Abstract
Hepatotropism is a prominent feature of hepatitis B virus (HBV) infection. Cell lines of nonhepatic origin do not independently support HBV replication. Here, we show that the nuclear hormone receptors, hepatocyte nuclear factor 4 and retinoid X receptor α plus peroxisome proliferator-activated receptor α, support HBV replication in nonhepatic cells by controlling pregenomic RNA synthesis, indicating these liver-enriched transcription factors control a unique molecular switch restricting viral tropism. In contrast, hepatocyte nuclear factor 3 antagonizes nuclear hormone receptor-mediated viral replication, demonstrating distinct regulatory roles for these liver-enriched transcription factors.
Hepatitis B virus (HBV) infects the hepatocytes of humans and primates, causing acute and chronic liver disease (1). It is an enveloped virus containing a 3.2-kb partially double-stranded DNA genome within its nucleocapsid (2). The viral genome is transcribed to produce the 3.5-, 2.4-, 2.1-, and 0.7-kb viral RNAs (1, 3). HBV DNA replication intermediates are synthesized inside nucleocapsids by the reverse transcription of the 3.5-kb pregenomic or core RNA using the viral polymerase (4). The reason viral tropism is restricted to hepatocytes is largely unknown, although it is assumed that infection is limited to the liver because of the tissue-restricted expression of the viral receptor. However, the observation that HBV transgenic mice display viral transcription and replication intermediates primarily in hepatocytes and kidney proximal convoluted tubules (5) indicates that liver-enriched transcription factors controlling viral RNA synthesis contribute to the hepatocyte-specific tropism of HBV. The observation that the activity of the nucleocapsid promoter is governed by multiple liver-enriched transcription factors (3, 6–8) suggested that these factors probably cooperate synergistically to control viral 3.5-kb pregenomic RNA transcription and viral replication, limiting HBV biosynthesis to cells of hepatic origin. However, the contribution of individual liver-enriched transcription factors to the restricted tissue specificity of HBV replication has not been examined because of the absence of suitable tissue culture systems. When viral RNA synthesis is controlled by the HBV promoters, replication is observed only in cell lines of hepatic origin (9–13). As the hepatoma cells used in these studies express multiple liver-enriched transcription factors, their individual roles in controlling viral replication cannot readily be examined.
In this study, a viral replication system has been developed using nonhepatoma cells where the effect of one or more liver-enriched transcription factors on HBV RNA synthesis and replication can be examined. Using this approach, it has been possible to demonstrate that the nuclear hormone receptors, hepatocyte nuclear factor 4 (HNF4) and retinoid X receptor α (RXRα) plus peroxisome proliferator-activated receptor α (PPARα), are the liver-enriched transcription factors that are essential for pregenomic RNA synthesis and viral replication. Hepatocyte nuclear factor 3 (HNF3) antagonizes the nuclear hormone receptor-mediated viral replication by inhibiting pregenomic RNA synthesis. Therefore, this replication system has identified roles for these liver-enriched transcription factors in controlling viral transcription and replication. Surprisingly, nuclear hormone receptors are the only essential liver-enriched transcription factors critical to pregenomic RNA synthesis and viral replication, indicating a previously unknown importance of these factors in the HBV life cycle and tissue-specific tropism of the virus.
Methods
Plasmid Constructions.
The steps in the cloning of the plasmid constructs used in the transfection experiments were performed by standard techniques (14). HBV DNA sequences in the greater-than-genome length constructions were derived from the plasmid pCP10, which contains two copies of the HBV genome (subtype ayw) cloned into the EcoRI site of pBR322 (15). The HBV DNA (4.1 kbp) construct that contains 1.3 copies of the HBV genome includes the viral sequence from nucleotide coordinates 1072–3182 plus 1–1990 (Fig. 1). This plasmid was constructed by cloning the NsiI/BglII HBV DNA fragment (nucleotide coordinates 1072–1990) into pUC13, generating pHBV(1072–1990). Subsequently, a complete copy of the 3.2-kbp viral genome linearized at the NcoI site (nucleotide coordinates 1375–3182 plus 1–1374) was cloned into the unique NcoI site (HBV nucleotide coordinate 1374) of pHBV(1072–1990), generating the HBV DNA (4.1 kbp) construct.
