavioral Assessment
A virtual optomotor system to quantify spatial vision was performed as previously described [26]. A rotating cylinder covered with a vertical sine wave grating gave virtual three-dimensional space on four computer monitors facing to form a square. Experimented mice standing unrestrained on a platform in the center of the square tracked the grating with reflexive head and neck movements. The spatial frequency of the grating was clamped at the viewing position by repeatedly recentering the cylinder on the head. Acuity was quantified by increasing the spatial frequency of the grating until an optomoter response could not be elicited. To obtain an internal control, the differences of spatial frequency between the right eye that received a transplant and the left eye that did not were evaluated.
Statistics
Data are expressed as means +/− SEM unless specified otherwise. Statistical comparisons between two groups were performed using a Student’s t test when appropriate. For multiple comparisons, analysis of variance (ANOVA) was employed followed by Dunnette’s post hoc tests. The acceptable level of significance was p < .05.
Results
Modulation of CHX10 Gives Rise to Photoreceptor Subtypes in hRSC Progeny
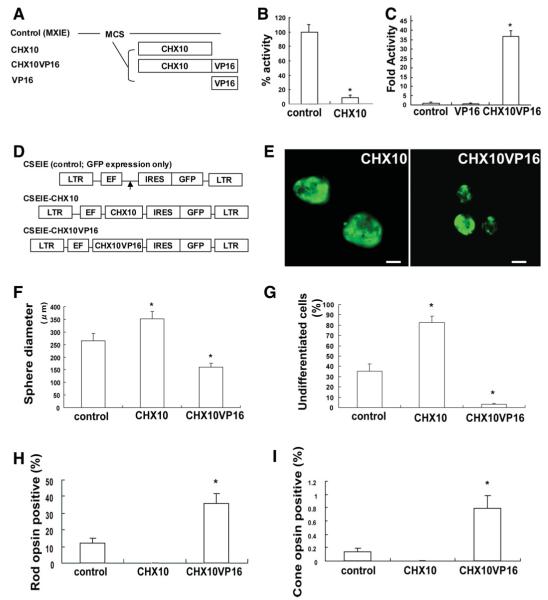
The expression of these genes was manipulated in hRSCs. The CHX10 gene is required for retinal progenitor proliferation and bipolar cell differentiation [15, 16, 27]. Further recent studies have demonstrated that the CHX10 protein targets and blocks photoreceptor-specific gene expression [18]. Moreover, the ability of Chx10 to drive bipolar cell genesis at the expense of rods is reversed if Chx10 is converted to an activator [16]. We hypothesized that modulation of CHX10 expression would increase the photoreceptor progeny of hRSCs, and designed CHX10VP16 [18] which encodes human CHX10 fused to the amino acids 410-490 of the VP16 activation domain [19] (Fig. 1A). To estimate its activity in vitro, CAT assays were performed using control, CHX10-, CHX10VP16-, and VP16-expressing vectors. Reporter transcription was significantly decreased in the presence of CHX10-expressing vector compared with control (Fig. 1B). Thus, CHX10 works as a transcriptional repressor. On the other hand, CHX10VP16 significantly activated reporter transcription compared with VP16 alone (Fig. 1C), indicating that CHX10VP16 acts as a functional CHX10 activator.
Figure 1.
Modulation of CHX10 expression is important for induction and early maturation of photoreceptors from human retinal stem cell (hRSC) progeny. (A): Schematic illustration of effecter vectors encoding CHX10, CHX10VP16 or VP16. CHX10VP16 encodes human CHX10 fused to amino acids 410-490 of the VP16 activation domain. All genes were introduced into the pMXIE expression vector. (B): The expression vector pMXIE-CHX10 represses activation. NG108 cells were cotransfected with equimolar amounts of control effector plasmid or pMXIE-CHX10 along with GAL4-HSF1 activator and HD4-pG5EC chloramphenicol acetyl transferase (CAT) reporter containing four homeodomain binding sites and five GAL4 DNA binding sites. One hundred percent CAT activity is taken as that obtained in the presence of control effector plasmid was set to 1.0. The y-axis indicates the percentage of reporter transcription with CHX10 transfection/reporter transcription with control transfection. *p < .05 indicates statistically significance with Student’s t test. (C): Transcription is activated by pMXIE-CHX10VP16. NG108 cells were cotransfected with equimolar amounts of control effector plasmid or pMXIE-CHX10VP16 along with HD4-pG5EC CAT reporter. The y-axis indicates fold activity of reporter transcription with VP16 or CHX10VP16 transfection/reporter transcription with control transfection set to 1.0. *p < .05 indicates statistically significance with analysis of variance (ANOVA) and a Dunnette’s multiple comparison test. (D): Schematic of replication-defective self-inactivating lentiviral vectors containing an internal ribosome entry site (IRES) sequence followed by enhanced green fluorescent protein (GFP) (CSEIE). CHX10 or CHX10VP16 cDNA were cloned into CSEIE, which directs the expression of the cloned genes together with GFP from the internal promoter, EF1α. Control vector expresses only GFP. (E): Human retinal stem cells-derived sphere transfected with CHX10 (left) and CHX10VP16 (right). Spheres ubiquitously express GFP, but some of the cells in the clonal sphere are pigmented, thus obscuring GFP and producing a mottled GFP appearance in the spheres. Scale bar: 100 μm. (F): Sphere diameters that are a proxy for total cell number generated by proliferation were measured in control, CHX10, or CHX10VP16-induced retinal stem cell (RSCs) colonies. We measured more than 30 spheres in each group in at least three independent experiments. Sphere diameters were significantly increased in CHX10-induced clonally-derived RSC colonies compared with control. On the other hand, sphere diameters were significantly decreased in CHX10VP16-induced clonally-derived RSC colonies (analysis of variance and Dunnette’s multiple comparison test, *p < .05). (G): PAX6/NESTIN double labeling cells, which indicate undifferentiated retinal cells, were measured in the in vitro differentiation assay with hRSC colonies transfected with control, CHX10,or CHX10VP16. With CHX10 transduction, most of hRSC progeny maintained an undifferentiated state. In contrast, CHX10VP16 transduction significantly decreased the number of undifferentiated cells (ANOVA and Dunnette’s multiple comparison test, *p < .05). The y-axis indicates the percentage of PAX6/NESTIN double labeling cells /total cell number after neomycin selection. (H,I): CHX10 transduction abolished photoreceptor cell differentiation, while CHX10VP16 transduction significantly increased rod (H) and cone (I) photoreceptor differentiation. Rho1D4 was used as a rod photoreceptor marker and human cone arrestin as a cone photoreceptor marker. CHX10 transduction abolished photoreceptor cell differentiation, whereas CHX10VP16 transduction significantly increased rod and cone photoreceptor differentiation (Student’s t test, *p < .05). The y-axis indicates the percentage of photoreceptor marker and GFP coexpressing cell number/GFP expressing cell number. Abbreviations: EF, elongation factor; GFP, green fluorescent protein; IRES, internal ribosome entry; LTR, long terminal repeat.
