Abstract
Ustilago maydis is a basidiomycete fungus that exhibits a yeast-like and a filamentous form. Growth of the fungus in the host leads to additional morphological transitions. The different morphologies are characterized by distinct nuclear movements. Dynein and α-tubulin are required for nuclear movements and for cell morphogenesis of the yeast-like form. Lis1 is a microtubule plus-end tracking protein (+TIPs) conserved in eukaryotes and required for nuclear migration and spindle positioning. Defects in nuclear migration result in altered cell fate and aberrant development in metazoans, slow growth in fungi and disease in humans (e.g. lissencephaly). Here we investigate the role of the human LIS1 homolog in U. maydis and demonstrate that it is essential for cell viability, not previously seen in other fungi. With a conditional null mutation we show that lis1 is necessary for nuclear migration in the yeast-like cell and during the dimorphic transition. Studies of asynchronous exponentially growing cells and time-lapse microscopy uncovered novel functions of lis1: It is necessary for cell morphogenesis, positioning of the septum and cell wall integrity. lis1-depleted cells exhibit altered axes of growth and loss of cell polarity leading to grossly aberrant cells with clusters of nuclei and morphologically altered buds devoid of nuclei. Altered septum positioning and cell wall deposition contribute to the aberrant morphology. lis1-depleted cells lyse, indicative of altered cell wall properties or composition. We also demonstrate, with indirect immunofluorescence to visualize tubulin, that lis1 is necessary for the normal organization of the microtubule cytoskeleton: lis1-depleted cells contain more and longer microtubules that can form coils perpendicular to the long axis of the cell. We propose that lis1 controls microtubule dynamics and thus the regulated delivery of vesicles to growth sites and other cell domains that govern nuclear movements.
Keywords: budding pattern, cell wall integrity, microtubule organization, morphology, nuclear migration, septum positioning
INTRODUCTION
The spatial organization of a cell during cell morphogenesis determines the proper site of growth and positioning of the nucleus and other organelles within the cellular space and involves interactions of the cell cortex with the cytoskeleton. The position of the nucleus is carefully orchestrated during several processes including fertilization, early embryonic development, neuronal migration, budding in yeast and hyphal growth in filamentous fungi (reviewed in Reinsch and Gönczy 1998, Schuyler and Pellman 2001, Dujardin and Vallee 2002, Morris 2003, Yamamoto and Hiraoka 2003, Pearson and Bloom 2004, Xiang and Fischer 2004). Defects in nuclear migration and spindle positioning can lead to aberrant development and altered cell fate in metazoans, slow growth in fungi, aberrant distribution of chromosomes and disease in humans (e.g. lissencephaly). A key issue is understanding how nuclear migration and spindle positioning are coordinated with cell morphogenesis.
Fungi exhibit a diversity of shapes during vegetative and reproductive growth, characterized by extensive and complex nuclear movements (see e.g. Raudaskoski 1972; Plamann et al. 1994; Xiang et al. 1994, 1995; Tinsley et al. 1996; Chiu et al. 1997; Inoue et al. 1998; Minke et al. 1999; Alberti-Segui et al. 2000; Liu et al. 2003; Finley et al. 2007). Cells can vary from spherical or ovoid in yeasts to cylindrical in hyphae and more unusual forms found in fruiting bodies and specialized structures formed during interaction of pathogenic and mycorrhizal fungi with their hosts. Many pathogenic fungi also exhibit dimorphism, adding a further level of complexity in fungal morphogenesis.
Nuclear migration and spindle positioning in diverse eukaryotes are governed by the microtubule cytoskeleton, the microtubule motor protein dynein, its regulator the dynactin complex and Lis1, a microtubule plus end tracking protein (+TIPs) and a highly conserved member of the WD40 protein family. Lis1 acts in the dynein/dynactin pathway (Xiang et al. 1995, Willins et al. 1997, Swan et al. 1999, Faulkner et al. 2000, Smith et al. 2000, Vallee et al. 2001, Lee et al. 2003, Sheeman et al. 2003, Cockell et al. 2004, Rehberg et al. 2005, Siller and Doe 2008). LIS1 was identified first in humans, where haploinsufficiency results in Mieller-Dieker lissencephaly, a syndrome characterized by altered neuronal migration in the cerebral cortex during brain development, resulting in a smooth brain and severe mental retardation (reviewed in Vallee et al. 2001, Morris 2003). Identification of NUDF, the LIS1 homolog in Aspergillus nidulans, and demonstration of a critical role in nuclear migration (Xiang et al. 1995) suggested a role for human LIS1 in nuclear translocation during neuronal migration (Xiang et al. 1995, Vallee et al. 2001, Morris 2003). A. nidulans nudF/lis1 null mutants exhibit slow growth and form tiny colonies, but polarized growth of the hypha is not affected (Xiang et al. 1995). The mutants exhibit altered development of the conidiophore (Xiang et al. 1995). S. cerevisiae pac1/lis1 null mutants do not exhibit altered morphology (Lee et al. 2003, Sheeman et al. 2003). Lis1 has not been characterized in other fungi.
Ustilago maydis is a basidiomycete fungus that infects maize (Zea mays L) and teosinte (Zea mays spp. mexicana or spp. parviglumis) and induces formation of tumors (reviewed in Banuett 1995). It exhibits two basic morphologies, a yeast-like and a filamentous form, and is capable of switching from one form to the other. This switch is crucial for pathogenicity (reviewed in Banuett 1995, 2002; Kahmann and Schirawski 2007; Klosterman et al. 2007). The repertoire of morphologies is increased by interaction with its hosts and includes development of appressoria, branching, and formation of clamp-like structures for nuclear migration and distribution (Snetselaar and Mims 1994, Banuett and Herskowitz 1996, Brachmann et al. 2003, Scherer et al. 2006, Doehlemann et al. 2008, Mendoza et al. 2009) and generation of distinct morphologies during teliospore (diploid spore) formation (Banuett and Herskowitz 1996). U. maydis is a particularly valuable experimental system for studies of morphogenesis because of the different forms exhibited during its life cycle and because it is amenable to molecular genetic and cell biological techniques.
The unicellular yeast-like form is haploid and divides by budding. Thus, as in S. cerevisiae, the position of the future division plane is determined at the time of bud formation and mechanisms have arisen to ensure proper segregation of the genetic material. The yeast-like form is elongated with slightly tapered ends and grows by tip extension (Banuett and Herskowitz 2002). It forms a bud once per cell cycle at the cell apex, slightly off center (Jacobs et al. 1994, Banuett and Herskowitz 2002). In the yeast-like form the nucleus occupies a central location throughout most of the cell cycle. This phase is characterized by an extensive array of cytoplasmic microtubules nucleated from a microtubule organizing center (MTOC) located at the base of the bud (Steinberg et al. 2001, Banuett and Herskowitz 2002, Straube et al. 2003). The yeast-like cell contains additional motile cytoplasmic MTOCs (Straube et al. 2003, Fink and Steinberg 2006). In G2 the nucleus migrates into the bud where nuclear division takes place (Holliday 1965, Steinberg et al. 2001, Banuett and Herskowitz 2002). Nuclear migration is accompanied by disassembly of the cytoplasmic array of microtubules, formation of a short intranuclear spindle and nucleation of astral microtubules from the spindle pole bodies (SPBs) (Steinberg et al. 2001, Banuett and Herskowitz 2002). These astral microtubules appear to contact the bud cortex (Banuett and Herskowitz 2002). Nuclear division in the bud creates a transient dikaryotic state and an anucleate mother cell. Subsequently one nucleus migrates to the mother cell and each nucleus locates in the cell middle (Steinberg et al. 2001, Banuett and Herskowitz 2002). Migration of the nucleus to the bud before nuclear division is a feature of basidiomycete yeasts and is not observed in ascomycete yeasts.
Nuclear movements also occur during the morphological switch from yeast-like to filamentous form and during growth in the host (Banuett and Herskowitz 1994a, 1996; Scherer et al. 2006). Little is known of the mechanisms that regulate nuclear movements in the two forms of U. maydis and how morphogenesis and nuclear positioning are coordinated. Recent work indicates that α-tubulin and dynein are necessary for nuclear migration and for cell morphogenesis of the yeast-like cell (Steinberg et al. 2001, Straube et al. 2001).
We identified the U. maydis homolog of human LIS1 in a screen for mutants altered in cell morphology. We demonstrate that Lis1 is essential for cell viability in contrast to its dispensability in S.cerevisiae and A. nidulans. We generated a conditional null mutation to determine its role in various cellular processes and present here the results of our comprehensive analysis. We demonstrate that in addition to a role in nuclear migration, as in other eukaryotes, Lis1 is necessary for cell morphogenesis, cell wall deposition and integrity, positioning of the septum and organization of the microtubule cytoskeleton. These novel functions of Lis1 have not been described previously in other fungi.
MATERIALS AND METHODS
Strains, media, and growth conditions
U. maydis strains used in these studies were FB1 (a1 b1), FB2 (a2 b2), FBD12 (a1/a2 b1/b2 ade+/ade− pan+/pan−) (Banuett and Herskowitz 1989), SG200 (a1 mfa2 bW1 bE2) (Regenfelder et al. 1997), FB1 Pcrg1∷lis1, SG200 Pcrg1∷lis1, and FBD12 lis+/Δlis1 (this work). Media were YEPS (Tsukuda et al. 1988), YEP 1% arabinose and YEP 1% glucose, UMC broth (U.maydis complete, Holliday 1974), charcoal agar (Holliday 1974) containing 1% arabinose or 1% glucose and charcoal broth (Banuett and Herskowitz 1994a). Media and conditions for induction of the lis1-depletion phenotype are described separately. U. maydis strains were grown at 28 C unless stated otherwise. Bacterial strains and growth conditions were as described by Chew et al. (2008).
DNA-mediated transformation of U. maydis
DNA-mediated transformation of U. maydis strains was as described by Chew et al. (2008). Transformants were selected on YEPS 1 M sorbitol supplemented with hygromycin B (200 μg/mL) or minimum medium (MM, Holliday 1974) 1 M sorbitol containing 0.6% arabinose and supplemented with hygromycin B (200 μg/mL), and purified on the same medium without sorbitol.
Induction of the lis1-depletion phenotype
For routine induction of the lis1-depletion phenotype, strains FB1 Pcrg1∷lis1 and SG200 Pcrg1∷lis1 were grown in MM 1% arabinose to OD6000.7–1.0 to allow expression of lis1. A 1 mL aliquot was washed with water, resuspended in the same volume of MM 1% glucose, and a 20–50 μL aliquot was inoculated into 50 mL MM supplemented with 1% glucose and grown to OD6000.9–1.0 to repress expression from the Pcrg1 promoter. Samples were removed at various intervals for analysis of cell morphology and nuclear number and position.
Similar procedures were followed for induction of the lis1-depletion phenotype in UMC glucose. For induction of the lis1-depletion phenotype in YEP glucose, a 4 mL culture in YEP 1% arabinose was grown to OD6000.2, a 1.0 mL aliquot washed with water, resuspended in the same volume of YEP 1% glucose, added to 4 mL YEP 1% glucose, grown overnight, diluted to OD6000.2 in YEP 1% glucose and incubated to OD6001.0. For growth on YEP glucose agar strains were grown as just described; a 1 mL aliquot was washed with water and resuspended in an equal volume of YEP 1% glucose. A total of 200 μL were removed, serially diluted in 10-fold increments and 3 μL each dilution spotted on YEP 1% glucose agar. Growth was monitored at 24, 48 and 72 h.
Enzymes and reagents
Monoclonal anti-α-tubulin antibody, thiabendazole, and Calcofluor white were from Sigma, secondary antibodies (TRITC or DyLight labeled goat-anti mouse) from Jackson ImmunoResearch and Sytox green from Molecular Probes/Invitrogen. All other enzymes and reagents were as described in Chew et al. (2008).
Plant inoculations and segregation analysis
An aliquot of a fresh overnight culture of strain FBD12 lis+/Δlis was inoculated into 5 d old maize seedlings (B164) as described in Banuett and Herskowitz (1996). Teliospores were removed from mature tumors, allowed to undergo meiosis on YEPS slabs and segregants analyzed as described by Banuett and Herskowitz (1989, 1994b).
