Abstract
Meiosis is a cellular differentiation process in which hundreds of genes are temporally induced. Because the expression of meiotic genes during mitosis is detrimental to proliferation, meiotic genes must be negatively regulated in the mitotic cell cycle. Yet, little is known about mechanisms used by mitotic cells to repress meiosis-specific genes. Here we show that the poly(A)-binding protein Pab2, the fission yeast homolog of mammalian PABPN1, controls the expression of several meiotic transcripts during mitotic division. Our results from chromatin immunoprecipitation and promoter-swapping experiments indicate that Pab2 controls meiotic genes post-transcriptionally. Consistently, we show that the nuclear exosome complex cooperates with Pab2 in the negative regulation of meiotic genes. We also found that Pab2 plays a role in the RNA decay pathway orchestrated by Mmi1, a previously described factor that functions in the post-transcriptional elimination of meiotic transcripts. Our results support a model in which Mmi1 selectively targets meiotic transcripts for degradation via Pab2 and the exosome. Our findings have therefore uncovered a mode of gene regulation whereby a poly(A)-binding protein promotes RNA degradation in the nucleus to prevent untimely expression.
Keywords: Gene Regulation, Polyadenylation, RNA-binding Protein, RNA Turnover, Yeast Genetics, RNA Exosome, S. pombe, Meiosis, Poly(A)-binding Protein
Introduction
Meiosis is a key differentiation process essential for the generation of genetically distinct individuals. During yeast meiosis, two cells of opposite mating types fuse and conjugate their DNA to form a diploid cell. This diploid cell undergoes DNA replication followed by two rounds of cell division to produce four haploid cells. Although many of the activities used to achieve cell division are common to both meiosis and mitosis, there are several features unique to meiosis. Importantly, the meiotic and mitotic cell cycles are mutually exclusive, and genes required for meiotic differentiation are solely expressed during meiosis. Such negative control of meiotic genes during mitosis suggests important regulatory systems to avoid inappropriate activation of meiosis. To date, however, the molecular mechanisms by which meiotic differentiation genes are suppressed during the mitotic cell cycle remain poorly understood.
In the fission yeast Schizosaccharomyces pombe, meiotic differentiation involves the temporal activation of hundreds of genes (1). Although previous studies have established the importance of transcriptional regulation during fission yeast meiosis (1, 2), recent results implicate post-transcriptional mechanisms of gene regulation, including pre-mRNA splicing and mRNA degradation. Accordingly, several meiotic genes are specifically spliced during meiosis, but remain unspliced during the mitotic cell cycle (3, 4). Selective mRNA turnover is another mechanism used by fission yeast to ensure the absence of meiosis-specific transcripts during the mitotic cell cycle. The rapid elimination of specific meiotic transcripts in mitotic cells requires the YTH-family RNA-binding protein, Mmi1 (5). Mmi1 promotes the destruction of specific meiotic transcripts via recognition of a cis-element termed “determinant of selective removal” (DSR).3 Although the mechanistic details of Mmi1-dependent degradation remain to be determined, genetic evidence suggest that Mmi1 cooperates with the exosome complex of 3′→5′ exonucleases (5).
The exosome is an evolutionarily conserved complex that functions as the primary RNA degradation system in the nucleus of eukaryotic cells (6–10). The exosome is involved in the processing and degradation of many RNA species, including ribosomal RNAs, small nuclear/small nucleolar RNAs, and transfer RNAs (11–14). The exosome is also implicated in the degradation of abnormal pre-mRNAs that result from processing defects (15–18). Structurally, the core exosome is a complex of 10 subunits that forms a ring structure (19) and contains a single 3′→5′ exonuclease subunit, Rrp44/Dis3 (20). Although the core exosome is present in both the nucleus and cytoplasm, specific proteins are associated exclusively to the nuclear exosome, including a second 3′→5′ exonuclease, Rrp6 (21). In contrast, adaptor proteins have been shown to be associated exclusively with the cytosolic form of the core exosome, such as Ski7 (22, 23). Studies have also shown that the exosome can be activated by short poly(A) tails catalyzed by a non-canonical polyadenylation complex called TRAMP (12, 24–26). Conversely, evidence also supports exosome-dependent degradation of transcripts polyadenylated via the canonical poly(A) polymerase (14, 27–29).
Interestingly, several noncoding RNAs are up-regulated during the initiation of fission yeast meiosis (30). One of the meiosis-specific noncoding RNAs is meiRNA, encoded by the sme2 gene (31). The expression of meiRNA at early meiosis is essential for meiotic differentiation, because sme2Δ cells cannot accomplish the first round of meiotic division (31). The RNA-binding protein Mei2 binds the meiRNA to form a nuclear dot structure at the onset of meiosis (31, 32). Because Mei2 has been shown to colocalize with Mmi1 in a meiRNA-dependent fashion during meiosis, it has been proposed that the Mei2-meiRNA ribonucleoprotein complex sequesters Mmi1, thereby preventing the ability of Mmi1 to bind DSR-containing transcripts and promote their elimination (5). Despite the crucial role of the meiRNA in meiotic differentiation, much remains unclear about how this noncoding RNA is negatively regulated during the fission yeast mitotic cycle.
In the course of studying the fission yeast homolog of the mammalian poly(A)-binding protein nuclear 1 (PABPN1), Pab2, we have recently performed a genome-wide study that uncovered a novel function for Pab2 in 3′ end processing of polyadenylated noncoding RNAs by the exosome (27). Further analysis of Pab2-regulated genes indicated that a substantial number of genes are associated with meiotic differentiation. In this study, we provide evidence that Pab2, Mmi1, and the nuclear exosome complex cooperate to rapidly eliminate several meiotic transcripts during mitosis, including the meiosis-specific noncoding meiRNA encoded by the sme2 gene. We also find that Pab2 is bound to polyadenylated meiotic transcripts and controls the expression of meiotic mRNAs independently of the meiRNA-Mei2 complex. Consistent with this finding, the critical requirement for meiRNA expression to progress into meiosis was bypassed in the absence of Pab2. Our findings are consistent with a polyadenylation-mediated mechanism of gene regulation in which Mmi1, Pab2, and the exosome promote the nuclear degradation of meiotic transcripts during mitosis.
EXPERIMENTAL PROCEDURES
Strains and Growth
All the S. pombe strains used in this study are listed in supplemental Table 1. Unless specified otherwise, YES medium was used to grow strains to log phase at 30 °C. Genes were deleted using the PCR-mediated gene disruption using 100-mer oligonucleotides with 80 nucleotides annealing to the target gene, as previously described (33). Gene knockouts were confirmed by colony PCR and RT-PCR.
