Abstract
Myosin 1b (Myo1b), a class I myosin, is a widely expressed, single-headed, actin-associated molecular motor. Transient kinetic and single-molecule studies indicate that it is kinetically slow and responds to tension. Localization and subcellular fractionation studies indicate that Myo1b associates with the plasma membrane and certain subcellular organelles such as endosomes and lysosomes. Whether Myo1b directly associates with membranes is unknown. We demonstrate here that full-length rat Myo1b binds specifically and with high affinity to phosphatidylinositol 4,5-bisphosphate (PIP2) and phosphatidylinositol 3,4,5-triphosphate (PIP3), two phosphoinositides that play important roles in cell signaling. Binding is not Ca2+-dependent and does not involve the calmodulin-binding IQ region in the neck domain of Myo1b. Furthermore, the binding site is contained entirely within the C-terminal tail region, which contains a putative pleckstrin homology domain. Single mutations in the putative pleckstrin homology domain abolish binding of the tail domain of Myo1b to PIP2 and PIP3 in vitro. These same mutations alter the distribution of Myc-tagged Myo1b at membrane protrusions in HeLa cells where PIP2 localizes. In addition, we found that motor activity is required for Myo1b localization in filopodia. These results suggest that binding of Myo1b to phosphoinositides plays an important role in vivo by regulating localization to actin-enriched membrane projections.
Keywords: Actin, Lipid-binding Protein, Myosin, Phosphatidylinositol, Phospholipid, Filopodia, Myosin I
Introduction
Class I myosins are single-headed members of the myosin superfamily that bind actin filaments and produce mechanical force by hydrolyzing ATP. Class I myosins consist of an N-terminal head or motor domain containing the ATP- and actin-binding sites, a neck region containing repeats of a light chain-binding region known as an IQ domain, and a C-terminal tail domain. Class I myosins are widely expressed in protozoans and metazoans. In mammals, there are eight class I myosins, Myo1a–h2 (1), which play roles in diverse cellular events such as membrane trafficking, formation of membrane protrusions, cell migration, and transcription in the nucleus (2).
The myosin I tail domain, a basic region referred to as the TH1 domain, is involved in membrane binding. Acanthamoeba myosin IC binds phosphatidylserine and phosphatidylinositol 4,5-bisphosphate (PIP2) and colocalizes with PIP2 in dynamic regions of the plasma membrane, including pseudopods, endocytic cups, and the base of filopodia (3). Vertebrate Myo1a, abundant in the brush border of the small intestine, also binds phosphatidylserine and PIP2 (4), suggesting that Myo1a tethers the core bundles of actin filaments in the microvilli directly to the membrane (5). The mammalian myosin I Myo1c, which mediates GLUT4 transport in adipocytes (6, 7) and adaptation in the specialized hair cells of the inner ear (8, 9), associates with phosphoinositides having phosphates at positions 4 and 5 of the inositol ring (10).
Vertebrate Myo1b is widely expressed in tissues such as the brain, heart, lung, kidney, and liver (11). Myo1b is kinetically slow, and the interaction of actin-Myo1b with ATP is biphasic, consisting of a fast phase followed by a slow phase (12, 13). In single-molecule studies, the interaction of Myo1b with actin can be separated into two mechanical phases; the first phase is thought to be associated with Pi release, and the second phase is presumably associated with ADP release (14). Moreover, like other class I myosins (15–18), Myo1b exhibits an ADP-induced conformational change.3 The results from kinetic, single-molecule, and structural studies suggest that Myo1b undergoes a conformational change before ADP release and predict that this step is load-dependent (12, 13). Single-molecule studies subsequently showed that the rate of Myo1b dissociation from actin is force-dependent (19). The results implicate Myo1b as a force-sensing motor protein that can cross-link load-bearing actin filaments. Such a protein is better able to maintain and control cortical tension rather than to transport cargo (12).
In fractionation studies of rat liver, Myo1b associates predominantly with the plasma membrane and endoplasmic reticulum (20). In normal rat kidney cells, Myo1b is concentrated in actin-enriched protrusions of the membrane such as ruffles and lamellipodia (21). When expressed, the tail domain localizes to the plasma membrane and associates with membrane fractions similar to full-length Myo1b, suggesting that the tail domain determines primarily the cellular localization (21). Myo1b is also associated with endosomes and lysosomes whose distribution and morphology are affected by Myo1b overexpression (22, 23).
Although Myo1b associates with membranes, whether it binds to membranes directly or indirectly, i.e. through a binding partner that binds to both Myo1b and membranes, is unknown. The specificity of Myo1b binding to membranes and whether it resembles that of the other mammalian class I myosins that have been studied to date remain unclear. Thus, in this study, we investigated the interaction of Myo1b with lipids and its specificity. In addition, we examined the roles of Myo1b-lipid binding in determination of Myo1b cellular distribution.
EXPERIMENTAL PROCEDURES
Construction of Recombinant cDNAs
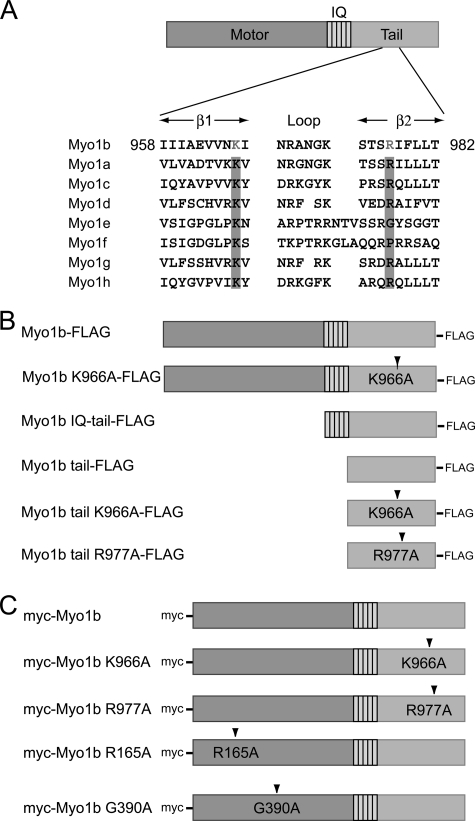
For expression in Sf9 insect cells, full-length Myo1b, the Myo1b IQ and tail domains (Myo1b IQ-tail; Asp706–Pro1107), or the tail domain only (tail; Val824–Pro1107) was amplified by PCR to contain a C-terminal FLAG tag using rat Myo1b cDNA as a template (a kind gift of Dr. Martin Bähler, University of Münster). As a control, the IQ and tail domains (Myo1c IQ-tail; Ala690–Arg1028) of mouse Myo1c were amplified by PCR to contain a C-terminal FLAG tag using enhanced GFP-mouse Myo1c (a kind gift of Dr. Thomas Friedman, NIDCD, National Institutes of Health). The PCR products were then cloned into the pFastBac Dual vector (Invitrogen) containing a calmodulin expression cassette (24). For expression in mammalian cells, Myo1b cDNAs in pFastBac Dual were amplified by PCR and ligated into pMyc (25), followed by verification of DNA by automatic sequencing. Point mutations were introduced by site-directed mutagenesis and verified by DNA sequencing. An espin/pcDNA3 construct (untagged) was kindly provided by Dr. James R. Bartles (Northwestern University, Evanston, IL).
