Structural insight into the oxidation-sensing mechanism of the antibiotic resistance of regulator MexR
The crystal structure of oxidized MexR, including the formation of interprotomer disulphide bonds, gives insight into the molecular mechanism of MexR-based oxidation-sensing and the regulation of antibiotic resistance in Pseudomonas aeruginosa.
Keywords: antibiotic resistance regulation, oxidative stress, Pseudomonas aeruginosa, redox sensing, thiol oxidation
Abstract
MexR functions as the primary regulator of the mexAB–oprM multidrug efflux expression in Pseudomonas aeruginosa. It has been shown that MexR senses oxidative stress by interprotomer disulphide bond formation between redox-active cysteines. This oxidation induces MexR to dissociate from the promoter DNA, thus activating the transcriptional expression of efflux pump genes. In this study, we present the crystal structure of MexR in its oxidized form at a resolution of 2.1 Å. This crystal structure reveals the mechanism by which oxidative signal allosterically derepresses the MexR-controlled transcription activation.
Introduction
The opportunistic human pathogen Pseudomonas aeruginosa is intrinsically resistant to many antibiotics and can acquire high levels of multidrug resistance (Poole, 2001). This Gram-negative bacterium possesses several multidrug efflux pump systems. The principal system contributing to intrinsic multidrug resistance is encoded by the mexAB–oprM operon (Poole, 2004). It has been shown that this multiprotein complex exports a wide variety of clinically used antibiotics, including quinolones, β-lactams, tetracycline, chloramphenicol and novobiocin, as well as macrolides and biocides, to the outer cellular space. This efflux system not only has an important role in the innate, multidrug-resistant phenotype exhibited by P. aeruginosa, but it also acts as a model system by which to understand the function and regulation of multidrug resistance in human pathogens in general. There have also been attempts to target this efflux system for the inhibition of its drug exportation function (Pagès et al, 2005; Blair & Piddock, 2009).
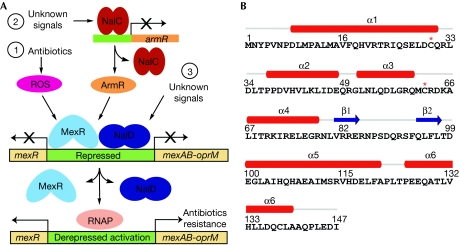
Over the past decade, extensive efforts have been made to study the regulation of the mexAB–oprM efflux system (Schweizer, 2003). Three regulatory genes, mexR, nalC and nalD, were identified as repressors of this efflux system. As shown in Fig 1A, MexR and NalD (Morita et al, 2006) repress the transcription of mexAB–oprM operon through binding directly to the promoter sites. NalC, a repressor of armR, also regulates this efflux pump expression but in an indirect manner (Cao et al, 2004). The overexpression of ArmR derepresses the MexR-controlled efflux pump by direct binding of ArmR to MexR, which prevents the binding of MexR to the promoter DNA (Wilke et al, 2008). As a result, P. aeruginosa exhibits increased multidrug resistance owing to the overproduction of efflux pumps. At present, the signal or inducer responsible for NalC activation is unknown.
Figure 1.
Pseudomonas aeruginosa MexR function and structure. (A) Transcriptional regulation of mexAB–oprM efflux expression. Three pathways are presented to mediate mexAB–oprM transcriptional expression. MexR uses two independent mechanisms in response to stress. (B) MexR protein sequence and secondary structure assignment. The two redox-active cysteines Cys 30 and Cys 62 are indicated by a red star. RNAP, RNA polymerase; ROS, reactive oxygen species.
MexR acts as the primary repressor of the mexAB–oprM multidrug efflux operon in P. aeruginosa (Poole & Srikumar, 2001). It belongs to the MarR family of transcriptional regulators that sense and respond to environmental changes and regulate bacterial resistance to various challenges (Ellison & Miller, 2006). In addition to the NalC/ArmR-mediated pathway, we have recently shown that MexR is redox active and can sense oxidative stress (Chen et al, 2008). This discovery solves a chemical puzzle: how can a small regulatory protein, such as MexR, recognize the presence of a wide range of antibiotics inside bacterial cells? The answer is simple: the protein senses oxidative stress generated from the bactericidal effect of various antibiotics (Kohanski et al, 2007). To the best of our knowledge, this discovery, for the first time, directly links oxidative regulation to multidrug efflux gene expression in P. aeruginosa.