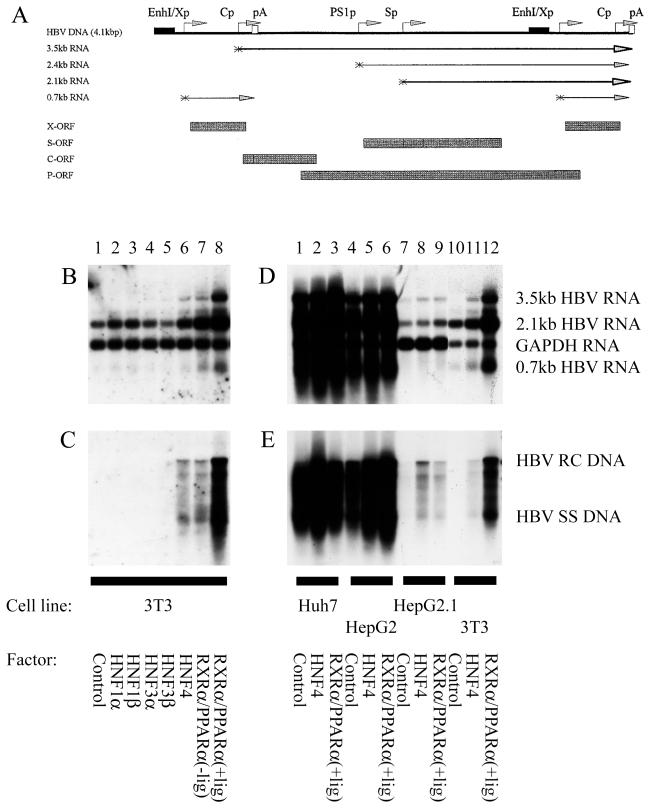
Figure 1.
Nuclear hormone receptors activate HBV replication in a nonhepatoma cell line. (A) Structure of the HBV DNA (4.1 kbp) construct used in transient transfection analysis. The 4.1-kbp greater-than-genome length HBV DNA sequence in this construct spans coordinates 1072–3182/1–1990 of the HBV genome (subtype ayw). The locations of the HBV 3.5-, 2.4-, 2.1-, and 0.7-kb transcripts are indicated. EnhI/Xp, enhancer I/X-gene promoter region; Cp, nucleocapsid or core promoter; pA, polyadenylation site; PS1p, presurface antigen promoter; Sp, major surface antigen promoter; X, X-gene; S, surface antigen gene; C, core gene; P, polymerase gene. (B–E) Cells were transiently transfected with the HBV DNA (4.1 kbp) construct and liver-enriched transcription factors. Mouse NIH 3T3 fibroblasts (3T3), human differentiated hepatoma cells (Huh7 and HepG2), and human dedifferentiated hepatoma cells (HepG2.1) were used for this analysis. (B and D) RNA (Northern) filter hybridization analysis of HBV transcripts. The glyceraldehyde 3-phosphate dehydrogenase (GAPDH) transcript was used as an internal control for RNA loading per lane. (C and E) DNA (Southern) filter hybridization analysis of HBV replication intermediates. HBV RC DNA, HBV relaxed circular DNA; HBV SS DNA, HBV single-stranded DNA. All-trans retinoic acid and clofibric acid at 1 μM and 1 mM, respectively, were used to activate the nuclear hormone receptors RXRα and PPARα (+lig).
The plasmid 4.1HNF4mut was derived by introducing clustered point mutations into the HBV DNA (4.1 kbp) construct using the Chameleon double-stranded, site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The HNF4mut mutation converted the 13-nt HNF4 recognition site sequence located between −28 and −16 (nucleotide coordinates 1757 and 1769) relative to the precore RNA initiation site from AGGTTAAAGGTCT to AGaTTgAAaGTtT. The sequence of the mutations introduced into the nucleocapsid promoter construct was verified by dideoxynucleotide sequencing (16).
Cells and Transfections.
The human hepatoma cell lines Huh7, HepG2, and HepG2.1 and the mouse NIH 3T3 fibroblast cell line were grown in RPMI 1640 medium and 10% FBS at 37°C in 5% CO2/air. Transfections for viral RNA and DNA analysis were performed as described (17) by using 10-cm plates, containing approximately 1 × 106 cells. DNA and RNA isolation was performed 3 days posttransfection. The transfected DNA mixture was composed of 10 μg of HBV DNA (4.1 kbp) plus 1.5 μg of the liver-enriched transcription factor expression vectors. The HNF1α, HNF1β, HNF3α, HNF3β, HNF4, RXRα, and PPARα expression vectors pMTHNF1α, pMTHNF1β, pCMVHNF3α, pCMVHNF3β, pCMVHNF4, pRS-hRXRα, and pCMVPPARα-G have been described (8, 18, 19). Controls were derived from cells transfected with HBV DNA (4.1 kbp) and the pCMV expression vector lacking a liver-enriched transcription factor cDNA insert (8). All-trans retinoic acid and clofibric acid at 1 μM and 1 mM, respectively, were used to activate the nuclear hormone receptors RXRα and PPARα.
Characterization of HBV Transcripts and Viral Replication Intermediates.