Protein Expression and Purification
The constructs were expressed in insect cells according to the manual provided for the Bac-to-Bac baculovirus expression system (Invitrogen). The infected insect cells were pelleted and homogenized in 50 mm Tris-HCl (pH 7.5), 300 mm KCl, 2 mm MgCl2, 1 mm EGTA, 2 mm ATP, and protease inhibitors. After centrifugation at 200,000 × g for 30 min, anti-FLAG M2-agarose (Sigma) was added to the supernatant and incubated for 1 h at 4 °C. The expressed protein was eluted with 100 μg/ml FLAG peptide in 30 mm HEPES (pH 7.5), 100 mm KCl, 2 mm MgCl2, and 1 mm EGTA. Purified protein containing 10% sucrose and 1 mm DTT was stored at −80 °C.
Lipid-Bead-Protein Pulldown Assay
The PIP BeadsTM sample pack (Echelon Biosciences Inc., Salt Lake City, UT) consisted of 50% slurries of eight different phosphoinositide-coated beads and control beads containing no phosphoinositide. In each case, 20 μl of the 50% slurry was washed by adding to 1 ml of binding buffer (30 mm HEPES (pH 7.5), 100 mm KCl, 2 mm MgCl2, and 0.25% IGEPAL CA-630) with 0.1 mm free calcium or 1 mm EGTA and then centrifuging at 100 × g for 5 min at 4 °C. Purified Myo1b in 250 μl of binding buffer (final concentration of 40 μg/ml) was incubated with the washed beads on a Labquake (Thermo Scientific, Asheville, NC) with gentle mixing for 90 min at 4 °C. The beads were collected at 100 × g for 5 min at 4 °C, followed by washing three times with 500 μl of binding buffer at 100 × g for 3 min each. The supernatant was removed, and the bound protein was eluted with 20 μl of 2× Laemmli sample buffer, followed by analysis by SDS-PAGE and Coomassie Blue staining.
Liposome Pulldown Assay
Myo1b IQ-tail (4 μm) or Myo1c IQ-tail (3.7 μm) was prespun at 353,000 × g (TL100 centrifuge and TLA100 rotor, Beckman Instruments) for 30 min at 25 °C in polycarbonate tubes (7 × 20 mm; Beckman centrifuge tubes 343775) to eliminate any aggregated protein. Then, Myo1b IQ-tail (final concentration of 25 nm) or Myo1c IQ-tail (final concentration of 50 nm) was incubated with various amounts (0–50 μm total lipid for Myo1b and 0–200 μm total lipid for Myo1c) of PolyPIPosomes (65% phosphatidylcholine, 30% phosphatidylethanolamine, and 5% phosphatidylinositol (PI), PIP2, or phosphatidylinositol 3,4,5-triphosphate (PIP3); Echelon Biosciences Inc.) in 200 μl of 30 mm HEPES (pH 7.5), 100 mm KCl, 2 mm MgCl2, 1 mm EGTA, and 0.1 mg/ml BSA for 20 min at room temperature in the same polycarbonate centrifuge tubes pretreated for 1 h with 300 μl of 100 μm phosphatidylcholine to lessen the possibility of absorption of the liposomes to the tubes. The protein-liposome complexes were centrifuged at 353,000 × g for 30 min at 25 °C. The pellets were resuspended in 12.5 μl of 2× Laemmli sample buffer, followed by analysis by SDS-PAGE and staining with Coomassie Brilliant Blue. The gels were scanned and analyzed using an Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, NE). The data are expressed as a percentage of bound Myo1b IQ-tail or Myo1c IQ-tail as a function of total lipid concentration, and the data were fit to hyperbolae with Prism. Klipid refers to the concentration of total lipid at which 50% of the protein is bound, whereas KPIP2 and KPIP3 are the concentrations of accessible PIP2 and PIP3, respectively ((total lipid × 0.05)/2), at which 50% of the protein is bound.
Immunofluorescence Microscopy
HeLa or COS-7 cells were transfected using FuGENE 6 (Roche Applied Science) and then replated on poly-l-lysine-coated glass coverslips. The cells were fixed in 4% formaldehyde and 0.05% glutaraldehyde 24 h after transfection and were permeabilized and blocked in 0.5% saponin, 5% goat serum, and 2.5% BSA in PBS for 1 h, followed by overnight incubation with the appropriate primary antibodies in 0.1% saponin, 5% goat serum, and 2.5% BSA in PBS. The primary antibodies used in this study were anti-Myc tag polyclonal antibody (Cell Signaling Technology, Danvers, MA), and anti-purified PIP2 mouse monoclonal antibody (Echelon Biosciences Inc.). The cells were then incubated with secondary antibodies (Alexa Fluor 488- and/or Alexa Fluor 594-conjugated goat anti-mouse or anti-rabbit) and/or rhodamine-phalloidin (Invitrogen). The cells were viewed with a Leica TCS SP5 AOBS 405 UV spectral confocal microscopy system. The localization patterns of the Myo1b mutants did not appear different over a wide range of expression levels; nevertheless, cells expressing similar amounts of protein were compared. Images were processed with Leica application suite advanced fluorescence imaging software and Adobe Photoshop.