We have observed that two cysteines in MexR, Cys 30 and Cys 62, which are located on helix α1 and in the loop between helices α3 and α4, respectively (Lim et al, 2002), are involved in the formation of interprotomer disulphide bonds under oxidative conditions in vitro and inside the microbe. This oxidation modification induces the dissociation of MexR from the promoter DNA and activation of the mexAB–oprM operon (Chen et al, 2008). However, this result leaves some doubts about the oxidation-sensing pathway and the detailed mechanism that leads to the dissociation of the oxidized MexR from the promoter DNA. An isolation and structural characterization of the oxidized MexR would firmly confirm the proposed oxidation pathway and reveal details about the activation mechanism.
Results And Discussion
Oxidized MexR construction and isolation
To obtain high-quality oxidized MexR crystals, different oxidants were tested carefully for the preparation of homogenous oxidized MexR on a large scale suitable for crystallography. Under mild conditions in vitro, MexR can be oxidized readily by oxidants such as hydrogen peroxide (H2O2), cumene hydroperoxide and glutathione disulphide (Chen et al, 2008); oxidized MexR can also be generated slowly through air-induced oxidation (supplementary Fig S1A online). However, we observed that 2,2′-dithiodipyridine was most effective for preparing disulphide-linked MexR dimer that can be readily isolated and crystallized. Typically, 10 μM MexR protomer was incubated with a stoichiometry mole ratio of 2,2′-dithiodipyridine for 30 min at room temperature (25°C). The oxidized MexR dimer was isolated from reduced MexR by ion-exchange chromatography. The purified fractions were confirmed by performing a 14% non-reduced SDS–polyacrylamide gel electrophoresis analysis and combined for crystallization experiments (supplementary Fig S1B online).
Crystallization of the oxidized MexR
We constructed a mutant, MexRC138S, to avoid potential side oxidation reactions on this residue, thus leaving only the two redox-active cysteines Cys 30 and Cys 62. In addition, five carboxy-terminal amino-acid residues were truncated to improve the quality of crystal diffraction. The structure of the oxidized MexR was determined at a resolution of 2.1 Å by molecular replacement using one protomer of the reduced MexR dimer (Lim et al, 2002) as the search model. Each asymmetric unit contains one oxidized MexR dimer. The final refined model contains residues 1–142 of each protomer, and shows good stereochemistry with 97% of residues in the most favourable region of the Ramachandran plot. Detailed parameters are summarized in supplementary Table S1 online.
Overall folding of the oxidized MexR
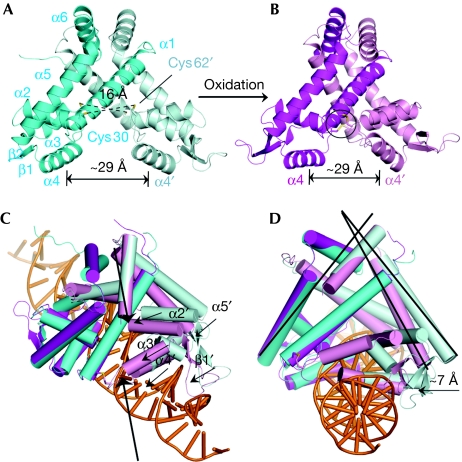
The overall oxidized MexR structure resembles the reduced apo-MexR (Figs 1B,2A,B), but its structural feature differs markedly in the details. In contrast to the four separate conformations observed in the reduced MexR (denoted AB, CD, EF and GH; Lim et al, 2002), the oxidized structure contains only one physiologically relevant dimer in each asymmetric unit. After oxidation, two interprotomer disulphide linkages, Cys30–Cys62′ and Cys30′–Cys62, are visualized clearly in the refined final model with good electron densities (Figs 2B,3A). Disulphide bond formation does not alter the spacing between the two DNA-binding helices α4 and α4′, which remains at ∼29 Å, similarly to that of one of the reduced apo-MexR structures, the CD dimer (Fig 2A,B). It should be noted that the CD dimer might represent the DNA-binding conformation (Lim et al, 2002); however, there are probably additional adjustments made when the protein actually binds to DNA. Superimpositions of all four structures in reduced state and oxidized MexR reveal that their DNA-binding domains have a similar orientation (supplementary Fig S2 online). The oxidized MexR was docked manually