Transfected cells from a single plate were divided equally and used for the preparation of total cellular RNA and viral DNA replication intermediates as described (20) with minor modifications. For RNA isolation (21), the cells were lysed in 1.8 ml of 25 mM sodium citrate, pH 7.0/4 M guanidinium isothiocyanate/0.5% (vol/vol) sarcosyl/0.1 M 2-mercaptoethanol. After addition of 0.18 ml of 2 M sodium acetate, pH 4.0, the lysate was extracted with 1.8 ml of water-saturated phenol plus 0.36 ml of chloroform-isoamyl alcohol (49:1). After centrifugation for 30 min at 3,000 rpm in a Sorval RT6000, the aqueous layer was precipitated with 1.8 ml of isopropanol. The precipitate was resuspended in 0.3 ml of 25 mM sodium citrate, pH 7.0/4 M guanidinium isothiocyanate/0.5% (vol/vol) sarcosyl/0.1 M 2-mercaptoethanol and precipitated with 0.6 ml of ethanol. After centrifugation for 20 min at 14,000 rpm in a microcentrifuge, the precipitate was resuspended in 0.3 ml of 10 mM Tris hydrochloride, pH 8.0/5 mM EDTA/0.1% (wt/vol) sodium lauryl sulfate and precipitated with 45 μl of 2 M sodium acetate plus 0.6 ml of ethanol.
For the isolation of viral DNA replication intermediates, the cells were lysed in 0.4 ml of 100 mM Tris hydrochloride, pH 8.0/0.2% (vol/vol) Nonidet P-40. The lysate was centrifuged for 1 min at 14,000 rpm in a microcentrifuge to pellet the nuclei. The supernatant was adjusted to 6.75 mM magnesium acetate plus 200 μg/ml DNase I and incubated for 1 h at 37°C to remove the transfected plasmid DNA. The supernatant was readjusted to 100 mM NaCl/10 mM EDTA/0.8% (wt/vol) sodium lauryl sulfate/1.6 mg/ml pronase and incubated for an additional 1 h at 37°C. The supernatant was extracted twice with phenol, precipitated with 2 vol of ethanol, and resuspended in 100 μl of 10 mM Tris hydrochloride, pH 8.0/1 mM EDTA. RNA (Northern) and DNA (Southern) filter hybridization analysis were performed with 10 μg of total cellular RNA and 30 μl of viral DNA replication intermediates, respectively, as described (14).
RNase protection assays were performed with the PharMingen Riboquant kit, and riboprobes were synthesized by using the Ambion Maxiscript kit as described by the manufacturers. Transcription initiation sites for the 3.5-kb HBV transcripts were examined by using 20 μg of total cellular RNA and a 333 (HBV coordinates 1990–1658)-nt-long 32P-labeled HBV riboprobe. As an internal control for the RNase protection analysis, a 32P-labeled mouse ribosomal protein L32 gene riboprobe spanning 101 nt of exon 3 was used (22). All riboprobes contained additional flanking vector sequences of 40–90 nt that are not protected by HBV RNA.
Results
HBV replication is primarily restricted to hepatocytes in natural infection (1). In cell culture, viral replication is restricted to transfected cells of hepatic origin (9–13) except when pregenomic RNA is expressed from a foreign promoter (23). This restriction presumably reflects the requirement for liver-enriched transcription factors for the synthesis of pregenomic RNA from the HBV nucleocapsid promoter (7, 8, 24). The importance of specific liver-enriched transcription factors in controlling pregenomic RNA synthesis and viral replication has not been defined because of the lack of appropriate replication systems. Analysis of viral transcription and replication in non-liver cells permits the role of specific liver-enriched transcription factors in controlling HBV replication to be examined.
A HBV DNA (4.1 kbp) construct that can encode the four HBV transcripts and support viral replication in hepatoma cells and the hepatocytes of transgenic mice was examined for its ability to support viral transcription and replication in mouse NIH 3T3 fibroblasts (Fig. 1) (5). In the absence of cotransfected liver-enriched transcription factor expression vectors, the 3.5-kb HBV RNA is not expressed, and viral replication is not apparent in transient transfection analysis (Fig. 1 B and C, lane 1). The 2.1-kb HBV RNA is abundantly expressed in the absence of liver-enriched transcription factors presumably because ubiquitous transcription factors primarily control the major surface antigen promoter activity in transient transfection analysis (25). Expression of the liver-enriched transcription factors, HNF1α, HNF1β, HNF3α, and HNF3β, does not stimulate transcription of the 3.5-kb HBV RNA and therefore does not activate viral replication (Fig. 1 B and C, lanes 2–5). C/EBPα, C/EBPβ, C/EBPδ, and HNF6 also did not support 3.5-kb HBV RNA synthesis or viral replication (results not shown). In contrast, HNF4 or RXRα plus PPARα stimulates transcription of the 3.5-kb HBV RNA, and this is associated with the synthesis of encapsidated viral replication intermediates (Fig. 1 B and C, lanes 6–8). These observations indicate that nuclear hormone receptors are a major determinant in controlling the transcription of the pregenomic RNA and therefore the ability of HBV to replicate in specific cell types.