RESULTS
Specific Binding of Myo1b to PIP2 and PIP3
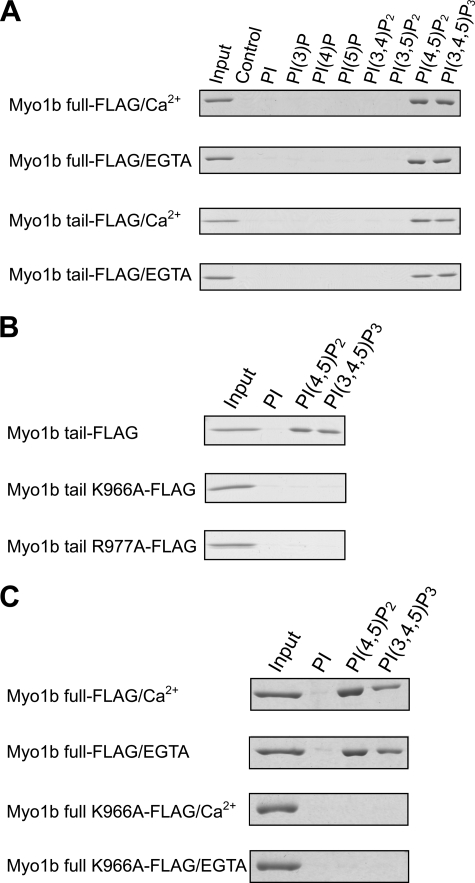
To determine whether Myo1b interacts with phosphoinositides directly, we used a pulldown assay involving beads coated with various phosphoinositides and purified expressed full-length Myo1b (Fig. 1B). As shown in Fig. 2, full-length Myo1b bound specifically to PIP2 and PIP3 in the presence and absence of Ca2+. This Ca2+-independent binding suggests that the calmodulin-binding IQ motifs in the neck domain are not involved in the interaction between Myo1b and phosphoinositides. To determine the phosphoinositide-binding domain of Myo1b, the tail domain without the IQ motifs was used in pulldown assays (Figs. 1B and 2A). The tail domain also showed specific binding to PIP2 and PIP3; binding was not dependent on Ca2+. Taken together, these results indicate that Myo1b interacts with specific lipids via its tail domain in a Ca2+-independent manner.
FIGURE 1.
Schematic diagram of rat Myo1b and constructs used in this study. A, rat Myo1b structure and alignment of the β1-loop-β2 motif of the PH domain of class I myosins. The rat Myo1b isoform used in this study consists of a motor domain, a neck domain with five IQ domains, and a tail domain. The tail domain of Myo1b contains the β1-loop-β2 motif of a putative PH domain in which conserved basic residues are highlighted. Conserved basic residues in other class I myosins are also highlighted. The GenBankTM accession numbers of the myosin isoforms are as follows: Myo1b, CAA48287; Myo1a, EDM16453; Myo1c, CAA52807; Myo1d, CAA50871; Myo1e, CAA52815; Myo1f, NP_001101546; Myo1g, NP_001128315; and Myo1h, NP_001158045. B, Myo1b constructs used in in vitro experiments. A FLAG tag was fused to the C terminus of full-length Myo1b, Myo1b IQ-tail (Asp706–Pro1107), or the Myo1b tail fragment only (Val824–Pro1107). Lys966, a conserved basic residue in the β1-loop-β2 motif, was replaced with alanine in full-length Myo1b K966A-FLAG and Myo1b tail K966A-FLAG. Also, Arg977 was replaced with alanine in Myo1b tail R977A-FLAG. C, constructs used for expression in mammalian cells. A Myc tag was fused at the N terminus to full-length wild-type Myo1b, the full-length Myo1b tail K966A mutant, or the full-length Myo1b tail R977A mutant. Mutations R165A and G390A reside in switches I and II, respectively, critical regions of the myosin motor domain.
FIGURE 2.
Lipid binding assays with epitope-tagged full-length wild-type Myo1b, Myo1b tail, or Myo1b tail mutants. Pulldown assays with lipid-coated beads were carried out in the presence of 0.1 mm free calcium or its absence (EGTA). A, Coomassie Blue-stained SDS-polyacrylamide gels show the amount of full-length Myo1b or Myo1b tail associating with various phosphoinositide (PI)-coated beads. 4% of the total protein used in the pulldown assays is shown as input. 21% of bound protein was loaded in the other lanes. Control lanes contain uncoated beads. B, FLAG-tagged Myo1b tail, Myo1b tail mutant K966A, or Myo1b tail mutant R977A was used in pulldown assays in the absence of calcium with beads coated with phosphoinositide, PIP2 (PI(4,5)P2), or PIP3 (PI(3,4,5)P3). Myo1b tail, but not the tail mutant, associated with PIP2 and PIP3. C, full-length Myo1b associates with PIP2 and PIP3 in a Ca2+-independent manner. The full-length Myo1b tail K966A mutant did not associate with PIP2 or PIP3 regardless of the Ca2+ concentration.
A Putative Pleckstrin Homology (PH) Domain Is the Binding Site of Phosphoinositides
Hokanson and Ostap (29) reported that Myo1c interacts with phosphoinositides through a PH domain and proposed that most mammalian class I myosins have a putative PH domain in the tail. Certain PH domains have a common secondary structure, β1-loop-β2, in which there are two conserved basic residues that are important for interacting with phosphoinositides (26, 27). The Myo1b tail domain has a putative PH domain containing a β1-loop-β2 structure, and the corresponding two basic residues are conserved (Lys966 and Arg977) (Fig. 1A). To determine whether these residues are involved in the binding of Myo1b to specific phosphoinositides, we constructed two Myo1b tail mutants, K966A and R977A (Fig. 1B), and examined the effect of these mutations on phospholipid binding using PIP bead pulldown assays. Fig. 2B shows that a single alanine mutation at Lys966 or Arg977 completely abolished binding to PIP2 and PIP3, suggesting that Lys966 and Arg977 within the putative PH domain play a critical role in the interaction of Myo1b with PIP2 and PIP3.
To test whether the IQ domain contributes to lipid binding, full-length Myo1b K966A (Fig. 1B) was used in pulldown assays with lipid-coated beads (Fig. 2C). Because the tail mutation eliminates lipid binding via the putative PH domain in the tail, any observed lipid binding could potentially result from the IQ domain, which was initially thought to contribute to lipid binding in the case of Myo1c (8, 28). Full-length Myo1b K966A does not bind PIP2 or PIP3 in the presence or absence of Ca2+, demonstrating that the putative PH domain is the sole lipid-binding domain and that neither the motor nor the IQ domain is involved in lipid binding.