To examine the effect of manipulating CHX10 and CHX10VP16 expression in hRSCs, we measured the size of clonal hRSC spheres following the transfer of bi-cistronic lentiviral vectors [11, 12], expressing these genes with green fluorescent protein (GFP) (Fig. 1D). Only all green spheres, which arose from a single green hRSC, were used for all further experiments. Sphere diameter is a proxy for total cell number generated by proliferation. Human adult retinal sphere size was significantly increased in CHX10-transduced clonal RSC colonies (352.0 ± 28.9 μm) compared with control (empty vector, 265.0 ± 29.1 μm, t = 2.12, p < .05). On the other hand, sphere size was decreased in CHX10VP16-transduced RSC clonal colonies (161.0 ± 15.2 μm) compared with control (t = 3.17, p < .05, Fig. 1E and 1F). This result is consistent with the finding that RSC spheres from Chx10orJ/orJ mutant mice are significantly smaller in diameter compared with their wild-type controls [28], confirming that CHX10 promotes retinal progenitor proliferation [15], and that CHX10VP16 decreases clonal RSC sphere proliferation.
To determine whether manipulation of CHX10 expression modifies the proliferation of the stem cell or the progenitor cell populations, the numbers of secondary spheres were compared after passaging the lentiviral-transduced clonal primary spheres. Spheres were bulk passaged to single cell suspensions, and 2,000 cells were plated per well and cultured for 2 weeks. The number of clonal secondary spheres is a direct reflection of the symmetrical divisions of stem cells in the primary clonal sphere, and the size of the sphere is attributed primarily to the progenitor population which comprises most of the cells in each sphere [4, 29]. There was no difference in secondary sphere number among control, CHX10-, and CHX10VP16-transduced hRSC spheres (F(2,6) = 0.44, p > .05, Supporting Fig. S2A). These results indicated that CHX10 directly enhances retinal progenitor proliferation, but not stem cell proliferation.
To examine the effect of modified CHX10 expression on the differentiation of cells in hRSC derived sphere, in vitro differentiation assays were carried out. Single hRSC sphere colonies were selected 7 days after transfection with control, CHX10-, or CHX10VP16-expressing lentiviral vectors with neomycin selection 3 days after virus infection (Supporting Fig. S1A). The transduced cells were then induced to differentiate in vitro for 3 weeks. In human retinal cells, PAX6/NESTIN double-labeling indicates undifferentiated cells, as PAX6 is expressed in retinal progenitor and mature amacrine cells, and NESTIN is expressed in retinal progenitor and mature Müller glial cells [4]. With increased CHX10 expression, most of the hRSC progeny maintained an undifferentiated state (80.2 ± 5.2%) compared with control (34.0 ± 6.9%) (t = 5.33, p < .05). In contrast, the expression of CHX10VP16 significantly decreased the number of undifferentiated cells (3.9 ± 0.7%) (t = 4.34, p < .05) (Fig. 1G, Supporting Fig. S2B). These results indicate that modulation of CHX10 may direct retinal stem cells progeny toward a differentiated state.
To estimate the effects on photoreceptor differentiation of modifying CHX10 activity, we used Rho1D4 as a rod photoreceptor marker, and human cone arrestin as a cone photoreceptor marker. We examined the differentiation of hRSC-derived colonies transfected with control (GFP), CHX10-, or CHX10VP16-expressing lentiviral vectors. With CHX10 transduction, no differentiated photoreceptors were detected. In contrast, CHX10VP16 transduction significantly increased the numbers of cells that differentiated into rod (35.9 ± 6.1%) (t = 3.52, p < .05, Fig. 1H) and cone photoreceptors (0.79 ± 0.19%) (t = 3.37, p < .05, Fig. 1I) compared with control (11.8 ± 3.2%, 0.14 ± 0.06%, respectively). Thus, CHX10VP16 transduction increases photoreceptor differentiation in hRSC progeny.
The Combination of CHX10VP16, OTX2 and CRX Strongly Induces Photoreceptor Differentiation of hRSC Progeny
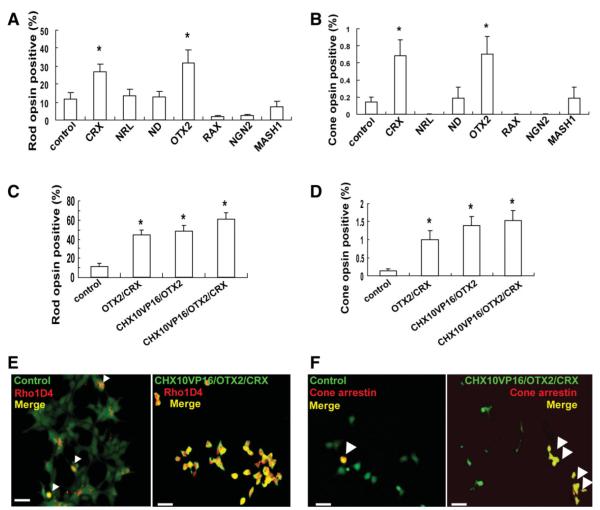
To test for the enhanced production of photoreceptor progeny from hRSC-derived cells, several retinal transcription factors were transferred into hRSC progeny, including CRX, NRL [30], NEUROD [31], OTX2, RAX [32], NEUROGENIN2 [33], and MASH1 [34, 35] (Supporting Fig. S1B). In the in vitro differentiation assay, photoreceptor differentiation was significantly promoted in hRSC progeny (F(7,32) = 8.06, p < .05) with OTX2 (31.5 ± 7.4%, p < .05) or CRX (26.8 ± 4.4%, p < .05), compared with control (11.8 ± 3.2%) (Fig. 2A). Similarly, cone photoreceptor differentiation was significantly increased in hRSC progeny (F(7,72) = 6.01, p < .05) with OTX2 (0.70 ± 0.20%, p < .05) or CRX (0.68 ± 0.19%, p < .05) transduction compared with control (0.14 ± 0.06%) (Fig. 2B). NRL, NEUROD, RAX, NGN2, or MASH1 did not affect rod nor cone photoreceptor differentiation (p > .05). Thus, OTX2 or CRX overexpression promotes photoreceptor induction from hRSC progeny in vitro.
Figure 2.