onto operator DNA (Fig 2C,D; supplementary Fig S3 online). This model suggests that the two DNA-binding helices α4 and α4′ can still interact with the major groove on the same face of the duplex DNA. In addition, the two winged-hairpin motifs (β1′–β2′) seem to be well positioned to interact properly with the DNA minor groove or phosphate backbone. However, the spacing for the DNA-binding domains is squeezed towards each other owing to the oxidative formation of the interprotomer disulphides. As shown in Fig 2C,D, several motifs in the unaligned protomer move towards DNA, such as helices α1′–α5′ and winged β1′–β2′. These overall motions prevent the oxidized MexR from binding to DNA. In local regions, this oxidation also leads to large structural changes in the helix–turn–helix (HTH) DNA-binding regulatory domain (α3′–α4′; Fig 3B). The rigid body rotation of helices α2′ and α3′, for example, results in a direct clash between helix α2′ and the DNA backbone (supplementary Fig S4 online). Overall, the newly formed disulphide bonds cause severe steric clashes with the DNA phosphate backbone, thereby significantly reducing the affinity of the oxidized MexR for DNA and locking MexR into a conformation such that high-affinity DNA binding cannot take place.
Figure 2.
Overall structural feature of oxidized MexR homodimer. (A) A representation of reduced apo-MexR CD dimer capable of DNA binding (PDB 1LNW). One protomer is shown in cyan and the other in pale cyan. The oxidizable cysteine residues Cys 30 and Cys 62 are shown as sticks and the distances between two corresponding sulphurs are indicated. The spacing of major-groove-binding motif α4 and α4′ is measured by the Cα-Cα distance for Arg 73 and Arg 73′. (B) Oxidized MexR homodimer with one protomer shown in magenta and the other in pink. The two interprotomer disulphide bonds are highlighted. (C) The modelling and structural implication of oxidized MexR binding to double stranded DNA showing that oxidized MexR adopts a conformation not suitable for DNA binding (the procedures to generate the model are described in the supplementary information online; supplementary Fig S3 online). Duplex DNA is adopted from PDB 1Z9C and coloured orange. The steric clashes are indicated with bold arrows, and motions of the motifs α1′–α5′ and β1′–β2′ in the unaligned protomer between dimer CD and oxidized MexR are shown by black arrows. (D) The side view of the docked model in (C) showing the movement of the DNA-binding domains in the oxidized MexR as compared with the reduced state. The estimated overall movement of the unaligned protomer in the oxidized MexR compared with that in the reduced dimer CD is indicated by the arrow. PDB, Protein Data Bank.
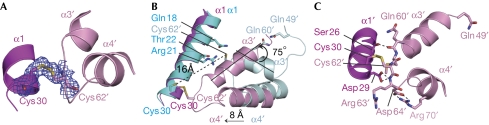
Figure 3.
A close view of the helix–turn–helix region in oxidized MexR. Magenta, pink, cyan and pale cyan each indicate a promoter. (A) The σA weighted 2Fo–Fc electron density map contoured at 1σ around the disulphide binding between Cys 30 and Cys 62′. (B) Close-up view of superimposition of oxidized MexR and reduced dimer CD showing conformational change around the HTH region before and after MexR oxidation. Atoms are coloured blue (nitrogen), red (oxygen) and yellow (sulphur) and the distance between Cys 30 and Cys 62′ is shown by black dashed lines. After oxidation, the rigid body rotation angle of helix α3′ and the movement of helix α4′ are shown. Key residues surrounding Cys 62′ in dimer CD structure (PDB 1LNW) are labelled. (C) Formation of new interactions in the oxidized MexR. The formation of the disulphide bond between Cys 30 and Cys 62′ is accompanied by newly formed interactions between the HTH motif and helix α1, including the hydrogen bonding of Ser 26 side chain and Gln 60′ backbone carbonyl, Asp 29 side chain and Arg 63′ backbone amide, and a salt bridge between the side chains of Asp 64′ and Arg 70′. HTH, helix–turn–helix.