Affinity of Myo1b IQ-Tail for Phosphoinositide
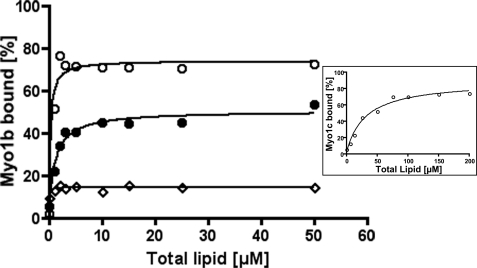
Binding of Myo1b IQ-tail to liposomes containing 5% PIP2 or PIP3 as a function of total lipid concentration is shown in Fig. 3. Although binding of Myo1b IQ-tail to PI was limited (open diamonds), Myo1b IQ-tail bound PIP2 tightly (open circles) and PIP3 less tightly (closed circles). An accurate KPIP2, the concentration of PIP2 at which 50% of the protein is bound, could not be determined but was estimated at 6 nm. The KPIP3 of Myo1b IQ-tail was determined to be 32 nm. More Myo1b (80%) associated with PolyPIPosomes containing 5% PIP2 than with PolyPIPosomes containing 5% PIP3 (50%). As the same Myo1b was used in each case, this difference in behavior cannot be attributed to the use of different batches of protein. Furthermore, the protein was prespun to eliminate any aggregates, and in the absence of PolyPIPosomes, little, if any (2–3%), Myo1b pelleted, indicating that the difference is also not due to the quality of the protein. One possibility is that the off-rate is higher for PIP3 versus PIP2 and that, during centrifugation, more Myo1b dissociates from PIP3 than from PIP2.
FIGURE 3.
Myo1b IQ-tail binding to liposomes containing PI, PIP2, or PIP3 as a function of lipid concentration. Pulldown assays of liposomes and Myo1b IQ-tail (25 nm) were performed with various concentrations of liposomes composed of 5% PI (◇), PIP2 (○), or PIP3 (●). Data are representative of three experiments for PIP2 and two experiments for PI and PIP3. The concentration at which 50% Myo1b IQ-tail bound PIP2 (KPIP2) was 6 nm, and the concentration at which 50% Myo1b IQ-tail bound PIP3 (KPIP3) was 32 nm. Inset, binding of 50 nm Myo1c IQ-tail to liposomes containing 5% PIP2 at various concentrations. The solid line is the best fit to a hyperbola yielding Klipid = 31.8 μm and KPIP2 = 0.8 μm.
The KPIP2 of Myo1c was also determined and was estimated to be 0.8 μm, which is close to previously reported values of 0.23 μm (29) and 0.53 μm (10). These results show that Myo1b has a higher affinity for PIP2 than for PIP3 and a much higher affinity (>100-fold) for PIP2 than does Myo1c.
Localization of Myo1b in Filopodia Requires Phosphoinositide Binding
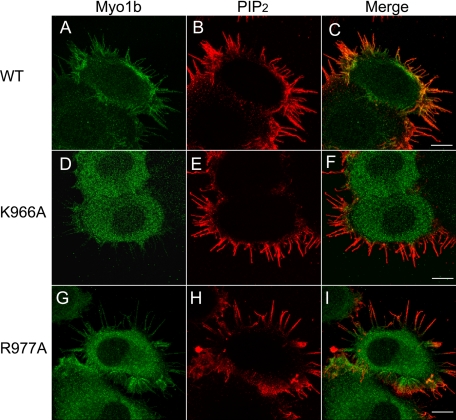
To examine the effect of a mutation in the putative PH domain that abolishes the interaction of Myo1b with phosphoinositides on the cellular distribution of Myo1b, we transfected HeLa cells with Myc-tagged full-length wild-type Myo1b or Myo1b K966A mutant (Fig. 1C), followed by staining with anti-Myc antibody. Myc-tagged wild-type Myo1b was found at the cell periphery and in filopodia (Fig. 4A), as reported previously for endogenous and Myc-tagged Myo1b (21) and GFP-tagged Myo1b (23, 30) in other cell types. The Myo1b K966A mutant localized diffusely throughout the cytoplasm and was not concentrated at the periphery or in filopodia (Fig. 4D). Transfected cells were also stained with anti-PIP2 antibodies (Fig. 4, B and E) in addition to anti-Myc antibody. Anti-PIP2 antibodies strongly stained filopodia and the cell periphery where wild-type Myo1b (Fig. 4C), but not Myo1b K966A (Fig. 4F), localized. These results suggest that phosphoinositide binding is required for localization of Myo1b in PIP2-enriched regions such as filopodia. Surprisingly, the R977A mutant localized diffusely in the cytoplasm like K966A but also localized to filopodia like the wild type (Fig. 4, G–I).
FIGURE 4.
Localization of Myo1b, Myo1b mutants, and PIP2 in HeLa cells. HeLa cells were transfected with full-length Myc-tagged wild-type Myo1b (A–C), full-length Myc-tagged Myo1b K966A (D–F), or full-length Myc-tagged Myo1b R977A (G–I) and stained with anti-Myc antibody (A, D, and G; green) and anti-PIP2 antibody (B, E, and H; red). C, F, and I are merged images of A and B, D and E, and G and H, respectively. PIP2 staining was observed at the cell periphery and in filopodia. Wild-type Myo1b, but neither of the mutants, colocalized with PIP2 at the periphery; however, like the wild type, some staining of filopodia was observed for R977A. Scale bars = 10 μm.
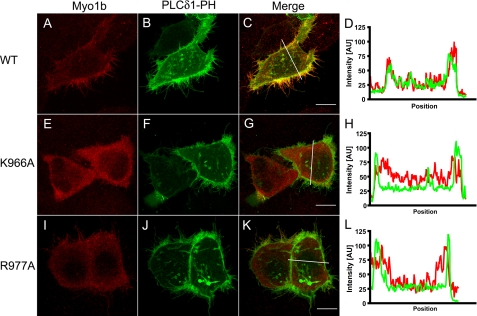
Myo1b and the Phospholipase Cδ1 (PLCδ1) PH Domain Colocalize at the Cell Periphery and in Filopodia
To monitor the Myo1b-PIP2 interaction in cells, HeLa cells were transfected with PLCδ1-PH-GFP (31), a PIP2-specific binding protein, and Myc-tagged full-length Myo1b constructs (Fig. 5). PLCδ1-PH-GFP was found at the cell periphery and in filopodia (Fig. 5, B and F), and wild-type Myo1b colocalized with PLCδ1-PH-GFP (Fig. 5, C and D). On the other hand, the Myo1b K966A mutant did not colocalize with PLCδ1-PH-GFP (Fig. 5, E–H). The Myo1b R977A mutant did not concentrate at the periphery and hence did not colocalize well with PLCδ1-PH (Fig. 5, I–L).