The ability of nuclear hormone receptors to control viral replication in various cell types was examined further (Fig. 1 D and E). The level of expression of the 2.1-kb HBV RNA was approximately 2- and 10-fold lower in the mouse NIH 3T3 fibroblasts and the dedifferentiated hepatoma cell line HepG2.1 (26), compared with the differentiated hepatoma cell lines Huh7 and HepG2, respectively (Fig. 1D). This indicates that the mouse NIH 3T3 fibroblasts and the differentiated hepatoma cells were probably transfected with similar efficiencies, as the level of the 2.1-kb HBV RNA depends primarily on ubiquitous transcription factors binding to the major surface antigen promoter. However, the transfection efficiency of the HepG2.1 cells may have been somewhat lower than the other cells. The lower level of the 2.1-kb HBV RNA in mouse NIH 3T3 fibroblasts and HepG2.1 cells compared with the differentiated hepatoma cell lines also may reflect the modest influence of liver-enriched transcription factors on the major surface antigen promoter activity.
It is apparent that transcription of the 3.5-kb HBV RNA and viral replication can occur in differentiated hepatoma cell lines without the ectopic expression of liver-enriched transcription factors (Fig. 1 D and E, lanes 1–6). In the dedifferentiated hepatoma cell line HepG2.1, transcription of the 3.5-kb HBV RNA and viral replication was stimulated by ectopic expression of nuclear hormone receptors (Fig. 1 D and E, lanes 7–9). In mouse NIH 3T3 fibroblasts, the ectopic expression of RXRα plus PPARα in the presence of their ligands stimulated expression of the 3.5-kb HBV RNA and viral replication to a level approximately one-third of that observed in the differentiated hepatoma cell lines Huh7 and HepG2 (Fig. 1 D and E, lanes 1–6 and 12). These observations indicate that nuclear hormone receptors may be the major positive modulators of viral replication in liver cells. Nuclear hormone receptors also were shown to activate transcription of the 3.5-kb HBV RNA and viral replication in diverse cell types including HeLa, 293T, SW1353, CV-1, and COS cells (results not shown). This finding suggests these cells of nonhepatic origin provide the ubiquitous transcription factors and basal transcription machinery necessary to mediate the nuclear hormone receptor-dependent synthesis of the 3.5-kb HBV RNA and viral replication. Increasing the amount of the nuclear hormone receptor expression vectors transfected with the HBV DNA (4.1 kbp) construct did not increase HBV replication to a major extent. This finding suggests that higher levels of nuclear hormone receptor expression do not produce an additional increase in viral replication intermediates.
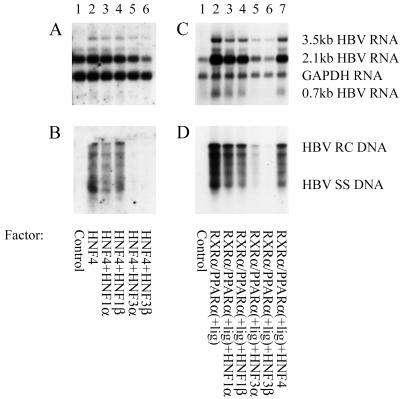
It has been shown previously that HNF3 increases transcription from the nucleocapsid promoter to approximately the same extent as nuclear hormone receptors using reporter gene constructs and transient transfection analysis (8, 19). Consequently, it might have been expected that HNF3 would have stimulated 3.5-kb HBV RNA synthesis and viral replication with similar efficiency to the nuclear hormone receptors. However, this is not the case, suggesting that measuring transcription from reporter gene constructs is not equivalent to measuring transcription from greater-than-genome length HBV constructs capable of supporting viral replication. As HNF3 did not stimulate 3.5-kb HBV RNA synthesis or viral replication, it was of interest to determine whether HNF3 could modulate the activity of the nuclear hormone receptors (Fig. 2). It is apparent that HNF3α and HNF3β reduced nuclear hormone receptor-mediated transcription of 3.5-kb HBV RNA 2- to 3-fold (Fig. 2 A and C). HNF4-mediated viral replication was essentially eliminated by ectopic expression of HNF3 (Fig. 2B, lanes 5 and 6). RXRα/PPARα-mediated viral replication was reduced approximately 8-fold by HNF3α and 25-fold by HNF3β (Fig. 2D, lanes 5 and 6). This result demonstrated that HBV replication was inhibited to a greater extent than 3.5-kb HBV RNA synthesis, and this inhibition of viral replication was specific to HNF3 as HNF1 failed to modulate HBV transcription or replication in mouse fibroblasts (Fig. 2).
Figure 2.