Microarray Analysis
The microarray data of pab2Δ cells were analyzed as previously described (27). A statistical test using significance analysis of microarrays (SAM) (34) identified 113 genes with a significant expression level increase in pab2Δ cells and low variation using a false discovery rate of 5%. The sme2 gene was not identified by SAM, although it displayed 11.4-, 2.8-, and 4.6-fold increases in the pab2Δ strain relative to wild-type cells in three biological replicates: the sme2 gene was not included, because the replicates represented variation higher than the threshold of SAM. Previously described algorithms (35, 36) were used to distinguish functional gene classes within the RNA species identified by SAM.
Real-time Quantitative PCR
Real-time quantitative PCR analysis using fission yeast total RNA was performed as described previously (37). Briefly, 1 μg of total RNA was treated with Promega DNase RQ1 and reverse transcribed using Qiagen Omniscript RT. cDNAs were diluted 100-fold and analyzed on an Eppendorf Realplex PCR instrument using Invitrogen Platinum SYBR Green mix. The -fold changes were calculated using the ΔΔCt method as previously described (37). Unless specified otherwise, -fold changes are relative to the wild-type strain and normalized to the nda2 mRNA.
ChIP
Chromatin immunoprecipitations (ChIPs) were performed as previously described (38) using a mouse monoclonal antibody (8WG16) specific to the C-terminal domain of the large subunit of RNA polymerase II. Quantification of the immunoprecipitated DNA was done by real-time quantitative PCR as described previously (38).
Promoter Swap Assay
Region 1–735 of the characterized meiRNA transcript (31) was inserted in the pJK148 vector (39) downstream of the adh1 promoter. The linearized plasmid was transformed into a sme2Δ strain (FBY165), and proper genomic integration was confirmed by PCR using genomic DNA. The resulting strain (FBY173) was deleted for pab2 using PCR-mediated disruption as described above. Total RNA from the FBY106, FBY165, FBY173, and FBY174 strains was analyzed by RT-PCR using primers specific for the rmt1 and meiRNA transcripts.
Northern Blot Analysis
Forty micrograms of total fission yeast RNA was resolved on a 1.25% agarose-formaldehyde gel and transferred onto nitrocellulose membrane by capillary diffusion. Membranes were cross-linked and hybridized overnight. The DNA probe used to detect meiRNA was generated by random labeling the PCR-amplified region 27–237 of the sme2 gene. Visualization and quantification of Northern blot signals were analyzed using a Typhoon Trio instrument. The -fold changes were calculated relative to the signal of meiRNA in the wild-type strain, and the signal was normalized to the 5 S rRNA.
Thermosensitive Strains Assay
FBY106, FBY107, FBY546, and FBY585 strains were grown to mid-log phase at 25 °C. The cells were then split into two flasks, one kept at 25 °C and the other shifted to the non-permissive temperature of 36 °C. The cells were grown for an additional hour before being harvested. The -fold changes were calculated relative to the WT strain for each temperature and normalized to the srp7 RNA.
RNA Immunoprecipitation
RNA immunoprecipitations were performed as described previously (27) using extracts prepared from an rrp6Δ strain that expresses a functional TAP-tagged version of Pab2 from its endogenous promoter (FBY347). A wild-type strain was used as a control for immunoprecipitation. RNA samples were treated with Promega DNase RQ1, and cDNA synthesis was primed with random hexamers (for srp7 RNA) and oligonucleotide d(T) (for polyadenylated meiotic RNA) using Qiagen Omniscript RT. The -fold changes were calculated relative to an untagged control strain and normalized to srp7 RNA.
Sporulation Assay
FBY200, FBY335, and FBY394 homothallic h90 cells were counted using a hemocytometer, and equal numbers of cells were spotted on minimal media and incubated at 30 °C for 6 days to induce sporulation. Cells spots were stained with iodine vapor, as described previously (40). In total, at least 600 cells for each strain from two independent experiments were counted manually to determine percentage of sporulation relative to wild-type h90 cells.
RESULTS
Meiotic Differentiation Genes Are Up-regulated in the Absence of Pab2
DNA microarrays were previously used to investigate the expression profile of fission yeast cells that have a deletion for the gene encoding Pab2 (27). A statistical analysis identified 113 and 85 genes that demonstrated increased and decreased RNA levels, respectively, in the pab2Δ strain (27). We used established computational algorithms (35, 36) to identify enriched functional gene classes within the differentially regulated RNA species. This analysis identified a significant number of gene classes associated with meiotic differentiation in the gene set that showed increased RNA levels in the pab2Δ strain. Accordingly, genes associated with DNA recombination (p value = 1.6e−10), reciprocal meiotic recombination (p = 8.7e−12), reproductive process (p = 6.3e−6), sexual reproduction (p = 6.3e−6), horsetail nuclear movement (p = 8.2e−8), meiotic sister chromatid segregation (p = 1.1e−6), synapsis (p = 5.7e−7), and meiotic sister chromatid cohesion (p = 2.9e−6) were up-regulated in pab2Δ cells. Overall, 32 genes previously demonstrated to be induced during fission yeast meiosis (1) were among the 113 up-regulated RNAs from pab2Δ cells (Table 1). Interestingly, of those 32 meiotic genes up-regulated in the pab2Δ strain, 17 (53%) were previously associated with the early phase of meiotic differentiation that includes premeiotic S phase and recombination (1). To validate this genome-wide data, we randomly selected seven meiotic genes within the list of Table 1 and performed quantitative PCR analysis to assess their expression levels in pab2Δ cells compared with a wild-type strain. As can be seen in Fig. 1, all of the tested meiotic genes demonstrated increased expression in the pab2Δ strain, whereas the adh1 and pgk1 housekeeping genes remained at baseline level. These results demonstrate that specific meiotic transcripts accumulate in pab2Δ cells during the mitotic cycle.
TABLE 1.