Transduction of OTX2 and CRX together with modulation of CHX10 produce the most potent induction of photoreceptor differentiation from human retinal stem cell progeny. (A,B): Results of the in vitro differentiation assay with clonal hRSC derived spheres transfected with control (green fluorescent protein [GFP]), CRX, NRL, NEUROD, OTX2, NEUROGENIN2, or MASH1-expressing lentiviral vectors. Rho1D4 was used as a rod photoreceptor marker and human cone arrestin as a cone photoreceptor marker. The y-axis indicates the percentage of photoreceptor marker and GFP coexpressing cell number/GFP expressing cell number. OTX2 or CRX transduction significantly increased the numbers of differentiated rod (A), and cone (B) photoreceptor (analysis of variance and a Dunnette’s multiple comparison test, *p < .05). (C,D): Photoreceptor differentiation was significantly promoted by coexpression of OTX2/CRX, CHX10VP16/OTX2, and CHX10VP16/OTX2/CRX compared with control ((C) rods, and (D) cones) (analysis of variance and Dunnette’s multiple comparison test, *p < .05). (E): Rho1D4 positive cells (red) or (F) human cone arrestin positive cells (red) coexpress GFP from the control (left) or from the CHX10VP16/OTX2/CRX0expression vector (right) as illustrated by the merged field (yellow, marks by arrowheads). Many more human retinal stem cell progeny differentiated into (E) rod or (F) cone photoreceptors with CHX10VP16/OTX2/CRX-transduction compared with control-GFP.
To determine whether photoreceptor differentiation from hRSC progeny could be further enhanced, we next examined the effect of the coexpression of OTX2/CRX, CHX10VP16/OTX2, or CHX10VP16/OTX2/CRX (Supporting Fig. S1C) in these cells. Overexpression of each gene was confirmed by PCR in double or triple expression constructs. In the in vitro differentiation assay, rod photoreceptor differentiation (Rho1D4 positive) was significantly promoted (F(3,31) = 8.07, p < .05) by the coexpression of OTX2/CRX (44.9 ± 5.0%, p < .05), CHX10VP16/OTX2 (48.7 ± 5.8%, p < .05), or CHX10VP16/OTX2/CRX (60.6 ± 7.3%, p < .05) compared with control (11.8 ± 3.2%) (Fig. 2C and 2E). Further PCR for other photoreceptor markers such as Rom1 (a rod photoreceptor outer segment protein), NRL, and recoverin showed similar enrichments were detected in differentiated retinal progeny after hRSC transfection with CHX10VP16/OTX2/CRX (data not shown). Similarly, cone photoreceptor differentiation (cone arrestin positive cells) significantly increased (F(3,36) = 6.87, p < .05) in the progeny of hRSC with coexpression of OTX2/CRX (0.99 ± 0.27%, p < .05), CHX10VP16/OTX2 (1.38 ± 0.27%, p < .05), or CHX10VP16/OTX2/CRX (1.54 ± 0.28%, p < .05) compared with control (0.14 ± 0.06%) (Fig. 2D and 2F). The photoreceptor-inducing activity of CHX10VP16/OTX2/CRX in hRSC progeny was significantly higher than the activity of CHX10VP16, OTX2, CRX, or OTX2/CRX alone. In comparison with CHX10VP16/OTX2, CHX10VP16/OTX2/CRX had a higher, but not a significantly higher, tendency for photoreceptor induction. These data indicate that the combination of CHX10VP16, OTX2, and CRX produced the greatest increase both in the proportion of rods and cones. CHX10VP16/OTX2/CRX-transfected hRSC progeny displayed the small-cell bodies characteristic of photoreceptors in culture [3, 4].
Interaction of CHX10, OTX2 and CRX in hRSC Progeny
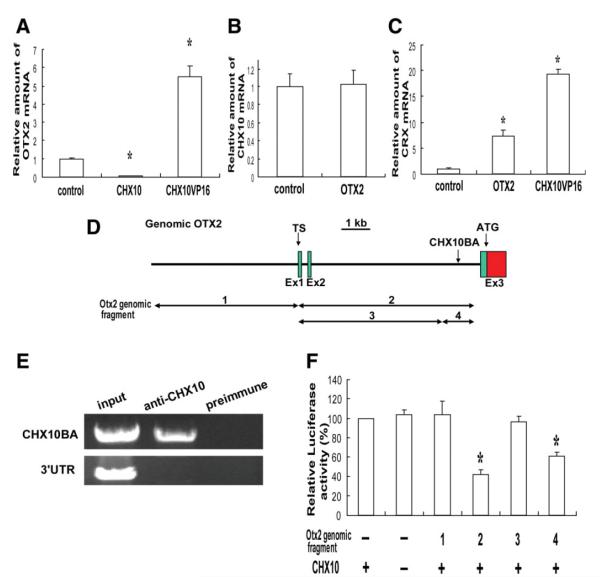
To examine the interactions between these transcription factors in photoreceptor differentiation from hRSC progeny, we performed RT-PCR analyses of OTX2, CHX10, and CRX mRNA levels. RNA was prepared from 3-day cultures of hRSC transfected with control, CHX10-, CHX10VP16-, and OTX2-expressing lentiviral vectors. OTX2 mRNA was decreased by CHX10 transduction (0.075 ± 0.01-fold, t = 25.79, p < .05). On the other hand, OTX2 mRNA was increased by CHX10VP16 transduction (5.5 ± 0.6-fold) compared with control (t = 13.72, p < .05, Fig. 3A). However, the levels of CHX10 mRNA were not affected by OTX2 transduction compared with controls (t = 0.19, p >.05, Fig. 3B). CRX mRNA levels were decreased by CHX10 (0.077 ± 0.02-fold, t = 8.44, p < .05), but on the other hand, were increased by OTX2 (7.4 ± 1.2-fold, t = 9.23, p < .05) or CHX10VP16 (19.4 ± 4.9-fold) (t = 33.21, p < .05) transduction compared with control (Fig. 3C). These results suggest that CHX10 suppresses OTX2 and CRX expression during photoreceptor differentiation from hRSC progeny.
Figure 3.
Molecular interaction of CHX10, OTX2, and CRX in human retinal stem cell (hRSC) progeny. (A–C): Real-time reverse transcription polymerase chain reaction (RT-PCR) analysis. OTX2, CHX10, or CRX mRNA levels were normalized to GAPDH mRNA (the control sample was set to 1.0). (A): OTX2 mRNA level was significantly downregulated in CHX10-transduced hRSC progeny, whereas OTX2 mRNA was increased in CHX10VP16 transduced progeny (analysis of variance and Dunnette’s multiple comparison test, *p < .05). (B): CHX10 expression is not affected in OTX2 transduced hRSC progeny. (C): CRX mRNA levels were significantly increased in OTX2 or CHX10VP16-transduced hRSC progeny (analysis of variance and a Dunnette’s multiple comparison test, *p < .05). (D): Partial genomic map (5′ region) for the human OTX2 gene. Exons (Ex1,2, and 3), transcriptional start sites (TS) and the initiator codon (ATG) are indicated. A putative CHX10-binding area (CHX10BA) is located in intron2. (E): Chromatin immunoprecipitation analysis. Anti-Chx10 antibody specifically precipitates the chromatin containing the end of intron two of OTX2 (CHX10BA), but not the 3′UTR region (control), from hRSC progeny. Preimmune serum does not precipitate these regions. (F): Luciferase assay. OTX2 genomic fragments 1-4 shown in (D) were cloned into a luciferase reporter and cotransfected into hRSC progeny with or without the CHX10 expression vector. The activity of the βactin minimal promoter-luciferase reporter without genomic OTX2 was taken as 100%. Suppression of the reporter activity by CHX10 required the CHX10 binding area (CHX10BA) present in fragments 2 and 4 (analysis of variance and a Dunnette’s multiple comparison test, *p < .05). Abbreviations: TS, transcriptional start sites.