FIGURE 5.
Cotransfection of Myo1b or Myo1b mutants with PLCδ1-PH-GFP in HeLa cells. HeLa cells were transfected with Myc-tagged wild-type Myo1b (A), Myo1b K966A (E) or Myo1b R977A (I) together with PLCδ1-PH-GFP (B, F, and J; green) and then stained with anti-Myc antibody (red). Merged images are shown in C, G, and K for A and B, E and F, and I and J, respectively. D, H, and L show fluorescence intensity along the white lines indicated in C, G, and K, respectively. PLCδ1-PH-GFP, a PIP2-specific binding protein, localized at the cellular periphery and in filopodia (B, F, and J) and colocalized with wild-type Myo1b (C and D). On the other hand, Myo1b K966A (E) did not colocalize with PLCδ1-PH-GFP either at the periphery or in filopodia (F–H). Myo1b R977A did not colocalize with PLCδ1-PH-GFP at the periphery. Scale bars = 10 μm.
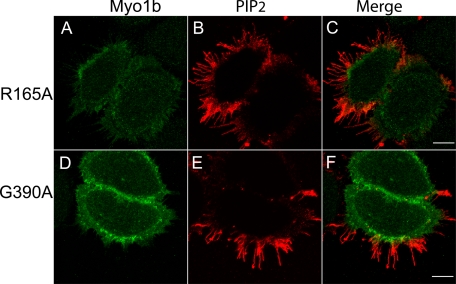
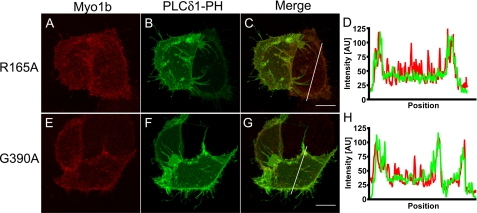
Motor Activity Is Required for Localization in Filopodia
We also examined whether motor activity is required for localization of Myo1b to filopodia, which contain an actin core. Arg165 and Gly390 of Myo1b are located in the switch I and switch II regions, respectively. These sites are strictly conserved among myosins, and replacement with alanine blocks ATP hydrolysis and isomerization, respectively. These myosins are held in the weak binding state in ATP (32, 33). We made Myc-tagged Myo1b constructs in which Arg165 and Gly390 were mutated to alanine to render the Myo1b motors inactive. These constructs were transfected into HeLa cells and stained with anti-Myc and anti-PIP2 antibodies. Both R165A (Fig. 6A) and G390A (Fig. 6D) were concentrated in the cell periphery but not in filopodia, where PIP2 is enriched (Fig. 6, B and E). These mutants were also cotransfected into HeLa cells with PLCδ-PH-GFP (Fig. 7). Both the R165A and G390A mutants colocalized with PLCδ1-PH-GFP at the cell periphery. These results indicate that, although these mutants should be freely diffusing and can associate with the cell periphery, motor activity is required for Myo1b to enter and/or remain in filopodia.
FIGURE 6.
Motor activity is required for localization of Myo1b in filopodia. Localization of Myc-Myo1b R165A (A) and Myc-Myo1b G390A (D) in HeLa cells is shown. HeLa cells were stained with anti-PIP2 antibody (B and E; red) in addition to anti-Myc antibody (A and D; green). C and F are merged images of A and B and of D and E, respectively. Both motor-dead Myo1b constructs were enriched at the cell periphery but were not present in filopodia. Scale bars = 10 μm.
FIGURE 7.
Motor activity is required for colocalization of Myo1b with PLCδ1-PH-GFP in filopodia. Myo1b R165A (A; red) and Myo1b G390A (E; red) were enriched at the cell periphery in HeLa cells that were cotransfected with PLCδ1-PH-GFP (B and F; green). D and H show fluorescence intensity along the white lines in C and G, respectively. PLCδ1-PH-GFP localized at the cell periphery and in filopodia (B and F), but motor-dead Myo1b was not present in filopodia. C and G are merged images of A and B and of E and F, respectively. Scale bars = 10 μm.
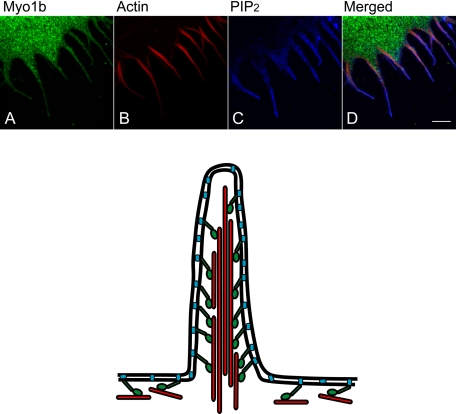
Localization of Myo1b Relative to PIP2 in Espin-induced Filopodia of COS-7 Cells
Because filopodia of HeLa cells are very thin, making it difficult to see the localization of Myo1b and PIP2 in detail, COS-7 cells were transfected with the actin-bundling protein espin, which induces actin-membrane protrusions (34). PIP2 localized at the membrane of espin-induced filopodia (Fig. 8C and supplemental Fig. S1, B and D), whereas Myo1b localized within the core (Fig. 8, A and D; and supplemental Fig. S1, A, C, G, and I). Actin filaments stained with phalloidin colocalized with Myo1b (Fig. 8, B and D; and supplemental Fig. S1, E, F, H, and I).
FIGURE 8.
Localization of Myo1b, PIP2, and actin in COS-7 cells with filopodia induced by espin. COS-7 cells were cotransfected with Myc-tagged wild-type Myo1b and untagged espin. Transfected cells were stained with anti-Myc antibody (A), rhodamine-phalloidin (B), and anti-PIP2 antibody (C). The localization of Myo1b, F-actin, and PIP2 is shown in green, red, and blue, respectively, in the merged image (D). PIP2 localized outside of Myo1b and actin in filopodia. Scale bar = 5 μm. A model of the localization of Myo1b, actin, and PIP2 in filopodia is shown below. Green, Myo1b; red, actin; blue, PIP2. Myo1b binds to actin through its head domain and to PIP2 through its tail domain.