Light microscopy
Cells were observed with Nomarski interference and epifluorescence optics with a BX61 Olympus microscope equipped with an ORCA CCD camera (Hamamatsu) and images captured with Simple PCI imaging software (Scientific Instruments, San Francisco, California) and processed with Adobe Photoshop CS.
Time-lapse microscopy
Pcrg1:lis1 and FB1 cells were grown in 4 mL MM 1% arabinose to OD6001.0. A 0.1 mL aliquot was washed with water, resuspended in MM 1% glucose and grown to OD6000.4–0.8. A 1–5 mL aliquot was removed and resuspended in 0.1–0.3 mL UMC 1% glucose at room temperature. A total of 45 μL melted UMC 1% glucose 0.7% agarose were placed into a 1 × 1 cm well, onto which 3 μL of the culture were layered, covered with a glass cover slip and sealed with parafilm. Images were captured every 30 s, with 2 s light exposure during capture time. Sequences were converted into movie files at 15 frames/s.
Cell wall, septum, and nuclear staining
Strains were grown in MM 1% glucose as described above, and an aliquot was treated with DAPI (4′,6-diamidino-2-phenylindole-dihydrochloride) at a final concentration of 2 μg/mL to stain the nucleus or 1% Congo red to stain the cell wall and septa, as described by Banuett and Herskowitz (2002). For Calcofluor white and Sytox green staining cells were grown as above; Calcofluor white was added to an aliquot of the culture to a final concentration of 1 μg/mL, incubated 15 min, fixed with 70% ethanol, washed with 1× PEM buffer, treated with extraction buffer (see Banuett and Herskowitz 2002), washed with 1× PBS buffer and resuspended in 0.1 mL 1× PBS buffer. Sytox green solution (Molecular Probes) was added to a final concentration of 0.25 μM.
Filament formation on charcoal agar
Strains SG200 and SG200 Pcrg1∷lis1 were grown in 4 mL UMC 1% arabinose or 1% glucose broth to OD6000.5–0.7, and 5–10 μL aliquots spotted on charcoal agar supplemented with 1% arabinose or 1% glucose. Plates were sealed with Parafilm and incubated at room temperature. Each strain was spotted on UMC arabinose and UMC glucose in parallel. Reactions were scored at 16, 24, 48, 72 and 96 h incubation. Samples were removed from the different spots and examined microscopically with Nomarski optics.
Nutritional induction of filaments in charcoal broth
To assay ability to form filaments under nitrogen-limiting conditions strains, SG200 and SG200 Pcrg1∷lis1 were grown overnight in 5 mL UMC 1% arabinose, washed with water, resuspended in 1 mL charcoal 1% glucose broth and processed as described by Banuett and Herskowitz (1994a). Aliquots were removed at the start of the incubation and at 3 h intervals for 16–24 h, stained with Congo red or DAPI as described above and examined microscopically with Nomarski and epifluorescence optics.
Growth on different media
To test the effect of different chemicals on growth of lis1-depleted cells, cultures of FB1 Pcrg1∷lis1 and SG200 Pcrg1∷lis1 and the parental strains (FB1 and SG200, respectively) were grown in MM 1% arabinose to OD6001.0, washed with water, serially diluted as indicated above and 3 μL each dilution for each strain spotted on these media: UMC 1% arabinose or 1% glucose agar containing 0, 2, 5 or 10 μg/mL thiabendazole (TBZ); 0.6 M KCl; Congo red (30 μg/mL) or Calcofluor white (40 μg/mL). Strains also were streaked on the above media. Growth was recorded at 24 h intervals for 5 d and photographed with a Nikon digital camera. Strains also were spotted on YEP 1% arabinose or YEP 1% glucose containing TBZ at 0, 2 or 5 μg/mL, and incubated as above.
For growth in liquid culture in the presence of thiabendazole, strains FB1 and FB1 Prcg:lis1 were grown in MM arabinose to OD6000.2–0.4 , resuspended in MM 1% glucose containing DMSO or TBZ at 2, 5 or 10 μg/mL, and grown to OD6001.0. Samples were removed for DAPI staining and microscopic observation.
Visualization of the microtubule cytoskeleton by indirect immunofluorescence
Strains FB1, FB1 Pcrg1∷lis1, SG200, SG200 Pcrg1∷lis1 and FBD12 were grown in MM 1% arabinose overnight to OD6000.2–0.4. A 1 mL aliquot adjusted to OD6000.15 was resuspended in MM 1% glucose and grown to OD6000.2–0.4. The cultures were monitored microscopically to assess budding in wild type strains or induction of the lis1 phenotype in the lis1 conditional mutants. Cells were fixed and processed as described by Banuett and Herskowitz (2002). The primary and secondary antibodies were as indicated above. The procedure was repeated with independent Pcrg1∷lis1 isolates in the FB1 and SG200 strain background.
Deletion of the lis1 ORF
The ORF was deleted in vitro according to Brachmann et al. (2004). Primers for amplification of a 1.5 kb fragment from the 5′ and 3′ regions were, respectively, FBU118 (5′GTTAATTAACCGTCGCCTGGTTGACCTGGC) and FBU119 (5′GAATGGCCATCTAGGCCTACCTCATCTTGAGAGCTAGTC), and FBU120 (5′GAATGGCCTGAGTGGCCTACAACGCTCGAGAACAAGTAG) and FBU121 (5′GTTAATTAACGGACCGTGCTCCAAGCTTGCTCG). FBU118 and FBU121 introduced a PacI restriction enzyme site (underlined) and FBU119 and FBU120 a modified SfiI restriction site (underlined) (see Brachmann et al. 2004). The amplified fragments were cloned into pCR2.1 TOPO II (Invitrogen) to generate, respectively, plasmids pCR5L1 and pCR3L1, which were treated with SfiI and NcoI. A 3.9 kb fragment from pCR5L1 and a 3.2 kb fragment from pCR3L1 were ligated with the 2.7 kb SfiI hygromycin cassette from plasmid pMF-1h (Brachmann et al. 2004) to generate pCR2.1 TOPOII containing the 5′lis1-hygB-3′lis1 targeting fragment that lacks the entire ORF (pCR-Δlis1). The 5.7 kb targeting fragment was released by treatment with restriction endonuclease PacI, introduced into strains a1 b1 (FB1), a2 b2 (FB2), and a1/a2 b1/b2 (FBD12), and transformants selected on YEPS hygromycin agar.
Deletion of the ORF was ascertained by PCR of genomic DNA from the transformants with primers FBU186 (5′CTGACCTTGCTGTTCGAGC) and FBU114 (Chew et al. 2008) or FBU187 (5′CTGAGCGAGTTGCACAGAG) and FBU115 (Chew et al. 2008), which are expected to generate a 2.3 kb fragment in the null mutant strains and no fragment in wild type strains. Primer FBU186 is upstream of FBU118; FBU187 is downstream of FBU121. The presence of the ORF was detected with ORF-specific primers FBU149 (5′GTACGTACGCATCTCTCGTGAGTCCGG) and FBU153 (5′GCGCGTTAATTAAGGGCGTCCAGATCTTGATGG), which are expected to amplify an approximately 1.4 kb fragment in wild type strains and no fragment in the null mutant strains.
Generation of a conditional lis1 mutation (Pcrg1∷lis1)
A 1.5 kb fragment from the lis1 5′ region was PCR amplified with primers FBU118 (described above) and FBU264 (5′GAATGGCCATCTAGGCCCTTGAGAGCTAGTCTTTGC). The entireORF (~ 1.4kb) was amplified with primers FBU261 (5′GAATGGCCTGAGTGGCCTAGCTCTCAAGATGAGCG) and FBU265 (5′GCGCGTTAATTAACTAGGGCGTCCAGATCTTG). FBU118 and FBU265 introduced a PacI site (underlined), and FBU261 and FBU264 a SfiI site (underlined). The amplified fragments were cloned into pCR2.1 TOPO to generate pCR-5L1M and pCR-L1ORF, respectively. The plasmids were digested with NcoI/SfiI, and the 3.9 kb and 2.9 kb fragment from pCR-5L1M and pCR-L1ORF, respectively, were ligated with a SfiI hygB-Pcrg1 cassette from plasmid pMF2-1h (Brachmann et al. 2004) to generate pCR2.1 TOPO-5′lis1-hyg-Pcrg1∷lis1 ORF (pCR-Pcrg1∷lis1) (Fig. 1B). The targeting fragment was released by restriction with PacI endonuclease and introduced into strains FB1 (a1 b1) and SG200 (a1 mfa2 bW1 bE2). Transformants were selected on MM 0.6% arabinose supplemented with hygromycin (200 μg/mL).
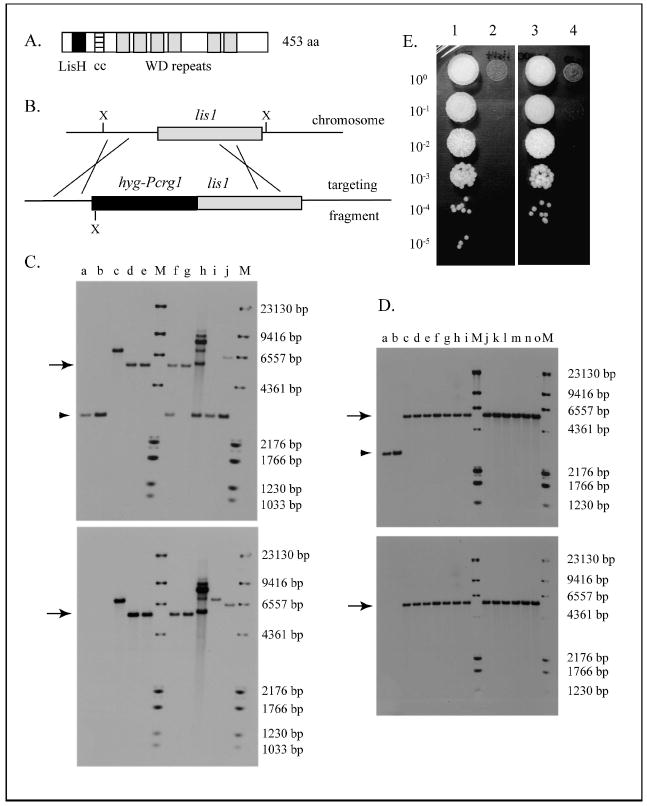
Fig. 1.
Ustilago maydis Lis1 protein and effect of its depletion on growth. A. Domain organization of the Ustilago maydis Lis1 protein. Lis H =Lis1 homology domain (amino acid residues 20–51); cc = coiled coil domain (aa residues 69–95); WD = tryptophan-aspartic acid repeats. Analysis was carried out with Expasy Prosite (Hulo et al. 2007, http://www.expasy.ch/prosite/) and Coils (Lupas et al. 1991, http://www.ch.embnet.org/software/COILS_form.html). B. Gene replacement of the wild type lis1 allele with the Pcrg1∷lis1 allele by homologous recombination. The targeting fragment was generated in vitro. X = XhoI. The XhoI fragment in the wild type allele is 3000 bp and in the Pcrg1∷lis1 allele 5544 bp because the hygB gene introduces a XhoI site, as indicated. C. Southern hybridization analysis of FB1 (a1 b1) transformants. a = FB1, b = FB2, c–j = independent transformants. Gene replacement occurred in strains d, e, f and g, as evidenced by the presence of the diagnostic 5544 bp band (arrow) and the absence of the wild type band (3000 bp, arrowhead). M = molecular weight markers consisting of a mixture of DIG-labeled MW marker II and VI (Roche). The top panel was probed with a 550 bp probe from the lis1 ORF region, and the bottom panel is the same membrane reprobed with a 872 bp probe from the hygB gene. D. Southern hybridization analysis of SG200 (a1 mfa2 bW1 bE2) transformants. a = FB1, b = SG200, c–o = independent transformants. Gene replacement occurred in all transformant strains analyzed (c–o) as evidenced by the presence of the diagnostic 5544 bp band (arrow) and the absence of the wild type band (arrowhead). M = molecular weight markers as in Panel C. Top and bottom panels were probed as indicated for Panel C. E. Growth of wild type and Pcrg1∷lis1 strains on YEP glucose. Strains were grown as indicated, serially diluted, spotted on YEP glucose medium and incubated at 28 C for 3 d. 1 = FB1 (a1 b1); 2 = FB1 Pcrg1∷lis1; 3 = SG200 (a1 mfa2 bW1 bE2); 4 = SG200 Pcrg1∷lis1.