HNF3 inhibits nuclear hormone receptor-activated HBV replication. (A–D) Mouse NIH 3T3 fibroblasts were transiently transfected with the HBV DNA (4.1 kbp) construct and liver-enriched transcription factors. (A and C) RNA (Northern) filter hybridization analysis of HBV transcripts. (B and D) DNA (Southern) filter hybridization analysis of HBV replication intermediates.
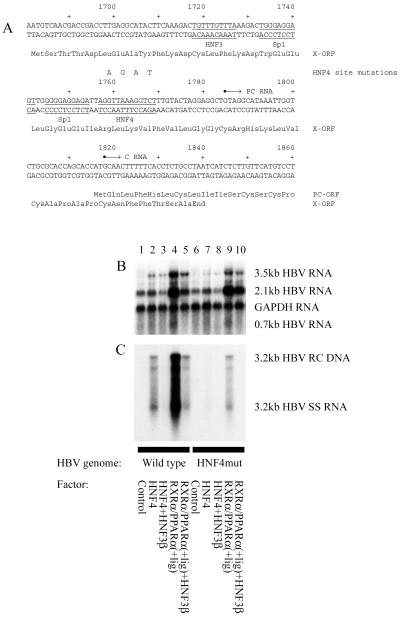
To investigate further the mechanism of modulation of viral replication by liver-enriched transcription factors, a HBV DNA (4.1 kbp) construct (Fig. 1A) with a 4-nt mutation in the nucleocapsid promoter proximal HNF4 binding site (HNF4mut) was examined for its ability to support viral transcription and replication in mouse fibroblasts (Fig. 3). The mutation blocks nuclear hormone receptor binding to the nucleocapsid promoter proximal HNF4 binding site (result not shown). The nuclear hormone receptor-dependent level of 3.5-kb HBV RNA expressed from the HNF4mut construct was slightly lower than that observed from the wild-type construct (Fig. 3B, lanes 2, 4, 7, and 9). Nuclear hormone receptor-dependent viral replication derived from the HNF4 mutant construct was greatly reduced compared with the wild-type construct, indicating the importance of the proximal HNF4 binding site in the nucleocapsid promoter for the control of viral replication (Fig. 3C, lanes 2, 4, 7, and 9). However, this HNF4 binding site is not the only site that can mediate nuclear hormone receptor-dependent HBV replication, as the HNF4mut HBV DNA (4.1 kbp) construct can still support HBV replication, albeit at a considerably lower level than the wild-type genome. This result indicates that although nuclear hormone receptors are essential transcription factors required for pregenomic RNA synthesis and viral replication in this system, they modulate HBV transcription from either the proximal nuclear hormone binding site in the core promoter or alternative binding sites present in the HBV genome. In addition, HNF3β decreased the level of viral replication from the HNF4 mutant construct in a manner similar to that observed for the wild-type HBV DNA (4.1 kbp) construct (Fig. 3C).
Figure 3.
The proximal nucleocapsid HNF4 binding site is a major determinant of nuclear hormone receptor-mediated HBV replication in mouse fibroblasts. (A) Sequence of the HBV core promoter region. The 4-nt HNF4 site mutation indicated above the wild-type sequence inhibits the binding of nuclear hormone receptors to the proximal HNF4 binding site (results not shown). The nucleotide substitutions do not alter the X-gene polypeptide sequence. The HNF3 and Sp1 binding sites are also indicated. (B and C) Mouse NIH 3T3 fibroblasts were transiently transfected with HBV DNA (4.1 kbp) constructs and liver-enriched transcription factors. The HBV HNF4mut DNA (4.1 kbp) construct (lanes 6–10) contained the 4-nt mutation (A) in the proximal HNF4 binding site of the core promoter. Both core promoter regions in this terminally redundant HBV construct (Fig. 1A) were mutated for this analysis, but similar results were obtained when only the upstream core promoter region was mutated (results not shown). (B) RNA (Northern) filter hybridization analysis of HBV transcripts. (C) DNA (Southern) filter hybridization analysis of HBV replication intermediates.