The oxidized conformation with disulphide bond
An allosteric mechanism for the oxidative formation of the disulphide bonds could be proposed on the basis of a comparison between reduced and oxidized MexR dimer structures. In reduced MexR, Gln 60′ at helix α3′ forms a hydrogen bond with Gln 49′ at helix α2′, which helps to keep the two redox-sensitive cysteines (Cys 30 and Cys 62′) apart by approximately 16 Å (Figs 2A,3B). A significant structural change is therefore necessary to bring them close enough to form a covalent disulphide bond. Cys 62 seems to be located in an environment more prone to oxidation. The side chain of Cys 62(′) can form direct hydrogen bonds with the side chains of Gln 18′, Thr 22′ and the backbone carbonyl of Arg 21 in different protomer structures of the reduced MexR dimer (Fig 3B; supplementary Fig S5A–H online). Two basic residues, Arg 21′ and Arg 63, are also in close vicinity, possibly to keep the thiol of Cys 62(′) in a de-protonated state. Upon oxidation, the hydrogen bonds to the other protomer might be disrupted, thus inducing a conformational change towards formation of the disulphide bond with Cys 30(′) from the other protomer. A biochemical study showed that the in vitro oxidation of Cys 62 was faster than that of Cys 30 (supplementary Fig S5K online), implying that oxidation of Cys 62 would probably first produce a sulphenic acid intermediate. The addition of an oxygen to the side-chain sulphur of residue Cys 62(′) might cause a steric clash with the surrounding residues, consequently favouring helix α3′ to rotate approximately 75° counterclockwise for this sulphenic acid intermediate of Cys 60, approaching residue Cys 30′ located in the other protein protomer (Fig 3B). This rigid body rotation is transmitted to the DNA-binding helix α4′ that moves 8 Å towards Cys 30 in the amino-terminal direction. After oxidation, the environment around the covalent disulphide bond is altered completely (Fig 3C). Tightly packed interactions and added stabilization are observed, including the side chain of Asp 29 (α1) to polypeptide amide of a flexible loop (between α3′and α4′), the side chain of Ser 26 to Asn 60′ carbonyl and the side-chain interaction between Asp 64′ and Arg 70′. All these structural elements ensure that helix α1 and the α3′–α4′ HTH contact each other properly to lock the oxidized MexR into a conformation that cannot bind to DNA.