Isolation of genomic DNA and Southern hybridization analysis
Genomic DNA was isolated as described by Chew et al. (2008). For Southern hybridization analysis of putative deletion and conditional mutant strains, genomic DNA from the transformants and the parental wild type strains (FB1, FB2, SG200 and FBD12) was digested with restriction endonuclease XhoI, fractionated and transferred to an Immobilon membrane (Millipore) as described by Chew et al. (2008). The membrane was probed with a 520 bp DIG-labeled PCR fragment from the lis1 5′ region (see below) or with a 550 bp DIG-labeled PCR fragment from the ORF region (see below) and stripped and reprobed with a 872 bp DIG-labeled PCR fragment from the HygB gene in pHL1 (Wang et al. 1988) (see below).
Labeling of DNA probes for Southern hybridization analysis
Probes were labeled with the PCR DIG-labeling kit (Roche); a 520 bp fragment from the lis1 5′ region with primers FBU75 (5′GTCTAGTTACGAGTCGAG) and FBU143 (5′TCACGAATCACGAATCGG); a 550 bp fragment from the lis1 ORF with primers FBU350 (5′CTGTCTGCAGCGCCATCGGC) and FBU351 (5′CCCTGGCGGTTTGGTCCGTC), and a 872 bp fragment from the hygB gene with primers FBU79 (5′GCGAGTACTTCTACACAG) and FBU80 (5′GCTTTCAGCTTCGATGTAGG). PCR fragment probes were gel purified with the QIAquick gel extraction kit (QIAGEN). Conditions for PCR were as described in Chew et al. (2008).
RESULTS
lis1 is essential for cell viability in U. maydis
lis1 was identified in a screen for mutants with altered cell morphology (F. Banuett unpubl). The U. maydis lis1 ORF has coding information for a protein of 453 amino acid residues that contains a LisH homology domain, a coiled-coil domain and six tryptophan-aspartic acid (WD) repeats (Fig. 1A). All three domains are present in other Lis1 homologs. U. maydis Lis1 exhibits 47–54% identity over its entire length to Lis1 in other eukaryotes, including human LIS1.
To determine the lis1 null phenotype, we attempted to generate a lis1 null mutation by a one-step gene replacement. We did not succeed in deleting the gene in haploid strains as shown by Southern hybridization analysis of transformants, suggesting that the gene is essential for cell viability. To determine whether the gene is indeed essential for cell viability, we generated a heterozygous Δlis1/lis1+ diploid strain by homologous recombination. A pure culture of this diploid was inoculated into 5 d old maize seedlings, tumors collected and teliospores (diploid spores) were induced to undergo meiosis. Analysis of the progeny indicated that all teliospores (30) segregated 2 live:2 dead (or very sick and dying progeny) (data not shown). These and additional observations (see below, Fig. 1E) indicate that U. maydis lis1 is essential for cell viability.
To circumvent inability to delete the gene, we generated a conditional null mutation in which the endogenous lis1 promoter was replaced with the carbon-regulatable Pcrg1 promoter from the crg1 gene (Fig. 1B, C, D). The Pcrg1 promoter is active in arabinose medium and inactive in glucose medium (Bottin et al.1996, Brachmann et al. 2004). The Pcrg1∷lis1 construct was introduced into haploid strains FB1 (a1 b1) and SG200 (a1 mfa2 bW1 bE2) and transformants selected on arabinose hygromycin medium. Gene replacement was confirmed by Southern hybridization analysis (Fig. 1C, D).
lis1-depleted cells exhibit altered morphology and defects in nuclear migration
Because lis1 was identified as a mutation conferring a cell morphology defect, we set out to determine the effect of depletion of lis1 on colony and cell morphology. We carried out a detailed analysis of cell morphology in asynchronous, exponentially growing cultures of lis1-depleted and wild type strains.
Colony and cell morphology
Pcrg1∷lis1 strains did not form colonies on YEP glucose medium (Fig. 1E; cf. 1, 2 and 3, 4), providing independent confirmation that lis1 is essential for cell viability. On MM glucose the strains formed tiny colonies that were at least 10–15 times smaller than those of the parental wild type strains and accumulated a greenish pigment (data not shown). In contrast, Pcrg1∷lis1 strains formed colonies indistinguishable in appearance and size from those of the parental wild type strains on MM arabinose (data not shown). The fact that Pcrg1∷lis1 strains did not form colonies on YEP glucose but were able to form tiny colonies on MM glucose likely suggests that the kinetics of lis1 depletion might be different under the growth conditions used.
To determine the effect of lis1 depletion on cell morphology of the yeast-like form, the Pcrg1∷lis1 and wild type parental strains were examined at various intervals after shift from arabinose to glucose medium with Nomarski optics. In MM arabinose, Pcrg1∷lis1 strains exhibited wild type cell morphology, indistinguishable from that of wild type strains grown under the same conditions (FIG. 2A1, B1).
Fig. 2.
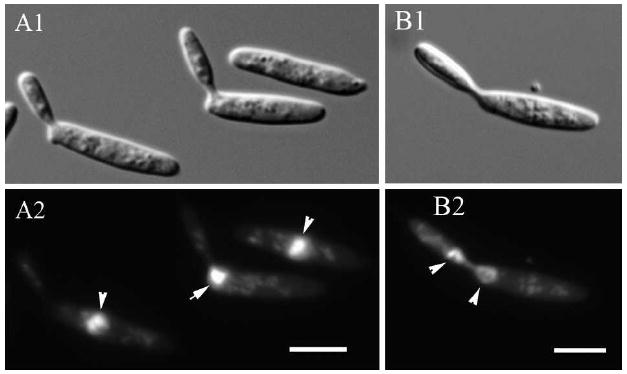
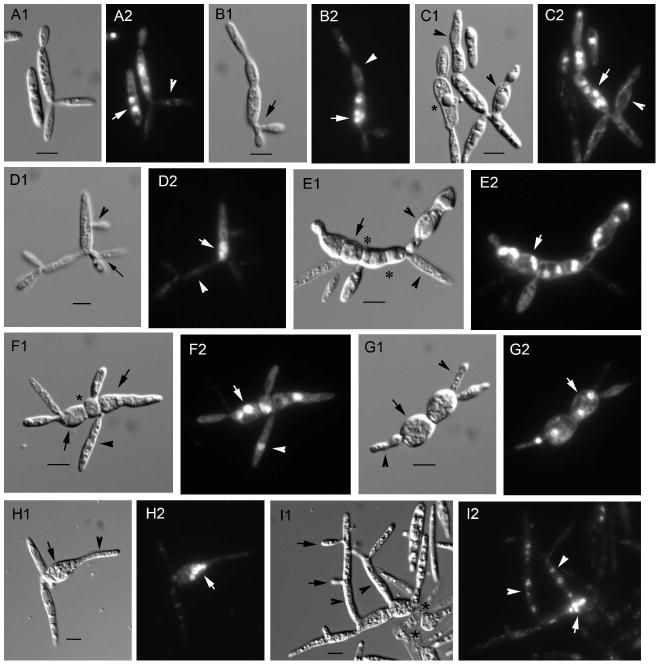
Nuclear position and migration in U. maydis wild type yeast-like cells. A. The nucleus locates in the cell center in unbudded cells and cells with small and medium-sized buds (arrowheads in A2). In large-budded cells, the nucleus locates near the neck region during migration to the bud (arrow in A2). B. Nuclear migration from the bud to the mother cell after nuclear division in the bud (arrowheads). A1, B1. Nomarski view of DAPI-stained cells in A2, B2, respectively. Bars = 5 μm.
In glucose medium, Pcrg1∷lis1 cells exhibited changes in cell morphology beginning 20–24 h after transfer from arabinose to glucose medium and became more dramatic after that time, as described below (Fig. 3, Supplemental table I). Cells with two buds of similar size or with two or more buds of different sizes were detected in the culture at 24–36 h (Fig. 3A1, B1, D1). Some of the buds arose laterally from the mother cell (Fig. 3D1 arrowhead) or from an immature bud (Fig. 3B1 arrow), not from the poles as they normally do (Fig. 2A, B). These observations suggest that the axis of growth is altered as lis1 is depleted. The phenotype was exacerbated with time; mother cells became wider in the middle, spherical or almost spherical (Fig. 3E1, F1, G1 arrows). These wider or spherical cells formed buds of two types, morphologically normal or aberrant (Fig. 3C1, E1, F1, G1 arrowheads), an observation supported by time-lapse video microscopy (see below). In some instances wider or spherical cells formed long appendages (Fig. 3H1, I1, arrowheads) and these in turn produced lateral buds (Fig. 1I arrows) resulting in clusters of grossly aberrant cells. These observations indicate that depletion of lis1 causes loss of cell polarity and cell morphology. Many cells in older cultures (> 72 h) lysed (Fig. 3I1 asterisks), suggesting that lis1-depleted cells have altered cell wall composition or properties. Others became vacuolated and collapsed (Fig. 3C1 asterisk), most likely due to absence of a nucleus (see below).
Fig. 3.
Nuclear position and migration in U. maydis lis1-depleted cells. Aliquots of asynchronous exponentially growing cultures of Pcrg1∷lis1 were examined at various times after shift from arabinose to glucose medium. Nuclear position was ascertained by staining with DAPI; cell morphology was analyzed with Nomarski optics. A1–I1. Nomarski view of the DAPI-stained cells in A2–I2, respectively. See text for description of what arrows, arrowheads and asterisks indicate. Bars = 5 μm.
Wild type cells grown under the same conditions for 96 h maintained wild type morphology and nuclear position throughout the growth period, except that budding decreased after 24 h growth (Supplemental table II), as observed for wild type strains grown in other media (F. Banuett unpubl).
Nuclear migration
Because lis1 is required for spindle position and nuclear migration in other eukaryotes, we determined nuclear position in the wild type and Pcrg1∷lis1 cells used above to analyze cell morphology. In wild type and Pcrg1∷lis1 cells grown in arabinose medium the nucleus was centrally located in the majority of cells (93%, n = 260) (Fig. 2A2 arrowheads). In a small fraction of the cells (7%, n = 310) the nucleus was near the neck region between mother and bud (Fig. 2A2 arrow), in the bud, or one nucleus in both mother and bud (Fig. 2B2 arrowheads), in agreement with observations of nuclear movements in wild type cells grown in YEPS medium (Steinberg et al. 2001, Banuett and Herskowitz 2002).
Pcrg1∷lis1 cells with two or more buds had two or more nuclei in the mother cell and none in the buds (n = 376) (Fig. 3A2, B2, C2, D2 arrows and arrowheads respectively). In some cases one of the buds contained a single nucleus (Fig. 3F2 arrowhead) but the majority of buds were devoid of nuclei at 24 and 48 h (Fig. 3A2, B2, C2, D2 arrowheads). These observations indicate that nuclear migration from mother to bud was impaired and suggest that nuclear division per se was not affected, at least in the first rounds of nuclear division. Wider or spherical cells also had two or more nuclei (Fig. 3E2, F2, G2 arrows). In wide or spherical cells with appendages, the mother cell was multinucleated (4–8 nuclei) (Fig. 3H2, I2 arrows) and in some cases the appendages also were multinucleated (Fig. 3I2 arrowheads), although most of them were devoid of nuclei.