To determine whether the CHX10 protein interacts with the OTX2 genomic region in vivo, a chromatin immunoprecipitation (ChIP) analysis was performed. Specific primers were used to detect the presence of several regions of OTX2 genomic DNA, including the CHX10-binding consensus sequence [19, 36]. Several primers, including the CHX10-binding consensus sequences, were studied over the entire OTX2 genomic region. Anti-CHX10 antibody, but not the pre-immune serum, specifically precipitated chromatin containing the OTX2 promoter region (CHX10 binding area, namely CHX10BA in Fig. 3D), but not the 3′UTR region from hRSC progeny (Fig. 3E). These results indicate that CHX10 interacts with OTX2 in hRSC progeny.
To evaluate OTX2 transcriptional regulation by CHX10 in hRSC progeny, we performed a luciferase reporter assay. The luciferase reporter was placed under the control of Otx2 5′ genomic region with or without the CHX10BA (fragment 1-4, Fig. 3D and 3F). The luciferase vector was cotransfected into hRSC progeny with or without the CHX10-expression vector. The activity of the luciferase vector without the genomic OTX2 fragment was taken as 100% (lane 1). Luciferase activity was significantly decreased (F(5,15) = 13.31, p < .05) when the CHX10BA was included in the OTX2 genomic DNA, as seen specifically with fragments two (42.4 ± 4.4%, p < .05) and four (61.2 ± 3.6%, p < .05) (Fig. 3F). These data indicate that CHX10-induced suppression of OTX2 expression required the end of intron two (CHX10BA). In addition, OTX2-fragment2 reporter activity was increased with the cotransfection of the CHX10VP16 expression vector.
Human Retinal Stem Cell Progeny Transfected With CHX10VP16/OTX2/CRX Adopted Photoreceptor Cell Fates More Effectively After In Vivo Transplantation and Contributed to Functional Recovery
To define the potential of retinal stem progeny for photoreceptor replacement in vivo, it is important to test their ability to integrate, migrate, and differentiate into appropriate cell types in the eye. To optimize photoreceptor differentiation from hRSC in vivo, the progeny of hRSCs transfected with CHX10VP16/OTX2/CRX were transplanted into the mouse eye. Control (including only GFP) or CHX10VP16/OTX2/CRX-transduced hRSC progeny were transplanted into the vitreous cavity of postnatal day 1 CD1 mice, as previously described [4, 24]. To suppress tissue rejection, cyclosporine [37] was administered intraperitoneally (i.p.) to animals every day beginning just before the transplantation surgery, and continuing until the hosts were killed. Host mice were sacrificed at 1, 3, or 5 weeks after transplantation, and the number of surviving hRSC progeny were counted at each time point.
A two-way ANOVA revealed significant effects of time (F(2,18) = 9.08, p < .05) and group (F(2,18) = 68.12, p < .05) on cell survival (Fig. 4D). Indeed, at all survival times after the transplant, the CHX10VP16/OTX2/CRX-transduced group had more human cells in the host mouse retina than did the control group, suggesting that the increase in photoreceptors produced by overexpressing the three transcription factors resulted in greater integration and/or early survival of the human photoreceptors. Multiple comparison tests revealed that the group that did not receive cyclosporineA showed a significant decrease in the numbers of cells surviving between 1 and 5 weeks after transplantation (post hoc Dunn’s correction, p < .05). However, the control vector and the CHX10VP16/OTX2/CRX-transduced groups treated with cyclosporine did not show significant differences in surviving cell numbers between week 1 and week 5 after transplantation (p >.05). Indeed, at 1 week after transplantation, similar numbers of human cells were seen in the host mouse eyes in the noncyclosporineA-treated and cyclosporineA-treated control vector groups (p >.05), but at 5 weeks after transplantation into the mouse eye, the transplanted hRSC progeny integrated significantly better with than without i.p. cyclosporinA treatment (p < .05).
Some control transfected hRSC progeny integrated after transplantation into various retinal layers and a few GFP-positive control vector cells also expressed photoreceptor markers. In contrast, hRSC progeny transfected with CHX10VP16/OTX2/CRX showed enhanced survival, and most integrated into the photoreceptor layer and expressed photoreceptor markers (Fig. 4A and 4B). In high magnification images, single donor hRSC integrated into the host retina showed photoreceptor morphology (Fig. 4C). The GFP protein is observed mostly in the inner and outer photoreceptor segments; the cell bodies containing the nucleus in the outer nuclear layer have very little cytoplasm, making it difficult to detect the GFP signal in the cell body, especially in low magnification images. The GFP in most transplanted cells was located in the outer segment region of photoreceptors. Rod photoreceptor differentiation (Rho1D4 positive) was significantly improved in hRSC progeny transfected with CHX10VP16/OTX2/CRX (91.3 ± 3.0% of GFP-positive cells) compared with control transfected cells 5 weeks transplantation (44.8 ± 3.8%, t = 9.70, p < .05, Fig. 4E). In addition, Rom1 staining of differentiated hRSC progeny in dissociated cell culture showed a similar enrichment with CHX10VP16/OTX2/CRX (data not shown). Cone photoreceptor differentiation (human cone arrestin positive) was also promoted in more hRSC progeny transfected with CHX10VP16/OTX2/CRX (2.5 ± 0.5%) than in control transfected cells (0.4 ± 0.4%) (t = 3.12, p < .05, Fig. 4F).
To evaluate whether transplanted hRSCs differentiated into functional photoreceptors in vivo, we used electrophysiologic and behavioral assays to assess visual function in the background of photoreceptor mutant mice 3 months after the transplantation. Human RSC progeny transfected with CHX10VP16/OTX2/CRX or with the control GFP vector were transplanted into the right eye of postnatal day 1 transducin mutant mice [38], which lack functional rod photoreceptors, and were treated with cyclosporine. Transducin mutant mice were chosen because the rod photoreceptors do not die in these mice, they simply do not function.
Since there are no functional rod photoreceptors in the transducin mutant mice, and because rod responses are detected only under low intensity light, the ERG [25] b-wave (bipolar) responses at low light intensities in dark adapted animals should be the best reflection of donor human photoreceptors that have integrated and functionally connected to host mouse bipolar cells. At the high flash intensities at which cone photoreceptors are activated, there was no difference between the groups. However, at the three lowest flash intensities tested (which progressively sample more rod photoreceptor activity), a repeatedly measured ANOVA demonstrated a significant interaction of group and flash intensity (F(4,60) = 20.37, p < .05, Fig. 4G). At the two lowest flash intensities (−3.2 and −2.8) the CHX10VP16/OTX2/CRX-treated group showed a higher response than the nontransplanted (post hoc Dunn’s correction, p < .05) and control GFP vector-treated groups (p < .05). A significant correlation of maximal b wave response (indicative of synaptic connections between donor human photoreceptors and the host mouse bipolar cells) and surviving human photoreceptor cell number in individual eyes was seen (r2 = 0.0372, p < .05, Fig. 4G inset). Control-hRSC transplanted eyes appeared to perform worse than noninjected control eyes, suggesting that the injection procedure itself might damage retina.