Comparison with the AmrR-bound MexR
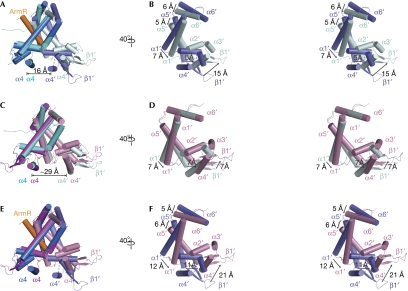
In addition to the ability of MexR to sense reactive oxygen species (ROS) as signals, a recent structural study shows that the binding of AmrR prevents MexR from recognizing the cognate DNA (Wilke et al, 2008). With the structures of the AmrR-bound and the oxidized MexR available, the two regulation mechanisms can now be compared. An overall alignment of the ArmR-bound MexR with the reduced MexR gives a core r.m.s.d. value of 2.8 Å2 (Fig 4A,B). This relatively large r.m.s.d. value indicates that a large conformational change takes place on ArmR binding (Fig 4B). To accommodate ArmR binding, the region in which MexR dimerization occurs must undergo significant conformational change, involving rigid body rotation and additional movements. The dimerization domain connects to the DNA-binding domain. On AmrR binding, the spacing between the DNA major-groove-binding helices of MexR is reduced greatly to 16 Å from 29 Å in the reduced MexR dimer. By contrast, the superimposition of oxidized MexR and reduced MexR results in a small r.m.s.d. value of 1.6 Å2, indicating a relatively small conformational change in the overall oxidized MexR (Fig 4C,D). Remarkably, oxidative formation of disulphide bonds does not much affect the orientation of the dimerization region, nor does it change the distance between the two α4–α4′ DNA major-groove-recognition helices when compared with that in the reduced form. Rather, oxidation directly induces a conformational change in the local HTH region to prevent oxidized MexR from binding to the promoter DNA. An overall structure-based alignment of antirepressor-bound MexR with oxidized MexR also demonstrates a big core r.m.s.d. value of 2.4 Å2 for 886 atoms (Fig 4E,F). Thus, the structure shown here, together with our previous biochemical and microbiological studies (Chen et al, 2008), confirm that there are two distinct cellular regulatory pathways that exist to negatively modulate MexR-controlled, multidrug-resistant efflux expression in P. aeruginosa (Fig 1A). In the oxidation-based regulation pathway, the transition from the oxidized state to the reduced state can be accomplished through reduction. Additional studies are necessary to probe the interesting question about the fate of the AmrR-bound MexR.
Figure 4.
Structural differences between ArmR-bound and oxidized MexR dimers. The positions of the DNA minor-groove-binding wing (β1′) and the helices, α1′, α2′, α3′, α4′, α5′ and α6′, in the unaligned protomers are labelled. All superimpositions were performed in PyMOL with one protomer aligned from each dimer. (A) Superimposition of ArmR-bound MexR and dimer CD with a core r.m.s.d. value of 2.8 Å2. The ArmR-bound MexR is shown with one protomer in marine and the other in slate, and the ArmR peptide in orange. The disordered loop between helices α3′ and α4′ in the ArmR-bound MexR is indicated by a dashed line. The distance between helices α4 and α4′ in the ArmR-bound MexR is 16 Å. (B) Stereoview of the unaligned protomer, as in (A), rotated 40° to the right. The conformational change of the DNA minor-groove-binding wing (β1′) and the dimerization helices, α1′, α5′ and α6′, is indicated with arrows. (C) Superimposition of the oxidized MexR and dimer CD to a core r.m.s.d. value of 1.6 Å2. The distance between helices α4 and α4′ in dimer CD is 29 Å. (D) Side view of the unaligned protomer as in (C) in stereo, rotated 40° to the right, revealing the conformational change due to the disulphide bonds formation. The conformational change of the DNA minor-groove-binding wing (β1′) and the dimerization helices, α1′ and α5′, is indicated with arrows. (E) Superimposition of the ArmR-bound and oxidized MexR dimers with a core r.m.s.d. value of 2.4 Å2. (F) Stereoview of the unaligned protomer as in (E), rotated 40° to the right, highlighting their significant conformational differences. The conformational change of the DNA minor-groove-binding wing (β1′) and the dimerization helices, α1′, α5′ and α6′, is indicated with arrows. cd, central distance.
Mechanism comparison with OxyR and OhrR
Several other bacterial transcription factors possess reactive cysteines that can be modified by ROS to regulate gene activation (Barford, 2004; Poole & Nelson, 2008). Escherichia coli OxyR and Xanthomonas campestris OhrR are examples of structurally well-studied thiol-based oxidation regulators (Choi et al, 2001; Newberry et al, 2007). Structurally, OxyR, OhrR and MexR share a common strategy for sensing peroxide through the formation of disulphide bonds between two distantly located cysteine residues. However, the respective mechanisms of ROS-induced disulphide bond formation are different. The oxidation of OxyR joins two distant regions in one domain, thereby producing the intraprotomer disulphide bond formation. The intraprotomer disulphide bond formation affects the mode of dimeric interaction in OxyR, thus changing its DNA-binding capability. Similarly, the OhrR protein also responds to oxidative stress, which incurs conformational change in the dimerization domains that propagate to the DNA recognition domain, resulting in an orientation of the winged-helix domains of the dimer, which is no longer capable of binding to DNA. The MexR protein uses a different allosteric mechanism of oxidation-induced conformational change. Oxidative formation of disulphide bonds in MexR does not sufficiently alter the orientation of dimerization domains or the spacing of the two DNA major-groove-binding domains. Instead, oxidation leads to a rigid body rotation of helices α2′ and α3′, which results in clashes of α2′ and the newly formed disulphide bond with the DNA backbone and which seems to be the primary reason for its attenuated DNA affinity.