Wider or spherical cells formed septa that divided the cell into smaller cells with one or two nuclei (n = 317) (Fig. 3E1, E2, F1, F2 asterisks), although some cells still retained more than two nuclei. Simultaneous staining of the cell wall with Calcofluor white and the nucleus with Sytox green confirmed these observations (n = 532) (data not shown). Similar observations were made when Pcrg1∷lis1 cells were grown in UMC glucose. Because of faster growth in this medium the terminal phenotype developed faster while the intermediate steps were not as easily detected under these growth conditions. Nonetheless the terminal phenotype was similar to that described above: Cells became enlarged and multinucleate and formed long projections that had no or a variable number of nuclei (data not shown). Taken together these observations indicate that U. maydis lis1 is necessary for nuclear migration in the yeast-like cell.
lis1-depleted cells exhibit altered septum positioning
In wild type cells, one, two or no septa were observed normally at the mother-bud neck region, depending on the stage of cell separation (n = 561) (Fig. 4A, C arrows). In large-budded cells, cell separation occurs in the region between the two septa, the fragmentation zone (O’Donnell and McLaughlin 1984, Banuett and Herskowitz 2002, Weinzierl et al. 2002). In lis1-depleted cells the number and location of septa were altered (n = 317); some had a single septum in the cell middle (Fig. 4E), others had two or more septa (Fig. 4D, J, K, L, M arrows) and still others had a septum close to one of the cell poles (Fig. 4F, G arrowhead), in most cases distal to the site of budding (Fig. 4F arrowhead). In addition, many of these cells with mislocalized septa had a normal doublet of septa separating the mother from one of its wild type-like buds (Fig. 4F, G asterisks). Formation of a septum in the cell middle appeared to be preceded by a constriction of the cell at that location, as suggested by the presence of peanut-shaped cells (data not shown). We also observed cells with thickened septa at the mother-bud junction (data not shown) or in the cell middle (Fig. 4M arrows [as]). In other cells a doublet of closely apposed septa formed in the cell middle (Fig. 4L arrowhead). In addition, cells undergoing rounding exhibited abnormal asymmetrical deposition of cell wall material on one of the lateral cell walls (Fig. 4I arrow) or over the entire cell surface (Fig. 4H, K arrowheads), compared to buds or appendages arising from the same cell (Fig. 4H, K), and many had thick striations of cell wall material parallel to the long axis of the cell (Fig. 4M arrows [cws]). Thickening of the cell wall, cell wall striations and mislocalized septa were not observed in wild type cells (Fig. 4A–C). Taken together, these observations indicate that lis1 is required for normal positioning of the septum, cell wall deposition and integrity.
Fig. 4.
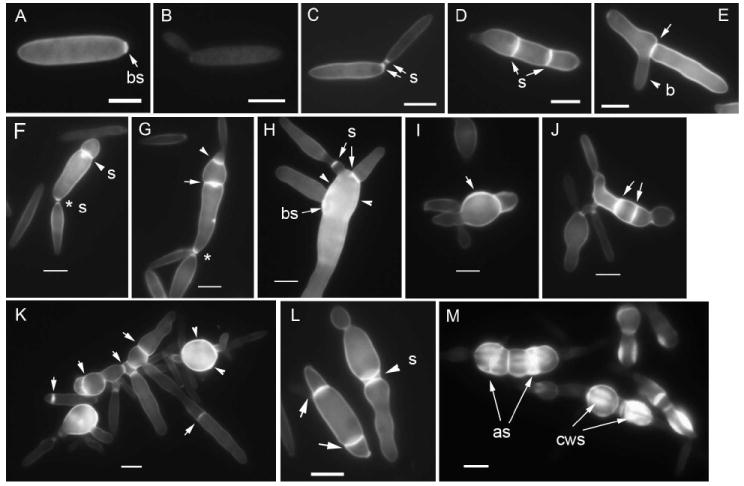
Septum positioning and cell wall deposition in U. maydis wild type and lis1-depleted cells. A–C. Wild type cells. A. Unbudded cell with chitin staining at the bud scar (arrow). B. Small budded cell showing faint overall cell wall staining. C. Large-budded cell showing two septa, one on the mother side, the other on the bud side of the neck, before cell separation. D–M. lis1-depleted cells. E. Cell with a septum (arrow) in the cell middle and a bud (b) emerging next to the septum. D, G, J, L. Cells with two aberrantly located septa (arrows); compare with normal location of septa (s) in C. F, G. Cells with a septum near one of the cell poles (arrowhead) and two normally positioned septa separating mother and bud (asterisks). I. Abnormal asymmetric cell wall deposition (arrow). H, K. Abnormal cell wall deposition over the entire cell surface (arrowheads, cf. A–C). L. Cell with a doublet of septa in the cell middle (arrowhead). M. Cells with aberrant septa (as) and cell wall striations (cws). Bars = 5 μm.
Analysis of cell morphology using time-lapse video microscopy
To document the temporal sequence of events leading to aberrant cell morphology in lis-depleted cells, we used time-lapse microscopy to compare growth of wild type and Pcrg1∷lis1 cells on glucose medium.
Wild type cells
Wild type haploid cells (a1 b1, FB1) were grown on UMC glucose agarose pads and analyzed with time-lapse video microscopy. Wild type cells exhibited the characteristic cigar shape under these growth conditions with an average length of 12.4 ± 1.85 μm and an average width of 2.9 ± 0.39 μm. The width of the neck remained constant at 0.89 ± 0.14 μm.
Wild type cells formed buds preferentially from one of the cell poles (Fig. 5; t = 0 to t = 6 h; Supplemental video I). At least 3–4 rounds of budding occurred from the same pole in a 6 h interval (Fig. 5) before budding switched to the opposite pole (not shown). The daughter cells formed buds preferentially from the distal end (the end farthest from the birth site), at least for the first two cycles of budding (Fig. 5; t = 3 to t = 6 h). The time from bud birth to bud separation averaged 102.3 min (1 h 42.3 min) (Fig. 5, Supplemental video I) and corresponded roughly to the doubling time of cells grown in rich medium in shaking culture (Holliday 1974, Banuett and Herskowitz 2002). Growth rate was 7.1 μm/h. Wild type cells did not exhibit cell lysis, shriveling or vacuolization as observed in lis1-depleted cells (see below) or the formation of lateral buds or simultaneous formation of buds from one or both cell poles.
Fig. 5.
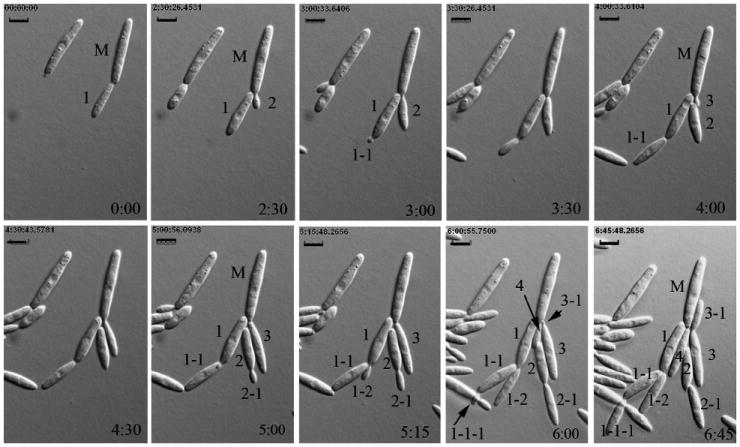
Time-lapse analysis of cell morphology and budding pattern in U. maydis wild type cells. Sequence of photomicrographs showing cell morphology of wild type cells through multiple cycles of budding of a mother cell and its daughters. The mother cell (M) forms four buds (1, 2, 3, 4) from the same cell pole and no buds from the opposite pole during this sequence (t = 0 to t = 6.75 h). Daughter cell 1 buds twice (1-1 at t = 3 h and 1-2 at t = 5:15 h) from the distal pole (opposite the birth site), and daughter 1-1 buds distally too (1-1-1 at t = 6 h). Daughter cell 2 also buds distally (2-1 at t = 5 h). Daughter cell 3 buds proximally (3-1 at t = 6 h). From bud initiation to bud separation is approximately 2 h. Arrows in the penultimate sequence point to emerging buds that are partially masked by other buds. Cell morphology remained unaltered in this and longer sequences. Time in hours is indicated on the bottom right hand of each panel. (See Supplemental video 1.) Bars = 5 μm.
lis1-depleted cells
Pcrg1∷lis1 cells first were grown in MM glucose broth in shaking culture for 36–48 h to begin induction of the lis1-depletion phenotype before transfer to an UMC glucose agarose pad for time-lapse video microscopy (Fig. 6A, B; Supplemental videos IIA, B; III). lis1-depleted mother cells were aberrantly shaped, mostly wider and longer than wild type cells and gave rise to two types of buds, morphologically aberrant and wild type-like (Fig. 6A). Growth rate of the wild type-like buds was similar to that of wild type cells (7.1 μm/h), and the time from bud emergence to bud separation was similar (1 h 42 min). These wild type-like cells became vacuolated and shriveled within 2 h of cell separation and no longer exhibited active vesicle motion (Fig. 6A). We propose that these vacuolated buds lack a nucleus and once separated from the mother cell are unable to carry out normal cellular functions. Aberrant buds were mostly carrot-shaped, with the broad part proximal to the neck region, and almost twice as long as wild type buds (22.6 μm) (Fig. 6A). The neck region separating the mother cell from the aberrant bud was wider than that in wild type cells (2.54 ± 0.11 μm vs. 0.89 ± 0.14 μm). The elongated buds formed buds from their tips. In 90% of the cases the aberrant bud did not separate from the mother cell and exhibited active movement of vesicles; in the cases when it did, it became vacuolated and shriveled and motion of vesicles ceased. Other buds remained short (7.6 μm), but interestingly growth appeared to switch to the base of the bud, which continued to increase in width.
Fig. 6.
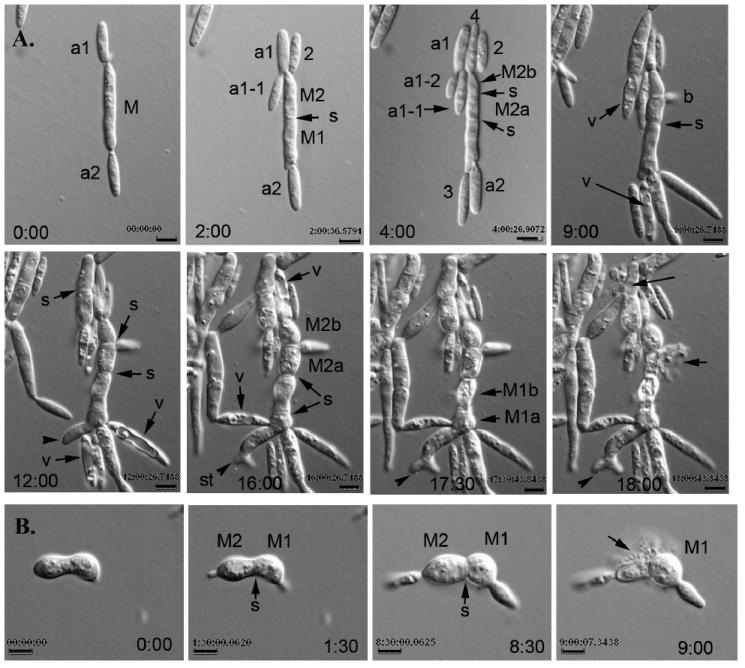
Time-lapse analysis of cell morphology in U. maydis lis1-depleted cells. A. Sequence of photomicrographs showing changes in cell morphology in lis1-depleted cells (see Supplemental videos IIA, B). A lis1-depleted mother cell budded multiple times from both cell poles with a preference for one of them (t = 2 to t = 9 h). The mother cell (M) widened and subsequently was divided into two cells (M1 and M2) (t = 2 h) and further subdivided by sequential septum formation into four cells (M1a, M1b, M2a, M2b) (t = 4 to t = 17:30 h). These cells eventually became spherical (t = 16 h); one of them formed a lateral bud next to the septum (b, t = 9). In many instances the morphologically aberrant mother cells gave rise to wild type-like buds, which on cell separation became vacuolated and stopped growing (t = 9 to t = 16 h, arrow [v]). Cell lysis was apparent, particularly in wider or spherical cells (t = 18 h, arrows). Lysis of one cell did not appear to affect the adjacent cells (cf. t = 17:30 and t = 18 h). One of the mother cells (M1a) produced an aberrantly shaped bud, which appeared to have a split tip (t = 12 to t = 18 h, arrowhead). On closer inspection the appearance of a split tip is due to the formation of two buds almost simultaneously: first a broad, slow-growing bud (0.49 μm/h) is formed, followed soon after by a second broad, faster growing one (1.47 μm/h) that emerges from the same birth site as the first one or nearby, giving the appearance of a split tip (t = 16 to t = 18 h). This abnormal tip growth appeared to be a common feature in many lis1-depleted cells. B. Sequence of photomicrographs showing cell lysis (see Supplemental video III). The peanut-shaped cell (t = 0 h) is divided into M1 and M2 by a septum (arrow) (t = 1:30 h). These two cells bud from their respective cell poles (t = 1:30 h). M2 lyses (t = 9 h, arrow) but its attached bud is not affected (cf. t = 8:30 and t = 9 h). Time in hours is indicated on the bottom right side of each panel (See Supplemental video III). Bars = 5 μm.