List of significantly up-regulated meiotic genes in pab2Δ cells
| Common name | Systematic ID | Description/function | Classification in meiosis cyclea | Mmi1-regulated |
|---|---|---|---|---|
| dic1 | SPBC646.17c | Dynein intermediate chain | Early | |
| mcp6 | SPBC582.06c | Meiosis specific coiled-coil protein | Early | |
| meu10 | SPCC1223.12c | GPI anchored protein | Early | |
| meu13 | SPAC222.15 | Meiotic expression up-regulated | Early | |
| meu32 | SPAP27G11.08c | Meiotic expression up-regulated | Early | |
| mug1 | SPCC11E10.03 | Dynactin complex subunit | Early | Yesc |
| mug10 | SPAC57A10.04 | Sequence orphan | Early | |
| mug4 | SPCC1393.07c | Sequence orphan | Early | |
| mug45 | SPBP8B7.04 | Sequence orphan | Early | |
| mug8 | SPAC32A11.01 | Conserved fungal protein | Early | |
| rdh54 | SPAC22F3.03c | ATP-dependent DNA helicase | Early | |
| rec10 | SPAC25G10.04c | Meiotic recombination protein | Early | Yesd |
| rec25 | SPAC17A5.18c | Meiotic recombination protein | Early | Yesc |
| rec8 | SPBC29A10.14 | Meiotic cohesin complex subunit | Early | Yesc |
| sfr1 | SPBC28F2.07 | Swi five-dependent recombination repair protein | Early | |
| sme2b | SPNCRNA.103 | Meiosis specific noncoding RNA | Early | |
| spo5 | SPBC29A10.02 | Meiotic RNA-binding protein 1 | Early | Yesc |
| ssm4 | SPAC27D7.13c | p150-Glued | Early | Yesc |
| gas4 | SPBC342.03 | GPI anchored protein | Middle | |
| mcp2 | SPCC1682.08c | RNA-binding protein | Middle | |
| mcp3 | SPAC1006.04c | Meiosis specific coiled-coil protein | Middle | |
| mde10 | SPAC17A5.04c | Peptidase family M12B | Middle | |
| meu1 | SPAC1556.06 | Meiotic expression up-regulated | Middle | Yesc |
| meu6 | SPBC428.07 | Lysine-rich protein | Middle | |
| mug28 | SPAC343.07 | Meiotically up-regulated gene | Middle | |
| sgo1 | SPBP35G2.03c | Shugoshin | Middle | |
| tht2 | SPAC23C4.07 | Meiotically up-regulated gene | Middle | |
| erg2 | SPAC20G8.07c | C-8 sterol isomerase | Unassigned | |
| mek1 | SPAC14C4.03 | Cds1/Rad53/Chk2 family protein kinase | Unassigned | Yesd |
| rik1 | SPCC11E10.08 | Silencing protein | Unassigned | |
| spn7 | SPBC19F8.01c | Septin | Unassigned | |
| SPAC29E6.07 | Sequence orphan | Unassigned | ||
| SPAC2G11.05c | BRO1 domain protein | Unassigned |
FIGURE 1.
Meiotic differentiation genes are up-regulated in pab2Δ cells. The expression levels of seven meiotic genes as well as of the adh1 and pgk1 housekeeping genes were analyzed using real-time quantitative PCR in the wild-type and pab2Δ strains. The expression levels are relative to the wild-type strain and normalized to the nda2 mRNA. The data and error bars represent the average and standard deviation from three independent experiments.
Post-transcriptional Control of Meiotic Genes by Pab2
The up-regulation of specific meiotic transcripts in the pab2Δ strain could be the consequence of a transcriptional derepression. To distinguish between transcriptional and post-transcriptional mechanisms, we assessed RNA polymerase II (pol II) occupancy by ChIP assays in wild-type and pab2Δ cells for two meiotic genes: sme2 and ssm4. Quantitative PCR analysis of the copurified DNA from RNA pol II precipitates was performed using primer sets located in the coding region of the sme2 and ssm4 genes. Whereas similar pol II densities were detected for the ssm4 gene, a slight but nonsignificant increase (as determined by Student's t test) was noted in the sme2 locus (Fig. 2A). However, this 1.5-fold increase in pol II density is in marked contrast to the 21-fold up-regulation of meiRNA level detected in the pab2Δ strain (Fig. 1). The absence of changes in RNA pol II occupancy detected at the sme2 and ssm4 genes between the wild-type and pab2Δ strains is not due to poor ChIP efficiency as demonstrated by the large dynamic range of our pol II ChIP assays after activation of nmt1 transcription (supplemental Fig. 1). These data suggest that the accumulation of meiRNA and ssm4 transcripts in pab2Δ cells is not the consequence of a transcriptional derepression.
FIGURE 2.

Post-transcriptional control of meiotic differentiation genes by Pab2. A, ChIP assays using an RNA pol II-specific antibody were analyzed using real-time quantitative PCR to assess abundance of RNA pol II on two early meiotic genes, sme2 and ssm4. The -fold enrichment of RNA pol II is relative to an untranscribed region. The data and error bars represent the average and standard deviation from at least five independent experiments. B, schematic representation of the adh1P-sme2 construct. C, total RNA prepared from wild-type (lane 4), sme2Δ (lane 1), as well as sme2Δ and sme2Δ pab2Δ strains that express the meiRNA from the adh1P-sme2 chimeric construct (lanes 2 and 3, respectively) were analyzed for meiRNA and rmt1 expression levels by RT-PCR.
To provide further evidence in favor of post-transcriptional gene regulation by Pab2, we replaced the meiosis-specific promoter of sme2 with the strong constitutive adh1 promoter and examined the expression of this chimeric construct upon deletion of pab2. The adh1 promoter was selected, because adh1 mRNA levels are not affected in the pab2Δ strain (27). The adh1P-sme2 chimeric construct (Fig. 2B) was chromosomally integrated as a single copy into sme2Δ and pab2Δ sme2Δ double mutant strains. The expression of meiRNA from the adh1P-sme2 chimeric construct in the sme2Δ strain was confirmed by RT-PCR analysis (Fig. 2C, compare lanes 1 and 2). Notably, the levels of meiRNA expressed from the adh1 promoter markedly increased in the absence of Pab2 (compare lanes 2 and 3). In contrast, levels of the housekeeping rmt1 transcript were not increased in the pab2Δ strain (compare lane 2 and 3). These results indicate that intrinsic features within the meiRNA account for its negative post-transcriptional regulation by Pab2.