DISCUSSION
Ca2+-independent Myo1b-Lipid Binding Is through the Tail Domain
In this study, we have shown direct binding of Myo1b through the putative PH domain in the tail domain to PIP2 and PIP3, two phosphoinositides that play major roles in cell signaling cascades that lead to important cellular events such as reorganization of the actin cytoskeleton. The results can be compared with those with the related mammalian myosin I, Myo1c. Hirono et al. (8) reported that Myo1c binds phosphoinositide through the IQ motif upon dissociation of calmodulin by calcium. On the other hand, Hokanson and Ostap (29) demonstrated that the Myo1c tail without the IQ domain is the major binding domain for lipid and that binding is not calcium/calmodulin-dependent. In the presence of calcium, the motor activity of Myo1b is inhibited, and this inhibition is reversed in the presence of exogenous calmodulin, suggesting that calcium induces dissociation of one or more calmodulins from the neck region of Myo1b (35); however, binding of full-length Myo1b to phosphoinositides is not Ca2+-dependent, and the tail domain without the IQ motifs binds both PIP2 and PIP3 (Fig. 2A). That the IQ domain plays no role in lipid binding in the case of Myo1b was further confirmed using the full-length Myo1b tail K966A mutant, which showed no binding to PIP2 or PIP3 in the presence or absence of Ca2+ (Fig. 2C).
Acanthamoeba MyoC (3) and chicken Myo1a (4) bind phosphatidylserine and PIP2; specificity for other phosphoinositides has not been investigated. Mouse Myo1c also binds to PIP2; however, binding is not specific for this particular phosphoinositide because Myo1c also binds to other phosphoinositides with phosphates at positions 4 and 5 of the inositol headgroup with a similar affinity (29). Mutation of the conserved basic residues (K872A and R903A) in the β1-loop-β2 motif of the putative PH domain of mouse Myo1c decreased its affinity for phosphoinositides by >8-fold, whereas the equivalent mutation in Acanthamoeba MyoC (R779A) resulted in only a 2–4-fold decrease in affinity for phosphatidylserine and PIP2.
Both of the homologous mutations in the β1-loop-β2 motif of Myo1b (K966A and R977A) completely abolished binding to PIP2 and PIP3 in vitro (Fig. 2B); moreover, the K966A mutant localized diffusely in the cytoplasm and lost the specific distribution at the cell periphery and in filopodia found for the wild type (Figs. 4 and 5). Although the R977A mutant also localized diffusely in the cytoplasm, it was found in filopodia (Figs. 4G and 5I). The difference in localization between the two tail mutants, neither of which binds PIP2 or PIP3 in vitro, can be explained by small differences in affinity for lipids between the two mutants with the affinity of R977A for PIP2 and/or PIP3 exceeding that of K966A. In the cell, these slightly different affinities for phosphoinositides could lead to differential localization.
Specificity and Affinity of Myo1b-Phosphoinositide Binding
Although Myo1b binds both PIP2 and PIP3 in vitro, its affinity for PIP2 exceeds that for PIP3 (Fig. 3). In addition, the cellular concentration of PIP3 is much less than that of PIP2 (36). Expression in HeLa cells of PLCδ1-PH-GFP, a specific PIP2-binding protein, demonstrates that PIP2 is concentrated at the cell periphery and in filopodia in which Myo1b colocalizes with PIP2 (Fig. 5), whereas Btk-PH-GFP (37), which binds specifically to PIP3, is found throughout the cytoplasm (supplemental Fig. S2). Together, the evidence from in vitro and in vivo studies indicates that Myo1b is more likely to associate with PIP2 than with PIP3 in HeLa cells. Upon stimulation, PIP2 and PIP3 levels dramatically increase in cells (36). Sharma et al. (38) reported that, in stimulated neutrophils, PIP2 and PIP3 localize to the leading edge of cells. Myo1b localization may also change upon stimulation involving an increase in PIP2 and PIP3.
The affinity of Myo1b for PIP2 is substantially higher than that determined for Myo1c with the same method (Fig. 3) or similar methods (10, 29). The high affinity of Myo1b for PIP2 might explain why Myo1b is more concentrated at the cell periphery compared with Myo1c (21).
Physiological Role of Myo1b-Lipid Binding
PIP2 plays an important role in actin reorganization in cellular protrusions. Many actin-binding proteins that regulate actin dynamics by capping, severing, branching, and bundling actin interact with and are activated by PIP2 (39). These actin-binding proteins are thought to function in membrane-cytoskeleton interactions (40). Reduction of cellular PIP2 by expression of PLCδ1-PH, which sequesters PIP2, or by targeting 5′-PIP2 phosphatase to the plasma membrane results in a decrease in plasma membrane-cytoskeletal adhesion energy (41). Activation of PLC by ionomycin and calcium that results in a decrease in the cellular PIP2 level leads to translocation of Myo1c from the plasma membrane to the cytoplasm (29). Myo1b may also be regulated by PIP2 given its localization at the membrane in regions exhibiting dynamic rearrangements of the actin cytoskeleton.
In HeLa and COS-7 cells, Myo1b localizes preferentially in filopodia, dynamic structures believed to sense the microenvironment and to determine the direction of cell movement. Filopodia are actin-filled membrane protrusions that function in diverse cellular processes such as cell migration, neurite outgrowth, and wound healing (42). The length of actin filaments in the core bundle is a consequence of actin polymerization and depolymerization, mediated by actin-binding proteins, which sever and cap filaments or bundle actin filaments. During the dynamic process of filopodial formation, the interaction of actin filaments with the membrane must be coordinated. The ability of Myo1b to sense tension between the membrane and the actin cytoskeleton (43) makes it an excellent candidate for regulating such actin-membrane interactions.
HeLa cells expressing motor-dead Myo1b or Myo1b unable to bind lipids have many filopodia, similar to that of untransfected cells and cells expressing wild-type Myo1b. Raposo et al. (22) demonstrated that overexpression of GFP-labeled wild-type Myo1b or Myo1b mutant in the motor domain affects the distribution of transferrin receptors in the hepatoma cell line BWTG3. Together, these results suggest that Myo1b mutants have a dominant-negative effect on endocytosis but not on filopodia.
Other class I myosins that bind phosphoinositides also localize to actin-enriched cell protrusions. Myo1a localizes to the lateral links found between the core bundle of actin filaments and the membrane in microvilli of the small intestine (5, 44, 45). Microvilli shed small vesicles into the intestinal lumen, and this activity is perturbed in Myo1a-deficient mice, suggesting a role for Myo1a in vesicle production (46). Myo1c is found in the brush border of the proximal tubules in kidney cells (47), in the stereocilia of the hair cells in the inner ear (48), and in membrane ruffles, where it supports vesicle fusion with the plasma membrane (7, 49, 50). In Myo1a knock-out mice, other class I myosins redistribute to the microvilli and may compensate for the lack of Myo1a (5, 51). Similarly, other class I myosins may also compensate for Myo1b in cells expressing mutant Myo1b.