The width of aberrant cells increased gradually and constantly. Septum placement in the medial section of these cells resulted in a beaded- or pear-shaped appearance, further contributing to shape distortion (Fig. 6A). These septa separated the contents of one cell from those of adjacent cells, as evidenced by the fact that when one cell lysed the adjacent cells remained intact, and cytoplasmic movement in the intact cells was uninterrupted (Fig. 6A, B). The width of cells before lysis was 3.5–8.8 μm, with an average of 5.5±1.5 μm. Cell lysis is indicative of altered cell wall properties or composition in lis1-depleted cells. Lateral buds formed adjacent and on either side of misplaced septa (Fig. 6A). Their growth rate varied from 0.95 μm/h to 7.1 μm/h. The growth rate of aberrant buds was 0.3–7.1 μm/h.
These observations lend support to our conclusions based on still pictures of cells grown in asynchronous exponential culture and indicate that lis1 is necessary for normal cell morphology, septum positioning, width of the neck region and integrity of the cell wall.
lis1-depletion alters filament formation on charcoal agar
On charcoal agar, strains carrying different a and b alleles, such as SG200 (a1 mfa2 bW1 bE2), form white mycelial colonies, whereas strains carrying identical a or b alleles, such as FB1 (a1 b1), form smooth grayish colonies (Banuett and Herskowitz 1989, Spellig et al. 1994, Regenfelder et al. 1997). To determine the effect of lis1 depletion on filament formation, we compared growth of SG200 Pcrg1∷lis1 and the parental strain SG200 (a1 mfa2 bW1 bE2) on charcoal agar. SG200 and SG200 Pcrg1∷lis1 were grown in UMC arabinose or glucose broth and spotted on charcoal agar containing arabinose or glucose. SG200 formed fuzzy colonies indicative of filament formation, regardless of the sugar in the medium. The reaction was detected beginning at 16 h incubation (Fig. 7A1, A3). The Pcrg1∷lis1 strain formed mycelial colonies, indistinguishable from those of the parental wild type strain, on arabinose medium (data not shown). If grown in arabinose medium and spotted on charcoal glucose agar it formed mycelial colonies (Fig. 7A2; cf. 7A1, A3), whose appearance changed after 48 h incubation; by 72 h filaments became brownish (Fig. 7B2, 7C2 cf. 7B1, 7C3 respectively) and no longer were visible after 96–120 h (data not shown). If grown in UMC glucose and spotted on charcoal glucose agar, no filaments formed (Fig. 7A4, 7B4 cf. 7A3, 7B3). These observations indicate that lis1 is necessary for filament formation on charcoal agar.
Fig. 7.
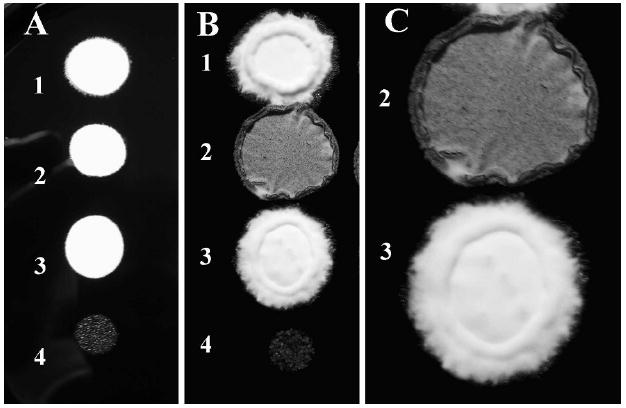
Filament formation on charcoal agar by U. maydis wild type and lis1-depleted strains. Strains SG200 and its isogenic derivative SG200 Pcrg1∷lis1 were grown as described, then spotted on charcoal glucose agar. The reaction was monitored beginning at 16 h and followed for 96 h. 1, 3 = SG200; 2, 4 = SG200 Pcrg1∷lis1. Strains (1) and (2) were grown in arabinose and strains (3) and (4) on glucose before spotting on charcoal glucose agar. A. 24 h. B. 72 h. C. Magnification of spots 2 and 3 in B.
lis1 is required for the filamentous morphology and for septum positioning in the filamentous form
Because inability to form filaments on charcoal agar might be due to a block in the transition from yeast-like to filamentous morphology or to a defect in filamentous growth per se (Banuett and Herskowitz 1994a), we analyzed cell morphology of wild type and lis1-depleted strains under two conditions. First, samples of the strains spotted on charcoal agar were analyzed with Normarski optics. The SG200 wild type parental and Pcrg1∷lis1 strains on arabinose charcoal medium formed filaments consisting of a long tip cell that contains cytoplasm, followed by short subapical compartments devoid of cytoplasm (Fig. 8A, B), as described for wild type strains (Banuett and Herskowitz 1994a, Spellig et al. 1994). The Pcrg1∷lis1 strain behaved similarly during the first 24–48 h growth after transfer from arabinose to glucose medium. After longer incubation on glucose charcoal agar the mutant formed aberrant filament-like structures; these were wider than wild type filaments, curved and branched (Fig. 8C, D cf. 8A, B).
Fig. 8.
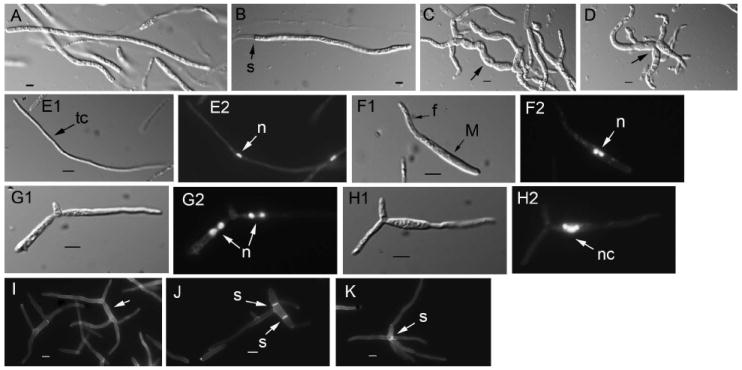
Cell morphology of filamentous structures in U. maydis wild type and lis1-depleted cells. A, B. Wild type filaments. Similar structures are formed by Pcrg1∷lis1 strains grown on charcoal arabinose medium. C, D. Filament-like structures formed by lis1-depleted cells. Arrows point to the wider, curved filament-like structures. Samples for the analysis in A–D were from a plate similar to that in Fig. 7. E–K. Strains were grown under nitrogen-limiting conditions. E. Wild type filament consisting of a long tip cell (E1, arrow) with a single nucleus (E2, arrow) (the strain is haploid but able to form filaments). E1. Nomarski view of DAPI-stained cell in E2. F, G and H. Filament-like structures formed by Pcrg1∷lis1 strains. F1. Initiation of filament formation. The mother cell contains two nuclei (F2 arrow). G and H show filament-like structures containing clusters of two nuclei in each appendage (G2 arrows) or multiple nuclei (H2 arrow) in an enlarged cell that gives rise to a filament-like structure devoid of nuclei. E1–H1. Nomarski view of the DAPI-stained cells in E2-H2, respectively. I, J and K. Congo red staining to reveal cell wall and septa. I. Multiple filament-like structures emanating from a central cell (arrow) that lacks septa. J. A mother cell with a filament-like structure and two septa (arrows) at right angles to the long axis of the cell. K. Filament-like structures with a septum (arrow) at their convergence. Bars = 5 μm.
We used an independent assay to determine the ability to transition from the yeast-like to the filamentous form and to examine the filamentous morphology as a function of time. We examined cell morphology of the wild type parental strain (SG200) and the lis1 conditional mutant (SG200 Pcrg1∷lis1) in MM glucose charcoal broth under nitrogen-limiting conditions. In this medium the parental wild type strain (SG200) initiated filament formation synchronously 3–4 h after inoculation; after 8 h incubation > 85% cells had responded and formed typical filaments (Fig. 8E1). The cultures became covered with a mat of white filaments after 16–24 h incubation (data not shown) as described by Banuett and Herskowitz (1994a). These filaments contained a single centrally located nucleus in the long (50–100 μm) apical cell (n = 100) (Fig. 8E2 arrow). The apical cell was separated from the mother cell by a septum, and if subapical compartments were present they lacked cytoplasm and nuclei (data not shown). Under the same growth conditions the Pcrg1∷lis1 strain initiated the transition as wild type strains do (Fig. 8F1) but the mother cell from which this filament arose contained two nuclei (n = 435), which is not observed in wild type cells undergoing the transition (Fig. 8F2 cf. 8E2). As time of incubation in glucose medium increased, multiple bud-like appendages arose from the mother cell. The appendages elongated and contained clusters of two or multiple nuclei (Fig. 8G2, H2 arrow). By 24 h the aberrant structures did not resemble the wild type filaments (Fig. 8I, J, K); they consisted of a central narrow cell from which 4–6 narrow filament-like structures or appendages arose. In the majority of cells (70–80%, n = 357), no septa separated these appendages from the central cell (Fig. 8I arrow). In the rest (20–30%), the central cell had two centrally located septa perpendicular to its long axis and a filament-like structure (Fig. 8J arrows) or, in rare cases, a curved septum at the junction of all the appendages (Fig. 8K arrow). These observations indicate that lis1 is required for maintenance of the normal filamentous morphology, for septum positioning and for nuclear migration under filament-inducing growth conditions.
The presence of multiple nuclei in the elongated and aberrantly shaped filament-like structures contrasts with the presence of a single nucleus in the filaments from the wild type strain and likely reflects a nuclear migration defect before the switch or might suggest additional roles of lis1 in controlling nuclear division under filament-inducing conditions in culture.
Organization of the microtubule cytoskeleton in wild type and lis1-depleted cells
Because microtubules are required for U. maydis cell morphology and nuclear position and because the phenotype of lis1-depleted cells resembles in some respects that conferred by α-tubulin depletion (Steinberg et al. 2001), we determined the organization of the microtubule cytoskeleton in wild type and lis1-depleted cells with an anti-α-tubulin monoclonal antibody and indirect immunofluorescence. In wild type haploid (FB1) and diploid (FBD12) strains an extensive array of cytoplasmic microtubules was present during most of interphase (Fig. 9A1), which was disassembled in late G2 when the nucleus migrated near the neck region and a short intranuclear spindle was assembled (Fig. 9B1 arrowhead), as described by Steinberg et al. (2001) and Banuett and Herskowitz (2002). The spindle migrated into the bud (Fig. 9B1 arrowhead) where mitosis occurred, and subsequently one of the nuclei migrated to the mother cell (data not shown). Astral microtubules could be detected during migration to the bud and appeared to contact the cell cortex of the bud (Fig. 9B1; arrow). The cytoplasmic network reassembled after the nuclei moved close to the cell poles but before they reached their destination in the cell center and before cytokinesis was complete (not shown), as described by Steinberg et al. (2001) and Banuett and Herskowitz (2002).
Fig. 9.
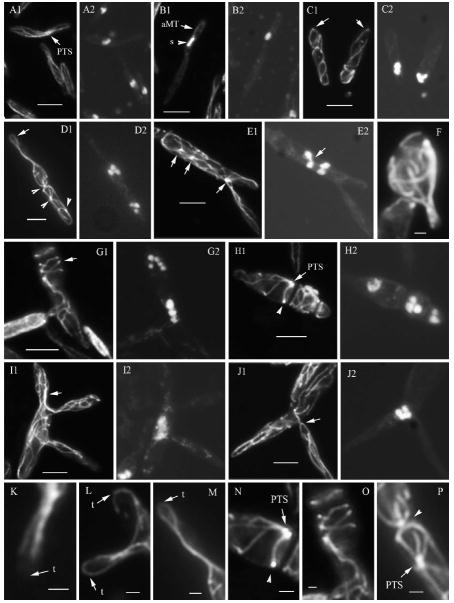
Microtubule organization in U. maydis wild type and lis1-depleted cells. Microtubules in wild type and lis1-depleted cells were visualized with an anti-α-tubulin antibody and indirect immunofluorescence. A, B, K. Wild type cells. C–J, L–P. lis1-depleted cells. A1–E1, F, G1–J1, K–P show tubulin staining, and panels A2–E2, G2–J2 the corresponding DAPI-stained cells. K. Microtubules at the cell tip (arrow) of a wild type cell. L, M. Microtubules at the cell tips (arrows) of lis1-depleted cells; M is an enlarged view of the tip of the cell in D1. N, P. Presumed PTS (arrows or arrowheads), N and P are enlarged views of a section of the cells in H1 and D1, respectively. O. An enlarged image of a section of the cell in G1. Bars: A–E, G–J = 5 μm; F, K–P = 1 μm.