The Nuclear Exosome Cooperates with Pab2 in the Regulation of Meiotic Transcripts
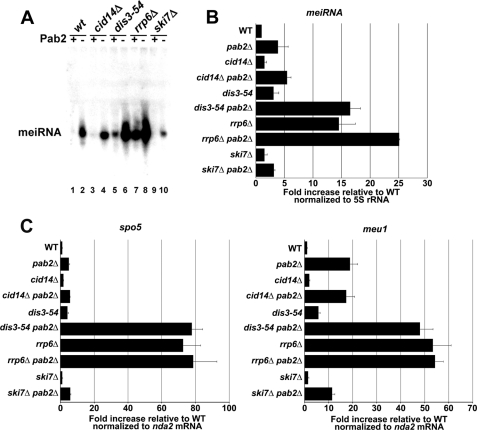
Most meiosis-specific transcripts are undetectable during normal mitotic growth in fission yeast (1). Because our data suggest that the up-regulation of meiotic genes in the pab2Δ strain results from a defect in a post-transcriptional mechanism, we reasoned that Pab2 may promote the rapid elimination of meiotic transcripts during the mitotic cell cycle. Accordingly, we have recently reported a significant overlap between the expression profile of pab2Δ cells and the expression profiles of dis3–54 and rrp6Δ strains (27), which are mutated in two exonucleases specific to the exosome. Precisely, 37 genes overlap between the up-regulated expression profiles of pab2Δ and the dis3–54 mutant, which possesses an allele that contains a mutation in the exonuclease domain of the exosome subunit Dis3/Rrp44 (41), and 37 genes overlap between the up-regulated expression profiles of pab2Δ and rrp6Δ mutants. Interestingly, we found a substantial number of meiotic genes within the list of up-regulated genes that overlap between the expression profiles of pab2Δ and dis3–54 mutants (14 of 37; supplemental Table 2) as well as between pab2Δ and rrp6Δ mutants (18 of 37; supplemental Table 3). To further examine the functional role of the exosome in the regulation of meiotic genes, we first analyzed the levels of the noncoding meiRNA in the dis3–54 and rrp6Δ strains. As can be seen in Fig. 3A, Northern blot analysis using total RNA prepared from pab2Δ (lane 2), dis3–54 (lane 5), and rrp6Δ (lane 7) strains clearly showed the accumulation of meiRNA. The meiRNA signal in each mutant was quantified relative to the meiRNA signal in the wild-type strain (Fig. 3B); quantification analyses indicated that the up-regulation of meiRNA was most robust in the rrp6Δ strain. The more robust up-regulation of meiRNA in pab2Δ as detected by qPCR (∼20-fold; Fig. 1) is likely due to the greater sensitivity of the PCR assay and the use of a different calibration RNA (nda2 mRNA versus 5 S rRNA). Strikingly, meiRNA levels in the pab2Δ dis3–54 double mutant were 4.3- and 5.5-fold greater than in the pab2Δ and dis3–54 single mutant strains, respectively (Fig. 3A, compare lanes 6 to 2 and 5; Fig. 3B). In contrast, meiRNA levels in the pab2Δ rrp6Δ double mutant were increased by 1.7-fold relative to the rrp6Δ single mutant strain (Fig. 3A, compare lanes 8 and 7; Fig. 3B). The deletion of ski7, which encodes a cytoplasmic-specific exosome-associated protein, did not cause the accumulation of meiRNA (Fig. 3A, lane 9), consistent with a nuclear degradation pathway. Taken together, these data support roles for Dis3 and Rrp6 in the nuclear degradation of the noncoding meiRNA during mitosis.
FIGURE 3.
Pab2 cooperates with the nuclear exosome in the negative regulation of meiotic genes. A, Northern blot analysis of total RNA prepared from various RNA degradation mutants in the presence (+) or absence (−) of Pab2. The meiRNA was detected using a sequence-specific probe. B, quantification of Northern blot experiments of meiRNA in various mutants of RNA degradation pathways. Expression levels are relative to the wild-type and normalized to the 5 S rRNA. The data and error bars represent the average and standard deviation from three independent experiments. C, real-time quantitative PCR analysis of spo5 and meu1 mRNAs in the same mutants described in A. The expression levels are relative to the wild-type strain and normalized to the nda2 mRNA. The data and error bars represent the average and standard deviation from three independent experiments.
The synthesis of a short 3′ poly(A) tail by the evolutionarily conserved TRAMP complex can promote the degradation of specific RNAs by the nuclear exosome (12, 24–26). To test whether the poly(A) polymerase activity of TRAMP was required for Pab2-dependent regulation of meiRNA, we deleted cid14, which encodes the single catalytic subunit of the fission yeast TRAMP complex (42, 43). Deletion of cid14 did not result in detectable levels of meiRNA (Fig. 3A, lane 3). Furthermore, meiRNA levels in a pab2Δ cid14Δ double mutant strain did not significantly change relative to the single pab2Δ mutant (Fig. 3B). These results indicate that the poly(A) polymerase activity of the TRAMP complex is not required to control meiRNA expression during mitosis.
We next assessed the role of the exosome in the post-transcriptional regulation of protein-coding meiotic genes by analyzing the expression levels of spo5 and meu1 mRNAs. As observed for meiRNA, deletion of ski7 or cid14 did not affect the expression levels of spo5 and meu1 transcripts (Fig. 3C). Consistent with data obtained for the meiRNA, a synergetic effect was noted between the pab2Δ and dis3–54 strains, whereas the rrp6Δ pab2Δ double mutant yielded similar expression levels as the rrp6Δ single mutant strain. Taken together, these results suggest that Pab2 cooperates with the nuclear exosome in the down-regulation of meiotic transcripts.
Pab2 Contributes to Mmi1-mediated Elimination of Meiotic Transcripts
The RNA-binding protein Mmi1 functions in the post-transcriptional destruction of specific meiotic mRNAs during fission yeast mitosis (5). To examine whether Pab2 and Mmi1 function in the same pathway that negatively controls meiotic genes, we used a previously described strain with a temperature-sensitive (ts) allele of mmi1 (5) to generate a pab2Δ mmi1-ts double mutant strain. The expression level of meiotic mRNAs was analyzed by qPCR using RNA prepared from wild-type, pab2Δ, mmi1-ts3, and pab2Δ mmi1-ts3 strains. Three common targets of Pab2 and Mmi1 were included in the analysis: spo5, ssm4, and meu1, as well as two targets of Pab2 that were not previously characterized as Mmi1 target mRNAs: rec10 and mek1 (Table 1) (5). As shown in Fig. 4, all five meiosis-specific transcripts, but not the adh1 control mRNA, accumulated after inactivation of Mmi1 using the mmi1-ts3 strain (compare RNA levels at permissive and non-permissive temperatures). These meiotic transcripts were also up-regulated in the pab2Δ strain, consistent with our microarray results (Table 1). Inactivation of Mmi1 in the context of a pab2 deletion (mmi1-ts3 pab2Δ) generally resulted in greater accumulation of meiotic mRNAs relative to either single mutant (Fig. 4; see non-permissive temperature of 36 °C). Yet, the up-regulation seen for some meiotic transcripts (ssm4, rec10, and mek1) in the mmi1-ts3 pab2Δ double mutant did not correspond to an additive effect between the pab2 deletion and Mmi1 inactivation, suggesting that Pab2 and Mmi1 contribute to the same pathway of gene regulation. This is in contrast to the meu1 and spo5 mRNAs, for which the up-regulation detected in the pab2Δ strain was 3.0- and 5.0-fold lower, respectively, than in the mmi1-ts3 strain (Fig. 4). Given the more important role of Mmi1 relative to Pab2 in the negative control of meu1 and spo5 expression, it is difficult to determine whether the 1.3-fold increase in meu1 and spo5 mRNAs in the mmi1-ts3 pab2Δ double mutant relative to the single mmi1-ts3 strain is biologically significant. Altogether, these results support a complex mechanism of meiotic RNA degradation that requires the coordination of Pab2 and Mmi1.