Conclusion
Our previous work on MgrA, a MarR family protein in the Gram-positive pathogen Staphylococcus aureus, has shown that oxidative stress acts as the signal to trigger the virulence and antibiotic resistance regulation (Chen et al, 2006). Recently, we have demonstrated that MgrA homologues exist in the Gram-negative pathogen P. aeruginosa, one of which has broad roles in redox sensing and regulation (Lan et al, 2010). MexR is not the exact homologue of MgrA, yet it also acts as a redox regulator that senses and responds to oxidative signals to mediate mexAB–oprM expression and antibiotics resistance (Chen et al, 2008). One of the main questions about the mechanistic regulation is how MexR senses and responds to oxidative stress. We show in this study that the oxidized MexR can be readily generated and purified, and structurally characterized by X-ray crystallography. The structural evidence and conformational features described in this study have led to the first complete molecular level picture of the oxidation-sensing regulation by MexR. The oxidative stress produced by many antibiotics under physiological conditions results in the oxidation of the redox-sensing cysteine to form a transiently active intermediate that is readily attacked by another cysteine distantly located in the second protomer to form an intermolecular cross-linkage. Disulphide bond formation induces a conformational change in the HTH DNA-binding domain, which abolishes MexR's capability of binding cognate operator, as a result of severe clashes of the newly formed disulphide bond with the DNA backbone. The repressor is released from the promoter, thus allowing transcriptional activation of the mexAB–oprM regulon to increase resistance to antibiotics.
Methods
Crystallization, data collection and structure solution. Published procedures were followed for the expression and purification of the C-terminal-truncated MexR (residues 1–142; Chen et al, 2008). Crystallization experiments were conducted using the hanging-drop vapour diffusion method at room temperature (25°C). The final optimized crystallization condition consists of 1 μl of oxidized MexR dimer at 5 mg/ml and 1 μl of precipitation solution containing 2.8 M sodium acetate trihydrate (pH 7.0). Crystals grown overnight were chosen for X-ray diffraction studies and flash-frozen in this buffer containing 20% glycerol as cryoprotectant. The X-ray data were collected at beamline 23ID-B (Advanced Photon Source, Argonne National Laboratory). For details about the determination of the structure see supplementary information online. For details on data collection and refinement statistics see supplementary Table S1 online. Coordinates and structure factors have been deposited in the Protein Data Bank with accession code 3MEX.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
This study was supported by the National Institutes of Health (NIAID R01 AI074658, to C.H.), the National Natural Science Foundation of China (NSFC; 20721002, to H.C.), the Bairen Program of the Chinese Academy of Sciences (C.Y.), the Major Research Plan of NSFC (90913010, to C.Y.), Key Project of Chinese National Programs for Fundamental Research and Development (2009CB918502, to C.Y.), and National Science and Technology Major Project ‘Key New Drug Creation and Manufacturing Program' (2009ZX09301-001, to C.Y.).
Footnotes
The authors declare that they have no conflict of interest.