In addition to class I myosins, other myosins are involved in actin-membrane interactions. Myosin VI, a minus-end-directed motor (52), binds PIP2 and dimerizes upon binding (53). In the renal brush border, myosin VI is required for parathyroid hormone-induced internalization of the sodium phosphate cotransporter and moves down along the actin bundle toward the base of microvilli (minus-end of the actin filament) together with the membrane-integrated transporter (54).
We have shown here that Myo1b localizes within filopodia adjacent to both PIP2 and actin filaments within filopodia (Fig. 8) and hypothesize that Myo1b associates with actin bundles and the plasma membrane and plays a role similar to that of Myo1a in microvilli. The requirement of motor activity for Myo1b for proper localization to filopodia (Figs. 6 and 7) supports this idea and suggests that, whereas a motor-independent mechanism is responsible for the location of Myo1b at the plasma membrane, Myo1b must actively move into filopodia. The ability of Myo1b to sense mechanical force and to change its motor properties depending on load (13, 19) presumably allows Myo1b to regulate actin-membrane dynamics in addition to anchoring actin filaments to the plasma membrane.
While this manuscript was under revision, a report appeared in which it was predicted, based on a computer program designed to recognize unstructured membrane-binding sites in protein sequences, that the tail of Myo1b binds lipids through an unstructured basic and hydrophobic (referred to as BH) region rather than through a highly defined tertiary structure such as a PH domain (55). No such BH region was identified in the tail of Myo1c, consistent with the previous identification of a PH domain in its tail (10). However, our studies provide experimental evidence clearly demonstrating that a PH domain exists in Myo1b and is responsible for the lipid binding of Myo1b both in vitro and in vivo.
During this same time, two other reports showed that a PH domain-like motif in the tail (and the motor domain) contributes to the localization of mammalian Myo1g to the plasma membrane (56, 57). Although not yet rigorously tested, the specificity of Myo1g for lipids appears distinct from that of Myo1c, suggesting, as our studies with Myo1b show, that different class I myosins have different affinities for phosphoinositides.
Supplementary Material
Acknowledgment
We thank our Boston Biomedical Research Institute colleague Lucia Rameh for helpful discussion.
This work was supported, in whole or in part, by National Institutes of Health Grants DC08793-01 and DC08793-01S1 (to L. M. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
C. P. Arthur, A. Lin, L. M. Coluccio, and R. A. Milligan, personal communication.
- Myo1
- myosin 1
- PI
- phosphatidylinositol
- PIP2
- phosphatidylinositol 4,5-bisphosphate
- PIP3
- phosphatidylinositol 3,4,5-triphosphate
- PH
- pleckstrin homology
- PLCδ1
- phospholipase Cδ1.
REFERENCES
- 1.Gillespie P. G., Albanesi J. P., Bähler M., Bement W. M., Berg J. S., Burgess D. R., Burnside B., Cheney R. E., Corey D. P., Coudrier E., de Lanerolle P., Hammer J. A., Hasson T., Holt J. R., Hudspeth A. J., Ikebe M., Kendrick-Jones J., Korn E. D., Li R., Mercer J. A., Milligan R. A., Mooseker M. S., Ostap E. M., Petit C., Pollard T. D., Sellers J. R., Soldati T., Titus M. A. (2001) J. Cell Biol. 155, 703–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coluccio L. M. (2008) in Myosins: A Superfamily of Molecular Motors (Coluccio L. M. ed) pp. 95–124, Springer, Dordrecht, The Netherlands [Google Scholar]
- 3.Brzeska H., Hwang K. J., Korn E. D. (2008) J. Biol. Chem. 283, 32014–32023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayden S. M., Wolenski J. S., Mooseker M. S. (1990) J. Cell Biol. 111, 443–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tyska M. J., Mackey A. T., Huang J. D., Copeland N. G., Jenkins N. A., Mooseker M. S. (2005) Mol. Biol. Cell 16, 2443–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bose A., Guilherme A., Robida S. I., Nicoloro S. M., Zhou Q. L., Jiang Z. Y., Pomerleau D. P., Czech M. P. (2002) Nature 420, 821–824 [DOI] [PubMed] [Google Scholar]
- 7.Bose A., Robida S., Furcinitti P. S., Chawla A., Fogarty K., Corvera S., Czech M. P. (2004) Mol. Cell. Biol. 24, 5447–5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirono M., Denis C. S., Richardson G. P., Gillespie P. G. (2004) Neuron 44, 309–320 [DOI] [PubMed] [Google Scholar]
- 9.Holt J. R., Gillespie S. K., Provance D. W., Shah K., Shokat K. M., Corey D. P., Mercer J. A., Gillespie P. G. (2002) Cell 108, 371–381 [DOI] [PubMed] [Google Scholar]
- 10.Hokanson D. E., Laakso J. M., Lin T., Sept D., Ostap E. M. (2006) Mol. Biol. Cell 17, 4856–4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruppert C., Kroschewski R., Bähler M. (1993) J. Cell Biol. 120, 1393–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coluccio L. M., Geeves M. A. (1999) J. Biol. Chem. 274, 21575–21580 [DOI] [PubMed] [Google Scholar]
- 13.Geeves M. A., Perreault-Micale C., Coluccio L. M. (2000) J. Biol. Chem. 275, 21624–21630 [DOI] [PubMed] [Google Scholar]
- 14.Veigel C., Coluccio L. M., Jontes J. D., Sparrow J. C., Milligan R. A., Molloy J. E. (1999) Nature 398, 530–533 [DOI] [PubMed] [Google Scholar]
- 15.Batters C., Arthur C. P., Lin A., Porter J., Geeves M. A., Milligan R. A., Molloy J. E., Coluccio L. M. (2004) EMBO J. 23, 1433–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jontes J. D., Milligan R. A. (1997) J. Cell Biol. 139, 683–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jontes J. D., Wilson-Kubalek E. M., Milligan R. A. (1995) Nature 378, 751–753 [DOI] [PubMed] [Google Scholar]