The organization of the microtubule cytoskeleton in Pcrg:lis1 strains grown in arabinose medium was similar to that in strains FB1 and FBD12 described above (data not shown). In early stages of growth in glucose medium, Pcrg1∷lis1 strains showed normal or almost normal cell morphology and organization of the microtubule cytoskeleton, yet these cells contained two nuclei (data not shown). These observations suggest that nuclear migration is altered before changes in the organization of the microtubule cytoskeleton can be detected, at least at this resolution. In contrast, after 24 h in glucose medium the strain exhibited an altered organization of the microtubule cytoskeleton (Fig. 9): lis1-depleted cells contained more and longer microtubules than wild type cells (Fig. 9F, I1, J1). In many instances microtubules curved around the cell tip and extended back toward the cell center (Fig. 9D1, L, M arrows; cf. 9K) or curved at the base of the bud (Fig. 9J1 arrow); this was not observed in wild type cells. Microtubules were undulated (Fig. 9C1, J1) and in extreme cases formed coils perpendicular to the long axis of the cell (Fig. 9G1, O). These coils were present throughout the cell or in a cell subcompartment. The aberrant microtubules extended into the buds where they also exhibited increased length, excessive numbers and undulations (Fig. 9I1, J1). Thick protruding bundles of microtubules also were observed (Fig. 9F), resulting in formation of aberrant blebs or extensions on the cell surface (Fig. 9F). Paired foci of strong tubulin staining material, likely corresponding to the paired tubulin structures (PTS), were randomly positioned in the cell; sometimes several such structures were present in one cell (Fig. 9D1 arrowheads; E1, H1, N, P arrows). In wild type cells, PTS are observed at the base of the bud (Fig. 9A1 arrow) during bud morphogenesis. They function as a MTOC that nucleates microtubules into the bud and mother cell (Steinberg et al. 2001, Straube et al. 2003). These observations indicate that lis1 controls microtubule organization, perhaps by controlling microtubule dynamics and position of the PTS.
Cells with abnormal microtubules were multinucleate, containing clusters of two, four or more nuclei at the base of the bud, in the cell middle, near the cell end distal to the bud or in one of the cell ends in unbudded cells (Fig. 9C2–E2,G2–J2), as described above.
During these studies we also observed cells with wild type-like morphology and arrangement of microtubules but lacking a nucleus, as described above. We hypothesized above that these cells arose from multinucleated mother cells and underwent normal cell separation; once separated from the mother cell they were unable to grow and form buds because they lacked a nucleus.
Treatment with microtubule depolymerizing drugs partially suppresses the nuclear migration phenotype of lis1-depleted cells
If aberrant microtubule organization is responsible for altered nuclear migration and cell morphology in lis1-depleted cells, we reasoned that interfering with microtubule polymerization might partially suppress the growth defect, cellular morphology and nuclear migration phenotypes of lis1-depleted cells. We thus streaked or spotted the wild type parental and Pcrg1∷lis1 strains on UMC arabinose or glucose medium containing thiabendazole (TBZ). On arabinose medium all strains grew vigorously in the presence of 0, 2 or 5 μg/mL TBZ (Fig. 10A1, 2, 3). On glucose medium with no TBZ the Pcrg1∷lis1 strain formed tiny colonies (Fig. 10A4), as described above. In the presence of 2 or 5 μg/mL TBZ, the Pcrg1∷lis1 strain formed colonies that were almost of similar size to those of the wild type parental strains (Fig. 10A5, 6; cf. 10A2, 3). In the presence of 10 μg/mL TBZ growth of the wild type strain was drastically reduced and colonies of the Pcrg1∷lis1 strain were of similar size to those of the wild type strain (data not shown). These observations indicate that interference with microtubule polymerization restores growth of the Pcrg1∷lis1 strain to almost wild type levels. Next, we examined the effect of TBZ on nuclear migration and cell morphology in asynchronous exponentially growing cultures of wild type and Pcrg1∷lis1 strains grown 39–45 h in MM glucose containing TBZ. Cells were stained with DAPI to ascertain nuclear position and number. In the absence of TBZ 98.2% (n = 286) of wild type cells contained a single nucleus (Fig. 10C). In the presence of 2 μg/mL or 5 μg/mL TBZ, 94.3% (n = 265) and 93.3% (n = 738) wild type cells respectively had a single nucleus (Fig. 10C). The remainder contained two nuclei. The average number of nuclei was, respectively, 1.02, 1.06 and 1.07 (Fig. 10B). Cells with more than two nuclei were not observed. In contrast, in the absence of TBZ, 20.6% (n = 165) lis1-depleted cells had a single nucleus; the majority were multinucleate (Fig. 10D). In the presence of 2 μg/ml or 5 μg/ml TBZ the percentage of uninucleated cells increased dramatically: 67.7% (n = 266) and 71.7% (n = 519), respectively (Fig. 10D). The average number of nuclei per cell under these conditions was, respectively, 2.25, 1.43 and 1.34 (Fig. 10B). In the absence of TBZ, cells with more than four nuclei were observed, whereas in the presence of TBZ they were absent (Fig. 10D). These observations indicate that depolymerization of microtubules partially suppresses the nuclear migration defect. Most of the cells with a single nucleus exhibited wild type-like cell morphology, indicating that there is also partial suppression of the cell morphology phenotype.
Fig. 10.
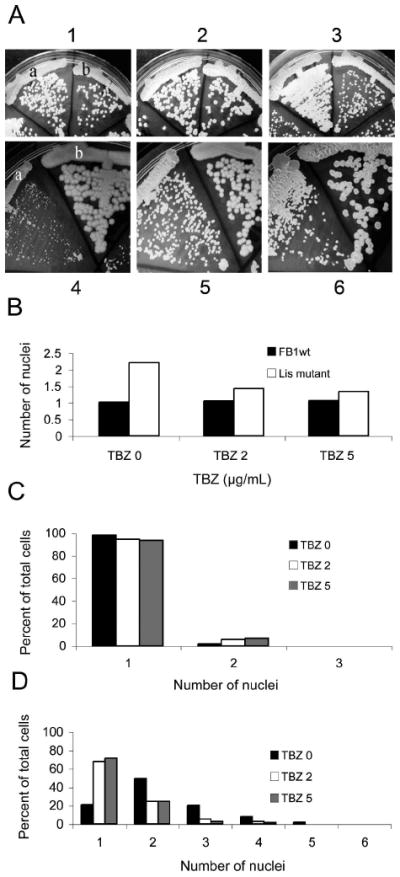
Effect of thiabendazole on growth and nuclear number in U. maydis wild type and lis1-depleted cells. A. Wild type and lis1-depleted cells were grown on UMC arabinose (1–3) or glucose (4–6) agar with 0 (1 and 4), 2 μg/mL (2 and 5), or 5 μg/mL (3 and 6) TBZ. Strains in 1–3: a = FB1 and b = FB1 Pcrg1∷lis1; 4–6, a = FB1 Pcrg1∷lis1 and b = FB1. Similar observations were made with independent Pcrg1∷lis1 isolates in the FB1 and SG200 strain backgrounds. B. Average number of nuclei in wild type (black bars) and lis1-depleted cells (white bars) grown in MM glucose broth containing DMSO, or 2 or 5 μg/mL TBZ for 39–45 h and stained with DAPI. (See Supplemental table III for cell and nuclear counts.) C. Percent of wild type cells with one or more nuclei (See Supplemental table III). DMSO (black bar); 2 μg/mL TBZ (white bar); and 5 μg/mL TBZ (gray bar). Cells were grown as described in B. D. Percent lis1-depleted cells with one or more nuclei (See Supplemental table III). Bars equal bars in C. Cells were grown as described in the legend to B.
In preliminary observations of the organization of the microtubule cytoskeleton in asynchronous exponentially growing cultures in the presence of TBZ at 0, 2 or 5 μg/mL, there was an increase in the number of cells with a wild type-like cell morphology and organization of the microtubule cytoskeleton (data not shown). Taken together, these observations support the notion that lis1 regulates organization of the microtubule cytoskeleton in U. maydis, perhaps by controlling microtubule dynamics.
DISCUSSION
In this study we characterized the homolog of the human LIS1 gene in the basidiomycete fungus Ustilago maydis and demonstrated, for the first time in fungi, that lis1 is essential for cell viability. We also showed that lis1 is necessary for nuclear migration as it is in other eukaryotes, including fungi and uncovered novel functions of lis1 in fungal cell morphogenesis, septum positioning, cell wall integrity and microtubule organization.
lis1 is essential for cell viability
This conclusion is based on three independent observations: inability to delete the gene in haploid strains, segregation analysis of meiotic progeny from a lis+/ Δlis1 heterozygous diploid and inability of lis1-depleted strains to form colonies on rich medium. The observations in U. maydis are in stark contrast with those in other fungi where lis1 is dispensable for cell viability (Xiang et al. 1995, Lee et al. 2003, Sheeman et al. 2003) but similar to those in metazoans where lis1 is essential for cell viability. In Drosophila, C. elegans and the mouse LIS1 null mutations result in embryonic lethality, indicating that LIS1 is required for an essential cellular function (Hirotsune et al. 1998, Swan et al. 1999, Dawe et al. 2001). The mechanism that leads to cell inviability in lis1-depleted Ustilago maydis cells is unknown and may differ from that in metazoan cells lacking Lis1.
lis1 is necessary for nuclear migration
Our work demonstrates that lis1 is required for nuclear migration in the yeast-like form. lis1-depleted cells contain clusters of nuclei. These multinucleated cells form buds devoid of nuclei, indicating that nuclear migration but not nuclear division is altered, at least in the first rounds of division. In some instances buds with a single nucleus are present. This nucleus could have migrated to the bud before lis1 depletion, or a low frequency of nuclear migration occurs in the absence of Lis1.
lis1 is also necessary for nuclear migration during the dimorphic switch: lis1-depleted cells undergoing the dimorphic transition contain clusters of two or more nuclei within aberrant filament-like structures. Because the nucleus does not normally divide in wild type cells during the switch (Banuett and Herskowitz 1994a) these observations most likely indicate a defect in nuclear migration before induction of the switch or might suggest additional roles of lis1 in controlling nuclear division under filament-inducing conditions.
The role of lis1 in nuclear migration and spindle positioning has been well documented in other fungi as well as in Drosophila, C. elegans, Dyctiostelium discoideum, and vertebrates (e.g. Xiang et al. 1995, Hirotsune et al. 1998, Swan et al. 1999, Dawe et al. 2001, Lee et al. 2003, Sheeman et al. 2003, Cockell et al. 2004, Rehberg et al. 2005, Siller and Coe 2008). Our results provide evidence, for the first time, that lis1 also is required for this function in a basidiomycete fungus.
Novel functions of lis1
Our work demonstrates, for the first time, a role for lis1 in fungal cell morphogenesis, septum localization and cell wall deposition and integrity: lis1-depleted cells exhibit altered axes of growth and cell polarity, aberrant thickened and mislocalized septa, uniformly or asymmetrically thickened walls, striations on the cell surface and a tendency to lyse. Altered cell wall deposition and lysis are indicative of altered cell wall properties or composition, a notion that is supported by the increased sensitivity of lis1-depleted cells to high salt concentration and to Congo red and Calcofluor white (M. Valinluck and F. Banuett unpubl obs); the latter two chemicals interfere with cell wall synthesis (Roncero and Duran, 1986).