FIGURE 4.
Pab2 contributes to the Mmi1-mediated pathway of meiotic gene regulation. Real-time quantitative PCR analysis of the indicated meiotic genes as well as the adh1 control gene in wild-type, pab2Δ, mmi1-ts3, and mmi1-ts3 pab2Δ strains at the permissive (25 °C) and non-permissive (36 °C) temperatures. The levels of transcripts are relative to the wild-type strain at each temperature and normalized to the srp7 RNA. The data and error bars represent the average and standard deviation from at least three independent experiments (*, p < 0.05; **, p < 0.01; and ***, p < 0.001; Student's t test).
Pab2 Regulates Meiotic mRNAs Independently of the Noncoding meiRNA
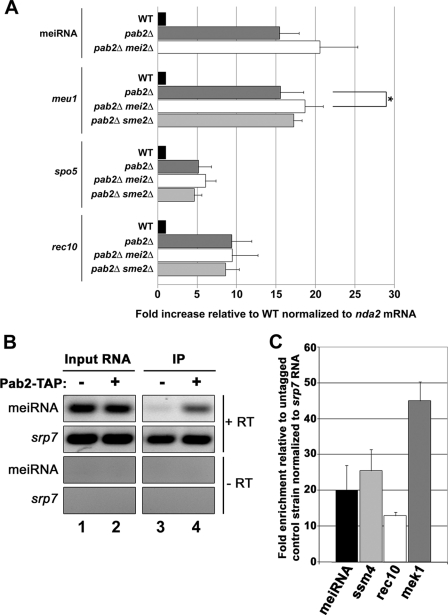
Our genome-wide analysis of gene expression changes in the pab2Δ strain revealed the up-regulation of the key meiosis-specific noncoding meiRNA, encoded by the sme2 gene (Table 1 and Figs. 1 and 3). It has been established that formation of the Mei2-meiRNA ribonucleoprotein complex during meiotic prophase sequesters Mmi1 to allow stable accumulation of meiotic mRNAs (5). To investigate whether the up-regulation of meiotic mRNAs in pab2Δ cells could be an indirect consequence of meiRNA accumulation that associates with Mei2, we analyzed the expression levels of meiotic mRNAs in wild-type, pab2Δ, pab2Δ mei2Δ, and pab2Δ sme2Δ strains. As can be seen in Fig. 5, the expression levels of meu1, spo5, and rec10 mRNAs were up-regulated in sme2Δ pab2Δ and mei2Δ pab2Δ double mutants compared with the wild-type strain, and similar to the pab2Δ single mutant. These results indicate that Pab2-dependent control of meiotic mRNAs is not mediated through inhibition of Mmi1 by the Mei2-meiRNA ribonucleoprotein.
FIGURE 5.
Regulation of meiotic mRNAs by Pab2 is direct and independent of the Mei2-meiRNA complex. A, real-time quantitative PCR analysis of the indicated meiotic genes in wild-type, pab2Δ, pab2Δ mei2Δ, and pab2Δ sme2Δ strains. The levels of transcripts are relative to the wild-type strain and normalized to the nda2 mRNA. The data and error bars represent the average and standard deviation from three independent experiments (*, p < 0.05; Student's t test). B, immunoprecipitation (IP) experiments showing that polyadenylated meiRNA, but not the cytoplasmic srp7 RNA, is enriched in Pab2-TAP precipitates. The meiRNA and srp7 cDNAs were synthesized using oligonucleotide d(T) and random primers, respectively. C, quantification using real-time quantitative PCR for the enrichment of meiotic RNAs in Pab2-TAP precipitates. The -fold changes are relative to an untagged control strain and normalized to the cytosolic srp7 RNA. The data and error bars represent the average and standard deviation from three independent experiments.
We then examined whether meiotic transcripts are bound by Pab2 in immunoprecipitation experiments using a functional TAP-tagged version of Pab2. Because meiotic transcripts are not detectable in mitotic cells, we performed these experiments in an rrp6Δ genetic background, as various meiotic transcripts accumulate in the absence of Rrp6 (Fig. 3). As can be seen in Fig. 5B, the noncoding meiRNA was clearly enriched in the Pab2-TAP precipitate compared with an untagged control strain (upper panel, compare lanes 3 and 4). In contrast, no enrichment was detected for the abundant cytosolic srp7 RNA (Fig. 5B, upper panel, compare lanes 3 and 4). Importantly, no signal was detected in the absence of reverse transcription (-RT), indicating that the observed amplification was not due to the presence of residual DNA in the immunoprecipitate (Fig. 5B, lower panels). We also quantified the enrichment of the noncoding meiRNA as well as other meiotic mRNAs by quantitative PCR. Meiotic transcripts were enriched from 12- to 45-fold in Pab2 precipitates relative to the control purification (Fig. 5C). These results indicate that Pab2 associates with polyadenylated meiotic transcripts during mitosis.