Visually guided behavior, that is the ultimate assay of visual function as it indicates that the signal derived from transplanted hRSC progeny can connect to the brain through synapses, was assessed. We used a virtual optomotor task [26] that enables spatial visual thresholds to be measured rapidly and without specific reinforcement training. These experiments were performed under low light illumination for evaluation of rod function. As a within-animal control in our transplantation model, we estimated the difference in spatial frequency resolution between the transplanted eye (right) and nontransplanted eye (left) in individual mice. All of the transplanted eyes showed better spatial frequency resolution than untransplanted eyes. The difference between the two eyes in the lowest spatial frequency detected behaviorally showed a significant positive correlation with human photoreceptor number derived from the transplanted hRSC progeny in individual mice (r2 = 0.9471, p < .05, Fig. 4H). Furthermore, CHX10VP16/OTX2/CRX-hRSCs transplanted eyes revealed better spatial vision compared with control transplanted eyes (t = 5.89, p < .05).
These data indicate that hRSC progeny expressing CHX10VP16/OTX2/CRX can integrate into the host mouse retina and differentiate into photoreceptors more efficiently than control hRSC progeny and promote significant functional electrophysiologic and behavioral recovery.
Discussion
To understand how intrinsic factors lead to the development of specific retinal cells from hRSCs, we analyzed the differentiation activity of several genes (CHX10, CRX, NRL, NEUROD, OTX2, RAX, NEUROGENIN2, and MASH1) that have been shown previously to be important for rodent photoreceptor development [15, 18, 20, 30-34, 39]. Understanding the activity of these genes in human-derived cells is critical for applications of human-retinal stem cell hRSC therapy. Overexpression of CHX10VP16, OTX2, or CRX alone each led hRSC progeny to differentiate into a photoreceptor subtype in vitro, but the overexpression of single genes still led to a rather small effect. However, it was reported previously that Otx2 or Crx overexpression induced efficient photoreceptor differentiation in mouse RSC progeny derived from the ciliary marginal zone [40, 41]. This discrepancy could be caused by differences in culture conditions or a species-specific difference between rodents and humans. Human RSC progeny might be more intrinsically restricted in their response to OTX2 or CRX alone. Crx overexpression in brain neural stem cells [42] or ES cells (data not shown) does not induce photoreceptor-specific markers in vitro, which indicates that other types of stem cells may be unable to respond to retinal transcription factors. RSCs may represent the optimal cell source for producing photoreceptors for transplantation into the eye.
RSCs are quite similar to brain stem cells, which actually produce a minority of neurons (less than 10% of all the differentiating progeny) in vitro. Most in vitro progeny of brain stem cells are glial cells, although in vivo stem cells certainly produce lots of glial progeny. A major focus in brain stem cell biology in the last 15 years has been to try to increase the numbers of neurons produced in vitro from adult brain stem cells. The most successful report with brain stem cells indicated that Pax6 overexpression substantially increased the numbers of neuronal progeny produced from brain stem cells [43]. This finding is consistent to our results that unmodified “hRSCs” yielded rather disappointing numbers of retinal cell types and little in the way of functional benefits, whereas the genetically modified cells did substantially better. The best transcriptional enhancement of photoreceptor development among hRSC progeny was achieved by overexpressing OTX2 and CRX, and converting CHX10 to an activator in vitro. A putative model for this differentiation pathway is shown in Figure 5A. We suggest that the CHX10VP16 blocks progenitor proliferation, thus causing cell cycle exit, and as a result, differentiation is promoted. In late-stage mouse progenitors, CHX10 drives bipolar cell differentiation at the expense of rod formation and CHX10VP16 does the opposite, and in both cases these effects are independent of any influence on proliferation [16]. Thus, in hRSCs, CHX10VP16 may promote photoreceptor formation through effects on both the cell cycle and differentiation. Because clonal hRSC-derived spheres include retinal progenitors that may have the competency to form only subsets of retinal cell types, OTX2 or CRX may induce only a subset of hRSC progeny to differentiate into photoreceptors. We speculate that CHX10VP16 increased the population of competent immature cells by blocking proliferation, and that coexpression of subtype specification factors such as OTX2 and/or CRX then may have biased these cells to adopt a photoreceptor cell fate. Alternatively, the new VP16 construct may have additional effects in RSC progeny besides lowering the level of CHX10. However, the reciprocal changes with CHX10 and CHX10VP16 overexpression are consistent with a simple interpretation of gain and loss of function effects through CHX10. Moreover, the similarity in decreasing retinal progenitor proliferation with CHX10VP16 (that is, smaller spheres) is consistent with the retinal progenitor proliferation deficit seen in the in vivo and in vitro data from CHX10 null mice [16, 28].
Figure 5.
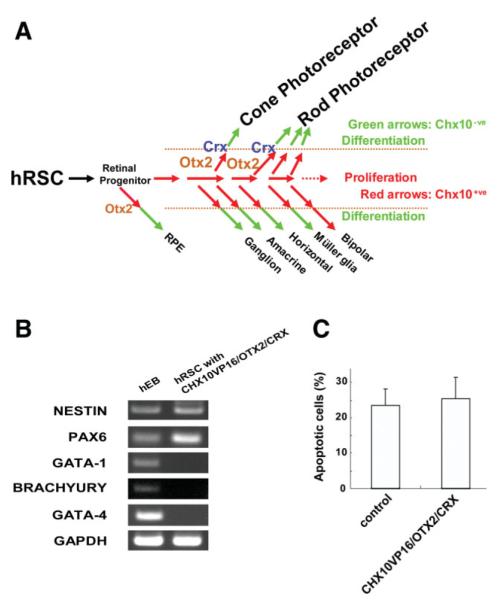
(A): Gene network model for photoreceptor differentiation from human retinal stem cells (hRSCs). CHX10VP16 increases the population of competent immature cells by blocking proliferation, and facilitates the coexpression of subtype specification factors such as OTX2 and/or CRX which serve to bias these cells to adopt a photoreceptor cell fate. (B): Reverse transcription polymerase chain reaction (RT-PCR) lineage analysis and apoptosis in CHX10VP16/OTX2/CRX transfected hRSC progeny. RT-PCR analysis of genes associated with neural (NESTIN, PAX6), mesodermal (GATA-1, BRACHYURY), and endodermal (GATA-4) identity in CHX10VP16/OTX2/CRX transfected hRSC progeny (right). Human embryoid body (hEB) samples were used as positive controls (left). The fate changes seen through transcriptional modulation are only within the retinal lineage. (C): Apoptosis in CHX10VP16/OTX2/CRX transfected hRSC progeny. Human retinal stem cell (hRSC) spheres transfected with control or CHX10VP16/OTX2/CRX were dissociated and subjected to immunocytochemistry for active caspase3 (an apoptotic cell marker). The apoptotic cell number was not significantly increased in CHX10VP16/OTX2/CRX-transfected hRSC progeny versus control (t = 0.24, p >.05). Abbreviations: hEB, human embryoid body; hRSC, human retinal stem cell; RPE, retinal progenitor.