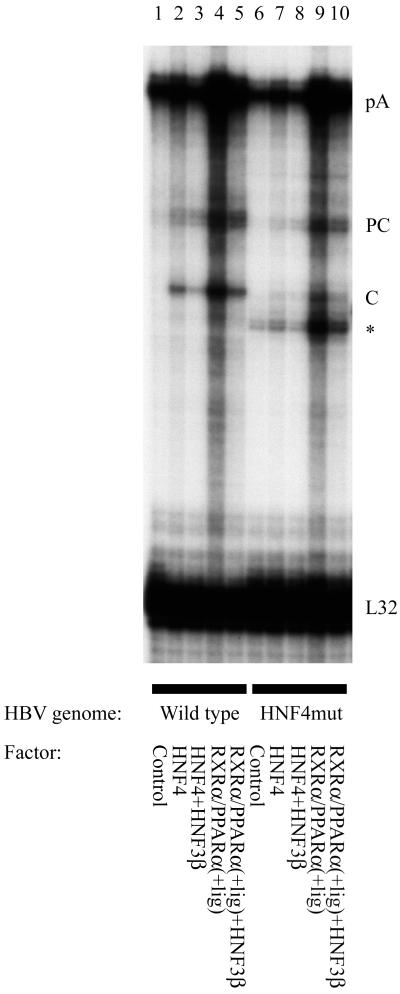
In an attempt to understand why the decrease in viral replication is greater than the decrease in the 3.5-kb HBV RNA, the sites of initiation of transcription from the core promoter were examined. This analysis was designed to determine whether the liver-enriched transcription factors modulated the relative abundance of precore and pregenomic or core RNA comprising the 3.5-kb HBV RNA (Fig. 4). The core RNA encodes the core and polymerase polypeptides and is the substrate for encapsidation and viral DNA synthesis (27–29). Therefore the level of the core RNA can directly affect the level of viral replication. In contrast, the precore RNA encodes hepatitis B e antigen (HBeAg), which has been shown to inhibit viral replication in a variety of systems (30–32). Consequently, the observed level of viral replication is likely to be positively influenced by increasing expression of the core RNA and negatively modulated by HBeAg encoded by the precore RNA.
Figure 4.
Effect of HNF3 and mutation of the proximal nuclear hormone binding site in the core promoter on the transcription initiation site of the 3.5-kb HBV RNA. Mouse NIH 3T3 fibroblasts were transiently transfected with HBV DNA (4.1 kbp) constructs and liver-enriched transcription factors. The HBV HNF4mut DNA (4.1 kbp) construct (lanes 6–10) contained the 4-nt mutation (Fig. 3A) in the proximal HNF4 binding site of the core promoter. Both core promoter regions in this terminally redundant HBV construct (Fig. 1A) were mutated for this analysis. RNase protection analysis was performed to map the transcription initiation sites of the HBV precore (PC) and pregenomic or core (C) transcripts. The HBV probe also protected a fragment (pA) derived from the 3′ end of all of the HBV RNAs that terminated at the HBV polyadenylation site. The protected fragment indicated with an asterisk is generated as a result of the cleavage of the pA-protected fragment at the site of the discontinuity between the wild-type probe and the HNF4mut containing HBV RNA. A riboprobe detecting the ribosomal gene L32 transcripts was included as an internal control.
Mutation of the proximal nucleocapsid promoter nuclear hormone receptor binding site results in a preferential decrease in the level of the core RNA compared with the precore RNA (Fig. 4, lane 2 vs. 7 and lane 4 vs. 9). This observation can account for the greater decrease in viral replication compared with the decrease in 3.5-kb HBV RNA synthesis, as the majority of the decrease in the 3.5-kb HBV RNA was because of the reduced synthesis of pregenomic RNA. Similarly, the majority of the decrease in the 3.5-kb HBV RNA synthesis from either the wild-type or the HNF4 mutant constructs resulting from the ectopic expression of HNF3 was because of the reduction in transcription from the core RNA initiation site (Fig. 4, lanes 2–5 and 7–10). The preferential inhibition of pregenomic RNA synthesis compared with precore RNA synthesis is associated with the larger decrease in viral replication relative to the change in total 3.5-kb HBV RNA synthesis. These observations indicate that the abundance of pregenomic RNA and its abundance relative to the precore RNA are important determinants controlling HBV replication level in this system.
Discussion
The development of a HBV replication system where pregenomic RNA synthesis and consequently viral replication are controlled by the ectopic expression of liver-enriched transcription factors in nonhepatoma cells has permitted the role of individual factors in this process to be analyzed. In this system, nuclear hormone receptors are the essential transcription factors required to synthesize pregenomic RNA and promote viral replication from wild-type viral genomes (Fig. 1). This is surprising as the level of expression of most genes is believed to be governed by the combinatorial activity of several transcription factors binding to the promoter region rather than a single transcription factor switching the promoter from an inactive to an active state. The dependence of HBV replication on nuclear hormone receptors suggests viral DNA synthesis may be restricted to cells expressing these transcription factors, and this requirement may be a major determinant of viral tropism. This possibility is supported by the observation that HNF4 expression is restricted to the liver, kidney, and intestine (33) and that PPARα synthesis is limited primarily to the liver, kidney, and heart (34). The limited tissue expression observed in HBV transgenic mice where transcription of pregenomic RNA and viral replication is largely restricted to the liver and the kidney proximal convoluted tubules is also consistent with an important role for these nuclear hormone receptors in determining viral tropism (5). Although nuclear hormone receptors may limit the tissues in which the HBV genome is transcribed, it is likely that a cell-surface hepatocyte-specific viral receptor restricts HBV infection to the liver. These two levels of restriction to viral replication probably limit HBV synthesis to the liver.