Deletion of Pab2 Can Rescue the Meiosis Arrest of sme2Δ Cells
Our data indicate that pab2Δ cells accumulate meiotic differentiation genes independently of the meiRNA-Mei2 complex (Fig. 5). This result raised the possibility that the deletion of pab2 could bypass the requirement for the formation of the Mei2-meiRNA dot during the initiation of meiosis. We therefore examined whether the meiotic differentiation arrest due to the absence of the Mei2 dot in sme2Δ cells (31) could be suppressed by deleting pab2. We used a homothallic strain (h90) that can switch between mating types, and therefore self-initiate meiotic differentiation. Wild-type, sme2Δ, and sme2Δ pab2Δ homothallic strains were generated to assess the importance of Pab2 as a negative regulator of meiotic differentiation. As can be seen by iodine staining of sporulated cells (Fig. 6A, lower panels), deleting pab2 rescued the meiotic arrest phenotype of sme2Δ cells. To confirm the iodine staining experiment, we manually counted the ascii population of each strain. As expected, sme2Δ cells sporulated at a frequency less than 1% (Fig. 6, A (panel b) and B), whereas a normal homothallic strain sporulated efficiently (Fig. 6, A (panel a) and B). Notably, the sme2Δ pab2Δ double mutant strain underwent meiosis and generated four-spore ascii at a 64% efficiency compared with wild-type cells (Fig. 6, A (panel c) and B). These results indicate that deletion of pab2 largely suppresses the meiotic arrest of sme2Δ cells and strengthen the idea that Pab2 functions in the negative regulation of meiotic differentiation during mitotic divisions.
FIGURE 6.
Pab2 is a negative regulator of meiotic differentiation. A, sporulation analysis of wild-type, sme2Δ, and pab2Δ sme2Δ homothallic strains. The spots were stained for spores using iodine vapors and cells from each strain were visualized by microscopy. B, the cells were manually counted to assess sporulation percentage relative to a wild-type homothallic strain. The data and error bars represent the average and standard deviation from two independent experiments. In total, at least 600 cells were counted for each strain.
DISCUSSION
Each eukaryotic cell is committed to either mitosis or meiosis. Accordingly, commitment to meiotic differentiation is accompanied by drastic and irreversible changes in global gene expression (1, 44, 45). Because the information encoded within the genome of most eukaryotic cells has the potential to switch from mitosis to meiosis, the expression of meiotic differentiation genes must be strongly repressed during the mitotic cycle. Here we report that Pab2, the fission yeast homolog of the mammalian PABPN1, functions as a negative regulator of meiosis-specific genes during the mitotic cycle. A polyadenylation-dependent mechanism has been previously established during the mitosis-meiosis decision in the Caenorhabditis elegans germ line. In this organism, a regulatory cytoplasmic polyadenylation complex stimulates the expression of meiotic genes (46, 47). Whereas this cytoplasmic pathway represents a positive regulatory step that reinforces commitment to the meiotic cycle in C. elegans, the Pab2-dependent mechanism described in this study is nuclear and represents a negative regulatory step to prevent untimely expression of meiotic genes. Our study therefore provides novel insights into the negative control of meiotic gene expression during fission yeast mitosis by demonstrating the important role of a poly(A)-binding protein in this process.
Post-transcriptional Gene Regulation of Meiotic Genes by Pab2
3′-End polyadenylation of mRNAs is generally thought to confer positive roles in eukaryotes, including nuclear export competence, stability, and efficient translational activity. However, the existence of an evolutionarily conserved mechanism in which the 3′ poly(A) tail can also target transcripts for degradation via the nuclear exosome complex of 3→5′ exonucleases has recently been established (24–26, 48, 49). Consistently, the data presented in this study suggest a polyadenylation-mediated mechanism of gene regulation in which Pab2 promotes the degradation of meiosis-specific transcripts during the mitotic cell cycle. Our attempts to determine changes in the half-life of meiotic transcripts that are regulated by Pab2 have been unsuccessful, however, because meiotic RNAs are undetectable during the mitotic cell cycle (Fig. 3) (5). This therefore suggests rapid co-transcriptional degradation of meiotic transcripts, similar to what has been seen for cryptic unstable transcripts that are also untraceable in normal growing cells (26, 50). Also similar to the pathway of cryptic unstable transcript decay, recent studies suggest that the exosome complex contributes to the degradation of fission yeast meiotic transcripts (51, 52). Consistently, we found several meiotic transcripts to be up-regulated in cells in which genes that encode exosome components were deleted or mutated (Fig. 3). Our data also go beyond previous findings and demonstrate the specific role of the nuclear exosome in this process. This conclusion is supported by results showing that the expression of meiotic genes was unaffected in the absence of the exosome-associated cytosolic factor, Ski7, whereas the nuclear exosome component Rrp6 was required for the elimination of meiotic transcripts (Fig. 3). The specific role of the nuclear exosome in the negative regulation of meiotic genes is consistent with the nuclear localization of Pab2 (53) and Mmi1 (5). Interestingly, genome-wide localization analyses in budding yeast indicate the recruitment of exosome components to meiotic intron-containing genes (54), suggesting that the role of the exosome in the negative regulation of meiotic gene expression is conserved between fission and budding yeasts.
Results of our RNA expression analyses using different meiotic genes revealed greater defects in the rrp6Δ strain compared with the dis3–54 mutant (Fig. 3). These results corroborate previous findings that demonstrate that depletion of Rrp6 or Dis3 can result in different RNA processing defects (11, 14, 20), suggesting that these exonucleases may have functionally distinct roles within the nuclear exosome complex. Consistently, electron micrographs of the purified Leishmania exosome suggest that Rrp6 and Dis3 are located on opposite sides of the exosome complex (55). Results also show that Rrp6 can perform RNA processing events independently of the core exosome in yeast (56). The enzymatic activity of Rrp6 may therefore be the main nuclease activity used to degrade meiotic transcripts during fission yeast mitosis. However, we cannot exclude that the differences in meiotic gene expression may be due to the use of a hypomorphic (dis3–54) versus a null (rrp6Δ) allele.
Our data revealed a functional relationship between Pab2 and the exosome, as indicated by the substantial number of meiotic genes that are present in the list of up-regulated genes that overlap between the expression profiles of pab2Δ, dis3–54, and the rrp6Δ mutant (supplemental Tables 2 and 3). Given that we have demonstrated direct physical interaction between Pab2 and the nuclear exosome (27) and shown that Pab2 is associated with meiotic transcripts (Fig. 5), these observations suggest a model in which Pab2 contributes to the rapid degradation of meiotic transcripts by promoting exosome-mediated decay. In this respect, our data support that meiotic gene regulation by Pab2 is more closely linked to Rrp6 than to Dis3: pab2Δ and dis3–54 mutants showed a synergetic effect, whereas the absence of Pab2 in rrp6Δ cells did not greatly increase meiotic gene expression compared with the rrp6Δ single mutant (Fig. 3).