Another possibility is that upregulation of OTX2 transcription levels by modulating CHX10 function might promote the photoreceptor cell lineage. We find that CHX10 directly binds to the OTX2 genomic locus in hRSC progeny and suppresses OTX2 expression, and that OTX2 mRNA levels are upregulated by CHX10VP16 expression. Furthermore, CRX mRNA levels were also upregulated by CHX10VP16 overexpression, and Otx2 was a direct upstream regulator of Crx in the mouse [20]. Indeed, in hRSC progeny overexpressing OTX2 and CRX, mRNA expression was upregulated. This type of disinhibitory and direct facilitatory regulatory network might amplify photoreceptor differentiation from hRSC progeny through feed-forward mechanisms. Otx2 and Chx10 were apparently coexpressed in the same single bipolar cells in the developing and adult retina [44]. Although this finding is not consistent with the simplest version of the present model, the suppression of Chx10 by Otx2 could be cell-type specific (that is, only in proliferating retinal precursors). Furthermore, Crx was reported to be expressed in bipolar cells along with Chx10 [45], and Crx expression was developmentally delayed in the Chx10-deficient mouse [46]. This later discrepancy can be explained easily, given that most Crx is expressed in photoreceptors. The delay of Crx expression in the Chx10 mutant mouse retinal may be an artifact of the delayed development of the retina in this mouse. More important, bipolar cells were almost completely absent in the smaller retina of the Chx10 mutant, which provides an alternative explanation for the lower levels of Crx—one of the cell types normally expressing Crx was missing.
Nevertheless, the fate changes seen through transcriptional modulation are only within the retinal lineage. RT-PCR analyses showed that neural lineage markers were maintained in human retinal stem and progenitor cells transfected with CHX10VP16/OTX2/CRX, whereas mesodermal and endodermal markers were not revealed (Fig. 5B). In addition, desmin (a muscle marker) and cytokeratin17 (an epithelial marker) were not detected in CHX10VP16/OTX2/CRX hRSC progeny by immunocytochemistry (data not shown). Thus, transfection of these genes did not change the retinal cell fates of the proliferating human retinal stem and progenitor cells. Furthermore, apoptotic cell number (assayed by immunostaining for active caspase3) was not affected in CHX10VP16/OTX2/CRX-transfected hRSC progeny as compared with control (Fig. 5C). This finding indicates that apoptosis was not promoted in CHX10VP16/OTX2/CRX gene-induced hRSC progeny. These data suggest that the transcriptional enrichment for photoreceptors from hRSC progenys is caused by a fate change within the retinal progeny rather than a selective survival effect.
With various candidate transplantable cells derived from hRSCs, embryonic retinal precursor cells [47] or human embryonic stem cells [48, 49], the problem of immunologic rejection remains to be resolved [50]. Although transcriptionally modified hRSC progeny appeared to integrate and survive well in the host mouse retina, they required immunologic suppression to avert severe immunologic rejection. Thus, the availability of an autologous stem cell source would offer a large advantage for future clinical therapy. In this respect, the use of autologous hRSCs after expansion and differentiation in culture may be an ideal therapy for human retinal disease. In addition to these cell-replacement therapies, another possibility is that inactive endogenous hRSCs may be stimulated by drugs or gene therapy. Grafted hRSC progeny should be considered for stem cell therapies given that they can successfully integrate without serious pathologic complications such as cancer. These cells do not appear to show pro-longed proliferation in the host animal as the proliferation marker (Ki67) was not detected in transplanted hRSC 5 weeks after surgery (data not shown). This result suggests that once RSC progeny enter into the retinal environment, they migrate and undergo proper differentiation without excessive proliferation or layer disruption. These more differentiated cells integrated as single cells in the outer nuclear layer, whereas the earlier precursor cells transplanted here tended to integrate more in clumps in the outer nuclear layer, perhaps because of their earlier differentiation state or because of proliferation of the donor cells in the outer nuclear layer in situ. The present genetically modified hRSC progeny may be in an optimal differentiation state for integration.
A sufficient number of hRSC photoreceptor progeny transplanted into the transducin mutant retina in vivo produced light responsiveness and made functional synaptic connections with rod bipolar cells, and could re-establish synaptic communication in the retina (and more important, with the brain) to improve spatial resolution. Excellent integration, differentiation, and function of single murine rod precursors selected on the basis of NRL expression has been reported [51]. Our previous report [4] indicated that the hRSCs transplanted to mouse retina show considerable GFP in the outer segments that is colocalized with Rom1, an outer segment marker. The preferential distribution of GFP protein to the segments is similar to what happens with the distribution of rhodopsin-most protein in the segments. We assume that MacLaren et al. [51] had higher expression of GFP in their rodent retinal precursor than we did in our human retinal stem cell progeny. Moreover, the small numbers (hundreds) of transplanted mouse rods were able to rescue a papillary light response in blind rd1-/- mice in these studies. We also see some rescue of vision (ERG and optomotor task) with similarly small numbers of transplanted donor human photoreceptors in transducin-/- mice. It may seem surprising that transplanted human rods and a very small number of transplanted human cones can improve behavior in on optomotor task that presumably assays cones function. However, in cone transducin knockout mouse, rods mediate visual behavior (at low-light intensities in dark-adapted animals) in the optomotor tasks at about a third the acuity of cones (as assessed in rod transducin knockout mice) (Prusky, unpublished data). Although we suggest that these functional results reveal the phototransduction function of the donor hRSC derived rods in the mouse eye, an alternative explanation might suggest a noncell-autonomous effect of the transplanted human cells on the survival of host cells and/or the preservation of early host developmental connections. Nevertheless, the selective improvement of ERG function at low-light intensities, where only (transplanted human rod) function should be sampled, speaks against a more general noncell-autonomous effect on the host mouse retina.
In summary, the present in vitro and in vivo results together demonstrate that appropriate modulation of retinal transcription increases the potential of hRSC progeny as substrates for the treatment of human photoreceptor disease.
Supplementary Material
Acknowledgments
We thank Dr. Hiroyuki Miyoshi for pCSII-EF plasmid, Dr. Janis Lem for transducin mutant mice, and Dr. Tamara Holowacz for help preparing the manuscript. This study was funded by: NIH, Canadian Institutes of Health Research, Lincy Foundation, Foundation Fighting Blindness of Canada, Canadian Stem Cell Network, Steinbach Foundation, and Japan Society for the Promotion of Science.