The dramatic alterations of cell morphology, septum localization and cell wall integrity in U. maydis lis1-depleted cells are in stark contrast with the normal cell morphology of S. cerevisiae pac1/lis1 null mutants. A. nidulans nudF/lis1 null mutants form tiny colonies due to slow growth but hyphae do not exhibit altered cell polarity or morphology (Xiang et al. 1995, Lee et al. 2003, Sheeman et al. 2003). Of note, A. nidulans nudA (dynein) mutants exhibit increased hyphal branching, indicative of establishment of new axes of growth in the absence of dynein (Liu and Morris 2000). They also form a septum near the tip of the germ tube, whereas in wild type cells the septum localizes to the base of the germ tube, near the spore body (Wolkow et al. 1996). Thus in A. nidulans, dynein plays a role in septum positioning in the germ tube and this requires dynein light chain (LC8) (Liu and Morris 2000, Liu et al. 2003). Dynein mutants in other fungi result in curled hyphal tips or spiral hyphae (Plamann et al. 1994, Inoue et al. 1998), suggesting a role in normal hyphal morphology. Altered cell wall deposition and cell lysis are observed in A. nidulans nudC mutants (Chiu et al. 1997). NUDC controls Lis1 protein levels and interacts with Lis1 in diverse eukaryotes (Chiu et al. 1997, Aumais et al. 2001, Helmstaedt et al. 2008). It is possible that different proteins in the dynein/dynactin pathway or proteins that regulate Lis1 levels or activity control differently in different fungi several aspects of cell morphology, septum localization and cell wall integrity.
Dynein and microtubules also are required for nuclear migration in U. maydis (Straube et al. 2001) as they are in other organisms. Of interest, depletion of U. maydis dynein and α-tubulin results in aberrant cell morphology reminiscent of that observed in lis1-depleted cells (Steinberg et al. 2001, Straube et al. 2001). Because Lis1 has been shown to be required for dynein retrograde transport in U. maydis (Lenz et al. 2006), the observed mislocalization of septa, altered cell wall deposition and aberrant cell morphology in lis1-depleted cells might reflect altered dynein function, resulting in altered transport of vesicles to the septal and growth regions. On the other hand, because several of the lis1-depletion phenotypes (altered filamentous cell morphology, mislocalized septa and altered cell wall properties) were not described for dynein-depleted cells (Straube et al. 2001), lis1 could control these cellular functions independently of dynein.
lis1 is required for organization of the microtubule cytoskeleton
In wild type cells, microtubules grow toward the cell cortex, pause briefly and undergo catastrophe. They do not curve around the tip or form unusual structures (Steinberg et al. 2001). During bud formation paired tubulin structures (PTS), a MTOC, at the base of the bud nucleate microtubules toward the bud and mother cell. PTS contain γ-tubulin and dynein (Straube et al. 2003).
We demonstrate that lis1 is required for organization of the microtubule cytoskeleton. lis1-depleted cells have mislocalized foci of strong tubulin staining, which we presume are displaced PTS; we did not corroborate whether these structures contain γ-tubulin and dynein. lis1-depleted cells also contain more and longer microtubules than wild type cells, which bend at the cell tips and grow backward and are undulated or form coils perpendicular to the long axis of the cell. These defects might result from altered dynamic instability of microtubules in the lis1 mutant, for example decreased rates of catastrophe or increased rates of growth. Lis1 has been shown to regulate rates of catastrophe or growth in other organisms (Sapir et al. 1997, Xiang et al. 2000, Han et al. 2001).
Interference with microtubule polymerization partially suppressed the nuclear migration, cell morphology and altered microtubule organization phenotype, lending support to the notion that microbutule dynamics are altered in the lis1-depleted cells.
Microtubule number and length were not altered in A. nidulans nudF/lis1 or S. cerevisiae pac1/lis1 mutants. Of note, dynein and dynactin mutants in A. nidulans, N. crassa and A. gossypii exhibit undulated or aberrant microtubules (Tinsley et al. 1996, Minke et al. 1999, Alberti-Segui et al. 2000, Liu et al. 2003). In both N. crassa and A. gossypii the aberrant microtubules occurred in the region of clustered nuclei, whereas anucleate hyphal regions contained normal microtubules oriented parallel to the long axis of the hypha. Nuclear clustering in A. gossypii and A. nidulans dynein mutants was suppressed by microtubule depolymerizing drugs or by mutations in α-tubulin (Willins et al. 1995, Alberti-Segui et al. 2000).
Because microtubules play a crucial role in cell morphogenesis in U. maydis, their aberrant organization in lis1-depleted cells likely derails delivery of landmark proteins that determine the site of polarized growth; consequently new axes of growth may arise, or of polarity factors and cell wall material resulting in aberrantly shaped cells with misplaced septa, and altered cell wall deposition.
Supplementary Material
Acknowledgments
We thank Michael Feldbrügge for a kind gift of plasmids for KO and conditional expression of U. maydis genes, Regine Kahmann for strain SG200 and anonymous reviewers for valuable comments and suggestions. This work was financially supported by NIH Grant 2S06 GM03119 to FB.
LITERATURE CITED
- Alberti-Segui C, Dietrich F, Altmann-Jöhl R, Hoepfner D, Philippsen P. Cytoplasmic dynein is required to oppose the force that moves nuclei toward the hyphal tip in the filamentous ascomycete Ashbya gossypii. J Cell Sci. 2001;114:975–986. doi: 10.1242/jcs.114.5.975. [DOI] [PubMed] [Google Scholar]
- Aumais JP, Tunstead JR, McNeil RS, Schaar BT, McConnell SK, Lin S-H, Clark GD, Yu-Lee L-y. NudC associates with Lis1 and the dynein motor at the leading pole of neurons. J Neurosci. 2001;21:1–7. doi: 10.1523/JNEUROSCI.21-24-j0002.2001. RC187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banuett F. Genetics of Ustilago maydis, a fungal pathogen that induces tumors in maize. Annu Rev Genet. 1995;29:179–208. doi: 10.1146/annurev.ge.29.120195.001143. [DOI] [PubMed] [Google Scholar]
- Banuett F. Pathogenic development in Ustilago maydis: a progression of morphological transitions that results in tumor formation and teliospore production. In: Osiewacz H, editor. Molecular biology of fungal development. New York: Basel: Marcel Dekker; 2002. pp. 349–398. [Google Scholar]
- Banuett F, Herskowitz I. Different a alleles of Ustilago maydis are necessary for maintenance of filamentous growth but not for meiosis. Proc Natl Acad Sci USA. 1989;86:5878–5882. doi: 10.1073/pnas.86.15.5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banuett F, Herskowitz I. Morphological transitions in the life cycle of Ustilago maydis and their genetic control by the a and b loci. Exp Mycol. 1994a;18:247–266. [Google Scholar]
- Banuett F, Herskowitz I. Identification of Fuz7, a U. maydis MEK/MAPKK homologue required for a-dependent and independent processes in the fungal life cycle. Genes Dev. 1994b;8:1367–1378. doi: 10.1101/gad.8.12.1367. [DOI] [PubMed] [Google Scholar]
- Banuett F, Herskowitz I. Discrete developmental stages during teliospore formation in the corn smut fungus Ustilago maydis. Development. 1996;122:2965–2976. doi: 10.1242/dev.122.10.2965. [DOI] [PubMed] [Google Scholar]
- Banuett F, Herskowitz I. Bud morphogenesis and the actin and microtubule cytoskeleton during budding in the corn smut fungus, Ustilago maydis. Fungal Genet Biol. 2002;37:149–170. doi: 10.1016/s1087-1845(02)00548-0. [DOI] [PubMed] [Google Scholar]
- Bottin A, Kämper J, Kahmann R. Isolation of a carbon source-regulated gene from Ustilago maydis. Mol Gen Genet. 1996;253:342–352. doi: 10.1007/pl00008601. [DOI] [PubMed] [Google Scholar]
- Brachmann A, Konig J, Julius C, Feldbrügge M. A reverse genetic approach for generating gene replacement mutants in Ustilago maydis. Mol Genet Genomics. 2004;272:216–226. doi: 10.1007/s00438-004-1047-z. [DOI] [PubMed] [Google Scholar]
- Brachmann A, Schirawski J, Müller P, Kahmann R. An unusual MAP kinase is required for efficient penetration of the plant surface by Ustilago maydis. EMBO J. 2003;22:2199–2210. doi: 10.1093/emboj/cdg198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew E, Aweiss Y, Lu C-y, Banuett F. Fuz1, a MYND domain protein, is necessary for cell morphogenesis in Ustilago maydis. Mycologia. 2008;100:31–46. doi: 10.3852/mycologia.100.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu Y-H, Xian X, Sawe AL, Morris NR. Deletion of nudC, a nuclear migration gene of Aspergillus nidulans, causes morphological and cell wall abnormalities and is lethal. Mol Biol Cell. 1997;8:1735–1749. doi: 10.1091/mbc.8.9.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockell MM, Baumer K, Gönczy P. lis-1 is required for dynein-dependent cell division processes in C. elegans embryos. J Cell Sci. 2004;117:4571–4582. doi: 10.1242/jcs.01344. [DOI] [PubMed] [Google Scholar]
- Dawe AL, Caldwell KA, Harris PM, Morris NR, Caldwell GA. Evolutionarily conserved nuclear migration genes required for early embryonic development in Caenorhabditis elegans. Dev Genes Evol. 2001;211:434–441. doi: 10.1007/s004270100176. [DOI] [PubMed] [Google Scholar]
- Doehlemann G, Wahl R, Horst RJ, Voll LM, Usadel B, Poree F, Stitt M, Pons-Kühnemann J, Sonnewald U, Kahmann R, Kämper J. Reprogramming a maize plant: transcriptional and metabolic changes induced by the fungal biotroph Ustilago maydis. Plant J. 2008;56:18–95. doi: 10.1111/j.1365-313X.2008.03590.x. [DOI] [PubMed] [Google Scholar]
- Dujardin DL, Vallee RB. Dynein at the cortex. Curr Opin Cell Biol. 2002;14:44–48. doi: 10.1016/s0955-0674(01)00292-7. [DOI] [PubMed] [Google Scholar]
- Faulkner NE, Dujardin DL, Tai C-Y, Vaughan KT, O’Connell CB, Wang Y-l, Vallee RB. A role for the lissencephaly gene LIS1 in mitosis and cytoplasmic dynein function. Nat Cell Biol. 2000;2:784–791. doi: 10.1038/35041020. [DOI] [PubMed] [Google Scholar]
- Fink G, Steinberg G. Dynein-dependent motility of microtubules and nucleation sites supports polarization of the tubulin array in the fungus Ustilago maydis. Mol Biol Cell. 2006;17:3242–3253. doi: 10.1091/mbc.E05-12-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley KR, Bouchonville KJ, Quick A, Berman J. Dynein-dependent nuclear dynamics affect morphogenesis in Candida albicans by means of the Bub2p spindle checkpoint. J Cell Sci. 2008;121:466–476. doi: 10.1242/jcs.015172. [DOI] [PubMed] [Google Scholar]
- Han G, Liu B, Zhang J, Zuo W, Morris NR, Xiang X. The Aspergillus cytoplasmic dynein heavy chain and NudF localize to microtubule ends and affect microtubule dynamics. Curr Biol. 2001;11:719–724. doi: 10.1016/s0960-9822(01)00200-7. [DOI] [PubMed] [Google Scholar]
- Helmstaedt K, Laubinger K, Vosskuhl K, Bayram O, Busch S, Hoppert M, Valerius O, Seiler S, Braus GH. The nuclear migration protein NUDF/LIS1 forms a complex with NUDC and BNFa at spindle pole bodies. Eukaryot Cell. 2008;7:1041–1052. doi: 10.1128/EC.00071-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsune S, Fleck MW, Gambello MJ, Bix GJ, Chen A, Clark GD, Ledbetter DH, McBain CJ, Wynshaw-Boris A. Graded reduction of Pafah1b1 (Lis1) activity results in neuronal migration defects and early embryonic lethality. Nat Genet. 1998;19:333–339. doi: 10.1038/1221. [DOI] [PubMed] [Google Scholar]