A Polyadenylation-associated Degradation Pathway Independent of the Poly(A) Polymerase Activity of TRAMP
The role of the poly(A)-binding protein Pab2 in the post-transcriptional elimination of meiotic transcripts suggest a polyadenylation-mediated mechanism of RNA decay. This is in fact consistent with the recently described role of Pab2 in promoting degradation of polyadenylated small nucleolar RNAs via the exosome (27). In eukaryotes, polyadenylation-mediated RNA decay has been largely associated to the activity of the TRAMP polyadenylation complex (12, 24–26). Unexpectedly, the specialized decay pathway described in this study functions independently of the polyadenylation activity catalyzed by the TRAMP complex, as our results indicate that the expression levels of meiotic transcripts are not affected in the cid14Δ strain (Fig. 3). This result echoes recent data demonstrating that the catalytic activity of TRAMP was not required to suppress the expression of crs1, which encodes a meiotic cyclin (51). Although the polyadenylation activity required for Pab2-mediated elimination of meiotic transcripts remains to be determined, Pab2/exosome-dependent degradation of RNAs polyadenylated by the classical poly(A) polymerase has been reported (27).
Mmi1 as a Specificity Factor for Pab2-mediated Decay of Meiotic RNAs
During fission yeast mitosis, Mmi1 functions as a trans-acting factor that eliminates meiotic transcripts that include a cis-element termed “determinant of selective removal” (DSR) (5). Because the RNA element bound by Pab2 is a widespread sequence (poly(A) tail), other factors are likely responsible for Pab2 recruitment and/or activation to selectively target meiotic RNAs for degradation. Several lines of evidence suggest that Mmi1 acts as the factor that selectively targets DSR-containing meiotic transcripts for degradation by Pab2 and the exosome: 1) Our genome-wide data revealed that a number of Pab2 target genes are also subjected to Mmi1-dependent regulation: mug1, rec25, spo5, ssm4, rec8, and meu1 (Table 1). In fact, most of the known Mmi1-regulated transcripts (5) are up-regulated in pab2Δ cells. Furthermore, our study identified novel Mmi1 target genes within the list of Pab2-controlled meiotic RNAs: the rec10 and mek1 mRNAs (Fig. 4), as well as the noncoding meiRNA (data not shown). 2) Our data using the pab2Δ mmi1-ts3 double mutant strain suggest that Pab2 and Mmi1 function in the same pathway that suppresses the expression of meiotic genes in mitosis. 3) Pab2 co-purifies with Mmi1 in immunoprecipitation assays (57). 4) Pab2 has recently been shown to promote the degradation of specific polyadenylated RNAs by the nuclear exosome complex (27). Because DSR elements are usually located near the 3′ end of meiotic transcripts (5), we speculate that binding of Mmi1 to DSR-containing transcripts alters normal 3′-end mRNA processing and triggers a polyadenylation-mediated decay pathway that involves Pab2 and the exosome. Consistent with such a model, the polyadenylation signal of the crs1 gene, a Mmi1-regulated meiotic transcript, was shown to be important for the negative regulation of crs1 expression in mitosis (51).
In summary, our study provides important new insights into mechanisms of meiotic gene regulation by the identification of a novel trans-acting factor, Pab2, which promotes the degradation of meiotic transcripts in mitosis. Our findings therefore unveiled a previously uncharacterized mode of gene regulation in which a poly(A)-binding protein targets transcripts for nuclear degradation.
Supplementary Material
Acknowledgments
We thank M. Yamamoto for providing the mmi1-ts strains and for communication of unpublished data. We thank members of the Bachand laboratory for helpful discussion and critical reading of the manuscript and S. Labbé for critical comments on the manuscript.
This work was supported in part by grants from Natural Sciences and Engineering Research Council of Canada (to F. B.) and Cancer Research UK (to J. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1, Tables 1–3, and references.
- DSR
- determinant of selective removal
- ChIP
- chromatin immunoprecipitation
- Pab2
- poly(A)-binding protein 2
- PABPN1
- poly(A)-binding protein nuclear 1
- pol II
- polymerase II
- ts
- temperature-sensitive
- SAM
- significance analysis of microarrays.
REFERENCES
- 1.Mata J., Lyne R., Burns G., Bähler J. (2002) Nat. Genet. 32, 143–147 [DOI] [PubMed] [Google Scholar]
- 2.Mata J., Bähler J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 15517–15522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Averbeck N., Sunder S., Sample N., Wise J. A., Leatherwood J. (2005) Mol. Cell 18, 491–498 [DOI] [PubMed] [Google Scholar]
- 4.Malapeira J., Moldón A., Hidalgo E., Smith G. R., Nurse P., Ayté J. (2005) Mol. Cell Biol. 25, 6330–6337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harigaya Y., Tanaka H., Yamanaka S., Tanaka K., Watanabe Y., Tsutsumi C., Chikashige Y., Hiraoka Y., Yamashita A., Yamamoto M. (2006) Nature 442, 45–50 [DOI] [PubMed] [Google Scholar]
- 6.Vanacova S., Stefl R. (2007) EMBO Rep. 8, 651–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parker R., Song H. (2004) Nat. Struct. Mol. Biol. 11, 121–127 [DOI] [PubMed] [Google Scholar]
- 8.Schmid M., Jensen T. H. (2008) Trends Biochem. Sci. 33, 501–510 [DOI] [PubMed] [Google Scholar]
- 9.Lebreton A., Séraphin B. (2008) Biochim. Biophys. Acta 1779, 558–565 [DOI] [PubMed] [Google Scholar]
- 10.Houseley J., LaCava J., Tollervey D. (2006) Nat. Rev. Mol. Cell Biol. 7, 529–539 [DOI] [PubMed] [Google Scholar]
- 11.Allmang C., Kufel J., Chanfreau G., Mitchell P., Petfalski E., Tollervey D. (1999) EMBO J. 18, 5399–5410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kadaba S., Krueger A., Trice T., Krecic A. M., Hinnebusch A. G., Anderson J. (2004) Genes Dev. 18, 1227–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitchell P., Petfalski E., Shevchenko A., Mann M., Tollervey D. (1997) Cell 91, 457–466 [DOI] [PubMed] [Google Scholar]