- 18.Whittaker M., Wilson-Kubalek E. M., Smith J. E., Faust L., Milligan R. A., Sweeney H. L. (1995) Nature 378, 748–751 [DOI] [PubMed] [Google Scholar]
- 19.Laakso J. M., Lewis J. H., Shuman H., Ostap E. M. (2008) Science 321, 133–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balish M. F., Moeller E. F., 3rd, Coluccio L. M. (1999) Arch. Biochem. Biophys. 370, 285–293 [DOI] [PubMed] [Google Scholar]
- 21.Ruppert C., Godel J., Müller R. T., Kroschewski R., Reinhard J., Bähler M. (1995) J. Cell Sci. 108, 3775–3786 [DOI] [PubMed] [Google Scholar]
- 22.Raposo G., Cordonnier M. N., Tenza D., Menichi B., Dürrbach A., Louvard D., Coudrier E. (1999) Mol. Biol. Cell 10, 1477–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salas-Cortes L., Ye F., Tenza D., Wilhelm C., Theos A., Louvard D., Raposo G., Coudrier E. (2005) J. Cell Sci. 118, 4823–4832 [DOI] [PubMed] [Google Scholar]
- 24.Perreault-Micale C., Shushan A. D., Coluccio L. M. (2000) J. Biol. Chem. 275, 21618–21623 [DOI] [PubMed] [Google Scholar]
- 25.Takamoto N., Komatsu S., Komaba S., Niiro N., Ikebe M. (2006) Arch. Biochem. Biophys. 456, 194–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cronin T. C., DiNitto J. P., Czech M. P., Lambright D. G. (2004) EMBO J. 23, 3711–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Isakoff S. J., Cardozo T., Andreev J., Li Z., Ferguson K. M., Abagyan R., Lemmon M. A., Aronheim A., Skolnik E. Y. (1998) EMBO J. 17, 5374–5387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang N., Lin T., Ostap E. M. (2002) J. Biol. Chem. 277, 42763–42768 [DOI] [PubMed] [Google Scholar]
- 29.Hokanson D. E., Ostap E. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 3118–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang N., Ostap E. M. (2001) Curr. Biol. 11, 1131–1135 [DOI] [PubMed] [Google Scholar]
- 31.Várnai P., Balla T. (1998) J. Cell Biol. 143, 501–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sasaki N., Sutoh K. (1998) Adv. Biophys. 35, 1–24 [PubMed] [Google Scholar]
- 33.Shimada T., Sasaki N., Ohkura R., Sutoh K. (1997) Biochemistry 36, 14037–14043 [DOI] [PubMed] [Google Scholar]
- 34.Loomis P. A., Zheng L., Sekerková G., Changyaleket B., Mugnaini E., Bartles J. R. (2003) J. Cell Biol. 163, 1045–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams R., Coluccio L. M. (1994) Cell Motil. Cytoskeleton 27, 41–48 [DOI] [PubMed] [Google Scholar]
- 36.Stephens L. R., Jackson T. R., Hawkins P. T. (1993) Biochim. Biophys. Acta 1179, 27–75 [DOI] [PubMed] [Google Scholar]
- 37.Rameh L. E., Arvidsson A., Carraway K. L., 3rd, Couvillon A. D., Rathbun G., Crompton A., VanRenterghem B., Czech M. P., Ravichandran K. S., Burakoff S. J., Wang D. S., Chen C. S., Cantley L. C. (1997) J. Biol. Chem. 272, 22059–22066 [DOI] [PubMed] [Google Scholar]
- 38.Sharma V. P., DesMarais V., Sumners C., Shaw G., Narang A. (2008) J. Leukocyte Biol. 84, 440–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yin H. L., Janmey P. A. (2003) Annu. Rev. Physiol. 65, 761–789 [DOI] [PubMed] [Google Scholar]
- 40.Sheetz M. P., Sable J. E., Döbereiner H. G. (2006) Annu. Rev. Biophys. Biomol. Struct. 35, 417–434 [DOI] [PubMed] [Google Scholar]
- 41.Raucher D., Stauffer T., Chen W., Shen K., Guo S., York J. D., Sheetz M. P., Meyer T. (2000) Cell 100, 221–228 [DOI] [PubMed] [Google Scholar]
- 42.Mattila P. K., Lappalainen P. (2008) Nat. Rev. Mol. Cell Biol. 9, 446–454 [DOI] [PubMed] [Google Scholar]
- 43.Nambiar R., McConnell R. E., Tyska M. J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 11972–11977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coluccio L. M., Bretscher A. (1989) J. Cell Biol. 108, 495–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsudaira P. T., Burgess D. R. (1979) J. Cell Biol. 83, 667–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McConnell R. E., Higginbotham J. N., Shifrin D. A., Jr., Tabb D. L., Coffey R. J., Tyska M. J. (2009) J. Cell Biol. 185, 1285–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wagner M. C., Blazer-Yost B. L., Boyd-White J., Srirangam A., Pennington J., Bennett S. (2005) Am. J. Physiol. Cell Physiol. 289, C120–C129 [DOI] [PubMed] [Google Scholar]
- 48.Gillespie P. G. (2004) Philos. Trans. R. Soc. Lond. B Biol. Sci. 359, 1945–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hagan G. N., Lin Y., Magnuson M. A., Avruch J., Czech M. P. (2008) Mol. Cell. Biol. 28, 4215–4226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang S., Lifshitz L., Patki-Kamath V., Tuft R., Fogarty K., Czech M. P. (2004) Mol. Cell. Biol. 24, 9102–9123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benesh A. E., Nambiar R., McConnell R. E., Mao S., Tabb D. L., Tyska M. J. (2010) Mol. Biol. Cell 21, 970–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wells A. L., Lin A. W., Chen L. Q., Safer D., Cain S. M., Hasson T., Carragher B. O., Milligan R. A., Sweeney H. L. (1999) Nature 401, 505–508 [DOI] [PubMed] [Google Scholar]
- 53.Spudich G., Chibalina M. V., Au J. S., Arden S. D., Buss F., Kendrick-Jones J. (2007) Nat. Cell Biol. 9, 176–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McDonough A. A. (2009) Am. J. Physiol. Cell Physiol. 297, C1331–C1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brzeska H., Guag J., Remmert K., Chacko S., Korn E. D. (2010) J. Biol. Chem. 285, 5738–5747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Olety B., Wälte M., Honnert U., Schillers H., Bähler M. (2010) FEBS Lett. 584, 493–499 [DOI] [PubMed] [Google Scholar]
- 57.Patino-Lopez G., Aravind L., Dong X., Kruhlak M. J., Ostap E. M., Shaw S. (2010) J. Biol. Chem. 285, 8675–8686 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.