References
- Barford D (2004) The role of cysteine residues as redox-sensitive regulatory switches. Curr Opin Struct Biol 14: 679–686 [DOI] [PubMed] [Google Scholar]
- Blair JMA, Piddock LJV (2009) Structure, function and inhibition of RND efflux pumps in Gram-negative bacteria: an update. Curr Opin Microbiol 12: 512–519 [DOI] [PubMed] [Google Scholar]
- Cao L, Srikumar R, Poole K (2004) MexAB–OprM hyperexpression in NalC-type multidrug-resistant Pseudomonas aeruginosa: identification and characterization of the nalC gene encoding a repressor of PA3720-PA3719. Mol Microbiol 53: 1423–1436 [DOI] [PubMed] [Google Scholar]
- Chen PR, Bae T, Williams WA, Duguid EM, Rice PA, Schneewind O, He C (2006) An oxidation-sensing mechanism is used by the global regulator MgrA in Staphylococcus aureus. Nat Chem Biol 2: 591–595 [DOI] [PubMed] [Google Scholar]
- Chen H, Hu J, Chen PR, Lan L, Li Z, Hicks LM, Dinner A, He C (2008) The Pseudomonas aeruginosa multidrug efflux regulator MexR uses an oxidation-sensing mechanism. Proc Natl Acad Sci USA 105: 13586–13591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HJ, Kim SJ, Mukhopadhyay P, Cho S, Woo JR, Storz G, Ryu SE (2001) Structural basis of the redox switch in the OxyR transcription factor. Cell 105: 103–113 [DOI] [PubMed] [Google Scholar]
- Ellison DW, Miller VL (2006) Regulation of virulence by members of the MarR/SlyA family. Curr Opin Microbiol 9: 153–159 [DOI] [PubMed] [Google Scholar]
- Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ (2007) A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130: 797–810 [DOI] [PubMed] [Google Scholar]
- Lan L, Murray TS, Kazmierczak BI, He C (2010) Pseudomonas aeruginosa OspR is an oxidative stress sensing regulator that affects pigment production, antibiotic resistance and dissemination during infection. Mol Microbiol 75: 76–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim D, Poole K, Strynadka NCJ (2002) Crystal structure of the MexR repressor of the mexAB–oprM multidrug efflux operon of Pseudomonas aeruginosa. J Biol Chem 277: 29253–29259 [DOI] [PubMed] [Google Scholar]
- Morita Y, Cao L, Gould VC, Avison MB, Poole K (2006) nalD Encodes a second repressor of the mexAB–oprM multidrug efflux operon of Pseudomonas aeruginosa. J Bacteriol 188: 8649–8654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newberry KJ, Fuangthong M, Panmanee W, Mongkolsuk S, Brennan RG (2007) Structural mechanism of organic hydroperoxide induction of the transcription regulator OhrR. Mol Cell 28: 652–664 [DOI] [PubMed] [Google Scholar]
- Pagès JM, Masi M, Barbe J (2005) Inhibitors of efflux pumps in Gram-negative bacteria. Trends Mol Med 11: 382–389 [DOI] [PubMed] [Google Scholar]
- Poole K (2001) Multidrug efflux pumps and antimicrobial resistance in Pseudomonas aeruginosa and related organisms. J Mol Microbiol Biotechnol 3: 255–264 [PubMed] [Google Scholar]
- Poole K (2004) Efflux pumps. In Pseudomonas, Vol I, Genomics, Life Style and Molecular Architecture, Ramos JL (ed), pp 635–674. New York: Kluwer Academic/Plenum [Google Scholar]
- Poole K, Srikumar R (2001) Multidrug efflux in Pseudomonas aeruginosa: components, mechanisms, and clinical significance. Curr Top Med Chem 1: 59–71 [DOI] [PubMed] [Google Scholar]
- Poole LB, Nelson KJ (2008) Discovering mechanisms of signaling-mediated cycteine oxidation. Curr Opin Chem Biol 12: 18–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer HP (2003) Efflux as a mechanism of resistance to antimicrobials in Pseudomonas aeruginosa and related bacteria: unanswered questions. Genet Mol Res 2: 48–62 [PubMed] [Google Scholar]
- Wilke MS, Heller M, Creagh AL, Haynes CA, Mclntosh LP, Poole K, Strynadka NCJ (2008) The crystal structure of MexR from Pseudomonas aeruginosa in complex with its antirepressor ArmR. Proc Natl Acad Sci USA 105: 14832–14837 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.