- Holliday R. Induced mitotic crossing-over in relation to genetic replication in asynchronously dividing cells of Ustilago maydis. Genet Res. 1965;6:104–120. doi: 10.1017/s0016672300003979. [DOI] [PubMed] [Google Scholar]
- Holliday R. Ustilago maydis. In: King RC, editor. Handbook of genetics. Vol. 1. New York: Plenum Press; 1974. pp. 575–595. [Google Scholar]
- Hulo N, Bairoch A, Bulliard V, Cerutti L, Cuche B, de Castro E, Lachaize C, Langendijk-Genevaux PS, Sigrist CJA. The 20 years of PROSITE. Nucleic Acids Res. 2007;36:D245–D249. doi: 10.1093/nar/gkm977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S, Turgeon BG, Yoder OC, Aist JR. Role of fungal dynein in hyphal growth, microtubule organization, spindle pole body motility and nuclear migration. J Cell Sci. 1998;111:1555–1566. doi: 10.1242/jcs.111.11.1555. [DOI] [PubMed] [Google Scholar]
- Jacobs CW, Mattichak SJ, Knowles JF. Budding patterns during the cell cycle of the maize smut pathogen Ustilago maydis. Can J Bot. 1994;72:1675–1680. [Google Scholar]
- Kahmann R, Schirawski J. Mating in the smut fungi: from a to b to the downstream cascades. In: Heitman J, Kronstad JW, Taylor J, Casselton LA, editors. Sex in Fungi: Molecular determination and evolutionary implications. Washington DC: ASM Press; 2007. pp. 377–387. [Google Scholar]
- Klosterman SJ, Perlin MH, Garcia-Pedrajas M, Covert SF, Gold SE. Genetics of morphogenesis and pathogenic development of Ustilago maydis. Adv Genet. 2007;57:1–47. doi: 10.1016/S0065-2660(06)57001-4. [DOI] [PubMed] [Google Scholar]
- Lee W-L, Oberle JR, Cooper JA. The role of the lissencephaly protein Pac1 during nuclear migration in budding yeast. J Cell Biol. 2003;160:355–364. doi: 10.1083/jcb.200209022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz JH, Schuchardt I, Straube A, Steinberg G. A dynein loading zone for retrograde endosome motility at microtubule plus-ends. EMBO J. 2006;25:2275–2286. doi: 10.1038/sj.emboj.7601119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Morris RN. A spindle pole body-associated protein, SNAD, affects septation and conidiation in Aspergillus nidulans. Mol Gen Genet. 2000;263:375–387. doi: 10.1007/s004380051181. [DOI] [PubMed] [Google Scholar]
- Liu B, Xiang X, Lee Y-RJ. The requirement of the LC8 dynein light chain for nuclear migration and septum positioning is temperature dependent in Aspergillus nidulans. Mol Microbiol. 2003;47:291–301. doi: 10.1046/j.1365-2958.2003.03285.x. [DOI] [PubMed] [Google Scholar]
- Lupas A, van Dyke M, Stock J. Predicting Coiled Coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Mendoza-Mendoza A, Berndt P, Djamei A, Weise C, Linne U, Marahiel M, Vranes M, Kämper J, Kahmann R. Physical-chemical plant-derived signals induce differentiation in Ustilago maydis. Mol Microbiol. 2009;71:895–911. doi: 10.1111/j.1365-2958.2008.06567.x. [DOI] [PubMed] [Google Scholar]
- Minke PF, Lee IH, Tinsley JH, Bruno KS, Plamann M. Neurospora crassa ro-10 and ro-11 genes encode novel proteins required for nuclear distribution. Mol Microbiol. 1999;32:1065–1076. doi: 10.1046/j.1365-2958.1999.01421.x. [DOI] [PubMed] [Google Scholar]
- Morris NR. Nuclear migration: from fungi to the mammalian brain. J Cell Biol. 2003;148:1097–1101. doi: 10.1083/jcb.148.6.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell KL, McLaughlin DJ. Postmeiotic mitosis, basidiospore development and septation in Ustilago maydis. Mycologia. 1984;76:486–502. [Google Scholar]
- Pearson CG, Bloom K. Dynamic microtubules lead the way for spindle positioning. Nat Rev Mol Cell Biol. 2004;5:481–492. doi: 10.1038/nrm1402. [DOI] [PubMed] [Google Scholar]
- Plamann M, Minke PF, Tinsley JH, Bruno K. Cytoplasmic dynein and centractin are required for normal nuclear distribution in filamentous fungi. J Cell Biol. 1994;127:139–149. doi: 10.1083/jcb.127.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raudaskoski M. Secondary mutations at the B beta incompatibility locus and nuclear migration in the basidiomycete Schizophyllum commune. Hereditas. 1972;72:175–182. doi: 10.1111/j.1601-5223.1972.tb01041.x. [DOI] [PubMed] [Google Scholar]
- Regenfelder E, Spellig T, Hartmann A, Lauenstein S, Bölker M, Kahmann R. G proteins in Ustilago maydis: transmission of multiple signals? EMBO J. 1997;16:1934–1942. doi: 10.1093/emboj/16.8.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehberg M, Kleylein-Sohn J, Faix J, Ho T-H, Schultz I, Gräf R. Dictyostelium LIS1 is a centrosomal protein required for microtubule/cell cortex interactions, nucleus/centrosome linkage and actin dynamics. Mol Cell Biol. 2005;16:2759–2771. doi: 10.1091/mbc.E05-01-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinsch S, Gönczy P. Mechanisms of nuclear positioning. J Cell Sci. 1998;11:2283–2295. doi: 10.1242/jcs.111.16.2283. [DOI] [PubMed] [Google Scholar]
- Roncero C, Duran A. Effect of Calcofluor white and Congo red on fungal cell wall morphogenesis: in vivo activation of chitin polymerization. J Bacteriol. 1985;162:1180–1185. doi: 10.1128/jb.163.3.1180-1185.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapir T, Elbaum M, Reiner O. Reduction of microtubule catastrophe events by Lis1, platelet activating factor acetylhydrolase subunit. EMBO J. 1997;16:6977–6984. doi: 10.1093/emboj/16.23.6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer M, Heimel K, Starke V, Kämper J. The Clp1 protein is required for clamp formation and pathogenic development of Ustilago maydis. Plant Cell. 2006;18:2388–2401. doi: 10.1105/tpc.106.043521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuyler SC, Pellman D. Search, capture and signal: games microtubules play and centrosomes play. J Cell Sci. 2001;114:247–255. doi: 10.1242/jcs.114.2.247. [DOI] [PubMed] [Google Scholar]
- Sheeman B, Carvalho P, Sagot I, Geisser J, Kho D, Hoyt MA, Pellman D. Determinants of S. cerevisiae dynein localization and activation: implications for the mechanism of spindle positioning. Curr Biol. 2003;13:364–372. doi: 10.1016/s0960-9822(03)00013-7. [DOI] [PubMed] [Google Scholar]
- Siller KH, Doe CQ. Lis1/dynactin regulates metaphase spindle orientation in Drosophila neuroblasts. Dev Biol. 2008;319:1–9. doi: 10.1016/j.ydbio.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DS, Niethammer M, Ayala R, Zhou Y, Gambellot JJ, Wynshaw-Boris A, Tsai L-H. Regulation of cytoplasmic dynein behaviour and microtubule organization by mammalian Lis1. Nat Cell Biol. 2000;2:767–775. doi: 10.1038/35041000. [DOI] [PubMed] [Google Scholar]
- Snetselaar KM, Mims CW. Light and electron microscopy of Ustilago maydis hyphae in maize. Mycol Res. 1994;98:347–355. [Google Scholar]
- Spellig T, Bölker M, Lottspeich F, Frank RW, Kahmann R. Pheromones trigger filamentous growth in Ustilago maydis. EMBO J. 1994;13:1620–1627. doi: 10.1002/j.1460-2075.1994.tb06425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg G, Wedlich-Söldner R, Brill M, Schulz I. Microtubules in the fungal pathogen Ustilago maydis are highly dynamic and determine cell polarity. J Cell Sci. 2001;114:609–622. doi: 10.1242/jcs.114.3.609. [DOI] [PubMed] [Google Scholar]
- Straube A, Brill M, Oakley BR, Horio T, Steinberg G. Microtubule organization requires cell cycle-dependent nucleation at dispersed cytoplasmic sites: polar and perinuclear microtubule organizing centers in the plant pathogen Ustilago maydis. Mol Biol Cell. 2003;14:642–657. doi: 10.1091/mbc.E02-08-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straube A, Enard W, Berner A, Wedlich-Söldner R, Kahmann R, Steinberg G. A split motor domain in a cytoplasmic dynein. EMBO J. 2001;20:5091–5100. doi: 10.1093/emboj/20.18.5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan A, Nguyen T, Suter B. Drosophila Lissencephaly-1 functions with Bic-D and dynein in oocyte determination and nuclear positioning. Nat Cell Biol. 1999;1:444–449. doi: 10.1038/15680. [DOI] [PubMed] [Google Scholar]
- Tinsley JH, Minke PF, Bruno KS, Plamann M. p150Glued, the largest subunit of the dynactin complex, is nonessential in Neurospora but required for nuclear distribution. Mol Biol Cell. 1996;7:731–742. doi: 10.1091/mbc.7.5.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukuda T, Carleton S, Fotheringham S, Holloman WK. Isolation and characterization of an autonomously replicating sequence from Ustilago maydis. Mol Cell Biol. 1988;8:3703–3709. doi: 10.1128/mcb.8.9.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee RB, Tai C-Y, Faulkner NE. LIS1: cellular function of a disease-causing gene. Trends Cell Biol. 2001;11:155–160. doi: 10.1016/s0962-8924(01)01956-0. [DOI] [PubMed] [Google Scholar]
- Wang J, Holden DW, Leong SA. Gene transfer system for the phytopathogenic fungus Ustilago maydis. Proc Natl Acad Sci USA. 1988;85:865–869. doi: 10.1073/pnas.85.3.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinzierl G, Leveleki L, Hassel A, Kost G, Wanner G, Bölker M. Regulation of cell separation in the dimorphic fungus Ustilago maydis. Mol Microbiol. 2002;45:219–231. doi: 10.1046/j.1365-2958.2002.03010.x. [DOI] [PubMed] [Google Scholar]
- Willins DA, Liu B, Xiang X, Morris NR. Mutations in the heavy chain of cytoplasmic dynein suppress the nudF nuclear migration mutation of Aspergillus nidulans. Mol Gen Genet. 1997;255:194–200. doi: 10.1007/s004380050489. [DOI] [PubMed] [Google Scholar]
- Willins DA, Xiang X, Morris NR. An alpha tubulin mutation suppresses nuclear migration mutations in Aspergillus nidulans. Genetics. 1995;141:1287–1298. doi: 10.1093/genetics/141.4.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolkow TD, Harris SD, Hamer JE. Cytokinesis in Aspergillus nidulans is controlled by cell size, nuclear positioning and mitosis. J Cell Sci. 1996;109:2179–2188. doi: 10.1242/jcs.109.8.2179. [DOI] [PubMed] [Google Scholar]
- Xiang X, Beckwith SM, Morris NR. Cytoplasmic dynein is involved in nuclear migration in Aspergillus nidulans. Proc Natl Acad Sci USA. 1994;91:2100–2104. doi: 10.1073/pnas.91.6.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang X, Fisher R. Nuclear migration and positioning in filamentous fungi. Fungal Genet Biol. 2004;41:411–419. doi: 10.1016/j.fgb.2003.11.010. [DOI] [PubMed] [Google Scholar]
- Xiang X, Han G, Winkelmann DA, Zuo W, Morris RN. Dynamics of cytoplasmic dynein in living cells and the effect of a mutation in the dynactin complex actinrelated protein Arp1. Curr Biol. 2000;10:603–606. doi: 10.1016/s0960-9822(00)00488-7. [DOI] [PubMed] [Google Scholar]
- Xiang X, Osmani AH, Osmani SA, Xin M, Morris NR. NudF, a nuclear migration gene in Aspergillus nidulans, is similar to the human LIS-1 gene required for neuronal migration. Mol Biol Cell. 1995;6:287–310. doi: 10.1091/mbc.6.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Hiraoka Y. Cytoplasmic dynein in fungi: insights from nuclear migration. J Cell Sci. 2003;116:4501–4512. doi: 10.1242/jcs.00835. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.