- 14.van Hoof A., Lennertz P., Parker R. (2000) Mol. Cell Biol. 20, 441–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bousquet-Antonelli C., Presutti C., Tollervey D. (2000) Cell 102, 765–775 [DOI] [PubMed] [Google Scholar]
- 16.Hilleren P., McCarthy T., Rosbash M., Parker R., Jensen T. H. (2001) Nature 413, 538–542 [DOI] [PubMed] [Google Scholar]
- 17.Libri D., Dower K., Boulay J., Thomsen R., Rosbash M., Jensen T. H. (2002) Mol. Cell Biol. 22, 8254–8266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torchet C., Bousquet-Antonelli C., Milligan L., Thompson E., Kufel J., Tollervey D. (2002) Mol. Cell 9, 1285–1296 [DOI] [PubMed] [Google Scholar]
- 19.Liu Q., Greimann J. C., Lima C. D. (2006) Cell 127, 1223–1237 [DOI] [PubMed] [Google Scholar]
- 20.Dziembowski A., Lorentzen E., Conti E., Séraphin B. (2007) Nat. Struct. Mol. Biol. 14, 15–22 [DOI] [PubMed] [Google Scholar]
- 21.Allmang C., Petfalski E., Podtelejnikov A., Mann M., Tollervey D., Mitchell P. (1999) Genes Dev. 13, 2148–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Araki Y., Takahashi S., Kobayashi T., Kajiho H., Hoshino S., Katada T. (2001) EMBO J. 20, 4684–4693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Hoof A., Staples R. R., Baker R. E., Parker R. (2000) Mol. Cell Biol. 20, 8230–8243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.LaCava J., Houseley J., Saveanu C., Petfalski E., Thompson E., Jacquier A., Tollervey D. (2005) Cell 121, 713–724 [DOI] [PubMed] [Google Scholar]
- 25.Vanácová S., Wolf J., Martin G., Blank D., Dettwiler S., Friedlein A., Langen H., Keith G., Keller W. (2005) PLoS Biol. 3, e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wyers F., Rougemaille M., Badis G., Rousselle J. C., Dufour M. E., Boulay J., Régnault B., Devaux F., Namane A., Séraphin B., Libri D., Jacquier A. (2005) Cell 121, 725–737 [DOI] [PubMed] [Google Scholar]
- 27.Lemay J. F., D'Amours A., Lemieux C., Lackner D. H., St-Sauveur V. G., Bähler J., Bachand F. (2010) Mol. Cell 37, 34–45 [DOI] [PubMed] [Google Scholar]
- 28.Grzechnik P., Kufel J. (2008) Mol. Cell 32, 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Milligan L., Torchet C., Allmang C., Shipman T., Tollervey D. (2005) Mol. Cell Biol. 25, 9996–10004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watanabe T., Miyashita K., Saito T. T., Yoneki T., Kakihara Y., Nabeshima K., Kishi Y. A., Shimoda C., Nojima H. (2001) Nucleic Acids Res. 29, 2327–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watanabe Y., Yamamoto M. (1994) Cell 78, 487–498 [DOI] [PubMed] [Google Scholar]
- 32.Yamashita A., Watanabe Y., Nukina N., Yamamoto M. (1998) Cell 95, 115–123 [DOI] [PubMed] [Google Scholar]
- 33.Bähler J., Wu J. Q., Longtine M. S., Shah N. G., McKenzie A., 3rd, Steever A. B., Wach A., Philippsen P., Pringle J. R. (1998) Yeast 14, 943–951 [DOI] [PubMed] [Google Scholar]
- 34.Tusher V. G., Tibshirani R., Chu G. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., Sherlock G. (2000) Nat. Genet. 25, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berriz G. F., King O. D., Bryant B., Sander C., Roth F. P. (2003) Bioinformatics 19, 2502–2504 [DOI] [PubMed] [Google Scholar]
- 37.Bachand F., Lackner D. H., Bähler J., Silver P. A. (2006) Mol. Cell Biol. 26, 1731–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lemieux C., Bachand F. (2009) Nucleic Acids Res. 37, 3418–3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keeney J. B., Boeke J. D. (1994) Genetics 136, 849–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Forsburg S. L., Rhind N. (2006) Yeast 23, 173–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murakami H., Goto D. B., Toda T., Chen E. S., Grewal S. I., Martienssen R. A., Yanagida M. (2007) PLoS ONE 2, e317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Win T. Z., Draper S., Read R. L., Pearce J., Norbury C. J., Wang S. W. (2006) Mol. Cell Biol. 26, 1710–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bühler M., Haas W., Gygi S. P., Moazed D. (2007) Cell 129, 707–721 [DOI] [PubMed] [Google Scholar]
- 44.Chu S., DeRisi J., Eisen M., Mulholland J., Botstein D., Brown P. O., Herskowitz I. (1998) Science 282, 699–705 [DOI] [PubMed] [Google Scholar]
- 45.Primig M., Williams R. M., Winzeler E. A., Tevzadze G. G., Conway A. R., Hwang S. Y., Davis R. W., Esposito R. E. (2000) Nat. Genet. 26, 415–423 [DOI] [PubMed] [Google Scholar]
- 46.Suh N., Jedamzik B., Eckmann C. R., Wickens M., Kimble J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 15108–15112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang L., Eckmann C. R., Kadyk L. C., Wickens M., Kimble J. (2002) Nature 419, 312–316 [DOI] [PubMed] [Google Scholar]
- 48.Chekanova J. A., Gregory B. D., Reverdatto S. V., Chen H., Kumar R., Hooker T., Yazaki J., Li P., Skiba N., Peng Q., Alonso J., Brukhin V., Grossniklaus U., Ecker J. R., Belostotsky D. A. (2007) Cell 131, 1340–1353 [DOI] [PubMed] [Google Scholar]
- 49.West S., Gromak N., Norbury C. J., Proudfoot N. J. (2006) Mol. Cell 21, 437–443 [DOI] [PubMed] [Google Scholar]
- 50.Davis C. A., Ares M., Jr. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 3262–3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McPheeters D. S., Cremona N., Sunder S., Chen H. M., Averbeck N., Leatherwood J., Wise J. A. (2009) Nat. Struct. Mol. Biol. 16, 255–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang S. W., Stevenson A. L., Kearsey S. E., Watt S., Bähler J. (2008) Mol. Cell Biol. 28, 656–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perreault A., Lemieux C., Bachand F. (2007) J. Biol. Chem. 282, 7552–7562 [DOI] [PubMed] [Google Scholar]
- 54.Moore M. J., Schwartzfarb E. M., Silver P. A., Yu M. C. (2006) Mol. Cell 24, 903–915 [DOI] [PubMed] [Google Scholar]
- 55.Cristodero M., Böttcher B., Diepholz M., Scheffzek K., Clayton C. (2008) Mol. Biochem. Parasitol. 159, 24–29 [DOI] [PubMed] [Google Scholar]
- 56.Callahan K. P., Butler J. S. (2008) Nucleic Acids Res. 36, 6645–6655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamanaka S., Yamashita A., Harigaya Y., Iwata R., Yamamoto M. (2010) EMBO J. 29, 2173–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.