Footnotes
Disclosure of Potential Conflicts of Interest The authors indicate no potential conflicts of interest.
REFERENCES
- 1.Cepko CL. The roles of intrinsic and extrinsic cues and bHLH genes in the determination of retinal cell fates. Curr Opin Neurobiol. 1999;9:37–46. doi: 10.1016/s0959-4388(99)80005-1. [DOI] [PubMed] [Google Scholar]
- 2.Livesey FJ, Cepko CL. Vertebrate neural cell-fate determination: Lessons from the retina. Nat Rev Neurosci. 2001;2:109–118. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- 3.Tropepe V, Coles BL, Chiasson BJ, et al. Retinal stem cells in the adult mammalian eye. Science. 2000;287:2032–2036. doi: 10.1126/science.287.5460.2032. [DOI] [PubMed] [Google Scholar]
- 4.Coles BL, Angenieux B, Inoue T, et al. Facile isolation and the characterization of human retinal stem cells. Proc Natl Acad Sci U S A. 2004;101:15772–15777. doi: 10.1073/pnas.0401596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahmad I, Tang L, Pham H. Identification of neural progenitors in the adult mammalian eye. Biochem Biophys Res Commun. 2000;270:517–521. doi: 10.1006/bbrc.2000.2473. [DOI] [PubMed] [Google Scholar]
- 6.Gao J, Cheon K, Nusinowitz S, et al. Progressive photoreceptor degeneration, outer segment dysplasia, and rhodopsin mislocalization in mice with targeted disruption of the retinitis pigmentosa-1 (Rp1) gene. Proc Natl Acad Sci U S A. 2002;99:5698–5703. doi: 10.1073/pnas.042122399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewis GP, Charteris DG, Sethi CS, et al. The ability of rapid retinal reattachment to stop or reverse the cellular and molecular events initiated by detachment. Invest Ophthalmol Vis Sci. 2002;43:2412–2420. [PubMed] [Google Scholar]
- 8.Johnson PT, Lewis GP, Talaga KC, et al. Drusen-associated degeneration in the retina. Invest Ophthalmol Vis Sci. 2003;44:4481–4488. doi: 10.1167/iovs.03-0436. [DOI] [PubMed] [Google Scholar]
- 9.Cicero SA, Johnson D, Reyntjens S, et al. Cells previously identified as retinal stem cells are pigmented ciliary epithelial cells. Proc Natl Acad Sci U S A. 2009;106:6685–6690. doi: 10.1073/pnas.0901596106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu S, Sunderland ME, Coles BL, et al. The proliferation and expansion of retinal stem cells require functional Pax6. Dev Biol. 2007;304:713–721. doi: 10.1016/j.ydbio.2007.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyoshi H, Blomer U, Takahashi M, et al. Development of a self-inactivating lentivirus vector. J Virol. 1998;72:8150–8157. doi: 10.1128/jvi.72.10.8150-8157.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tahara-Hanaoka S, Sudo K, Ema H, et al. Lentiviral vector-mediated transduction of murine CD34(-) hematopoietic stem cells. Exp Hematol. 2002;30:11–17. doi: 10.1016/s0301-472x(01)00761-5. [DOI] [PubMed] [Google Scholar]
- 13.Inoue T, Hojo M, Bessho Y, et al. Math3 and NeuroD regulate amacrine cell fate specification in the retina. Development. 2002;129:831–842. doi: 10.1242/dev.129.4.831. [DOI] [PubMed] [Google Scholar]
- 14.Hatakeyama J, Kageyama R. Retinal cell fate determination and bHLH factors. Semin Cell Dev Biol. 2004;15:83–89. doi: 10.1016/j.semcdb.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 15.Burmeister M, Novak J, Liang MY, et al. Ocular retardation mouse caused by Chx10 homeobox null allele: Impaired retinal progenitor proliferation and bipolar cell differentiation. Nat Genet. 1996;12:376–384. doi: 10.1038/ng0496-376. [DOI] [PubMed] [Google Scholar]
- 16.Livne-Bar I, Pacal M, Cheung MC, et al. Chx10 is required to block photoreceptor differentiation but is dispensable for progenitor proliferation in the postnatal retina. Proc Natl Acad Sci U S A. 2006;103:4988–4993. doi: 10.1073/pnas.0600083103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dorval KM, Bobechko BP, Ahmad KF, Bremner R. Transcriptional activity of the paired-like homeodomain proteins CHX10 and VSX1. J Biol Chem. 2005;280:10100–10108. doi: 10.1074/jbc.M412676200. [DOI] [PubMed] [Google Scholar]
- 18.Dorval KM, Bobechko BP, Fujieda H, et al. CHX10 Targets a Subset of Photoreceptor Genes. J Biol Chem. 2006;281:744–751. doi: 10.1074/jbc.M509470200. [DOI] [PubMed] [Google Scholar]
- 19.Walker S, Greaves R, O’Hare P. Transcriptional activation by the acid domain of Vmw65 requires the integrity of the domain and involves additional determinants distinct from those necessary for TFIIB binding. Mol Cell Biol. 13:5223–5244. doi: 10.1128/mcb.13.9.5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishida A, Furukawa A, Koike C, et al. Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat Neurosci. 2003;6:1255–1263. doi: 10.1038/nn1155. [DOI] [PubMed] [Google Scholar]
- 21.Furukawa T, Morrow EM, Li T, et al. Retinopathy and attenuated circadian entrainment in Crx-deficient mice. Nat Genet. 1999;23:466–470. doi: 10.1038/70591. [DOI] [PubMed] [Google Scholar]
- 22.Bremner R, Cohen BL, Sopta M, et al. Direct transcriptional repression by pRB and its reversal by specific cyclins. Mol Cell Biol. 1995;15:3256–3265. doi: 10.1128/mcb.15.6.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shang Y, Hu X, DiRenzo J, et al. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 24.Young MJ, Ray J, Whiteley SJ, et al. Neuronal differentiation and morphological integration of hippocampal progenitor cells transplanted to the retina of immature and mature dystrophic rats. Mol Cell Neurosci. 2000;16:197–205. doi: 10.1006/mcne.2000.0869. [DOI] [PubMed] [Google Scholar]
- 25.Tremblay F, Abdel-Majid R, Neumann PE. Electroretinographic oscillatory potentials are reduced in adenylyl cyclase type I deficient mice. Vision Res. 2002;42:1715–1725. doi: 10.1016/s0042-6989(02)00113-x. [DOI] [PubMed] [Google Scholar]
- 26.Prusky GT, Alam NM, Beekman S, Douglas RM. Rapid quantification of adult and developing mouse spatial vision using a virtual optomotor system. Invest Ophthalmol Vis Sci. 2004;45:4611–4616. doi: 10.1167/iovs.04-0541. [DOI] [PubMed] [Google Scholar]