Nuclear hormone receptor-dependent viral transcription and replication is modulated by HNF3 as a result of the effect of these factors on pregenomic RNA synthesis (Figs. 2–4). This finding suggests that the level of viral replication in a cell depends not only on nuclear hormone receptors but on their relative abundance compared with HNF3. Therefore, alterations in the relative ratios or activities of these transcription factors within the hepatocyte occurring under various physiological or pathophysiological conditions also may be expected to affect viral replication. These conditions may include maturity-onset diabetes of the young (MODY1) where the HNF4 activity is altered (35), inflammation where the level of the PPARα ligand, leukotriene B4, is increased (36), and treatment with the fibrate class of hypolipidemic drugs, which increases PPARα activity (37, 38).
The analysis of the HNF4 mutant HBV construct (4.1 kbp) demonstrated that the nuclear hormone receptor activation of viral replication was mediated primarily through the nucleocapsid promoter proximal HNF4 binding site (Fig. 3). However, comparison of the properties of this construct with the wild-type construct demonstrated that HNF3 repressed pregenomic RNA synthesis independently of whether the proximal nucleocapsid HNF4 binding site was occupied by a nuclear hormone receptor (Fig. 3). The inhibition of nuclear hormone receptor-activated pregenomic RNA synthesis by HNF3 directly reduced viral replication in both cases.
These studies demonstrate that HBV replication using only the regulatory elements of the virus can occur in cells that normally do not support viral replication. This replication has been achieved by the ectopic expression of liver-enriched transcription factors, permitting RNA synthesis of viral genes that would not normally be expressed in a nonhepatoma cell line. This approach may be generally applicable and permit the replication of other DNA viruses that display cell type-restricted replication to be observed in a variety of normally nonpermissive cell lines if the appropriate tissue-enriched transcription factors are known. Various steps in the viral life cycle in addition to the relationship between transcription and replication may also be amenable to study in more detail using this approach. In the case of HBV, this will include the role of HNF3 in modulating transcription initiation site selection.
Acknowledgments
We are grateful to Dr. Eric F. Johnson (The Scripps Research Institute) for the plasmids pCMVHNF4 and pCMVPPARα-G, Dr. Ronald M. Evans (The Salk Institute) for the plasmid pRS-hRXRα, Dr. Robert Costa (University of Illinois, Chicago) for the plasmids pCMVHNF3α and pCMVHNF3β, Dr. Riccardo Cortese (Instituto di Ricerche di Biologia Molecolare, Rome, Italy) for the plasmid pB1.1 (rat HNF1α cDNA), and Dr. Gerald R. Crabtree (Stanford University, Stanford, CA) for the plasmid 28–1 (mouse HNF1β cDNA). We thank Dr. Eric F. Johnson, Dr. Kevin Sullivan, and Dr. Anneke K. Raney for many helpful discussions and critical reading of this manuscript. This work was supported by Public Health Service Grant AI30070 from the National Institutes of Health. This is publication number 13320-CB from The Scripps Research Institute.
Abbreviations
- HBV
hepatitis B virus
- RXRα
retinoid X receptor α
- PPARα
peroxisome proliferator-activated receptor α
- HNF
hepatocyte nuclear factor
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.041479698.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.041479698
References
- 1.Raney A K, McLachlan A. In: Molecular Biology of the Hepatitis B Virus. McLachlan A, editor. Boca Raton, FL: CRC; 1991. pp. 1–37. [Google Scholar]
- 2.Ganem D, Varmus H E. Annu Rev Biochem. 1987;56:651–693. doi: 10.1146/annurev.bi.56.070187.003251. [DOI] [PubMed] [Google Scholar]
- 3.Yen T S B. Semin Virol. 1993;4:33–42. [Google Scholar]
- 4.Will H, Reiser W, Weimer T, Pfaff E, Buscher M, Sprengle R, Cattaneo R, Schaller H. J Virol. 1987;61:904–911. doi: 10.1128/jvi.61.3.904-911.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guidotti L G, Matzke B, Schaller H, Chisari F V. J Virol. 1995;69:6158–6169. doi: 10.1128/jvi.69.10.6158-6169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang P, McLachlan A. Virology. 1994;202:430–440. doi: 10.1006/viro.1994.1359. [DOI] [PubMed] [Google Scholar]
- 7.Johnson J L, Raney A K, McLachlan A. Virology. 1995;208:147–158. doi: 10.1006/viro.1995.1138. [DOI] [PubMed] [Google Scholar]
- 8.Raney A K, Johnson J L, Palmer C N A, McLachlan A. J Virol. 1997;71:1058–1071. doi: 10.1128/jvi.71.2.1058-1071.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sureau C, Romet-Lemonne J-L, Mullins J I, Essex M. Cell. 1986;47:37–47. doi: 10.1016/0092-8674(86)90364-8. [DOI] [PubMed] [Google Scholar]
- 10.Tsurimoto T, Fujiyama A, Matsubara K. Proc Natl Acad Sci USA. 1987;84:444–448. doi: 10.1073/pnas.84.2.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sells M A, Chen M-L, Acs G. Proc Natl Acad Sci USA. 1987;84:1005–1009. doi: 10.1073/pnas.84.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang C, Jeng K, Hu C, Lo S J, Su T, Ting L-P, Chou C-K, Han S, Pfaff E, Salfeld J, et al. EMBO J. 1987;6:675–680. doi: 10.1002/j.1460-2075.1987.tb04807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yaginuma K, Shirakata Y, Kobayashi M, Koike K. Proc Natl Acad Sci USA. 1987;84:2678–2682. doi: 10.1073/pnas.84.9.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 15.Dubois M F, Pourcel C, Rousset S, Chany C, Tiollais P. Proc Natl Acad Sci USA. 1980;77:4549–4553. doi: 10.1073/pnas.77.8.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McLachlan A, Milich D R, Raney A K, Riggs M G, Hughes J L, Sorge J, Chisari F V. J Virol. 1987;61:683–692. doi: 10.1128/jvi.61.3.683-692.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raney A K, Easton A J, Milich D R, McLachlan A. J Virol. 1991;65:5774–5781. doi: 10.1128/jvi.65.11.5774-5781.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raney A K, Zhang P, McLachlan A. J Virol. 1995;69:3265–3272. doi: 10.1128/jvi.69.6.3265-3272.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Summers J, Smith P M, Huang M, Yu M. J Virol. 1991;65:1310–1317. doi: 10.1128/jvi.65.3.1310-1317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 22.Dudov K P, Perry R P. Cell. 1984;37:457–468. doi: 10.1016/0092-8674(84)90376-3. [DOI] [PubMed] [Google Scholar]
- 23.Junker M, Galle P, Schaller H. Nucleic Acids Res. 1987;15:10117–10132. doi: 10.1093/nar/15.24.10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.López-Cabrera M, Letovsky J, Hu K-Q, Siddiqui A. Virology. 1991;183:825–829. doi: 10.1016/0042-6822(91)91019-d. [DOI] [PubMed] [Google Scholar]
- 25.Raney A K, McLachlan A. J Gen Virol. 1997;78:3029–3038. doi: 10.1099/0022-1317-78-11-3029. [DOI] [PubMed] [Google Scholar]
- 26.Raney A K, Milich D R, Easton A J, McLachlan A. J Virol. 1990;64:2360–2368. doi: 10.1128/jvi.64.5.2360-2368.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ou J-H, Bao H, Shih C, Tahara S M. J Virol. 1990;64:4578–4581. doi: 10.1128/jvi.64.9.4578-4581.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nassal M, Junker-Niepmann M, Schaller H. Cell. 1990;63:1357–1363. doi: 10.1016/0092-8674(90)90431-d. [DOI] [PubMed] [Google Scholar]
- 29.Bartenschlager R, Schaller H. EMBO J. 1992;11:3413–3420. doi: 10.1002/j.1460-2075.1992.tb05420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buckwold V E, Xu Z C, Chen M, Yen T S B, Ou J H. J Virol. 1996;70:5845–5851. doi: 10.1128/jvi.70.9.5845-5851.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guidotti L G, Matzke B, Pasquinelli C, Shoenberger J M, Rogler C E, Chisari F V. J Virol. 1996;70:7056–7061. doi: 10.1128/jvi.70.10.7056-7061.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scaglioni P P, Melegari M, Wands J R. J Virol. 1997;71:345–353. doi: 10.1128/jvi.71.1.345-353.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sladek F M, Zhong W, Lai E, Darnell J E., Jr Genes Dev. 1990;4:2353–2365. doi: 10.1101/gad.4.12b.2353. [DOI] [PubMed] [Google Scholar]
- 34.Issemann I, Green S. Nature (London) 1990;347:645–650. doi: 10.1038/347645a0. [DOI] [PubMed] [Google Scholar]
- 35.Yamagata K, Furuta H, Oda N, Kaisaki P J, Menzel S, Cox N J, Fajans S S, Signorini S, Stoffel M, Bell G I. Nature (London) 1996;384:458–460. doi: 10.1038/384458a0. [DOI] [PubMed] [Google Scholar]
- 36.Devchand P R, Keller H, Peters J M, Vazquez M, Gonzalez F J, Wahli W. Nature (London) 1996;384:39–43. doi: 10.1038/384039a0. [DOI] [PubMed] [Google Scholar]
- 37.Schoonjans K, Staels B, Auwerx J. J Lipid Res. 1996;37:907–925. [PubMed] [Google Scholar]
- 38.Forman B M, Chen J, Evans R M. Proc Natl Acad Sci USA. 1997;94:4312–4317. doi: 10.1073/pnas.94.9.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]