- 27.Rowan S, Cepko CL. Genetic analysis of the homeodomain transcription factor Chx10 in the retina using a novel multifunctional BAC transgenic mouse reporter. Dev Biol. 2004;271:388–402. doi: 10.1016/j.ydbio.2004.03.039. [DOI] [PubMed] [Google Scholar]
- 28.Coles BL, Horsford DJ, McInnes RR, van der Kooy D. Loss of retinal progenitor cells leads to an increase in the retinal stem cell population in vivo. Eur J Neurosci. 2006;23:75–82. doi: 10.1111/j.1460-9568.2005.04537.x. [DOI] [PubMed] [Google Scholar]
- 29.Tropepe V, Hitoshi S, Sirard C, et al. Direct neural fate specification from embryonic stem cells: A primitive mammalian neural stem cell stage acquired through a default mechanism. Neuron. 2001;30:65–78. doi: 10.1016/s0896-6273(01)00263-x. [DOI] [PubMed] [Google Scholar]
- 30.Mears AJ, Kondo M, Swain PK, et al. Nrl is required for rod photoreceptor development. Nat Genet. 2001;29:447–452. doi: 10.1038/ng774. [DOI] [PubMed] [Google Scholar]
- 31.Morrow EM, Furukawa T, Lee JE, Cepko CL. NeuroD regulates multiple functions in the developing neural retina in rodent. Development. 1999;126:23–36. doi: 10.1242/dev.126.1.23. [DOI] [PubMed] [Google Scholar]
- 32.Kimura A, Singh D, Wawrousek EF, et al. Both PCE-1/RX and OTX/CRX interactions are necessary for photoreceptor-specific gene expression. J Biol Chem. 2000;275:1152–1160. doi: 10.1074/jbc.275.2.1152. [DOI] [PubMed] [Google Scholar]
- 33.Akagi T, Inoue T, Miyoshi G, et al. Requirement of multiple basic helix-loop-helix genes for retinal neuronal subtype specification. J Biol Chem. 2004;279:28492–28498. doi: 10.1074/jbc.M400871200. [DOI] [PubMed] [Google Scholar]
- 34.Hatakeyama J, Tomita K, Inoue T, Kageyama R. Roles of homeobox and bHLH genes in specification of a retinal cell type. Development. 2001;128:1313–1322. doi: 10.1242/dev.128.8.1313. [DOI] [PubMed] [Google Scholar]
- 35.Tomita K, Nakanishi S, Guillemot F, Kageyama R. Mash1 promotes neuronal differentiation in the retina. Genes Cells. 1996;1:765–774. doi: 10.1111/j.1365-2443.1996.tb00016.x. [DOI] [PubMed] [Google Scholar]
- 36.Percin E Ferda, Ploder LA, Yu JJ, et al. Human microphthalmia associated with mutations in the retinal homeobox gene CHX10. Nat Genet. 2000;25:397–401. doi: 10.1038/78071. [DOI] [PubMed] [Google Scholar]
- 37.DiLoreto D, Jr., del Cerro C, del Cerro M. Cyclosporine treatment promotes survival of human fetal neural retina transplanted to the sub-retinal space of the light-damaged Fischer 344 rat. Exp Neurol. 1996;140:37–42. doi: 10.1006/exnr.1996.0112. [DOI] [PubMed] [Google Scholar]
- 38.Calvert PD, Krasnoperova NV, Lyubarsky AL, et al. Phototransduction in transgenic mice after targeted deletion of the rod transducin alpha - subunit. Proc Natl Acad Sci U S A. 2000;97:13913–13918. doi: 10.1073/pnas.250478897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Furukawa T, Morrow EM, Cepko CL. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell. 1997;91:531–541. doi: 10.1016/s0092-8674(00)80439-0. [DOI] [PubMed] [Google Scholar]
- 40.Akagi T, Mandai M, Ooto S, et al. Otx2 homeobox gene induces photoreceptor-specific phenotypes in cells derived from adult iris and ciliary tissue. Invest Ophthalmol Vis Sci. 2004;45:4570–4575. doi: 10.1167/iovs.04-0697. [DOI] [PubMed] [Google Scholar]
- 41.Jomary C, Jones SE. Induction of functional photoreceptor phenotype by exogenous Crx expression in mouse retinal stem cells. Invest Ophthalmol Vis Sci. 2008;49:429–437. doi: 10.1167/iovs.07-0812. [DOI] [PubMed] [Google Scholar]
- 42.Haruta M, Kosaka M, Kanegae Y, et al. Induction of photoreceptor-specific phenotypes in adult mammalian iris tissue. Nat Neurosci. 2001;4:1163–1164. doi: 10.1038/nn762. [DOI] [PubMed] [Google Scholar]
- 43.Hack MA, Sugimori M, Lundberg C, et al. Regionalization and fate specification in neurospheres: The role of Olig2 and Pax6. Mol Cell Neurosci. 2004;25:664–678. doi: 10.1016/j.mcn.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 44.Baas D, Bumsted KM, Martinez JA, et al. The subcellular localization of Otx2 is cell-type specific and developmentally regulated in the mouse retina. Brain Res Mol Brain Res. 2000;78:26–37. doi: 10.1016/s0169-328x(00)00060-7. [DOI] [PubMed] [Google Scholar]
- 45.Bibb LC, Holt JK, Tarttelin EE, et al. Temporal and spatial expression patterns of the CRX transcription factor and its downstream targets. Critical differences during human and mouse eye development. Hum Mol Genet. 2001;10:1571–1579. doi: 10.1093/hmg/10.15.1571. [DOI] [PubMed] [Google Scholar]
- 46.Rutherford AD, Dhomen N, Smith HK, Sowden JC. Delayed expression of the Crx gene and photoreceptor development in the Chx10-deficient retina. Invest Ophthalmol Vis Sci. 2004;45:375–384. doi: 10.1167/iovs.03-0332. [DOI] [PubMed] [Google Scholar]
- 47.Chacko DM, Rogers JA, Turner JE, Ahmad I. Survival and differentiation of cultured retinal progenitors transplanted in the subretinal space of the rat. Biochem Biophys Res Commun. 2000;268:842–846. doi: 10.1006/bbrc.2000.2153. [DOI] [PubMed] [Google Scholar]
- 48.Lamba DA, Karl MO, Ware CB, Reh TA. Efficient generation of retinal progenitor cells from human embryonic stem cells. Proc Natl Acad Sci U S A. 2006;103:12769–12774. doi: 10.1073/pnas.0601990103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lamba DA, Gust J, Reh TA. Transplantation of human embryonic stem cell-derived photoreceptors restores some visual function in Crx-deficient mice. Cell Stem Cell. 2009;4:73–79. doi: 10.1016/j.stem.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Drukker M, Katz G, Urbach A, et al. Characterization of the expression of MHC proteins in human embryonic stem cells. Proc Natl Acad Sci U S A. 2002;99:9864–9869. doi: 10.1073/pnas.142298299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.MacLaren RE, Pearson RA, MacNeil A, et al. Retinal repair by transplantation of photoreceptor precursors. Nature. 2006;444:203–207. doi: 10.1038/nature05161. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.